Abstract
Aberrant glycosylation has been extensively reported in cancer, with fundamental changes in the glycosylation patterns of cell–surface and secreted proteins largely occurring during cancer progression. As such, serum glycan and glycopeptide biomarkers have been discovered using mass spectrometry and proposed for cancer detection. Here, we report for the first time potential serum N-glycan and glycopeptide biomarkers for Philippine lung cancer patients. The N-glycan and glycoprotein profiles of a cohort (n = 26 patients, n = 22 age- and gender-matched) of lung cancer patients were analyzed and compared to identify potential N-glycan and glycopeptide serum biomarkers using nano-QToF-MS/MS and ultra-high-performance liquid chromatography coupled with triple quadrupole mass spectrometry dynamic multiple monitoring methods, respectively. Statistical analyses identified differential N-glycan and glycopeptide abundances. The N-glycans were mostly sialylated and sialofucosylated branched structures. The glycopeptides involved proteins in complement and coagulation cascades (padj= 6.418 × 10–4), innate immunity (padj= 6.094 × 10–3), acute inflammatory response (padj= 6.404 × 10–5), defense response (padj= 2.082 × 10–4), complement activation pathways (padj= 1.895 × 10–2), and immunoglobulin-mediated immune response pathways (padj= 4.818 × 10–2). Biomarker models were constructed using serum N-glycans [area under the curve (AUC) = 0.775; 95% CI: 0.617–0.931] and glycopeptides (AUC = 0.959; 95% CI: 0.85–1.0), with glycopeptides having higher accuracies than N-glycans. The results suggest that in the Philippine lung cancer patient sera, specific N-glycans and site-specific glycans are differentially expressed between cases and controls. This report represents the first serum glycan and glycopeptide biomarkers of Philippine lung cancer patients, further demonstrating the utility of mass spectrometry-based glycomic and glycoproteomic methods.
Introduction
Cancer incidence and mortality are growing rapidly worldwide, reflecting several factors including aging, population growth, increasing cancer risk factors, and socioeconomic development. According to the GLOBOCAN 2020 database of 185 countries and 36 cancers, there will be 19.3 million new cases and 10 million cancer deaths worldwide.1 Of these, 2,093,876 cases (11.6%) and 1,761,007 (18.4%) deaths for both sexes will be due to lung cancer. Additionally, lung cancer is the leading cause of cancer death among men in 93 countries and among women in 28 countries. There are two main types of lung cancer—small cell lung cancer and the more common non-small cell lung cancer (NSCLC), which accounts for approximately 85% of all lung cancer cases.2
Several risk factors contribute to the pathogenesis of NSCLC such as cigarette smoking,3 secondhand or passive smoking,4 alcohol consumption,5 and the presence of susceptibility genes such as TP536 and epidermal growth factor receptor T790M sequence variation.7 Recently, the effect of race-ethnicity in cancer trends have been reported as well, showing unique cancer biomarkers due to racial and ethnic differences.8−12 As such, it is important to identify race-specific cancer biomarkers for a specific population.
Protein glycosylation is one of the most complicated and most common post-translational modifications. It is involved in many cellular interactions such as host–pathogen interactions, cell differentiation and trafficking, and intra- and intercellular signaling.13 Protein glycosylation is a step-wise process that starts at the endoplasmic reticulum and continues in the Golgi apparatus to achieve the diversity and complexity of final glycan structures through biosynthetic steps involving glycosyltransferases and glycosidases. In cancer, these glycan-processing enzymes are overexpressed, resulting in enhanced expression of related glycan structures. For example, the enzymes Alpha1-6FucT, B4GALT2, MAN1A2, and MAN2A1 are overexpressed in lung cancer tissue samples.14 Likewise, high-mannose, fully galactosylated, and fucosylated N-glycans are also overexpressed in lung cancer tissues.15 Furthermore, altered glycosylation is also correlated with the other hallmarks of the disease such as enhanced proliferation, angiogenesis potential, apoptosis and tumor suppression, replicative immortality, and metastatic potential.16 Aberrant glycosylation has been well documented in cancer, with fundamental changes in the glycosylation patterns of cell surface and excreted proteins during cancer progression. Growing evidence further supports the role of glycosylation during tumor progression and cancer cell proliferation, invasion, metastasis, and angiogenesis.17
The utility of serum biomarkers using liquid chromatography–tandem mass spectrometry (LC–MS/MS) has been demonstrated previously, with methods focusing on N-glycans15,18 and glycopeptides19−22 responding to cancer progression. In particular, distinct glycans including Hex4HexNAc5Fuc1, Hex5HexNAc6Fuc1, NeuAc2, and Gal4 have been previously shown to provide discriminating area under the curve (AUC) (AUC = 0.74, 95% CI: 0.68–0.80) in a primarily Caucasian cohort (n = 100 cases, 199 controls for discovery set; n = 108 cases, 216 controls for test set).18 Furthermore, comparison of glycans on isolated IgG from lung cancer patients and healthy volunteers showed overexpression of Hex3HexNAc4Fuc1 and underexpression of Hex5HexNAc5NeuAc1, Hex5HexNAc5NeuAc2, and Hex5HexNAc5Fuc1NeuAc2.23
In this study, the N-glycan and glycoprotein profiles of a cohort (n = 26 patients, n = 22 age- and gender-matched volunteers) of Philippine lung cancer patients were analyzed and compared using LC–MS/MS to identify potential N-glycan and glycopeptide serum biomarkers. This report represents the first attempt to characterize differences in serum protein N-glycosylation of Philippine lung cancer patients and to identify N-glycan and glycopeptide biomarkers that specifically target this population.
Materials and Methods
Ethical Statement and Clinical Sample Collection
This research study was approved by the Lung Center of the Philippines Institutional and Ethics Review Board and was performed in accordance with the institutional guidelines/regulations (LCPIERB; ethics approval number: LCP-CS-003-2018). Before recruitment, informed consent was obtained from all participants and/or their legal guardians. A total of 26 TTF1 (thyroid transcription factor-1)-positive24 confirmed non-small cell lung adenocarcinoma patients from the Lung Center of Philippines, Quezon City, Philippines, and 22 age- and gender-matched control individuals were enrolled in the current study from June 2018 to June 2021. All enrolled individuals were Filipino up to second degree of consanguinity, aged 18–85 years old, and had neither TB nor HIV. Case individuals were chemotherapy—as well as radiotherapy—naïve. Control individuals, on the other hand, were physically assessed to be healthy by a physician, had no smoking history for 2 years, had no history of malignancies, were non-diabetic, and did not have serious illness in the past 6 months. Complete clinical and demographic information of the cohort can be found in the Supporting Information, Table 1. Venous blood samples were collected in tubes containing clot activator and were centrifuged to separate the serum. The collected serum samples were then stored at −70 °C ultra-low temperature freezer and shipped to the University of California Davis for LC–MS/MS analyses.
N-Glycan Sample Preparation and LC–MS/MS Analysis
Serum N-glycans were prepared and analyzed as previously described, with modifications.25 Serum samples and quality control, a 1:1 mixture of human serum (Sigma-Aldrich) and RNAse B, were resolubilized in N-glycan release solution (100 mM NH4HCO3, 5 mM DTT) and then boiled for 1 min in a water bath. Afterward, 2 μL peptide-N-glycosidase F (PNGase F) was added, and the samples were microwaved (20 W, 10 min, 33 °C; Discover Proteomics) followed by overnight incubation (18 h, 37 °C) to release covalently linked N-glycans. These were then cleaned up using porous graphitized carbon-solid phase extraction plates and eluted with 40% ACN + 0.05% TFA. N-glycans were analyzed as previously described25,26 using an Agilent 6520 chip-QToF mass spectrometer (Santa Clara, CA). N-glycan samples were reconstituted in 25 μL of Milli-Q water, and 5 μL of the resulting solution was used for injection into the nano-LC-MS/MS system. Separation was performed using an Agilent PGC-Chip II with a 40 nL enrichment and 43 mm × 75 μm analytical column (particle size 5 μm) and a binary solvent system composed of mobile phase A (3% v/v acetonitrile and 0.1% v/v formic acid in water) and mobile phase B (90% v/v acetonitrile and 1% v/v formic acid in water). The gradient sequence for separation used was as follows: 0–2.5 min, 1% B; 2.5–20 min, 16% B; 20–35 min, 58% B; 35–40 min, 100% B; 40–50 min, 100% B; 50.01–65 min, 0% B with a flow rate of 0.3 μL/min. Tandem MS spectra were acquired via collision-induced dissociation (CID), with spectra measured at 0.8 s per spectrum in the positive ion mode.
Analysis of the N-glycan data was performed using MassHunter Qualitative Analysis Software B.07.00 (Agilent Technologies). MassHunter was used to automatically identify the N-glycan structures by matching of the monoisotopic masses obtained against our in-house database consisting of known N-glycan compositions and their exact masses, using a quality score >30 and mass accuracy <20 ppm.25,27 Additionally, N-glycan hits were subsequently verified through their corresponding MS/MS spectra. The relative abundance of each glycan in a sample was determined using the peak area of all glycans from extracted ion chromatograms in MassHunter. N-glycan structure and compositions are reported based on the number of each type of residue, HexaHexNAcbFuccNeuAcd, where Hexa refers to the number of hexose (i.e., mannose, galactose), HexNAcb refers to the number of N-acetylhexosamine (i.e., N-acetylglucosamine), Fucc refers to the number of fucose, and NeuAcd refers to the number of neuraminic acid (i.e., sialic acid) residues. The N-glycans were subsequently classified using an in-house classification system: high-mannose (HexaHexNAcb, where b = 2 denotes the chitobiose core and “a” refers solely to mannose residues), undecorated (HexaHexNAcb, where “a” refers to both mannose and galactose residues), fucosylated (HexaHexNAcbFucc, where c ≥ 1), sialylated (HexaHexNAcbNeuAcd, where d ≥ 1), or sialofucosylated (HexaHexNAcbFuccNeuAcd, where both c and d ≥ 1). Relative abundances of N-glycan groups (high-mannose, undecorated, fucosylated, sialylated, and sialofucosylated glycans) were calculated by adding the relative abundance of each individual glycan belonging to a specific glycan group.
Glycopeptide Sample Preparation and LC–MS/MS Analysis
Serum glycopeptides were prepared and analyzed as previously described, with modifications.19−21,28 Ten microliters (10 μL) each of serum samples, glycoprotein standards—immunoglobulin M (IgM) and alpha-2-macroglobulin (A2MG)—and standard serum (Sigma-Aldrich) were resolubilized in 50 μL buffer (50 mM NH4CO3), reduced with 40 μL dithiothreitol (25 mM, 60 °C, 50 min), alkylated with 20 μL iodoacetamide (90 mM, 30 min), and then digested with 60 μL trypsin (0.067 μg/μL) overnight at 37 °C. After trypsin digestion, 20 μL of internal standard (peptide sequence: RPAIAINNPYVPR; 9 μg/mL) and 10 μL of 18% formic acid were added to quench the reaction. To determine the linearity of the procedure, dilutions of digested standard serum samples were prepared (50–1500 μg/mL). Digested glycopeptides and peptides were analyzed in an Agilent 1290 infinity LC system coupled to an Agilent 6495 triple-quadrupole (QqQ) mass spectrometer (Santa Clara, CA) using a previously described dynamic multiple reaction monitoring method (dMRM).19−21,28 2 μL of each sample was injected into an Agilent Eclipse plus C18 column (rapid resolution high definition [RRHD] 1.8 μm, 2.1 × 100 mm) coupled with a C18 guard column (RRHD 1.8 μm, 2.1 × 5 mm). Separation was performed using a binary solvent system composed of mobile phase A (3% v/v acetonitrile and 0.1% v/v formic acid in water) and mobile phase B (90% v/v acetonitrile and 0.1% v/v formic acid in water). The gradient sequence for separation used was as follows: 0–2.5 min, 1% B; 2.5–20 min, 16% B; 20–35 min, 58% B; 35–40 min, 100% B; 40–50 min, 100% B; and 50.01–65 min, 0% B with a flow rate of 0.5 mL/min. The dMRM method applied predetermined CID energies to detect and quantify 505 specific glycopeptide transitions.28 The dMRM results were analyzed using MassHunter Quantitative Analysis Software B.08.00 (Agilent Technologies). Data was exported as integrated areas, and glycopeptide abundance was normalized to corresponding peptide concentrations to reduce the variability caused by over- or underexpression of proteins. Glycopeptides are reported as follows: UniProt Protein name_Asparagine glycosite_N-glycan composition; for example, IgM-Asn46_Hex5HexNAc4Fuc1NeuAc1 refers to immunoglobulin M at glycosite asparagine 46 with N-glycan composition of IgM-Asn46_Hex5HexNAc4Fuc1NeuAc1.
Statistical Analysis
Statistical analyses were performed using the MetaboAnalyst 5.0 platform.29 Before statistical analysis, missing values were imputed with 1/5th of the analyte minima. The resulting values were log10-transformed and mean-centered to transform and scale the data. Differential analyses were done using the “Statistical Analysis [one factor]” module in MetaboAnalyst 5.0. Orthogonal Projections to Latent Structure Discriminant Analysis (OPLS-DA) was performed based on the R package ropls(30) as implemented in MetaboAnalyst, to generate the score plot and identify important features using variable importance in projection (VIP) scores. Biomarker models were calculated using the “Biomarker Analysis” module in MetaboAnalyst 5.0. Multivariate receiver operating characteristic (ROC) curves were generated using support vector machine (SVM) classification method and feature ranking methods. Biomarker models incorporating significantly different N-glycan, glycopeptide, and both N-glycan and glycopeptide analytes were generated using a linear SVM algorithm.
To identify the pathway involvement of differentially glycosylated serum glycoproteins, the protein IDs were mapped to different Reactome, KEGG, and gene ontology pathways using g:profiler.31
Results and Discussion
Serum N-Glycan Biomarkers of Lung Cancer in Philippine Patients
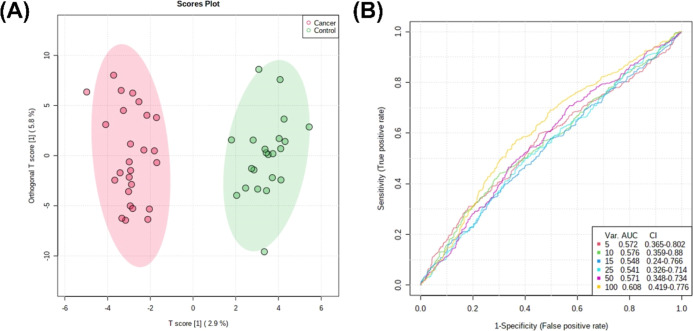
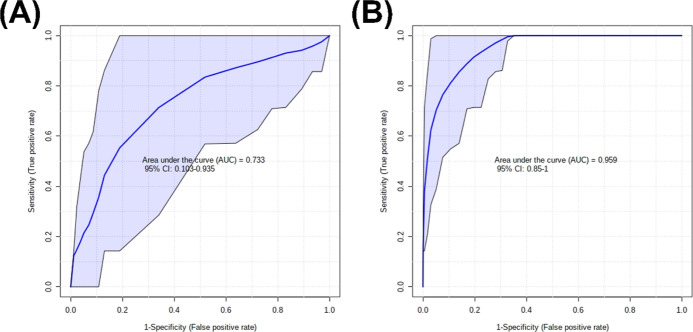
N-Glycans obtained from the sera of lung cancer patients and healthy controls were primarily sialylated only (>50%) and sialofucosylated (>20%) (Supporting Information Figure 1). Orthogonal partial least square discriminant analysis (OPLS-DA) of the relative N-glycan abundances between cancer patients and healthy volunteers suggests separation between the two groups (Figure 1a). Moreover, ROC-curve analyses also suggest good accuracy, with biomarker models using 100 N-glycans having AUC = 0.608 (95% CI: 0.419–0.776) (Figure 1b).
Figure 1.
Biomarker analysis of serum N-glycan abundance comparing lung cancer patients and healthy volunteers. (A) Scores plot showing distribution of cancer patients (red) and healthy volunteers (green) across components 1 and 2 given the OPLS-DA model for serum N-glycans. (B) ROC curve analysis of serum N-glycans.
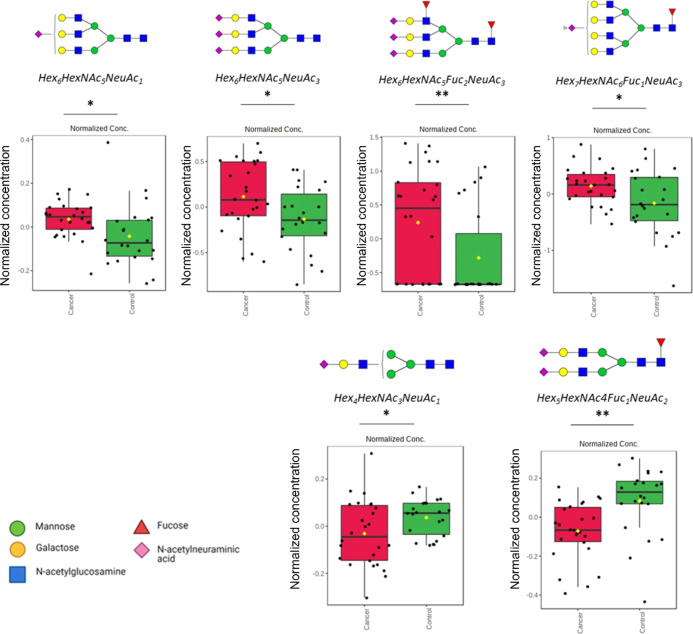
Further statistical test was performed to identify the highly discriminating N-glycans. Multiple t-tests (p < 0.05) show that these identified N-glycans were significantly different (Figure 2; Supporting Information, Figure 2). Specifically, significantly overexpressed N-glycans were identified as having the compositions Hex6HexNAc5NeuAc1, Hex6HexNAc5NeuAc3, Hex6HexNAc5Fuc2NeuAc3, and Hex7HexNAc6Fuc1NeuAc3. On the other hand, significantly underexpressed N-glycans were identified as Hex4HexNAc3NeuAc1 and Hex5HexNAc4Fuc1NeuAc2.
Figure 2.
Relative abundances of significantly over- and underexpressed N-glycans (reported as Hex_HexNAc_Fuc_NeuAc) in lung cancer sera compared to healthy volunteers, identified using OPLS-DA; *p < 0.05; **p < 0.01.
Interestingly, majority of the N-glycans that were significantly different between cancer and healthy sera were fucosylated, sialylated, and sialofucosylated. We observed highly branched sialylated and sialofucosylated structures to be overexpressed in cancer sera as well as a significant decrease in the abundance of biantennary structures. Comparison of serum N-glycan biomarkers with reported tissue N-glycan biomarkers in lung cancer tissues coincides with decrease in fully galactosylated N-glycans observed in lung cancer tissues—Hex5HexNAc4, Hex5HexNAc4NeuAc1, and Hex5HexNAc4Fuc1.15
Serum Glycopeptide Biomarkers of Lung Cancer in Philippine Patients
Multiple reaction monitoring (MRM) method involves selecting a predetermined precursor ion, fragmenting it, and detecting and quantifying a predetermined fragment ion. High selectivity, sensitivity, and robustness is obtained due to the non-scanning nature of the method and selectivity steps in filtering out background ions.32 However, monitoring analytes throughout the whole chromatographic period poses a limit on the number of analytes that can be monitored simultaneously. A more recent approach, dMRM, alleviates this limit by monitoring analytes only at specific retention times.19,20 Serum glycopeptides were analyzed using a previously established dMRM method consisting of 505 transitions representing 505 glycoforms of 62 unique serum glycoproteins.19−21
The dMRM results were extremely reproducible, producing 85 transitions having less than 20% CV in a 5-day batch run (Supporting Information, Figure 3). Furthermore, the dMRM method was very quantitative, with high linear range of the internal standard (R2 = 0.9963) in the 50–1500 μg/mL range (Supporting Information, Figure 4). Glycopeptides obtained from sera of lung cancer patients and control suggested marked differences in the glycopeptide profiles (Supporting Information, Figure 5).
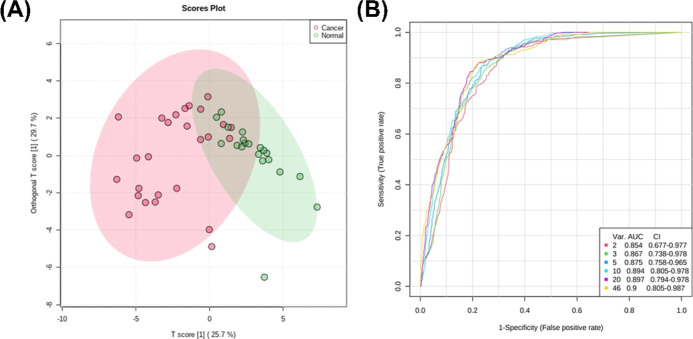
OPLS-DA of the peptide-normalized glycopeptide abundances between cancer patients and controls suggested good separation between the two groups (Figure 3a). ROC-curve analyses suggest excellent accuracy with biomarker models using 20 glycopeptides having AUC = 0.897 (95% CI: 0.794–0.978) (Figure 3b).
Figure 3.
Biomarker analysis of serum glycopeptides comparing lung cancer patients and healthy volunteers. (A) Score plot showing distribution of cancer patients (red) and healthy volunteers (green) across components 1 and 2 given by the OPLS-DA model for serum glycopeptides. (B) ROC curve analysis of serum glycopeptides.
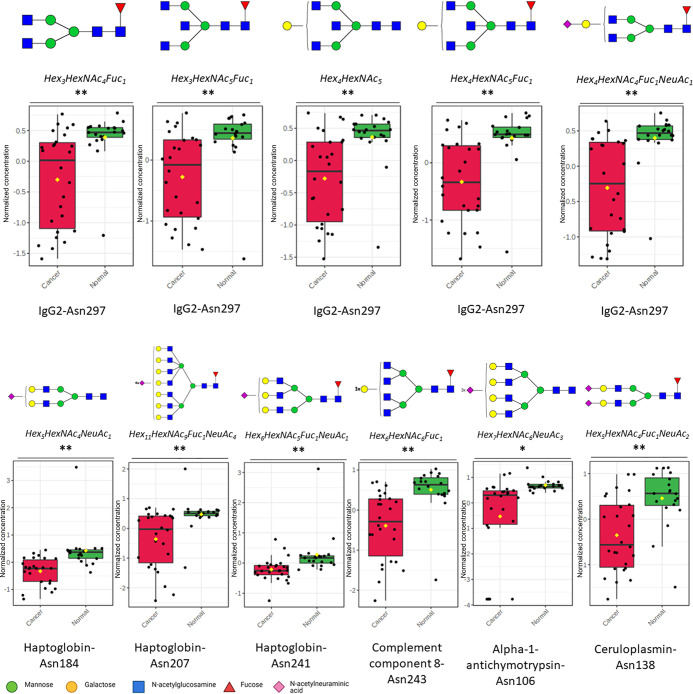
Multiple t-tests (FDR < 0.05) show that these identified glycopeptides were significantly different. Specifically, the glycopeptides TRFE-Asn630_Hex6HexNAc5NeuAc3, TRFE-Asn630_Hex6HexNAc5Fuc1NeuAc3, A1AT-Asn271_Hex5HexNAc4NeuAc1, A1AT-Asn107_Hex5HexNAc4Fuc1NeuAc2, A1AT-Asn107_Hex6HexNAc5Fuc1NeuAc3, CO3-Asn85_Hex7HexNac2, IgM-Asn46_Hex4HexNAc3Fuc1NeuAc1, IgM-Asn46_Hex5HexNAc5NeuAc1, IgM-Asn209_Hex5HexNAc4NeuAc1, IgM-Asn46_Hex5HexNAc4Fuc1NeuAc1, and IgM-Asn46_Hex5HexNAc5Fuc1NeuAc1 were significantly overexpressed in cancer patients (Figure 4). These glycopeptide compositions correspond to changes in serum glycosylation of TRFE (serotransferrin), A1AT (alpha-1-antitrypsin), CO3 (complement C3), and IgM (immunoglobulin M).
Figure 4.
Relative abundances of significantly overexpressed glycopeptides (reported as Protein_Asn glycosite-N-Glycan) in lung cancer; **q < 0.01; ***q < 0.001.
Likewise, the glycopeptides HPT-Asn184_Hex5HexNAc4NeuAc1, HPT_Asn241_Hex6HexNAc5Fuc1NeuAc1, HPT-Asn207_Hex11HexNac9Fuc1NeuAc4, CO8B-Asn243_Hex6HexNAc6Fuc1, CERU-Asn138_Hex5HexNAc4Fuc1NeuAc2, AACT-Asn106_Hex7HexNAc6NeuAc3, IgG2-Asn297_Hex4HexNAc5Fuc1, IgG2-Asn297_Hex4HexNac4Fuc1NeuAc1, IgG2-Asn297_Hex4HexNAc5, IgG2-Asn297_Hex3HexNAc5Fuc1, and IgG2-Asn297_Hex3HexNAc4Fuc1 were significantly underexpressed in cancer patients (Figure 5). These underexpressed glycopeptide changes correspond to glycosylation of HPT (Haptoglobin), CO8B (complement component C8 beta chain), CERU (ceruloplasmin), AACT (alpha-1-antichymotrpysin), and IgG2 (immunoglobulin G2).
Figure 5.
Relative abundances of significantly underexpressed glycopeptides (Protein_Asn glycosite-Glycan) in lung cancer; *q < 0.05; **q < 0.01.
Comparison of the significant glycopeptides with significant N-glycans yielded interesting results. The overexpressed triantennary sialylated N-glycan Hex6HexNAc5NeuAc3 was also overexpressed in the glycopeptide TRFE-Asn630_Hex6HexNAc5NeuAc3. Triantennary N-glycans were similarly overexpressed in the glycopeptides TRFE-Asn630_Hex6HexNAc5NeuAc3, TRFE-Asn630_Hex6HexNAc5Fuc1NeuAc3, and A1AT-Asn107_Hex6HexNAc5Fuc1NeuAc3. On the other hand, the underexpressed biantennary sialofucosylated N-glycan Hex5HexNac4Fuc1NeuAc2 was also underexpressed CERU-Asn138_Hex5HexNAc4Fuc1NeuAc2 as well as overexpressed in Asn107_Hex5HexNAc4Fuc1NeuAc2. These results show the difference in quantifying global glycosylation (in glycomics) and glycoprotein/glycosite-specific glycosylation (in glycoproteomics).
Interestingly, quantification of these glycoproteins using the same dMRM method suggests significant differences in serum glycoprotein expression as well (Supporting Information, Table 2). The glycoproteins TRFE (q = 3 × 10–6), A1AT (q = 0.003353), IgM (q = 0.000825), and IgG2 (q = 0.028177) were found to be significantly underexpressed, while HPT was significantly overexpressed (q = 0.002399) in serum cancer samples compared to healthy volunteers. This result suggests that not only do serum protein expression vary between disease states but also glycosylation of specific protein glycosites differ as well.
Due to the high abundance of protein glycosylation in serum, N-glycans and glycoproteins serve as prime candidate biomarkers.33 These glycoconjugates serve critical roles in cellular functions, including chemical signaling, cell differentiation and proliferation, and protein–protein interactions.34,35 Interestingly, as broad as the functions of these biomolecules are in critical biological processes, aberrantly glycosylated proteins are highly associated with several human diseases, especially cancer.36
Using pathway enrichment analysis,31 the glycoproteins we identified with significantly different glycosylation were found to be involved in complement and coagulation cascades (padj= 6.418 × 10–4), innate immune system (padj= 6.094 × 10–3), acute inflammatory response (padj= 6.404 × 10–5), defense response (padj= 2.082 × 10–4), complement activation pathway (padj= 1.895 × 10–2), and immunoglobulin-mediated immune response pathway (padj= 4.818 × 10–2) (Supporting Information, Figure 6). Most notably, the glycoproteins IgM, IgG2, and complement components 3 and 8 were found to be critical in these pathways. In addition, the glycoproteins we identified with significantly different glycosylation have been previously implicated in cancer. Changes in serotransferrin (TRFE) glycosylation have been implicated in cholangiocarcinoma37 and ovarian cancer.22 Moreover, alpha-1 antitrypsin (A1AT) deficiency is considered as a risk factor for lung cancer development in non-smokers.38 The components of the complement pathway were also suggested as potential drug target in lung cancer.39,40 CO3 has been shown as a prognostic factor in NSCLC.41 The result for CO3 was very interesting, in that it was the only significantly different glycopeptide containing high-mannose glycosylation in all the N-glycan and glycopeptide analytes detected in the study. Furthermore, the abundance of high-mannose N-glycans was negligible in the sera of healthy volunteers (Figure 4). Increased abundance of high-mannose N-glycans has been correlated with cancer progression in tissues and tumor models of lung cancer,15 cholangiocarcinoma,42 and breast cancer.43 The heterotropic production of ceruloplasmin (CERU) was also reportedly higher in invasive lung adenocarcinoma compared to adenocarcinoma in situ, suggesting poor prognostic outcome.44 In our results, the glycosylation of IgM and IgG were significantly overexpressed and underexpressed, respectively. Traditionally, immunoglobulins are thought of as produced only by B immune cells, but recent studies have suggested that cancer cells also produce tumor-derived immunoglobulins during cancer progression.45,46
Serum Biomarker Models for Philippine Lung Cancer Patients Using Significantly Different N-Glycan and Glycopeptide Analytes
Using these results, several biomarker models incorporating N-glycans and glycopeptides were developed (Figure 6). The model features were selected based on LASSO (least absolute shrinkage and selection operator) frequencies as calculated from MetaboAnalyst 5.0 Biomarker module.29 The N-glycan model (AUC = 0.775; 95% CI: 0.617–0.931) was composed of the N-glycans Hex6HexNAc5NeuAc1, Hex6HexNAc5NeuAc3, Hex6HexNAc5Fuc2NeuAc1, Hex7HexNAc6Fuc1NeuAc3, Hex4HexNAc3NeuAc1, and Hex5HexNAc4Fuc1NeuAc2. A biomarker model using glycopeptides (AUC = 0.959; 95% CI: 0.85–1.0) was proposed as well using the following: TRFE-Asn630_Hex6HexNAc5NeuAc3, A1AT-Asn271_Hex5HexNAc4NeuAc1, CO3-Asn85_Hex7HexNac2, CO8B-Asn243_Hex6HexNAc6Fuc1, and A1AT-Asn107_Hex5HexNAc4NeuAc2. The biomarker models using glycopeptides (AUC = 0.959; 95% CI: 0.85–1.0) had higher accuracy than the N-glycan model (AUC = 0.775; 95% CI: 0.617–0.931). These biomarker models illustrate the higher specificity obtained and utility in using glycopeptides for cancer serum diagnostics compared to N-glycans.
Figure 6.
ROC curves of serum biomarker models for lung cancer patients using (A) N-glycan and (B) glycopeptides.
The N-glycans and glycopeptides included in the biomarker models were primarily sialylated, fucosylated, and sialofucosylated. This result highlights the relevance of fucosylation and sialylation in cancer progression.13,17,47−49 Several glycosyltransferases had been associated as cancer biomarkers.13 UDP-N-acetyl-D-glucosamine:N-acetylglucosamine transferase V (GlcNAcT-V), which catalyzes β1-6 branching of N-glycans, has been observed in breast carcinoma.50 Sialyltransferases are another example of glycosyltransferases that are abnormally expressed in cancers and are therefore implicated in carcinogenesis, progression, and metastasis.51−53 Overexpression of α2-3 sialyltransferase III (ST3Gal-III) in pancreatic cancer has been implicated in pancreatic tumor progression. Overexpression of α2-6 sialyltransferase I (ST6GalNAc-I) was related to poor patient survival in colorectal carcinoma patients.54 Similarly, a prior study of glycosylation gene expression changes in lung cancer patients showed significantly upregulated expressions of MAN1A2, MAN2A1, MGAT2, MGAT4B, B4GALT2, FUT2, FUT3, FUT6, and FUT8, while several enzymes, MAN1A1, MAN1C1, MAN2A2, MGAT1, MGAT3, and FUT1 were significantly down-regulated in lung tissue samples of lung cancer patients.14,15
Conclusions
In summary, the N-glycan and glycoprotein profiles of a cohort (n = 26 patients, n = 22 age- and gender-matched volunteers) of Philippine lung cancer patients were analyzed and compared using mass spectrometry techniques to identify potential N-glycan and glycopeptide serum biomarkers. Although no dramatic differences were observed in the total N-glycan compositions of cancer and healthy volunteers, we identified several N-glycan compositions that were significantly different (p < 0.05). This result suggests that in lung cancer patient sera, only specific glycoproteins are differentially expressed and the overall N-glycan composition may not be significantly different. To test this hypothesis, further quantification of the differences between lung cancer and volunteer serum glycopeptides using dMRM gave greater number of highly significantly different glycopeptides (FDR < 0.05) and a better discrimination between the groups (AUC = 0.9 for 46 glycopeptides). Using the information from significantly different N-glycan and glycopeptide analytes, biomarker models were constructed and proposed to discriminate between cancer patients and healthy volunteer, using serum N-glycans (AUC = 0.775; 95% CI: 0.617–0.931) and glycopeptides (AUC = 0.959; 95% CI: 0.85–1.0). To develop serum biomarker models for lung cancer patients, we utilized differentially abundant N-glycans and glycopeptides. These results could open potential diagnostic platforms specifically targeting lung cancer in the Philippine population. One possible limitation of the study is the small sample size; as such, it will be interesting to observe similar changes in cancer stages with significantly larger sample size. Additional validation studies of the identified serum biomarkers are still needed for its use in the development of potential lung cancer diagnostic platforms for the Philippine population.
Data Availability
Mass spectrometry data for glycomics (doi:10.25345/C5R20S05Z, MSV000089041) and glycoproteomics (doi:10.25345/C5VT1GS9C, MSV000089040) are available on MassIVE data repository upon reasonable request.
Acknowledgments
The authors would like to acknowledge the contributions of Dr. Ruby King, Dr. Treah May Suacillo-Sayo, Mary Suzette Angeles, Allysa Marantan, Jennifer Panuelos, Perlita Apelado, Abbie Bautista, Kim Fernandez, Patrick Moreno, Dennis Macapagal, and Shelley Bridget Tomacas for their contributions in clinical trial enrolment and sample collection.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05111.
Representative chromatograms of serum N-glycans from a Philippine lung cancer patient and healthy volunteer; MS/MS spectra of significantly different N-glycans; validation of dMRM method reproducibility; overlaid LC–MS chromatograms of standard digested sera; coefficients of variance (%CVs) of glycopeptide responses that are un-normalized, ISTD-normalized, and peptide-normalized; peptide-normalized glycopeptide response was used to normalize the data; linearity of dMRM method; overlaid LC–MS chromatograms of standard diluted digested sera; linear range and linearity of the response of the internal standard curve; comparative heatmap plot of individual patient and volunteer glycopeptide profiles of the top 15 discriminating glycopeptides (Protein_Asn glycosite-Glycan) from OPLS-DA; pathway enrichment analysis of differentially glycosylated serum glycoproteins, showing enrichment in the innate immune system, acute inflammatory response, defense response, complement activation and cascade, and immunoglobulin-mediated immune response; demographic and clinical information of the Philippine lung cancer cohort; and significantly different peptide compositions (FDR < 0.05) between Philippine lung cancer patients and healthy volunteers (PDF)
This research was supported by the Philippine Commission on Higher Education (CHED) through the Philippine-California Advanced Research Institute (PCARI-IHITM 2017–18) grant.
The authors declare no competing financial interest.
Supplementary Material
References
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca-Cancer J. Clin. 2021, 71, 209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Navada S.; Lai P.; Schwartz A. G.; Kalemkerian G. P. Temporal Trends in Small Cell Lung Cancer: Analysis of the National Surveillance, Epidemiology, and End-Results (SEER) Database. J. Clin. Oncol. 2006, 24, 7082. 10.1200/jco.2006.24.18_suppl.7082. [DOI] [Google Scholar]
- Alberg A. J.; Brock M. V.; Samet J. M. Epidemiology of Lung Cancer: Looking to the Future. J. Clin. Oncol. 2005, 23, 3175–3185. 10.1200/JCO.2005.10.462. [DOI] [PubMed] [Google Scholar]
- Taylor R.; Najafi F.; Dobson A. Meta-Analysis of Studies of Passive Smoking and Lung Cancer: Effects of Study Type and Continent. Int. J. Epidemiol. 2007, 36, 1048–1059. 10.1093/ije/dym158. [DOI] [PubMed] [Google Scholar]
- Freudenheim J. L.; Ritz J.; Smith-Warner S. A.; Albanes D.; Bandera E. V.; van den Brandt P. A.; Colditz G.; Feskanich D.; Goldbohm R. A.; Harnack L.; Miller A. B.; Rimm E.; Rohan T. E.; Sellers T. A.; Virtamo J.; Willett W. C.; Hunter D. J. Alcohol Consumption and Risk of Lung Cancer: A Pooled Analysis of Cohort Studies. Am. J. Clin. Nutr. 2005, 82, 657–667. 10.1093/ajcn/82.3.657. [DOI] [PubMed] [Google Scholar]
- Hwang S.-J.; Lozano G.; Amos C. I.; Strong L. C. Germline P53 Mutations in a Cohort with Childhood Sarcoma: Sex Differences in Cancer Risk. Am. J. Hum. Genet. 2003, 72, 975–983. 10.1086/374567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.-I.; Jeong J.; Lee C. W. Association between EGFR Mutation and Ageing, History of Pneumonia and Gastroesophageal Reflux Disease among Patients with Advanced Lung Cancer. Eur. J. Cancer 2019, 122, 101–108. 10.1016/j.ejca.2019.09.010. [DOI] [PubMed] [Google Scholar]
- Aldrighetti C. M.; Niemierko A.; Van Allen E.; Willers H.; Kamran S. C. Racial and Ethnic Disparities Among Participants in Precision Oncology Clinical Studies. JAMA Netw. Open 2021, 4, e2133205 10.1001/jamanetworkopen.2021.33205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno D. S.; Hess L. M.; Li X.; Su E. W.; Zhu Y. E.; Patel M. Racial Disparities in Biomarker Testing and Clinical Trial Enrollment in Non-Small Cell Lung Cancer (NSCLC). J. Clin. Oncol. 2021, 39, 9005. 10.1200/JCO.2021.39.15_suppl.9005. [DOI] [Google Scholar]
- Cheng T.-Y. D.; Cramb S. M.; Baade P. D.; Youlden D. R.; Nwogu C.; Reid M. E. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J. Thorac. Oncol. 2016, 11, 1653–1671. 10.1016/j.jtho.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero S.; López-Cortés A.; Indacochea A.; García-Cárdenas J. M.; Zambrano A. K.; Cabrera-Andrade A.; Guevara-Ramírez P.; González D. A.; Leone P. E.; Paz-y-Miño C. Analysis of Racial/Ethnic Representation in Select Basic and Applied Cancer Research Studies. Sci. Rep. 2018, 8, 13978. 10.1038/s41598-018-32264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne U.; Jadhav T.; Putcha B.-D. K.; Samuel T.; Soni S.; Shanmugam C.; Suswam E. A. Molecular Biomarkers of Colorectal Cancer and Cancer Disparities: Current Status and Perspective. Curr. Colorectal Cancer Rep. 2016, 12, 332–344. 10.1007/s11888-016-0338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meany D. L.; Chan D. W. Aberrant Glycosylation Associated with Enzymes as Cancer Biomarkers. Clin. Proteomics 2011, 8, 7. 10.1186/1559-0275-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi M. T.; Dracheva T.; Rotunno M.; Figueroa J. D.; Liu H.; Dasgupta A.; Mann F. E.; Fukuoka J.; Hames M.; Bergen A. W.; Murphy S. E.; Yang P.; Pesatori A. C.; Consonni D.; Bertazzi P. A.; Wacholder S.; Shih J. H.; Caporaso N. E.; Jen J. Gene Expression Signature of Cigarette Smoking and Its Role in Lung Adenocarcinoma Development and Survival. PLoS One 2008, 3, e1651 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Taylor S. L.; Stroble C.; Nguyen U. T.; Parker E. A.; Song T.; Lebrilla C. B.; Rom W. N.; Pass H.; Kim K.; Kelly K.; Miyamoto S. Differential N-Glycosylation Patterns in Lung Adenocarcinoma Tissue. J. Proteome Res. 2015, 14, 4538–4549. 10.1021/acs.jproteome.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajaria B. N.; Patel P. S. Glycosylation: A Hallmark of Cancer?. Glycoconjugate J. 2017, 34, 147–156. 10.1007/s10719-016-9755-2. [DOI] [PubMed] [Google Scholar]
- Pinho S. S.; Reis C. A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Stroble C.; Dai J.; Barnett M.; Taguchi A.; Goodman G. E.; Miyamoto S.; Gandara D.; Feng Z.; Lebrilla C. B.; Hanash S. Serum Glycans as Risk Markers for Non-Small Cell Lung Cancer. Cancer Prev. Res. 2016, 9, 317–323. 10.1158/1940-6207.CAPR-15-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Q.; Ruhaak L. R.; Stroble C.; Parker E.; Huang J.; Maverakis E.; Lebrilla C. B. A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J. Proteome Res. 2015, 14, 5179–5192. 10.1021/acs.jproteome.5b00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Kailemia M. J.; Merleev A. A.; Xu G.; Serie D.; Danan L. M.; Haj F. G.; Maverakis E.; Lebrilla C. B. Site-Specific Glycosylation Quantitation of 50 Serum Glycoproteins Enhanced by Predictive Glycopeptidomics for Improved Disease Biomarker Discovery. Anal. Chem. 2019, 91, 5433–5445. 10.1021/acs.analchem.9b00776. [DOI] [PubMed] [Google Scholar]
- Miyamoto S.; Stroble C. D.; Taylor S.; Hong Q.; Lebrilla C. B.; Leiserowitz G. S.; Kim K.; Ruhaak L. R. Multiple Reaction Monitoring for the Quantitation of Serum Protein Glycosylation Profiles: Application to Ovarian Cancer. J. Proteome Res. 2018, 17, 222–233. 10.1021/acs.jproteome.7b00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova R.; Royle L.; Radcliffe C. M.; Abd Hamid U. M.; Evans R.; Arnold J. N.; Banks R. E.; Hutson R.; Harvey D. J.; Antrobus R.; Petrescu S. M.; Dwek R. A.; Rudd P. M. Ovarian Cancer Is Associated with Changes in Glycosylation in Both Acute-Phase Proteins and IgG. Glycobiology 2007, 17, 1344–1356. 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. R.; Nguyen U. T.; Stroble C.; Taylor S. L.; Taguchi A.; Hanash S. M.; Lebrilla C. B.; Kim K.; Miyamoto S. Enrichment Strategies in Glycomics-Based Lung Cancer Biomarker Development. Proteomics: Clin. Appl. 2013, 7, 664–676. 10.1002/prca.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H.; Kim H. S.; Kim B. J.; Han B.; Choi D. R.; Kwon J. H. Prognostic Impact of TTF-1 Expression in Non-Squamous Non-Small-Cell Lung Cancer: A Meta-Analysis. J. Cancer 2018, 9, 4279–4286. 10.7150/jca.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Xie Y.; Wong M.; Barboza M.; Lebrilla C. B. Comprehensive Structural Glycomic Characterization of the Glycocalyxes of Cells and Tissues. Nat. Protoc. 2020, 15, 2668–2704. 10.1038/s41596-020-0350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R.; Taylor S. L.; Miyamoto S.; Kelly K.; Leiserowitz G. S.; Gandara D.; Lebrilla C. B.; Kim K. Chip-Based NLC-TOF-MS Is a Highly Stable Technology for Large-Scale High-Throughput Analyses. Anal. Bioanal. Chem. 2013, 405, 4953–4958. 10.1007/s00216-013-6908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronewitter S. R.; An H. J.; de Leoz M. L.; Lebrilla C. B.; Miyamoto S.; Leiserowitz G. S. The Development of Retrosynthetic Glycan Libraries to Profile and Classify the Human Serum N-Linked Glycome. Proteomics 2009, 9, 2986–2994. 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena J.; Tang X.; Zhou Q.; Harvey D.; Barajas-Mendoza M.; Jin L.; Maezawa I.; Zivkovic A. M.; Lebrilla C. B. Glycosylation alterations in serum of Alzheimer’s disease patients show widespread changes in N -glycosylation of proteins related to immune function, inflammation, and lipoprotein metabolism. Alzheimer’s Dement.: Diagn. Assess. Dis. Monit. 2022, 14, e12309 10.1002/dad2.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.; Chong J.; Zhou G.; de Lima Morais D. A.; Chang L.; Barrette M.; Gauthier C.; Jacques P.-É.; Li S.; Xia J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenot E. A.; Roux A.; Xu Y.; Ezan E.; Junot C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- Raudvere U.; Kolberg L.; Kuzmin I.; Arak T.; Adler P.; Peterson H.; Vilo J. G. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Q.; Lebrilla C. B.; Miyamoto S.; Ruhaak L. R. Absolute Quantitation of Immunoglobulin G and Its Glycoforms Using Multiple Reaction Monitoring. Anal. Chem. 2013, 85, 8585–8593. 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X.; Hood B. L.; Sun M.; Conrads T. P.; Day R. S.; Weissfeld J. L.; Siegfried J. M.; Bigbee W. L. Lung Cancer Serum Biomarker Discovery Using Glycoprotein Capture and Liquid Chromatography Mass Spectrometry. J. Proteome Res. 2010, 9, 6440–6449. 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J.; Elliott D. J. Hallmarks of Glycosylation in Cancer. Oncotarget 2016, 7, 35478–35489. 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Chen S.; Li Q.; Sheng Y.; Alvarez M. R.; Reyes J.; Xu G.; Solakyildirim K.; Lebrilla C. B. Glycan-Protein Cross-Linking Mass Spectrometry Reveals Sialic Acid-Mediated Protein Networks on Cell Surfaces. Chem. Sci. 2021, 12, 8767. 10.1039/D1SC00814E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake P. M.; Cho W.; Li B.; Prakobphol A.; Johansen E.; Anderson N. L.; Regnier F. E.; Gibson B. W.; Fisher S. J. Sweetening the Pot: Adding Glycosylation to the Biomarker Discovery Equation. Clin. Chem. 2010, 56, 223–236. 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamnongkan W.; Lebrilla C. B.; Barboza M.; Techasen A.; Loilome W.; Sithithaworn P.; Khuntikeo N.; Pairojkul C.; Chamadol N.; Thanan R.; Yongvanit P. Discovery of Serotransferrin Glycoforms: Novel Markers for Diagnosis of Liver Periductal Fibrosis and Prediction of Cholangiocarcinoma. Biomolecules 2019, 9, 538. 10.3390/biom9100538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Durán M.; Ruano-Ravina A.; Parente-Lamelas I.; Abal-Arca J.; Leiro-Fernández V.; Montero-Martínez C.; Pena C.; Castro-Añón O.; Golpe-Gómez A.; González-Barcala F. J.; Martínez C.; Guzmán-Taveras R.; Provencio M.; Mejuto-Martí M. J.; Fernández-Villar A.; Barros-Dios J. M. Alpha-1 Antitrypsin Deficiency and Lung Cancer Risk. J. Thorac. Oncol. 2015, 10, 1279–1284. 10.1097/JTO.0000000000000609. [DOI] [PubMed] [Google Scholar]
- Kleczko E. K.; Kwak J. W.; Schenk E. L.; Nemenoff R. A. Targeting the Complement Pathway as a Therapeutic Strategy in Lung Cancer. Front. Immunol. 2019, 10, 954. 10.3389/fimmu.2019.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.; Liu Q.; Li T.; Liao Q.; Zhao Y. Role of the Complement System in the Tumor Microenvironment. Cancer Cell Int. 2019, 19, 300. 10.1186/s12935-019-1027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.; He S.; He L.; Chen J.; Cheng X.; Zhang G.; Zhu B. Complement Component 3 Is a Prognostic Factor of Non-Small Cell Lung Cancer. Mol. Med. Rep. 2014, 10, 811–817. 10.3892/mmr.2014.2230. [DOI] [PubMed] [Google Scholar]
- Park D. D.; Phoomak C.; Xu G.; Olney L. P.; Tran K. A.; Park S. S.; Haigh N. E.; Luxardi G.; Lert-itthiporn W.; Shimoda M.; Li Q.; Matoba N.; Fierro F.; Wongkham S.; Maverakis E.; Lebrilla C. B. Metastasis of Cholangiocarcinoma Is Promoted by Extended High-Mannose Glycans. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 7633–7644. 10.1073/pnas.1916498117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leoz M. L. A.; Young L. J. T.; An H. J.; Kronewitter S. R.; Kim J.; Miyamoto S.; Borowsky A. D.; Chew H. K.; Lebrilla C. B. High-Mannose Glycans Are Elevated during Breast Cancer Progression. Mol. Cell. Proteomics 2011, 10, M110.002717. 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka R.; Shiba-Ishii A.; Nakano N.; Togayachi A.; Sakashita S.; Sato Y.; Minami Y.; Noguchi M. Heterotopic Production of Ceruloplasmin by Lung Adenocarcinoma Is Significantly Correlated with Prognosis. Lung Cancer 2018, 118, 97–104. 10.1016/j.lungcan.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Isaeva O. I.; Sharonov G. V.; Serebrovskaya E. O.; Turchaninova M. A.; Zaretsky A. R.; Shugay M.; Chudakov D. M. Intratumoral Immunoglobulin Isotypes Predict Survival in Lung Adenocarcinoma Subtypes. J. Immunother. Cancer 2019, 7, 279. 10.1186/s40425-019-0747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Lin D.; Peng H.; Huang Y.; Huang J.; Gu J. Cancer-Derived Immunoglobulin G Promotes Tumor Cell Growth and Proliferation through Inducing Production of Reactive Oxygen Species. Cell Death Dis. 2013, 4, e945 10.1038/cddis.2013.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Olio F.; Malagolini N.; Trinchera M.; Chiricolo M. Mechanisms of Cancer-Associated Glycosylation Changes. Front. Biosci., Landmark Ed. 2012, 17, 670–699. 10.2741/3951. [DOI] [PubMed] [Google Scholar]
- Dall’Olio F.; Chiricolo M. Sialyltransferases in Cancer. Glycoconjugate J. 2001, 18, 841–850. 10.1023/a:1022288022969. [DOI] [PubMed] [Google Scholar]
- Honma R.; Kinoshita I.; Miyoshi E.; Tomaru U.; Matsuno Y.; Shimizu Y.; Takeuchi S.; Kobayashi Y.; Kaga K.; Taniguchi N.; Dosaka-Akita H. Expression of Fucosyltransferase 8 Is Associated with an Unfavorable Clinical Outcome in Non-Small Cell Lung Cancers. Oncology 2015, 88, 298–308. 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- Handerson T.; Camp R.; Harigopal M.; Rimm D.; Pawelek J. β1,6-Branched Oligosaccharides Are Increased in Lymph Node Metastases and Predict Poor Outcome in Breast Carcinoma. Clin. Cancer Res. 2005, 11, 2969–2973. 10.1158/1078-0432.CCR-04-2211. [DOI] [PubMed] [Google Scholar]
- Burchell J.; Poulsom R.; Hanby A.; Whitehouse C.; Cooper L.; Clausen H.; Miles D.; Taylor-Papadimitriou J. An 2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology 1999, 9, 1307–1311. 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- Picco G.; Julien S.; Brockhausen I.; Beatson R.; Antonopoulos A.; Haslam S.; Mandel U.; Dell A.; Pinder S.; Taylor-Papadimitriou J.; Burchell J. Over-Expression of ST3Gal-I Promotes Mammary Tumorigenesis. Glycobiology 2010, 20, 1241–1250. 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchi M. A.; Hebbar M.; Hornez L.; Harduin-Lepers A.; Peyrat J. P.; Delannoy P. Multiplex Reverse Transcription Polymerase Chain Reaction Assessment of Sialyltransferase Expression in Human Breast Cancer. Cancer Res. 1998, 58, 4066–4070. [PubMed] [Google Scholar]
- Schneider F.; Kemmner W.; Haensch W.; Franke G.; Gretschel S.; Karsten U.; Schlag P. M. Overexpression of Sialyltransferase CMP-Sialic Acid:Galbeta1,3GalNAc-R Alpha6-Sialyltransferase Is Related to Poor Patient Survival in Human Colorectal Carcinomas. Cancer Res. 2001, 61, 4605–4611. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data for glycomics (doi:10.25345/C5R20S05Z, MSV000089041) and glycoproteomics (doi:10.25345/C5VT1GS9C, MSV000089040) are available on MassIVE data repository upon reasonable request.