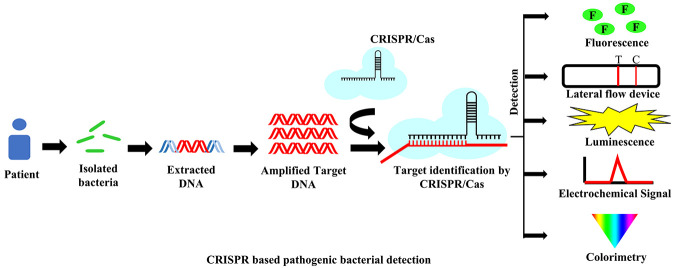
Abstract
Methods enabling rapid and on-site detection of pathogenic bacteria are a prerequisite for public health assurance, medical diagnostics, ensuring food safety and security, and research. Many current bacteria detection technologies are inconvenient and time-consuming, making them unsuitable for field detection. New technology based on the CRISPR/Cas system has the potential to fill the existing gaps in detection. The clustered regularly interspaced short palindromic repeats (CRISPR) system is a part of the bacterial adaptive immune system to protect them from intruding bacteriophages. The immunological memory is saved by the CRISPR array of bacteria in the form of short DNA sequences (spacers) from invading viruses and incorporated with the CRISPR DNA repeats. Cas proteins are responsible for triggering and initiating the adaptive immune function of CRISPR/Cas systems. In advanced biological research, the CRISPR/Cas system has emerged as a significant tool from genome editing to pathogen detection. By considering its sensitivity and specificity, this system can become one of the leading detection methods for targeting DNA/RNA. This technique is well applied in virus detection like Dengue, ZIKA, SARS-CoV-2, etc., but for bacterial detection, this CRISPR/Cas system is limited to only a few organisms to date. In this review, we have discussed the different techniques based on the CRISPR/Cas system that have been developed for the detection of various pathogenic bacteria like L. monocytogenes, M. tuberculosis, Methicillin-resistant S. aureus, Salmonella, E. coli, P. aeruginosa, and A. baumannii.
1. Introduction
Rapid, sensitive, specific, and on-site detection of pathogenic bacteria is critical in clinical diagnosis, treatment, surveillance of foodborne disease, and biological research.1−3 It helps to gather clinical information in order to provide appropriate treatment and prevent the spread of disease.4 According to the World Health Organization’s standard, an ideal pathogen diagnostic test should be assured: affordable, sensitive, specific, easy-to-use, rapid, without large equipment, and delivered to the user.5 In order to detect nucleic acid signatures of pathogens, a vast array of detection methods have emerged based on PCR/qPCR, isothermal amplification-based detection assays, and next-generation sequencing.6−8 To improve sensitivity, affordability, simplicity, and rapidity there are various advanced nucleic acid detection techniques developed so far. One of the latest and advanced methods is clustered regularly interspaced short palindromic repeats (CRISPR) associated systems (CRISPR/Cas),9,10 which have recently gained great importance and attention in nucleic acid analysis and detection. In order to achieve higher sensitivity, the CRISPR/Cas system is frequently associated with polymerase chain reaction (PCR) and with isothermal nucleic acid amplification techniques like NASBA,11 RCA,12−14 SDA,15,16 LAMP,17 RPA,18 and EXPAR.19 The combination of CRISPR/Cas with advanced isothermal amplification technologies is promoting the development of novel optical and electrochemical biosensing devices.
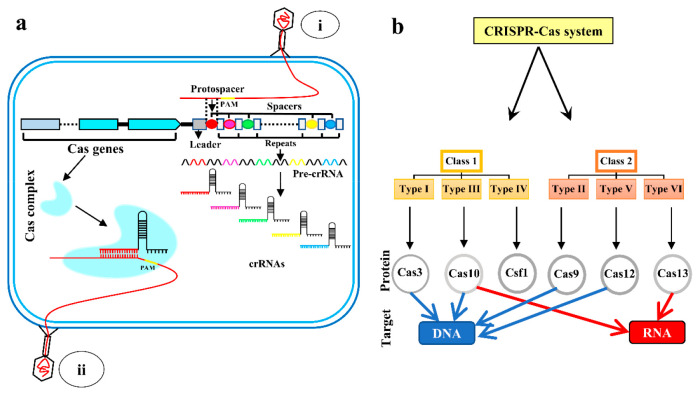
CRISPR/Cas systems provide adaptive protection to bacteria and archaea against invading foreign nucleic acids.9,10 The CRISPR/Cas system in bacteria recognizes and degrades foreign genetic elements generally from viruses.20 These systems are primarily guided by an RNA called guide-RNA (gRNA) or CRISPR RNA (crRNA), which recognizes the target and directs Cas proteins to locate and cleave invading DNA sequences.21 This system works through three steps: adaptation or spacer-acquisition,22 crRNA processing,23 and interference.24 In the spacer acquisition step, when new foreign DNA/RNA is introduced in the bacterial cell, a short piece of the DNA/RNA segment, called protospacer in the immediate upstream vicinity of a protospacer adjacent motif (PAM) present in the foreign DNA/RNA, is excised out by the help of the Cas1–Cas2 complex. This protospacer was then inserted as a new spacer into the bacterial genomic region of the CRISPR array (Figure 1a) where all the acquired spacer resides.22,25
Figure 1.
Basic understanding of CRISPR/Cas system and its classification. (a) CRISPR loci consisted of short repeats (gray boxes) interspaced by various short spacers (colored circular). CRISPR loci are flanked by a cluster of CRISPR-associated (Cas) genes (blue arrows) that encode proteins responsible for the different stages of the CRISPR/Cas system in adaptive immunity. When a bacteriophage (i) enters the cell, a small piece of invader DNA (protospacer) is added to the host chromosome’s CRISPR array as a new spacer (red circle). Transcription takes place across the CRISPR array to generate pre-crRNA, which is then processed into short mature crRNAs. Each crRNA sequence contains one spacer and one flanking repetition. The effector complexes formed by the assembly of the crRNAs and Cas proteins serve as antisense guides for the effector complexes. When the same bacteriophage (ii) will attack the bacteria again, the existing crRNA having the spacer sequences complementary to the portion of the bacteriophage genome will degrade the phage genome by the CRISPR/Cas system. (b) There are two classes and six types of CRISPR/Cas systems where Cas3, Cas9, and Cas12 target the DNA, Cas13 targets RNA, and Cas10 targets both DNA and RNA.
The second step is crRNA biogenesis, which occurs when pre-CRISPR RNA (pre-crRNA) is transcribed by RNA polymerase (RNAP) from the CRISPR array region, then cleaved by specific endoribonucleases into small mature crRNA (Figure 1a). Each crRNA contains one complementary sequence of a spacer.26,27 The final step is interference,24 which entails sequence-specific targeting and cleavage of foreign DNA/RNA having a protospacer that is complementary to the spacer sequence in crRNA. To commence crRNA-mediated DNA binding, a protospacer adjacent motif (PAM) must be present in the immediate vicinity of a protospacer sequence.28 crRNAs recognize and produce complementary base pairs unique to foreign RNA or DNA, resulting in the cleavage of the crRNA-foreign nucleic acid complex29,30 (Figure 1a).
CRISPR/Cas systems are classified according to their utilization of specific Cas enzymes and methods of interference. In accordance with recent publications, CRISPR/Cas systems can be categorized into two classes: class 1 and 2, six types: types I–VI, and numerous subtypes.31 Class 1 comprises types I, III, and IV; and Class 2 includes types II, V, and VI (Figure 1b). Each type is distinguished by discrete effector module configurations that include various signature proteins.32 The most widely used toolbox for nucleic acid detection belongs to the class 2 system that contains Cas9, Cas12, Cas13, and Cas14.33 Cas9 (type II) and Cas12 (type V) target DNA, while Cas13 (type VI) targets RNA34 and Cas14 targets ssDNA.33 CRISPR/Cas systems, specifically Cas9 (type II), have become a popular tool for transcription regulation, genome editing, and in situ DNA/RNA detection in recent years.35 Cas12 and Cas13 effectors have a unique property called “collateral cleavage”.36 In the presence of a target or reporter DNA/RNA, these Cas effectors are activated and can do collateral (nonspecific) cleavage on any single-stranded DNA/RNA present in the near vicinity.37 The advantage of this collateral cleavage is that it can easily be detected by fluorescence reporters tagged in single-stranded DNA/RNA. This has recently displayed remarkable potential in developing novel biosensing technologies for nucleic acid detection.37 This technology is widely harnessed for the detection of viral diseases, such as specific high-sensitivity enzymatic reporter unlocking (SHERLOCK)38−40 to detect Zika and Dengue38 and DNA endonuclease-targeted CRISPR trans reporter (DETECTR)41,42 for rapid and specific detection of HPV and SARS-CoV-2 in humans. Though the futuristic developments of CRISPR/Cas-based viral detection techniques are expanding rapidly in biosensing, this technique is limited to very few bacterial pathogens to date. This limited use for bacterial pathogens may be because there is a well established gold-standard detection technique for pathogenic bacteria and establishing these emerging techniques in practice will be time-consuming. Also, the viral rate of mutation is relatively much higher than bacteria, which provides more priority to develop new methods for viral detection and lesser focus on bacterial detection. In this review, we have discussed the different CRISPR/Cas systems as biosensors used for the detection of bacterial pathogens like L. monocytogenes, M. tuberculosis, Methicillin-resistant S. aureus, Salmonella, E. coli, P. aeruginosa, and A. baumannii.
2. Detection of L. monocytogenes Using CRISPR/Cas System
Listeria monocytogenes is one of the most virulent foodborne pathogens43 and can be found in a variety of foods like milk, milk products, eggs, poultry, and meat.44 The FDA upholds a zero-tolerance policy for L. monocytogenes since it has a low infectious dose and high mortality rate. In healthy people, it can cause invasive listeriosis. In young, elderly, or immunocompromised people, it can cause septicemia, meningitis, and infections related to the central nervous system. Infections in pregnant women can be fatal and can result in spontaneous abortion or fetal death.45 The slow growth rate of L. monocytogenes is challenging for the conventional culture and plating-based detection methods, which can take up to 7 days to yield results.46
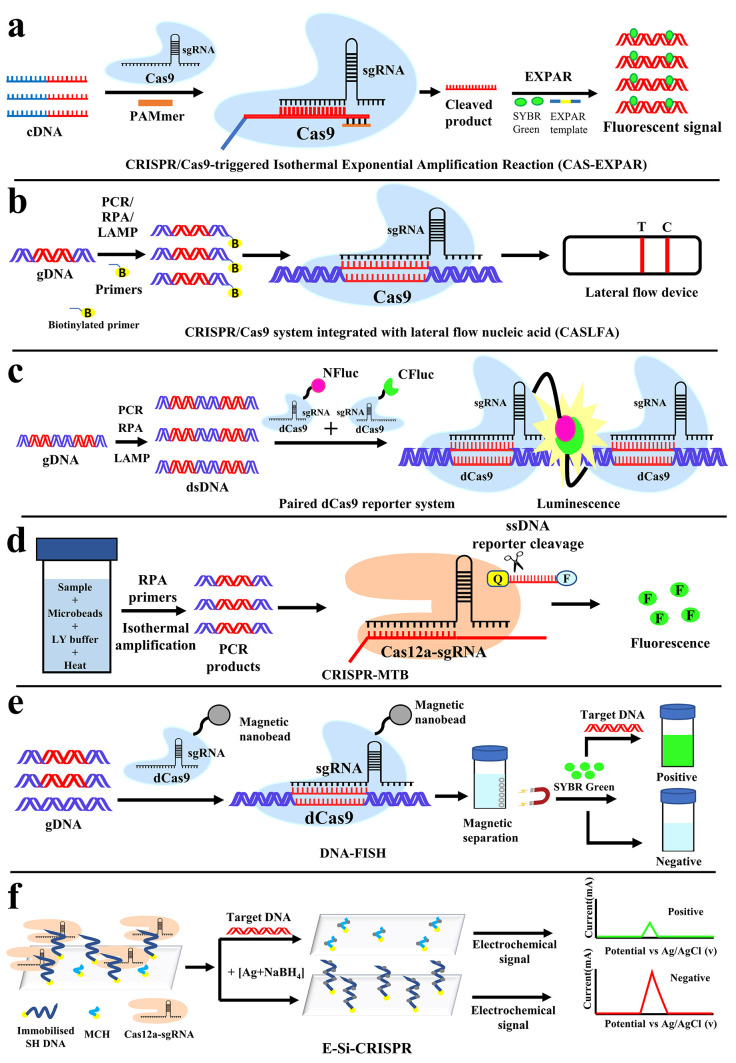
CRISPR/Cas9-triggered isothermal exponential amplification reaction (CAS-EXPAR) based detection against Listeria monocytogenes was developed by Huang et al.47 Here the hemolysin (hly) gene of L. monocytogenes was used as the target sequence. It utilizes the target-specific nicking activity of Cas9 and nicking endonuclease (NEase)-mediated amplification. From bacteria, RNA was isolated and cDNA was generated. cDNAs were cleaved by Cas9 with the help of specific sgRNA and PAMmers. These cleaved products are now subjected to EXPAR-mediated amplification by EXPAR templates and without exogenous primers. Finally, the amplified products were detected by fluorescence using SYBR green (Figure 2a). This method combines the benefits of Cas9/sgRNA site-specific cleavage and EXPAR fast amplification kinetics. This process is reported to be highly specific in discriminating single-nucleotide mismatch. Reprogrammable cleavage activity of Cas9/sgRNA is also beneficial for targeting various other pathogens. The merit of this method does not require exogenous primers for amplification. Therefore, the chances of nonspecific amplification followed by false positivity could be minimized. The sensitivity of this technique was reported to be 0.82 amol (Table 1) of synthetic ssDNA, but in bacteria, this technique was verified with 1.25 and 2.5 μg of total RNA. This method may have a problem to detect long targets, as EXPAR is not efficient for long DNA or RNA targets.
Figure 2.
Detection of pathogen nucleic acids with a CRISPR/Cas-based assay to detect L. monocytogenes, M. tuberculosis, and MRSA. Schematic illustration of (a) CAS-EXPAR and (b) CASLFA for the detection of L. monocytogenes, (c) paired dCas9 (PC) reporter system, and (d) CRISPR-MTB for the detection of M. tuberculosis. (e) CRISPR-mediated DNA-FISH and the (f) E-Si-CRISPR technique for the detection of Methicillin-resistant Staphylococcus aureus (MRSA).
Table 1. List of CRISPR/Cas Systems Used for Detection of Bacterial Pathogens.
| bacteria | system name | target nucleic acid for extraction | target | effector Cas | amplification method | sensitivity | read out | time | ref |
|---|---|---|---|---|---|---|---|---|---|
| L. monocytogenes | Cas-EXPAR | RNA | hemolysin (hly) gene | Cas9 | EXPAR | 0.82 amol (0.82 × 10–9 nmol) | fluorescence detection | ≈1 h | Huang et al. 201847 |
| L. monocytogenes | CASLFA | DNA | hemolysin (hly) gene | dCas9 | PCR/Isothermal amplification | 150 copies | lateral flow nucleic acid (LFA) | 40 min–1 h | Wang et al. 202048 |
| M. tuberculosis | Paired dCas9 | DNA | 16s rRNA | dCas9 | PCR | 5 × 10–5 nmol/mL | luminescence | 1 h | Zhang et al. 201749 |
| M. tuberculosis | CRISPR-MTB | DNA | IS6110 | Cas12a | RPA | 50 CFU/mL | qPCR instrument | ≈1.5 h | Ai et al. 201950 |
| Methicillin-resistant S. aureus (MRSA) | DNA-FISH | DNA | mecA gene | dCas9 | Amplification free | 10 CFU/mL | fluorescence detection | ≈ 0.5 h | Guk et al. 201751 |
| Methicillin-resistant S. aureus (MRSA) | E-Si-CRISPR | DNA | mecA gene | Cas12a | Amplification free | 3.5 fM (3.5 × 10–9 nmol/mL) | electrochemical | 1.5 h | Suea-Ngam et al. 202152 |
| Salmonella sp. | APC-Cas | -- | whole bacteria | Cas13a | Isothermal amplification | 1 CFU | fluorescence detection | 2 h20 min | Shen et al. 202053 |
| Salmonella sp. | CRISPR/Cas12a-powered dual-mode biosensor | DNA | invA gene | Cas12a | PCR | 1 CFU/mL | naked eye/colorimetric/photothermal | 1 h | Ma et al. 202154 |
| E. coli O157:H7 | CRISPR/Cas9 triggered SDA-RCA | DNA | hemolysin A (hlyA) gene | Cas9 | SDA + RCA | 40 CFU/mL | fluorescence detection | 2 h | Sun et al. 202055 |
| P. aeruginosa | CRISPR/Cas and loop mediated isothermal amplification (CIA) | DNA | acyltransferase gene | Cas12a | LAMP | 1 CFU/mL | naked eye detection | 50 min | Mukama et al. 202037 |
| A. baumannii | multiplex PCR integrated with CRISPR/Cas system | DNA | β-lactamase genes | Cas12a | Multiplex PCR | 50 CFU/mL | fluorescence detection | 2 h | Wang et al. 202156 |
Another method to detect L. monocytogenes was developed by Wang et al.48 based on CRISPR/Cas9 system integrated with lateral flow nuclic acid (CASLFA). Here also, the hly gene was used as a target. This technique is termed as the DNA unwinding-based hybridization assay with a lateral flow device for simple and easy detection by the naked eye. Genomic DNA from bacteria was subjected to amplification (by PCR or any isothermal reaction) with gene-specific biotinylated primers. Then biotinylated amplicons are incubated with target specific sgRNA and dCas9 to form a complex (Cas9/sgRNA-biotinylated amplicons) without cleaving the targets. This complex, when applied to a lateral flow device, bound with an AuNP-DNA probe (gold nanoparticle bound with complementary DNA sequence of target gene/sgRNA) and combined with immobilized streptavidin in the test line (T). Accumulation of AuNP-generated color band occurred on the test line (T). The excess unbound AuNP-DNA probe will form a control line (C) by binding with the precoated DNA probes with their control line hybridization region (Figure 2b). There are two kinds of DNA probes reported by the authors: DNA unwinding and sgRNA anchor-based.48 The DNA unwinding probe has a specific DNA sequence for individual target. Therefore, for every target, there is a need to generate a new probe. The sgRNA-based probe has a target sequence that has DNA sequences specific to the crRNA region. The advantage of the sgRNA-based probe is that it can detect multiple targets as this is specific to a portion of sgRNA but not the target DNA. This is a simple and rapid method for the detection of genetic targets by the naked eye. This detection limit was reported as low as 150 copies (Table 1) of bacterial targets. As reported, this method can detect the gene target with almost no background signal interference and can be completed based on a cheap and portable tool kit within 40 min in point of care.48
3. CRISPR/Cas-Mediated Detection of M. tuberculosis
Tuberculosis (TB) is the leading cause of death among infectious diseases.57 Due to the difficulty in its diagnosis, it was anticipated that 40% of the cases failed to be identified and reported.50
Zhang et al.49 have developed a novel and sensitive detection method to detect M. tuberculosis using CRISPR/dCas9. The Mtb 16S rRNA gene was used as a target sequence. In this method, the luciferase gene was split into N- and C-terminal halves (NFluc and CFluc) and fused each with separate dCas9 termed as a paired-dCas9 (PC) reporter system. Two guide RNAs (sgRNA) were also employed which are complementary to the upstream and downstream proximal segments (∼44bp) of a target DNA. Upstream and downstream sgRNAs are separately mixed with NFluc-Cas9 and CFluc-Cas9, respectively, to achieve higher efficiency and specificity. The two halves can initiate heterodimerization to rebuild the intact enzyme when target DNA containing the two segments in proximity is detected and bound by the corresponding sgRNAs followed by a pair of dCas9 (Figure 2c). Luminescence is generated from the catalytic activity of luciferase. The PC reporter system was reported to be sensitive up to 5 × 10–5 nmol/mL (Table 1) but also observed that when the target concentration was beyond 6 × 10–3 nmol/mL detection signals decreased due to inefficient pairing between two halves of luciferase.49 This method requires two target sites, for separate sgRNAs, spanning approximately 44bp, which could limit the selection of target region for other pathogens.
The other method to detect M. tuberculosis by CRISPR-MTB was developed by Ai et al.50 Here MTB-specific insertion sequence IS6110 of ∼1.5 kb in length was used as the target sequence. In this method, optimization of the extraction process was done based on a combination strategy of bead beating, chemical lysis, and heating to ensure the higher efficiency of DNA extraction. As the amplification technique is RPA, there was no special need for any thermal-cycler. This method is a combination of RPA reaction with a CRISPR/Cas12a system (Figure 2d). Fluorescence signal can be detected by target activated reporter cleavage of Cas12a trans cleavage activity. The sensitivity of this method was reported to be 2–5 copies/μL or 50 CFU/mL (Table 1). It was reported that the extraction efficiency was found to be high and the extraction time was reduced with fewer centrifugation steps over the column-based traditional method. This method also was tested on a variety of sample types such as sputum, BALF, CSF, and pus. The drawback of this detection will be that it will fail to detect MTB strains that lack IS6110 genomic segments.
4. Methicillin-Resistant S. aureus (MRSA) Detection by CRISPR/Cas System
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important multidrug-resistant human pathogens, causing severe life-threatening diseases.58 MRSA infections are four times more likely than methicillin-susceptible S. aureus (MSSA), and it causes serious morbidity and mortality worldwide.59,60 Traditional culture-based identification methods for MRSA are time-consuming, and conventional techniques like MALDI-TOF,61 RT-PCR,62S. aureus protein A (spa) typing,63 multilocus sequence typing (MLST),64 and pulsed-field gel electrophoresis (PFGE)65 are labor intensive and require a high level of professional expertise. Therefore, MRSA detection requires simplified detection procedures that are faster, less labor-intensive, and highly specific.
Kyeonghye Guk and colleagues recently introduced a CRISPR-mediated DNA-FISH.51 This CRISPR-mediated DNA-FISH was developed to detect methicillin-resistant Staphylococcus aureus (MRSA) by targeting the gene mecA. This technique involves dCas9 to specifically recognize the target gene without cleavage activity and SYBR Green as a fluorescent probe. The genomic DNA of the target organism was isolated and treated with dCas9/sgRNA for 15 min at room temperature to bind the dCas9 with the target. After hybridization, the dCas9/sgRNA complex was isolated using Ni-NTA magnet beads, and nontarget unbound DNA was removed by washing. SYBR green was added to detect the presence of bound DNA as a target (Figure 2e). In clinical isolates, this method can detect as low as 10 CFU/mL within 30 min (Table 1). This approach is both quick and sensitive. This method does not require any amplification which is an advantage that reduces the detection time and complexity. The combination of dCas9/sgRNA and SYBR green as a fluorescent probe makes for labeling a reasonably straightforward and inexpensive approach. This method of detection has great potential to be used easily in patient point of care.
Suea-Ngam et al. have developed another amplification-free method for the detection of MRSA.66 Here also, the mecA gene was chosen as the target. In this method of detection, the silver metallization technology was combined with the CRISPR/Cas to create a novel silver-enhanced E-CRISPR biosensor (E-Si-CRISPR) for MRSA detection. In the presence of target DNA the Cas12a–gRNA complex cleaves the ssDNA at random sites, destroying the electrode’s ssDNA surface layer (Figure 2f). The trans-cleavage mechanism fails in the absence of the target. The degree of silver deposition during the succeeding silver metallization stage is proportional to the quantity of ssDNA left and thus proportional to the initial amount of target DNA. Square wave voltammetry was used to read the final electrochemical signal (Figure 2f). The detection and quantification limits were found to be 3.5 and 10 fM (Table 1). This new electrochemical CRISPR/Cas biosensor, based on silver metallization, was stated to be highly selective, sensitive, and without DNA amplification cycles. As reported, this amplification-free detection method could yield results within 1.5 h.66 This method is innovative in the aspect of its unique readout of results through electrochemical signals. One drawback can be perceived that, contrary to the other conventional methods, the positive signals are lower than the negative signals in this method, which could be inconvenient.
5. Detection of Salmonella by CRISPR/Cas System
Food poisoning by Salmonella species is the second most prevalent cause of food poising followed by severe gastroenteritis and bacteremia worldwide.67 To date, traditional biochemical culture, immunological testing, and molecular biological approaches (PCR/real-time PCR) have been used to detect Salmonella.68 These procedures are time-consuming, have low specificity, and require expensive laboratory equipment.69
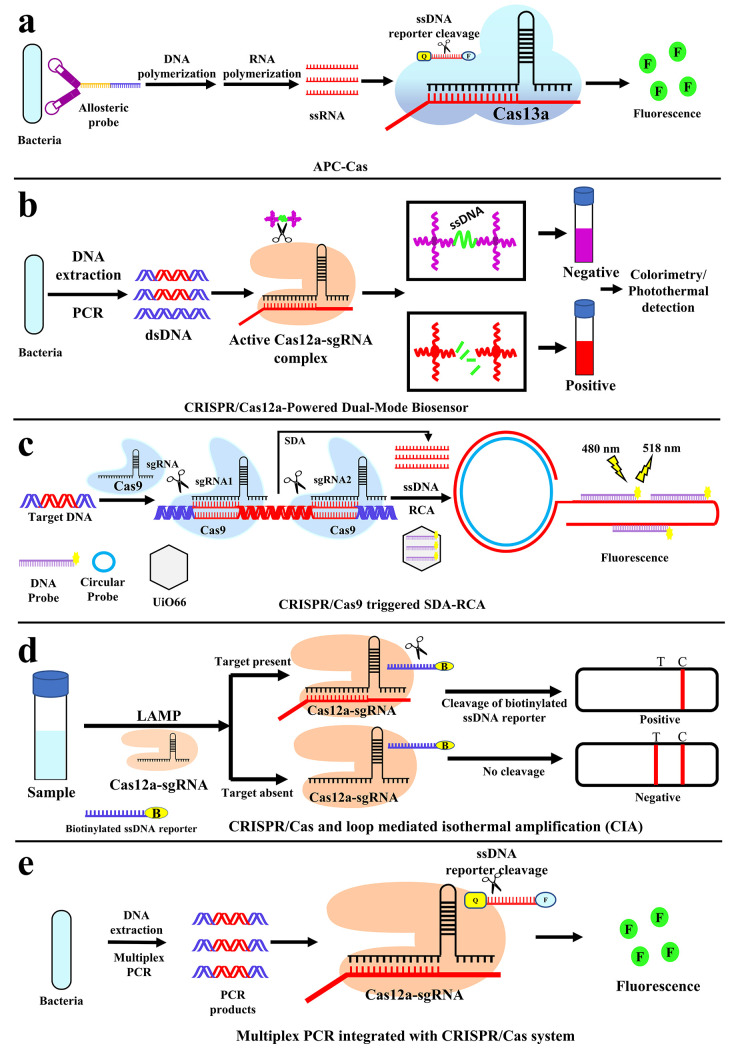
Detection of Salmonella enteritidis using a unique allosteric probe (AP) with a combination of CRISPR/Cas13a (APC-Cas)is developed by Shen et al.,53 where whole bacteria were used as a target. The allosteric probe (AP) comprises of three functional domains (Figure 3a): aptamer domain for target pathogen identification (purple), primer binding site domain (blue), and T7 promoter domain (yellow). A phosphate group was added to the 3′ end of AP to prevent self-extension and make the DNA molecule resistant to enzymatic hydrolysis. The aptamer domain of AP can specifically recognize and engage with the target pathogen that is Salmonella enteritidis. The hairpin structure of AP will unfold and flip to its active configuration, allowing primers to anneal to the exposed primer binding site domain. The AP then acts as a template for the production of double-stranded DNA (dsDNA) with the help of DNA polymerase, followed by the displacement of the target pathogen for the next catalytic cycle (primary amplification) because of the polymerase extension reaction. T7 RNA polymerase is then utilized to identify the T7 promoter sequence on the created dsDNA and perform amplification via transcription to generate a large number of single-stranded RNAs (ssRNAs) (secondary amplification). Finally, the crRNA is intended to contain two areas, a guide sequence that is complementary to the transcripted ssRNA, and the repeat sequence that is required for crRNA to attach the Cas13a enzyme. When the above ssRNAs hybridize with Cas13a/crRNA, Cas13a/collateral crRNA’s cleavage capacity is activated, allowing numerous RNA reporter probes to be cleaved (tertiary amplification), resulting in the amplification of fluorescence signals (Figure 3a). This procedure does not require bacterial isolation, nucleic acid extraction, and a washing step. It is cost-effective, very sensitive up to 1 CFU (Table 1), and can be done in a relatively short period.53 Designing an allosteric probe for different bacteria could be a challenging task for this method of detection.
Figure 3.
Detection of pathogen nucleic acids with CRISPR/Cas-based assay to detect S. enteritidis, Salmonella, E. coli, P. aeruginosa, and A. baumannii. Schematic representation of (a) allosteric probe-induced catalysis and CRISPR/Cas13a (APC-Cas) system for the detection of S. enteritidis, (b) CRISPR/Cas12a-powered dual-mode biosensor for colorimetric and photothermal-based detection of Salmonella, (c) CRISPR/Cas9-triggered SDA-RCA for the detection of E. coli O157:H7, (d) CIA-based LFB for the detection of P. aeruginosa, and (e) simultaneous detection of multiple genes with Multiplex PCR integrated with the CRISPR/Cas system for A. baumannii.
In order to detect Salmonella, Ma et al.54 have developed gold-nanoparticles (AuNPs)-based method termed as CRISPR/Cas12a-powered dual-mode biosensor. The target DNA was Invasion gene A (invA), a virulence gene of Salmonella.70 This method involves DNA extraction as well as PCR amplification of target sequence. The designed biosensor is built on the trans-cleavage activity of the CRISPR/Cas12a. The AuNPs probe is coated with DNA, and a linker ssDNA hybridizes with AuNPs-DNA probe pairs. In the absence of target amplicons, the linker ssDNA remains intact, and aggregated AuNPs are maintained with a purple color (Figure 3b). Upon recognition of the target amplicons with designed crRNA, the trans-cleavage of CRISPR/Cas12a is activated and the linker ssDNA is cut off, and the AuNPs are dispersed in solution. The dispersed AuNP solution exhibits a red color, and the change can be detected by the naked eye or colorimetrically or photothermally (Figure 3b). The detection limit for this technique was reported as 1 CFU/mL (Table 1). This method was just used to detect the bacteria in milk samples. There is a need to explore with other food samples. This technique was the first to explore the gold-based nanoparticles as a probe.54
6. CRISPR/Cas-Mediated Detection of E. coli
Escherichia coli O157:H7 is one of the most common causes of hemorrhagic colitis.71,72E. coli O157:H7 can be found in water as well as other food sources such as milk, juice, fruits, and vegetables. Infections that are severe enough can lead to hemorrhagic colitis, hemolytic uremic syndrome, and even death.73,74
A CRISPR/Cas9 triggered SDA–RCA method on the UiO66 platform was developed by Sun et al.55 to detect Escherichia coli O157:H7. The method employs the target sequence of gene hemolysin A (hlyA). Nanoparticle (UiO66) and Two amplification methods: strand displacement amplification (SDA) and rolling circle amplification (RCA) are used for this method. After isolation of DNA from the bacteria, the pair of CRISPR/Cas9 (by sgRNA1 and sgRNA2) recognized and cleaved the two proximal regions of the target DNA. Primary amplification by SDA synthesis and extending at the nicked position results in short–ssDNA indefinitely. This short–ssDNA was the template for secondary amplification by RCA, which generates long–ssDNA having repetitive sequences complementary to the fluorescence-labeled DNA probe (Figure 3c). This probe in bound form with long–ssDNA can be detected with 480 and 518 nm of excitation and emission wavelength. Unbound probes are absorbed in the UiO66 where fluorescence is quenched. In the presence of the target sequence, short (by SDA) followed by long ssDNA (by RCA) will be generated. The fluorescence probes will leave UiO66 and hybridize with the long–ssDNA, resulting in a fluorescence signal. As a result, the fluorescence intensity can be used to detect the target DNA quantitatively (Figure 3c). It is reported that this technique can detect low amounts of E. coli O157:H7 (40 CFU/mL) with high sensitivity and a wide detection range under mild response conditions (Table 1).
7. CRISPR/Cas-Mediated Detection of Pseudomonas aeruginosa
Pseudomonas aeruginosa is a multidrug-resistant, highly infectious opportunistic human pathogenic bacteria with a large and complex genome.75 Its widespread distribution in nature indicates a high level of genetic and physiological flexibility in response to environmental changes.76 In 2017, the World Health Organization designated P. aeruginosa as a critical pathogen that poses a serious threat to human health, necessitating the development of new treatments.77
Mukama et al. have developed a method to detect P. aeruginosa(37) based on CRISPR/Cas and loop mediated Isothermal Amplification (CIA). The acyltransferase gene from P. aeruginosa was used as a target. Here the samples were directly used for loop-mediated isothermal amplification (LAMP) for the target gene. Products of LAMP were incubated with CRISPR/Cas12, to activate the collateral cleavage of the biotinylated ssDNA reporter, followed by a run on a strip for final results (Figure 3d). In the absence of a target, the gold nanoparticle-streptavidin (AuNP-SA) complex binds with the biotin of the ssDNA reporter. Then the whole complex (AuNP-SA-ssDNA) binds with complementary DNA to the ssDNA reporter immobilized in the test (T) line which results in a visible colored band (Figure 3d). But, in the presence of a target, the reporter DNA was cleaved by CRISPR/Cas12, so there will not be any formation of the AuNP-SA-ssDNA complex, hence no visible band on the test line. In the control line (C), only AuNP-SA is bound with immobilized antibodies to streptavidin. That means in the presence of any P. aeruginosa, there will be only one band in the control and no visible band in the test line (Figure 3d). Whereas, if the sample is negative for P. aeruginosa then the visible band will be there in the T as well as in the C line in the strip. This method was reported to be a fast, accurate, robust, and inexpensive technique with a detection limit of 1 CFU/mL (Table 1). The best feature of this approach is that it allows for naked-eye detection. That means this detection technique has the potential to apply in the patient point of care. On the other hand, the method of detection is unconventional too. In general, we are accustomed with a positive sample with a positive band in the test line, but here the positive sample is associated with a negative band in the test line, which may have some inconvenience in the hand of technicians. Additionally, we cannot eliminate the possibility of false positivity due to various reasons like degradation of ssDNA reporter, AuNP-SA, or complementary DNA in the test line.
8. CRISPR/Cas-Mediated Detection of A. baumannii
The rapid emergence of multidrug-resistant A. baumannii has posed a severe threat to worldwide public health.78 In humans, it can be an opportunistic pathogen that affects immunocompromised persons and is becoming more common as a hospital-borne (nosocomial) infection.79 Detection of A. baumannii based on the CRISPR/Cas system was developed by Wang et al.56 This method was integrated with multiplex PCR where simultaneously many genes of β-lactamase, responsible for antibiotic resistance, were detected. Extracted genomic DNA from bacteria was used for the multiplex PCR reaction. When the target gene is present, the system will amplify the target and then initiate Cas12a’s nonspecific ssDNA trans cleavage activity. The ssDNA reporter, conjugated with fluorophore and quencher, was cleaved after the Cas12a-crRNA-DNA assembly, resulting in an increase in fluorescence signals (Figure 3e). Different crRNAs were used to detect different genes. The detection limit was reported to be 50 CFU/mL (Table 1). Integration of multiplex PCR provided an added advantage where multiple targets can be detected at once, provided the individual target specific crRNA needs to be developed.
9. Discussion
All the methods discussed above based on CRISPR/Cas-based approaches for bacterial detection are distinct in their own way, employing various Cas enzymes and techniques. According to the ASSURED standards of WHO, all the above-described methods may not fully qualify all the standards, whereas some might be more affordable and others might be more sensitive or have less bulky equipment, or be easy to use in point of care. Likewise, amplification free methods, such as DNA-FISH51 and E-Si-CRISPR,66 need less time and fewer components, making them relatively inexpensive. Methods like CASLFA,48 APC-Cas,53 CIA,37 and CRISPR-MTB50 can also be conducted utilizing the cost-effective isothermal amplification technique.80 The CASLFA,48 CIA,37 and CRISPR/Cas12a-Powered Dual-Mode Biosensor54 have naked eye readout capabilities requiring no expensive equipment. The APC-Cas53 technique also does not need bacterial isolation or DNA extraction, which contributes to its low cost. The turnaround times for the methods described above ranged from 30 to 140 min. The CRISPR-mediated DNA-FISH for Methicillin-resistant Staphylococcus aureus (MRSA) detection51 has the quickest turnaround time of 30 min as it is an amplification free method. In terms of the analytical sensitivity of the CRISPR/Cas assays, the limit of detection (LoD) was reported in different units: copies/mL, CFU/mL, moles, and molarity. The majority of methods reported the LoD in CFU/mL, with the lowest reported LoD being 1 CFU/mL for both CRISPR-Cas12a-powered dual-mode biosensor54 and CIA37 (Table 1). APC-Cas53 and CASLFA48 was reported to be 1 CFU and 150 copies (Table 1). Between dCas949 and E-Si-CRISPR,52 where both the LoD were reported in molar concentration, the lowest being for E-Si-CRISPR52 that is up to 3.5 fM (Table 1). The LoD of CAS-EXPAR47 was estimated to 0.82 amol. Expressing the sensitivity with different units is misleading; therefore, it is hard to compare among methods. In the case of the bacterial detection method, it would have been helpful for the readers, if the sensitivity could have been reported to the standard unit that is CFU/mL.
The CRISPR-based methods reduced the need for large equipment, which is a notable feature that has significant possibilities for field implementation, particularly for controlling epidemic outbreaks in resource-limited areas. CRISPR/Cas systems make it easier to create a wide range of readout signals from fluorescence to naked eye detection. Methods based on LFA such as CASLFA48 and CIA37 might have higher utility at the patient point of care. Though CRISPR/Cas-based pathogen detection technologies have exceptional sensitivity and specificity, there are yet many scopes for future advancements. Due to the concerning inherent off-target impact of CRISPR-based detection, improved specificity is the utmost requirement for practical detection approaches.36 For example, outside of the PAM-proximal 5–12 bp seed areas, Cas9-mediated cleavage is very tolerant to mismatches,81 and dCas9 off-target binding is random,82 which can weaken the analytical specificity and sensitivity. In the past few years, Cas9 variants with reduced off-target cleavage, such as SpCas9-HF1,83 eSpCas9 (1.1),84 and HypaCas9,85 have been developed, thereby providing potential future solutions for the off-target effect of Cas9. Therefore, for Cas enzymes, future research should be concentrated on high-fidelity nucleic acid detection to minimize off-targeting. A more user-friendly one-step diagnostic that comprises pathogen nucleic acid release, preamplification, CRISPR/Cas-induced reaction, and signal readout should be developed in the future. For example, several simple formats, such as paper-based biosensors with visual readout (NASBACC,86 SHERLOCK,40 DETECTR87), pathogen detection without nucleic acid extraction (HUDSON),88 and a single-tube test combining isothermal amplification and Cas-mediated reaction, have been developed,39,89 which might be combined for more simplicity, affordability, and user-friendliness. More equipment-free approaches for signal readout, such as lateral flow assay39 or naked-eye view89 under light, should be introduced, as they might be easier at the point of care. Currently, the CRISPR-based detection system must be stored and delivered in a cold chain, which is an inconvenience in many remote areas. The NASBACC38 detection system with freeze-dried reagents and the SHERLOCK40 system with freeze-dried and paper spotting reagents showed long-term storage and transport. Therefore, more storage and transportation strategies for the CRISPR-based reaction kit should be developed.
10. Conclusion
CRISPR/Cas-mediated detection system is a very powerful and advanced technique with high specificity and sensitivity. Therefore, this CRISPR/Cas technique could be of high potential for early diagnosis in the present emerging scenario of antibiotic resistance. In most of the techniques discussed above for pathogenic bacterial detection, there are different amplification techniques like PCR, LAMP, RCA, and SDA integrated along with the CRISPR/Cas system. Previously, only positive amplification was sufficient for detection, but due to the high rate of false positivity, reliance toward only amplification to detect pathogens accurately becomes untrustworthy. Therefore, target amplification followed by CRISPR/Cas as biosensor-mediated detection of pathogenic bacteria can make the process more robust, reliable, sensitive, and specific.
Acknowledgments
J.C. acknowledges the Swami Vivekananda Merit-cum-Means Scholarship from Govt. of West Bengal. Authors are thankful to Raiganj University for all the suport. The authors extend their appreciation to the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) for funding and supporting this work through Research Group no. RG-21-09-89.
Author Contributions
H.S. and J.C. conceptualize the idea and prepared the original draft. S.M.R., A.A.C., S.K., and H.A.R. reviewed and edited the manuscript.
The authors declare no competing financial interest.
References
- Qiao Z.; Fu Y.; Lei C.; Li Y. Advances in Antimicrobial Peptides-Based Biosensing Methods for Detection of Foodborne Pathogens: A Review. Food Control 2020, 112, 107116. 10.1016/j.foodcont.2020.107116. [DOI] [Google Scholar]
- Velusamy V.; Arshak K.; Korostynska O.; Oliwa K.; Adley C. An Overview of Foodborne Pathogen Detection: In the Perspective of Biosensors. Biotechnol. Adv. 2010, 28 (2), 232–254. 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Sassolas A.; Leca-Bouvier B. D.; Blum L. J. ChemInform Abstract: DNA Biosensors and Microarrays. ChemInform. 2008, 39 (17), 1. 10.1002/chin.200817270. [DOI] [PubMed] [Google Scholar]
- Broadhurst M. J.; Brooks T. J. G.; Pollock N. R. Diagnosis of Ebola Virus Disease: Past, Present, and Future. Clin. Microbiol. Rev. 2016, 29 (4), 773–793. 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosack C. S.; Page A. L.; Klatser P. R. A Guide to Aid the Selection of Diagnostic Tests. Bull. World Health Organ. 2017, 95 (9), 639–645. 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler O.; Glynn B.; Kurg A. Nucleic Acid Detection Technologies and Marker Molecules in Bacterial Diagnostics. Expert Rev. Mol. Diagn. 2014, 14 (4), 489–500. 10.1586/14737159.2014.908710. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Chen F.; Li Q.; Wang L.; Fan C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115 (22), 12491–12545. 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Matthijs G.; Souche E.; Alders M.; Corveleyn A.; Eck S.; Feenstra I.; Race V.; Sistermans E.; Sturm M.; Weiss M.; Yntema H.; Bakker E.; Scheffer H.; Bauer P. Guidelines for Diagnostic Next-Generation Sequencing. Eur. J. Hum. Genet. 2016, 24 (1), 2–5. 10.1038/ejhg.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R.; Fremaux C.; Deveau H.; Richards M.; Boyaval P.; Moineau S.; Romero D. A.; Horvath P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science (80-.) 2007, 315 (5819), 1709–1712. 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Jinek M.; Chylinski K.; Fonfara I.; Hauer M.; Doudna J. A.; Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science (80-.) 2012, 337 (6096), 816–821. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic Acid Sequence-Based Amplification. Nature. 1991, 350 (6313), 91–92. 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Nilsson M.; Malmgren H.; Samiotaki M.; Kwiatkowski M.; Chowdhary B. P.; Landegren U. Padlock Probes: Circularizing Oligonucleotides for Localized DNA Detection. Science (80-.) 1994, 265 (5181), 2085–2088. 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- Fire A.; Xu S. Q. Rolling Replication of Short DNA Circles. Proc. Natl. Acad. Sci. U. S. A. 1995, 92 (10), 4641–4645. 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Lu C. H.; Liu X.; Freage L.; Willner I. Amplified and Multiplexed Detection of DNA Using the Dendritic Rolling Circle Amplified Synthesis of DNAzyme Reporter Units. Anal. Chem. 2014, 86 (3), 1614–1621. 10.1021/ac4033033. [DOI] [PubMed] [Google Scholar]
- Walker G. T.; Little M. C.; Nadeau J. G.; Shank D. D. Isothermal in Vitro Amplification of DNA by a Restriction Enzyme/DNA Polymerase System. Proc. Natl. Acad. Sci. U. S. A. 1992, 89 (1), 392–396. 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. T.; Fraiser M. S.; Schram J. L.; Little M. C.; Nadeau J. G.; Malinowski D. P. Strand Displacement Amplification - an Isothermal, in Vitro DNA Amplification Technique. Nucleic Acids Res. 1992, 20 (7), 1691–1696. 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28 (12), 63e–63. 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O.; Williams C. H.; Stemple D. L.; Armes N. A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4 (7), e204. 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J.; Van Ness L. K.; Galas D. J. Isothermal Reactions for the Amplification of Oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (8), 4504–4509. 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir M. A. B.; Shabbir M. Z.; Wu Q.; Mahmood S.; Sajid A.; Maan M. K.; Ahmed S.; Naveed U.; Hao H.; Yuan Z. CRISPR-Cas System: Biological Function in Microbes and Its Use to Treat Antimicrobial Resistant Pathogens. Ann. Clin. Microbiol. Antimicrob. 2019, 18 (1), 1–9. 10.1186/s12941-019-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M.; Jiang F.; Taylor D. W.; Sternberg S. H.; Kaya E.; Ma E.; Anders C.; Hauer M.; Zhou K.; Lin S.; Kaplan M.; Iavarone A. T.; Charpentier E.; Nogales E.; Doudna J. A. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science (80-.) 2014, 343 (6176), 2–18. 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau J. E.; Dupuis M. È.; Villion M.; Romero D. A.; Barrangou R.; Boyaval P.; Fremaux C.; Horvath P.; Magadán A. H.; Moineau S. The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature. 2010, 468 (7320), 67–71. 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Newsom S.; Parameshwaran H. P.; Martin L.; Rajan R. The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies. Front. Cell. Infect. Microbiol. 2021, 10, 1–10. 10.3389/fcimb.2020.619763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H.; Garneau J. E.; Moineau S. CRISPR/Cas System and Its Role in Phage-Bacteria Interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- McGinn J.; Marraffini L. A. Molecular Mechanisms of CRISPR–Cas Spacer Acquisition. Nat. Rev. Microbiol. 2019, 17 (1), 7–12. 10.1038/s41579-018-0071-7. [DOI] [PubMed] [Google Scholar]
- Puhan M. A.; Chandra D.; Mosenifar Z.; Ries A.; Make B.; Hansel N. N.; Sciurba F.; Sinai C.; Angeles L.; Centre H. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Sci. 2008. August 2017, 37 (4), 784–790. 10.1126/science.1159689.Small. [DOI] [Google Scholar]
- Carte J.; Wang R.; Li H.; Terns R. M.; Terns M. P. Cas6 Is an Endoribonuclease That Generates Guide RNAs for Invader Defense in Prokaryotes. Genes Dev. 2008, 22 (24), 3489–3496. 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T.; Gasiunas G.; Young J.; Bigelyte G.; Silanskas A.; Cigan M.; Siksnys V. Rapid Characterization of CRISPR-Cas9 Protospacer Adjacent Motif Sequence Elements. Genome Biol. 2015, 16 (1), 1–13. 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K.; Babu K.; Sundaresan R.; Rajan R.; Sashital D. G. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol. Cell 2017, 68 (1), 15–25. 10.1016/j.molcel.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. Y.; Doudna J. A. Chemistry of Class 1 CRISPR-Cas Effectors: Binding, Editing, and Regulation. J. Biol. Chem. 2020, 295 (42), 14473–14487. 10.1074/jbc.REV120.007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P.; Makarova K. S.; Zetsche B.; Zhang F.; Koonin E. V.; van der Oost J. Diverse Evolutionary Roots and Mechanistic Variations of the CRISPR-Cas Systems. Science 2016, 353 (6299), 1. 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- Koonin E. V.; Makarova K. S. Origins and Evolution of CRISPR-Cas Systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374 (1772), 20180087. 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S.; Wolf Y. I.; Iranzo J.; Shmakov S. A.; Alkhnbashi O. S.; Brouns S. J. J.; Charpentier E.; Cheng D.; Haft D. H.; Horvath P.; Moineau S.; Mojica F. J. M.; Scott D.; Shah S. A.; Siksnys V.; Terns M. P.; Venclovas Č.; White M. F.; Yakunin A. F.; Yan W.; Zhang F.; Garrett R. A.; Backofen R.; van der Oost J.; Barrangou R.; Koonin E. V. Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18 (2), 67–83. 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.; Upadhyay D. J.; Srivastava A. Next-Generation Molecular Diagnostics Development by CRISPR/Cas Tool: Rapid Detection and Surveillance of Viral Disease Outbreaks. Front. Mol. Biosci. 2020, 7, 1. 10.3389/fmolb.2020.582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Li S.; Wang J.; Liu G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37 (7), 730–743. 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Wang M.; Zhang R.; Li J. CRISPR/Cas Systems Redefine Nucleic Acid Detection: Principles and Methods. Biosens. Bioelectron. 2020, 165 (June), 112430. 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukama O.; Wu J.; Li Z.; Liang Q.; Yi Z.; Lu X.; Liu Y.; Liu Y.; Hussain M.; Makafe G. G.; Liu J.; Xu N.; Zeng L. An Ultrasensitive and Specific Point-of-Care CRISPR/Cas12 Based Lateral Flow Biosensor for the Rapid Detection of Nucleic Acids. Biosens. Bioelectron. 2020, 159, 112143. 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- Gootenberg J. S.; Abudayyeh O. O.; Lee J. W.; Essletzbichler P.; Dy A. J.; Joung J.; Verdine V.; Donghia N.; Daringer N. M.; Freije C. A.; Myhrvold C.; Bhattacharyya R. P.; Livny J.; Regev A.; Koonin E. V.; Hung D. T.; Sabeti P. C.; Collins J. J.; Zhang F. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356 (6336), 438–442. 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S.; Abudayyeh O. O.; Kellner M. J.; Joung J.; Collins J. J.; Zhang F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a and Csm6. Science 2018, 360 (6387), 439–444. 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M. J.; Koob J. G.; Gootenberg J. S.; Abudayyeh O. O.; Zhang F. SHERLOCK: Nucleic Acid Detection with CRISPR Nucleases. Nat. Protoc. 2019, 14 (10), 2986–3012. 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S.; Ma E.; Harrington L. B.; Da Costa M.; Tian X.; Palefsky J. M.; Doudna J. A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360 (6387), 436–439. 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P.; Deng X.; Yu G.; Fasching C. L.; Servellita V.; Singh J.; Miao X.; Streithorst J. A.; Granados A.; Sotomayor-Gonzalez A.; Zorn K.; Gopez A.; Hsu E.; Gu W.; Miller S.; Pan C. Y.; Guevara H.; Wadford D. A.; Chen J. S.; Chiu C. Y. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38 (7), 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. Identification, Subtyping and Virulence Determination of Listeria Monocytogenes, an Important Foodborne Pathogen. J. Med. Microbiol. 2006, 55 (6), 645–659. 10.1099/jmm.0.46495-0. [DOI] [PubMed] [Google Scholar]
- Amagliani G.; Brandi G.; Omiccioli E.; Casiere A.; Bruce I. J.; Magnani M. Direct Detection of Listeria Monocytogenes from Milk by Magnetic Based DNA Isolation and PCR. Food Microbiol. 2004, 21 (5), 597–603. 10.1016/j.fm.2003.10.008. [DOI] [Google Scholar]
- Ferreira V.; Wiedmann M.; Teixeira P.; Stasiewicz M. J. Listeria Monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77 (1), 150–170. 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- Gasanov U.; Hughes D.; Hansbro P. M. Methods for the Isolation and Identification of Listeria Spp. and Listeria Monocytogenes: A Review. FEMS Microbiol. Rev. 2005, 29 (5), 851–875. 10.1016/j.femsre.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Huang M.; Zhou X.; Wang H.; Xing D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90 (3), 2193–2200. 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- Wang X.; Xiong E.; Tian T.; Cheng M.; Lin W.; Wang H.; Zhang G.; Sun J.; Zhou X. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Lateral Flow Nucleic Acid Assay. ACS Nano 2020, 14 (2), 2497–2508. 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Qian L.; Wei W.; Wang Y.; Wang B.; Lin P.; Liu W.; Xu L.; Li X.; Liu D.; Cheng S.; Li J.; Ye Y.; Li H.; Zhang X.; Dong Y.; Zhao X.; Liu C.; Zhang H. M.; Ouyang Q.; Lou C. Paired Design of DCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2017, 6 (2), 211–216. 10.1021/acssynbio.6b00215. [DOI] [PubMed] [Google Scholar]
- Ai J. W.; Zhou X.; Xu T.; Yang M.; Chen Y.; He G. Q.; Pan N.; Cai Y.; Li Y.; Wang X.; Su H.; Wang T.; Zeng W.; Zhang W. H. CRISPR-Based Rapid and Ultra-Sensitive Diagnostic Test for Mycobacterium Tuberculosis. Emerg. Microbes Infect. 2019, 8 (1), 1361–1369. 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guk K.; Keem J. O.; Hwang S. G.; Kim H.; Kang T.; Lim E. K.; Jung J. A Facile, Rapid and Sensitive Detection of MRSA Using a CRISPR-Mediated DNA FISH Method, Antibody-like DCas9/SgRNA Complex. Biosens. Bioelectron. 2017, 95 (April), 67–71. 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Suea-Ngam A.; Howes P. D.; Demello A. J. An Amplification-Free Ultra-Sensitive Electrochemical CRISPR/Cas Biosensor for Drug-Resistant Bacteria Detection. Chem. Sci. 2021, 12 (38), 12733–12743. 10.1039/D1SC02197D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.; Zhou X.; Shan Y.; Yue H.; Huang R.; Hu J.; Xing D. Sensitive Detection of a Bacterial Pathogen Using Allosteric Probe-Initiated Catalysis and CRISPR-Cas13a Amplification Reaction. Nat. Commun. 2020, 11 (1), 1–10. 10.1038/s41467-019-14135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.; Peng L.; Yin L.; Liu G.; Man S. CRISPR-Cas12a-Powered Dual-Mode Biosensor for Ultrasensitive and Cross-Validating Detection of Pathogenic Bacteria. ACS Sensors. 2021, 6 (8), 2920–2927. 10.1021/acssensors.1c00686. [DOI] [PubMed] [Google Scholar]
- Sun X.; Wang Y.; Zhang L.; Liu S.; Zhang M.; Wang J.; Ning B.; Peng Y.; He J.; Hu Y.; Gao Z. CRISPR-Cas9 Triggered Two-Step Isothermal Amplification Method for E. Coli O157:H7 Detection Based on a Metal-Organic Framework Platform. Anal. Chem. 2020, 92 (4), 3032–3041. 10.1021/acs.analchem.9b04162. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Guo Y.; Zhang L.; Yang Y.; Yang S.; Yang L.; Chen H.; Liu C.; Li J.; Xie G. Integration of Multiplex PCR and CRISPR-Cas Allows Highly Specific Detection of Multidrug-Resistant Acinetobacter Baumannii. Sensors Actuators, B Chem. 2021, 334, 129600. 10.1016/j.snb.2021.129600. [DOI] [Google Scholar]
- Kamariza M.; Keyser S. G. L.; Utz A.; Knapp B. D.; Ealand C.; Ahn G.; Cambier C. J.; Chen T.; Kana B.; Huang K. C.; Bertozzi C. R. Toward Point-of-Care Detection of Mycobacterium Tuberculosis : A Brighter Solvatochromic Probe Detects Mycobacteria within Minutes. JACS Au. 2021, 1 (9), 1368–1379. 10.1021/jacsau.1c00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo V. F.; Oliveira A. G.; Nishio E. K.; Perugini M. R. E.; Andrade C. G. T. J.; Silveira W. D.; Durán N.; Andrade G.; Kobayashi R. K. T.; Nakazato G. Antibacterial Activity of Extracellular Compounds Produced by a Pseudomonas Strain against Methicillin-Resistant Staphylococcus Aureus (MRSA) Strains. Ann. Clin. Microbiol. Antimicrob. 2013, 12 (1), 1–8. 10.1186/1476-0711-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekema D. J.; Pfaller M. A.; Schmitz F. J.; Smayevsky J.; Bell J.; Jones R. N.; Beach M. Survey of Infections Due to Staphylococcus Species: Frequency of Occurrence and Antimicrobial Susceptibility of Isolates Collected in the United States, Canada, Latin America, Europe, and the Western Pacific Region for the SENTRY Antimicrobial Surveillanc. Clin. Infect. Dis. 2001, 32, S114. 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- Boucher H.; Miller L. G.; Razonable R. R. Serious Infections Caused by Methicillin-Resistant Staphylococcus Aureus. Clin. Infect. Dis. 2010, 51, S183. 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- Wolters M.; Rohde H.; Maier T.; Belmar-Campos C.; Franke G.; Scherpe S.; Aepfelbacher M.; Christner M. MALDI-TOF MS Fingerprinting Allows for Discrimination of Major Methicillin-Resistant Staphylococcus Aureus Lineages. Int. J. Med. Microbiol. 2011, 301 (1), 64–68. 10.1016/j.ijmm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Hagen R. M.; Seegmüller I.; Navai J.; Kappstein I.; Lehn N.; Miethke T. Development of a Real-Time PCR Assay for Rapid Identification of Methicillin-Resistant Staphylococcus Aureus from Clinical Samples. Int. J. Med. Microbiol. 2005, 295 (2), 77–86. 10.1016/j.ijmm.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Harmsen D.; Claus H.; Witte W.; Rothgänger J.; Claus H.; Turnwald D.; Vogel U. Typing of Methicillin-Resistant Staphylococcus Aureus in a University Hospital Setting by Using Novel Software for Spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41 (12), 5442–5448. 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M. C.; Day N. P. J.; Davies C. E.; Peacock S. J.; Spratt B. G. Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus Aureus. J. Clin. Microbiol. 2000, 38 (3), 1008–1015. 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchan S.; Kaufmann M. E.; Deplano A.; De Ryck R.; Struelens M.; Zinn C. E.; Fussing V.; Salmenlinna S.; Vuopio-Varkila J.; El Solh N.; Cuny C.; Witte W.; Tassios P. T.; Legakis N.; Van Leeuwen W.; Van Belkum A.; Vindel A.; Laconcha I.; Garaizar J.; Haeggman S.; Olsson-Liljequist B.; Ransjo U.; Coombes G.; Cookson B. Harmonization of Pulsed-Field Gel Electrophoresis Protocols for Epidemiological Typing of Strains of Methicillin-Resistant Staphylococcus Aureus: A Single Approach Developed by Consensus in 10 European Laboratories and Its Application for Tracing the Spre. J. Clin. Microbiol. 2003, 41 (4), 1574–1585. 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suea-Ngam A.; Howes P. D.; Stanley C. E.; Demello A. J. An Exonuclease I-Assisted Silver-Metallized Electrochemical Aptasensor for Ochratoxin A Detection. ACS Sensors. 2019, 4 (6), 1560–1568. 10.1021/acssensors.9b00237. [DOI] [PubMed] [Google Scholar]
- Bell R. L.; Jarvis K. G.; Ottesen A. R.; Mcfarland M. A.; Brown E. W. Recent and Emerging Innovations in Salmonella Detection: A Food and Environmental Perspective. Microb. Biotechnol. 2016, 9 (3), 279–292. 10.1111/1751-7915.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama W. B.; Dudley E. G.; Doores S.; Cutter C. N. Commercially Available Rapid Methods for Detection of Selected Food-Borne Pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56 (9), 1519–1531. 10.1080/10408398.2013.775567. [DOI] [PubMed] [Google Scholar]
- Gao S.; Liu J.; Li Z.; Ma Y.; Wang J. Sensitive Detection of Foodborne Pathogens Based on CRISPR-Cas13a. J. Food Sci. 2021, 86 (6), 2615–2625. 10.1111/1750-3841.15745. [DOI] [PubMed] [Google Scholar]
- Lou L.; Zhang P.; Piao R.; Wang Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9 (July), 1–12. 10.3389/fcimb.2019.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N. T.; Plunkett G.; Burland V.; Mau B.; Glasner J. D.; Rose D. J.; Mayhew G. F.; Evans P. S.; Gregor J.; Kirkpatrick H. A.; Pósfai G.; Hackett J.; Klink S.; Boutin A.; Shao Y.; Miller L.; Grotbeck E. J.; Davis N. W.; Lim A.; Dimalanta E. T.; Potamousis K. D.; Apodaca J.; Anantharaman T. S.; Lin J.; Yen G.; Schwartz D. C.; Welch R. A.; Blattner F. R. Genome Sequence of Enterohaemorrhagic Escherichia Coli O157:H7. Nature 2001, 409 (6819), 529–533. 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Varshney M.; Li Y.; Srinivasan B.; Tung S. A Label-Free, Microfluidics and Interdigitated Array Microelectrode-Based Impedance Biosensor in Combination with Nanoparticles Immunoseparation for Detection of Escherichia Coli O157:H7 in Food Samples. Sensors Actuators, B Chem. 2007, 128 (1), 99–107. 10.1016/j.snb.2007.03.045. [DOI] [Google Scholar]
- Ross S. A.; Lane J. A.; Kilcoyne M.; Joshi L.; Hickey R. M. Defatted Bovine Milk Fat Globule Membrane Inhibits Association of Enterohaemorrhagic Escherichia Coli O157:H7 with Human HT-29 Cells. Int. Dairy J. 2016, 59, 36–43. 10.1016/j.idairyj.2016.03.001. [DOI] [Google Scholar]
- Saeedi P.; Yazdanparast M.; Behzadi E.; Salmanian A. H.; Mousavi S. L.; Nazarian S.; Amani J. A Review on Strategies for Decreasing E. Coli O157:H7 Risk in Animals. Microb. Pathog. 2017, 103, 186–195. 10.1016/j.micpath.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Tacconelli E.; Carrara E.; Savoldi A.; Harbarth S.; Mendelson M.; Monnet D. L.; Pulcini C.; Kahlmeter G.; Kluytmans J.; Carmeli Y.; Ouellette M.; Outterson K.; Patel J.; Cavaleri M.; Cox E. M.; Houchens C. R.; Grayson M. L.; Hansen P.; Singh N.; Theuretzbacher U.; Magrini N.; Aboderin A. O.; Al-Abri S. S.; Awang Jalil N.; Benzonana N.; Bhattacharya S.; Brink A. J.; Burkert F. R.; Cars O.; Cornaglia G.; Dyar O. J.; Friedrich A. W.; Gales A. C.; Gandra S.; Giske C. G.; Goff D. A.; Goossens H.; Gottlieb T.; Guzman Blanco M.; Hryniewicz W.; Kattula D.; Jinks T.; Kanj S. S.; Kerr L.; Kieny M. P.; Kim Y. S.; Kozlov R. S.; Labarca J.; Laxminarayan R.; Leder K.; Leibovici L.; Levy-Hara G.; Littman J.; Malhotra-Kumar S.; Manchanda V.; Moja L.; Ndoye B.; Pan A.; Paterson D. L.; Paul M.; Qiu H.; Ramon-Pardo P.; Rodríguez-Baño J.; Sanguinetti M.; Sengupta S.; Sharland M.; Si-Mehand M.; Silver L. L.; Song W.; Steinbakk M.; Thomsen J.; Thwaites G. E.; van der Meer J. W.; Van Kinh N.; Vega S.; Villegas M. V.; Wechsler-Fördös A.; Wertheim H. F. L.; Wesangula E.; Woodford N.; Yilmaz F. O.; Zorzet A. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18 (3), 318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Pang Z.; Raudonis R.; Glick B. R.; Lin T. J.; Cheng Z. Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37 (1), 177–192. 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Sauvage S.; Hardouin J. Exoproteomics for Better Understanding Pseudomonas Aeruginosa Virulence. Toxins (Basel) 2020, 12 (9), 571. 10.3390/toxins12090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D.; Nielsen T. B.; Bonomo R. A.; Pantapalangkoor P.; Luna B.; Spellberg B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30 (1), 409–447. 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang Z.; Chen Y.; Hua X.; Yu Y.; Ji Q. A Highly Efficient CRISPR-Cas9-Based Genome Engineering Platform in Acinetobacter Baumannii to Understand the H2O2-Sensing Mechanism of OxyR. Cell Chem. Biol. 2019, 26 (12), 1732. 10.1016/j.chembiol.2019.09.003. [DOI] [PubMed] [Google Scholar]
- Gadkar V. J.; Goldfarb D. M.; Gantt S.; Tilley P. A. G. Real-Time Detection and Monitoring of Loop Mediated Amplification (LAMP) Reaction Using Self-Quenching and De-Quenching Fluorogenic Probes. Sci. Reports 2018, 8 (1), 5548. 10.1038/s41598-018-23930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Scott D. A.; Kriz A. J.; Chiu A. C.; Hsu P. D.; Dadon D. B.; Cheng A. W.; Trevino A. E.; Konermann S.; Chen S.; Jaenisch R.; Zhang F.; Sharp P. A. Genome-Wide Binding of the CRISPR Endonuclease Cas9 in Mammalian Cells. Nat. Biotechnol. 2014, 32 (7), 670–676. 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C.; Arslan S.; Singh R.; Thorpe J.; Adli M. Genome-Wide Analysis Reveals Characteristics of off-Target Sites Bound by the Cas9 Endonuclease. Nat. Biotechnol. 2014, 32 (7), 677–683. 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B. P.; Pattanayak V.; Prew M. S.; Tsai S. Q.; Nguyen N. T.; Zheng Z.; Joung J. K. High-Fidelity CRISPR–Cas9 Nucleases with No Detectable Genome-Wide off-Target Effects. Nat. 2016, 529 (7587), 490–495. 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M.; Gao L.; Zetsche B.; Scott D. A.; Yan W. X.; Zhang F. Rationally Engineered Cas9 Nucleases with Improved Specificity. Science (80-.) 2016, 351 (6268), 84–88. 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S.; Dagdas Y. S.; Kleinstiver B. P.; Welch M. M.; Sousa A. A.; Harrington L. B.; Sternberg S. H.; Joung J. K.; Yildiz A.; Doudna J. A. Enhanced Proofreading Governs CRISPR–Cas9 Targeting Accuracy. Nat. 2017, 550 (7676), 407–410. 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; Daringer N. M.; Bosch I.; Dudley D. M.; O’Connor D. H.; Gehrke L.; Collins J. J. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165 (5), 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Mustafa M. I.; Makhawi A. M. Sherlock and Detectr: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J. Clin. Microbiol. 2021, 59 (3), 1. 10.1128/JCM.00745-20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Myhrvold C.; Freije C. A.; Gootenberg J. S.; Abudayyeh O. O.; Metsky H. C.; Durbin A. F.; Kellner M. J.; Tan A. L.; Paul L. M.; Parham L. A.; Garcia K. F.; Barnes K. G.; Chak B.; Mondini A.; Nogueira M. L.; Isern S.; Michael S. F.; Lorenzana I.; Yozwiak N. L.; MacInnis B. L.; Bosch I.; Gehrke L.; Zhang F.; Sabeti P. C. Field-Deployable Viral Diagnostics Using CRISPR-Cas13. Science (80-.) 2018, 360 (6387), 444–448. 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Wang R.; Wang D.; Wu J.; Li J.; Wang J.; Liu H.; Wang Y. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91 (19), 12156–12161. 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]