Abstract
Multidimensional analyses have demonstrated the presence of a unique tumor microenvironment (TME) in liver cancer. Tumor‐associated macrophages (TAMs) are among the most abundant immune cells infiltrating the TME and are present at all stages of liver cancer progression, and targeting TAMs has become one of the most favored immunotherapy strategies. In addition, macrophages and liver cancer cells have distinct origins. At the early stage of liver cancer, macrophages can provide a niche for the maintenance of liver cancer stem cells. In contrast, cancer stem cells (CSCs) or poorly differentiated tumor cells are key factors modulating macrophage activation. In the present review, we first propose the origin connection between precursor macrophages and liver cancer cells. Macrophages undergo dynamic phenotypic transition during carcinogenesis. In this course of such transition, it is critical to determine the appropriate timing for therapy and block specific markers to suppress pro‐tumoral TAMs. The present review provides a more detailed discussion of transition trends of such surface markers than previous reviews. Complex crosstalk occurs between TAMs and liver cancer cells. TAMs play indispensable roles in tumor progression, angiogenesis, and autophagy due to their heterogeneity and robust plasticity. In addition, macrophages in the TME interact with other immune cells by directing cell‐to‐cell contact or secreting various effector molecules. Similarly, tumor cells combined with other immune cells can drive macrophage recruitment and polarization. Despite the latest achievements and the advancements in treatment strategies following TAMs studies, comprehensive discussions on the communication between macrophages and cancer cells or immune cells in liver cancer are currently lacking. In this review, we discussed the interactions between TAMs and liver cancer cells (from cell origin to maturation), the latest therapeutic strategies (including chimeric antigen receptor macrophages), and critical clinical trials for hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA) to provide a rationale for further clinical investigation of TAMs as a potential target for treating patients with liver cancer.
Keywords: hepatocellular carcinoma, intrahepatic cholangiocarcinoma, tumor‐associated macrophages, immunotherapy
Abbreviations
- ABTAA
Tie2‐Activating Antibody
- ABA
Ang2‐Blocking Antibody
- AD
Alzheimer's disease
- AGER
advanced glycosylation end product‐specific receptor
- Αkg
α‐ketoglutarate
- ApoE
apolipoprotein E
- B7‐H1
B7 homolog 1
- BACE1
β‐site amyloid precursor protein‐cleaving enzyme 1
- BM
bone marrow
- CA1P
combretastatin A‐1 phosphate
- CAFs
cancer‐associated fibroblasts
- CAR‐M
chimeric antigen receptor macrophage
- CAR‐T
chimeric antigen receptor T cell
- CCL2
C‐C motif chemokine ligand 2
- CCL5
C‐C motif chemokine ligand 5
- CCL20
C‐C motif chemokine ligand 20
- CCL17
C‐C motif chemokine ligand 17
- CCL22
C‐C motif chemokine ligand 22
- CCR2
C‐C motif chemokine receptor 2
- CeO2NPs
cerium oxide nanoparticles
- cGAS
cyclic GMP‐AMP synthase
- CHCCA
hepatocellular cholangiocarcinoma
- Clec4F
C‐type lectin domain family 4 member F
- CSCs
Cancer stem cells
- CSF1
colony‐stimulating factor‐1
- CSF1R
colony‐stimulating factor 1 receptor
- CTLA‐4
cytotoxic T‐lymphocyte antigen 4
- CX3CR
C‐X3‐C motif chemokine receptor
- CXCL2
C‐X‐C motif chemokine ligand 2
- CXCL9
C‐X‐C motif chemokine ligand 9
- CXCL10
C‐X‐C motif chemokine ligand 10
- DCs
dendritic cells
- ECs
endothelial cells
- ECT2
epithelial cell transforming 2
- EMPs
erythromyeloid progenitors
- EMT
epithelial mesenchymal transition
- ERK
extracellular regulated protein kinase
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- FAK
focal adhesion kinase
- FAO
Fatty acid oxidation
- FAS
factor related apoptosis
- FASL
factor related apoptosis ligand
- GATA
glutamyl‐tRNA amidotransferase, subunit A
- GM‐CSF
colony‐stimulating factor
- G‐MDSCs
granulocytic myeloid‐derived suppressor cells
- GPC3
Glypican‐3
- HCC
hepatocellular carcinoma
- HDAC6
histone deacetylase 6
- HGF
hepatocyte growth factor
- HLA
human leukocyte antigen
- HLA‐DR
human leukocyte antigens DR
- HLA‐E
human leukocyte antigen E
- HLA‐G
human leukocyte antigen G
- HME
human macrophage metalloelastase
- HMGB1
high mobility group box 1
- HSCs
hepatic stellate cells
- iCCA
intrahepatic cholangiocarcinoma
- ICIs
immune‐checkpoint inhibitors
- ID3
inhibitor of differention‐3
- IFNγ
interferon‐gamma
- IGF
insulin growth factor
- IGF‐R
insulin growth factor receptor
- IL‐1β
interleukin 1β
- IL‐1Rinterleukin 1 receptor;IL‐6
interleukin 1 receptor;IL‐6interleukin 6
- IL‐10
interleukin 10
- IL‐12
interleukin 12
- IL‐13/34
interleukin 13/34
- IL21interleukin 21;iNOS
interleukin 21;iNOSinducible nitric oxide synthase
- LOXL4
lysyl oxidase‐like 4
- IRF1
interferon regulatory factor 1
- KCs
Kupffer cells
- M1 TAMs
M1 type tumor‐associated macrophages
- M2 TAMs
M2 type tumor‐associated macrophages
- MACRO
macrophage receptor with collagenous
- MAPK
mitogen‐activated protein kinase
- MCP‐1
Monocyte chemoattractant protein‐1
- MDSCs
myeloid‐derived immunosuppressive cells
- MEK
mitogen‐activated protein kinase kinase
- MHC‐II
major histocompatibility complex type II
- MIF
migration inhibitory factor
- miRNAs
microRNAs
- MMP9
matrix metallopeptidase 9
- MoMϕs
monocyte‐derived macrophages
- mTOR
mechanistic target of rapamycin
- NK cells
natural killer cells
- NOS2
nitric oxide synthase
- NOX2
NADPH oxidase 2
- PCR
polymerase chain reaction
- PD‐1
programmed cell death protein 1
- PD‐L1
programmed cell death protein 1 ligand 1
- PFKFB3
6‐phosphofructo‐2‐kinase/fructose‐2,6‐biphosphatase 3
- PGE2
prostaglandin E2
- PI3Kγ
Phosphoinositide 3‐kinase gamma
- POSTN
Periostin
- PPAR
peroxisome proliferators‐activated receptors
- PKM2
pyruvate kinase M2
- pMacs
pre‐macrophages
- p‐STAT‐3
phospho‐signal transducer and activator of transcription 3
- REDD1
Regulated in development and DNA damage response 1
- RelB
Reticuloendotheliosis viral oncogene homolog B
- RIG‐I
retinotic acid‐inducible gene I; , regulation of Igh‐1b 1
- RIPK3
receptor‐interacting protein kinase 3
- RNA‐Seq
RNA Sequencing
- ROS
reactive oxygen species
- SALL4
Sal‐like protein‐4
- S100A9
S100 calcium‐binding protein A9
- SIRT1
sirtuin 1
- SIRT4
sirtuin 4
- SLC7A11
solute carrier family 7 member 11
- SPON2
matricellular protein spondin2
- STING
stimulator of interferon genes
- TANs
tumor‐associated neutrophils
- TAMs
tumor‐associated macrophages
- TAZ
tafazzin
- TCA
tricarboxylic acid cycle
- TEMs
Tie2‐expressing monocytes
- TFH cells
T follicular helper cells
- TGF‐β
transforming growth factor‐β
- Th1
type 1 T helper cells
- Tim‐3
T cell immunoglobulin and mucin‐containing molecule 3
- Tim‐4
T‐cell immunoglobulin and mucin domain containing 4
- TKIs
tyrosine kinase inhibitors
- TLR
Toll‐like receptor
- TLR2
toll‐like receptor 2
- TME
tumor microenvironment
- TNF
tumor necrosis factor
- TNF‐α
tumor necrosis factor α
- TNFR1
tumor necrosis factor receptor 1
- TNFSF
tumor necrosis factor superfamily
- TRAF2
tumor necrosis factor receptor‐associated factor 2
- Treg
regulatory T
- TREM‐1
triggering receptor expressed on myeloid cells‐1
- TSP1
thrombospondin‐1
- T‐VEC
Talimogene laherparepvec
- TWEAK
Tumor necrosis factor‐like weak inducer of apoptosis
- YAP
yes‐associated protein 1
- VEGF
vascular endothelial growth factor
1. BACKGROUND
Emerging evidence has shown that the TME plays a pivotal role in driving cancer progression and governing the response to standard‐of‐care therapies [1]. Multiple components coexist and interact in the TME, including tumor‐associated macrophages (TAMs), CD4+ and CD8+ T cells, dendritic cells (DCs), natural killer (NK) cells, tumor‐related endothelial cells (ECs), abnormal tumor vasculature, cancer‐associated fibroblasts (CAFs), and myeloid‐derived immunosuppressive cells (MDSCs). As a dynamic system orchestrated by multiple cellular and non‐cellular components, each cellular component in the tumor immune microenvironment represents a potential target for reprogramming the TME (Figure 1).
FIGURE 1.
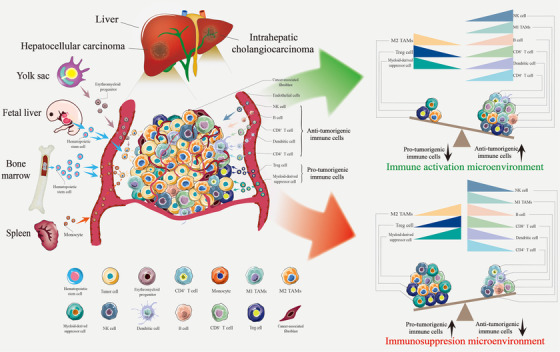
TAMs in the liver cancer immune microenvironment. TAMs constitute the core cell population of the tumor immune microenvironment in liver cancer. BM and embryos are the most critical sources of TAMs, followed by the spleen. The immunosuppressive cell population (based on TAMs) facilitates tumor progression and suppresses the immune response. In contrast, increasing infiltration of immune cell populations, including M1 and cytotoxic T lymphocytes, is associated with a better clinical prognosis. Abbreviations: TAMs, tumor‐associated macrophages; M1 TAMs, M1 type tumor‐associated macrophages; M2 TAMs, M2 type tumor‐associated macrophages; BM, bone marrow; NK cell, natural killer cell
Clinical studies and experimental mouse models have indicated that TAMs are particularly abundant among innate and adaptive immune cells recruited to the tumor milieu [2, 3, 4]. TAMs typically exhibit distinct functional phenotypes. M1‐like TAMs exert pro‐inflammatory and anti‐tumor activities, whereas M2‐like TAMs exert anti‐inflammatory and tumor‐promoting effects. M2 TAMs can promote cancer initiation, suppress antitumor immunity, stimulate angiogenesis, and enhance tumor cell invasion, motility and intravasation [5, 6] (Figure 1). However, the classification of macrophages is far more complex than previously thought, and the markers on the surface of macrophages are constantly in flux as cancer progresses [7, 8]. Rather than simply classifying macrophages, in the present review, we provide a comprehensive discussion of the subpopulation and the dynamic transformation of markers on their surfaces. Specific macrophage subsets assume distinct roles in cancer progression and antitumor immunity [9]. Traditional flow cytometry and histological methods to define TAMs appear to be limited because of the inability to capture the full diversity of the cells or distinguish them from other cell populations. With the development of single‐cell technology and spatial transcriptomics, determining the origin and spatial distribution of TAMs and identifying specific macrophage markers to design targeted and personalized medicine is essential for preventing and treating hepatic malignancies [10, 11].
Very few systematic reviews have discussed the interactions between the origin of macrophages and cancer cells in liver cancer. We first propose a relationship between precursor macrophages and liver cancer stem cells. This potential interaction in cellular evolution contributes to a better understanding of how macrophages can promote early‐stage liver cancer progression and develop appropriate treatment strategies. Therefore, the interactions of cell origins cannot be ignored. Moreover, it is unclear whether macrophage precursors interact with other immune cell precursors, such as lymphoid cell precursors. Additional research is required to test such hypotheses.
In the past, associated research on TAMs mainly has focused on how TAMs interact with cancer cells and their phenotype switching [12, 13, 14, 15, 16]. The present review is a more in‐depth discussion than previous reviews. We described the effects of macrophages on cancer cells and summarized the regulation of macrophages by cancer cells. Moreover, we discussed the regulation of macrophage metabolism, including glucose metabolism, lipid metabolism, and amino acid metabolism, which are also essential key research directions of our research group [15, 17, 18]. The liver is vital for metabolic homeostasis; therefore, more metabolomic data on liver TAMs are warranted to confirm that macrophage metabolism regulates liver cancer progression.
The role of the formation of an immunosuppressive microenvironment is not limited to TAMs alone. Studies on the relationship between macrophages and CD8+ T cells are currently widespread [19, 20]. However, studies on the relationships between macrophages and other immune cells, such as CD4+ T cells, B cells, Tregs, and MDSCs, are still lacking. The reciprocal regulation between TAMs and these cells is an important and novel research direction.
Finally, we divided the macrophage‐targeted therapy for liver cancer into three categories based on published studies and summarized the latest treatment strategies and clinical trials compared to those of the past. Combination therapy is the primary treatment strategy for liver cancer [21]. We explored studies on combined therapy with multi‐target inhibitors such as sorafenib or lenvatinib and evaluated the efficacy of immune checkpoint inhibitors (ICIs) such as anti‐programmed cell death protein 1 (PD‐1) [22, 23, 24]. A critical hypothesis is whether macrophage metabolism disorder is the core resistance mechanism to such drugs. To support such conclusions, more studies on liver cancer are required to explore drug resistance targets around macrophage metabolism.
In summary, in the present review, we illustrated the potential origin connection between TAMs and liver cancer cells, the dynamic transitions of TAMs, and the latest insights into therapeutic strategies used with TAMs in the liver field, with particular emphasis on interactions between these and other cells in the liver cancer microenvironment, which could promote the clinical translation of macrophage‐based combination therapy.
2. POTENTIAL ORIGIN CONNECTION BETWEEN TAMs AND LIVER CANCER CELLS
TAMs are one of the most abundant infiltrative immune cells in tumor stroma and play a pivotal role in inflammation. They can be divided into two categories according to their source: tissue‐resident macrophages and monocyte‐derived macrophages (MoMϕs) [10]. In the normal liver, macrophages mainly exist in the form of tissue‐resident macrophages, namely Kupffer cells (KCs). Increasing evidence shows that tissue‐resident macrophages may be derived from erythromyeloid progenitors (EMPs) that express the macrophage colony‐stimulating factor 1 receptor (CSF1R) in the yolk sac or fetal liver [25]. During liver cancer progression, tissue‐resident macrophages are stimulated by pro‐tumorigenic factors, which causes them to undergo a phenotypic switch and eventually become TAMs. Recent data have confirmed that EMPs can produce pre‐macrophages (pMacs) and differentiate into KCs in a chemokine‐receptor‐dependent manner [26]. The sources of KCs are also diverse, contributing to the significant heterogeneity in the resident cells of liver tissues. When becoming malignant, macrophages undergo sequential stages as EMPs, monocyte progenitors, mature monocytes, KCs, and TAMs. Therefore, unraveling the mystery of this heterogeneity is essential for targeted macrophage therapy [27, 28] (Figure 2).
FIGURE 2.
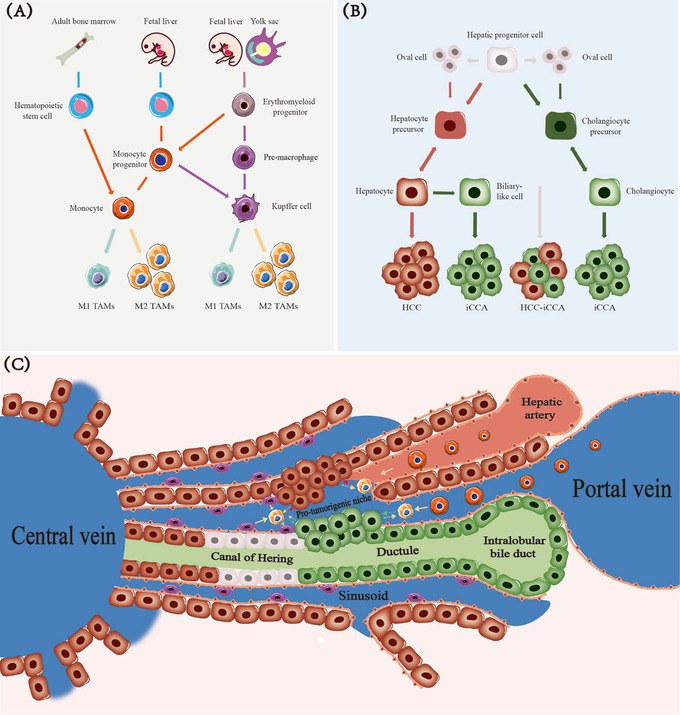
Potential origin connection between TAMs and liver cancer cells. (A) Erythroid progenitor cells can differentiate into pMacs and monocyte progenitors. Hematopoietic stem cells are also a source of monocyte precursor cells. pMacs and monocyte progenitors differentiate into tissue‐resident macrophages (KCs in the liver) during growth in the body. Such cells maintain themselves by self‐renewal and can bind to peripheral blood monocytes to serve as two essential sources of TAMs when stimulated by certain cancer factors. (B) Liver progenitor cells have strong differentiation potential. Oval cells may be a subset of liver progenitor cells. Liver cancer and cholangiocarcinoma usually originate from mature hepatocytes and bile duct cells formed by precursor cells. Mixed cell carcinomas are primarily derived directly from progenitor cells. (C) The hepatic progenitor cells located in the canals of Hering have a subtle connection with TAM development during mature hepatocytes formations and subsequent transformation into hepatocarcinoma cells. TAMs can enable cancer cells to form a pre‐tumor niche. The development of cancer cells recruits a large number of mononuclear‐derived macrophages and activates tissue‐resident macrophages. Abbreviations: pMacs, pre‐macrophages; KCs, Kupffer cells; TAMs, tumor‐associated macrophages; M1 TAMs, M1 type tumor‐associated macrophages; M2 TAMs, M2 type tumor‐associated macrophages; HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; HCC‐iCCA, hepatocellular carcinoma and intrahepatic cholangiocarcinoma
Another main line of macrophage genesis is very complex and lengthy. Embryonic hematopoietic stem cells that colonize the fetal liver (embryonic period) and migrate to the bone marrow (BM, adult period) can serve as a source of monocytes. MoMϕs penetrate tumor tissues through the blood and differentiate into TAMs during liver cancer [29, 30] (Figure 2). Furthermore, splenic monocytes represent a secondary source of TAMs, which can play a significant role in the inflammatory response following acute injury [31, 32].
Liver stem/progenitor cells are known as hepatic progenitor cells in humans and oval cells in rodents. We previously conducted multiple studies on liver progenitor cells [33, 34]. Clinical and pathological analyses of combined hepatocellular cholangiocarcinoma (CHCCA) have revealed the characteristics of progenitor cells [35]. These findings suggest that CHCCA originates directly from liver progenitor cells. The current consensus is that mature hepatocytes and bile duct cells derived from hepatic progenitor cells or precursor cells transform into hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA), respectively, in the presence of cancer‐stimulating factors [36, 37]. Intriguingly, adult hepatocytes and bile duct cells can dedifferentiate into precursor cells, ultimately transforming into cancer cells with progenitor cell markers [38]. Moreover, hepatocytes can transdifferentiate into bile duct‐like cells and evolve into iCCAs [39] (Figure 2).
Recently, a significant discovery revealed that tissue‐resident macrophages could provide a pro‐tumorigenic niche for early cancer [10]. Based on our previous liver cancer research, we proposed that KCs combined with hepatic stellate cells (HSCs) may facilitate the dissolution of the basement membrane. This process could allow residual hepatic cancer stem cells in the canals of Hering to move into the adjacent hepatic lobe and differentiate into cancer cells [34]. Macrophages undergoing malignant transformation can regulate liver precursor cells before maturation and provide a unique niche for maintaining CSCs and controlling their behavior from the time of their origin.
CSCs or poorly differentiated tumor cells in the microenvironment act as crucial factors in modulating macrophage activation (Figure 2). Recently, the discovery of oncofetal reprogramming of the tumor ecosystem revealed that reprogramming of embryonic HCC ECs promotes the production of immunosuppressive macrophages [40]. Furthermore, a novel subgroup of cells with a resident CXCR5−PD‐1−BTLA−CD69high phenotype has been identified. These protumorigenic T follicular helper (TFH) cells can create conditions for M2b macrophage polarization through the interleukin 21 (IL21)‐ interferon‐gamma (IFNγ) pathway [41]. Such findings suggest that cancer cells and other immune cells can influence macrophages during the early stages of liver cancer.
As more TAM sources are characterized, the diversity and complexity of macrophages in the TME will be better understood. In addition, both macrophage differentiation and cancer cell development are inextricably linked. We discuss these processes in depth in this review, offering the tantalizing possibility of using therapies targeting the recruitment of primitive macrophage subsets and their communication with hepatic progenitors and progeny.
3. TAMs ARE RECRUITED AND ACTIVATED IN THE LIVER CANCER NICHE
TAMs are recruited and activated by different chemokines in the immuno‐inflammatory microenvironment of liver cancer and differentiate into particular polarized forms associated with specific pathological conditions. Macrophages can be divided into two different polarization states according to the state and function of macrophage activation: classically activated M1 macrophages and alternatively activated M2 macrophages [42]. Both polarization forms are interconvertible under specific circumstances and in the presence of certain stimuli. M1 macrophages usually play pro‐inflammatory roles and secrete large amounts of pro‐inflammatory cytokines. Classical macrophage activation can occur when cells are stimulated with: 1) lipopolysaccharide, a component of the cell wall of gram‐negative bacteria, 2) IFN‐γ, released by NK cells and type 1 T helper (Th1) cells, 3) tumor necrosis factor (TNF), 4) granulocyte‐macrophage colony‐stimulating factor (GM‐CSF), and 5) Toll‐like receptor (TLR) ligands [43]. Activated M1 macrophages secrete certain interleukins, chemokines and TNF‐α to elicit pro‐inflammatory effects and can also exert cytotoxic effects by activating nitric oxide synthase (NOS2) or inducible nitric oxide synthase (iNOS) to produce NO and release reactive oxygen species (ROS). In addition, M1‐type macrophages, which highly express major histocompatibility complex type II (MHC‐II), can regulate and promote Th‐1 type cellular immune responses by presenting antigens to T cells [44].
In contrast, M2 macrophages usually perform functions opposite to those of M1 macrophages. The cytokines IL‐4, IL‐10, IL‐13 and transforming growth factor‐β (TGF‐β) are secreted by Th2 cells and tumor cells, and CSF1 and prostaglandin E2 (PGE2) can induce the alternative activation of macrophages resulting in the M2 polarized phenotype. M2 macrophages can secrete several complex immunosuppressive factors, cytokines, and growth factors; regulate Th‐2 type immune responses; promote tumor cell growth, and participate in tumor angiogenesis [45].
Furthermore, owing to their high heterogeneity, M2‐type macrophages are usually divided into four subtypes: M2a, M2b, M2c, and M2d [46]. The phenotypic heterogeneity and plasticity of the M2 macrophage subtypes have been described in diseases such as atherosclerosis [47]. Table 1 presents a detailed summary of the surface markers of human and mouse macrophages. Research on the specific subtypes of M1/M2 TAMs in liver cancer has been inconclusive. Therefore, it is urgent and necessary to classify the unique phenotypes exhibited by M1/M2 TAMs and propose precise targeted therapies for disparate patients with various stages of cancer. For example, the results of single‐cell RNA sequencing have indicated that microglia‐derived TAMs are predominant in newly diagnosed glioblastoma. In patients with tumor recurrence, monocyte‐derived TAMs outnumber microglia‐derived TAMs [48]. Such findings indicate that tumor intervention strategies may be distinct at multiple stages.
TABLE 1.
Stimuli, Secretion and Markers of macrophage in Human and Mouse
| M1 | M2 | ||||||
|---|---|---|---|---|---|---|---|
| Markers | Markers | ||||||
| Stimuli | Secretion | Surface | Intracellular | Stimuli | Secretion | Surface | Intracellular |
| ATP | CCL4 | CCR5(CD195) | IRF5 | CSF‐1 | Arg‐1 | B7‐H4 | IRF4 |
| GM‐CSF | CCL5 | CD11b(ITGAM) | NOS2(iNOS) | IL‐1R | CCL1 | CCR2(CD192) | STAT6 |
| IFNγ | CCL8 | CD14 | STAT1 | IL‐4 | CCL2(MCP1) | CCR5(CD195) | Ym1* |
| LPS | CCL10 | CD16(FCγR3A) | IL‐6 | CCL5 | CD11b(ITGAM) | ||
| TLR | CCL11 | CD32 | IL‐10 | CCL17 | CD14 | ||
| TNF | CXCL1 | CD40 | IL‐13 | CCL18 | CD40 | ||
| CXCL2(MIP‐2a) | CD64(FCGR1A) | PEG2 | CCL22 | CD68(SCARD1) | |||
| CXCL3 | CD68(SCARD1) | TGF‐β | CCL24 | CD115(CSF1R) | |||
| CXCL5 | CD80(B7‐1) | CXCL10 | CD163 | ||||
| CXCL8 | CD86(B7‐2) | CXCL16 | CD169(Siglec‐1) | ||||
| CXCL9 | CD115(CSF1R) | EGF | CD206(MRC1) | ||||
| CXCL10 | CD169(Siglec‐1) | Fizz1* | CD209(DC‐SIGN) | ||||
| Galectin‐3 | CD204 | Galectin‐3 | CD369(Dectin‐1) | ||||
| IFNγ | F4/80* | IDO | F4/80* | ||||
| IL‐1α | Galectin‐3 | IL‐4 | FcεR1 | ||||
| IL‐1β | IL‐1R | IL‐10 | Galectin‐3 | ||||
| IL‐6 | Ly‐6C* | IL‐12 | Ly‐6C* | ||||
| IL‐12 | Ly‐6G* | MMP9 | Ly‐6G* | ||||
| IL‐18 | Mer (MerTK) | PDGFβ | MACRO | ||||
| IL‐23 | MHC II | PPARγ | MHC II | ||||
| TNFα | TLR2(CD282) | TGFβ | PD‐L1(CD274) | ||||
| TLR4(CD284) | TNFα | PD‐L2 | |||||
| PD‐L1(CD274) | VEGF | Tie | |||||
| PD‐L2 | Ym1* | TLR1 | |||||
| TLR7 | |||||||
| TLR8 | |||||||
Only Mouse
Dynamic phenotypic transitions accompany macrophage recruitment and activation. Quiescent KCs originate from yolk sac‐derived CSF1R+ EMPs, which are usually CD11blow, F4/80high, CD68+ and Ly6C− [49, 50]. The predominant hepatic macrophages are located under the sinusoid endothelium and are involved in scavenging dying cells, pathogens, and molecules. In one study, the inhibitor of differention‐3 (ID3)+ progenitors were found to infiltrate the fetal liver during embryogenesis and are essential for giving rise to self‐maintaining KCs [26] (Figure 3).
FIGURE 3.

Dynamic phenotypic transition of TAMs during liver cancer progression. (A) KCs are unique macrophages in the liver and are part of the mononuclear phagocyte system. Those KCs in the liver sinusoids can phagocytize foreign antigens, antigen‐antibody complexes, and cell debris and also secrete cytokines, which is unfavorable for cancer initiation. DCs and HSCs present in the space of Disse cooperate with other immune cells such as KCs to maintain the liver immune microenvironment. (B) The immune microenvironment of liver cancer (HCC or iCCA) is constantly changing. The resident macrophages inside the tumor can alter their phenotype following stimulation. Furthermore, numerous circulating monocytes in the peripheral blood, including Tie+ monocytes, are recruited to the tumor milieu. Prior to converting pro‐tumoral TAMs, CD11b and Ly6C are highly expressed, while F4/80 is low. Nevertheless, TAMs, which suppresses the immune response, have higher F4/80 and lower CD11b and Ly6C levels of expression. HSCs in the space of Disse are also activated and may synergize with TAMs to promote liver cancer progression. Abbreviations: KCs, Kupffer cells; DCs, dendritic cells; HSCs, hepatic stellate cells; HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; Clec4F, C‐type lectin domain family 4 member F; Tim‐4, T‐cell immunoglobulin and mucin domain‐containing 4; CX3CR, C‐X3‐C motif chemokine receptor; CSF1R, colony‐stimulating factor 1 receptor; CCR2, C‐C motif chemokine receptor 2; MHC‐II, major histocompatibility complex class II; GATA, glutamyl‐tRNA amidotransferase, subunit A; MoMFs, monocyte‐derived macrophages; MACRO, macrophage receptor with collagenous
MoMϕs are usually CD11b+, F4/80low, CSF1R+, C‐C motif chemokine ligand 2 (CCL2)+and Ly6C− [51, 52]. These cells may be derived from CX3CR1+CD117+Lin− progenitor cells in the BM [53]. Regarding mouse models of liver disease, hepatic MoMϕs are divided into two main subpopulations according to their Ly6C‐expression levels: Ly6Chigh and Ly6Clow MoMϕs [54]. Similar to pro‐inflammatory M1 macrophages, Ly6Chigh MoMϕs can exacerbate inflammation and fibrogenesis [55]. A previous report indicated that recruited CCR2+Ly6Chigh monocytes replaced embryonic precursor cells and differentiated into mature anti‐inflammatory macrophages. This finding showed that Ly6Chigh MoMϕs had similar characteristics to M1 TAMs, while M2 TAMs usually showed low expression of Ly6C. Therefore, Ly6C is a significant dynamic surface marker during the evolution of TAMs [56] (Figure 3).
In addition, macrophages expressing CD11bhigh, F4/80low and Ly6Chigh may constitute an early macrophage phenotype in the dynamic TAM environment in the liver [8, 57]. Moreover, the surface markers of M2‐like TAMs appear to be consistent with those of CD11blow, F4/80high and Ly6Clow MoMϕs. These three markers exhibit dynamic phenotypic transitions in the TAM environment, which suggests that surface markers can be used to track the distribution of TAMs to determine the progression of liver cancer [40, 58]. Although a schematic waterfall model of TAMs development has also been described, we further summarized the published research on liver cancer and generated a more detailed map of dynamic phenotypic transitions in macrophages during carcinogenesis [57] (Figure 3).
The transcriptional profiles of the human liver have been reported with the help of single‐cell RNA sequencing. Researchers have segregated intrahepatic CD68+ macrophages into two distinct populations: the CD68+ macrophage receptor with collagenous (MARCO)+ immunoregulatory phenotype and the CD68+ MARCO− proinflammatory phenotype [59, 60]. Human CD68+ MARCO+ cells are transcriptionally similar to the mice population participating in maintaining immune tolerance and suppressing inflammation. This observation indicated that a subtype of suppressive TAMs express MARCO. In contrast, CD68+ MARCO− cells are strongly associated with inflammation. This macrophage subpopulation is characterized by the enriched expression of inflammatory genes [59]. This classification is consistent with the characteristics of our defined macrophage subpopulation, which affects inflammation during development (Figure 3).
A recent study has demonstrated that glutamyl‐tRNA amidotransferase, subunit A 6 (GATA6) macrophages could migrate toward sites of injury and repair focal lesions rapidly when the human body suffers peritoneal injury. However, abdominal adhesions can occur as a side effect of this otherwise beneficial repair process [61]. The rapid response of macrophages plays a decisive role in adhesion. However, whether GATA6 macrophages can serve as a source of TAMs that participate in liver cancer progression remains unclear.
Tie2‐expressing monocytes (TEMs) play an important role in tumor angiogenesis, and selective elimination of these TEMs through suicide genes has become a novel treatment strategy [62, 63]. Some research groups have developed a mathematical framework for systematically estimating the roles of TEMs in the M1 and M2 macrophage phenotypes during the growth of vascularized tumor lesions [64]. TEMs have been identified as a distinct TAM subpopulation influencing tumor angiogenesis, vascular remodeling and monocyte differentiation [65].
Although previous findings have revealed the recruitment of macrophages and dynamic switching of surface markers, these processes vary across cancer types. In addition, convincing evidence on how human macrophages recruit and find suitable surface markers is required to formulate strategies based on targeting macrophages.
4. M1 TAMs AND LIVER CANCER
Classically activated (or inflammatory) macrophages exhibit anti‐cancer properties. In HCC, M1 can inhibit tumor progression through various mechanisms [66, 67, 68]. Most current research is focused on regulating genes or proteins in cancer cells that can induce the polarization or infiltration of macrophages. The matricellular protein spondin2 (SPON2) activates RhoA and Rac1 and increases F‐actin reorganization through SPON2‐α4β1 integrin signaling to promote the infiltration of M1‐like macrophages [69]. High sirtuin1 (SIRT1) expression in hepatoma carcinoma cells regulates M1 polarization via the NF‐κB pathway [70]. In addition, an increase in retinoic acid‐inducible gene I (RIG‐I) expression can promote the M1 polarization of mouse peritoneal macrophages via the RIG‐I/MAVS/TRAF2/NF‐κB pathway, thereby inducing apoptosis of HCC cells [71]. Furthermore, monocytes overexpressing IL‐12 can downregulate phospho‐signal transducer and activator of transcription 3 (p‐STAT‐3) and c‐Myc to directionally differentiate into M1 and inhibit HCC growth [72]. However, M1 also shows a positive correlation with cancer, and this abnormal mechanism is relatively rare. For instance, M1 macrophages secrete IL‐1β to activate hepatoma carcinoma cells and induce programmed cell death protein 1 ligand 1(PD‐L1) expression through the transcription factors IRF1 and NF‐κB [73](Figure 4). Therefore, M1 TAMs and M2 TAMs are not always mutually exclusive; on the contrary, the two types of cells often coexist in the TME. Consequently, the two types of macrophages cannot be considered entirely distinct macrophage populations. The function favored by the mixed TAMs phenotype depends on the balance between macrophage activation and suppression and the immune microenvironment.
FIGURE 4.
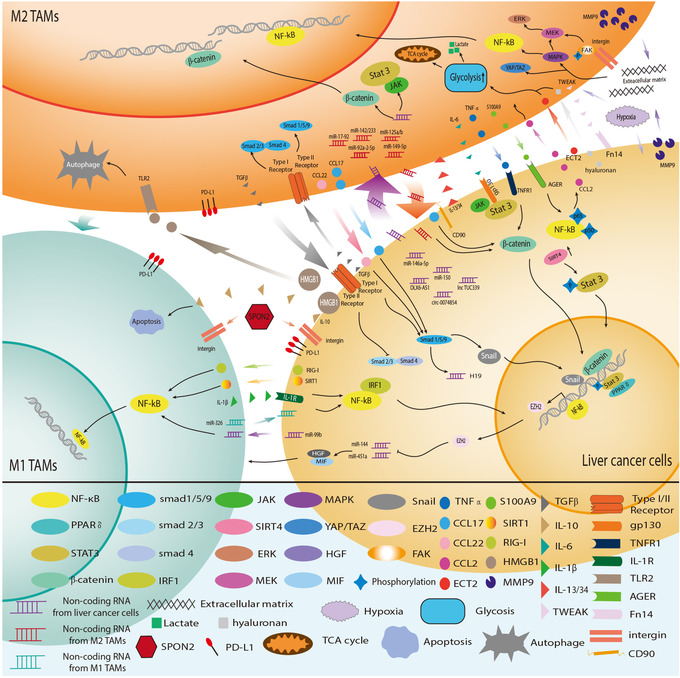
Crosstalk between TAMs and liver cancer cells. An extensive and intricate network exists between tumor cells and TAMs. M1‐type macrophages exhibit anti‐tumor effects through various mechanisms, with abundant crosstalk between M2 macrophages and tumor cells, which is closely related to tumor progression. Tumor cells and HCC‐related macrophages coordinate to adapt to the immune environment of HCC. Abbreviations: TAMs, Tumor‐associated macrophages; M1 TAMs, M1 type Tumor‐associated macrophages; M2 TAMs, M2 type Tumor‐associated macrophages; HCC, hepatocellular carcinoma; PPAR, peroxisome proliferators‐activated receptors; IRF1, interferon regulatory factor 1; SIRT4, sirtuin 4; ERK, extracellular regulated protein kinase; MEK, mitogen‐activated protein kinase kinase; MAPK, mitogen‐activated protein kinase; YAP, yes‐associated protein 1; TAZ, tafazzin; HGF, hepatocyte growth factor; MIF, migration inhibitory factor; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; FAK, focal adhesion kinase; TNF‐α, tumor necrosis factor α; CCL2, C‐C motif chemokine ligand 2; CCL17, C‐C motif chemokine ligand 17; CCL22, C‐C motif chemokine ligand 22; ECT2, epithelial cell transforming 2; S100A9, S100 calcium binding protein A9; SIRT1, sirtuin 1; RIG‐I, retinotic acid‐inducible gene I; HMGB1, high mobility group box 1;MMP9, matrix metallopeptidase 9;TGF‐β, transforming growth factor‐β; IL‐6, interleukin 6;IL‐10, interleukin 10; IL‐1β, interleukin 1β; IL‐13/34, interleukin 13/34; TWEAK, tumor necrosis factor‐like weak inducer of apoptosis; TNFR1, tumor necrosis factor receptor 1; IL‐1R, interleukin 1 receptor; TLR2, toll‐like receptor 2; SPON2, matricellular protein spondin2; PD‐L1, programmed cell death protein 1 ligand 1; AGER, advanced glycosylation end product‐specific receptor; TCA, tricarboxylic acid cycle
M1 TAMs‐derived exosomes surface‐modified with IL4RPep‐1(named IL4R‐Exo si/mi) reportedly suppress the IL‐4 receptor of M2 TAMs and decrease the levels of M2 cytokines. Such findings indicated that IL4R‐Exo(si/mi) induces the M1 TAMs phenotype and enhances antitumor immunity [74]. Multiple studies have shown that M1 macrophages are positively associated with response to anti‐PD‐1 therapy. Persistent DNA damage contributes to the cyclic GMP‐AMP synthase (cGAS)‐stimulator of interferon genes (STING) signaling pathway in M1 TAMs. Activated M1 TAMs induce T‐lymphocyte infiltration, which could enhance the response to anti‐PD‐1 [75]. Inhibition of p38 kinase phosphorylation and downstream Creb1/Klf4 activity in bone marrow‐derived macrophages by regorafenib reverses M2 polarization and enhances synergistic antitumor effect with anti‐PD‐1 [76]. In a recent study, human leukocyte antigens DR (HLA‐DR)high CD86high glycolytic phenotype macrophages were shown to represent the primary cellular source of PD‐L1 in human HCC tumors. Inhibiting the critical glycolytic enzyme pyruvate kinase M2 (PKM2) or PD‐L1 blockade liberated the inherent antitumor capability of PKM2+ glycolytic macrophages by producing antitumorigenic cytokines such as IL‐12p70. In addition, patients with more PD‐L1+PKM2+ glycolytic TAMs demonstrated a poorer prognosis [77].
In a phase I Hodgkin lymphoma clinical trial, using PI3Kδ/γ inhibitor RP6530 could switch macrophages to the M1 phenotype. PI3Kδ/γ inhibition is an effective therapeutic strategy for Hodgkin lymphoma [78]. The higher the CD137 level in the serum and the higher the M1 density in the matrix in 50 patients with advanced HCC who received sintilimab (anti‐PD‐1) plus IBI305 (a bevacizumab biosimilar), the longer the median progression‐free survival and median overall survival [24]. β‐site amyloid precursor protein‐cleaving enzyme 1 (BACE1) inhibitor MK‐8931 potently reprogramed M2 TAMs into M1 TAMs via inhibiting IL‐6‐STAT3 signaling and activated macrophage phagocytosis in cancer cells [79]. Encouragingly, MK‐8931 has been tested in clinical trials for Alzheimer's disease (AD) treatment and suggested that M1‐type antitumor macrophages were positively associated with a favorable prognosis [80]. Interfering different signaling pathways in macrophages or blocking receptors on the surface of M2 TAMs to convert them into M1 TAMs is an attractive clinical translational strategy.
5. M2 TAMs AND LIVER CANCER
5.1. M2‐TAMs contribute to cancer cell stemness in liver cancer
In hepatic tumors, the essential characteristics of aggressiveness are associated with achieving stemness. CSCs are self‐renewing cells that can facilitate tumor initiation and enhance immune resistance. Cancer cells with these biological properties may positively correlate with tumor development and metastasis. It is particularly important to clarify the crosstalk mechanisms between TAMs and CSCs in HCC.
Compared with tumor cells, the interaction between CSCs and TAMs plays a more central role in tumorigenesis, progression, and metastasis formation. Inflammatory microenvironment‐related secreted S100 calcium‐binding protein A9 (S100A9) is highly expressed in TAMs. S100A9 can promote the stemness of hepatoma carcinoma cells by activating the NF‐κB signaling pathway. Intriguingly, HCC cells treated with S100A9 can recruit more macrophages via chemokine ligand 2 [81]. Macrophage‐induced long noncoding RNA H19 upregulation enhances stemness and promotes tumorigenesis, confirming that macrophage‐induced H19 is significantly correlated with HCC progression [82]. Besides, cytokines (TNF‐α, IL‐6, and TGF‐β) and low levels of microRNAs (miRNAs, such asmiR‐125a and miR‐125b) derived from M2 TAMs can also promote cancer stemness and CSC expansion [13, 83‐85](Figure 4). The intervention of CSCs by targeting TAMs is a novel tumor immunotherapy strategy.
The interaction between TAMs and CSCs is bidirectional. Periostin (POSTN; a member of the fasciclin family secreted by CSCs) may significantly promote the recruitment of M2 TAMs in iCCA [86]. CSCs in iCCA release multiple molecules, such as IL13, OA and IL34, which can guide macrophage precursors to the M2‐like cancer‐promoting phenotype [87]. In conclusion, further understanding of the biology of CSCs and elucidating the interaction between CSCs and TAMs in various stages of tumor growth are the keys to mitigating liver cancer progression.
5.2. M2 TAMs facilitate cancer cell proliferation, aggressiveness, and metastasis
Although most of the current related research have focused on tumor‐derived exosomes, the presence of TAM‐derived exosomes is necessary for tumor progression and metastasis [88, 89]. Alternatively, activated (M2) macrophages can secrete the cytokine CCL22 to enhance tumor invasion and induce epithelial‐mesenchymal transition (EMT) via Smad2/3 and Smad1/5/8 activation and Snail upregulation [90]. In addition, CCL17 secreted by M2 macrophages is closely related to tumor stemness and EMT in the TGF‐β1 and Wnt/β‐catenin signaling pathways [91]. In vitro data have indicated that M2‐TAMs orchestrate the immune microenvironment of iCCA by secreting various cytokines, such as TNF‐α, ICAM‐1, IL‐6, and modulating the EMT of cancer cells [92](Figure 4). In an animal model of ICC, tumor cells were shown to accelerate THP‐1 cells (human acute monocytic leukemia cell line) differentiation into M2 macrophages, and the polarized macrophages could secrete IL‐10 to promote the growth, invasion and EMT of liver cancer cells [93].
The accumulation of KCs‐derived ROS and paracrine TNF causes mitochondrial dysfunction and induces oxidative stress, which could lead to the formation of premalignant lesions [94]. TNF has been positively associated with bile duct cell proliferation and carcinogenesis. Blocking the ROS/TNF/JNK axis may be an effective therapeutic strategy for attenuating the growth of iCCA tumors [94]. Intriguingly, enhanced communication between TAMs and tumor‐associated cells also promoted cancer invasion and metastasis. After co‐culturing tumor‐associated neutrophils (TANs) with macrophages, oncostatin M and interleukin‐11 were both expressed at higher levels than in the corresponding individual cultures. Crosstalk between TANs and TAMs was shown to enhance the proliferation and aggression of ICC via STAT3 signaling [95].
The above studies suggest that macrophages play exciting roles in tumorigenesis. Therefore, an attractive therapeutic strategy for liver cancer could be to block communication between M2 TAMs and liver cancer cells, such as using noncoding RNA inhibitors and TAMs receptor inhibitors.
5.3. M2 TAMs stimulate angiogenesis in liver cancer
Angiogenesis is essential for hepatocarcinogenesis and metastasis due to the hypervascular nature of most HCC tumors. Angiogenesis in HCC is a multidimensional process orchestrated by hepatoma carcinoma cells and a repertoire of tumor‐associated stromal cells, including TAMs and their bioactive products. The tumor‐microvessel density correlated positively with macrophage counts, which revealed a crucial role for TAMs in early‐stage HCC neovascularization [96]. In recent years, the monocyte/macrophage subpopulation, characterized by the expression of the tyrosine kinase receptor Tie‐2, has attracted attention. TEMs mainly aggregate in the perivascular area of tumor tissues and participate in HCC angiogenesis [97].
Human macrophage metalloelastase (HME) and vascular endothelial growth factor (VEGF) have been implicated in tumor angiogenesis. The balance between HME and VEGF gene expression can significantly affect tumor angiogenesis [98, 99]. For instance, cytokines derived from ICC cells can induce macrophage differentiation into M2‐TAMs, with increased vessels and VEGF expression [100].
Macrophage populations and phenotypes are positively correlated with angiogenesis and clinical prognosis in liver cancer. CCR2+ TAMs are more abundant at the edge of highly vascularized HCC, while the absence of CCR2+ TAM infiltration attenuates pathogenic vascularization [101]. A case‐controlled study showed that CD14+ inflammatory macrophages secreted large amounts of IL‐23 after stimulation by hepatitis virus‐infected hepatocytes, accompanied by the upregulation of IL‐23 receptors and intense macrophage‐associated angiogenesis [102]. In addition, CD14+ CD16+ monocytes from patients with liver cancer express high levels of angiogenic factor‐related genes (epiregulin, VEGF‐A and CXCL3) and predict the tissue invasive character of iCCA [103].
Hypoxic TAMs acquire angiogenic and immunosuppressive properties. Regulated in development and DNA damage response 1 (REDD1), a negative regulator of the mechanistic target of rapamycin (mTOR), was significantly upregulated in hypoxic TAMs. This inhibitor can hinder glycolysis and the vascular‐hyperactivation response in TAMs. Thwarting glycolysis in REDD1‐knockout TAMs may lead to abnormal angiogenesis [104].
The pro‐angiogenic properties of TAMs and vasculogenic mimicry in TME are fundamental reasons for the poor prognosis of tumor patients. The accumulation of macrophages was shown to correlate with the emergence of resistance to anti‐VEGF therapy in a preclinical model [105]. The escape from VEGF‐directed treatment may be due to the downregulation of VEGFR‐1 and VEGFR‐3 and the upregulation of angiogenic‐promoting genes. Such a key finding suggests that using VEGF blockade combined with macrophage blockade (such as CSF1 or CCR2 inhibitors) could enhance the anti‐VEGF therapeutic response.
5.4. TAM‐associated autophagic progress in liver cancer
Substantial evidence has demonstrated that autophagy plays an essential role in cell stress response, thereby maintaining internal environment stability. Listeria‐based HCC vaccines induce autophagy in TAMs via the Toll‐like receptor 2 (TLR2)/Myd88/NF‐κB pathway [106]. The induction of autophagy led to macrophage repolarization from the M2 to the M1 phenotype and recruited mounting anti‐tumor cytokines. Although ICIs have achieved promising therapeutic outcomes in liver cancer, ICIs still have a low response rate. Combined with PD‐1 blockade, this vaccine induced a robust antitumor response and reshaped the tumor immune microenvironment [106]. Additionally, TAMs can induce autophagy in HCC cells and attenuate the toxic effects of oxaliplatin. This autophagy‐mediated, drug‐resistance mechanism provides a new therapeutic strategy [107]. Other findings have unveiled the autophagy‐associated tumor necrosis factor receptor‐associated factor 2 (TRAF2) degradation and RelB/p52 activation can initiate TAM reprogramming to M1 macrophages, as observed in the presence of baicalin [108]. Moreover, autophagy in macrophages is triggered by the high‐mobility group box 1/Toll‐like receptor 2/NADPH oxidase 2 (HMGB1/TLR2/NOX2) autophagy axis, where hepatoma‐derived HMGB1 can skew TAMs to the M2‐like phenotype to support HCC growth [109] (Figure 4).
Autophagy is critical for controlling macrophage production and polarization at different stages. Modulating autophagy in TAMs could be a promising strategy for inhibiting liver cancer growth and progression.
5.5. Liver cancer cells enhance the M2 phenotype by releasing cytokines and exosomes
Tumor‐derived extracellular vesicles are essential mediators of cell‐to‐cell communication during tumorigenesis. Macrophages can release cytokines and exosomes, which can in turn affect tumor cells. Similarly, liver cancer cells secrete different factors into their local environment, promoting macrophage polarization.
Emerging evidence suggests that liver cancer cell‐derived exosomes facilitate cancer progression. Exosomal Sal‐like protein‐4 (SALL4) binds to the promoter region of miR‐146a‐5p and upregulates its expression in HCC‐derived exosomes. In addition, SALL4 plays a key role in T cell exhaustion [110]. Liver tumor‐derived lncRNAs (such as TUC339), circRNAs (such as hsa_circ_0074854), and miRNAs (such as miR150) are implicated as critical signaling mediators that orchestrate macrophage M1/M2 polarization [111, 112, 113](Figure 4).
Cholangiocarcinoma cells can also secrete multiple cytokines to guide macrophages into the tumor milieu. Tumor necrosis factor‐like weak inducer of apoptosis (TWEAK), a chemical messenger, and its receptor Fn14 are overexpressed in iCCA cohorts. TWEAK/Fn14 expression correlated positively with CAF proliferation. TWEAK derived from TAMs can bind to the Fn14 receptor on iCCA cells and increase the Monocyte chemoattractant protein‐1 (MCP‐1) secretion. MCP‐1 can recruit more monocytes to enter the tumor and transform into TAMs [114](Figure 4).
In this section, we emphasized on how tumor cells regulate TAMs by releasing cytokines and exosomes. However, these studies are mostly limited to the animal level. Consequently, more robust evidence is required to facilitate the translation of basic research into clinical practice. For example, in a phase III clinical trial against melanoma, Talimogene laherparepvec (T‐VEC), a herpes simplex virus type 1‐derived oncolytic immunotherapy, could selectively replicate within tumors and recruit macrophages to enhance systemic antitumor immune responses [115].
5.6. Liver cancer cells modulate the metabolic reprogramming of TAMs
Previous studies have highlighted the role of metabolic reprogramming in macrophage activation. This process of switching from a quiescent to an activated state can direct macrophage differentiation and regulate the function of these immune cells. In both mice and humans, glycolytic metabolism is involved in the classical activation of M1 TAMs, whereas mitochondrial oxidative phosphorylation is restricted to alternative activation of M2 TAMs [116]. Thus, glycolysis upregulation in specific macrophages may cause them to acquire an anti‐tumor phenotype. Nevertheless, macrophage metabolism is more complex than previously thought. Previous reports have shown that elevated glycolysis can regulate PD‐L1 levels and lead to M2‐type polarization [117, 118]. Recent data demonstrated a population of macrophages displaying an HLA‐DRhighCD86high PD‐L1+ glycolytic phenotype in HCC tumors. Intrinsic glycolytic metabolism confers PD‐L1+macrophages with anti‐tumorigenic properties [77]. Thus, targeting glycolytic metabolism in macrophages abrogated the PD‐L1‐mediated immune privilege. Tumor‐derived soluble factors, including hyaluronan fragments, can modulate glycolysis in peritumoral monocytes by up‐regulating 6‐phosphofructo‐2‐kinase/fructose‐2,6‐biphosphatase 3 (PFKFB3). This key glycolytic enzyme orchestrates cellular metabolism and induces PD‐L1 expression to attenuate cytotoxic T lymphocyte responses in tumor tissues [119]. Thus, there is an urgent need to understand the glycolytic status of macrophages and their precursor monocytes at different stages and to target critical glycolytic enzymes. In addition, the culture supernatant of HCC cells can activate the Wnt2b/β‐catenin/c‐Myc signaling pathway and augment the glycolysis of M2 TAMs [120]. The process can be inhibited by the TLR9 agonist, suggesting that targeting TLR9 might be a potential therapeutic strategy. Exosomal PKM2 derived from HCC cells induces metabolic reprogramming in monocytes, phosphorylates STAT3 and upregulates differentiation‐associated transcription factors. These data showed that PKM2 could promote monocyte‐to‐macrophage differentiation and remodel the TME [17]. The development of small‐molecule inhibitors similar to PKM2 may prevent monocyte differentiation into tumor‐promoting macrophages. Although essential, identifying tumor metabolites or vital glycolytic enzymes in macrophages would be a complex and challenging task.
Lipid metabolic reprogramming in TAMs is indispensable for macrophage polarization and hepatocarcinogenesis. Downregulation of receptor‐interacting protein kinase 3 (RIPK3) in TAMs reduced ROS levels and promoted fatty acid metabolism via PPAR activation, contributing to the accumulation and polarization of M2 TAMs [18]. Fatty acid oxidation (FAO) blockade reverses the immunosuppressive activity of TAMs, which also appears to provide a potential strategy for inhibiting tumor progression in HCC.
Several studies have provided mechanistic insights into amino acid metabolism in TAMs, and most of these studies have focused on how amino acid changes in tumor cells affect TAMs. For instance, an amine oxidase involved in extracellular matrix remodeling, lysyl oxidase‐like 4 (LOXL4), is upregulated during liver carcinogenesis in mice concomitantly fed a choline‐deficient, L‐amino acid‐defined diet. LOXL4 from neoplastic cells may facilitate macrophage infiltration into the liver and accelerate tumor growth. Glutamine metabolism is essential for macrophage activation, and α‐ketoglutarate (Αkg) production via glutaminolysis provides synergistic support for the activation [121]. Investigators in our hospital demonstrated that solute carrier family 7 member 11 (SLC7A11) led to intratumoral TAMs and MDSCs accumulation by increasing colony‐stimulating factor‐1 (CSF1) expression through Αkghypoxia‐inducible factor 1 subunit alpha (HIF1α) cascade [15].
Tryptophan‐derived microbial metabolites have been reported to mediate anti‐tumor immunity and activate aryl hydrocarbon receptors in TAMs, suggesting a potential therapeutic strategy [122]. How amino acid metabolism in macrophages affects the direction of polarization and tumor progression remains unclear; therefore, further research is warranted.
In conclusion, extensive studies have been conducted on glucose metabolism, lipid metabolism and amino acid metabolism in TAMs. From the perspective of macrophage metabolism, M1 macrophages are characterized by glycolytic metabolism, iNOS expression, and proinflammatory cytokine production. The energy distribution of M2 macrophages is characterized by the increased expression of genes related to improved glucose and fatty acid uptake, transport, and oxidation. Modulating cellular metabolic remodeling to facilitate tumor progression is a novel and promising approach. Therefore, more reliable studies on the metabolic properties, dependencies, and adaptations of TAMs are warranted.
5.7. Liver cancer cells could polarize TAM by affecting the mechanical environment
Most hepatocarcinogenesis is based on fibrotic or cirrhotic livers. Patients with HCC and severe cirrhosis usually have a worse prognosis and shorter median survival times. Increased liver stiffness plays a deleterious role in HCC progression. In addition, the degree of matrix stiffness is used to evaluate the histopathological characteristics of HCC [123]. The results of most related studies have shown that increased matrix stiffness can promote the macrophage polarization to the M2 phenotype, and the effects of biomechanical cues on HCC progression remain largely unexplored. Research has shown that a 3D gel‐like microenvironment induces adhesion and differentiation of monocytes via MAPK‐NF‐κβ activation [124]. Activation of the integrin β5‐FAK‐MEK1/2‐ERK1/2 pathway facilitates matrix stiffness‐mediated HIF‐1α and LOXL2 expression in polarized macrophages [125]. The high expression of Nogo‐B in TAMs of patients with HCC is closely correlated with yes‐associated protein 1(YAP)/ tafazzin (TAZ)‐mediated M2 polarization [126]. In summary, high matrix stiffness promotes cancer cell proliferation and resistance to chemotherapeutic agents, regulates angiogenesis, and enhances stemness. The effect of M2 polarization on HCC reflects the above processes. Thus, it is a very innovative point to explore how matrix stiffness affects macrophage polarization and functions, such as cytokine secretion and phagocytosis.
6. SYNERGISTIC REGULATION BETWEEN TAMs AND OTHER IMMUNE CELLS
Communication between macrophages and other immune cells involves intricate exposure to different microenvironments. TAMs express an array of effector molecules that inhibit immune response in liver cancer. These immunosuppressing molecules include cytokines, chemokines, enzymes and cell‐surface receptors, which are mainly ligands and receptors expressed by the target immune effector cells (Figure 5).
FIGURE 5.
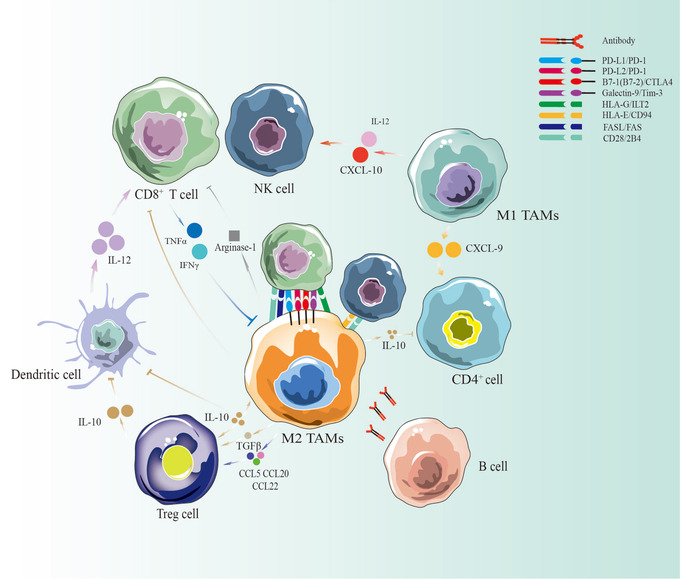
TAMs coordinate with other immune cells in liver cancer. TAMs may be considered a double‐edged sword in the immune microenvironment of liver cancer. M2 macrophages and Tregs can secrete IL‐10 to inhibit DCs, T cells and NK cells. They can also secrete arginase and directly contact each other to inhibit the immune response. M1 macrophages can secrete IL‐12 and CXCL10 to recruit more T cells and NK cells, whereas CXCL9 derived from M1 macrophages can stimulate CD4+ T cells. B cells participate in anti‐tumor‐related macrophage activities by secreting antibodies. Abbreviations: TAMs, tumor‐associated macrophages; M1 TAMs, M1 type tumor‐associated macrophages; M2 TAMs, M2 type tumor‐associated macrophages; HCC, hepatocellular carcinoma; NK cell, natural killer cell; IL‐10, interleukin 10; IL‐12, interleukin 12; CCL5, C‐C motif chemokine ligand 5; CCL20, C‐C motif chemokine ligand 20; CCL22, C‐C motif chemokine ligand 22; TNF‐α, tumor necrosis factor α; TGF‐β, transforming growth factor‐β; IFNγ, interferon‐gamma; CXCL9, C‐X‐C motif chemokine ligand 9; CXCL10, C‐X‐C motif chemokine ligand 10; PD‐L1, programmed cell death protein 1 ligand 1; PD‐1, programmed cell death protein 1; CTLA‐4, cytotoxic T‐lymphocyte antigen 4; HLA‐E, human leukocyte antigen E; HLA‐G, human leukocyte antigen G; ILT2, Ig‐like transcript 2; FAS, factor‐related apoptosis; FASL, factor‐related apoptosis ligand
6.1. TAMs and CD8+ T cells
6.1.1. TAMs interact with CD8+ T cells through direct contact
TAMs can express HLA molecules, such as HLA‐G and HLA‐E, to disable cytotoxic antitumor immune response by interacting with the costimulatory T cells markers Ig‐like transcript 2 (ILT2) and CD94, respectively [127]. In addition to HLA molecules, macrophages can also express ligands for the inhibitory receptors PD‐1 and PD‐2, cytotoxic T‐lymphocyte antigen 4 (CTLA‐4), and T cell immunoglobulin and mucin‐containing molecule 3 (Tim‐3) [128, 129, 130]. These inhibitory ligands induce T cell apoptosis or functional inactivation. Selective CD28 blockade causes 2B4 upregulation in specific CD8+ T cells, which can facilitate the control of antigen‐specific CD8+ T cell responses and functions [131]. Studies on liver metastasis revealed that FasL+CD11b+F4/80+ MoMϕs can mediate apoptosis in activated antigen‐specific Fas+CD8+ T cells [132].
In addition to the molecules mentioned above, many undiscovered mechanisms can mediate direct interactions between macrophages and CD8+ T cells. Therefore, targeting surface markers between TAMs and CD8+ T cells is a promising strategy for reversing the immunosuppressive microenvironment in liver cancer treatment.
6.1.2. TAMs indirectly exacerbate CD8+ T cells apoptosis and inhibit cytotoxic functions
Generally, macrophages are influenced by tumor cells after entering the TME and are transformed into a state that promotes tumor growth and inhibits T cell function. Therefore, TAMs are one of the leading factors that induce T‐cell dysfunction.
The TME is typically a low‐oxygen, low‐glucose, and high‐lactate milieu due to uncontrolled cancer cell proliferation and immune cell dysregulation. Some research has revealed that HIF1α induces increased expression of triggering receptor expressed on myeloid cells‐1 (TREM‐1) in TAMs in a hypoxic HCC environment. Furthermore, TREM‐1+ TAMs induced CD8+ T cell apoptosis and impaired cytotoxic functions by reducing the release of granzyme B and perforin [133]. Intriguingly, this process is independent of the PD‐L1/PD‐1 axis. In addition, TAMs have been found to express arginase‐1 and suppress the function of activated (but not resting) CD8+ T cells [134]. These results indicated that TAMs could also release small molecule secretions to indirectly repress CD8+ T cells.
TAMs are major immunosuppressive components in liver cancer. Nevertheless, reprogramming of tumor‐surveillance phenotypes of macrophages can enhance CD8+ T cell activity. For instance, a low dose of type‐I IFN can effectively reconstitute MoMϕs into CD169high macrophages. These macrophages exhibited significantly enhanced phagocytic and CD8+ T cell activation [135]. In a concurrent study, microRNA‐206 facilitated the infiltration of CD8+ T cells by inducing M1 polarization [20].
In conclusion, future research should explore macrophage remodeling to prevent M2 TAMs from affecting the infiltration of CD8+ T cells. Moreover, attenuating the expression of PD‐L1 on the surface of macrophages is a significant strategy for CD8+ T cell accumulation. [19]. However, the mechanism whereby TAMs exacerbate CD8+ T cell suppression remains unclear. Recently, a study on other cancers demonstrated that the depletion of RNA N6‐adenosine methyltransferase in TAMs induced CD8+ T cell dysfunction [136]. Thus, targeting molecules in TAMs that cause CD8+ T cell suppression represents an attractive immunotherapeutic strategy.
6.2. TAMs and regulatory T (Treg) cells are mutual accomplices
Similar to TAMs, Treg cells are essential for maintaining a suppressive immune microenvironment and contributing to tumor immune escape. Previous findings demonstrated that Tregs could mediate fatty acid synthesis in M2 TAMs [137]. In addition, Tregs indirectly but selectively maintain metabolic fitness, mitochondrial integrity, and cell survival in M2‐like TAMs. Treg cells can mediate the metabolic remodeling of TAMs and influence the direction of macrophage polarization to promote tumor progression [137]. In contrast, CCL5, CCL20, and CCL22 derived from TAMs were responsible for Tregs induction [138, 139]. In epithelial ovarian cancer, exosomes from TAMs, such as miR‐29a‐3p and miR‐21‐5p, can mediate the interaction between TAMs and T ‐cells by suppressing the STAT3 signaling pathway [140]. Regardless of the type of cancer, TAMs and Tregs act as accomplices to maintain an intratumoral immunosuppressive microenvironment.
TAMs and Tregs can also cooperate to maintain the immunosuppressive microenvironment [133]. Therefore, while targeting TAMs, Tregs also seem to be affected. However, the current understanding is insufficient, and further work in this area is merited.
6.3. TAMs and other immune cells
Tumor‐infiltrating lymphocytes can secrete TNF‐α and IFNγ, which inhibit TAMs. In turn, IL‐10 and TGF‐β produced by M2 TAMs and Tregs are pivotal chemokines that block the differentiation and maturation of T‐cells, DCs and CD4+ T cells [141, 142]. Cytokines secreted by M1 TAMs contribute to the infiltration and activation of CTLs, NK cells, and CD4+ cells [143, 144]. The CSF1 receptor signaling pathway mediates CD11b+Gr‐1lowLy6Chigh MDSC infiltration while recruiting CD11b+F4/80+ TAMs. Therefore, TAMs and MDSCs usually appear together. Intriguingly, owing to the compensatory appearance of MDSCs, the elimination of TAMs alone cannot inhibit tumor progression in iCCA [145]. As C‐X‐C motif chemokine ligand 2 (CXCL2) is a known chemoattractant for MDSCs, the researchers observed significant upregulation of CAF‐derived CXCL2 in mouse liver cancer tumors when compared with adjacent liver using the chemokine array and quantitative polymerase chain reaction (PCR). Another potential mechanism of TAM blockade–mediated MDSC accumulation is due to their enhanced survival. A distinct apolipoprotein E (ApoE) MDSC subset was uncovered by single‐cell RNA Sequencing (RNA‐Seq) analysis. Both TAMs and MDSCs are immunosuppressive cells in the TME. Eliminating TAMs may lead to the compensatory emergence of MDSCs. ApoE as well as cathepsin D (ctsd) and cathepsin B (ctsb), which mediate MDSC death via interrupted autophagy and endoplasmic reticulum stress, were downregulated in MDSCs. Thus, ApoE agonist GW3965 combined with TAMs blockade have tumor‐suppressive effects in the murine liver cancer model [145]. Moreover, RGX‐104, an LXR/ApoE agonist, was shown to significantly decrease MDSC levels in patients in a phase I human clinical trial [146]. Therefore, the effectiveness of such combinations of MDSCs blockade and TAMs blockade in the treatment of human liver cancers requires further evaluation.
In summary, M2 TAMs and other pro‐tumorigenic immune cells such as Tregs and MDSCs constitute the main drivers of the immunosuppressive microenvironment. They can directly or indirectly inhibit the function of immune cells. In contrast, M1 TAMs mediate T cell infiltration and enhance tumor immunity by reshaping the tumor immune microenvironment. Balance the interactions of macrophages with other immune cell types is key for treating malignant tumors.
7. EMERGING STRATEGIES FOR TARGETING TAMs IN LIVER CANCER
Over the past decade, experimental and clinical research results have indicated that the destruction or redifferentiation of TAMs may be a viable therapeutic strategy for patients with liver cancer [45, 147, 148]. We have divided TAMs‐related therapeutic strategies into three approaches:1) cutting off the source and eliminating the production of M2 TAMs, 2) remodeling M2 TAMs to M1 TAMs, and 3) blocking communication between M2 TAMs and liver cancer cells (Figure 6). Furthermore, we summarize the most cutting‐edge research on these treatment strategies and provide sufficient and robust evidence to support promising strategies for limiting liver cancer growth and progression (Table 2).
FIGURE 6.
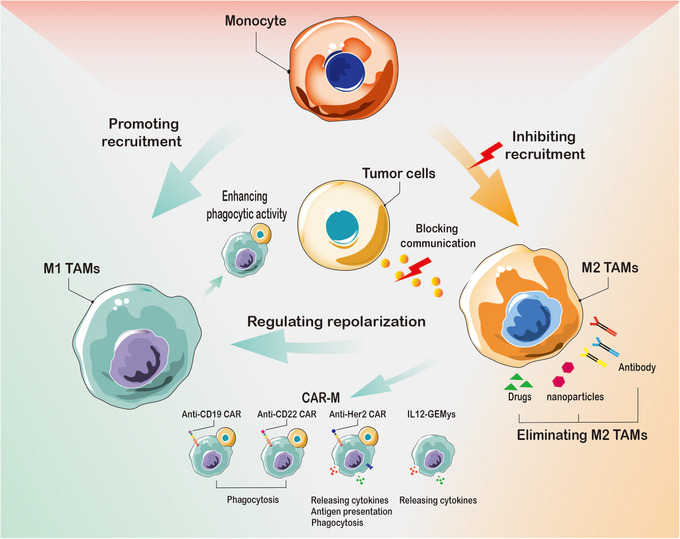
Therapeutic strategies with TAMs in liver cancer. Targeting TAMs can be carried out from three perspectives: 1. Cutting off the source and eliminating the production of M2 TAMs, including inhibiting the transition of monocytes to M2 TAMs and eliminating specific pro‐tumoral tissue‐resident macrophages in liver cancer. 2. Remodeling M2 TAMs to M1 TAMs and CAR‐M. 3. Blocking the communication between M2 TAMs and liver cancer cells. Abbreviations: TAMs, tumor‐associated macrophages; M1 TAMs, M1 type tumor‐associated macrophages; M2 TAMs, M2 type tumor‐associated macrophages. CAR‐M, chimeric antigen receptor macrophage; GEMys, genetically engineered myeloid cells. IL12, interleukin 12
TABLE 2.
Preclinical agents and clinical trials targeting TAMs for HCC treatment
| Treatment strategy | Drug name | Combinational therapy | Cancer type | Results | Clinical trial number or Reference |
|---|---|---|---|---|---|
| Inhibiting the transition of monocytes to M2 TAMs | |||||
| Blocking CSF‐1/CSF1R | Cabiralizumab | Anti‐PD1‐mAb: Nivolumab | HCC | Clinical trial for HCC patients(ongoing) | NCT04050462 |
| Chiauranib | N/A | HCC | Clinical trial for HCC patients | NCT03245190 | |
| GW2580 or AZD7507 | N/A | iCCA | Inhibit iCCA growth in xenografts model | [151] | |
| Anti‐CSF1R:AFS98 | Anti‐Ly6G:1A8 and anti‐PD‐1: G4 | iCCA | Potentiate anti‐PD‐1 therapy and attenuate iCCA progression | [145] | |
| SNDX‐6532 | Durvalumab | iCCA | Clinical trial for iCCA patients(ongoing) | NCT04301778 | |
| CCL2/CCR5 antagonist | BMS‐813160 | Anti‐PD1‐mAb: Nivolumab | HCC | Clinical trial for HCC patients(ongoing) | NCT04123379 |
| Cenicriviroc | N/A | HCC | Reverse liver damage and steatosis | [152] | |
| CCR2 antagonist | 747 | Low‐dose sorafenib | HCC | Anti‐tumor and enhance the efficacy of sorafenib | [154] |
| RDC018 | N/A | HCC | Inhibit HCC growth and metastasis | [153] | |
| CCL2 antibody | 2H5 | N/A | iCCA | Downregulate macrophage accumulation and suppress tumor growth | [114] |
| C/EBPα saRNA | MTL‐CEBPA | Sorafenib | HCC | Clinical trial for HCC patients (ongoing) | NCT02716012/[155] |
| MTL‐CEBPA | Pembrolizumab | HCC/iCCA | Clinical trial for solid tumor patients (ongoing) | NCT04105335 | |
| CCL5 blockade | aPKCɩ‐siRNA | Gemcitabine | iCCA | Inhibited TAM infiltration and iCCA progression | [156] |
| Remodeling M2 TAMs to M1 TAMs | |||||
| Tyrosine kinase inhibitors | Sorafenib | N/A | HCC | Inhibit tumor growth and improve survival | [165] |
| Pan‐PI3K inhibitor | SF1126 | Anti‐PD1‐mAb: Nivolumab | HCC | Clinical trial for HCC patients | NCT03059147 |
| PI3Kγ inhibitor | TG100‐115 | Sorafenib | HCC | Higher levels of antitumor efficiency | [170] |
| GSK2636771 | N/A | iCCA | MATCH Screening Trial for solid tumor patients(ongoing) | NCT02465060 | |
| RIPK3 inhibitor | GSK872 | N/A | HCC | Dampened HCC tumorigenesis | [18] |
| Natural compound | Baicalin | N/A | HCC | Suppress tumor progression | [108] |
| CSF‐1R inhibitor | PLX3397 | Anti‐PD‐L1 | HCC | Suppress tumor progression | [167] |
| C‐Met and EGFR inhibitor | Norcantharidin | N/A | HCC | Suppress tumor progression | [169] |
| Agonistic anti‐CD40 | Clone FGK4.5 | Anti‐PD‐1 antibodies: Clone 29F.1A12 | iCCA | Increase the infiltration of immune cells and inhibit iCCA growth | [171] |
| Tie2 inhibitor | Regorafenib | N/A | HCC | Clinical trial for HCC patients | NCT04476329 |
| Regorafenib | Nivolumab | HCC | Clinical trial for HCC patients(ongoing) | NCT04170556 | |
| Regorafenib | PD‐1 inhibitor | HCC | Clinical trial for HCC patients(ongoing) | NCT05048017 | |
| Regorafenib | N/A | HCC | Improves overall survival in HCC patients with sorafenib treatment | [176] | |
| TLR7 agonist | RO7119929 | Tocilizumab | HCC/iCCA | Clinical trial for HCC/iCCA patients(ongoing) | NCT04338685 |
| GS‐9620 | Anti‐SIGLEC‐3 mAb | HCC | Reducing the risk of HCC in chronic hepatitis patients | [172] | |
| Eliminating specific pre‐tumoral tissue‐resident macrophages in liver cancer | |||||
| Microtubule polymerization inhibitor | CA1P | Sorafenib | HCC | Suppress tumor growth and enhance the effect of PD‐1 sorafenib | [157] |
| Macrophage depletion drug | Zoledronic acid | Sorafenib | HCC | Suppress tumor growth and enhance the effect of PD‐1 sorafenib | [158] |
| Lipclod | N/A | iCCA | Deplete phagocytic macrophages and inhibit iCCA growth | [151] | |
| Zoledronic acid | Sorafenib | HCC | Clinical trial for HCC patients(ongoing) | NCT01259193 | |
| Tyrosine kinase inhibitors | Lenvatinib | Anti‐mouse PD‐1 antibody | HCC | Suppress tumor growth and enhance the effect of PD‐1 antibody | [159] |
| Antioxidant agent | CeO2NPs | N/A | HCC | Eliminating existent TAMs and Suppress tumor progression | [160] |
| Blocking the communication between M2 TAMs and liver cancer cells | |||||
| A2A antagonist | SCH58261 | Anti‐GM‐CSF antibodies | HCC | Eliminating existent TAMs and Suppress tumor progression | [14] |
| CD47‐SIRPα blocking | Anti‐human‐SIRPα | N/A | HCC | Clinical trial for HCC patients | NCT02868255 |
| Anti‐CD47‐Ab | TACE or Tocilizumab | HCC | Suppress tumor growth and enhance the effect of TACE | [184] | |
| Anti‐CD47‐Ab | Doxorubicin | HCC | Suppress tumor growth and enhance the effect of doxorubicin | [187] | |
| CD47mAb400 | N/A | HCC | Suppress tumor progression | [188] | |
| Anti‐CD47‐Ab | B6H12.2 | iCCA | Promotes macrophage phagocytosis and suppress iCCA growth | [186] | |
| CAR‐M | |||||
| CAR macrophages | CT‐0508 | N/A | HCC | Clinical trial for Her2 overexpressing patients(ongoing) | NCT04660929 |
Abbreviations: A2A, adenosine A2a receptor; CAR‐M, chimeric antigen receptor macrophage; CCL2, C‐C motif chemokine ligand 2; CCL5, C‐C motif chemokine ligand 5; CCR2, C‐C motif chemokine receptor 2; CCR5, C‐C motif chemokine receptor 5; C/EBPα, CCAAT/enhancer‐binding protein alpha; CSF1, colony‐stimulating factor‐1; CSF1R, colony‐stimulating factor 1 receptor; EGFR, epidermal growth factor receptor; HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; IL6, interleukin 6; PD1, programmed cell death protein 1; PI3Kγ, Phosphoinositide 3‐kinase gamma; RIPK3, receptor‐interacting protein kinase 3; saRNA, small activating RNA; SIGLEC‐3, sialic acid binding Ig like lectin 3; SIRPα, signal regulatory protein alpha; TACE, transcatheter arterial chemoembolization; TAMs, tumor‐associated macrophages; TLR7, toll‐like receptor 7.
7.1. Cutting off the source and eliminating the production of M2 TAMs
7.1.1. Inhibiting the transition of monocytes to M2 TAMs
Circulating monocytes are the primary source of infiltrating macrophages in tumors, and the accessibility of mononuclear cells in clinical practice (which enables a treatment strategy based on blocking the source of monocytes) is particularly significant. Blocking the CSF1/CSF1R axis is the most established method of reducing TAM survival [149]. This ligand‐receptor pair is indispensable for TAM differentiation and survival [150]. The CSFR1 inhibitors GW2580 and AZD7507 were shown to prevent the recruitment of peripheral monocytes in iCCA [151]. TAM blockade alone does not inhibit tumor progression, with a compensatory increase of granulocytic myeloid‐derived suppressor cells (G‐MDSCs). Combined treatment with antibodies against CSF1R and Ly6G or ApoE agonist GW3965 could reduce tumor volumes and increase the efficacy of anti‐PD‐1 in the YAP/AKT driven murine iCCA model.
Along with the CSF1/CSF1R axis, a growing body of evidence indicates that the CCL2/CCR2 axis‐mediated macrophage infiltration is a potential immunotherapeutic target [152]. CCL2 derived from HSCs can promote macrophage accumulation and modulate TAM polarization. Blocking the CCL2/CCR2 axis with a CCR2 antagonist impaired the accumulation of blood inflammatory monocytes and suppressed murine liver tumor growth [153]. Furthermore, a natural CCR2 antagonist, 747, could recruit more CD8+ cytotoxic T cells and enhance the therapeutic effect of sorafenib by blocking tumor‐infiltrating macrophage‐mediated immunosuppression [154]. In cholangiocarcinoma, monocyte chemoattractant protein‐1(MCP‐1, also known as CCL‐2) directed the trafficking of CCR2+monocytes to the tumor niche, and an anti‐MCP‐1 antibody attenuated the aggregation of circulating macrophages [114].
C/EPBα, a transcription factor, can enhance the function of MDSCs and M2 macrophages. In a phase I/Ib multicenter clinical study, C/EBPα saRNA (MTL‐CEBPA) treatment played an indispensable role in hampering aggregation and reversing the suppressive activities of MDSCs and TAMs, and combination therapy with MTL‐CEBPA and a PD‐1 antibody significantly abrogated tumor progression [155]. Moreover, some researchers have used 10X Genomics single‐cell RNA sequencing technologies to determine that M2 TAMs infiltration was associated with iCCA progression and to measure the expression of macrophage markers and aPKCɩ in 70 human tumor samples at various stages. Furthermore, the study presented a novel combination therapy strategy with cationic liposome‐mediated co‐delivery of gemcitabine and aPKCɩ‐siRNA, which significantly attenuated macrophage recruitment [156].
None of the antibodies and inhibitors above can selectively act on M2 TAMs and affect the antitumor activity of M1 TAMs. Therefore, M2‐selective clearance antibodies and inhibitors need to be developed in the future.
7.1.2. Eliminating specific pro‐tumoral tissue‐resident macrophages in liver cancer
Specific pro‐tumoral tissue‐resident macrophages accumulate close to tumor cells early during tumor formation and provide a pro‐tumorigenic niche and induce a potent regulatory T cell response that protects tumor cells from adaptive immunity [10]. In liver cancer, KCs represent the vast majority of tissue‐resident macrophages. In the presence of carcinogenic factors, these cells will gradually transform into TAMs to promote cancer progression.
A previous report showed that combretastatin A‐1 phosphate (CA1P), a microtubule polymerization inhibitor, exerted remarkable antitumor activity against HCC cells and TAMs. CA1P could induce TAM apoptosis by inhibiting the Wnt/β‐catenin pathway and down‐regulating Treg levels. The authors also indicated that combination therapy with CA1P and sorafenib therapy was most likely to achieve treatment effects in drug‐resistant patients [157]. Although emerging clinical evidence and preclinical findings have revealed that sorafenib increases overall survival and improves outcomes in patients with HCC, macrophages may exert a pro‐tumor role under sorafenib treatment [22]. However, zoledronic acid combined with sorafenib could significantly inhibit tumor progression in two xenograft nude mouse models [158]. Other multi‐targeted kinase inhibitors, such as lenvatinib, have been reported to decrease the tumor macrophage population and increase the number of CD8+ cells [159]. That research also indicated that combination treatment with lenvatinib and an anti‐PD‐1 antibody notably enhanced antitumor activity.
Recent research showed that treating rats with HCC with cerium oxide nanoparticles (CeO2NPs) reduced the macrophage infiltration into the liver, inhibited the expression of inflammatory response‐related genes, and improved the survival of Wistar rats [160]. Moreover, the macrophage scavenger liposomal clodronate (Lipclod) can selectively inhibit Wnt signaling and the growth of iCCA in bilateral xenografts of human cholangiocarcinoma cells SNU‐1079, CC‐LP‐1 or WITT1 [151].
Tissue‐resident macrophages play a pivotal role in shaping the liver tumor immune microenvironment, making them a crucial target in the prevention and treatment of early liver cancer lesions. Nonetheless, the lack of a comprehensive understanding of macrophage molecular and functional diversity makes modulating them extremely difficult. Therefore, using single‐cell sequencing to trace how such TAM lineages contribute to the TME and liver cancer progression should be explored in future research. [10, 59, 161, 162].
7.2. Remodeling M2 TAMs to M1 TAMs
Preclinical experimental data suggest that targeting macrophage repolarization can be beneficial in cancer immune therapy [163]. Sorafenib is a classic multikinase inhibitor approved for treating systemic HCC. Recent data have shown that sorafenib inhibits the macrophage‐induced growth of hepatoma cells [164]. Sorafenib regulates the functions of M2 TAMs and hampers HCC growth mediated by the insulin growth factor (IGF)/insulin growth factor receptor (IGF‐R) signaling axis [165]. Further study on targeting macrophages with sorafenib has shown that the multi‐kinase inhibitors stimulate proinflammatory cytokine production in macrophages and activate hepatic NK cells in cytokine‐ and NF‐κB‐dependent manners [166]; providing robust evidence that tyrosine kinase inhibitors (TKIs) can be used to reverse macrophage polarization and induce anti‐tumor immune responses.
RIPK3‐mediated lipid metabolic reprogramming is correlated with tumorigenesis, macrophage accumulation, and polarization in the tumor milieu. The RIPK3 inhibitor, GSK872, or FAO blockade, was shown to remodel the immune activity of TAMs and markedly abrogate tumor progression [18]. A natural compound, baicalin, has been reported to directly induce TAM reprogramming to M1‐like macrophages through autophagy‐associated activation of Reticuloendotheliosis viral oncogene homolog B (RelB)/p52. The authors demonstrated the tumor‐suppressive effect of baicalin in skewing macrophages away from M2‐like macrophages toward the M1‐like phenotype for treating HCC [108]. Recently, it was reported that blocking the CSF1/CSF1R signaling by PLX3397 reduced tumor growth and reduced M2 phenotype polarization [167]. Targeting miRNAs could also be an effective therapeutic strategy for regulating macrophage polarization. MiR‐98 may play a vital role in shifting the polarization of TAMs towards the M1 phenotype [168]. Norcantharidin, a common anticancer drug, modulates macrophage polarization through miR‐214 to exert an anti‐HCC effect [169].
Phosphoinositide 3‐kinase gamma (PI3Kγ) regulates the immunosuppressive properties of TAMs. PI3Kγ inhibitors, such as TG100‐115, can induce the expression of MHC‐II and proinflammatory cytokines, skewing macrophages to the M1 phenotype while reducing immunosuppressive molecules and inhibiting tumor progression [170].
Multiple stages of the immune response are mediated by the tumor necrosis factor superfamily (TNFSF) ligands and their receptors, such as the CD40/CD40L pair. CD40 functions as a crucial communication medium connecting innate and adaptive immunity and is a significant trigger for monocyte to differentiate into M1 macrophages and DC cells. The CD40‐mediated activation of macrophages and DCs in iCCA has been shown to ameliorate responses to checkpoint inhibitors [171].
TLR can trigger the release of pro‐inflammatory cytokines in response to bacteria or viruses infection. Intratumoral injection of TLR agonists can increase monocyte recruitment and infiltration and induce repolarization, preventing TAM polarization to the M2 phenotype [172, 173]. Clinical trials have also been conducted to study the efficacy of TLR agonists, such as RO7119929, against liver cancer.
The infiltration of pro‐angiogenic TAMs that express the angiopoietin receptor Tie2 could serve as a diagnostic marker for HCC [97]. Transient expression of a dominant‐negative Tie2 ectodomain (sTie2) can block Ang2‐mediated Tie2 signaling and may be a promising approach for tumor treatment [174]. In addition, data from a study of other cancers showed that Ang2‐Binding and Tie2‐Activating Antibody (ABTAA) induced polarization of TAM toward an anti‐tumor phenotype and promoted tumor vascular normalization more effectively than treatment with Ang2‐Blocking Antibody (ABA) alone. These findings demonstrate that ABTAA is a promising therapeutic for inhibiting tumor growth and reorchestrating the TME [175]. Regorafenib is an oral small‐molecule TKI that potently blocks multiple protein kinases. Interestingly, Tie2 may be a tempting target for regorafenib. Preclinical data and some clinical findings have confirmed that regorafenib could effectively inhibit liver cancer progression [176].
7.2.1. A novel cellular immunotherapy: chimeric antigen receptor macrophage (CAR‐M) treatment
Clinical trials have significantly advanced cancer treatment in adoptive cell therapy, such as chimeric antigen receptor T cell (CAR‐T) therapy, especially for hematologic neoplasms. Recent clinical trials of CAR‐M therapy have opened up a new field for using macrophages to treat solid tumors (Table 2). After modification via genetic engineering technology, macrophages can target specific antigens, such as CD19, CD22, and Her2, to recognize tumor cells [177]. CAR‐M cells can phagocytize tumor cells, secrete pro‐inflammatory cytokines to change the tumor immune microenvironment, present tumor antigens to T cells and activate the immune response [178]. Studies on solid tumors have revealed that highly active CD3‐based CARs on the surface of macrophages can phagocytose and destroy targeted tumor cells in a Syk‐dependent manner instead of soluble opsonizing factors [177, 179]. Ad5f35, a chimeric adenoviral vector, can induce a durable M1 phenotype after macrophage transduction in humanized mouse models. Intriguingly, this group of macrophages did not convert to M2 macrophages upon stimulation with cytokines or a tumor‐conditioned medium, which confirmed that CAR‐Ms could elicit an inflammatory microenvironment, improve antitumor T cell activity, and significantly abrogate solid tumor progression [177].
CAR‐M technologies are constantly evolving. The targets recognized by CAR‐M are primarily expressed in other cancer types. Nevertheless, due to the heterogeneity of liver cancer, exploring liver tumor cell‐specific targets and engineering macrophages in liver TME remains challenging. It is also necessary to pay attention to the therapy's potential off‐target toxicity and immunogenicity [180, 181, 182]. If the results of associated studies are to be translated into clinical practice, safer, more reliable and efficient CAR‐M technologies should be developed. In addition, more research is warranted to confirm whether CAR‐M combined with CAR‐T, multi‐targeted kinase inhibitors, and ICIs can enhance tumor inhibitory effects.
7.3. Blocking the communication between M2 TAMs and liver cancer cells
The relationships between tumor cells and macrophages are diverse. Many of the relationships involve the modulation of signaling pathways that contribute to homeostasis in non‐malignant organs. In addition, intervention with M2 TAMs should not affect macrophages not associated with malignancy. Mechanistic data have demonstrated that tumor‐derived adenosine, an ATP‐AMP metabolite, with autocrine GM‐CSF can accelerate suppressive tumor‐infiltrating macrophage proliferation via the PI3K/AKT pathway. Hence, using adenosine receptor antagonists or anti‐GM‐CSF antibodies to inhibit macrophage accumulation may be a novel immunotherapy strategy [14].
CD47 is an immunoglobulin‐like protein known to interact with its receptor on macrophages, SIRPα, and participates in phagocyte‐mediated tumor clearance [183].CD47 expression is modulated by several mechanisms. For instance, IL‐6 secreted by TAMs can upregulate CD47 expression in hepatoma cells through the STAT3 pathway [184]. Moreover, histone deacetylase 6 (HDAC6) mediates thrombospondin‐1 (TSP1) expression, and TSP1 binds the CD47 receptor to block the CD47‐SIRPα‐mediated anti‐phagocytosis of the macrophage in a spontaneous mouse HCC model [185]. Thus, blocking CD47‐SIRPα signaling is a potential strategy for enhancing the ability of macrophages to phagocytose tumor cells and inducing antitumor responses [184, 186‐188].
Combination therapy of CD47 blockade and other target inhibitors presents new insights to improve HCC treatment. Glypican‐3 (GPC3) is a characteristic antigen of hepatocellular carcinoma. Bispecific antibodies to CD47 and Glypican‐3 (GPC3) have more potent antitumor effects and lower toxicity than monotherapy in humanized mice [189]. Furthermore, a preclinical study showed that CD47 blockade with CD47 antibody is associated with a good prognosis. CD47‐blocking increased the time‐to‐progression of metastatic tumors and prolonged survival in a murine splenic injection model of hepatic micrometastatic pancreatic ductal adenocarcinoma [190].
8. CONCLUSION AND PERSPECTIVES
Over the past decades, extensive research has brought forth the revolutionary idea that hepatic immune cell populations based on TAMs play central roles in initiating and perpetuating liver cancer. In the specific immune microenvironment of hepatic neoplasms, TAMs serve functions that differ from those in other organs, including the regulation of signal‐transduction mechanisms, the change of self‐metabolism patterns in response to the context of the internal environment, and the spectrum of secreted factors. Whether starting from the communication between TAMs and cancer cells, the corresponding metabolic changes, or the biological stress of TAMs themselves, there seems to be more room for exploring the field of TAMs in liver cancer. In clinical research, TAMs are closely related to postoperative tumor progression and treatment. Experts have concluded that the TAMs population is a significant clinical prognostic indicator. Many clinical trials are still exploratory and are in an early stage. Some studies, such as using in vitro‐cultured macrophages in patients with advanced cancer (who have failed standard therapy) have been conducted. After intravenous or intraperitoneal injection of these cultured macrophages into 15 patients, the disappearance of ascites and inflammatory reactions occurred in individual patients. It is noteworthy that there were no other adverse reactions except for low‐grade fever and abdominal discomfort. Although lacking significant regression of the primary tumor site and clinical efficacy, these studies provide valuable information for the development of human macrophage‐based therapies.
Although preclinical and clinical studies on TAMs have provided encouraging results, the use of macrophages as a targeted therapy for liver cancer still faces some challenges. First, most TAM research has been limited to animal models. There is considerable heterogeneity between mouse models and humans in terms of pathogenesis and responses to drug therapy. To facilitate the introduction of TAMs applications in the clinical arena, it is necessary to explore inhibitors targeting human TAMs and the immune microenvironment in patients with liver cancer. Second, due to the diversity of the origin of macrophages and heterogeneity after differentiation, TAMs show diverse characteristics at different stages. It appears that using specific blocking agents, such as CCL2 antibodies alone, is insufficient to overcome liver malignancies. Additionally, pan‐macrophage therapeutic approaches may also have adverse effects, such as CSF1 blockade targeting all macrophages, leading to systemic toxicity. Therefore, determining an optimal time for treatment and more precise therapeutic targets of TAMs during ongoing treatment are tasks worth exploring. Third, the checkpoint blockade on the surface of macrophages is currently limited to PD‐L1. Several novel immune checkpoints are expressed on the surface of macrophages, such as SIRPα and Tim‐3. Additional preclinical and clinical trials are required to support other ICIs combined with classic therapies. Finally, eliminating TAMs seems to lead to the compensatory emergence of other immunosuppressive cells. Thus, TAM elimination would also need to compensate for other immunosuppressive cells, such as Tregs and MDSCs, which cause tolerance to targeted TAMs alone.
In this review, we have elaborated on the origin of TAMs, the communication of TAMs with surrounding cells, and the latest treatment progress, which provides more options and substantial evidence for treating patients with liver cancer by targeting macrophages. In the future, pharmaceuticals targeting macrophages in the specific immune environment of the liver as well as more stable, safe and efficient immune combination therapy could facilitate the further development of immunotherapy for liver cancer.
DECLARATIONS
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
KC collected the related papers and was a major contributor to the writing of the manuscript. NC prepared the figures and tables. JHZ and XY drafted the paper and revised it critically for important intellectual content. HFL and WGZ provided the final approval for the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGMENTS
We thank several professional English editors (American and Chinese Journal Experts) for assistance in improving the quality of the language. We thank Professor Huifang Liang and Wanguang Zhang for their valuable scientific advice and substantial support. This work was supported by The National Natural Science Foundation of China (No. 8187111473 to Wanguang Zhang; No. 8217113337 to Wanguang Zhang).
Cheng K, Cai N, Zhu J, Yang X, Liang H, Zhang W. Tumor‐associated macrophages in liver cancer: From mechanisms to therapy. Cancer Commun. 2022;42:1112–1140. 10.1002/cac2.12345
Kun Cheng and Ning Cai contributed equally.
Contributor Information
Huifang Liang, Email: lianghuifang1997@126.com.
Wanguang Zhang, Email: wgzhang@tjh.tjmu.edu.cn.
REFERENCES
- 1. Hiam‐Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer. 2021;21(6):345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, et al. Clinical significance of tumor‐associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol. 2012;30(28):3525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seton‐Rogers S. Taming TAMs in brain metastases. Nat Rev Cancer. 2022;22(1):2–3. [DOI] [PubMed] [Google Scholar]
- 4. Sun L, Kees T, Almeida AS, Liu B, He XY, Ng D, et al. Activating a collaborative innate‐adaptive immune response to control metastasis. Cancer Cell. 2021;39(10):1361–74.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sunakawa Y, Stintzing S, Cao S, Heinemann V, Cremolini C, Falcone A, et al. Variations in genes regulating tumor‐associated macrophages (TAMs) to predict outcomes of bevacizumab‐based treatment in patients with metastatic colorectal cancer: results from TRIBE and FIRE3 trials. Ann Oncol. 2015;26(12):2450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang X, Wang J, Lu D, Xu X. Targeting tumor‐associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther. 2021;6(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bian Z, Gong Y, Huang T, Lee CZW, Bian L, Bai Z, et al. Deciphering human macrophage development at single‐cell resolution. Nature. 2020;582(7813):571–6. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–45.e20. [DOI] [PubMed] [Google Scholar]
- 9. De Simone G, Andreata F, Bleriot C, Fumagalli V, Laura C, Garcia‐Manteiga JM, et al. Identification of a Kupffer cell subset capable of reverting the T cell dysfunction induced by hepatocellular priming. Immunity. 2021;54(9):2089–100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casanova‐Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, et al. Tissue‐resident macrophages provide a pro‐tumorigenic niche to early NSCLC cells. Nature. 2021;595(7868):578–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, et al. Cross‐tissue single‐cell landscape of human monocytes and macrophages in health and disease. Immunity. 2021;54(8):1883–900.e5. [DOI] [PubMed] [Google Scholar]
- 12. Wang D, Li X, Li J, Lu Y, Zhao S, Tang X, et al. APOBEC3B interaction with PRC2 modulates microenvironment to promote HCC progression. Gut. 2019;68(10):1846–57. [DOI] [PubMed] [Google Scholar]
- 13. Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, et al. Tumor‐associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147(6):1393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J, Wang Y, Chu Y, Li Z, Yu X, Huang Z, et al. Tumor‐derived adenosine promotes macrophage proliferation in human hepatocellular carcinoma. J Hepatol. 2021;74(3):627–37. [DOI] [PubMed] [Google Scholar]
- 15. He Q, Liu M, Huang W, Chen X, Zhang B, Zhang T, et al. IL‐1β‐Induced elevation of solute carrier family 7 member 11 promotes hepatocellular carcinoma metastasis through Up‐regulating programmed death ligand 1 and colony‐stimulating factor 1. Hepatology. 2021;74(6):3174–93. [DOI] [PubMed] [Google Scholar]
- 16. Sun R, Zhang Z, Bao R, Guo X, Gu Y, Yang W, et al. Loss of SIRT5 promotes bile acid‐induced immunosuppressive microenvironment and hepatocarcinogenesis. J Hepatol. 2022. [DOI] [PubMed] [Google Scholar]
- 17. Hou PP, Luo LJ, Chen HZ, Chen QT, Bian XL, Wu SF, et al. Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol Cell. 2020;78(6):1192–206.e10. [DOI] [PubMed] [Google Scholar]
- 18. Wu L, Zhang X, Zheng L, Zhao H, Yan G, Zhang Q, et al. RIPK3 orchestrates fatty acid metabolism in tumor‐associated macrophages and hepatocarcinogenesis. Cancer Immunol Res. 2020;8(5):710–21. [DOI] [PubMed] [Google Scholar]
- 19. Chen J, Lin Z, Liu L, Zhang R, Geng Y, Fan M, et al. GOLM1 exacerbates CD8(+) T cell suppression in hepatocellular carcinoma by promoting exosomal PD‐L1 transport into tumor‐associated macrophages. Signal Transduct Target Ther. 2021;6(1):397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu N, Wang X, Steer CJ, Song G. MicroRNA‐206 promotes the recruitment of CD8(+) T cells by driving M1 polarisation of Kupffer cells. Gut. 2022. Aug;71(8):1642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng AL, Hsu C, SL Chan, Choo SP, Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72(2):307–19. [DOI] [PubMed] [Google Scholar]
- 22. Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor‐associated neutrophils recruit macrophages and T‐regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology. 2016;150(7):1646–58.e17. [DOI] [PubMed] [Google Scholar]
- 23. Adachi Y, Kamiyama H, Ichikawa K, Fukushima S, Ozawa Y, Yamaguchi S, et al. Inhibition of FGFR Reactivates IFNγ signaling in tumor cells to enhance the combined antitumor activity of lenvatinib with Anti‐PD‐1 antibodies. Cancer Res. 2022;82(2):292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang W, Gong C, Peng X, Bi X, Sun Y, Zhou J, et al. Serum concentration of CD137 and tumor infiltration by M1 macrophages predict the response to sintilimab plus bevacizumab biosimilar in advanced hepatocellular carcinoma patients. Clin Cancer Res. 2022. Mar 11:clincanres.3972.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laviron M, Boissonnas A. Ontogeny of tumor‐associated macrophages. Front Immunol. 2019;10:1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, et al. Specification of tissue‐resident macrophages during organogenesis. Science. 2016;353(6304):aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Franklin RA, Li MO. Ontogeny of tumor‐associated macrophages and its implication in cancer regulation. Trends Cancer. 2016;2(1):20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature. 2015;518(7540):547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kieusseian A, Brunet de la Grange P, Burlen‐Defranoux O, Godin I, Cumano A. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 2012;139(19):3521–30. [DOI] [PubMed] [Google Scholar]
- 30. Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446(7139):1056–61. [DOI] [PubMed] [Google Scholar]
- 31. Cortez‐Retamozo V, Etzrodt M, Newton A, Rauch PJ, Chudnovskiy A, Berger C, et al. Origins of tumor‐associated macrophages and neutrophils. Proc Natl Acad Sci U S A. 2012;109(7):2491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shand FH, Ueha S, Otsuji M, Koid SS, Shichino S, Tsukui T, et al. Tracking of intertissue migration reveals the origins of tumor‐infiltrating monocytes. Proc Natl Acad Sci U S A. 2014;111(21):7771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong HH, Xiang S, Chen XP, Liang HF, Zhang W, Jing K, et al. The epithelial‐mesenchymal transition promotes transdifferentiation of subcutaneously implanted hepatic oval cells into mesenchymal tumor tissue. Stem Cells Dev. 2009;18(9):1293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong HH, Xiang S, Liang HF, Li CH, Zhang ZW, Chen XP. The niche of hepatic cancer stem cell and cancer recurrence. Med Hypotheses. 2013;80(5):666–8. [DOI] [PubMed] [Google Scholar]
- 35. Shibahara J, Hayashi A, Misumi K, Sakamoto Y, Arita J, Hasegawa K, et al. Clinicopathologic characteristics of hepatocellular carcinoma with reactive Ductule‐like components, a subset of liver cancer currently classified as combined hepatocellular‐cholangiocarcinoma with stem‐cell features, typical subtype. Am J Surg Pathol. 2016;40(5):608–16. [DOI] [PubMed] [Google Scholar]
- 36. Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Molecular pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. Clin Cancer Res. 2016;22(2):291–300. [DOI] [PubMed] [Google Scholar]
- 37. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152(4):745–61. [DOI] [PubMed] [Google Scholar]
- 38. Chen Y, Wong PP, Sjeklocha L, Steer CJ, Sahin MB. Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology. 2012;55(2):563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nishikawa Y, Doi Y, Watanabe H, Tokairin T, Omori Y, Su M, et al. Transdifferentiation of mature rat hepatocytes into bile duct‐like cells in vitro. Am J Pathol. 2005;166(4):1077–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma A, Seow JJW, Dutertre CA, Pai R, Blériot C, Mishra A, et al. Onco‐fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183(2):377–94.e21. [DOI] [PubMed] [Google Scholar]
- 41. Chen MM, Xiao X, Lao XM, Wei Y, Liu RX, Zeng QH, et al. Polarization of tissue‐resident TFH‐Like cells in human hepatoma bridges innate monocyte inflammation and M2b macrophage polarization. Cancer Discov. 2016;6(10):1182–95. [DOI] [PubMed] [Google Scholar]
- 42. Li Z, Wu T, Zheng B, Chen L. Individualized precision treatment: Targeting TAM in HCC. Cancer Lett. 2019;458:86–91. [DOI] [PubMed] [Google Scholar]
- 43. Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18(9):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour‐associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rőszer T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediators Inflamm. 2015;2015:816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Colin S, Chinetti‐Gbaguidi G, Staels B. Macrophage phenotypes in atherosclerosis. Immunol Rev. 2014;262(1):153–66. [DOI] [PubMed] [Google Scholar]
- 48. Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, et al. Single‐cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci. 2021;24(4):595–610. [DOI] [PubMed] [Google Scholar]
- 49. Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17(5):306–21. [DOI] [PubMed] [Google Scholar]
- 50. Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow‐derived monocytes give rise to self‐renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol. 2016;13(3):316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wen Y, Lambrecht J, Ju C, Tacke F. Hepatic macrophages in liver homeostasis and diseases‐diversity, plasticity and therapeutic opportunities. Cell Mol Immunol. 2021;18(1):45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–7. [DOI] [PubMed] [Google Scholar]
- 54. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wehr A, Baeck C, Heymann F, Niemietz PM, Hammerich L, Martin C, et al. Chemokine receptor CXCR6‐dependent hepatic NK T Cell accumulation promotes inflammation and liver fibrosis. J Immunol. 2013;190(10):5226–36. [DOI] [PubMed] [Google Scholar]
- 56. Bain CC, Bravo‐Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15(10):929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McGrath KE, Frame JM, Palis J. Early hematopoiesis and macrophage development. Semin Immunol. 2015;27(6):379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor‐associated macrophage and monocyte transcriptional landscapes reveal cancer‐specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35(4):588–602.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9(1):4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. La Fleur L, Botling J, He F, Pelicano C, Zhou C, He C, et al. Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res. 2021;81(4):956–67. [DOI] [PubMed] [Google Scholar]
- 61. Zindel J, Peiseler M, Hossain M, Deppermann C, Lee WY, Haenni B, et al. Primordial GATA6 macrophages function as extravascular platelets in sterile injury. Science. 2021;371(6533):eabe0595. [DOI] [PubMed] [Google Scholar]
- 62. Larionova I, Kazakova E, Gerashchenko T, Kzhyshkowska J. New angiogenic regulators produced by TAMs: Perspective for targeting tumor angiogenesis. Cancers (Basel). 2021;13(13):3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lewis CE, De Palma M, Naldini L. Tie2‐expressing monocytes and tumor angiogenesis: regulation by hypoxia and angiopoietin‐2. Cancer Res. 2007;67(18):8429–32. [DOI] [PubMed] [Google Scholar]
- 64. Mahlbacher G, Curtis LT, Lowengrub J, Frieboes HB. Mathematical modeling of tumor‐associated macrophage interactions with the cancer microenvironment. J Immunother Cancer. 2018;6(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Turrini R, Pabois A, Xenarios I, Coukos G, Delaloye JF, Doucey MA. TIE‐2 expressing monocytes in human cancers. Oncoimmunology. 2017;6(4):e1303585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bai ZZ, Li HY, Li CH, Sheng CL, Zhao XN. M1 macrophage‐derived exosomal MicroRNA‐326 suppresses hepatocellular carcinoma cell progression via mediating NF‐κB signaling pathway. Nanoscale Res Lett. 2020;15(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67. Zhao J, Li H, Zhao S, Wang E, Zhu J, Feng D, et al. Epigenetic silencing of miR‐144/451a cluster contributes to HCC progression via paracrine HGF/MIF‐mediated TAM remodeling. Mol Cancer. 2021;20(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang L, Hu YY, Zhao JL, Huang F, Liang SQ, Dong L, et al. Targeted delivery of miR‐99b reprograms tumor‐associated macrophage phenotype leading to tumor regression. J Immunother Cancer. 2020. Sep;8(2):e000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang YL, Li Q, Yang XM, Fang F, Li J, Wang YH, et al. SPON2 promotes M1‐like macrophage recruitment and inhibits hepatocellular carcinoma metastasis by distinct integrin‐Rho GTPase‐hippo pathways. Cancer Res. 2018;78(9):2305–17. [DOI] [PubMed] [Google Scholar]
- 70. Zhou B, Yang Y, Li C. SIRT1 inhibits hepatocellular carcinoma metastasis by promoting M1 macrophage polarization via NF‐κB pathway. Onco Targets Ther. 2019;12:2519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou B, Li C, Yang Y, Wang Z. RIG‐I promotes cell death in hepatocellular carcinoma by inducing M1 Polarization of perineal macrophages through the RIG‐I/MAVS/NF‐κB pathway. Onco Targets Ther. 2020;13:8783–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Q, Cheng F, Ma TT, Xiong HY, Li ZW, Xie CL, et al. Interleukin‐12 inhibits the hepatocellular carcinoma growth by inducing macrophage polarization to the M1‐like phenotype through downregulation of Stat‐3. Mol Cell Biochem. 2016;415(1‐2):157–68. [DOI] [PubMed] [Google Scholar]
- 73. Zong Z, Zou J, Mao R, Ma C, Li N, Wang J, et al. M1 Macrophages induce PD‐L1 expression in hepatocellular carcinoma cells through IL‐1β signaling. Front Immunol. 2019;10:1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gunassekaran GR, Poongkavithai Vadevoo SM, Baek MC, Lee B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL‐4 receptor inhibit tumor growth by reprogramming tumor‐associated macrophages into M1‐like macrophages. Biomaterials. 2021;278:121137. [DOI] [PubMed] [Google Scholar]
- 75. Ma H, Kang Z, Foo TK, Shen Z, Xia B. Disrupted BRCA1‐PALB2 interaction induces tumor immunosuppression and T‐lymphocyte infiltration in HCC through cGAS‐STING pathway. Hepatology. 2022; 10.1002/hep.32335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ou DL, Chen CW, Hsu CL, Chung CH, Feng ZR, Lee BS, et al. Regorafenib enhances antitumor immunity via inhibition of p38 kinase/Creb1/Klf4 axis in tumor‐associated macrophages. J Immunother Cancer. 2021;9(3):e001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lu LG, Zhou ZL, Wang XY, Liu BY, Lu JY, Liu S, et al. PD‐L1 blockade liberates intrinsic antitumourigenic properties of glycolytic macrophages in hepatocellular carcinoma. Gut. 2022. Feb 16:gutjnl‐2021‐326350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Locatelli SL, Careddu G, Serio S, Consonni FM, Maeda A, Viswanadha S, et al. Targeting cancer cells and tumor microenvironment in preclinical and clinical models of hodgkin lymphoma using the dual PI3Kδ/γ inhibitor RP6530. Clin Cancer Res. 2019;25(3):1098–112. [DOI] [PubMed] [Google Scholar]
- 79. Zhai K, Huang Z, Huang Q, Tao W, Fang X, Zhang A, et al. Pharmacological inhibition of BACE1 suppresses glioblastoma growth by stimulating macrophage phagocytosis of tumor cells. Nat Cancer. 2021;2(11):1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Panza F, Lozupone M, Solfrizzi V, Sardone R, Piccininni C, Dibello V, et al. BACE inhibitors in clinical development for the treatment of Alzheimer's disease. Expert Rev Neurother. 2018;18(11):847–57. [DOI] [PubMed] [Google Scholar]
- 81. Wei R, Zhu WW, Yu GY, Wang X, Gao C, Zhou X, et al. S100 calcium‐binding protein A9 from tumor‐associated macrophage enhances cancer stem cell‐like properties of hepatocellular carcinoma. Int J Cancer. 2021;148(5):1233–44. [DOI] [PubMed] [Google Scholar]
- 82. Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, et al. Macrophages‐induced long noncoding RNA H19 up‐regulation triggers and activates the miR‐193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310–22. [DOI] [PubMed] [Google Scholar]
- 83. Chen Y, Wen H, Zhou C, Su Q, Lin Y, Xie Y, et al. TNF‐α derived from M2 tumor‐associated macrophages promotes epithelial‐mesenchymal transition and cancer stemness through the Wnt/β‐catenin pathway in SMMC‐7721 hepatocellular carcinoma cells. Exp Cell Res. 2019;378(1):41–50. [DOI] [PubMed] [Google Scholar]
- 84. Fan QM, Jing YY, Yu GF, Kou XR, Ye F, Gao L, et al. Tumor‐associated macrophages promote cancer stem cell‐like properties via transforming growth factor‐beta1‐induced epithelial‐mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014;352(2):160–8. [DOI] [PubMed] [Google Scholar]
- 85. Wang Y, Wang B, Xiao S, Li Y, Chen Q. miR‐125a/b inhibits tumor‐associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. 2019;120(3):3046–55. [DOI] [PubMed] [Google Scholar]
- 86. Zeng J, Liu Z, Sun S, Xie J, Cao L, Lv P, et al. Tumor‐associated macrophages recruited by periostin in intrahepatic cholangiocarcinoma stem cells. Oncol Lett. 2018;15(6):8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Raggi C, Correnti M, Sica A, Andersen JB, Cardinale V, Alvaro D, et al. Cholangiocarcinoma stem‐like subset shapes tumor‐initiating niche by educating associated macrophages. J Hepatol. 2017;66(1):102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, et al. The miR‐92a‐2‐5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p‐AKT/β‐catenin signaling. Cell Death Differ. 2020;27(12):3258–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato‐carcinoma cells and inhibit proliferation. J Immunol. 2013;191(12):6250–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62(3):607–16. [DOI] [PubMed] [Google Scholar]
- 91. Zhu F, Li X, Chen S, Zeng Q, Zhao Y, Luo F. Tumor‐associated macrophage or chemokine ligand CCL17 positively regulates the tumorigenesis of hepatocellular carcinoma. Med Oncol. 2016;33(2):17. [DOI] [PubMed] [Google Scholar]
- 92. Sun D, Luo T, Dong P, Zhang N, Chen J, Zhang S, et al. M2‐polarized tumor‐associated macrophages promote epithelial‐mesenchymal transition via activation of the AKT3/PRAS40 signaling pathway in intrahepatic cholangiocarcinoma. J Cell Biochem. 2020;121(4):2828–38. [DOI] [PubMed] [Google Scholar]
- 93. Yuan H, Lin Z, Liu Y, Jiang Y, Liu K, Tu M, et al. Intrahepatic cholangiocarcinoma induced M2‐polarized tumor‐associated macrophages facilitate tumor growth and invasiveness. Cancer Cell Int. 2020;20(1):586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yuan D, Huang S, Berger E, Liu L, Gross N, Heinzmann F, et al. Kupffer cell‐derived tnf triggers cholangiocellular tumorigenesis through JNK due to chronic mitochondrial dysfunction and ROS. Cancer Cell. 2017;31(6):771–89.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhou Z, Wang P, Sun R, Li J, Hu Z, Xin H, et al. Tumor‐associated neutrophils and macrophages interaction contributes to intrahepatic cholangiocarcinoma progression by activating STAT3. J Immunother Cancer. 2021;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fujita N, Nishie A, Aishima S, Kubo Y, Asayama Y, Ishigami K, et al. Role of tumor‐associated macrophages in the angiogenesis of well‐differentiated hepatocellular carcinoma: pathological‐radiological correlation. Oncol Rep. 2014;31(6):2499–505. [DOI] [PubMed] [Google Scholar]
- 97. Matsubara T, Kanto T, Kuroda S, Yoshio S, Higashitani K, Kakita N, et al. TIE2‐expressing monocytes as a diagnostic marker for hepatocellular carcinoma correlates with angiogenesis. Hepatology. 2013;57(4):1416–25. [DOI] [PubMed] [Google Scholar]
- 98. Gorrin‐Rivas MJ, Arii S, Mori A, Takeda Y, Mizumoto M, Furutani M, et al. Implications of human macrophage metalloelastase and vascular endothelial growth factor gene expression in angiogenesis of hepatocellular carcinoma. Ann Surg. 2000;231(1):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non‐coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR‐140. Am J Physiol Cell Physiol. 2020;318(3):C649–c63. [DOI] [PubMed] [Google Scholar]
- 100. Hasita H, Komohara Y, Okabe H, Masuda T, Ohnishi K, Lei XF, et al. Significance of alternatively activated macrophages in patients with intrahepatic cholangiocarcinoma. Cancer Sci. 2010;101(8):1913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bartneck M, Schrammen PL, Möckel D, Govaere O, Liepelt A, Krenkel O, et al. The CCR2(+) Macrophage Subset Promotes Pathogenic Angiogenesis for Tumor Vascularization in Fibrotic Livers. Cell Mol Gastroenterol Hepatol. 2019;7(2):371–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zang M, Li Y, He H, Ding H, Chen K, Du J, et al. IL‐23 production of liver inflammatory macrophages to damaged hepatocytes promotes hepatocellular carcinoma development after chronic hepatitis B virus infection. Biochim Biophys Acta Mol Basis Dis. 2018;1864(12):3759–70. [DOI] [PubMed] [Google Scholar]
- 103. Subimerb C, Pinlaor S, Lulitanond V, Khuntikeo N, Okada S, McGrath MS, et al. Circulating CD14(+) CD16(+) monocyte levels predict tissue invasive character of cholangiocarcinoma. Clin Exp Immunol. 2010;161(3):471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wenes M, Shang M, Di Matteo M, Goveia J, Martín‐Pérez R, Serneels J, et al. Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 2016;24(5):701–15. [DOI] [PubMed] [Google Scholar]
- 105. Dalton HJ, Pradeep S, McGuire M, Hailemichael Y, Ma S, Lyons Y, et al. Macrophages facilitate resistance to anti‐VEGF therapy by altered VEGFR expression. Clin Cancer Res. 2017;23(22):7034–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Xu G, Feng D, Yao Y, Li P, Sun H, Yang H, et al. Listeria‐based hepatocellular carcinoma vaccine facilitates anti‐PD‐1 therapy by regulating macrophage polarization. Oncogene. 2020;39(7):1429–44. [DOI] [PubMed] [Google Scholar]
- 107. Fu XT, Song K, Zhou J, Shi YH, Liu WR, Shi GM, et al. Tumor‐associated macrophages modulate resistance to oxaliplatin via inducing autophagy in hepatocellular carcinoma. Cancer Cell Int. 2019;19:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tan HY, Wang N, Man K, Tsao SW, Che CM, Feng Y. Autophagy‐induced RelB/p52 activation mediates tumour‐associated macrophage repolarisation and suppression of hepatocellular carcinoma by natural compound baicalin. Cell Death Dis. 2015;6(10):e1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shiau DJ, Kuo WT, Davuluri GVN, Shieh CC, Tsai PJ, Chen CC, et al. Hepatocellular carcinoma‐derived high mobility group box 1 triggers M2 macrophage polarization via a TLR2/NOX2/autophagy axis. Sci Rep. 2020;10(1):13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yin C, Han Q, Xu D, Zheng B, Zhao X, Zhang J. SALL4‐mediated upregulation of exosomal miR‐146a‐5p drives T‐cell exhaustion by M2 tumor‐associated macrophages in HCC. Oncoimmunology. 2019;8(7):1601479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes‐Mediated Macrophage M2 Polarization. Int J Nanomedicine. 2021;16:2803–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC‐Derived exosomal lncRNA TUC339. Int J Mol Sci. 2018;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liu Y, Zhao L, Li D, Yin Y, Zhang CY, Li J, et al. Microvesicle‐delivery miR‐150 promotes tumorigenesis by up‐regulating VEGF, and the neutralization of miR‐150 attenuate tumor development. Protein Cell. 2013;4(12):932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dwyer BJ, Jarman EJ, Gogoi‐Tiwari J, Ferreira‐Gonzalez S, Boulter L, Guest RV, et al. TWEAK/Fn14 signalling promotes cholangiocarcinoma niche formation and progression. J Hepatol. 2021;74(4):860–72. [DOI] [PubMed] [Google Scholar]
- 115. Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–8. [DOI] [PubMed] [Google Scholar]
- 116. Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30(1):36–50. [DOI] [PubMed] [Google Scholar]
- 117. Wang F, Zhang S, Vuckovic I, Jeon R, Lerman A, Folmes CD, et al. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab. 2018;28(3):463–75.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Huang SC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, et al. Metabolic reprogramming mediated by the mTORC2‐IRF4 signaling axis is essential for macrophage alternative activation. Immunity. 2016;45(4):817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen DP, Ning WR, Jiang ZZ, Peng ZP, Zhu LY, Zhuang SM, et al. Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3‐PD‐L1 axis in human hepatocellular carcinoma. J Hepatol. 2019;71(2):333–43. [DOI] [PubMed] [Google Scholar]
- 120. Jiang Y, Han Q, Zhao H, Zhang J. Promotion of epithelial‐mesenchymal transformation by hepatocellular carcinoma‐educated macrophages through Wnt2b/β‐catenin/c‐Myc signaling and reprogramming glycolysis. J Exp Clin Cancer Res. 2021;40(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tan HY, Wang N, Zhang C, Chan YT, Yuen MF, Feng Y. Lysyl oxidase‐like 4 fosters an immunosuppressive microenvironment during hepatocarcinogenesis. Hepatology. 2021;73(6):2326–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan‐derived microbial metabolites activate the aryl hydrocarbon receptor in tumor‐associated macrophages to suppress anti‐tumor immunity. Immunity. 2022;55(2):324–40.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Thompson SM, Wang J, Chandan VS, Glaser KJ, Roberts LR, Ehman RL, et al. MR elastography of hepatocellular carcinoma: Correlation of tumor stiffness with histopathology features‐Preliminary findings. Magn Reson Imaging. 2017;37:41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bhattacharya A, Agarwal M, Mukherjee R, Sen P, Sinha DK. 3D micro‐environment regulates NF‐κβ dependent adhesion to induce monocyte differentiation. Cell Death Dis. 2018;9(9):914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Xing X, Wang Y, Zhang X, Gao X, Li M, Wu S, et al. Matrix stiffness‐mediated effects on macrophages polarization and their LOXL2 expression. Febs j. 2021;288(11):3465‐77. [DOI] [PubMed] [Google Scholar]
- 126. Zhao X, Wang X, You Y, Wen D, Feng Z, Zhou Y, et al. Nogo‐B fosters HCC progression by enhancing Yap/Taz‐mediated tumor‐associated macrophages M2 polarization. Exp Cell Res. 2020;391(1):111979. [DOI] [PubMed] [Google Scholar]
- 127. Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)‐E complexed with HLA class I signal sequence‐derived peptides by CD94/NKG2 confers protection from natural killer cell‐mediated lysis. J Exp Med. 1998;187(5):813‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nebbia G, Peppa D, Schurich A, Khanna P, Singh HD, Cheng Y, et al. Upregulation of the Tim‐3/galectin‐9 pathway of T cell exhaustion in chronic hepatitis B virus infection. PLoS One. 2012;7(10):e47648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wei Z, Zhang X, Yong T, Bie N, Zhan G, Li X, et al. Boosting anti‐PD‐1 therapy with metformin‐loaded macrophage‐derived microparticles. Nat Commun. 2021;12(1):440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hua GY, Wang P, Takagi K, Shimozato O, Yagita H, Okigaki T, et al. Expression of a soluble form of CTLA4 on macrophage and its biological activity. Cell Res. 1999;9(3):189–99. [DOI] [PubMed] [Google Scholar]
- 131. Liu D, Krummey SM, Badell IR, Wagener M, Schneeweis LA, Stetsko DK, et al. 2B4 (CD244) induced by selective CD28 blockade functionally regulates allograft‐specific CD8+ T cell responses. J Exp Med. 2014;211(2):297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Yu J, Green MD, Li S, Sun Y, Journey SN, Choi JE, et al. Liver metastasis restrains immunotherapy efficacy via macrophage‐mediated T cell elimination. Nat Med. 2021;27(1):152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wu Q, Zhou W, Yin S, Zhou Y, Chen T, Qian J, et al. Blocking triggering receptor expressed on myeloid Cells‐1‐Positive Tumor‐Associated macrophages induced by hypoxia reverses immunosuppression and anti‐programmed Cell death ligand 1 resistance in liver cancer. Hepatology. 2019;70(1):198–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Xiong H, Mittman S, Rodriguez R, Moskalenko M, Pacheco‐Sanchez P, Yang Y, et al. Anti‐PD‐L1 treatment results in functional remodeling of the macrophage compartment. Cancer Res. 2019;79(7):1493–506. [DOI] [PubMed] [Google Scholar]
- 135. Liao J, Zeng DN, Li JZ, Hua QM, Huang CX, Xu J, et al. Type I IFNs repolarized a CD169(+) macrophage population with anti‐tumor potentials in hepatocellular carcinoma. Mol Ther. 2022;30(2):632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)‐adenosine methyltransferase Mettl14 in tumor‐associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39(7):945–57.e10. [DOI] [PubMed] [Google Scholar]
- 137. Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, et al. Treg cells promote the SREBP1‐dependent metabolic fitness of tumor‐promoting macrophages via repression of CD8(+) T cell‐derived interferon‐γ. Immunity. 2019;51(2):381–97.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF‐β‐miR‐34a‐CCL22 signaling‐induced Treg cell recruitment promotes venous metastases of HBV‐positive hepatocellular carcinoma. Cancer Cell. 2012;22(3):291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wei Y, Lao XM, Xiao X, Wang XY, Wu ZJ, Zeng QH, et al. Plasma cell polarization to the immunoglobulin G phenotype in hepatocellular carcinomas involves epigenetic alterations and promotes hepatoma progression in mice. Gastroenterology. 2019;156(6):1890–904.e16. [DOI] [PubMed] [Google Scholar]
- 140. Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor‐associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6(12):1578–92. [DOI] [PubMed] [Google Scholar]
- 141. Li L, Sun P, Zhang C, Li Z, Zhou W. MiR‐98 suppresses the effects of tumor‐associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL‐10. Biochimie. 2018;150:23–30. [DOI] [PubMed] [Google Scholar]
- 142. Kuipers H, Muskens F, Willart M, Hijdra D, van Assema FB, Coyle AJ, et al. Contribution of the PD‐1 ligands/PD‐1 signaling pathway to dendritic cell‐mediated CD4+ T cell activation. Eur J Immunol. 2006;36(9):2472–82. [DOI] [PubMed] [Google Scholar]
- 143. Petty AJ, Li A, Wang X, Dai R, Heyman B, Hsu D, et al. Hedgehog signaling promotes tumor‐associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. J Clin Invest. 2019;129(12):5151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bloemendaal FM, Koelink PJ, van Schie KA, Rispens T, Peters CP, Buskens CJ, et al. TNF‐anti‐TNF immune complexes inhibit IL‐12/IL‐23 secretion by inflammatory macrophages via an Fc‐dependent mechanism. J Crohns Colitis. 2018;12(9):1122–30. [DOI] [PubMed] [Google Scholar]
- 145. Loeuillard E, Yang J, Buckarma E, Wang J, Liu Y, Conboy C, et al. Targeting tumor‐associated macrophages and granulocytic myeloid‐derived suppressor cells augments PD‐1 blockade in cholangiocarcinoma. J Clin Invest. 2020;130(10):5380–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Tavazoie MF, Pollack I, Tanqueco R, Ostendorf BN, Reis BS, Gonsalves FC, et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172(4):825–40.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xia Y, Rao L, Yao H, Wang Z, Ning P, Chen X. Engineering macrophages for cancer immunotherapy and drug delivery. Adv Mater. 2020;32(40):e2002054. [DOI] [PubMed] [Google Scholar]
- 148. Quail DF, Joyce JA. Molecular pathways: deciphering mechanisms of resistance to macrophage‐targeted therapies. Clin Cancer Res. 2017;23(4):876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ao JY, Zhu XD, Chai ZT, Cai H, Zhang YY, Zhang KZ, et al. Colony‐stimulating factor 1 receptor blockade inhibits tumor growth by altering the Polarization of tumor‐associated macrophages in hepatocellular carcinoma. Mol Cancer Ther. 2017;16(8):1544–54. [DOI] [PubMed] [Google Scholar]
- 150. Stanley ER, Chitu V. CSF‐1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol. 2014;6(6):a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Boulter L, Guest RV, Kendall TJ, Wilson DH, Wojtacha D, Robson AJ, et al. WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J Clin Invest. 2015;125(3):1269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ambade A, Lowe P, Kodys K, Catalano D, Gyongyosi B, Cho Y, et al. Pharmacological inhibition of CCR2/5 signaling prevents and reverses alcohol‐induced liver damage, steatosis, and inflammation in mice. Hepatology. 2019;69(3):1105–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour‐infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66(1):157‐67. [DOI] [PubMed] [Google Scholar]
- 154. Yao W, Ba Q, Li X, Li H, Zhang S, Yuan Y, et al. A natural CCR2 antagonist relieves tumor‐associated macrophage‐mediated immunosuppression to produce a therapeutic effect for liver cancer. EBioMedicine. 2017;22:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Hashimoto A, Sarker D, Reebye V, Jarvis S, Sodergren MH, Kossenkov A, et al. Upregulation of C/EBPα inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin Cancer Res. 2021;27(21):5961–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yang T, Deng Z, Xu L, Li X, Yang T, Qian Y, et al. Macrophages‐aPKC(ɩ)‐CCL5 Feedback Loop Modulates the Progression and Chemoresistance in Cholangiocarcinoma. J Exp Clin Cancer Res. 2022;41(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mao J, Wang D, Wang Z, Tian W, Li X, Duan J, et al. Combretastatin A‐1 phosphate, a microtubule inhibitor, acts on both hepatocellular carcinoma cells and tumor‐associated macrophages by inhibiting the Wnt/β‐catenin pathway. Cancer Lett. 2016;380(1):134–43. [DOI] [PubMed] [Google Scholar]
- 158. Zhang W, Zhu XD, Sun HC, Xiong YQ, Zhuang PY, Xu HX, et al. Depletion of tumor‐associated macrophages enhances the effect of sorafenib in metastatic liver cancer models by antimetastatic and antiangiogenic effects. Clin Cancer Res. 2010;16(13):3420–30. [DOI] [PubMed] [Google Scholar]
- 159. Kato Y, Tabata K, Kimura T, Yachie‐Kinoshita A, Ozawa Y, Yamada K, et al. Lenvatinib plus anti‐PD‐1 antibody combination treatment activates CD8+ T cells through reduction of tumor‐associated macrophage and activation of the interferon pathway. PLoS One. 2019;14(2):e0212513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Fernández‐Varo G, Perramón M, Carvajal S, Oró D, Casals E, Boix L, et al. Bespoken nanoceria: An effective treatment in experimental hepatocellular carcinoma. Hepatology. 2020;72(4):1267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single‐cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–308.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Nalio Ramos R, Missolo‐Koussou Y, Gerber‐Ferder Y, Bromley CP, Bugatti M, Núñez NG, et al. Tissue‐resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell. 2022;185(7):1189–207.e25. [DOI] [PubMed] [Google Scholar]
- 163. Wei CY, Zhu MX, Zhang PF, Huang XY, Wan JK, Yao XZ, et al. PKCα/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti‐PD1 resistance in hepatocellular carcinoma. J Hepatol. 2022. Jul;77(1):163–76. [DOI] [PubMed] [Google Scholar]
- 164. Hage C, Hoves S, Strauss L, Bissinger S, Prinz Y, Pöschinger T, et al. Sorafenib induces pyroptosis in macrophages and triggers natural killer cell‐mediated cytotoxicity against hepatocellular carcinoma. Hepatology. 2019;70(4):1280–97. [DOI] [PubMed] [Google Scholar]
- 165. Sprinzl MF, Puschnik A, Schlitter AM, Schad A, Ackermann K, Esposito I, et al. Sorafenib inhibits macrophage‐induced growth of hepatoma cells by interference with insulin‐like growth factor‐1 secretion. J Hepatol. 2015;62(4):863–70. [DOI] [PubMed] [Google Scholar]
- 166. Sprinzl MF, Reisinger F, Puschnik A, Ringelhan M, Ackermann K, Hartmann D, et al. Sorafenib perpetuates cellular anticancer effector functions by modulating the crosstalk between macrophages and natural killer cells. Hepatology. 2013;57(6):2358–68. [DOI] [PubMed] [Google Scholar]
- 167. Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumour‐associated macrophage trafficking by the osteopontin‐induced colony‐stimulating factor‐1 signalling sensitises hepatocellular carcinoma to anti‐PD‐L1 blockade. Gut. 2019;68(9):1653–66. [DOI] [PubMed] [Google Scholar]
- 168. Li L, Sun P, Zhang C, Li Z, Cui K, Zhou W. MiR‐98 modulates macrophage polarization and suppresses the effects of tumor‐associated macrophages on promoting invasion and epithelial‐mesenchymal transition of hepatocellular carcinoma. Cancer Cell Int. 2018;18:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lu S, Gao Y, Huang X, Wang X. Cantharidin exerts anti‐hepatocellular carcinoma by miR‐214 modulating macrophage polarization. Int J Biol Sci. 2014;10(4):415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Li G, Zhao L. Sorafenib‐loaded hydroxyethyl starch‐TG100‐115 micelles for the treatment of liver cancer based on synergistic treatment. Drug Deliv. 2019;26(1):756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Diggs LP, Ruf B, Ma C, Heinrich B, Cui L, Zhang Q, et al. CD40‐mediated immune cell activation enhances response to anti‐PD‐1 in murine intrahepatic cholangiocarcinoma. J Hepatol. 2021;74(5):1145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Tsai TY, Huang MT, Sung PS, Peng CY, Tao MH, Yang HI, et al. SIGLEC‐3 (CD33) serves as an immune checkpoint receptor for HBV infection. J Clin Invest. 2021;131(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Bao D, Zhao J, Zhou X, Yang Q, Chen Y, Zhu J, et al. Mitochondrial fission‐induced mtDNA stress promotes tumor‐associated macrophage infiltration and HCC progression. Oncogene. 2019;38(25):5007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Tanaka S, Sugimachi K, Yamashita Yi Y, Ohga T, Shirabe K, Shimada M, et al. Tie2 vascular endothelial receptor expression and function in hepatocellular carcinoma. Hepatology. 2002;35(4):861–7. [DOI] [PubMed] [Google Scholar]
- 175. Park JS, Kim IK, Han S, Park I, Kim C, Bae J, et al. Normalization of tumor vessels by Tie2 activation and Ang2 inhibition enhances drug delivery and produces a favorable tumor microenvironment. Cancer Cell. 2016;30(6):953–67. [DOI] [PubMed] [Google Scholar]
- 176. Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet. 2017;389(10064):56–66. [DOI] [PubMed] [Google Scholar]
- 177. Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38(8):947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Mukhopadhyay M. Macrophages enter CAR immunotherapy. Nat Methods. 2020;17(6):561. [DOI] [PubMed] [Google Scholar]
- 179. 30th annual meeting and associated programs of the society for immunotherapy of cancer (SITC 2015). J Immunother Cancer. 2015;3(Suppl 2):O1‐p453. [PubMed] [Google Scholar]
- 180. Wang S, Yang Y, Ma P, Zha Y, Zhang J, Lei A, et al. CAR‐macrophage: An extensive immune enhancer to fight cancer. EBioMedicine. 2022;76:103873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kang M, Lee SH, Kwon M, Byun J, Kim D, Kim C, et al. Nanocomplex‐mediated in vivo programming to chimeric antigen receptor‐M1 macrophages for cancer therapy. Adv Mater. 2021;33(43):e2103258. [DOI] [PubMed] [Google Scholar]
- 182. Zhang W, Liu L, Su H, Liu Q, Shen J, Dai H, et al. Chimeric antigen receptor macrophage therapy for breast tumours mediated by targeting the tumour extracellular matrix. Br J Cancer. 2019;121(10):837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138(2):271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Chen J, Zheng DX, Yu XJ, Sun HW, Xu YT, Zhang YJ, et al. Macrophages induce CD47 upregulation via IL‐6 and correlate with poor survival in hepatocellular carcinoma patients. Oncoimmunology. 2019;8(11):e1652540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yang HD, Kim HS, Kim SY, Na MJ, Yang G, Eun JW, et al. HDAC6 suppresses Let‐7i‐5p to elicit TSP1/CD47‐mediated anti‐tumorigenesis and phagocytosis of hepatocellular carcinoma. Hepatology. 2019;70(4):1262–79. [DOI] [PubMed] [Google Scholar]
- 186. Vaeteewoottacharn K, Kariya R, Pothipan P, Fujikawa S, Pairojkul C, Waraasawapati S, et al. Attenuation of CD47‐SIRPα signal in cholangiocarcinoma potentiates tumor‐associated macrophage‐mediated phagocytosis and suppresses intrahepatic metastasis. Transl Oncol. 2019;12(2):217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lo J, Lau EY, So FT, Lu P, Chan VS, Cheung VC, et al. Anti‐CD47 antibody suppresses tumour growth and augments the effect of chemotherapy treatment in hepatocellular carcinoma. Liver Int. 2016;36(5):737–45. [DOI] [PubMed] [Google Scholar]
- 188. Xiao Z, Chung H, Banan B, Manning PT, Ott KC, Lin S, et al. Antibody mediated therapy targeting CD47 inhibits tumor progression of hepatocellular carcinoma. Cancer Lett. 2015;360(2):302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Du K, Li Y, Liu J, Chen W, Wei Z, Luo Y, et al. A bispecific antibody targeting GPC3 and CD47 induced enhanced antitumor efficacy against dual antigen‐expressing HCC. Mol Ther. 2021;29(4):1572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Michaels AD, Newhook TE, Adair SJ, Morioka S, Goudreau BJ, Nagdas S, et al. CD47 blockade as an adjuvant immunotherapy for resectable pancreatic cancer. Clin Cancer Res. 2018;24(6):1415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


