Abstract
Accurate and rapid diagnosis of coronavirus disease 2019 (COVID-19) from chest CT scans is of great importance and urgency during the worldwide outbreak. However, radiologists have to distinguish COVID-19 pneumonia from other pneumonia in a large number of CT scans, which is tedious and inefficient. Thus, it is urgently and clinically needed to develop an efficient and accurate diagnostic tool to help radiologists to fulfill the difficult task. In this study, we proposed a deep supervised autoencoder (DSAE) framework to automatically identify COVID-19 using multi-view features extracted from CT images. To fully explore features characterizing CT images from different frequency domains, DSAE was proposed to learn the latent representation by multi-task learning. The proposal was designed to both encode valuable information from different frequency features and construct a compact class structure for separability. To achieve this, we designed a multi-task loss function, which consists of a supervised loss and a reconstruction loss. Our proposed method was evaluated on a newly collected dataset of 787 subjects including COVID-19 pneumonia patients, other pneumonia patients, and normal subjects without abnormal CT findings. Extensive experimental results demonstrated that our proposed method achieved encouraging diagnostic performance and may have potential clinical application for the diagnosis of COVID-19.
Keywords: COVID-19, deep supervised autoencoder, multi-view features, multi-task learning
1. Introduction
Since the coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in December 2019, in Wuhan, China [1], it has brought tremendous panic all around the world. The challenges related to the shortage of medical resources and staffs in the global medical system posed by COVID-19 remain intractable. As of January 29, over one hundred million cases were confirmed in the world [2]. Given the facts that COVID-19 is highly contagious and no effective vaccine is routinely used in clinical practice, strict prevention and early diagnosis remain the most effective way to fight the outbreak of COVID-19.
Reverse-transcription polymerase chain reaction (RT-PCR) has become standard of care in the diagnosis of COVID-19 [3]. However, its inherent disadvantage, that is false negative, can limit the clinical applicability to early diagnose the disease [4], [5]. A delayed diagnosis can increase the risk of viral transmission, which is not conducive to the epidemic prevention and control. Therefore, a more sensitive diagnostic tool is urgently needed. Computed tomography (CT) has played a vital role in the screening, diagnosis and evaluation of treatment response of COVID-19 [6], [7], [8]. More importantly, previous studies have reported that some patients have typical chest CT scan findings and symptoms for COVID-19 but their initial RT-PCR results are negative [5], [9], [10]. In this context, CT was considered as a clinical diagnostic tool in China and helped us to screen out and isolate suspected cases. In this way, a lot of cases were timely diagnosed and the spread of virus had been substantially avoided. Although CT has high sensitivity, it has pitfalls such as a relatively low specificity. The typical imaging findings of COVID-19 pneumonia are bilateral and peripheral ground-glass and consolidative opacities [6], [11]. However, other lung diseases may also present the aforementioned imaging manifestations [9], [12]. Moreover, accurately differentiating COVID-19 pneumonia from other pneumonia in a large number of CT examinations is a tedious and inefficient work, which could compromise the accuracy. In this context, it is urgently and clinically needed to find an efficient and accurate diagnostic tool to help the radiologists to fulfill this difficult task.
With the state-of-the-art data analysis strategy, artificial intelligence (AI) technologies, especially convolutional neural networks (CNNs), have achieved remarkable success in medical imaging analysis. Numerous studies have shown great potential in automatic diagnosis of COVID-19 from medical images [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. According to the difference of inputs and screening classifiers, we categorize pioneering methods used for classifying COVID-19 into four classes. The first class is the feature engineering-based approaches that manually annotate the infection areas, quantify radiomic features, and then train machine learning classifiers based on the features [17], [18]. A preliminary study conducted by Fang et al. reported that some image biomarkers had strong predictive power for screening out COVID-19 pneumonia from CT images [17]. Subsequently, they built a radiomic signature by combining clinical features and radiomic features to improve the diagnostic performance [18]. However, the number of data samples used in their studies is relatively small, which requires further validation by larger prospective multicenter studies.
The second class is the transfer learning-based methods that take a series of CT slices as input, employ the state-of-the-art pre-trained models as backbones to generate features and perform slice-wise decisions [19], [20], [25]. Li et al. used the segmented lung regions as input and employed RestNet50 architecture, which won the first place in multi-intelligence tasks [26], as the backbone to extract features for differentiating COVID-19 pneumonia from community acquired pneumonia and other lung diseases [19]. Similarly, Bai el al. used EfficientNet architecture, which achieved state-of-the-art accuracy on ImageNet and CIFAR-100 datasets [27], to distinguish COVID-19 from other pneumonia with CT images [20]. Their high predictive performance partially benefited from carefully preprocessed and selected data, such as manual corrections and annotations.
The third class is the 3D CNNs-based methods that directly take 3D CT images as input and train their proposed 3D CNNs to identify COVID-19 pneumonia [21], [22], [28], [29]. Wang et al. applied a 3D connected component algorithm [30] for lesion localization in an unsupervised manner, and then took 3D CT images with the corresponding 3D lesion masks into a CNN model to generate the probabilities of COVID-19 positive and COVID-19 negative [21]. Although it is no longer required to manually annotate the COVID-19 lesions on CT images, the segmentations they gained are still imperfect. To improve the accuracy of automatic identification of infected regions, Ouyang et al. used an established segmentation model to automatically extract the lung regions with infection lesions and put them into a dual-sampling attention network to focus on diagnosing COVID-19 pneumonia [22]. This work can avoid errors that may be caused by intermediate processes, but the reliability of their network needs to be trained with a large amount of data.
The forth class is the representation learning-based methods that learn latent representation from CT images to diagnose COVID-19 pneumonia [23], [24]. Kang et al. explored multiple features from infected lesions and designed a multi-view representation-based framework for diagnosis of COVID-19 [23]. Han et al. employed an attention-based deep multiple instance learning to obtain bag representations and transformed them into final prediction by using two fully connected layers [24]. Since they are highly predictive and well interpretable, representation learning-based approaches have great potential in diagnosing COVID-19. Although significant advancements have achieved, the diagnostic methods remain underexplored.
In this study, we proposed a classification framework towards identify COVID-19 pneumonia rapidly and accurately from other pneumonia and normal subjects. It is worth noting that automatic and accurate segmentation of COVID-19 or other pneumonia lesions from CT images is extremely challenging due to complex and changeable manifestations of pneumonia lesions. Since there is a large contrast difference between the lesion and the normal tissue in the lungs, we employed a morphological technique to detect lesions as an alternative for segmentation of lesions. To further demonstrate the superiority of the alternative, we took subjects without abnormal CT findings as normal group into consideration for the triple classification task. Thus, we only performed lung parenchyma segmentation from 3D CT images using a pre-trained 3D U-Net model [31]. Then, the lung parenchyma was texturized by using 3D wavelet transform to capture multiple different frequency subbands. We extracted the multiple features including gray features and texture features from the subbands with different frequencies, which were considered as a multi-view feature set. Based on multi-view learning and supervised autoencoder [32], we designed a deep supervised autoencoder (DSAE) to map the original features into a latent space, aiming to learn informative and structured representations. A series of experiments on a newly collected dataset from multiple institutions were conducted to evaluate our proposed method, and the results showed that our method could achieve encouraging diagnostic performance. Our main contributions are summarised as follows:
-
1)
We employed a morphological technique, namely 3D wavelet transform, to decompose the original image with its lung mask into multiple frequency subbands, and then the features extracted from the subbands constituted a multi-view features set for diagnosis of COVID-19.
-
2)
We proposed a DSAE network to map the original features into a latent space to learn the latent representation.
-
3)
We developed a multi-task loss function to make the latent representations more informative and structured.
-
4)
We evaluated the performance of the proposed DSAE on a newly collected dataset from multiple institutions and provided clinical insights for diagnosis of COVID-19.
The remainder of this paper is organized as follows. Materials and methods are introduced in Section 2. Experiments and results are presented in Section 3. A discussion of our works is provided in Section 4. Finally, a brief conclusion to this study is provided in Section 5.
2. Materials and Methods
This retrospective study was approved by our Medical Ethical Committee (Approved Number.2020024), which waived the requirement of informed consent of patients.
2.1. Dataset
In this study, we retrospectively included three groups. First, 317 confirmed COVID-19 cases were collected from nine institutions in Hunan Province, China (152 women, 165 men; mean age, 45.33 years 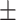 19.41 [SD]; age range, 1-84 years), named as COVID-19 group. The inclusion criteria for COVID-19 group were : 1) patients were confirmed as COVID-19; 2) patients underwent CT scanning before or upon admission; and 3) patients had abnormal CT findings. Second, 248 cases of non-COVID-19 pneumonia were also collected from our institution (103 women, 145 men; mean age, 49.32 years
19.41 [SD]; age range, 1-84 years), named as COVID-19 group. The inclusion criteria for COVID-19 group were : 1) patients were confirmed as COVID-19; 2) patients underwent CT scanning before or upon admission; and 3) patients had abnormal CT findings. Second, 248 cases of non-COVID-19 pneumonia were also collected from our institution (103 women, 145 men; mean age, 49.32 years 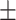 19.49 [SD]; age range, 0-90 years), named as non-COVID-19 group. The inclusion criteria for non-COVID-19 group were: 1) patients were identified as community acquired pneumonia (CAP); and 2) patients had imaging manifestations showing viral pneumonia but without accurate diagnosis. Lastly, 222 cases without abnormal imaging findings were retrieved from the picture archiving and communication system (PACS) as a comparison (114 women, 108 men; mean age, 27.18 years
19.49 [SD]; age range, 0-90 years), named as non-COVID-19 group. The inclusion criteria for non-COVID-19 group were: 1) patients were identified as community acquired pneumonia (CAP); and 2) patients had imaging manifestations showing viral pneumonia but without accurate diagnosis. Lastly, 222 cases without abnormal imaging findings were retrieved from the picture archiving and communication system (PACS) as a comparison (114 women, 108 men; mean age, 27.18 years 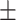 18.90 [SD]; age range, 0-57 years), named as normal group. The CT images of all included cases were retrieved from PACS and anonymized for further investigation. Note that we only included cases who had CT images with slice less than 5
18.90 [SD]; age range, 0-57 years), named as normal group. The CT images of all included cases were retrieved from PACS and anonymized for further investigation. Note that we only included cases who had CT images with slice less than 5 and non-contrast enhanced CT images. For patients had multiple CT scans, we only included the first CT scan images. Finally, a total of 787 subjects were used in this study. According to the data collection dates, we divided all subjects into a primary cohort of 529 cases and a validation cohort of 258 cases. The gender, age, and institution distributions of the two cohorts are presented in Table 1.
and non-contrast enhanced CT images. For patients had multiple CT scans, we only included the first CT scan images. Finally, a total of 787 subjects were used in this study. According to the data collection dates, we divided all subjects into a primary cohort of 529 cases and a validation cohort of 258 cases. The gender, age, and institution distributions of the two cohorts are presented in Table 1.
TABLE 1. Clinical Characteristics of Patients in the Primary and Validation Cohorts.
| Primary cohort (n = 529) | Validation cohort (n = 258) | ||||||
|---|---|---|---|---|---|---|---|
| COVID-19 | Non-COVID-19 | Normal | COVID-19 | Non-COVID-19 | Normal | ||
| Gender | Male | 119 [54.34] | 85 [49.71] | 71 [51.08] | 46 [46.94] | 60 [77.92] | 37 [44.58] |
| Female | 100 [45.66] | 86 [50.29] | 68 [48.92] | 52 [53.06] | 17 [22.08] | 46 [55.42] | |
Age (mean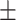 std, years) std, years)
|
45.39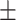 16.46 16.46 |
49.12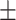 19.84 19.84 |
30.46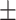 14.07 14.07 |
45.18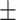 17.59 17.59 |
49.76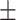 18.86 18.86 |
21.67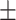 15.60 15.60 |
|
0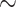 20 20 |
9 [4.11] | 17 [9.94] | 31 [22.30] | 4 [4.08] | 4 [5.19] | 40 [48.19] | |
20 40 40 |
66 [30.14] | 24 [14.04] | 73 [52.52] | 36 [36.73] | 21 [27.27] | 29 [34.94] | |
40 60 60 |
99 [45.20] | 70 [40.94] | 35 [25.18] | 36 [36.73] | 25 [32.47] | 14 [16.87] | |
60 80 80 |
41 [18.72] | 55 [32.16] | 0 | 20 [20.42] | 21 [27.27] | 0 | |
80 100 100 |
4 [1.83] | 5 [2.92] | 0 | 2 [2.04] | 6 [1.30] | 0 | |
| Institution | |||||||
| The Second Xiangya Hospital | 11 | 171 | 139 | 0 | 77 | 83 | |
| The First People Hospital of Changde | 14 | 0 | 0 | 0 | 0 | 0 | |
| The First People Hospital of Huaihua | 1 | 0 | 0 | 0 | 0 | 0 | |
| The First People Hospital of Yueyang | 51 | 0 | 0 | 0 | 0 | 0 | |
| The First People Hospital of Changsha | 34 | 0 | 0 | 98 | 0 | 0 | |
| The Second People Hospital of Chenzhou | 10 | 0 | 0 | 0 | 0 | 0 | |
| The Central Hospital of Zhuzhou | 12 | 0 | 0 | 0 | 0 | 0 | |
| The Central Hospital of Shaoyang | 75 | 0 | 0 | 0 | 0 | 0 | |
| The First Affiliated Hospital University of South China | 9 | 0 | 0 | 0 | 0 | 0 | |
| Total | 219 [41.40] | 171 [32.32] | 139 [26.28] | 98 [37.98] | 77 [28.84] | 83 [32.17] | |
Note that data are presented as mean  SD, or n [%].
SD, or n [%].
2.2. Data Preprocessing
In this study, all CT images of each patient were first reconstructed into a three-dimensional image using dcm2nii package [33]. Then, each image was preprocessed with an U-Net model [31], which is widely used in medical image segmentation [34], [35], [36], to extract the lung parenchyma. To overcome the difference between the varying thickness of samples, the volumetric data of lung parenchyma were resampled to 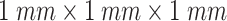 voxel resolution by the B-spline interpolation. After that, each segmented volume was texturized by using 3D wavelet transform (3D-WT) to capture eight different frequency subbands (Fig. 1A). Each frequency subband was treated as a view image. The 3D-WT provides a spatial and frequency representation of the original signal. For a wavelet decomposition, the 3D-WT can be denoted by a tensor product
voxel resolution by the B-spline interpolation. After that, each segmented volume was texturized by using 3D wavelet transform (3D-WT) to capture eight different frequency subbands (Fig. 1A). Each frequency subband was treated as a view image. The 3D-WT provides a spatial and frequency representation of the original signal. For a wavelet decomposition, the 3D-WT can be denoted by a tensor product
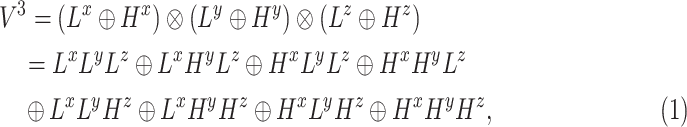
|
where  and
and 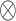 represent a space direct sum and a convolution operation, respectively.
represent a space direct sum and a convolution operation, respectively. 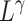 and
and 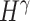 represent the low- and high-pass filters along the
represent the low- and high-pass filters along the 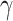 -axis, where
-axis, where 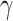
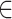
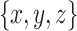 .
.
Fig. 1.
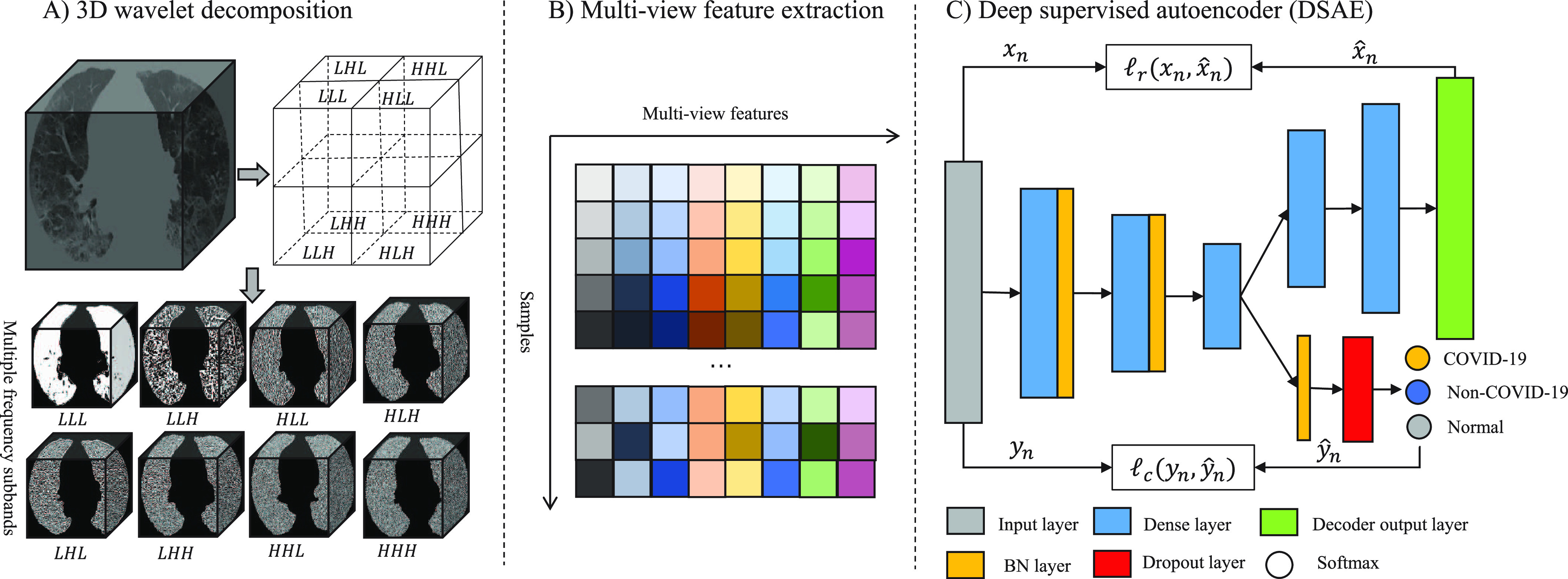
Overview of the proposed diagnostic framework. 3D U-Net model is first used to segment the lung parenchyma first from chest CT images, and then 3D-WT is used to decompose each segmented volume into 8 different frequency subbands, as shown in A); B) the features extracted from multiple frequency subbands are considered as multi-view feature set; and C) shows that deep supervised autoencoder we proposed jointly optimize the supervised loss and reconstruction loss to learn the informative and structured latent representation.
2.3. Multi-View Feature Extraction
Gray features, with a total of 18 features, mainly consist of the first-order statistics which describe the distribution of voxels within the volume of interest (VOI), such as entropy, energy, maximum, mean, and so on. Texture features are extracted from gray level co-occurrence matrix (GLCM, 24 features), gray level dependence matrix (GLDM, 14 features), gray level run length matrix (GLRLM, 16 features), gray level size zone matrix (GLSZM, 16 features) and neighboring gray tone difference matrix (NGTDM, 5 features). Thus, there are 93 radiomics features extracted from each subband. A total of 744 radiomic features were extracted from eight frequency subbands for each subject and designed as multi-view features in this study.
2.4. Deep Supervised Autoencoder and Representation Learning
The presence of heterogeneity in multiple frequency subbands may provide additional information for the diagnosis of COVID-19, and a multi-view learning-based approach [37], [38] was used in this study. To effectively exploit these features from multiple frequency subbands, a DSAE was proposed to learn the latent representation by multi-task learning.
2.4.1. Deep Supervised Autoencoder
Autoencoder is an artificial neural network designed to learn latent data representations in an unsupervised manner, which can optimally reconstruct the original data [39]. Therefore, autoencoder has been demonstrated the capacity of reducing dimensionality [40], [41] and mining latent fetures [42]. To learn latent representations with class structure, we proposed a DSAE framework for this diagnostic task. The structure of our proposed DSAE shown in Fig. 1C consists of three components: a) an encoder, which learns the latent representations from the original input; b) a decoder, which reconstructs the input from the latent representations; and c) a supervisor, which structures the latent representations and discriminates disease types. For our settings, the encoder has three hidden layers with 256, 128, and 16 neurons, respectively, and the last hidden layer serves as the representation layer. On the contrary, the decoder is regarded as the reverse operation of the encoder. It has two hidden layers with 128 and 256 neurons, and the output of the decoder has the same size as the input layer of the encoder. The supervisor is behind the representation layer followed by a batch normalization layer, a dropout layer with a drop rate of 0.5, and a classification output layer. More formally, we defined the following notations. Let the training samples be 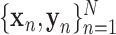 , where
, where 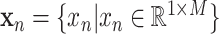 is a multi-view feature set (
is a multi-view feature set (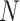 and
and 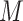 represent the number of samples and multi-view features, respectively.) and
represent the number of samples and multi-view features, respectively.) and 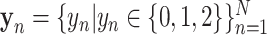 is the corresponding label set. Generally,
is the corresponding label set. Generally, 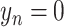 indicates the non-COVID-19 patient,
indicates the non-COVID-19 patient, 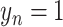 indicates the COVID-19 patient, and
indicates the COVID-19 patient, and 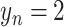 indicates the normal subject.
indicates the normal subject.
2.4.2. Representation Learning for Multi-View Features
To discover latent high-level representation for each subject, the multi-view features were used as input and encoded into a low-dimensional space. Then, the latent representation was reconstructed to the original dimension of the input. The reconstruction error was minimized through back propagation to learn two stable mappings, that is, 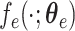 in encoding path and
in encoding path and 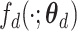 in decoding path, where
in decoding path, where 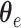 and
and 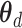 indicate the parameters of the two paths. Let
indicate the parameters of the two paths. Let 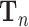 denote the learned latent representation and
denote the learned latent representation and 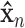 be the decoded output. Thus, they can be formulated as
be the decoded output. Thus, they can be formulated as

|

|
Our proposed aotoencoder learns the latent representation by minimizing the mean squared error (MSE) loss function between the inputs and outputs. The reconstruction loss is defined as
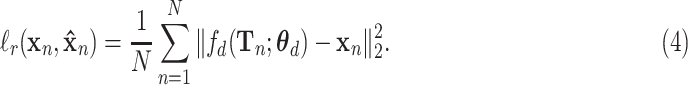
|
2.4.3. Structure for Latent Representation
To make the learned latent representation of these different pneumonia diseases well structured, a supervised block was introduced in the representation layer. The advantage is that it enables the network to better learn latent representations associated with pneumonia diseases. Batch normalization [43] and dropout [44] strategies were introduced into the supervised block to reduce overfitting issues. And the softmax layer was used to predict the subject class. The output probability can be computed as

|
where 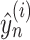 denotes the probability of
denotes the probability of 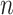 th sample for class
th sample for class 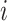 .
. 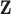 is the output vector in last fully connected layer and
is the output vector in last fully connected layer and 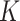 is the number of classes. For this supervised task, we minimized the cross-entropy loss function defined as (6) to enforce the compactness for the same type of disease and to present boundaries between COVID-19 pneumonia and others.
is the number of classes. For this supervised task, we minimized the cross-entropy loss function defined as (6) to enforce the compactness for the same type of disease and to present boundaries between COVID-19 pneumonia and others.
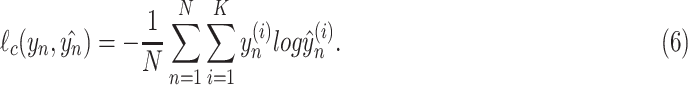
|
To take informativeness and separability into consideration, two tasks are jointly trained with the following multi-task loss:
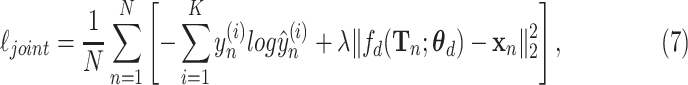
|
where 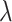 is a balance factor between the two tasks. In this study, the supervised loss is served as a major task to distinguish COVID-19 pneumonia from others, and the reconstruction loss is used as an auxiliary task to learn latent representation.
is a balance factor between the two tasks. In this study, the supervised loss is served as a major task to distinguish COVID-19 pneumonia from others, and the reconstruction loss is used as an auxiliary task to learn latent representation.
3. Experiments and Results
3.1. Experimental Settings
We conducted multiple experiments on the CT images to evaluate the proposed pipeline. Since the original features extracted from multi-view CT images are quite different, a preprocessing step of standardized features is essential for training the model. Thus, the widely used  -score standardization was employed and computed as
-score standardization was employed and computed as
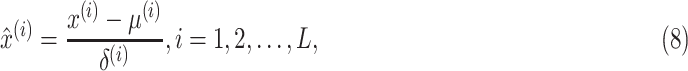
|
where 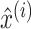 is the standardization feature of feature
is the standardization feature of feature 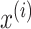 and
and 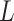 denotes the number of features.
denotes the number of features. 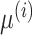 and
and 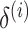 are mean value and standard deviation of the feature
are mean value and standard deviation of the feature 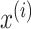 , respectively. For the training procedure, Adam [45] was used as an optimizer with an initial learning of 0.001, which was reduced by half after each 20 epochs. The batch size was set to 8, and the maximum number of epochs was set to 500. To avoid the overfitting problem, we used an early stopping strategy that the training would be terminated if the validation loss does not decrease within 50 epochs. Furthermore, we used a 5-fold cross-validation technique on the primary cohort to determine the factor
, respectively. For the training procedure, Adam [45] was used as an optimizer with an initial learning of 0.001, which was reduced by half after each 20 epochs. The batch size was set to 8, and the maximum number of epochs was set to 500. To avoid the overfitting problem, we used an early stopping strategy that the training would be terminated if the validation loss does not decrease within 50 epochs. Furthermore, we used a 5-fold cross-validation technique on the primary cohort to determine the factor 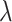 in (7) from the range [0,1] with an interval of 0.05. We found that the overall accuracy was the best when
in (7) from the range [0,1] with an interval of 0.05. We found that the overall accuracy was the best when 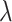 = 0.75. Hence,
= 0.75. Hence, 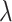 was fixed at 0.75 in the following experiments.
was fixed at 0.75 in the following experiments.
To clarify and compare the fairness, we used the standardized data as input for all experimental methods. We compared the proposed method with radiomics-based methods and a deep neural network (DNN). The radiomisc-based methods first used the minimum redundancy maximum relevance (mRMR) [46] algorithm to select features, and then the selected features were entered into the logistic regression (LR) [47], random forest (RF) [48] and support vector machine (SVM) [49] classifier to separately build a radiomic signature for the diagnosis task. The DNN is the remaining parts of the proposed DSAE excluding the decoder. For each of these methods, we performed ten experiments on CT images and reported the mean and standard deviation. Diagnostic performance was evaluated using overall accuracy in a triple classification task. Furthermore, we used a one-vs-rest strategy, treating each class as a positive in turn and the rest as negatives, to evaluate the performance with respect to accuracy (ACC), sensitivity (SEN), specificity (SPE) and F1-score (F1) metrics, which can be formulated as
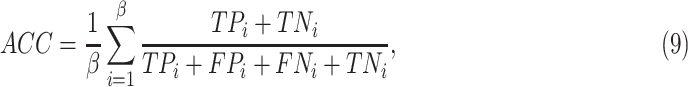
|

|

|
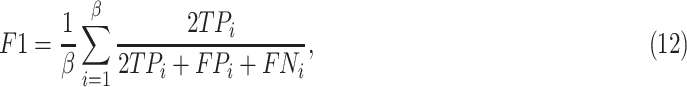
|
where 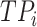 ,
, 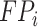 ,
, 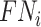 , and
, and 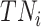 denote the number of true positives, false positives, false negatives and true negatives at
denote the number of true positives, false positives, false negatives and true negatives at 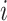 th experiment, respectively.
th experiment, respectively. 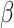 indicates the number of experiments, which is equal to 10 in this study.
indicates the number of experiments, which is equal to 10 in this study.
3.2. Diagnostic Power of Different Frequency Features
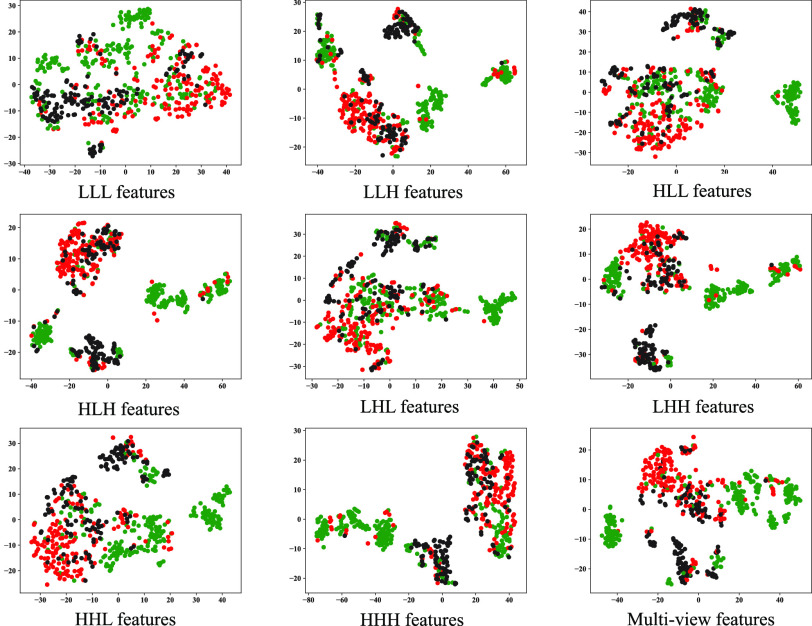
To investigate the diagnostic power of different frequency features, we first used a visualization technique called t-distributed stochastic neighbor embedding (t-SNE) [50]. Fig. 2 shows different distributions of the 8 types of original features and their fused multi-view features. For quantitative analysis, we conducted five-fold cross-validation experiments on the primary cohort for each type of features. Table 2 shows the overall accuracy of the triple classification task. Tables 3, 4, 5, and 6 show the diagnostic performance of one-vs-rest in terms of mean accuracy, sensitivity, specificity and F1-score, respectively. We can first observe that different frequency features have large performance gaps for all methods. For example, the features extracted from high-frequency subbands have better predictive performances than those extracted from low-frequency subbands for COVID-19 patients. However, low-frequency subbands have a strong predictive power for normal subjects. As expected, the high-pass filter can detect lesions with large gradient changes, while the low-pass filter can detect normal tissues with smooth gradient changes. Noteworthily, the features from different frequency subbands have different diagnostic power and they can be regarded as multiple views to complement each other for enhancing diagnostic power. As shown in Tables 2, 3, 4, 5, and 6, the approaches using multi-view features (i.e., eight different frequency features) have better predictive performance than those using individual type of features.
Fig. 2.
Visualization of 8 original features and multi-view features using t-SNE technique, which is particularly suitable for visualization of high-dimensional data. Green, red and black represent COVID-19, non-COVID-19 and normal cases, respectively.
TABLE 2. Mean Overall Accuracy of the Proposed Method and Compared Methods of the Triple Classification Task on the Primary Cohort.
| Methods | Multi-view | LLL | LLH | LHL | LHH | HLL | HLH | HHL | HHH |
|---|---|---|---|---|---|---|---|---|---|
| LR | 80.51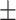 0.63 0.63 |
75.41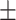 0.79 0.79 |
73.72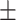 0.91 0.91 |
77.79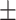 0.49 0.49 |
74.14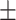 0.81 0.81 |
74.24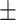 0.86 0.86 |
74.14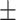 0.52 0.52 |
75.90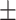 0.64 0.64 |
74.46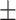 1.26 1.26 |
| RF | 84.16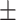 0.98 0.98 |
75.01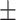 1.01 1.01 |
80.47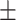 0.79 0.79 |
76.59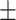 0.95 0.95 |
78.94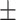 0.87 0.87 |
74.23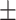 1.04 1.04 |
79.04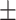 0.67 0.67 |
79.36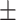 0.81 0.81 |
78.22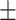 0.61 0.61 |
| SVM | 81.06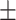 1.21 1.21 |
75.67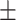 0.75 0.75 |
75.46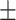 1.14 1.14 |
77.19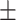 0.77 0.77 |
74.80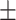 0.91 0.91 |
75.11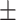 1.01 1.01 |
74.56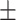 0.68 0.68 |
75.88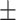 1.57 1.57 |
74.48 1.22 1.22 |
| DNN | 83.34 0.93 0.93 |
75.93 1.01 1.01 |
79.28 0.85 0.85 |
77.77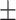 1.23 1.23 |
77.43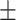 1.00 1.00 |
77.62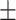 1.21 1.21 |
78.86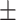 0.96 0.96 |
78.65 1.26 1.26 |
77.58 1.03 1.03 |
| DSAE | 86.44 0.70 0.70 |
78.99 1.20 1.20 |
81.19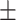 0.55 0.55 |
81.10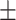 0.91 0.91 |
80.58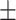 0.74 0.74 |
80.59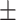 0.89 0.89 |
81.15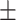 1.08 1.08 |
81.64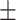 0.84 0.84 |
80.62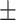 0.86 0.86 |
Note that LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH represent different frequency features extracted from the multiple frequency subbands.
TABLE 3. Mean Accuracy of the Proposed Method and Compared Methods Based on the Primary Cohort.
| Method | Class | Multi-view | LLL | LLH | LHL | LHH | HLL | HLH | HHL | HHH |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | COVID-19 | 89.62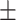 1.55 1.55 |
82.74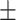 0.94 0.94 |
84.52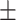 0.58 0.58 |
86.24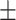 0.63 0.63 |
86.34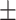 1.07 1.07 |
84.27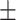 0.83 0.83 |
87.07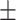 0.45 0.45 |
86.71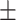 0.62 0.62 |
87.41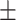 0.71 0.71 |
| Non-COVID-19 | 84.24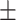 1.41 1.41 |
81.13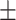 0.79 0.79 |
78.79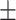 1.33 1.33 |
81.98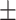 0.43 0.43 |
78.58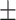 0.64 0.64 |
80.26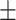 0.76 0.76 |
78.75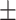 0.67 0.67 |
81.10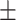 0.58 0.58 |
78.62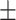 1.00 1.00 |
|
| Normal | 88.19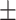 1.18 1.18 |
86.94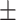 0.47 0.47 |
84.14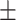 0.71 0.71 |
87.36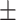 0.67 0.67 |
83.35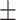 0.72 0.72 |
83.94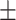 0.76 0.76 |
82.46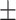 0.45 0.45 |
83.99 0.75 0.75 |
82.89 1.01 1.01 |
|
| RF | COVID-19 | 93.95 0.47 0.47 |
82.51 0.98 0.98 |
92.19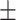 0.64 0.64 |
87.09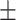 0.61 0.61 |
91.98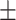 0.67 0.67 |
85.63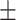 0.94 0.94 |
92.27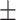 0.55 0.55 |
91.91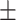 0.54 0.54 |
91.55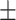 0.53 0.53 |
| Non-COVID-19 | 86.26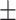 1.22 1.22 |
80.47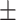 1.01 1.01 |
82.93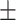 0.73 0.73 |
80.33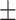 0.68 0.68 |
81.49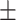 0.80 0.80 |
79.81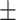 1.19 1.19 |
81.83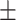 0.61 0.61 |
82.27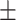 0.78 0.78 |
82.08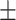 0.60 0.60 |
|
| Normal | 88.83 1.45 1.45 |
87.04 0.75 0.75 |
85.82 0.51 0.51 |
85.77 0.83 0.83 |
84.41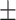 0.59 0.59 |
83.03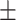 0.84 0.84 |
83.97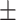 0.79 0.79 |
84.54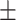 0.89 0.89 |
82.82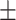 0.88 0.88 |
|
| SVM | COVID-19 | 90.63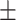 2.01 2.01 |
83.33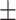 0.77 0.77 |
87.20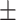 0.69 0.69 |
86.69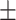 0.63 0.63 |
87.92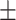 0.85 0.85 |
86.09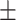 0.89 0.89 |
88.94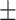 0.50 0.50 |
88.22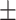 1.01 1.01 |
88.09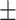 0.95 0.95 |
| Non-COVID-19 | 84.44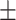 1.27 1.27 |
80.76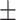 0.96 0.96 |
78.81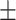 1.22 1.22 |
81.16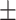 0.62 0.62 |
78.36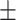 0.62 0.62 |
79.47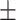 1.07 1.07 |
77.47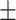 0.60 0.60 |
79.94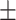 1.26 1.26 |
78.17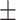 1.34 1.34 |
|
| Normal | 88.30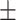 1.32 1.32 |
87.26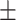 0.50 0.50 |
84.92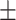 1.09 1.09 |
86.53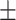 0.76 0.76 |
83.33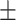 0.89 0.89 |
84.65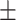 0.62 0.62 |
82.71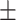 0.60 0.60 |
83.60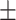 1.17 1.17 |
82.71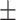 0.76 0.76 |
|
| DNN | COVID-19 | 93.74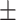 0.51 0.51 |
83.53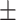 0.69 0.69 |
89.99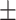 0.44 0.44 |
87.73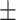 1.12 1.12 |
90.30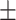 0.88 0.88 |
87.03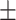 0.88 0.88 |
91.74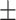 0.59 0.59 |
90.58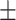 0.91 0.91 |
90.43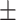 0.71 0.71 |
| Non-COVID-19 | 85.23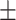 0.92 0.92 |
80.60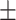 0.77 0.77 |
81.98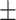 0.46 0.46 |
81.68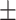 0.72 0.72 |
79.96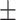 0.96 0.96 |
80.45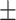 1.01 1.01 |
80.79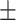 0.79 0.79 |
81.23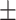 1.10 1.10 |
80.18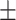 0.88 0.88 |
|
| Normal | 88.16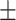 0.62 0.62 |
88.14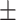 0.91 0.91 |
85.03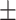 0.60 0.60 |
85.82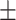 0.97 0.97 |
82.74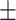 1.12 1.12 |
84.88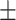 1.01 1.01 |
83.12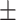 0.94 0.94 |
83.16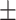 1.23 1.23 |
83.29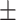 0.77 0.77 |
|
| DSAE | COVID-19 | 94.61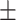 0.44 0.44 |
85.65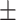 0.92 0.92 |
91.70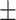 0.52 0.52 |
89.90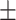 0.52 0.52 |
92.34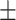 0.44 0.44 |
90.51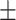 0.45 0.45 |
92.61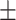 0.42 0.42 |
92.82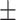 0.57 0.57 |
93.46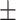 0.38 0.38 |
| Non-COVID-19 | 87.60 0.61 0.61 |
82.91 0.90 0.90 |
83.95 0.71 0.71 |
83.80 0.94 0.94 |
81.87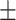 1.10 1.10 |
82.99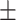 0.68 0.68 |
82.80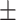 0.65 0.65 |
83.25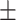 0.76 0.76 |
83.67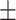 0.59 0.59 |
|
| Normal | 90.38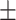 0.58 0.58 |
89.77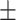 0.40 0.40 |
87.52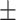 0.80 0.80 |
88.15 0.67 0.67 |
86.09 1.24 1.24 |
86.81 0.67 0.67 |
86.26 0.59 0.59 |
86.20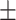 0.73 0.73 |
85.82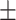 0.66 0.66 |
Note that LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH represent different frequency features extracted from the multiple frequency subbands.
TABLE 4. Mean Sensitivity of the Proposed Method and Compared Methods Based on the Primary Cohort.
| Method | Class | Multi-view | LLL | LLH | LHL | LHH | HLL | HLH | HHL | HHH |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | COVID-19 | 87.33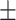 1.86 1.86 |
78.50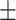 1.65 1.65 |
79.52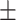 1.69 1.69 |
84.03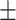 0.87 0.87 |
84.90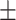 1.45 1.45 |
83.22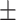 1.04 1.04 |
86.32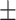 0.42 0.42 |
84.57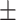 1.35 1.35 |
85.24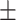 1.25 1.25 |
| Non-COVID-19 | 73.29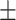 2.67 2.67 |
65.77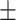 2.05 2.05 |
65.53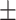 1.77 1.77 |
68.59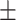 0.93 0.93 |
66.53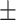 1.36 1.36 |
67.07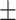 2.32 2.32 |
66.86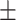 1.39 1.39 |
70.83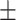 1.36 1.36 |
67.53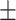 2.35 2.35 |
|
| Normal | 80.79 3.01 3.01 |
83.00 2.08 2.08 |
75.15 1.93 1.93 |
79.70 1.75 1.75 |
66.92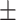 2.16 2.16 |
69.47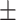 1.75 1.75 |
64.07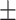 1.10 1.10 |
68.82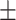 2.26 2.26 |
66.54 1.74 1.74 |
|
| RF | COVID-19 | 89.86 0.74 0.74 |
79.03 1.33 1.33 |
88.84 0.80 0.80 |
82.87 0.61 0.61 |
88.30 0.85 0.85 |
82.62 1.44 1.44 |
89.24 1.04 1.04 |
88.94 0.71 0.71 |
87.41 0.90 0.90 |
| Non-COVID-19 | 79.37 2.05 2.05 |
65.64 2.28 2.28 |
73.64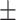 1.87 1.87 |
71.63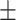 2.50 2.50 |
71.87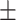 2.20 2.20 |
67.62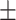 2.29 2.29 |
72.32 2.03 2.03 |
73.85 1.04 1.04 |
73.17 1.04 1.04 |
|
| Normal | 82.99 3.10 3.10 |
80.52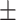 2.17 2.17 |
76.08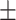 1.53 1.53 |
73.52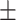 2.84 2.84 |
73.18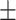 2.15 2.15 |
69.22 1.94 1.94 |
71.71 1.40 1.40 |
71.63 2.63 2.63 |
70.47 1.66 1.66 |
|
| SVM | COVID-19 | 86.67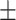 2.26 2.26 |
77.63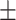 1.29 1.29 |
80.11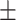 1.39 1.39 |
82.00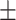 0.95 0.95 |
85.03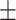 1.37 1.37 |
82.58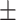 1.38 1.38 |
86.55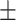 1.03 1.03 |
82.37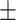 0.99 0.99 |
84.09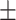 1.17 1.17 |
| Non-COVID-19 | 75.27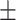 2.96 2.96 |
67.21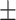 1.73 1.73 |
71.20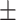 1.89 1.89 |
68.99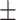 1.41 1.41 |
74.43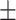 2.39 2.39 |
71.85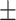 1.59 1.59 |
76.66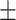 1.88 1.88 |
75.42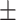 2.03 2.03 |
71.40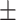 2.74 2.74 |
|
| Normal | 81.99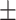 3.14 3.14 |
83.65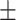 1.23 1.23 |
74.08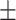 2.77 2.77 |
80.10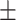 2.09 2.09 |
59.49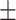 2.17 2.17 |
67.85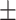 2.20 2.20 |
53.51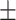 1.60 1.60 |
66.69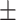 3.62 3.62 |
63.70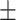 2.56 2.56 |
|
| DNN | COVID-19 | 89.85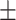 0.41 0.41 |
79.46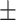 1.42 1.42 |
84.61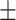 0.81 0.81 |
84.06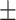 2.24 2.24 |
84.95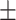 1.38 1.38 |
85.20 1.44 1.44 |
87.53 0.91 0.91 |
84.64 1.46 1.46 |
85.81 1.18 1.18 |
| Non-COVID-19 | 74.35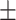 2.09 2.09 |
65.61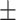 2.72 2.72 |
71.10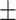 1.88 1.88 |
68.91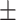 1.78 1.78 |
65.43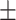 3.03 3.03 |
62.84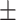 2.61 2.61 |
68.04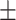 3.00 3.00 |
67.74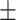 1.11 1.11 |
67.98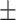 2.38 2.38 |
|
| Normal | 85.11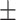 2.48 2.48 |
84.16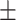 2.26 2.26 |
78.12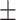 1.50 1.50 |
78.49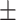 1.89 1.89 |
77.50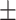 2.36 2.36 |
78.82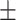 2.92 2.92 |
74.52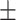 3.11 3.11 |
78.39 2.83 2.83 |
74.10 2.27 2.27 |
|
| DSAE | COVID-19 | 91.22 1.02 1.02 |
79.21 1.97 1.97 |
88.82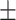 1.06 1.06 |
85.04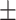 1.32 1.32 |
89.52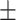 0.78 0.78 |
87.14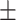 1.31 1.31 |
89.78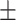 1.09 1.09 |
88.26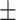 1.48 1.48 |
89.56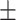 0.92 0.92 |
| Non-COVID-19 | 80.32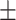 1.77 1.77 |
72.55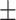 3.58 3.58 |
75.27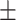 2.24 2.24 |
76.07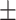 2.20 2.20 |
75.91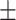 2.89 2.89 |
74.91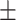 2.33 2.33 |
76.27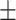 2.77 2.77 |
77.88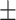 2.45 2.45 |
78.18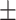 2.72 2.72 |
|
| Normal | 85.63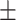 1.84 1.84 |
87.22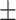 1.60 1.60 |
77.58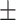 3.58 3.58 |
80.34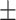 2.52 2.52 |
70.37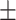 3.28 3.28 |
75.27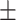 3.08 3.08 |
71.69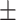 2.64 2.64 |
73.49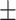 2.46 2.46 |
72.39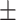 1.99 1.99 |
Note that LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH represent different frequency features extracted from the multiple frequency subbands.
TABLE 5. Mean Specificity of the Proposed Method and Compared Methods Based on the Primary Cohort.
| Method | Class | Multi-view | LLL | LLH | LHL | LHH | HLL | HLH | HHL | HHH |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | COVID-19 | 91.22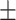 1.78 1.78 |
85.86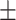 1.59 1.59 |
88.14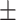 0.94 0.94 |
87.86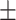 0.72 0.72 |
87.41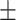 1.23 1.23 |
85.09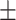 0.82 0.82 |
87.72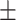 0.92 0.92 |
88.21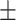 0.99 0.99 |
89.00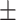 0.93 0.93 |
| Non-COVID-19 | 89.63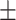 1.14 1.14 |
88.63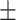 0.85 0.85 |
85.21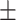 1.32 1.32 |
88.51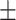 0.58 0.58 |
84.43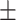 0.96 0.96 |
86.74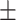 0.52 0.52 |
84.54 0.63 0.63 |
86.16 0.83 0.83 |
84.12 0.84 0.84 |
|
| Normal | 90.85 1.34 1.34 |
88.48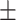 1.02 1.02 |
87.52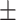 0.69 0.69 |
90.20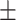 0.59 0.59 |
89.32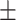 0.94 0.94 |
89.27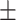 0.99 0.99 |
89.03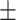 0.75 0.75 |
89.48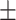 0.99 0.99 |
88.86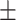 1.03 1.03 |
|
| RF | COVID-19 | 96.88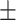 0.56 0.56 |
84.92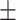 1.21 1.21 |
94.62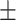 0.83 0.83 |
90.17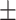 0.92 0.92 |
94.60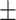 0.82 0.82 |
87.74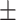 1.11 1.11 |
94.43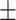 0.41 0.41 |
94.03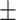 0.74 0.74 |
94.47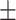 1.05 1.05 |
| Non-COVID-19 | 89.68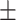 1.12 1.12 |
87.62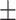 0.73 0.73 |
87.46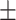 0.68 0.68 |
84.74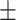 0.92 0.92 |
86.17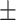 0.92 0.92 |
85.68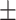 0.98 0.98 |
86.51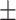 0.98 0.98 |
86.39 0.95 0.95 |
86.52 0.81 0.81 |
|
| Normal | 91.09 1.10 1.10 |
89.53 0.77 0.77 |
89.40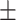 0.40 0.40 |
90.27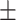 0.96 0.96 |
88.54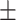 0.70 0.70 |
88.00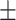 1.25 1.25 |
88.54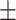 1.10 1.10 |
89.35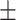 0.50 0.50 |
87.37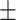 0.97 0.97 |
|
| SVM | COVID-19 | 93.39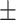 2.43 2.43 |
87.46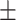 1.56 1.56 |
92.27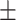 0.72 0.72 |
90.03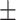 1.05 1.05 |
89.98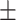 1.22 1.22 |
88.66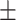 0.97 0.97 |
90.72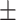 6.23 6.23 |
92.40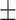 1.31 1.31 |
91.02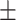 1.34 1.34 |
| Non-COVID-19 | 88.96 0.99 0.99 |
87.37 1.02 1.02 |
82.60 1.22 1.22 |
87.15 0.85 0.85 |
80.39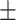 0.89 0.89 |
83.26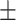 1.19 1.19 |
78.03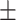 0.76 0.76 |
82.27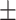 1.33 1.33 |
81.60 1.50 1.50 |
|
| Normal | 90.64 1.39 1.39 |
88.68 0.79 0.79 |
88.97 1.02 1.02 |
88.92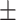 0.52 0.52 |
91.91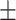 0.89 0.89 |
90.80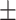 0.62 0.62 |
93.17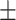 0.98 0.98 |
89.76 0.76 0.76 |
89.57 0.95 0.95 |
|
| DNN | COVID-19 | 96.51 0.86 0.86 |
86.48 1.25 1.25 |
93.07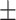 0.70 0.70 |
90.41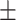 1.51 1.51 |
94.17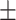 1.07 1.07 |
88.41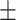 1.40 1.40 |
94.77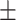 0.93 0.93 |
94.75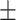 0.82 0.82 |
93.71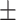 0.99 0.99 |
| Non-COVID-19 | 90.47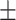 1.14 1.14 |
87.78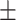 1.86 1.86 |
87.33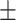 0.74 0.74 |
87.36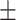 1.25 1.25 |
86.97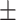 1.17 1.17 |
88.98 1.17 1.17 |
86.98 0.87 0.87 |
87.84 1.39 1.39 |
86.15 0.96 0.96 |
|
| Normal | 89.24 0.87 0.87 |
89.63 0.99 0.99 |
87.41 0.98 0.98 |
88.47 1.11 1.11 |
84.76 1.48 1.48 |
87.08 1.28 1.28 |
86.02 1.24 1.24 |
84.84 0.73 0.73 |
86.45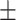 0.89 0.89 |
|
| DSAE | COVID-19 | 97.00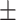 0.73 0.73 |
90.18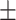 1.72 1.72 |
93.75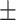 0.46 0.46 |
93.34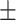 1.02 1.02 |
94.33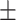 0.38 0.38 |
92.88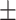 0.64 0.64 |
94.59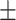 0.75 0.75 |
95.98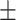 0.98 0.98 |
96.21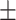 0.99 0.99 |
| Non-COVID-19 | 91.06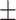 0.77 0.77 |
87.83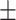 1.72 1.72 |
88.08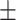 1.10 1.10 |
87.47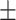 1.64 1.64 |
84.75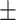 1.69 1.69 |
86.76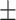 1.44 1.44 |
85.76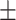 1.06 1.06 |
85.73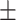 1.39 1.39 |
86.26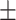 0.99 0.99 |
|
| Normal | 92.04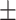 0.84 0.84 |
90.71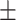 0.48 0.48 |
90.93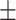 1.07 1.07 |
90.96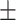 0.90 0.90 |
91.56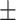 1.64 1.64 |
90.86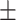 0.96 0.96 |
91.19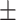 1.10 1.10 |
90.61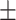 0.95 0.95 |
90.50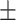 1.01 1.01 |
Note that LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH represent different frequency features extracted from the multiple frequency subbands.
TABLE 6. Mean F1-Score of the Proposed Method and Compared Methods Based on the Primary Cohort.
| Method | Class | Multi-view | LLL | LLH | LHL | LHH | HLL | HLH | HHL | HHH |
|---|---|---|---|---|---|---|---|---|---|---|
| LR | COVID-19 | 87.32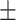 1.96 1.96 |
78.84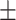 1.07 1.07 |
80.88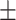 0.83 0.83 |
83.40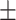 0.72 0.72 |
83.68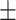 1.29 1.29 |
81.35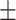 0.95 0.95 |
84.59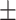 0.49 0.49 |
83.98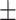 0.76 0.76 |
84.78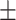 0.85 0.85 |
| Non-COVID-19 | 74.91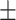 2.27 2.27 |
69.07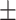 1.53 1.53 |
66.30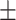 1.94 1.94 |
70.30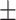 0.78 0.78 |
66.44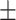 0.92 0.92 |
68.47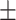 1.52 1.52 |
66.82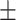 1.05 1.05 |
70.60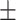 0.98 0.98 |
66.84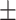 1.63 1.63 |
|
| Normal | 77.88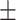 2.31 2.31 |
76.73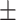 0.77 0.77 |
71.03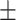 1.54 1.54 |
76.54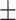 1.41 1.41 |
67.51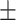 1.61 1.61 |
69.12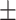 1.32 1.32 |
65.35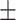 0.82 0.82 |
68.92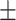 1.71 1.71 |
66.93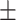 1.67 1.67 |
|
| RF | COVID-19 | 92.39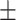 0.60 0.60 |
78.77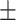 1.17 1.17 |
90.33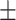 0.78 0.78 |
84.05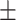 0.76 0.76 |
90.03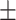 0.86 0.86 |
82.55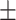 1.19 1.19 |
90.42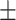 0.73 0.73 |
90.00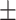 0.69 0.69 |
89.45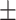 0.65 0.65 |
| Non-COVID-19 | 78.72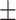 1.84 1.84 |
68.26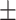 1.86 1.86 |
73.41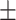 1.20 1.20 |
69.97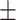 1.23 1.23 |
71.31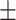 1.38 1.38 |
68.14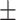 1.93 1.93 |
71.71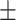 1.23 1.23 |
72.68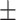 1.14 1.14 |
72.35 0.93 0.93 |
|
| Normal | 79.43 2.70 2.70 |
76.38 1.47 1.47 |
73.53 1.14 1.14 |
72.73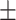 1.81 1.81 |
70.84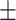 1.32 1.32 |
67.79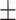 1.25 1.25 |
69.97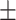 1.20 1.20 |
70.60 1.97 1.97 |
68.02 1.49 1.49 |
|
| SVM | COVID-19 | 88.34 2.47 2.47 |
79.24 0.89 0.89 |
83.77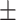 0.96 0.96 |
83.52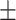 0.72 0.72 |
85.31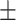 1.03 1.03 |
82.99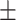 1.06 1.06 |
86.57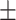 0.68 0.68 |
85.15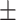 1.21 1.21 |
85.28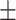 1.11 1.11 |
| Non-COVID-19 | 75.54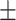 2.19 2.19 |
69.10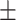 1.60 1.60 |
68.30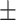 1.59 1.59 |
70.07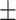 1.05 1.05 |
68.80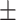 1.13 1.13 |
69.12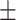 1.47 1.47 |
68.55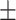 1.15 1.15 |
70.62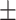 1.71 1.71 |
67.75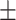 2.05 2.05 |
|
| Normal | 78.36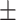 2.36 2.36 |
77.33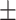 0.68 0.68 |
71.56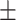 2.46 2.46 |
75.45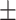 1.67 1.67 |
64.64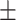 2.00 2.00 |
69.52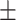 1.48 1.48 |
61.42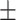 0.82 0.82 |
67.51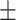 3.19 3.19 |
65.50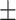 1.91 1.91 |
|
| DNN | COVID-19 | 92.15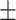 0.54 0.54 |
79.79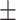 0.83 0.83 |
87.39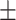 0.52 0.52 |
84.92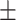 1.46 1.46 |
87.80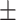 1.16 1.16 |
84.37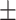 1.03 1.03 |
89.70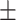 0.73 0.73 |
88.03 1.22 1.22 |
88.05 0.81 0.81 |
| Non-COVID-19 | 76.25 1.52 1.52 |
68.27 1.61 1.61 |
71.68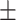 0.97 0.97 |
70.61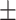 0.99 0.99 |
67.46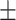 2.08 2.08 |
67.06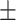 1.89 1.89 |
69.22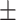 2.05 2.05 |
69.62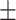 1.60 1.60 |
68.60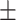 1.51 1.51 |
|
| Normal | 78.99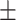 1.30 1.30 |
78.75 1.62 1.62 |
73.00 0.91 0.91 |
74.20 1.51 1.51 |
70.17 1.74 1.74 |
73.05 1.76 1.76 |
69.59 1.75 1.75 |
70.78 2.30 2.30 |
69.67 1.51 1.51 |
|
| DSAE | COVID-19 | 93.24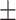 0.56 0.56 |
81.87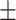 1.09 1.09 |
89.76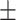 0.67 0.67 |
87.36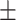 0.63 0.63 |
90.55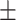 0.57 0.57 |
88.27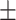 0.64 0.64 |
90.86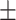 0.59 0.59 |
90.93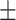 0.78 0.78 |
91.82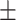 0.42 0.42 |
| Non-COVID-19 | 80.51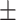 1.06 1.06 |
73.00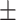 1.65 1.65 |
75.07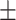 1.24 1.24 |
75.03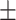 1.30 1.30 |
72.81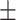 1.65 1.65 |
73.78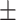 1.07 1.07 |
73.79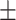 1.30 1.30 |
74.80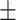 1.18 1.18 |
75.31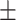 1.24 1.24 |
|
| Normal | 82.17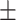 1.08 1.08 |
81.61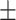 0.88 0.88 |
76.16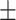 1.74 1.74 |
77.89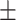 1.39 1.39 |
72.31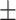 2.25 2.25 |
74.62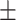 1.44 1.44 |
72.89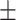 1.24 1.24 |
73.37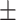 1.48 1.48 |
72.53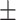 1.16 1.16 |
Note that LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH represent different frequency features extracted from the multiple frequency subbands.
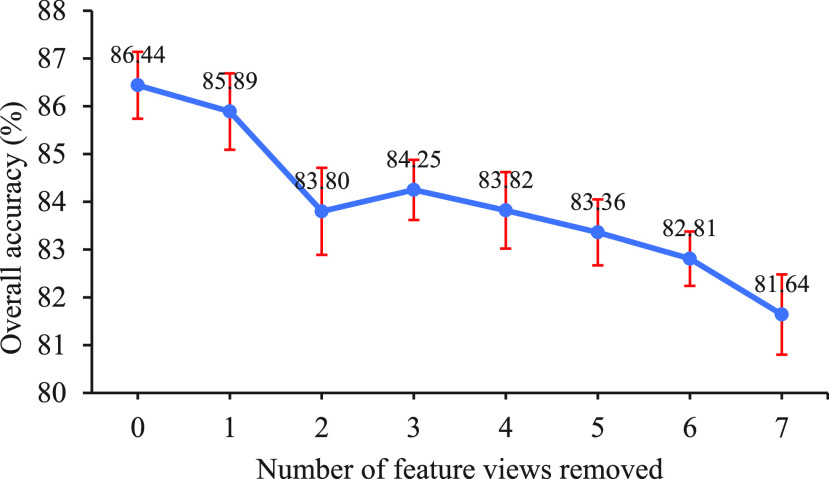
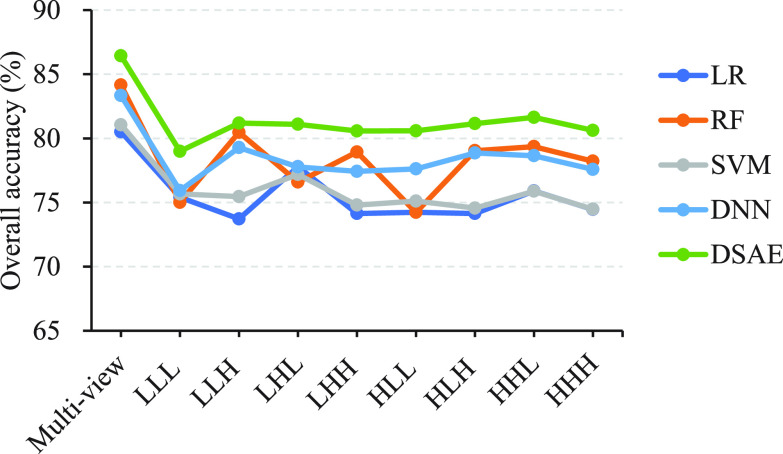
To further demonstrate the strong diagnostic power of multi-view features, we conducted experiments to explore the overall accuracy of our proposed model under different combinations of feature views. For simplicity, we randomly removed the feature views one by one and performed 10 five-fold cross-validation experiments on the primary cohort. Fig. 3 reveals the performance trend of our proposed method as the number of feature views removed varies from 0 to 7. We can see the overall accuracy becomes significantly lower as the number of feature views removed increases, which strongly supports the need to combine feature views of eight different frequencies. Fig. 4 shows the performance trends of our proposed method and the comparaed methods across different frequency features. It reveals that the performance of the methods using multi-view features is better than that using individual frequency features.
Fig. 3.
Overall accuracy of the proposed method (DSAE) with the number of feature views removed. The order of randomly removing feature views one by one is HLL, LLL, LHH, HHH, LHL, HLH, and LLH.
Fig. 4.
Overall accuracy for multi-view features and different frequency features across different methods.
3.3. Efficacy and Discovery of Latent Representations
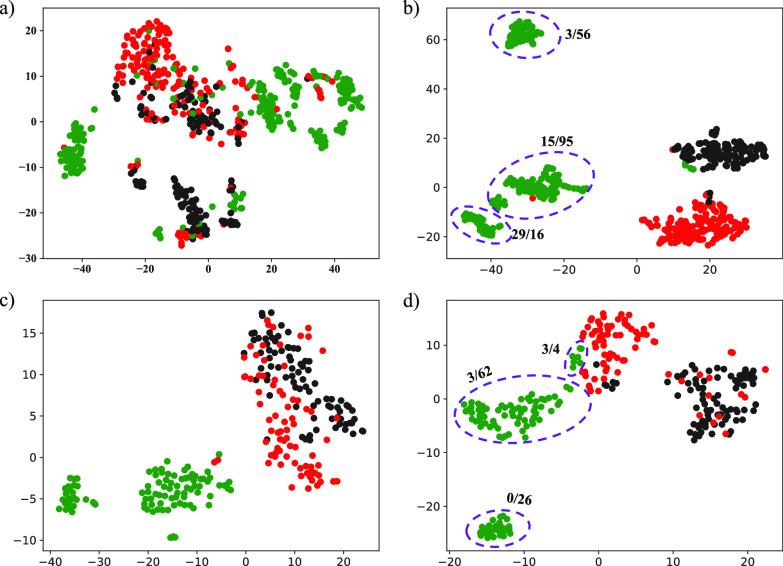
To demonstrate the effectiveness of latent representations, we visualized the learned features in representation layer and original multi-view features in both primary and validation cohorts. Fig. 5 shows the distributions of the original multi-view features and the latent representations and vividly illustrates that the latent representations are more informative and structured compared to the original multi-view features. More specifically, the visualization results in Fig. 5a) show that the underlying class structure is not well revealed for the original multi-view features, while Fig. 5b) indicates that the latent representations learned from original multi-view features and classes are more informative and well-structured. As expected, Fig. 5c) and Fig. 5d) also illustrate a similar situation in the validation cohort. As shown in Tables 2, 3, 4, 5, and 6, we can also observe that the performance with latent representations is better than that without latent representations. For example, the DSAE achieved an overall accuracy of 86.44 percent, which is 3.10 percent higher than the DNN without learning latent representations.
Fig. 5.
Visualization of the original multi-view features and the latent representations in the primary cohort and validation cohort. For the primary cohort, the class structure of the original multi-view features in a) is poor, while the structure of the learned latent representations in b) is much better and consistent with classes. A similar situation is observed in the validation cohort, as shown in c) and d). Green, red and black represent COVID-19, non-COVID-19 and normal cases, respectively. Numerical symbol (n/m) around the purple circle indicates that n is the number of severe cases and m is the number of non-server cases.
In addition, we discovered the following two phenomena from Fig. 5. One is that there is a certain margin between the COVID-19 patients and the others in the original multi-view features (Fig. 5a) and Fig. 5c)). This indicates that the feature extraction method we designed is beneficial to distinguishing COVID-19 patients from others. The other is that the learned latent representations have an internal class structure in COVID-19 patients (Fig. 5b) and Fig. 5d)). This means that COVID-19 patients can be further classified into multiple sub-clusters using our proposed method. We will further discuss this in Section 4.
3.4. Comparison With Other Methods
Fig. 4 and Table 2 show the overall accuracy of the triple classification task of the proposed method and compared methods. Obviously, the proposed method achieved the best overall accuracy up to 84.66 percent by multi-view features learning. Compared to the radiomics-based methods, our latent representation-based approach improved the overall accuracy by 2.28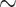 5.93 percent in multi-view features learning. To further demonstrate the effectiveness of our proposed method, we used the DNN model for different frequency features and multi-view features to directly learn the mapping from the original features to the class labels. The experimental results show that the performance of DNN model is comparable to that of radiomics-based methods, but much worse than that of our proposed DSAE model. This indicates that the DSAE model can achieve a better predictive performance by multi-task learning.
5.93 percent in multi-view features learning. To further demonstrate the effectiveness of our proposed method, we used the DNN model for different frequency features and multi-view features to directly learn the mapping from the original features to the class labels. The experimental results show that the performance of DNN model is comparable to that of radiomics-based methods, but much worse than that of our proposed DSAE model. This indicates that the DSAE model can achieve a better predictive performance by multi-task learning.
Tables 3, 4, 5, and 6 also show the performance of the proposed method and comparison methods on the three diagnostic tasks. Compared with other methods, our proposed method overall achieved the best performance in each diagnostic task across different frequency features and multi-view features. More precisely, when identifying COVID-19 from others, our proposed method achieved the best performance in multi-view features learning, with an accuracy of 94.61 percent, a sensitivity of 91.22 percent, a specificity of 97.00 percent, and a F1-score of 93.24 percent. When distinguishing non-COVID-19 from others, the proposed method achieved the best performance in multi-view features learning, with an accuracy of 87.60 percent, a sensitivity of 80.32 percent, a specificity of 91.06 percent and a F1-score of 80.51 percent. Similarly, it achieved the best diagnostic performance, an accuracy of 93.38 percent, a sensitivity of 85.63 percent, a specificity of 92.04 percent and a F1-score of 82.17 percent, in distinguishing normal subjects from COVID-19 and non-COVID-19 patients. As seen from the performance differences of three diagnostic tasks, our proposed model has the strongest ability to identify COVID-19 subjects, followed by normal and non-COVID-19 subjects.
3.5. Independent Validation
To further validate our proposed method, we performed independent validation experiments based on the validation cohort presented in Table 1. More specifically, we used the primary cohort as training data to obtain more generalized models and used validation cohort to evaluate the performance. Fig. 6A) shows the confusion matrix of our proposed method, and Fig. 6B) shows the overall accuracy of our proposed method and the compared methods of the triple classification task on the validation cohort. It was observed that our proposed method achieves the best overall accuracy with a value of 89.53 percent in independent validation cohort. Although our proposed DSAE achieves promising performance, there are still 10 COVID-19 cases, 14 non-COVID-19 cases and 3 normal cases that are misdiagnosed. We will further discuss the underlying reason of misclassification in the next section. Moreover, Table 7 presents the corresponding diagnostic performance in terms of accuracy, sensitivity, specificity and F1-score under the one-vs-rest strategy. As shown in Table 7, our proposed method has a consistent pattern across the three binary classification tasks and achieves the best diagnostic performance compared with other methods. Although the diagnostic performance of our proposed method is similar to that of the RF-based approach on the primary cohort, our proposed DSAE model performs better on the validation cohort. For example, the accuracy of our proposed DSAE model is 1.55, 7.76, and 6.20 percent higher in distinguishing each category than that of the RF-based approach, respectively. Moreover, the radiomics-based methods are less sensitive in distinguishing normal subjects from other patients, while our proposed method and DNN-based method are more sensitive.
Fig. 6.
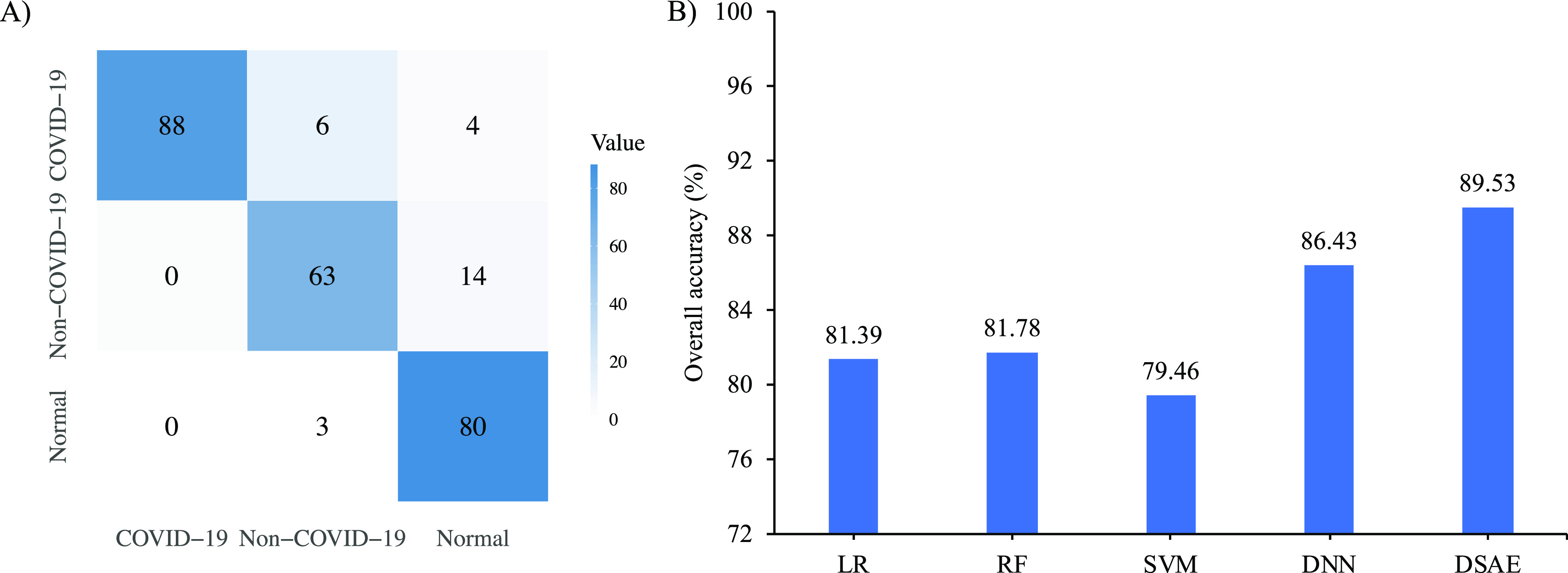
Diagnostic performance of triple classification task on the validation cohort. A) is the confusion matrix of the proposed method; and B) is the overall accuracy of the proposed method and compared methods.
TABLE 7. Diagnostic Performance of the Proposed Method and Compared Methods on the Validation Cohort.
| Method | Class | ACC(%) | SEN(%) | SPE(%) | F1(%) |
|---|---|---|---|---|---|
| LR | COVID-19 | 92.25 | 96.94 | 89.38 | 90.48 |
| Non-COVID-19 | 86.82 | 87.01 | 86.74 | 79.76 | |
| Normal | 83.72 | 57.83 | 96.00 | 69.57 | |
| RF | COVID-19 | 94.57 | 96.94 | 93.14 | 93.14 |
| Non-COVID-19 | 83.33 | 76.62 | 86.18 | 73.29 | |
| Normal | 85.66 | 68.67 | 93.71 | 75.50 | |
| SVM | COVID-19 | 92.64 | 92.86 | 92.50 | 80.55 |
| Non-COVID-19 | 85.66 | 88.31 | 84.53 | 78.61 | |
| Normal | 82.95 | 59.04 | 94.29 | 69.01 | |
| DNN | COVID-19 | 95.74 | 88.78 | 100.00 | 94.05 |
| Non-COVID-19 | 87.21 | 75.32 | 92.26 | 77.85 | |
| Normal | 89.92 | 93.98 | 88.00 | 85.71 | |
| DSAE | COVID-19 | 96.12 | 89.80 | 100.00 | 94.62 |
| Non-COVID-19 | 91.09 | 81.82 | 95.03 | 84.56 | |
| Normal | 91.86 | 96.39 | 89.71 | 88.40 |
To further prove the diagnosis power of the purposed methods, other radiomics features such as shape-based features (14 features), and gray and texture features (93 features, named as original features) extracted directly from original images were also used to distinguish the COVID-19 pneumonia from others using the proposed DSAE method. As shown in Table 8, the DSAE method using shape features achieves the lowest overall accuracy with an value of 46.51 percent, which means that only using shape-based features are difficult to screen out COVID-19 pneumonia from others. The method using gray and texture features achieves a moderate performance with an overall accuracy of 77.13 percent. However, the method using our designed multi-view features achieves encouraging performance with an overall accuracy of 89.53 percent. This demonstrates that the multi-view feature we designed has a strong diagnostic power for distinguishing the COVID-19 pneumonia from others. Moreover, we implemented multiple works [19], [20], [23], [25] to compare the performance. The comparison performance is shown in Table 8. We found that the representation learning-based method [23] performed better than the transfer learning-based methods [19], [20], [25]. More precisely, the overall accuracy of the literature [23] is 10.85 percent higher than that of the literature [25], which suggests that the radiomic features have a strong predictive power for identifying COVID-19. However, our proposed method is based on multi-view representation learning and achieves better performance compared with these methods.
TABLE 8. Diagnostic Performance on the Validation Cohort When Distinguishing COVID-19 From Other Subjects.
| Method | Class | ACC (%) | SEN (%) | SPE (%) | F1 (%) | Overall accuracy(%) |
|---|---|---|---|---|---|---|
| Li et al. [19] | COVID-19 | 76.36 | 63.27 | 84.38 | 67.03 | 63.95 |
| Non-COVID-19 | 79.07 | 65.39 | 85.00 | 65.39 | ||
| Normal | 72.48 | 63.42 | 76.71 | 59.43 | ||
| Shah et al. [25] | COVID-19 | 79.85 | 59.18 | 92.50 | 69.05 | 71.32 |
| Non-COVID-19 | 80.22 | 79.49 | 80.56 | 70.86 | ||
| Normal | 82.56 | 78.05 | 84.66 | 73.99 | ||
| Bai et al. [20] | COVID-19 | 70.93 | 38.78 | 90.63 | 50.33 | 62.02 |
| Non-COVID-19 | 75.19 | 75.64 | 75.00 | 64.84 | ||
| Normal | 77.91 | 76.83 | 78.41 | 68.85 | ||
| Kang et al. [23] | COVID-19 | 94.57 | 88.78 | 98.13 | 92.55 | 82.17 |
| Non-COVID-19 | 83.72 | 54.55 | 96.13 | 66.67 | ||
| Normal | 86.05 | 100.00 | 79.43 | 82.18 | ||
| DSAE (Shape) | COVID-19 | 57.36 | 36.74 | 70.00 | 39.56 | 46.51 |
| Non-COVID-19 | 65.89 | 50.65 | 72.38 | 46.99 | ||
| Normal | 69.77 | 54.22 | 77.14 | 53.57 | ||
| DSAE (Original) | COVID-19 | 87.21 | 68.37 | 98.75 | 80.24 | 77.13 |
| Non-COVID-19 | 82.17 | 72.73 | 86.19 | 70.89 | ||
| Normal | 84.88 | 91.56 | 81.71 | 79.58 | ||
| DSAE (Multi-view) | COVID-19 | 96.12 | 89.80 | 100.00 | 94.62 | 89.53 |
| Non-COVID-19 | 91.09 | 81.82 | 95.03 | 84.56 | ||
| Normal | 91.86 | 96.39 | 89.71 | 88.40 |
4. Discussion
The COVID-19 pandemic has infected more than 14 million patients worldwide and quickly become a major global health threat [2]. Computer-aided diagnosis systems are playing an increasingly important role in the diagnosis and monitoring of COVID-19, which can reduce the burden of radiologists and help them make clinical decisions. In this study, we proposed an efficient and accurate diagnosis system for automatically differentiating COVID-19 pneumonia from other pneumonia and normal subjects. Compared with previous studies [17], [18], [20], [51] which require manual or semi-automatic lesions annotation from CT images, our proposed method only needs an easy-to-implement preprocessing step, that is, the use of AI model to automatically segment the lung regions from CT images. It also does not require select key slices to represent a full 3D CT scan, but only uses 3D-WT technique to decompose the 3D lung images into multiple frequency subbands for exploiting multi-view features. More importantly, compared to the CNN-based methods, our proposed method can achieve promising performance on a limited amount of data.
Leveraging the complementarity of multiple views, multi-view representation learning is capable of learning more informative and compact representations for improving predictive performance as proved in this study (see Table 2 and Fig. 5). Moreover, visualization of latent representations in Fig. 5b) and Fig. 5d) has revealed an internal class structure in COVID-19 subjects. We further retrospectively investigated severity of COVID-19 (non-severe and severe) based on clinical assessment criteria. Statistical results presented in Fig. 5b) and Fig. 5d) show that non-severe and severe are not completely separated, but there are three types of structures with a high, medium, and low presence probability of severe subjects. The reason for this difference may be that the severity assessment criteria are not fully derived from CT imaging. In fact, CT evaluation has little reference value in clinical classification [52].
Automatically identifying patients with abnormal CT findings and further screening out COVID-19 pneumonia from other pneumonia is urgently needed in the clinical practice. Inspired by the clinical requirements, we proposed a classification system to address the task. Despite the promising performance, 14 cases with non-COVID-19 pneumonia were identified as normal cases (see Fig. 6A)). After carefully reviewed the misdiagnosed cases by radiologists, 9 cases had small and low-density lesions, which were too subtle to detect. This circumstance was also presented in COVID-19 group. Among 10 misdiagnosed cases characterized by our system in COVID-19 group, 4 cases with small and low-density lesions were identified as normal cases. This means that the sensitivity of our system requires further improvement. Other 6 misdiagnosed cases in COVID-19 group had non-typical imaging manifestations due to the potential of a relatively later stage of the disease. Although we only used the first CT scans of each case, the interval time varied between onset of symptoms and first CT scans. We stated it as one of our limitations. Three cases with normal CT findings were misdiagnosed as non-COVID-19 pneumonia. All the 3 failure cases had false lesions due to the relatively higher density of the posterior lung. It is worth noting that none failure cases in normal and non-COVID-19 group were identified as COVID-19 pneumonia, indicating a substantial inherent difference between COVID-19 group and other two groups. In clinical practice, it is easy to identify normal and abnormal lung CT findings. Therefore, we further investigated the binary classification, that is, differentiating COVID-19 pneumonia from non-COVID-19 pneumonia. On independent validation cohort, the diagnostic results showed that only two cases with small and low-density lesions from COVID-19 group were identified as non-COVID-19 cases, and three cases in non-COVID-19 group were identified as COVID-19 cases (see Fig. 7A)). Specifically, our proposed DSAE achieved encouraging predictive performance with an accuracy of 97.14 percent, a sensitivity of 96.10 percent, a specificity of 97.37 percent and an F1-score of 96.73 percent (see Fig. 7B)). As expected, the performance of binary classification task outperformed that of triple classification task. Actually, the triple classification task is naturally more difficult than the binary classification task.
Fig. 7.
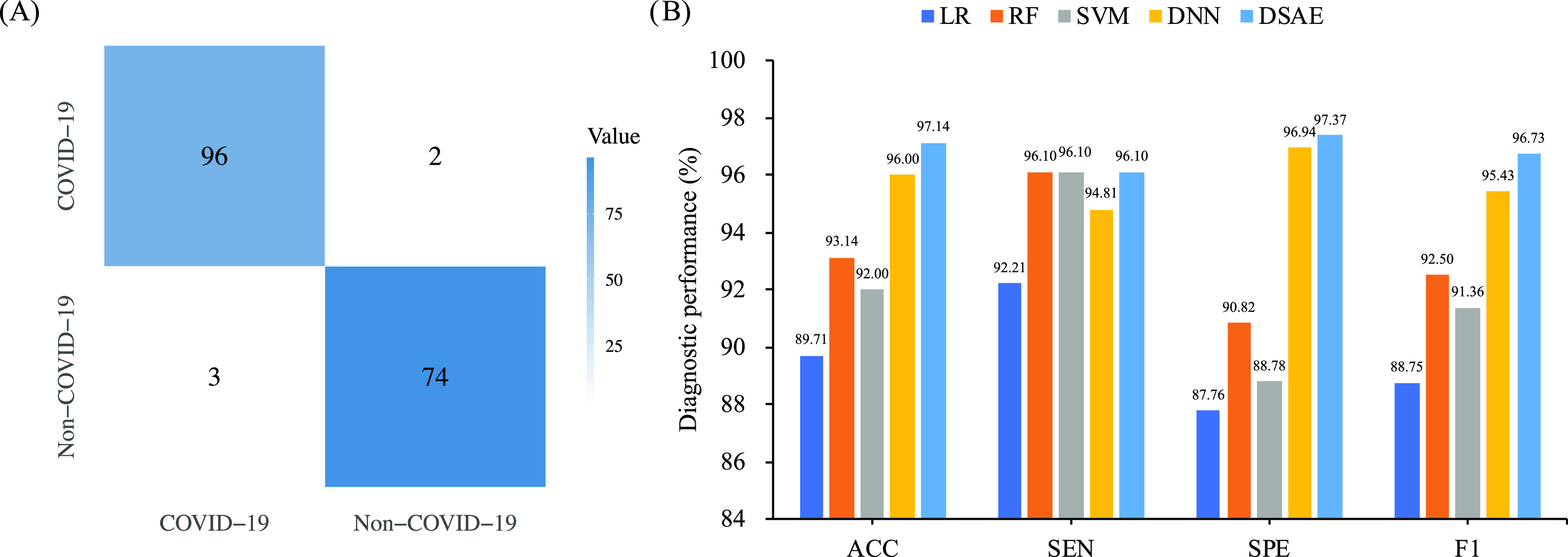
Diagnostic performance of binary classification task (COVID-19 versus non-COVID-19) on the validation cohort. A) is the confusion matrix of the proposed method; and B) is the diagnostic performance of the proposed method and compared methods.
Training a deep learning model with high generalization, especially in multiple classification tasks, may require the use of more samples. One of the biggest advantages of using deep learning approach is the ability to automatically learn the latent features associated with pneumonia diseases. However, it still lacks interpretability and can’t extract quantitative features as the same as radiomic features. Therefore, we used radiomic features from the multiple frequency subbands as the multi-view features for the diagnosis of COVID-19 in this study. Moreover, we further investigated whether our approach was influenced by age. Considering the size and imbalance of samples, we divided the validation cohort into three groups based on age. Table 9 shows the diagnostic performance based on three age groups. We found that our proposed method yielded consistently good performance across age groups and was more sensitive to COVID-19 over the age of 40. Therefore, our model is not affected by age differences when distinguishing COVID-19 from non-COVID-19 pneumonia.
TABLE 9. Diagnostic Performance of Different Age Groups of the Proposed Method on the Validation Cohort When Distinguishing COVID-19 From Non-COVID-19 Subjects.
| Age group | ACC (%) | SEN (%) | SPE (%) | F1 (%) |
|---|---|---|---|---|
| 0-40 | 95.39 | 92.00 | 97.50 | 93.88 |
| 40-60 | 98.36 | 96.00 | 100.00 | 97.96 |
| 60-100 | 97.96 | 100.00 | 95.46 | 98.18 |
Our study still comprises some limitations. First, only radiomic features are used, the deep learning features may have the potential to identify COVID-19 pneumonia. Next work will collect more data and use the CNN-based methods to automatically learn the latent features associated with pneumonia diseases. Second, we only included the first CT scans, thus the longitudinal CT changes were not investigated. Whether our model has the same performance in identifying different stages of COVID-19 pneumonia from other pneumonia is unclear. Moreover, only early-stage patients of the pandemic are considered in this study, the patients infected with SARS-CoV-2 variants are further investigated. Future work will focus on collecting more data on patients after virus mutations to validate the performance of the model. Third, our deep learning model only integrated chest CT features without involving the clinical information such as symptoms, exposure history, and so on. A recent research has reported that combining CT imaging and clinical information can improve the diagnostic performance of AI model [51]. We will further validate it in future work. Finally, the interpretability of the deep learning system remains unclear, and the clinical meaning of the feature learned by the system is difficult to explain. Actually, we have investigated the visualization of the original multi-view features and the latent representations in two cohorts to mine the inherent mechanism. However, the further investigation is needed in the future work.
5. Conclusion
In conclusion, we proposed an easy-to-use diagnostic method based on multi-view representation learning, which used 3D CT images to rapidly screen out COVID-19 from other pneumonia and normal subjects without abnormal CT findings. Our proposed diagnostic model achieved an overall accuracy of 89.54 percent in the triple classification task. When only considering to distinguish COVID-19 from non-COVID-19 pneumonia, the model had a more generalization performance with an accuracy of 97.14 percent, a sensitivity of 96.10 percent, a specificity of 97.37 percent and an F1-score of 96.73 percent. Comprehensive results have demonstrated that our proposed method has great potential in accurately and rapidly screening out COVID-19 pneumonia, which is beneficial to fight the current disease outbreak.
Acknowledgments
This work was supported in part by Key Emergency Project of Pneumonia Epidemic of novel coronavirus infection under Grant 2020SK3006, Emergency Project of Prevention and Control for COVID-19 of Central South University under Grant 160260005, Foundation from Changsha Scientific and Technical bureau, China under Grant kq2001001 National Natural Science Foundation of China under Grants 61802442, 61877059, Natural Science Foundation of Hunan Province under Grant 2019JJ50775, 111 Project under Grant B18059, the Hunan Provincial Science and Technology Program under Grant 2018WK4001, the Hunan Provincial Science and Technology Innovation Leading Plan under Grant 2020GK2019, the Science and Technology Innovation Program of Hunan Province under Grant 2020SK53423, and Clinical Research Center for Medical Imaging In Hunan Province under Grant 2020SK4001. Jianhong Cheng and Wei Zhao are with equal contribution.
Biographies

Jianhong Cheng received the BS degree from Liaoning Technical University, Fuxin, China, in 2014, the MS degree in software engineering from Central South University, Changsha, China, in 2017. He is currently working toward the PhD degree in the School of Computer Science and Engineering, Central South University, Changsha, China. His research interests include machine learning, deep learning, and medical image analysis.

Wei Zhao received the PhD degree in imaging and nuclear medicine from Fudan University, China. He is a radiologist of The Second Xiangya Hospital. His research interests include chest CT imaging, radiomics, and deep learning.

Jin Liu (Member, IEEE) received the PhD degree in computer science from Central South University, China, in 2017. He is currently a lecturer at the School of Computer Science and Engineering, Central South University, Changsha, Hunan, China. His current research interests include medical image analysis, machine learning, and bioinformatics.

Xingzhi Xie received the bachelor’s degree in clinical medicine from Central South University, China. She is currently working toward the graduate degree in imaging and nuclear medicine of The Second Xiangya Hospital. Her research interests include CT imaging, radiomics, and deep learning.

Shangjie Wu received the bachelor’s degree of clinical medicine from the Central South University, China, the master’s degree of DME from Fudan University, China, and the PhD degree from Central South University, China. She is postdoctoral fellow at the Cancer Research Institute of Central South University, China. She is visiting scholar at Imperial College London, U.K. Her research interests include basic and clinical research of pulmonary vascular diseases, Venous Thrombus Embolism, pulmonary hypertension and Evidence-based Medicine.

Liangliang Liu received the MS degree from Henan University, China, in 2014, and the PhD degree from the School of Computer Science and Engineering, Central South University, Changsha, China, in 2020. He worked as a visiting scholar with Tulane University, New Orleans, Louisiana, from 2019 to 2020. He is an distinguished professor with the College of Information and Management Science, Henan Agricultural University, Zhengzhou, China, since 2021. His research interests include machine learning, deep learning, and medical image analysis.

Hailin Yue received the BS degree from NanJing XiaoZhuang University, Nanjing, China, in 2019. He is currently working toward the MS degree in the School of Computer Science and Engineering, Central South University, Changsha, China. His research interests include machine learning, deep learning, and medical image analysis.

Junjian Li received the BS degree from the Hefei University of Technology, Hefei, China, in 2019. He is currently working toward the MS degree in the School of Computer Science and Engineering, Central South University, Changsha, China. His research interests include unsupervised learning, self-supervised learning, and medical image analysis.

Jianxin Wang (Senior Member, IEEE) received the BS and MS degrees in computer science and application from the Central South University of Technology, China, and the PhD degree in computer science and technology from Central South University, China. Currently, he is the dean and a professor in the School of Computer Science and Engineering, Central South University, Changsha, Hunan, China. He is also a leader in Hunan Provincial Key Lab on Bioinformatics, Central South University, Changsha, Hunan, China. His current research interests include algorithm analysis and optimization, parameterized algorithm, bioinformatics and computer network. He has published more than 200 papers in various International journals and refereed conferences. He has been on numerous program committees and NSFC review panels, and served as editors for several journals such as the IEEE/ACM Transactions Computational Biology and Bioinformatics (TCBB), the International Journal of Bioinformatics Research and Applications, the Current Bioinformatics, the Current Protein & Peptide Science, the Protein & Peptide Letters.

Jun Liu is the director of Radiology Department of The Second Xiangya Hospital. He is also the leader of 225 subjects in Hunan Province, National member of the Neurology Group of the Chinese Society of Radiology, National Committee of the Neurology Group of the Radiological Branch of the Chinese Medical Association. His research interests include brain functional imaging, radiomics, and deep learning.
Funding Statement
This work was supported in part by Key Emergency Project of Pneumonia Epidemic of novel coronavirus infection under Grant 2020SK3006, Emergency Project of Prevention and Control for COVID-19 of Central South University under Grant 160260005, Foundation from Changsha Scientific and Technical bureau, China under Grant kq2001001 National Natural Science Foundation of China under Grants 61802442, 61877059, Natural Science Foundation of Hunan Province under Grant 2019JJ50775, 111 Project under Grant B18059, the Hunan Provincial Science and Technology Program under Grant 2018WK4001, the Hunan Provincial Science and Technology Innovation Leading Plan under Grant 2020GK2019, the Science and Technology Innovation Program of Hunan Province under Grant 2020SK53423, and Clinical Research Center for Medical Imaging In Hunan Province under Grant 2020SK4001.
Contributor Information
Jianhong Cheng, Email: jianhong_cheng@csu.edu.cn.
Wei Zhao, Email: wei.zhao@csu.edu.cn.
Jin Liu, Email: liujin06@mail.csu.edu.cn.
Xingzhi Xie, Email: xingzhixie123@csu.edu.cn.
Shangjie Wu, Email: wushangjie@csu.edu.cn.
Liangliang Liu, Email: liuhau@126.com.
Hailin Yue, Email: yuehailin@mail.csu.edu.cn.
Junjian Li, Email: 194712147@mail.csu.edu.cn.
Jianxin Wang, Email: jxwang@mail.csu.edu.cn.
Jun Liu, Email: junliu123@csu.edu.cn.
References
- [1].WHO, “Novel coronavirus-China,” Website, Accessed: Jan. 12, 2020, 2020. [Online]. Available: https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/
- [2].WHO, “Coronavirus disease (COVID-19) pandemic,” Website, Accessed: Jan. 29, 2021, 2020. [Online]. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- [3].CDC, “CDC diagnostic tests for COVID-19,” Website, Accessed: Aug. 5, 2020, 2020. [Online]. Available: https://www.cdc.gov/coronavirus/2019-ncov/lab/testing.html
- [4].Ai T. et al. , “Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. E32–E40, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xie X., Zhong Z., Zhao W., Zheng C., Wang F., and Liu J., “Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: Relationship to negative RT-PCR testing,” Radiology, vol. 296, no. 2, pp. E41–E45, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhao W., Zhong Z., Xie X., Yu Q., and Liu J., “Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study,” Amer. J. Roentgenol., vol. 214, no. 5, pp. 1072–1077, 2020. [DOI] [PubMed] [Google Scholar]
- [7].Colombi D. et al. , “Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia,” Radiology, vol. 296, no. 2, pp. E86–E96, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao W., Zhong Z., Xie X., Yu Q., and Liu J., “CT scans of patients with 2019 novel coronavirus (COVID-19) pneumonia,” Theranostics, vol. 10, no. 10, 2020, Art. no. 4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bai H. X. et al. , “Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT,” Radiology, vol. 296, no. 2, pp. E46–E54, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fang Y. et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bernheim A. et al. , “Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection,” Radiology, vol. 295, no. 3, 2020, Art. no. 200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu M., Zeng W., Wen Y., Zheng Y., Lv F., and Xiao K., “COVID-19 pneumonia: CT findings of 122 patients and differentiation from influenza pneumonia,” Eur. Radiol., vol. 30, no. 10, pp. 5463–5469, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y.-D., Satapathy S. C., Zhang X., and Wang S.-H., “COVID-19 diagnosis via DenseNet and optimization of transfer learning setting,” Cogn. Comput., Springer, 2021. [Online]. Available: https://doi.org/10.1007/s12559-020-09776-8 [DOI] [PMC free article] [PubMed]
- [14].Wong H. Y. F. et al. , “Frequency and distribution of chest radiographic findings in COVID-19 positive patients,” Radiology, vol. 296, no. 2, pp. E72–E78, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang S.-H., Nayak D. R., Guttery D. S., Zhang X., and Zhang Y.-D., “COVID-19 classification by CCSHNet with deep fusion using transfer learning and discriminant correlation analysis,” Inf. Fusion, vol. 68, pp. 131–148, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang L., Lin Z. Q., and Wong A., “COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Sci. Rep., vol. 10, no. 1, pp. 1–12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fang M. et al. , “CT radiomics can help screen the coronavirus disease 2019 (COVID-19): A preliminary study,” Sci. China Inf. Sci., vol. 63, no. 7, 2020, Art. no. 172103. [Google Scholar]
- [18].Chen H. J. et al. , “Machine learning-based CT radiomics model distinguishes COVID-19 from other viral pneumonia,” 2020. [Online]. Available: https://doi.org/10.21203/rs.3.rs-32511/v1 [DOI] [PMC free article] [PubMed]
- [19].Li L. et al. , “Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy,” Radiology, vol. 296, no. 2, pp. E65–E71, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bai H. X. et al. , “Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT,” Radiology, vol. 296, no. 3, pp. E156–E165, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X. et al. , “A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2615–2625, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [22].Ouyang X. et al. , “Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2595–2605, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [23].Kang H. et al. , “Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2606–2614, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [24].Han Z. et al. , “Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2584–2594, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [25].Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., and Mehendale N., “Diagnosis of COVID-19 using CT scan images and deep learning techniques,” Emergency Radiol., vol. 28, pp. 497–505, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2016, pp. 770–778. [Google Scholar]
- [27].Tan M. and Le Q., “EfficientNet: Rethinking model scaling for convolutional neural networks,” in Proc. 36th Int. Conf. Mach. Learn., 2019, pp. 6105–6114. [Google Scholar]
- [28].Zhang Y.-D., Satapathy S. C., Liu S., and Li G.-R., “A five-layer deep convolutional neural network with stochastic pooling for chest CT-based COVID-19 diagnosis,” Mach. Vis. Appl., vol. 32, no. 1, pp. 1–13, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang X. et al. , “Diagnosis of COVID-19 pneumonia via a novel deep learning architecture,” J. Comput. Sci. Technol., 2021. [Online]. Available: https://jcst.ict.ac.cn/EN/10.1007/s11390–020-0679-8 [DOI] [PMC free article] [PubMed]
- [30].Liao F., Liang M., Li Z., Hu X., and Song S., “Evaluate the malignancy of pulmonary nodules using the 3-D deep leaky noisy-or network,” IEEE Trans. Neural Netw. Learn. Syst., vol. 30, no. 11, pp. 3484–3495, Nov. 2019. [DOI] [PubMed] [Google Scholar]
- [31].Hofmanninger J., Prayer F., Pan J., Röhrich S., Prosch H., and Langs G., “Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem,” Eur. Radiol. Exp., vol. 4, no. 1, pp. 1–13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Le L., Patterson A., and White M., “Supervised autoencoders: Improving generalization performance with unsupervised regularizers,” in Proc. 32nd Int. Conf. Neural Inf. Process. Syst., 2018, vol. 31, pp. 107–117. [Google Scholar]
- [33].Li X., Morgan P. S., Ashburner J., Smith J., and Rorden C., “The first step for neuroimaging data analysis: DICOM to NIfTI conversion,” J. Neurosci. Methods, vol. 264, pp. 47–56, 2016. [DOI] [PubMed] [Google Scholar]
- [34].Cheng J., Liu J., Liu L., Pan Y., and Wang J., “Multi-level glioma segmentation using 3D U-net combined attention mechanism with atrous convolution,” in Proc. IEEE Int. Conf. Bioinf. Biomed., 2019, pp. 1031–1036. [Google Scholar]
- [35].Liu L., Cheng J., Quan Q., Wu F.-X., Wang Y.-P., and Wang J., “A survey on U-shaped networks in medical image segmentations,” Neurocomputing, vol. 409, pp. 244–258, 2020. [Google Scholar]
- [36].Cheng J., Liu J., Jiang M., Yue H., Wu L., and Wang J., “Prediction of egfr mutation status in lung adenocarcinoma using multi-source feature representations,” in Proc. IEEE Int. Conf. Acoust. Speech Signal Process., 2021, pp. 1350–1354. [Google Scholar]
- [37].Zhang Y. and Yang Q., “A survey on multi-task learning,” IEEE Trans. Knowl. Data Eng., early access, Mar. 31, 2021. [Online]. Available: https://doi.org/10.1109/TKDE.2021.3070203
- [38].Sun S., “A survey of multi-view machine learning,” Neural Comput. Appl., vol. 23, no. 7/8, pp. 2031–2038, 2013. [Google Scholar]
- [39].Vincent P., Larochelle H., Lajoie I., Bengio Y., and Manzagol P.-A., “Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion,” J. Mach. Learn. Res., vol. 11, no. Dec, pp. 3371–3408, 2010. [Google Scholar]
- [40].Wang Y., Yao H., and Zhao S., “Auto-encoder based dimensionality reduction,” Neurocomputing, vol. 184, pp. 232–242, 2016. [Google Scholar]
- [41].Cheng J. et al. , “Multimodal disentangled variational autoencoder with game theoretic interpretability for glioma grading,” IEEE J. Biomed. Health Inform., 2021. [Online]. Available: https://doi.org/10.1109/JBHI.2021.3095476 [DOI] [PubMed]
- [42].Lu C., Wang Z.-Y., Qin W.-L., and Ma J., “Fault diagnosis of rotary machinery components using a stacked denoising autoencoder-based health state identification,” Signal Process., vol. 130, pp. 377–388, 2017. [Google Scholar]
- [43].Ioffe S. and Szegedy C., “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” in Proc. 32nd Int. Conf. Mach. Learn., 2015, pp. 448–456. [Google Scholar]
- [44].Srivastava N., Hinton G., Krizhevsky A., Sutskever I., and Salakhutdinov R., “Dropout: A simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res., vol. 15, no. 1, pp. 1929–1958, 2014. [Google Scholar]
- [45].Wang Y., Liu J., Xiang Y., Wang J., Chen Q., and Chong J., “Mage: Automatic diagnosis of autism spectrum disorders using multi-atlas graph convolutional networks and ensemble learning,” Neurocomputing, 2021. [Online]. Available: https://doi.org/10.1016/j.neucom.2020.06.152
- [46].Cheng J., Liu J., Yue H., Bai H., Pan Y., and Wang J., “Prediction of glioma grade using intratumoral and peritumoral radiomic features from multiparametric MRI images,” IEEE/ACM Trans. Comput. Biol. Bioinf., 2020. [Online]. Available: https://doi.org/10.1109/TCBB.2020.3033538 [DOI] [PubMed]
- [47].Peng C.-Y. J., Lee K. L., and Ingersoll G. M., “An introduction to logistic regression analysis and reporting,” J. Educ. Res., vol. 96, no. 1, pp. 3–14, 2002. [Google Scholar]
- [48].Liaw A. et al. , “Classification and regression by randomforest,” R News, vol. 2, no. 3, pp. 18–22, 2002. [Google Scholar]
- [49].Burges C. J., “A tutorial on support vector machines for pattern recognition,” Data Mining Knowl. Discov., vol. 2, no. 2, pp. 121–167, 1998. [Google Scholar]
- [50].Maaten L. V. D. and Hinton G., “Visualizing data using t-SNE,” J. Mach. Learn. Res., vol. 9, no. Nov, pp. 2579–2605, 2008. [Google Scholar]
- [51].Mei X. et al. , “Artificial intelligence–enabled rapid diagnosis of patients with COVID-19,” Nat. Med., vol. 26, no. 8, pp. 1224–1228, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zheng C., “Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19),” Radiology, vol. 295, no. 3, pp. 715–721, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]