Abstract
Candida auris is a nosocomial fungal pathogen of prime importance due to its global emergence and rapid spread in healthcare facilities worldwide. One important concern is that routine, conventional methods fail to identify C. auris. While molecular and protein-based assays accurately detect/identify C. auris, these methods are time-consuming, expensive, and require expertise. Therefore, the objective of the present study was to assess the potential use of a novel chromogenic medium, CHROMagar™ Candida Plus, as an economical alternative to expensive and laborious diagnostic tests. We compared CHROMagar™ Candida Plus with the standard enrichment (salt Sabouraud Dulcitol broth) medium to test the recovery efficiency of C. auris from surveillance samples. We also tested CHROMagar™ Candida Plus for its ability to distinguish C. auris from other yeast species. One hundred surveillance samples were cultured on CHROMagar™ Candida Plus and Dulcitol broth and incubated at 37 °C and 40 °C, respectively. Additionally, 32 Candida and yeast species were cultured on CHROMagar™ Candida Plus at 37 °C for three days to rule out any close resemblance to C. auris. Of 100 surveillance samples tested, 69 yielded presumptive positive C. auris exhibiting creamy pink colonies with a blue halo on CHROMagar™ Candida Plus within three days of incubation, and MALDI-TOF MS confirmed all by day 4. On the other hand, 69 of 100 surveillance samples yielded turbidity in Dulcitol broth by days 3–14 with final MALDI identification by days 5 to 17. Both media failed to identify one sample each, resulting in assay sensitivity and specificity of 99% and 97%, respectively. Of Candida and yeast species tested, 75–80% of C. metapsilosis and C. orthospilosis were misidentified as C. auris. However, previous studies indicated that these species are rarely detected in surveillance screening of C. auris. Naganishia diffluens also resembled C. auris, although it required different temperature growth (30 °C). In conclusion, CHROMagar™ Candida Plus provides rapid presumptive identification of C. auris. It would be another valuable tool in surveillance efforts to control the spread of C. auris in healthcare.
Keywords: Candida auris, MALDI-TOF MS, Chromogenic, Surveillance
Introduction
Candida auris is the newly discovered member of the Candida/Clavispora clade that presents a serious global health threat. Even though C. auris was first identified from ear discharge of a patient in Japan in 2009, a retrospective study revealed that its earliest occurrence dated back to 1996 in South Korea, where it was misidentified as C. haemulonii [1, 2]. Whole genome sequencing has clustered C. auris isolates into four clades corresponding to the regions of origin and spread. These clades are South Asia (Clade I), East Asia (Clade II), Africa (Clade III) and South America (Clade IV) [3]. Recently, a fifth clade of C. auris was discovered in Iran, separated by > 200,000 SNPs from the other four clades [4, 5]. Over the past decade, several healthcare-associated outbreaks of C. auris have been a major concern [6–10]. In 2016, the Centers for Disease Control and Prevention (CDC) issued a global alert for C. auris, recommending surveillance and infection control protocols. After worldwide alerts from different organizations, C. auris cases have been tracked in all continents except Antarctica [11]. C. auris is troublesome in clinical settings due to high transmission rates and multi-drug resistance [3, 9, 10, 12]. Prolonged colonization on the skin and indoor surfaces and tolerance to healthcare disinfectants further increases the risks of C. auris hospital outbreaks [9, 12]. Colonization not only increases the risk of transmission but also elevates the risk of invasive infection especially in immunocompromised patients [13]. Invasive infections of C. auris often have high mortality rates ranging from 40 to 60% [13, 14].Therefore, the early diagnosis of this “superbug” is crucial to reduce the rate of transmission as well as the progression to invasive infection. Many commercially available phenotype-based methods such as API ID 32 C system (version 4.0 database, Biomerieux), BD Phoenix (BD Diagnostics, Sparks, MD, USA) and RapID Yeast Plus (Remel, Thermo Fisher Scientific, Lenexa, KS, USA) misidentify C. auris [10, 12, 15]. Molecular methods like C. auris real-time PCR, Sanger sequencing of the Internal Transcribed Spacer (ITS) and D1/D2 regions of the ribosomal gene, protein-based technologies like matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can identify C. auris accurately [12, 15–17]. Although these technologies provide reliable detection of C. auris, the instruments and reagents, along with the maintenance, are expensive for low-complexity laboratories with limited resources. Therefore, low-complexity laboratories depend on culture-based methods such as selective or differential media for the identification of C. auris [15]. Welsh et al. have established salt Sabouraud Dulcitol broth as a selective medium for recovery of C. auris from clinical and environmental samples [18]. Many high-complexity laboratories, including regional, and state public health laboratories in the USA, have successfully utilized salt Sabouraud Dulcitol broth for recovery of C. auris from surveillance samples. However, we observed that this culture method requires two weeks incubation before calling it negative [17]. Chromogenic isolation media provide better detection rates of pathogenic Candida spp. in mixed cultures than traditional media due to the production of species-specific color based on enzymatic reactions [19, 20]. However, currently used commercial chromogenic media were designed to target major Candida pathogenic spp. prior to C. auris outbreaks, and these media do not clearly distinguish C. auris from all other Candida spp. [21–23]. In this study, we compared CHROMagar™ Candida Plus with the salt Sabouraud Dulcitol broth for presumptive identification of C. auris from surveillance samples collected from patients from healthcare facilities in northeast region of the United States. We also evaluated the potential of the CHROMagar™ Candida Plus (CHROMagar, Paris, France) for presumptive identification of C. auris from other Candida and yeast species.
Materials and Methods
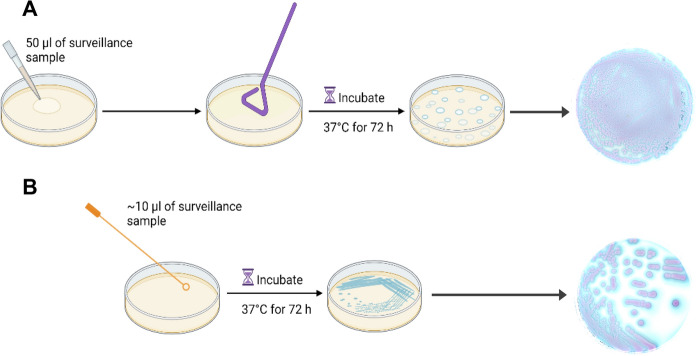
A total of 100 surveillance samples were collected as swabs from a single colonization site (axilla, groin, or nares) or as a composite swab (axilla/groin or nares/axilla/groin) from patients in health care facilities from the northeast region of the United States [16]. These swabs were submitted to the Wadsworth Center Mycology Laboratory (WCML) within 24 h of sample collection in the BD ESwab Liquid Amies Collection and Transport System (Becton, Dickinson, Franklin Lakes, NJ, USA) at ambient temperature. A total of 32 Candida and other yeast species procured from the Wadsworth Center’s Mycology Culture Collection Repository were also part of this investigation. For C. auris recovery from surveillance samples, ESwabs containing 1 ml liquid Amies medium were vortexed for 30 s, and 200 µl of the ESwab liquid was inoculated in salt Sabouraud Dulcitol broth (henceforth, Dulcitol broth) and incubated at 40 °C for 2 weeks in a shaking incubator at 150 RPM [18]. CHROMagar™ Candida Plus plates were prepared from the dry powder as per the manufacturer’s instructions and stored at 4 °C in the dark with a shelf life of 4 weeks. Approximately 10 µl and 50 µl of surveillance samples were also inoculated on CHROMagar™ Candida Plus medium using spread (Fig. 1A) and streak (Fig. 1B) methods, and plates were incubated at 37 °C for 24 to 72 h. The choice of two volumes of surveillance samples was to ensure isolation of individual colonies of presumptive C. auris on CHROMagar™ Candida Plus. Colonies recovered from CHROMagar™ Candida Plus and Dulcitol broth were streaked on Sabouraud dextrose agar, followed by MALDI-TOF MS (Bruker, Bremen, Germany) identification. Since colony color is a critical factor for presumptive identification of C. auris on CHROMagar™ Candida Plus, other Candida (21) and pathogenic yeast (11) spp. were simultaneously streaked on this medium and incubated at 37 °C for 24–72 h. Colony images were taken from the front and back of the plate using Canon EOS 80D Camera through Ortery Photosimile software and processed using Photoshop 22.4.2.
Fig. 1.
Workflow of surveillance sample testing using CHROMagar™ Candida Plus medium. A 50 µl of surveillance sample was inoculated on a CHROMagar™ Candida Plus plate and spread evenly using a disposable spreader. B 10 µl of surveillance sample was also inoculated on another CHROMagar™ Candida Plus plate and streaked with a 10 µl yellow plastic loop to obtain single colonies. Plates were incubated at 37 ˚C for 72 h and then imaged using a Canon EOS 80D Camera through Ortery Photosimile software and processed using Photoshop 22.4.2. Workflow template was created with BioRender.com
Results and Discussion
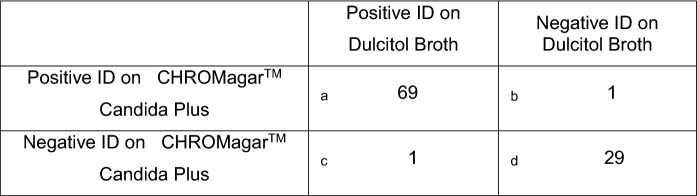
Of the 100 surveillance swabs tested, 69 yielded positive colonies at 72 h with a creamy pink color with a blue halo on CHROMagar™ Candida Plus, indicating the presence of C. auris. According to the manufacturer’s instruction manual, C. auris yields a light blue color colony with a blue halo on CHROMagar™ Candida Plus at 48 h [24]. However, we observed that an additional day of incubation allowed the development of creamy pink color along with the blue halo, thus helping the distinction of C. auris from other Candida spp. studied in this investigation. All suspected colonies were confirmed to be C. auris by MALDI-TOF MS, providing 100% agreement with the phenotypic results. These results also suggested that the unique creamy pink color with a blue halo produced by C. auris on CHROMagar™ Candida Plus can be utilized as presumptive identification of C. auris from surveillance samples. The spread and streak plate methods were both helpful in the isolation of single colonies of C. auris from surveillance samples with a moderate number of C. auris. The streak plate method was useful for the isolation of single colony from surveillance samples with high load (> 1000 CFU/50 µl) of C. auris, while the spread plate method was useful for the isolation of a single colony from surveillance samples with a low load (< 10 CFU/50 µl) of C. auris. A combination of spread and streak plate methods yielded 69 of 100 surveillance samples positive for C. auris on CHROMagar™ Candida Plus, which were also positive in Dulcitol broth. Both media failed to identify one sample each resulting in assay sensitivity of 99% and specificity of 97% (Table 1).
Table 1.
Method comparison for C. auris identification (ID) using CHROMagar™ Candida Plus and Dulcitol broth.
a = true positives
b = false positives
c = false negatives
d = true negatives
Sensitivity of CHROMagar™ Candida Plus = a/a + c = 99%
Specificity of CHROMagar™ Candida Plus = d/d + b = 97%
PPV (Positive Predictive Value) = a/a + b = 99%
NPV (Negative Predictive Value) = d/c + d = 97%
Next, we compared the time taken to presumptively identify C. auris on CHROMagarTM Candida Plus to that of Dulcitol broth. CHROMagarTM Candida Plus yielded results in 3 days for all positive surveillance samples irrespective of the presence of a high (> 1000 CFU/ 50 µl) or low (< 10 CFU/ 50 µl) load of C. auris. In contrast, it took anywhere from 3 to 14 days for the same surveillance samples to become turbid in Dulcitol broth, with timing dictated by how high or low a load of C. auris was present in each surveillance sample. All presumptive positive C. auris on CHROMagarTM Candida Plus were confirmed by MALDI-TOF MS by day 4, while the final MALDI-TOF MS identification of turbid surveillance samples from Dulcitol broth ranged from 5 to 17 days (Fig. 2).
Fig. 2.
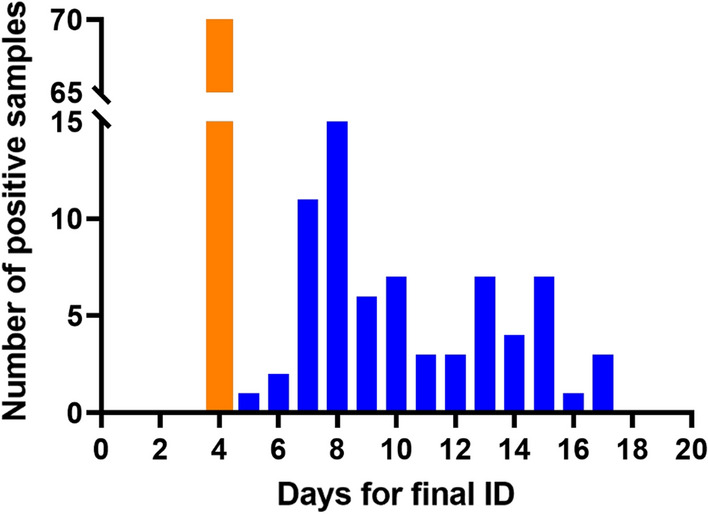
Comparison of days required for final identification of C. auris using CHROMagar™ Candida Plus and Dulcitol broth. Presumptive identification on CHROMagar™ Candida Plus for all C. auris positive surveillance samples took 3 days followed by final identification by MALDI-TOF MS on the fourth day (orange bar). Final identification with Dulcitol broth and MALDI-TOF MS had a broad range from day 5 to day 17 (blue bars)
Dulcitol broth relies on the enrichment of C. auris at higher temperatures (40 °C), higher salinity (10%), and dulcitol as a carbon source [18]. Additionally, other yeasts are not grown but are viable in Dulcitol broth, hence confirmation by MALDI-TOF MS requires an additional step of streaking 10 µl of turbid broth onto Sabouraud dextrose agar and incubation at 40 °C for at least 48 h for the isolation of single colonies followed by streaking those single colonies on Sabouraud dextrose agar at 30 °C for 24 h before MALDI-TOF MS identification. This additional step increases the processing time for Dulcitol broth cultures. On the other hand, CHROMagar™ Candida Plus not only allows the rapid growth of Candida and yeast spp. but can successfully distinguish C. auris from other yeasts based on color change within 3 days of incubation. Thus, CHROMagar™ Candida Plus would be a useful tool in the context of hospital outbreaks where faster detection would prove effective in curbing the spread. This is the first publication directly comparing Dulcitol broth, a selective medium for growing C. auris, against CHROMagar™ Candida Plus, a differential medium. Our results indicate that CHROMagar™ Candida Plus provides rapid presumptive identification of C. auris from surveillance samples. These results are encouraging and provide an additional culture-based method to identify this deadly fungal pathogen from surveillance samples.
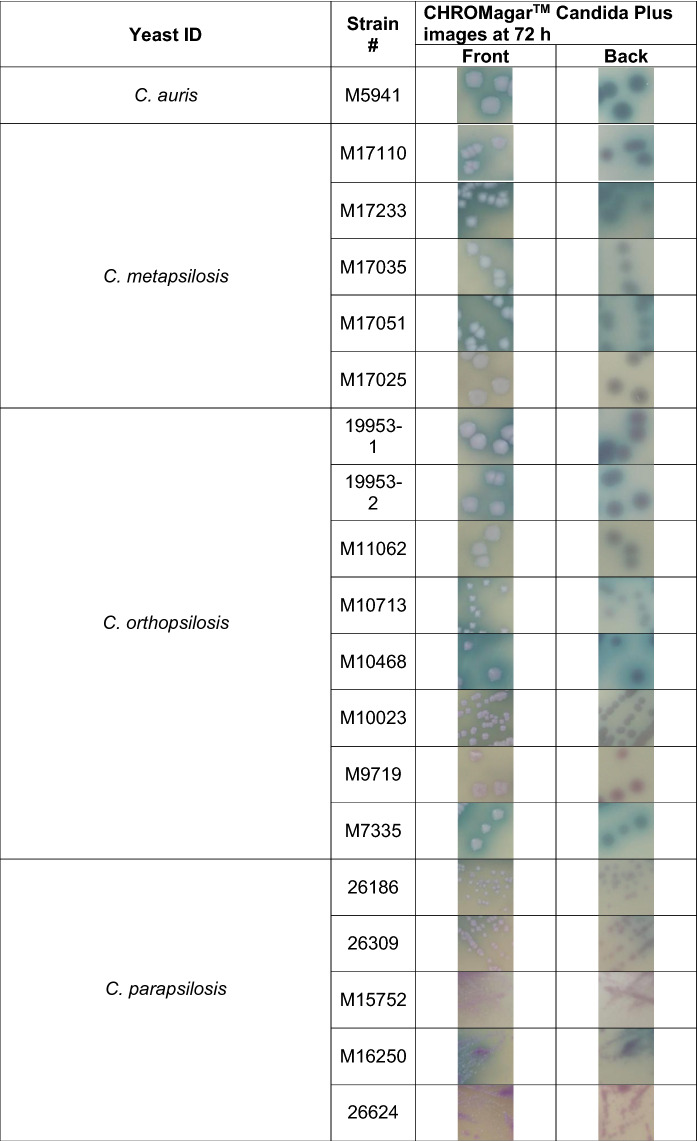
Before advocating the use of CHROMagar™ Candida Plus for presumptive identification of C. auris from surveillance samples, we wanted to observe the pigmentation from different Candida and yeast spp. to rule out any close resemblance to C. auris. To achieve this objective, an additional 32 Candida and yeast species either recovered from surveillance samples [16], or identified as part of reference testing, were used (Table 2). The majority of Candida and yeast species were incubated at 37 °C, except for Candida famata, Naganishia diffluens, and Rhodotorula mucilaginosa, which were incubated at 30 °C. The poor growth of these species at temperatures above 30 °C has been observed previously [25]. C. auris colonies were creamy pink with a blue halo post 72 h incubation at 37 °C. Of the 32 comparator species tested, Naganishia diffluens most closely resembled C. auris (Table 2). However, its inability to grow at 37 °C precludes the probability of confusing one for another. Another important distinction was the texture of colonies; Naganishia diffluens colonies were mucoid, unlike the creamy colonies of C. auris (Table 2). Recently, while performing surveillance studies, we noted that C. orthopsilosis and C. metapsilosis resembled C. auris on CHROMagar™ Candida Plus. This led us to investigate an extensive panel of C. parapsilosis species complex (Table 3). Of the C. parapsilosis species complex tested, a high percentage (75–80%) of C. metapsilosis and C. othropsilosis indeed resembled C. auris in color and texture and would have easily been misidentified as C. auris (Table 3). However, since these species are rarely the cause of Candida infection and colonization [16], the similarity between these species on CHROMagar™ Candida Plus should not hinder the presumptive identification of C. auris. Recently, Borman et al. reported that Candida diddensiae, a rare yeast that resembled C. auris on CHROMagar™ Candida Plus. Therefore, we looked closely at all the C. diddensiae isolates available in Mycology Culture Collection Repository, but we found no resemblance to any of the C. auris clades tested (Table 2).
Table 2.
Colony images of Candida and other yeast spp. at 72 h on CHROMagar™ Candida Plus
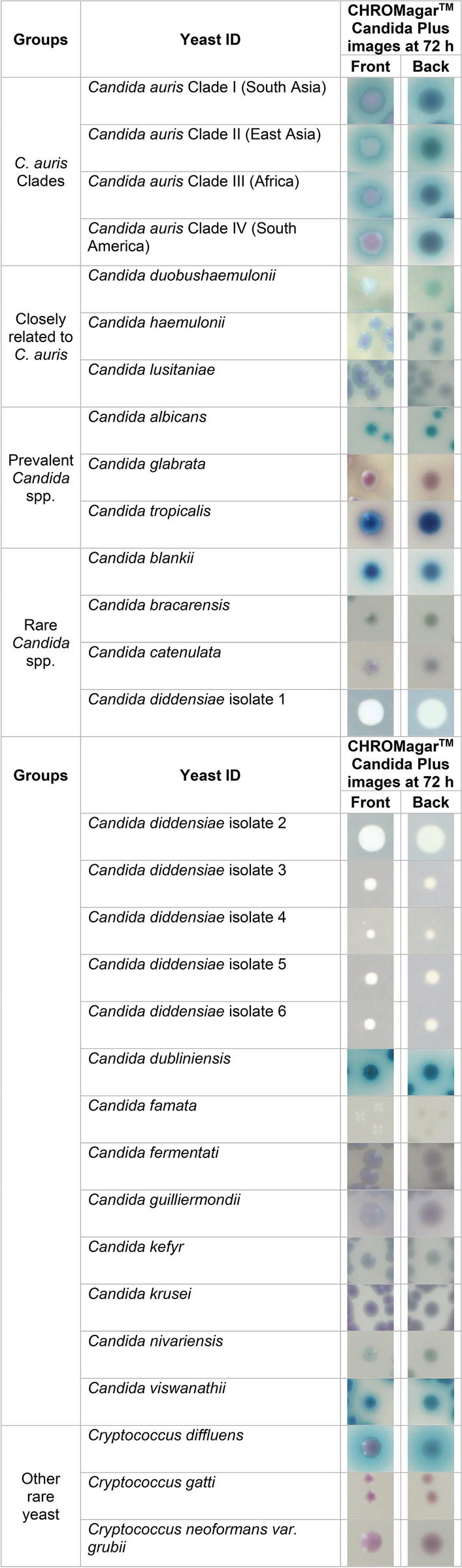
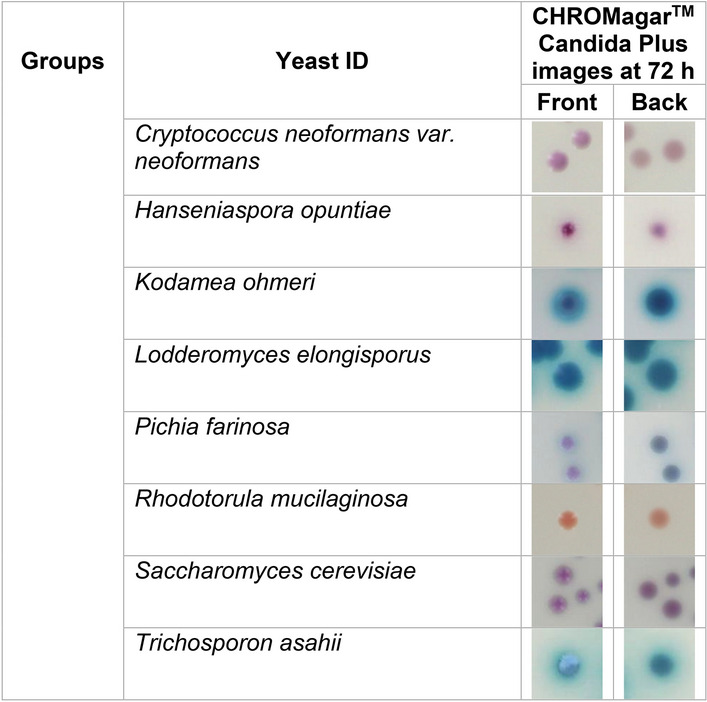
Table 3.
Colony images of Candida parapsilosis species complex on CHROMagar™ Candida Plus
In conclusion, our findings indicate that CHROMagar™ Candida Plus is a valuable medium for presumptive identification of C. auris from yeast isolates recovered from clinical specimens, and it can also serve as a rapid surveillance screening tool for assessing C. auris colonization from patients in the healthcare setting. A CHROMagar™ Candida Plus medium, a valuable culture-based tool, can easily be adapted by both the low- and high-complexity laboratories for the presumptive identification of C. auris. The quick identification supported by this tool will boost the effort to curb the spread of C. auris, which is a critical and immediate need considering the significant rise of C. auris infection/colonization in combination with the emergence of COVID-19 [26–29].
Acknowledgements
We want to acknowledge the Wadsworth Center (WC) Tissue Culture & Media Cores for providing various media for the culture of Candida species. We thank CHROMagar, Paris, France, and their US distributors for providing dehydrated chromogenic media for differentiation of Candida species. We also thank Dr. Kimberlee McClive-Reed for the critical reading and editing of the manuscript. This work was supported partly by the funds from the WC, the New York State Department of Health (NYSDOH), the National Institutes of Health (1R21AI156573-01A1), and the Centers for Disease Control and Prevention (CDC) grant number NU50CK000516. The contents of this study are solely the authors’ responsibility and do not necessarily represent the official view of the NYSDOH or the CDC.
Author Contributions
AM was responsible for surveillance sample testing, Candida spp. culture and imaging, data analysis, and draft preparation. YZ helped with Candida spp. culture and imaging, review, and editing. VC provided expertise with the study design and critically reviewed the manuscript. SC conceived and designed the study, edited, and critically reviewed the manuscript.
Declarations
Conflict of Interest
The authors declare that they have no financial or non-financial conflicts of interest.
Footnotes
Handling Editor: Ferry Hagen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Satoh K, et al. Candida auris sp nov, a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Oh BJ, et al. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol. 2011;49(1):98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart SR, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow NA, et al. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safari F, et al. A Chronic autochthonous fifth clade case of Candida auris otomycosis in Iran. Mycopathologia. 2022;187(1):121–127. doi: 10.1007/s11046-021-00605-6. [DOI] [PubMed] [Google Scholar]
- 6.Schelenz S, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuichard-Gysin D, et al. Candida auris - recommendations on infection prevention and control measures in Switzerland. Swiss Med Wkly. 2020;150:w20297. doi: 10.4414/smw.2020.20297. [DOI] [PubMed] [Google Scholar]
- 8.Sathyapalan DT, et al. Evaluating the measures taken to contain a Candida auris outbreak in a tertiary care hospital in South India: an outbreak investigational study. BMC Infect Dis. 2021;21(1):425. doi: 10.1186/s12879-021-06131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13(5):e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kathuria S, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and Iis antifungal susceptibility profile variability by vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol. 2015;53(6):1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Tracking Candida auris. 2019 [cited 2021; Available from: https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html.
- 12.Du H, et al. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taori SK, et al. Candida auris outbreak: mortality, interventions and cost of sustaining control. J Infect. 2019;79(6):601–611. doi: 10.1016/j.jinf.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Arensman K, et al. Clinical outcomes of patients treated for Candida auris infections in a multisite health system, Illinois, USA. Emerg Infect Dis. 2020;26(5):876–880. doi: 10.3201/eid2605.191588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis EK, Chaturvedi S, Chaturvedi V. So many diagnostic tests, so little time: Review and preview of Candida auris testing in clinical and public health laboratories. Frontiers in Microbiol. 2021;12:1–13. doi: 10.3389/fmicb.2021.757835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, et al. Laboratory analysis of an outbreak of Candida auris in New York from 2016 to 2018: impact and lessons learned. J Clin Microbiol. 2020;58(4):1. doi: 10.1128/JCM.01503-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leach L, Zhu Y, Chaturvedi S. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol. 2018;56(2):123. doi: 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welsh RM, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017;55(10):2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry JD. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin Microbiol Rev. 2017;30(2):449–479. doi: 10.1128/CMR.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadeem SG, Hakim ST, Kazm SU. Use of CHROMagar Candida for the presumptive identification of Candida species directly from clinical specimens in resource-limited settings. Libyan J Med. 2010;5(1):2144. doi: 10.3402/ljm.v5i0.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borman AM, Fraser M, Johnson EM. CHROMagarTM Candida Plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med Mycol. 2021;59(3):253–258. doi: 10.1093/mmy/myaa049. [DOI] [PubMed] [Google Scholar]
- 22.Mulet Bayona JV, et al. Evaluation of a novel chromogenic medium for Candida spp identification and comparison with CHROMagar Candida for the detection of Candida auris in surveillance samples. Diagn Microbiol Infect Dis. 2020;98(4):115168. doi: 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed] [Google Scholar]
- 23.de Jong AW, et al. Performance of two novel chromogenic media for the identification of multidrug-resistant Candida auris compared with other commercially available formulations. J Clin Microbiol. 2021;59(4):1–9. doi: 10.1128/JCM.03220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CHROMagarTM. Candida Plus Data Sheet, CHROMagarTM, Editor. CHROMagarTM The Chromogenic Media Pioneer. 2020
- 25.Barnett JA, Payne RW, Yarrow D. Yeasts: characteristics and identification. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 26.Nori P, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021;42(1):84–88. doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villanueva-Lozano H, et al. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin Microbiol Infect. 2021;27(5):813–816. doi: 10.1016/j.cmi.2020.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajni E, et al. A High Frequency of Candida auris blood stream infections in coronavirus disease 2019 patients admitted to intensive care units, northwestern India: A case control ctudy. Open Forum Infect Dis. 2021;8(12):ofab452. doi: 10.1093/ofid/ofab452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janniger EJ, Kapila R. Public health issues with Candida auris in COVID-19 patients. New Jersey: World Med Health Policy; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]