SUMMARY
Kinetochore assembly on centromeres is central for chromosome segregation and defects in this process cause mitotic errors and aneuploidy. Besides the well-established protein network, emerging evidence suggests the involvement of regulatory RNA in kinetochore assembly; however, it has remained elusive about the identity of such RNA, let alone its mechanism of action in this critical process. Here, we report CCTT, a previously uncharacterized long non-coding RNA (lncRNA) transcribed from the arm of human chromosome 17, that plays a vital role in kinetochore assembly. We show that CCTT highly localizes to all centromeres via the formation of RNA-DNA triplex and specifically interacts with CENP-C to help engage this blueprint protein in centromeres, and consequently, CCTT loss triggers extensive mitotic errors and aneuploidy. These findings uncover a non-centromere-derived lncRNA that recruits CENP-C to centromeres and shed critical lights on the function of centromeric DNA sequences as anchor points for kinetochore assembly.
Keywords: centromere, lncRNA, CENP-C recruitment, mitotic defects, aneuploidy
eTOC blurb
Zhang et al. identify a non-centromere-derived long non-coding RNA, CCTT, that plays a vital role in kinetochore assembly at centromeres in trans. The authors show that CCTT localizes to all centromeres via the formation of RNA-DNA triplex and specifically interacts with CENP-C for its recruitment to centromeres.
INTRODUCTION
Accurate chromosome segregation during cell division depends on highly delicate centromere-kinetochore interactions. Centromeres of most eukaryotes consist of a large stretch of tandemly repeated DNA elements (Fukagawa and Earnshaw, 2014; McKinley and Cheeseman, 2016). Centromere function seems to be defined by the incorporation of a unique histone H3 variant CENP-A in centromeric nucleosomes (Black and Cleveland, 2011; McKinley and Cheeseman, 2016; Logsdon et al., 2019). CENP-A initiates the assembly of the inner kinetochore network, known as the constitutive centromere-associated network (CCAN) (Weir et al., 2016; Yan et al., 2019), which then connects to the outer kinetochore network consisting of KNL1, Mis12 and Ndc80 (KMN) for microtubule attachment (Varma and Salmon, 2012).
Among CCAN, CENP-C is the first one to bind to centromeres and then provides a blueprint for kinetochore assembly by facilitating the recruitment of other CCAN components (Nagpal et al., 2015; McKinley et al., 2015; Klare et al., 2015; Weir et al., 2016). It is well-established that CENP-A is responsible for CENP-C recruitment through its carboxy-terminal tail that directly binds to two specific domains in the CENP-C protein, the central domain (CD) and the c-motif domain (CM) (Carroll et al., 2010; Guse et al., 2011; Fachinetti et al., 2013; Kato et al., 2013). Puzzling, however, are the more recent observations that neither deletion of CENP-A (Fachinetti et al., 2013) nor blocking the interaction between CENP-A and CENP-C (Fachinetti et al., 2013; Guo et al., 2017) completely diminished CENP-C recruitment to centromeres. These observations suggest the existence of an additional unknown mechanism(s) for CENP-C recruitment.
Recent evidence suggests that both centromeric transcription and resulting RNAs (or cenRNAs) play a critical role in kinetochore assembly (Fukagawa and Earnshaw, 2014; Talbert and Henikoff, 2018; Sullivan and Sullivan, 2020). The transcription process may facilitate nucleosome remodeling to help CENP-A incorporation (Quénet and Dalal, 2014), whereas cenRNAs may mediate the localization of several centromere-associated proteins (Ferri et al., 2009; Quénet and Dalal, 2014; Blower, 2016; McNulty et al., 2017; Ling and Yuen, 2019). However, no sequence specificity appears to be required for cenRNAs to bind centromere proteins tested to date (Du et al., 2010; Jambhekar et al., 2014; Liu et al., 2015; Blower, 2016). Importantly, the functional requirement for cenRNAs remains enigmatic due to the misinterpretation of data on targeting a putative cenRNA in human cells (Quénet and Dalal, 2018). Therefore, compared to the well-established protein-protein interaction networks, both the involvement of the transcripts produced from non-centromere regions and their mechanism of action in kinetochore assembly remain unexplored.
We herein report a previously uncharacterized non-centromere-derived lncRNA transcribed from chromosome 17 (Chr. 17), which provides a key missing link in CENP-C recruitment to centromeres. This lncRNA, now renamed as CCTT, interacts with both CENP-C through specific RNA-protein interactions and cenDNA possibly via RNA-DNA triplex formation. Importantly, loss of CCTT induces chromosomal instability and aneuploidy by regulating CENP-C level at centromeres, suggesting that CCTT is required for faithful chromosome segregation during mitosis. These findings unveil a regulatory lncRNA with a key role in kinetochore assembly and chromosomal stability in human cells.
RESULTS
Binding of a non-centromere-derived lncRNA CCTT to CENP-C
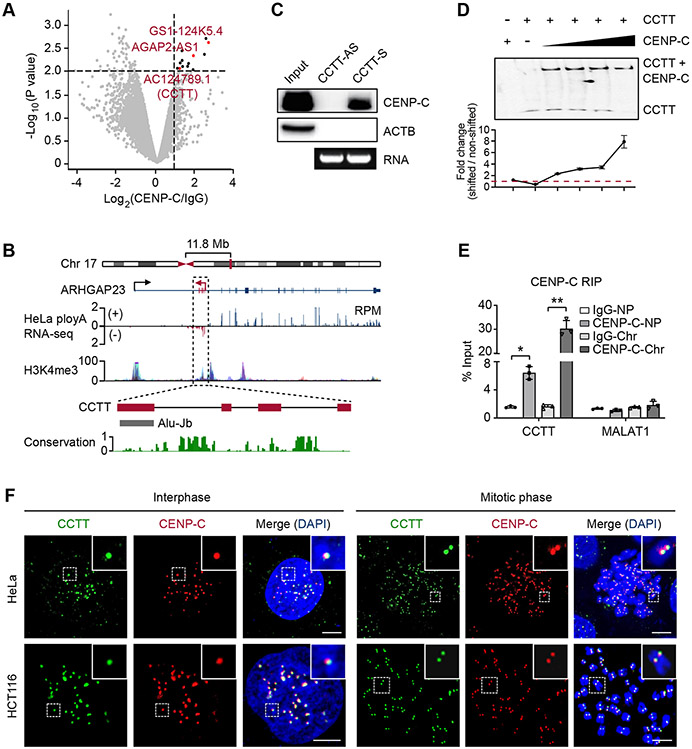
Given the central role of CENP-C in providing a blueprint for kinetochore assembly, it has been a focal point in understanding how CENP-C is recruited to centromeres. A previous study suggests the involvement of certain regulatory RNAs in CENP-C recruitment, because RNase treatment diminished the localization of CENP-C at centromeres and CENP-C appeared to have the capacity to bind RNA with high affinity (Wong et al., 2007). Although cenRNAs might fulfill such a role, their specificity and functional requirement have been controversial (Wong et al., 2007; Quénet and Dalal, 2014; Rošić et al., 2014; McNulty et al., 2017). We therefore sought an unbiased approach to enrich CENP-C-interacting RNAs by performing RNA immunoprecipitation sequencing (RIP-seq) with whole cell lysate of HeLa cells. This led to the identification of a small set of RNAs significantly enriched with anti-CENP-C antibody relative to control IgG (fold-change > 2, p < 0.01) (Figure 1A). Besides a few mRNAs identified, we noted three annotated lncRNAs (AGAP2-AS1, GS1-124K5.4, and AC124789.1), which we further validated by RIP-qPCR (Figure S1A). We reasoned that a functional CENP-C-interacting RNA would be present with sufficient quantity in the nucleus. By quantifying their relative levels in the cytoplasm versus the nucleus, we found that only AC124789.1 was selectively enriched in the nucleus, like the well-characterized nuclear lncRNA NEAT1 (Figure S1B). Based on its specific binding to CENP-C and additional functional properties (see below), we renamed this previously uncharacterized RNA as CENP-C targeting transcript (CCTT).
Figure 1. CENP-C Interacts with A New LncRNA CCTT.
(A) Volcano plot of CENP-C-binding transcripts in HeLa cells. Transcripts with over 2-fold enrichment relative to IgG (p < 0.01) are highlighted as red (lncRNAs) or black dots (mRNAs).
(B) Diagram of the CCTT locus. CCTT consists of 4 exons, with the last one harboring an Alu-Jb element. The coverage of ENCODE polyA RNA-seq and H3K4me3 ChIP-seq signals within the CCTT locus is prevalent in multiple human cell lines. The conservation score in 30 mammals is calculated by PhastCons.
(C) RNA pulldown with biotinylated sense or antisense CCTT transcripts from HeLa whole cell lysate followed by western blot analysis of CENP-C. The agarose gel shows the corresponding RNA of sense or antisense CCTT.
(D) CCTT interacted with CENP-C. Top: EMSA imaging of the biotinylated CCTT binding to increasing concentrations (2.5, 3.5, 4.5, 6 μM) of purified recombinant full-length CENP-C proteins. Gels were visualized and quantified by ChemiImager analysis. Bottom: quantification of the EMSA results. For each condition, the fraction of shifted RNA in each lane versus the amount of non-shifted RNA was plotted. Results are presented as the mean and SD of triplicate determinations. The dashed line indicates the shifted signal is same with the non-shifted signal.
(E) CENP-C RIP followed by qPCR analysis of CCTT in nucleoplasm (NP) and chromatin (Chr) fractions of HeLa cells. MALAT1 was used as a negative control. *p < 0.05; **p < 0.01 (mean ± SD, n = 3 per group).
(F) The co-localization of CCTT and CENP-C at centromeres. CCTT FISH (green) and CENP-C IF (red) analysis of HeLa (top) and HCT116 (bottom) cells in interphase and mitotic phases. Scale bars, 5 μm.
See also Figures S1-S2.
CCTT contains four exons with the last one harboring an Alu element and is an anti-sense transcript encoded in the first intron of the ARHGAP23 protein-coding gene in Chr. 17 (Figure 1B). Interestingly, this RNA is conserved only in primates, beginning to diverge from more basal primates (i.e., tarsiers and prosimians) where their centromere sequences also become diversified (Figure S1C; also see Melters et al., 2013). These observations suggest that this RNA may have been co-evolved with primate cenDNA. We performed 5’ and 3’ rapid amplification of cDNA ends (RACE) and confirmed the authentic CCTT sequence of 730 nt in HeLa cells (Figure S1D). Primary CCTT transcript appears to have been efficiently spliced as indicated by a single band of ~730 nt in northern blot performed in both HeLa and HCT116 cells (Figure S1E). CCTT is quite abundant in HeLa cells, estimated to have ~2,000 copies per cell (Figure S1F), which is close to or exceeding the amount of many highly abundant lncRNAs (Figure S1G). In addition, three lines of evidence support the annotation of CCTT as a lncRNA: 1) little association with ribosomes based on analyzing the existing ribosome profiling data (Wang et al., 2014) (Figure S1H), 2) bioinformatics analysis using four different algorithms – CNCI (Sun et al., 2013), CPAT (Wang et al., 2013), CPC (Kong et al., 2007), and PhyloCSF (Lin et al., 2011) – to show the low protein-coding potential (Figure S1I), and 3) the inability to translate into putative micropeptides from two short open reading frames (ORFs) in CCTT (Figure S1J). Collectively, these observations indicate that CCTT is a lncRNA generated from a non-centromeric locus.
Quantitative localization of CCTT on centromeres
We next characterized the interaction of CCTT with CENP-C in vitro and within cells. By using biotinylated CCTT (schemed in Figure S1D), we performed RNA pulldown followed by western blot to show that CCTT, but not its anti-sense version, efficiently captured CENP-C from HeLa whole cell extracts (Figure 1C). Moreover, RNA pulldown coupled with mass spectrometry (MS) identified 20 CCTT-binding protein candidates, and notably, CENP-C was the only component of currently annotated centromere-associated proteins (Figure S1K). We further used this biotinylated CCTT and full-length CENP-C expressed in bacteria (Figure S2A) to perform the electrophoretic mobility shift assay (EMSA). Results showed progressive shifts with increasing amounts of recombinant CENP-C (Figure 1D). In contrast, anti-sense CCTT failed to bind to CENP-C (Figure S2B). Cell fractionation followed by RIP-qPCR showed that CCTT was largely associated with chromatin (Chr), relative to the nucleoplasm (NP) (Figure 1E).
To determine the spatial relationship between CCTT and CENP-C within cells, we performed CCTT fluorescence in situ hybridization (FISH) in comparison with CENP-C localization by immunostaining and found that CCTT precisely colocalized with CENP-C in interphase HeLa and HCT116 cells (Figures 1F, left and S2C). By preparing chromosome spreads from mitotically arrested cells, we found CCTT signals on centromeres of all chromosomes, precisely colocalized with CENP-C (Figures 1F, right and S2C). Taken together, these data demonstrate that this non-centromere-produced lncRNA specifically interacts with CENP-C at all centromeres throughout the cell cycle.
Requirement of CCTT for CENP-C assembly on centromeres
Given the specific interaction between CCTT and CENP-C, we next investigated the function of CCTT in CENP-C assembly on centromeres. We used CRISPR/Cas9-mediated genome editing to generate conditional CCTT knockout HeLa cells (note that HeLa cells carry 3 copies of CCTT, Figure S2D). For this purpose, we first generated a heterozygous CCTT+/− cell line (Figures S2E and S2F), which slightly reduced CCTT expression (Figure S2F), and then inserted two LoxP sites between exon 1 of CCTT (CCTTfloxed/−) followed by infection with adenovirus-Cre (Ad-Cre) to convert CCTTfloxed/− to CCTT−/− cells. We verified CCTT ablation in CCTT−/− cells by RT-PCR, RNA FISH, and northern blot (Figures S2F right bottom, S2G-S2H). Importantly, the mRNA level of ARHGAP23, the parental gene of CCTT, was unchanged in CCTT−/− cells (Figure S2I), thus excluding any contribution of ARHGAP23 to CCTT function.
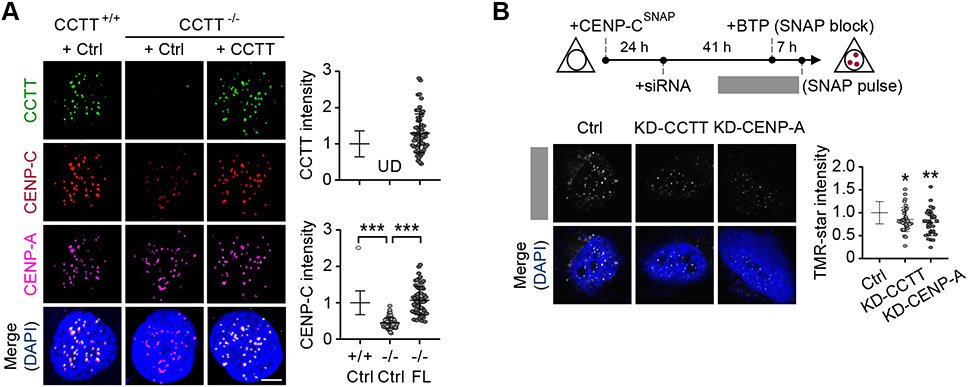
CCTT deletion resulted in ~40% reduction of CENP-C signals on centromeres without affecting CENP-C expression at the RNA level and this reduction could be rescued by re-expressing exogenous CCTT in CCTT−/− cells (Figures 2A, S3A, and S3B). Interestingly, we found only a slight increase of CCTT levels in centromeres, despite ~180-fold increase in CCTT expression relative to the endogenous level (Figures 2A and S3A). This implies that the available CCTT-binding sites in cenDNA have been saturated, thus preventing further incorporation of overexpressed CCTT. To further validate the contribution of CCTT to CENP-C assembly on centromeres, we performed CENP-C ChIP-qPCR. We used siRNA to knockdown CENP-A as a positive control, and to keep CCTT depletion under the same experimental conditions, we similarly used siRNA to knockdown CCTT (Figure S3C). We observed that transient depletion of both CCTT and CENP-A significantly decreased CENP-C on centromeres (Figure S3D).
Figure 2. CCTT Recruits CENP-C to Centromeres.
(A) Re-expression of CCTT rescued CENP-C abundance at centromeres in CCTT−/− cells. Representative images of CCTT FISH (green), CENP-C (red) and CENP-A (pink) IF analyses of HeLa cells (left) and the quantifications of CCTT (top) or CENP-C (bottom) signals by IMARIS (right) are shown. n = 61 for CCTT+/+ cells, n = 72 for CCTT−/− cells, and n = 61 for CCTT−/− cells transfected with full-length (FL) CCTT. ***p < 0.001; UD, undetected (mean ± SD of three biological replicates). Scale bar, 5 μm. Each point represents an averaged fluorescence of all centromeres within a cell.
(B) CCTT knockdown (KD-CCTT) decreased the deposition of new CENP-C at centromeres. Top: The strategy to determine the role of CCTT in CENP-C recruitment. Bottom: TMR-star staining (white) represents the newly deposited CENP-C during 7-hours chasing period. The TMR-star signals were quantified by IMARIS. n = 34 for Ctrl cells, n = 34 for KD-CCTT cells, n = 32 for KD-CENP-A cells. The cells collected from one batch experiment. *p < 0.05; **p < 0.01 (mean ± SD of three biological replicates). Scale bar, 5 μm. Each point represents an averaged TMR-star signals of all centromeres within a cell.
See also Figures S2-S3.
Prior to our current work, it is well established that CENP-A and CENP-B are both involved in CENP-C abundance at centromeres (Carroll et al., 2010; Guse et al., 2011; Fachinetti et al., 2013; Kato et al., 2013; Fachinetti et al., 2015), raising the possibility that CCTT depletion-induced reduction of CENP-C might result from reduced CENP-A. To test this possibility, we depleted CCTT by CCTT ASO, which takes advantage of the relatively long-lasting knockdown effect of ASO in targeting nuclear RNA compared to siRNA. This enabled us to monitor both short and long-term impact of CCTT depletion on CENP-A levels on centromeres. Significant CCTT reduction was observed after CCTT ASOs treatment (ASO-CCTT #1 or #2, see Figures S1D and S3E). Although no significant decrease of CENP-A on centromeres was found at 48 hours after CCTT ASO treatment, while at 96 hours a ~65% reduction of CENP-A was observed (Figure S3F), which is consistent with the reported function of CENP-C in stabilizing CENP-A nucleosomes at centromeres (Falk et al., 2015). We further ruled out the potential dosage effect of CCTT depletion in short versus long-term ASO treatments by mimicking short-term CCTT depletion with Ad-Cre to treat CCTTfloxed/− cells for 48 hours and similarly observed CENP-C reduction without significant change in CENP-A on centromeres (Figure S3G). RIP-qPCR further showed that CENP-A does not bind CCTT (Figure S3H). Together, these data demonstrate that the reduction of CENP-A is a downstream event following CENP-C decrease in response to CCTT depletion, suggesting that CCTT directly modulates CENP-C assembly on centromeres in a CENP-A independent manner.
Critical role of CCTT in CENP-C recruitment
Because CCTT depletion reduced CENP-C at centromeres and given the established dynamics of CENP-C during the G1 and G2 phases (Hemmerich et al., 2008), we hypothesized that CCTT may play a direct role in facilitating the recruitment of newly synthesized CENP-C to centromeres. To test this hypothesis, we employed a SNAP-based labeling strategy (Jansen et al., 2007; Bodor et al., 2012). We first expressed a SNAP-tagged CENP-C (CENP-CSNAP), blocked the existing pool of CENP-CSNAP by pre-incubation with a nonfluorescent substrate (bromothenylpteridine, BTP), and labeled newly synthesized CENP-CSNAP with a fluorescent substrate (tetramethylrhodamine; TMR) of SNAP (Figure 2B, top). We first confirmed no TMR-star signal immediately after adding BTP (Figures S3I and S3J). By comparing the TMR-star signals in control and CCTT knockdown cells, we found the deposition of new CENP-C at centromeres was significantly compromised (Figure 2B). CENP-A depletion served as a positive control (Figures 2B). These data revealed CCTT-dependent recruitment of CENP-C to centromeres.
Genomic mapping of CCTT binding to centromeres
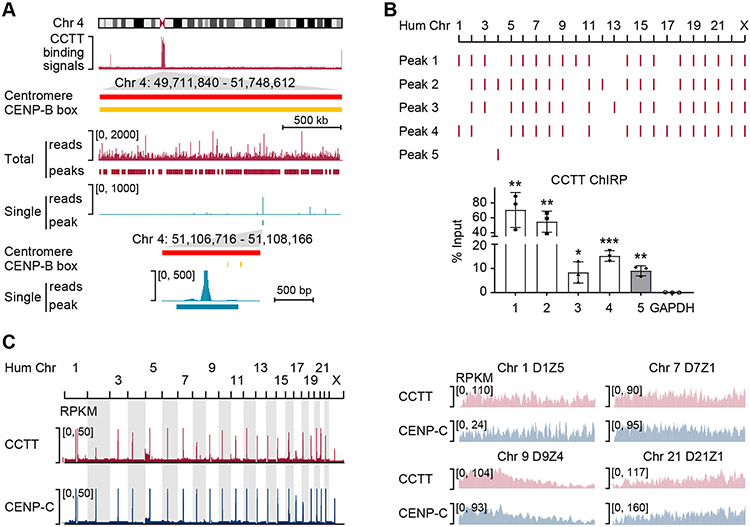
As we detected no interaction between CCTT and CENP-A (see Figure S3H), this raised a possibility that CCTT is directly associated with centromeric α-satellite DNA. To test this possibility and to investigate the CCTT binding profile at the genome-wide scale, we developed a new method by modifying chromatin isolation by RNA purification followed by deep-sequencing (ChIRP-seq) (Chu et al., 2011) via treating cells with 4’-aminomethyltrioxsalen (AMT, a psoralen derivative), a crosslinker that helps stabilize the interactions between nucleic acids (AMT-ChIRP-seq; Figure S4A). This method could explore the interactions between RNA and DNA with minimal impact on the interactions bridged by proteins (Shen et al., 1977; Mondal et al., 2015; Wang et al., 2020). We designed eight complementary DNA oligonucleotides to tile the non-Alu part of CCTT, affinity-purified CCTT-bound DNA, and mapped enriched DNA sequences onto the reference human genome (hg38; note that this version includes centromere models, which provide the approximate repeat number and order for each centromere (Miga et al., 2014)). We called peaks with the SICER algorithm by using input reads as background and setting stringent parameters previously optimized for analyzing the human CENP-A ChIP-seq data (Zang et al., 2009; Lacoste et al., 2014).
Among 26,044 CCTT-binding peaks, 10,895 (41.83%) mapped onto the centromere regions, corresponding to predominant peaks in all 23 chromosomes (Figure S4B). This is further exemplified on Chr. 4 (Figure 3A), showing that the peak density (peak number per Mb) within the centromere region is 36.6-fold higher than that occupying elsewhere on the chromosome (183 versus 5 peaks per Mb). Most peaks contained multi-mapped reads that were evenly distributed among individual genomic locations. A small fraction of peaks (n = 3,023) that contained uniquely mapped reads were also found in specific centromere regions (see a specific example in Figure 3A).
Figure 3. CCTT Binds to CenDNA.
(A) CCTT AMT-ChIRP-seq signals as well as SICER peaks across the centromere of Chr. 4. The middle two tracks separately show all mapped reads and deduced peaks (red) as well as single-mapped reads and deduced peaks (blue). The bottom panel further highlights the peak based on single-mapped reads in reference to annotated CENP-B boxes (yellow).
(B) AMT-ChIRP-qPCR validation of representative peaks deduced from genome-wide CCTT binding profile by AMT-ChIRP-seq. Four representative peaks (1-4) were based on total reads which were detectable in multiple centromeres, and the last peak (5) was deduced based on single reads uniquely localized in Chr. 4. The AMT-ChIRP-qPCR results are shown at the bottom. GAPDH served as a negative control. *p < 0.05; **p < 0.01; ***p < 0.001 (mean ± SD, n = 3 per group).
(C) Comparison between genome-wide binding profiles of CCTT and CENP-C. Mapped reads of CCTT AMT-ChIRP-seq and CENP-C ChIP-seq across all human chromosomes (left) and at different HORs within 4 representative centromeres (right) of HeLa cells. RPKM: reads per kb per million mapped reads.
See also Figure S4.
To validate the AMT-ChIRP-seq results, we performed AMT-ChIRP-qPCR on HeLa cells. We randomly selected 5 peaks: peaks 1-4 containing multi-mapped reads and peak 5 in Chr. 4 containing only uniquely mapped reads (Figure 3B, top). We then designed PCR primers flanking each of these peaks for AMT-ChIRP-qPCR and found an 8~70-fold enrichment relative to input (Figure 3B). Finally, using previously reported CENP-C ChIP-seq datasets from HeLa cells, we found a significant overlap between CCTT and CENP-C binding sites across the entire genome (Figure 3C, left and Figure S4C; Pearson correlation = 0.75), as exemplified by higher order repeats (HORs) on 4 representative centromeres (Figure 3C, right) as well as on the whole centromere region of Chr. 19 (Figure S4D). Collectively, these results demonstrate that CCTT specifically interacts with cenDNA at all centromeres.
Evidence for CCTT binding to cenDNA via the formation of RNA-DNA triplex
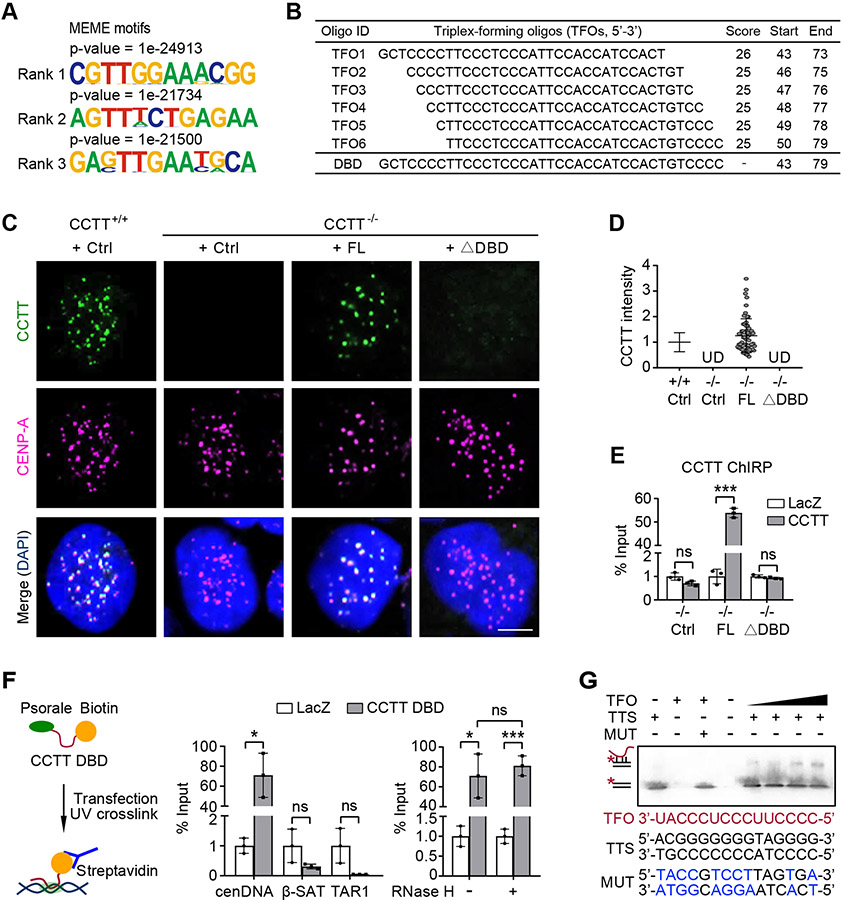
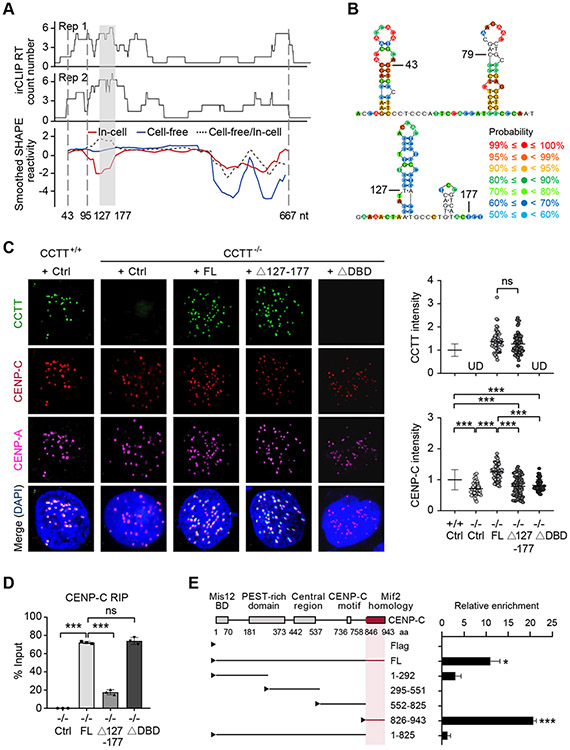
CCTT may interact with DNA either directly or via an accessory factor. Given the fact that CCTT only interacts with CENP-C among centromere-enriched proteins (see Figure S1K), we explored the possibility that CCTT might directly interact with cenDNA. To determine the propensity, we examined enriched motifs associated with CCTT-bound peaks and found that the top three overrepresented motifs exhibit a GA-rich preference (Figure 4A). It has been shown that lncRNAs could bind to specific DNA sequences through the formation of RNA-DNA triplex (Wang and Chang, 2011; Mondal et al., 2015; Li et al., 2016; Li and Fu, 2019). A feature associated with triplex formation is purine-rich sequences in duplex DNA, to which pyrimidine-rich single-stranded RNA binds via Hoogsteen hydrogen bonding (Beal et al., 1991; Escudé et al., 1993). All these top three motifs appear to fulfill the requirement for triplex formation. Moreover, these motifs are enriched in the centromere regions by using the FIMO software (Grant et al., 2011) to map the whole genome (Figure S4E). We next utilized the Triplexator software (Buske et al., 2012) to predict the triplex-forming oligos (TFOs) within the CCTT sequence and identified a single putative DNA binding domain (DBD) in CCTT, consisting of 6 highly overlapped TFOs that span the pyrimidine-rich (43-79 nt) sequences near the 5’ end of CCTT (Figure 4B).
Figure 4. CCTT Possibly Forms RNA-DNA Triplex with CenDNA.
(A) Top three GA-rich motifs enriched in CCTT peaks using MEME.
(B) Predicted 6 CCTT TFOs (triplex-forming oligos) using Triplexator, all of which correspond to a DBD (DNA binding domain) located between 43-79 nt in CCTT.
(C) The exogenous FL CCTT but not the DBD-deleted (ΔDBD) mutant localized at centromeres of CCTT−/− HeLa cells, shown by CCTT FISH (green) and CENP-A IF (pink) analyses. Scale bar, 5 μm.
(D) The quantification of the data in C by IMARIS (n = 65 for CCTT+/+ cells, n = 61 for CCTT−/− cells transfected with FL CCTT). UD, undetected (mean ± SD of three biological replicates). Each point represents an averaged CCTT signals of all centromeres within a cell.
(E) Confirmation of CCTT localization at centromeres by ChIRP-qPCR in CCTT−/− HeLa cells complemented with Ctrl, FL CCTT, or ΔDBD CCTT. LacZ served as a negative control. ***p < 0.001; ns, no significant difference (mean ± SD, n = 3 per group).
(F) The DBD was sufficient to mediate CCTT localization at centromeres. Left: A diagram showing the use of psoralen and biotin-labeled CCTT DBD to capture cenDNA in HeLa cells. Middle: Quantified results of CCTT DBD captured cenDNA in comparison with β-satellite (β-SAT) and telomere-associated repeat sequences (TAR1). Right: CCTT DBD that captures cenDNA is resistant to RNase H. LacZ served as a negative control. *p < 0.05; ***p < 0.001; ns, no significant difference (mean ± SD, n = 3 per group).
(G) Triplex formation between CCTT and cenDNA. EMSA imaging of 10 μM biotinylated dsDNA oligos (TTS) or mutant ones (MUT) binding to increasing concentrations (0.25, 0.5, 2, 4 μM) of CCTT TFO. The mutant nucleotides are shown in blue.
See also Figure S4.
Next, we tested the functional requirement of the DBD in CCTT knockout cells complemented with either full-length or mutant (deletion of 43-79 nt, ΔDBD) exogeneous CCTT. FISH analysis showed that the exogenous full-length CCTT localized at centromeres, but the ΔDBD mutant failed to do so (Figures 4C and 4D). Moreover, ChIRP-qPCR analysis showed that the re-expressed full-length CCTT, but not the ΔDBD mutant, restored cenDNA enrichment in CCTT ablated cells, demonstrating that the 43-79 nt sequence in CCTT is indispensable for CCTT binding to centromeres (Figure 4E). To assess the sufficiency of the DBD domain in binding to centromeres, we transfected HeLa cells with a biotinylated CCTT-DBD oligoribonucleotide carrying a psoralen moiety at its 5’ end (Figure 4F, left). This biotinylated CCTT-DBD enriched cenDNA ~70-fold relative to the control lacZ RNA (Figure 4F, middle). As expected, this CCTT-DBD RNA did not capture other repetitive DNAs, such as β-satellites and telomere TAR1, highlighting its specificity in interaction with cenDNA (Figure 4F, middle). Furthermore, the interaction between CCTT-DBD and cenDNA was resistant to RNase H, supporting the possibility that CCTT interacts with cenDNA via the formation of RNA-DNA triplex rather than R-loop (Figure 4F, right).
Furthermore, we used the Triplex Domain Finder (TDF) software (Kuo et al., 2019) to analyze our CCTT AMT-ChIRP-seq data to predict GA-rich triplex target DNA sites (TTSs). Strikingly, 68.36% of centromeric CCTT-bound peaks showed similar TTS sequences. To verify the direct interaction between CCTT and predicted TTS, we performed EMSA with synthetic 15 nt CCTT-TFO RNA sequences and a biotinylated TTS DNA motif. The EMSA results showed a retarded mobility of double-stranded TTS DNA after incubation with single-strand CCTT-TFO RNA (Figure 4G), suggesting RNA-DNA triplex formation between CCTT-TFO and double-strand TTS, but not with the mutant DNA (Figure 4G). Altogether, these results strongly suggest that CCTT may directly bind to cenDNA through the formation of RNA-DNA triplex.
Distinct domains of CCTT involved in binding CENP-C and cenDNA
To map the region within CCTT responsible for direct binding to CENP-C, we performed infrared crosslinking and immunoprecipitation followed by RNA sequencing (irCLIP-seq) (Zarnegar et al., 2016) and identified a potential CENP-C binding region that spans 95-177 nt of CCTT (Figure 5A). To complement the irCLIP-seq result, we also globally mapped CCTT secondary structure by using selective 2’-hydroxyl acylation analysis by primer extension and mutational profiling (SHAPE-MaP) (Siegfried et al., 2014; Smola et al., 2015a; Smola et al., 2015b) under cell-free conditions as well as within HeLa cells. The results showed that 43-79 nt of CCTT, the determinant for RNA-DNA triplex formation, has continuous single-stranded propensity, thus making it accessible for DNA binding (Figure 5B, top) and that 127-177 nt of CCTT is predicted to fold into a two-stem-loop structure (Figure 5B, bottom), and this region is protected by proteins in cells as shown by a significant decrease in SHAPE reactivity in the in-cell state compared with the cell-free state (Figure 5A), indicating that this region may be responsible for interaction with CENP-C.
Figure 5. CCTT Uses Distinct Domains to Interact with CENP-C and CenDNA.
(A) The CENP-C binding domain of CCTT deduced by irCLIP (top two tracks from two biological replicates) and SHAPE-MaP (bottom track) in cell-free (blue line) or in-cell state (red line). The ratio of cell-free smoothened SHAPE reactivities relative to in-cell reactivities is indicated by the dotted line. The gray-shaded area (127-177 nt) indicates the region with the highest ratio of cell-free versus in-cell SHAPE reactivities, which is co-incident with the highest CENP-C binding detected by irCLIP. The sequence between the two vertical dotted lines (43-95 nt) corresponds to the region with most single-strandness predicted by SHAPE-MaP.
(B) The deduced secondary structures of the predicted single-strand region (top) and CENP-C binding domain (bottom) of CCTT. Color-coded probability scores at individual nucleotide positions are based on the SHAPE-MaP data and predicate the confidence probability of secondary structure. The predicated secondary structure contains paired and unpaired base, and the higher scores imply the more reliable structure in this position.
(C) Exogenous FL CCTT, but not the mutants depleted of the CENP-C binding domain (Δ127-177) or the DNA binding domain (ΔDBD), restored the abundance of CENP-C at centromeres in CCTT−/− HeLa cells, shown by CCTT FISH (green), CENP-C (red) and CENP-A (pink) IF analyses (left). The quantifications of CCTT (top) or CENP-C (bottom) signals by IMARIS (n = 77 for CCTT+/+ cells, n = 46 for CCTT−/− cells, n = 58 for CCTT−/− cells transfected with FL CCTT, n = 65 for CCTT−/− cells transfected with Δ127-177 CCTT, n = 51 for CCTT−/− cells transfected with ΔDBD) are shown (right). ***p < 0.001; ns, no significant difference; UD, undetected (mean ± SD of three biological replicates). Scale bar, 5 μm. Each point represents an averaged CCTT or CENP-C signals of all centromeres within a cell.
(D) Quantified results of captured CCTT by CENP-C RIP in CCTT−/− HeLa cells complemented with Ctrl, FL CCTT, Δ127-177 CCTT, or ΔDBD CCTT. ***p < 0.001; ns, no significant difference (mean ± SD, n = 3 per group).
(E) Mapping the specific CCTT binding domain in CENP-C by CENP-C RIP-qRT-PCR. Left: Annotated CENP-C domains in literature and various Flag-tagged CENP-C truncated mutants tested by the CCTT capture assay. The deduced CCTT binding domain in CENP-C is highlighted in red. Right: Quantification of the RIP-qRT-PCR data. *p < 0.05; ***p < 0.001 (mean ± SD, n = 3 per group).
See also Figure S5.
To validate these findings, we expressed full-length, Δ127-177, or ΔDBD CCTT in CCTTfloxed/− cells (Figure S5A). After deleting endogenous CCTT, we performed CCTT FISH and CENP-C immunostaining and found that although Δ127-177 CCTT normally localized to centromeres, its ability to direct CENP-C recruitment to centromeres was compromised (Figure 5C). Similarly, the ΔDBD CCTT failed to rescue CENP-C levels at centromeres compared to what the full-length CCTT did in CCTT−/− cells (Figure 5C). CENP-C RIP-qPCR analysis further showed that Δ127-177 CCTT was much less efficient than full-length CCTT in binding to CENP-C, and as expected, the interaction of ΔDBD CCTT and CENP-C was unaffected (Figure 5D). We additionally mapped the domain in CENP-C responsible for interacting with CCTT by expressing different Flag-tagged truncations of CENP-C in HeLa cells (Figures 5E and S5B). By RIP with an anti-Flag antibody, we identified a key domain between 826-943 aa responsible for specific CCTT binding (Figure 5E), which is dispensable for interactions with other previously established proteins, including CENP-A, CENP-L/N, CENP-HIKM, and the Mis12 complex (Weir et al., 2016). Taken together, these results suggest that CCTT bridges CENP-C and cenDNA through two separate domains.
Functional requirement of CCTT for mitosis
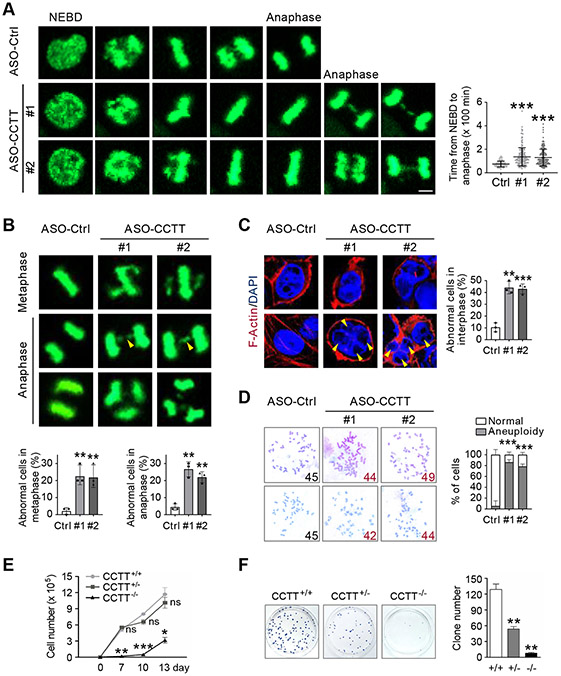
We finally explored the function of CCTT during mitosis. To avoid potential indirect effects accumulated in CCTT knockout cell lines, we examined CCTT function by using CCTT ASOs. Live-cell imaging of HeLa cells stably expressing histone 2B (H2B)-GFP revealed that CCTT knockdown resulted in a marked prolongation of the mitotic phase from nuclear envelope breakdown (NEBD) to anaphase (Figure 6A, left), which typically lasted ~75 minutes, and upon CCTT knockdown, increased to ~180 minutes (Figure 6A, right). Furthermore, we observed significant mitotic errors in CCTT knockdown cells (Figure 6B, top), as indicated by a ~10-fold increase in the frequency of misaligned chromosomes in metaphase (Figure 6B, bottom left) and a ~5-fold elevation in the frequency of chromosome bridges and multipolar-spindles in anaphase (Figure 6B, bottom right). Consequently, CCTT knockdown cells displayed a significantly higher frequency of binuclei and micronuclei in interphase (Figure 6C).
Figure 6. CCTT Is Required for Accurate Mitosis.
(A) CCTT knockdown led to prolonged mitosis of HeLa cells. Left: representative time-lapse microscopic images of ASO-Ctrl and ASO-CCTT HeLa cells expressing histone H2B-GFP during mitosis. Right: Quantification of CCTT expression by qRT-PCR. **p < 0.01 (mean ± SD, n = 3 per group). Left bottom: Quantification of time from nuclear envelope breakdown (NEBD) to anaphase. n = 90 for ASO-Ctrl, n = 180 for ASO-CCTT #1, n = 173 for ASO-CCTT #2. ***p < 0.001 (mean ± SD). Each point represents a cell. Scale bar, 5 μm.
(B) CCTT knockdown caused mitotic errors in metaphase and anaphase HeLa cells. Top: Representative images of mitotic errors by time-lapse assay, including alignment defects (white arrowheads), chromosome bridges (yellow arrowheads), and multipolar spindles. Bottom: Quantification of the percentage of abnormal cells in metaphase (top) and anaphase (bottom). n = 90 for ASO-Ctrl, n = 180 for ASO-CCTT #1, n = 173 for ASO-CCTT #2. **p < 0.01 (mean ± SD of three biological replicates). Scale bar, 5 μm.
(C) CCTT knockdown induced abnormal nuclei in interphase HeLa cells. Left: Binuclei (white arrowheads) and micronuclei (yellow arrowheads) were detected. Cells were stained with F-Actin (red) and DAPI (blue). Right: Quantification of the percentage of abnormal cells. n = 94 for ASO-Ctrl, n = 69 for ASO-CCTT #1, n = 65 for ASO-CCTT #2. **p < 0.01; ***p < 0.001 (mean ± SD of three biological replicates). Scale bars, 5 μm.
(D) CCTT knockdown caused aneuploidy in HCT116 cells. Left: Representative images of chromosomes and abnormal numbers are highlighted in red. Right: Quantification of the percentage of aneuploid cells. n = 28 for ASO-Ctrl, n = 36 for ASO-CCTT #1, n = 35 for ASO-CCTT #2. **p < 0.01; ***p < 0.001 (mean ± SD of three biological replicates).
(E) Cell growth curve of CCTT+/+, CCTT+/−, and CCTT−/− HeLa cells. Cell numbers were counted at 0, 7, 10, 13 days after CCTT inducible knockout. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference (mean ± SD, n = 3 per group).
(F) CCTT+/+, CCTT+/−, and CCTT−/− HeLa cells were cultured for colony formation. After 3 weeks, clones were visualized by crystal violet and the numbers were quantified. **p < 0.01 (mean ± SD, n = 3 per group).
See also Figure S6.
As mitotic errors invariably lead to aneuploidy (lost or gain chromosome), we directly tested this possibility on HCT116 cells, which have a relatively stable karyotype, and indeed found that the level of aneuploidy was elevated from the baseline of ~5% to ~80% after CCTT knockdown (Figure 6D). All these phenotypes were mirrored by CENP-C knockdown (Figures S6A-S6E), suggesting that CCTT deficiency-induced mitotic errors and chromosomal instability likely resulted from compromised CENP-C abundance during mitosis. As expected, CCTT knockout induced cell immortality, as shown by severely retarded cell growth and compromised colony formation (Figures 6E and 6F). Notably, the heterozygous cells with a ~50% decrease in CCTT expression (see Figure S2F) did not immediately impact cell fitness (Figure 6E) while displayed impaired cell growth in the long-term colony formation assay (Figure 6F).
We further explored the mechanism by which aberrant CCTT led to mitotic defects and aneuploidy. During cell division, the kinetochore unattached to microtubules provides a platform for the assembly of mitotic checkpoint complex (MCC), which inhibits the E3 ligase activity of the anaphase-promoting complex/cyclosome (APC/C), thereby preventing cells from exiting mitosis. The spindle assembly checkpoint (SAC) must be precisely activated and then silenced to enable cell cycle progression, defects of which are known to induce aneuploidy (Corbett, 2017). Given the newly elucidated function of CCTT in kinetochore assembly, we hypothesized that CCTT reduction may affect the SAC during mitosis. By performing immunostaining for Bub1 and BubR1, two critical factors for the assembly of the MCC, we found that in CCTT-depleted HeLa cells, both Bub1 and BubR1 were inefficiently localized to kinetochores (Figures S6F and S6G), indicating that incomplete CENP-C recruitment resulted in a series of compromised events during kinetochore assembly, including impaired SAC activation, together leading to mitotic failure.
DISCUSSION
In this study, we uncover a previously unannotated lncRNA CCTT generated from a non-centromeric region in Chr. 17 that specifically targets to repetitive cenDNA in all chromosomes. We show that CCTT binds to CENP-C, the kinetochore assembly blueprint protein, via RNA-protein interactions and to cenDNA via RNA-DNA interactions, possibly through the formation of RNA-DNA triplex. These findings suggest that CCTT is the long-sought RNA component critical for bringing together a blueprint for kinetochore assembly.
It has been well established that CENP-A binding defines functional centromeres and initiates kinetochore assembly (Black and Cleveland, 2011). CENP-A then recruits CENP-C to centromeres in mid-G1 followed by other CCAN deposition during S/G2 (Prendergast et al., 2011; McKinley et al., 2015; Weir et al., 2016; Hoffmann et al., 2016). Importantly, CENP-A is not the only factor responsible for CENP-C recruitment, as deletion of the interaction domains in CENP-C or CENP-A fails to fully diminish CENP-C localization at centromeres (Fachinetti et al., 2013; Guo et al., 2017). This is in line with the observations that even when endogenous CENP-A declines to below 1%, ~30% CENP-C is still associated with centromeres (Fachinetti et al., 2013) and that CENP-C does not always colocalize with CENP-A within the centromere domain (Kyriacou and Heun, 2018).
The possibility for the involvement of RNA in kinetochore assembly is initially supported by RNase A treatment that results in significant reduction of CENP-C on metaphase centromeres (Wong et al., 2007). α-satellite RNAs transcribed from cenDNA have been long suspected to play a role, but these RNAs lack the specificity in RNA-protein interactions and their functional requirement has remained controversial (Wong et al., 2007; Quénet and Dalal, 2014; Rošić et al., 2014; McNulty et al., 2017). In our study, we retrieved CENP-C-binding RNAs by CENP-C RIP-seq and did not find any α-satellite RNA sequence, and more importantly, we demonstrated that CCTT is functionally required for kinetochore assembly, thus fulfilling a long-suspected RNA involved in this critical biological process (Talbert and Henikoff, 2018).
Our in vitro experiments suggest that CCTT seems to target cenDNA via the formation of RNA-DNA triplex. Interestingly, the characterized CCTT DNA-binding motif is GA-rich instead of AT-rich, the latter of which is more prevalently distributed in human centromeres. However, a previous study clearly showed that GA-rich sequences are also frequently presented in human centromeres, and more importantly, these GA-rich sequences have the propensity to form a non-B hairpin DNA structure (Catasti et al., 1994) and/or a four-stranded i-motif (Garavis et al., 2015). These structures may favor the formation of RNA-DNA triplex. Future studies to precisely map such triplex formation regions within centromeres will be required to substantiate this hypothesis. Interestingly, a mutant CCTT that failed to bind to DNA can still interact with CENP-C, suggesting that CCTT may first bind soluble CENP-C and then facilitate CENP-C recruitment to cenDNA via triplex formation.
Given its essential function for CENP-C recruitment, it is not surprising that we detected significant mitotic errors, which led to severe aneuploidy, in CCTT-depleted cells. As chromosome stability is frequently linked to tumorigenesis, we predict that altered CCTT expression might be characteristic of certain cancer types. Our current mechanistic insights thus pave the path for future studies to understand the function of CCTT in cancer etiology and/or progression.
Limitations of the Study
Our results suggest that CCTT functions to facilitate CENP-C recruitment during kinetochore assembly. Through the SNAP-tag labeling scheme, we provide initial evidence that CCTT is required for the recruitment of newly synthesized CENP-C to centromeres. Because CENP-C is also known for its dynamic association/dissociation with the established centromeres in the interphase, it remains to be determined whether CCTT shows a similar dynamic regulation as CENP-C. Accordingly, our current data could not differentiate between the following two possibilities: 1) CENP-C and CCTT may first bind and then be recruited together to cenDNA or 2) CCTT may first bind to cenDNA, thereby creating a “landing pad” for subsequent CENP-C recruitment. Future experiments are needed to address these possibilities.
The putative function of CCTT as a structural component of the kinetochore would predict that the requirement of CCTT for CENP-C recruitment could not be bypassed by simply increasing the supply of CENP-C to “rescue” CCTT depletion-induced phenotype in chromosome segregation. Unfortunately, we are precluded from carrying out this seemly simple experiment to test the prediction, as CENP-C overexpression is also known to induce mitotic defects, thus complicating data interpretation. The comprehensive understanding of CCTT in kinetochore assembly thus awaits future insights from high resolution kinetochore structure.
STAR*METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yan Teng (tengyan@bmi.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
The accession number for RIP-seq, AMT-ChIRP-seq, irCLIP-seq, and SHAPE-MaP from HeLa cells is GEO: GSE149534.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Cell Lines
Human cell lines including uterine cancer cells HeLa, colon cancer cells HCT116 were purchased from the American Type Culture Collection (ATCC; https://www.atcc.org).
Bacterial Strains
E. coli DH5α competent cells were procured from Biomed (#BC116) and E. coli DL21 (DE3) chemically competent cells were procured from Transgen (#CD601-02) and were grown in LB culture at 37°C.
METHOD DETAILS
Cell Lines and Culture Conditions
Human uterine cancer cells HeLa (CCL-2, ATCC), colon cancer cells HCT116 (CCL-247, ATCC) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, HYCLONE) high glucose supplemented with 10% fetal bovine serum (FBS, GIBCO). All cell lines were maintained at 37°C with 5% CO2.
ASOs, siRNAs and Adenovirus-Cre Treatment
ASOs (antisense oligonucleotides) and siRNAs were introduced using jetPRIME (polyplus). ASOs against CCTT: #1 GCTACCAGAAGGGAGCACCA, #2 CCAGAT GTCTTCAGCTCCAA; siRNAs against CCTT: CCAGAUGUCUUCAGCUCCA, CENP-C: UUGACUUUCUACCUUGAAGG, CENP-A: AAAGGAGAUCCGAAAGCU UCA, CENP-B: CCAACAAGCUGUCUCCCUA were purchased from Ribobio. For the CCTT inducible knockout, CCTTfloxed/− HeLa cells were treated with adenovirus-Cre for 12 hours.
Plasmid Construction
Full-length and various mutant CCTT were cloned into the pcDNA3.1(+) vector for overexpression. Two ORFs within CCTT sequences were cloned with GFP into the pcDNA3.1(+) vector for the in vitro translation assay. Full-length CENP-C proteins were cloned into the pET-28a vector for expression and purification for the protein-RNA EMSA assay. Full-length and truncated CENP-C proteins were cloned into the CMV10-Flag vector for the RIP assay. The primers were shown in Table S1.
Subcellular Fractionation Analysis
The distribution of RNAs in the nuclear and cytosolic fractions of HeLa cells was assessed using the PARIS Kit (Life Technologies) following the manufacturer’s instruction.
RACE
The 5’ and 3’ RACE was performed using the FirstChoice RLM-RACE RNA Ligase Mediated RACE Kit (Ambion) following the manufacturer’s instruction. RNA was extracted from HeLa cells. Primers used for 5’ and 3’ RACE were designed based on the known sequence information and are listed in Table S1.
RNA Isolation and qRT-PCR
Total RNA from each cultured cell line with different treatments was extracted with TRIzol Reagent (Invitrogen) according to the manufacturer’s instruction. For qRT-PCR, cDNA synthesis was carried out using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO). qPCR was performed using SYBR Green Real-time PCR Master Mix (TOYOBO) and a StepOnePlus real-time PCR system (Applied Biosysterms 7500 Fast). The relative expression of different sets of genes was quantified to GAPDH mRNA. Primer sequences for qRT-PCR and RT-PCR are listed in Table S1.
Northern Blot
Total RNA was isolated from HeLa and HCT116 cells using TRIzol Reagent (Invitrogen), and 10 μg total RNA was diluted in 2 x RNA Loading Dye (Invitrogen) and denatured at 65°C. Samples were separated on a 1.2% agarose, 6.6% formamide, 1 x TBE gel containing ethidium bromide. After imaging the rRNA subunits (18s and 28s), RNA was transferred to a Hybond N+ nylon membrane and crosslinked to the membrane by baking at 80°C for 2 hours. The membrane was prehybridized in DIG Easy Hyb (Roche) and hybridized with 100 ng/mL digoxigenin-labelled probe (sequences are 1-404 nt within CCTT) overnight at 68°C, followed by two washes with 2 x SSC/0.1% SDS and one wash with 0.2 x SSC/0.1% SDS. Digoxigenin-probe hybridization was detected using the DIG-High Prime DNA Labeling and Detection Protocol (Roche). After addition of CDP-Star chemiluminescent substrate, the probe signal was visualized using an Image Quant LAS 4000 mini (GE healthcare). A single-stranded RNA probe was generated using the DIG Northern Starter Kit (Roche) according to the manufacturer’s instruction.
Measurement of CCTT Copy Numbers
A serial dilution of the linearized plasmid pcDNA3.1-CCTT was used qRT-PCR to generate a standard curve for CCTT. The copy number of the diluted plasmid pcDNA3.1-CCTT was calculated by DNA/RNA Copy Number Calculator from website (http://endmemo.com/bio/dnacopynum.php). To measure the CCTT copy number in HeLa cells, total RNA extracted from 2~3 x 106 cells was reverse transcribed into cDNAs for qPCR analysis, and the copy number could be quantitated from the standard curve.
In Vitro Translation Assay
Conserved open reading frames (ORFs) within CCTT sequences were predicted by Pfam (http://pfam.xfam.org) and shown in Table S2. The ORFs (ORF1: 38-373 nt and ORF2: 223-417 nt) were cloned with GFP into the pcDNA3.1(+) vector for fused expression. Total RNA and proteins were extracted for RT-PCR and western blot using anti-GFP antibody (BioVision).
CCTT Knockout by CRISPR/Cas9
HeLa cells were infected with lentivirus by pLenti-U6-spgRNA v2.0-CMV-Puro-P2A-3Flag-spCas9 (H7548, Obio) plasmid transfection for stable expression of Cas9. First, we deleted a CCTT allele in Cas9-expressing HeLa cells. Two gRNA-coding sequences (5’-GATGGGGCTAGCAGAGGCCTTGG-3’ and 5’-GGCTGCCTTCACCCCCACCAAGG-3’) were located upstream and downstream of the CCTT locus, respectively. After transfection with two sgRNAs transcribed using the GeneArt™ Precision gRNA Synthesis Kit (Thermo) using Lipofectamine 2000 Reagent (Thermo), a 2585-bp DNA fragment spanning full-length CCTT was deleted. If the other allele was intact, a heterozygous deletion was achieved (CCTT+/−). 3 pairs of primers were used to screen CCTT+/− cells (Figures S2E-S2F). Products of primers 1 (391 bp) and 2 (715 bp) are specific for the wildtype allele and products of primers 3 are specific for the CCTT full-length deletion allele with a 3.5 minutes extension step in PCR reaction (3164 bp for wildtype allele and ~588 bp for deletion allele, marked by 1 kb ladder and DNA Marker I, respectively). Next, we engineered the other allele in CCTT+/− cells. The gRNA-coding sequence (5’-CCCACTCCTGAATCTAGGAGGAG-3’) was located upstream of exon 1. The homologous arms (HA) of the donor vector are indicated as HA-L (724 bp) and HA-R (752 bp). In the donor sequence, two LoxP sites flanked exon 1. After transfection with sgRNA and donor sequences, two LoxP sites were integrated at the CCTT locus (CCTTfloxed/−). Finally, upon adenovirus-Cre treatment, a 339-bp DNA fragment containing exon 1 was deleted, resulting in two deletion alleles (CCTT−/−). Primers for genotyping are shown in Table S1. The knockout efficiency was validated by RT-PCR, FISH, and northern blot, as shown in Figures S2F-S2H. We constructed 4 independent clones to confirm CCTT conditional knockout.
Western Blot
Western blot was performed as described previously (Hao et al., 2020). Cells were collected and lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5% Tween-20, 0.5% NP-40, 2 mM PMSF, 2.5 mM β-glycerophosphate (all from Sigma-Aldrich) and protease inhibitor cocktail tablet (complete Mini, Roche Diagnostics). Equal amounts of protein were resolved by 10% Tris-Glycine gel (SDS-PAGE). Proteins were blotted onto a nitrocellulose membrane, blocked with blocking buffer (1xTBS, 0.1% Tween-20) with 5% w/v nonfat dry milk and incubated with the corresponding primary/secondary antibodies: anti-CENP-C (1:1000, Abcam), anti-GFP (1:1000, BioVision), anti-Actin (1:5000, Abcam). After incubation with peroxidase conjugated secondary antibodies (1:5000, Abcam), blots were developed with Supersignal West Pico Chemiluminescent Substrate (Pierce) and exposed to film (SAGECREATION, MiNiChemi).
RNA FISH
We used PCR to amplify an Alu-deleted part of the CCTT sequence (1-404 nt) from HeLa cells using T7 primers. RNA probes were prepared by T7 (Promega) with the PCR purification products as a template and were labelled with Alexa Fluor 488 using the ULYSIS Nucleic Acid Labeling Kit (Invitrogen) according to the manufacturer’s instruction. To perform FISH on interphase cells, unsynchronized cells cultured in chamber slides were washed with PBS and fixed with 4% formaldehyde diluted in DEPC-PBS for 10 minutes at room temperature. FISH probes were combined as indicated, applied to interphase cells or metaphase spreads on glass slides, and sealed with a coverslip. Samples and probes were codenatured at 75°C for 2 minutes, followed by sealing with rubber cement and overnight hybridization at 37°C with yeast tRNA blocking in a humidified chamber. Slides were washed in 0.4 x SSC at 72°C for 2 minutes and in 2 x SSC, 0.05% Tween-20 at room temperature for 30 seconds. Slides were rinsed in water, stained with DAPI, and mounted in ProLong Gold anti-fade mounting solution.
Immunofluorescence and Immuno-RNA FISH
For immunofluorescence, cells seeded on coverslips or chamber slides were fixed with 4% formaldehyde diluted in PBS for 10 minutes and washed with PBS. Cells were then permeabilized with 0.3% Triton X-100, PBS for 5 minutes, washed with PBS, and blocked with Block (0.1% Triton X-100, 2.5% FBS, 0.2 M glycine, PBS). Primary antibodies were diluted in the Triton Block and applied to cells for 1 hour at room temperature or overnight at 4°C, followed by three 10-minute washes with 0.1% Triton X-100, PBS. The following primary antibodies were used in the Triton Block: CENP-A (MBL), CENP-C (MBL), and CREST (kind gift from Li lab) with a 1:1,000 dilution; Bub1 (Abcam) with a 1:500 dilution; BubR1 (Abcam) with a 1:50 dilution. Fluorescence-conjugated secondary antibodies (Invitrogen) were diluted 1:200 in Triton Block and applied to cells for 1 hour at room temperature, followed by three 10-minute washes with 0.1% Triton X-100, PBS and staining with DAPI. Cells were mounted in ProLong Gold anti-fade mounting solution and imaged as described.
For immunofluorescence combined with RNA FISH, the immunofluorescence procedure was performed as described followed by fixation in 4% formaldehyde diluted in DEPC-PBS for 10 minutes at room temperature. Samples were then processed for RNA FISH as described.
SNAP-Tag Labeling
SNAP labeling was conducted as described previously (Jansen et al., 2007; Bodor et al., 2012) with modifications. For mammalian expression, CENP-CSNAP plasmid is a kind of gift from Guohong Li. Human gene CENP-C in full length was cloned into pSNAP26m (N9172, New England Biolabs) using the standard molecular biology methods. HeLa cells were transfected CENP-CSNAP before related siRNAs treatment. In the quench-chase-pulse experiment, SNAP-tag was quenched with SNAP-Cell Block (S9106S, New England Biolabs). Newly synthesized CENP-CSNAP was labelled 7 hours after release with SNAP-Cell TMR-star (S9105S, New England Biolabs). Cells were fixed at the indicated times and processed for TMR-star signal detection with confocal microscopy (LSM880, Zeiss).
RIP-Seq
RIP-seq was carried out as previously described (Zhang et al., 2013) with modifications. HeLa cells (1 x 107) were harvested, resuspended in lysis buffer (25 mM Tris pH7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, 1 mM PMSF, and 2 mM VRC, with Protease Inhibitor Cocktail, Roche) and then lysed on ice for 30 minutes. After incubation with anti-CENP-C (2 μg, Abcam) or anti-IgG (Santa Cruz) antibodies, the beads were washed once in lysis buffer and four times in washing buffer (50 mM Tris pH7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), followed by elution with extraction buffer C (100 mM Tris pH6.8, 4% SDS, 12% β-mercaptoethanol, 20% glycerol) at room temperature for 10 minutes. One-third of the RIP materials was used for immunoblotting and the other two-thirds were used for RNA extraction. Each RNA sample was treated with DNase I (Ambion, DNA-freeTM kit). We adhered to the protocols supplied with NEB Next® UltraTM Directional RNA Library Prep Kit for Illumina®.
Biotinylated RNA Pulldown Assay
Biotinylated RNA pulldown assays were performed as previously described (Tsai et al., 2010; Klattenhoff et al., 2013) with modifications. Linearized CCTT RNAs (sequences are full length of CCTT) were in vitro transcribed with Biotin RNA Labeling Mix (Roche). T7 RNA polymerase (Roche) was applied followed by RNase-free DNase I (Ambion) treatment and purification with NucleoSpin RNA Clean-up XS (Macherey Nagel). Then, 3 μg biotinylated RNA was heated for 5 minutes at 90°C in RNA capture buffer (20 mM Tris-HCl pH 7.5, 1 M NaCl, 1 mM EDTA) and slowly cooled down to room temperature. We harvested 2 x 107 HeLa cells, resuspended them in Pierce® IP lysis buffer (Thermo Fisher Scientific) with anti-RNase, protease/phosphatase inhibitor cocktail and then lysed them on ice for 30 minutes. Streptavidin-conjugated magnet beads C1 (Invitrogen) were prepared according to the manufacturer's instructions. After centrifugation at 20,000 rcf for 20 minutes, the supernatant was transferred to a new tube and precleared by applying 30 μL prepared Streptavidin C1 Dynabeads for 1 hour at 4°C. Folded RNAs were added and incubated for 3 hours at 4°C, followed by addition of 30 μL of the prepared Streptavidin Dynabeads and incubation for 3 hours at 4°C. The RNA-binding protein complexes were washed sequentially with NT2 buffer (twice), NT2-high salt buffer containing 500 mM NaCl (twice), NT2-high salt buffer containing 1 M NaCl (twice), NT2-KSCN buffer containing 750 mM KSCN (twice) and DEPC-PBS (once) for 5 minutes at 4°C, and they were eluted with 2 mM D-biotin in PBS. The eluted protein complexes were analyzed by western blot with anti-CENP-C (MBL) and anti-CENP-A (MBL) antibodies. For mass spectrometric analysis, the protein-RNA complexes were subjected to 4~12% NuPAGE Bis-Tris gels (Life Technologies) followed by silver staining with SilverQuest Silver Staining Kit (Life Technologies).
Protein Purification and EMSA
The protein used in the EMSA was expressed in E.coli DL21 (DE3) by using the pET-28a expression vector. After transfection, the bacteria were incubated until reaching 0.6 at OD600. Then, the recombinant protein was induced by adding 1 mM IPTG for overnight at 16°C. Next, Ni-NTA Fast Start Kit (#30600, Qiagen) was used to purify the recombinant CENP-C. Using this kit, the His-tagged CENP-C was purified in the presence of high salt, resolved by 10% Tris-Glycine gel (SDS-PAGE) and stained with Coomassie Brilliant Blue. Full-length CCTT was labeled with biotin through in vitro transcription assays (sequences are full length of CCTT). EMSA was conducted as previously reported (Xing et al., 2017). The 0.7 pmol biotinylated CCTT was used for each reaction. The secondary structure of CCTT was formed by heating at 95°C for 5 minutes following by a slow cool down to room temperature. The indicated amount of purified protein (2.5, 3.5, 4.5, 6 μM) and biotinylated CCTT was incubated in binding buffer (100 mM HEPES pH7.5, 200 mM KCl, 10 mM MgCl2, 10 mM DTT) for 30 minutes at room temperature. The mixtures were subjected to electrophoresis on a 6% nondenaturing polyacrylamide 0.5 x TBE gel. The gel was then transferred to a nylon membrane and imaged by incubation with a Chemiluminescent Nucleic Acid Detection Module Kit (#89880, Thermo).
RNA-DNA Triplex EMSA
Triplex was performed as previously described (Mondal et al., 2015; Kalwa et al., 2016) with modifications. For the initial centromeric double-stranded oligonucleotide TTS hybridization, the complementary biotin-labeled DNA single strand was incubated in hybridization buffer (10 mM Tris-HCl pH7.5, 50 mM NaCl, and 10 mM MgCl2) for 5 minutes at 85°C and then cooled down to room temperature. To remove secondary structures present in CCTT TFO, RNA oligonucleotides were heated at 95°C for 5 minutes followed by quickly cooling them down on ice. Triplex formation was carried out in a 20-μL reaction volume with reaction mixtures consisting of nuclease free water, labeled dsDNA oligonucleotides (0.4 pmol), and different concentrations of CCTT TFO in hybridization buffer for 1 hour at 37°C. 10 μM labeled dsDNA corresponding to the CCTT-targeted centromere peak sequence or a mutated version were incubated with increasing concentrations (0.025, 0.5, 2, 4 μM) of CCTT ssRNA. RNase H treatment was performed after triplex formation at 37°C for 30 minutes. The reaction was monitored by EMSA on 6% polyacrylamide TBE gels. The gel was then transferred to a nylon membrane and imaged by incubation using a Chemiluminescent Nucleic Acid Detection Module Kit (#89880, Thermo).
ChIP and qPCR Analysis
1~1.2 x 107 cultured cells exposed to each treatment were washed with PBS, crosslinked in 1% formaldehyde for 10 minutes, and quenched by addition of 0.25 M glycine. The cells were then washed with cold PBS and harvested by scraping. Pelleted cells were incubated in ChIP lysis buffer (50 mM Tris pH 8.1, 10 mM EDTA, 1% SDS) for 10 minutes before being sonicated eight times for 10 seconds to shear DNA to an average fragment size of approximately 300~500 bp. Samples were diluted ten times in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris pH8.1, 167 mM NaCl) and precleared with previously blocked Protein A/G beads (Life Technologies) for 3 hours at 4°C. The beads were blocked with BSA and herring sperm DNA. Precleared samples were incubated overnight with 2-μg-specific antibodies at 4°C. Next, 2 μL of the Protein A/G beads were added and incubated for another 4 hours at 4°C followed by washing the beads in ChIP lysis buffer once and in high salt wash buffer (1% Triton X-100, 0.1% deoxycholate, 50 mM Tris pH8.0, 500 mM NaCl, 5 mM EDTA), LiCl immune complex wash buffer (0.25 M LiCl, 0.5% igepal, 0.5% deoxycholate, 10 mM Tris pH8.0, 1 mM EDTA) and 1 x TE Buffer (10 mM Tris pH8.0, 1 mM EDTA) at 4°C. We then added 100 μL of elution buffer and incubated the sample for 15 minutes. The elution process was repeated once, followed by the addition of 8 μL 5 M NaCl and incubation overnight at 65°C to reverse the cross-link. After treatment with 5 μL RNase A (20 mg/mL) and 5 μL proteinase K (20 mg/mL), the recovered DNA and soluble chromatin (input) were purified with phenol/chloroform and analyzed by qPCR using a LightCycler 480 (Roche). The primers used are shown in Table S1. The anti-CENP-C (Abcam) and anti-CENP-A (MBL) antibodies were used.
AMT-ChIRP
We developed the AMT-ChIRP by modifying previously reported ChIRP (chromatin isolation by RNA purification) (Chu et al., 2011) with the crosslinker 4’-aminomethyltrioxsalen (AMT, a psoralen derivative, Sigma), which allows the fixation of nucleic acid interaction by ultraviolet light without protein crosslinking (Shen et al., 1977; Mondal et al., 2015; Wang et al., 2020). Briefly, antisense DNA probes against CCTT were designed using the ChIRP Probe Designer tool. The 3’-end Biotin-TEG-modified probes were synthesized by Invitrogen (Thermo). HeLa cells were incubated with 0.5 mg/mL AMT solution for 10 minutes and chilled on ice. The cells were placed on ice and irradiated in a tissue culture dish for 10 minutes from a distance of 2.5 cm with 365 nm ultraviolet light. The cross-linked cells were lysed with lysis buffer (50 mM Tris-Cl pH7.5, 10 mM EDTA, 1% SDS, protease inhibitors, and 2 U/mL RNasin Ribonuclease Inhibitor). The lysates were sonicated with a Bioruptor (Diagenode) at 4°C using the high setting with pulse intervals of 20 seconds on and 20 seconds off for a total of 15 minutes. The sonicated cell lysates were hybridized with the mixture of biotinylated DNA probes against human CCTT in hybridization buffer (50 mM Tris-Cl pH7.5, 750 mM NaCl, 1% SDS, 1 mM EDTA, 15% formamide, protease inhibitors and 2 U/mL RNasin Ribonuclease Inhibitor) for 4 hours at 37°C. The binding complexes were then recovered using streptavidin-conjugated magnet beads C1 (Invitrogen) for 45 minutes at 37°C. The C1 beads were washed 5 times in 1 mL wash buffer (2 x NaCl and sodium citrate (SSC) (diluted from 20 x SSC Invitrogen stock), 0.5% SDS, PMSF) for 5 minutes at 37°C. DNA was eluted with elution buffer (50 mM NaHCO3, 1% SDS). Each sample of beads was resuspended in 150 μL of DNA Elution Buffer (50 mM NaHCO3, 1% SDS) with RNases (10 μL RNase A (10 mg/mL) and 10 μL RNase H (10 U/μL) per mL of DNA Elution Buffer), and the DNA input was resuspended in 140 μL. Samples were incubated at 37°C for 30 minutes with shaking. Beads and supernatant were separated on a DynaMag-2 magnetic strip. The previous step was repeated. All the supernatant (should be ~300 μL) was collected, and 15 μL proteinase K (20 mg/mL) was added. The samples were incubated at 50°C for 45 minutes with shaking. DNA samples were transferred to phase-lock gel tubes and treated according to the protocols from Qiagen. We adhered to the protocols supplied with the NEB Next® Ultra™ II DNA Library Prep Kit for Illumina®.
For in vivo triplex capture assays, HeLa cells were transfected with 10 μM 5’-6C-psoralen- and 3’-biotin-modified RNA oligonucleotides using INTERFERin®-RNA transfection reagent (Polyplus). After 24 hours, isolated nuclei were irradiated for 10 minutes with UVA (365 nm), and the cross-linked cells from 2 x 106 cells were processed as described for ChIRP. In the case of the RNase H control reaction, the supernatants were treated with 15 units of RNase H for 20 minutes at 37°C before addition of the streptavidin-magnetic beads.
irCLIP-Seq
irCLIP-seq was performed as previously described (Xue et al., 2009). Briefly, 2 x 108 HeLa cells were irradiated at 400 mJ with 254 nm UV light. The cross-linked cells were lysed, and 15 μg of anti-CENP-C antibody was applied to pull down specific protein-RNA complexes. After micrococcal nuclease treatment, alkaline phosphatase treatment, and pre-adenylated 3’ DNA linker (IRdye-800CW-labelled) ligation, the immunoprecipitated complexes were fractionated on a 4~12% NuPAGE Bis-Tris gel and transferred to a nitrocellulose membrane. The IRdye-800CW marked CENP-C-specific smear bands were cut and treated with proteinase K (20 mg/mL) prior to the extraction of respective RNA by phenol and chloroform. The extracted RNA was further ligated with a 5’ RNA linker and converted into a library as we previously described for deep sequencing. The possible CENP-C binding regions of all ncRNAs according to irCLIP-seq are shown in Table S3.
SHAPE-MaP
HeLa cells were grown, and RNA was extracted and modified, both in cell and ex vivo, as described previously (Smola et al., 2015). In brief, in-cell modification was carried out by treating HeLa cells in fresh growth medium with 1M7 (10 mM final) and incubating them at 37°C for 5 minutes before RNA isolation. For ex vivo analysis, total cellular RNA was gently extracted from cells into RNA folding buffer (100 mM HEPES pH8.0, 100 mM NaCl, and 10 mM MgCl2), incubated at 37°C for 20 minutes, and subjected to SHAPE modification with 1M7. From each sample of RNA from HeLa in-cell and cell-free probing experiments, 1~3 μg was subjected to mutational profiling (MaP) reverse transcription with primers specific to CCTT, followed by CCTT-specific PCR amplification (Siegfried et al., 2014). The cDNA generated was buffer exchanged over Illustra microspin G-50 columns (GE Healthcare). Output cDNA (5 μL) was used as a template for 50-μL PCR reactions (Q5 Hot-start polymerase, NEB) with primers generated to amplify CCTT and add adapter sequences (1 x Q5 reaction buffer, 250 nM each primer, 200 μM dNTPs, 3% DMSO, 0.02 U/μL Q5 Hot-start polymerase). PCR was performed using a touchdown format and high-throughput sequencing library construction.
Cell Synchronization
For mitotic synchronization, HeLa cells were synchronized by thymidine-nocodazole arrest and shaken off. Cells were incubated in thymidine-containing (2 mM) medium for 18 hours, released into fresh medium for 3 hours and treated with nocodazole (100 ng/mL) for 11 hours.
Time-Lapse Imaging
HeLa cells stably expressing histone H2B-GFP (HeLa-GFP-H2B) were seeded on an eight-chambered cover glass (Merck Millipore) in DMEM (Hyclone). For the time-lapse imaging, five 2.5-μm-separated z-planes covering the entire volume of mitotic cells were collected every 5 minutes for 12 hours using a 20 x lens objective on an inverted fluorescence microscope (Nikon Eclipse Ti-E) with an UltraView spinning-disc confocal scanner unit (Perkin Elmer). The temperature of the imaging medium was kept at 37°C. Image sequences were viewed using Volocity software, and the cell behaviors were analyzed manually. Normal HeLa cells take about 25 to 30 minutes to go through the prometaphase and then enter the metaphase. So, if the abnormal chromosomal configuration continued more than ~50 minutes after cells entered the prometaphase, we defined these cells as aberrant metaphase.
Chromosome Spreads and Karyotyping
To analyze the metaphase spreads, HCT116 cells were treated with 0.02 mg/mL colcemid for 18 hours, harvested with trypsin, washed with PBS, and incubated in hypotonic solution (0.4% KCl) for 10 minutes at 37°C. Cells were then resuspended in fixation buffer (3:1 mix of methanol:glacial acetic acid) and spread on slides cooled on ice. Karyotype analysis were performed by crystal violet staining for 20 minutes, washed with ddH2O, dried, and imaged under microscopy (Imager A2, Zeiss).
Cell Growth Assay
We seeded 5 x 103 CCTT+/+, CCTT+/− or CCTT−/− HeLa cells in 24-well plates and the cell numbers were counted at the designated time points (0, 7, 10, 13 days) after CCTT inducible knockout.
Colony Formation Assay
We seeded 1 x 102 CCTT+/+, CCTT+/− or CCTT−/− HeLa cells in 10-cm dishes and then cultured them at 37°C for about 3 weeks. Crystal violet staining was performed for 20 minutes at room temperature, and the number of clones per dish were counted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Ribosome Profiling Data Analysis
We downloaded the ribosome profiling data and input sequencing data in HeLa cells from SRA databases (Ribo-seq: SRR944657 and input: SRR944659). The ribosome profiling sequencing reads were stripped of adaptor sequences using Trimmomatic, and reads shorter than 28 bases were discarded before removing reads aligning to rRNA sequences using Bowtie2 with default parameters. Additionally, using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), sequences were discarded if they contained a base with a quality score of below 20. We obtained the read counts of genes using FeatureCounts and then normalized the read counts to RPKM using edgeR.
Centromere Quantification
Centromere quantification of CCTT, CENP-C, CENP-A, and TMR-star intensities were conducted with maximum intensity projections of images and were determined using IMARIS (Bitplane) Surface model. The average fluorescence value of approximately 40 centromeres in each individual cell and 30~60 cells were quantified in each sample. The results have been repeated by three independent experiments.
RIP-Seq Data Analysis
After obtaining RIP-seq data, we first used Trimmomatic to perform sliding window trimming, cutting the average quality once within the 4-bp window falling below 20 and removing the trimmed read if it had a length less than 80. SortMeRNA (Kopylova et al., 2012) was used to filter ribosomal RNAs with default parameters. We mapped the filtered reads to the genome (genome version hg38) using STAR (Dobin et al., 2013) with parameters --outFilterScoreMinOverLread 0.1, --outFilterMatchNminOverLread 0.1, --outFilterMismatchNmax 4 and --outFilterMultimapNmax 8. After obtaining the mapping files, we removed the duplicate reads using Picard (http://broadinstitute.github.io/picard/) with parameters MarkDuplicates and REMOVE_DUPLICATES = true. We calculated the gene read counts using FeatureCounts (Liao et al., 2014) and converted them to RPKM using the edgeR (Robinson et al., 2010) package in R language. Limma were used to determine the differentially expressed genes between CENP-C and IgG RIP-seq datasets. Only genes with a fold change greater than 2 and a p-value less than 0.01 were retained for further analysis. The wig files of each RIP-seq data set from the sense strand and antisense strand were obtained using bamCoverage from DeepTools with a parameter --binSize 20, --smoothLength 100, --ignoreDuplicates and --normalizeUsingRPKM.
ChIP-Seq Data Analysis
First, we obtained CENP-C (SRR2061974) and input (SRR2061983) raw reads from the SRA database and then used Fastq-dump to convert SRA files to Fastq files. Next, Trimmomatic, a mapping process and bigwig files were generated as described for the ChIRP-seq data analysis. Finally, peak calling between IPs and input was performed with the SICER pipeline using the same parameters described for the ChIRP-seq analysis.
AMT-ChIRP-Seq Data Analysis
Two CCTT AMT-ChIRP-seq libraries were combined before processing. Combined AMT-ChIRP-seq and input datasets were performed, followed by adapter trimming and filtering low-quality reads with Trimmomatic using the following parameters: MINLEN: 80, ILLUMINACLIP: 2:30:10 and SLIDINGWINDOW: 4:20.
Filtered pair-end reads were merged using PEAR software with default parameters. The merged reads were then mapped to a hg38 reference genome (including centromere models) using BWA-MEM with default parameters. BamCoverage in deeptools was used to obtain the bigwig files using the parameters --normalizeUsingRPKM --binSize 10 --ignoreDuplicates.
Enrichment peaks for AMT-ChIRP-seq data were determined using SICER software (Xu et al., 2014) with the input experiment as background. The parameters were the same as those used for previous processing of the human CENP-A ChIP-seq datasets (Nechemia-Arbely et al., 2019). Only the high-quality peaks (raw read count number > 20 and fold change > 2) were retained for further analysis. Peaks were determined as centromeric peaks if they had overlapping sites (BEDTools: intersect) in the genome previously annotated as centromere regions. To obtain the peak enriched motifs, the MEME suite was used to discover the motifs using only the peaks with a fold change greater than 5. These high quality peaks were also used to predict CCTT-bound DNA regions using the triplex format with TDF (Triplex Domain Finder, Kuo et al., 2019), allowing a maximal error rate = 20% and minimum triplex length = 14.
Triplex TFOs Prediction
Triplexator is a computational framework (Buske et al., 2012) for in silico prediction of triplex structures within specific genomic regions. Triplex formation is governed by sequence-specific binding rules. The triplex-forming oligonucleotides (TFO) are located in the region of the single-stranded nucleotide (RNA) capable of forming Hoogsteen (or reverse) bonds with the duplex (Escude et al., 1993). The triplex target site (TTS) is defined as the polypurine-polypyrimidine tract of a duplex (DNA) that is able to accommodate the TFO. We searched TFOs using CCTT RNA as single-stranded input and TTSs using centromere satellite sequences as double-stranded input, separately. A maximal error rate = 20% was used.
irCLIP-Seq Data Analysis
After obtaining the irCLIP-seq raw datasets, we first used fastx_collapser to remove PCR duplicates with default parameters. Then, we clipped 5’ barcodes and 3’ adaptors using fastx_clipper, and only the clipped reads with a length greater than 15 bp were retained for further analysis. We implemented the STAR alignment algorithm to map reads to the genome (genome version hg38) with the parameters --outFilterScoreMinOverLread 0.1, --outFilterMatchNminOverLread 0.1, --outFilterMismatchNoverLmax 0.08 and --outFilterMultimapNmax 8. In addition, we only considered uniquely mapped reads, and we used the parameter –q 30 in samtools to filter the reads. Next, we obtained the RT stop location for each read, and we annotated the RT stop to the transcripts. The transcript annotation file was obtained from Gencode (Version 25). Next, we obtained significantly cross-linked sites in CCTT regions according to the following steps: (i) irCLIP RT-stops within CCTT were extended ±25 nt surrounding the regions; (ii) the height of each nucleotide position was the number of reads overlapping that position; (iii) the background frequency was computed after randomly placing the same number of extended reads within CCTT for 100 iterations; (iv) the FDR was calculated for each height according to the method described previously (Yeo et al., 2009); (v) only loci with a height with an FDR < 0.05 were assigned as significantly cross-linked sites. Finally, we combined the two significantly cross-linked replicate irCLIP-seq regions together, and nucleotides 127-177 of CCTT were defined as the CENP-C-binding region.
SHAPE-MaP Data Analysis
ShapeMapper was used for processing the in-cell and cell-free SHAPE-MaP datasets separately. Only nucleotides with an uniquely mapped read-depth greater than 10 were retained for modification detection. The distribution of mutation rates in the SHAPE-modified sample should be distinctly greater than that in the denaturing control and no-reagent control sample. After obtaining the SHAPE reactivity information, DeltaShAPE was used to determine the significantly protein bound regions by comparing the difference between the in-cell and cell-free conditions. First, the absolute change in SHAPE reactivity was calculated for each nucleotide along the transcript of interest. The raw absolute difference was then smoothed by calculating the 50-nt sliding median. The regions were predicted as the significantly protein bound regions meeting the following three criteria: (i) a Z-factor for a nucleotide greater than zero, indicating that the 95% confidence intervals of the measurements in the two conditions do not overlap; (ii) a standard score greater than one standard deviation from the mean ΔSHAPE; and (iii) three of five nucleotides in a sliding window meeting both the Z-factor and standard score criteria. Finally, based on the SHAPE-MaP reactivity data, we predicted the RNA secondary structure using the RNAstructure web server with default parameters.
Statistical Analysis
Statistical comparisons between two groups were carried out using the unpaired Student’s t-test when a normal distribution could not be assumed. GraphPad Prism was used to analyze the statistical significance. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference.
Supplementary Material
Table S1. Primers and Probes Used in the Study, Related to Star Methods
Table S3. The Possible CENP-C Binding Regions of All ncRNAs According to irCLIP-seq, Related to Figure 5
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Bub1 | Abcam | Cat# ab227228 |
| Rabbit monoclonal anti-BubR1 | Abcam | Cat# ab254326 |
| Mouse monoclonal anti-CENP-A | Abcam | Cat# ab13939; RRID: AB_300766 |
| Mouse monoclonal anti-CENP-A | MBL | Cat# D115-3; RRID: AB_591074 |
| Rabbit monoclonal anti-CENP-C | Abcam | Cat# ab193666 |
| Guinea pig monoclonal anti-CENP-C | MBL | Cat# PD030; RRID: AB_10693556 |
| Mouse monoclonal anti-Flag | Sigma | Cat# F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-GAPDH | Abcam | Cat# ab37168; RRID: AB_732652 |
| Mouse monoclonal anti-GFP | BioVision | Cat# 3992-100; RRID: AB_1121019 |
| Normal rabbit IgG antibody | Cell Signaling Technology | Cat# 2729; RRID: AB_1031062 |
| Mouse monoclonal anti-β-actin | Abcam | Cat# ab8226; RRID: AB_306371 |
| CREST antiserum | kind gift from Li lab | N/A |
| Bacterial and Virus Strains | ||
| E. coli DH5α | Biomed | Cat# BC116 |
| E. coli DL21 (DE3) | Transgen | Cat# CD601-02 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SNAP-Cell® Block (bromothenylpteridine, BTP) | New England BioLabs | Cat# S9106S |
| SNAP-Cell® TMR-star (tetramethylrhodamine) | New England BioLabs | Cat# S9105S |
| 4’-aminomethyltrioxsalen (AMT) | Sigma | Cat# A4330 |
| 6 x His-CENP-C | This paper | N/A |
| Critical Commercial Assays | ||
| Chemiluminescent Nucleic Acid Detection Module Kit | Thermo | Cat# 89880 |
| DIG Northern Starter Kit | Roche | Cat# 12039672910 |
| FirstChoice RLM-RACE RNA Ligase Mediated RACE Kit | Ambion | Cat# AM1700 |
| GeneArt™ Precision gRNA Synthesis Kit | Thermo | Cat# A29377 |
| His-Tag Protein Purification Kit | Qiagen | Cat# 31314 |
| PARIS Kit | Life Technologies | Cat# AM1921 |
| Deposited Data | ||
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| CCTT AMT-ChIRP-seq data | This paper | GEO: GSE149534 |
| CCTT irCLIP-seq data | This paper | GEO: GSE149534 |
| CCTT SHAPE-MaP data | This paper | GEO: GSE149534 |
| CENP-C RIP-seq data | This paper | GEO: GSE149534 |
| Experimental Models: Cell Lines | ||
| HCT116 | ATCC | Cat# CCL-247 |
| HeLa | ATCC | Cat# CCL-2 |
| Oligonucleotides | ||
| PCR primers, siRNAs, ASOs, sgRNAs target sequences, and probes | This paper | See Table S1 and Star Methods |
| Recombinant DNA | ||
| CENP-CSNAP | gift from Li lab | N/A |
| CMV10-Flag-CENP-C full length | This paper | N/A |
| CMV10-Flag-CENP-C 1-292 aa | This paper | N/A |
| CMV10-Flag-CENP-C 295-551 aa | This paper | N/A |
| CMV10-Flag-CENP-C 552-825 aa | This paper | N/A |
| CMV10-Flag-CENP-C 826-943 aa | This paper | N/A |
| CMV10-Flag-CENP-C 1-825 aa | This paper | N/A |
| pcDNA3.1-CCTT full length | This paper | N/A |
| pcDNA3.1-CCTT ORF1-GFP | This paper | N/A |
| pcDNA3.1-CCTT ORF2-GFP | This paper | N/A |
| pcDNA3.1-CCTT ΔAlu | This paper | N/A |
| pcDNA3.1-CCTT Δ43-79 (ΔDBD) | This paper | N/A |
| pcDNA3.1-CCTT Δ127-177 | This paper | N/A |
| pcDNA3.1-GFP | This paper | N/A |
| pET-28a-CENP-C full length | This paper | N/A |
| pLenti-U6-spgRNA v2.0-CMV-Puro-P2A-3Flag-s pCas9 | Obio | Cat# H7548 |
| Software and Algorithms | ||
| BEDTools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtiebio.sourceforge.net/bowtie2/indexs.html |
| BWA-MEM | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| CNCI | Sun et al., 2013 | http://www.bioinfo.org/software/cnci/ |
| CPAT | Wang et al., 2013 | http://rna-cpat.sourceforge.net/ |
| CPC | Kong et al., 2007 | http://cpc.cbi.pku.edu.cn |
| DeepTools | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| deltaSHAPE | Smola et al., 2015 | https://weekslab.com/software/ |
| DNA/RNA Copy Number Calculator | N/A | http://endmemo.com/bio/dnacopynum.php/ |
| edgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Fastq-dump | N/A | https://ncbi.github.io/sra-tools/fastq-dump.html |
| FASTX-Toolkit | N/A | http://hannonlab.cshl.edu/fastx_toolkit/ |
| FeatureCounts | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| GraphPad Prism | GraphPad Software | http://www.graphpad.com/scientifificsoftware/prism/ |
| IMARIS | Bitplane | N/A |
| Limma | Ritchie et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/limma.html |
| MEME | Bailey et al., 2009 | http://meme-suite.org |
| PEAR | Zhang et al., 2013 | https://cme.h-its.org/exelixis/web/software/pear/ |
| Pfam | El-Gebali et al., 2019 | http://pfam.xfam.org |
| PhyloSCF | Lin et al., 2011 | https://github.com/mlin/PhyloCSF/ |
| Picard | N/A | http://broadinstitute.github.io/picard/ |
| RNAstructure web server | DiChiacchio and Mathews, 2016 | https://rna.urmc.rochester.edu/RNAstructureWeb/ |
| ShapeMapper | Busan and Weeks, 2018 | https://help.rc.ufl.edu/doc/ShapeMapper |
| SICER | Xu et al., 2014 | http://gensoft.pasteur.fr/docs/SICER/1.1/SICER-README.pdf |
| SortMeRNA | Kopylova et al., 2012 | http://bioinfo.lifl.fr/RNA/sortmerna/ |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| TDF | Kuo et al., 2019 | https://www.regulatory-genomics.org/tdf/basic-introduction/ |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Triplexator | Buske et al., 2012 | http://abacus.qfab.org/tools/triplexator/manual.html |
| UCSC | N/A | http://genome.ucsc.edu/ |
| Volocity Demo (x 64) | Volocity Software | N/A |
HIGHLIGHTS.
Chromosome 17-derived lncRNA, CCTT, specifically localizes on all centromeres
CCTT interacts with CENP-C to facilitate its recruitment to centromeres
CCTT targets centromeres likely via the formation of RNA-DNA triplex on cenDNA
Depletion of CCTT induces severe mitotic defects and aneuploidy
ACKNOWLEDGMENTS
We thank A. Desai, K. D. Corbett and M. F. Liu for helpful discussions, Center for Big Data Research in Health (http://bigdata.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences for support with the data analysis and computing resource. This work was supported by grants from the Ministry of Science and Technology (2018YFA0801104) and the National Natural Science Foundation of China (81772952) to Y.T., the National Natural Science Foundation of China (31630093, 82030011) to X.Y., the National Key R&D Program of China (2018YFA0106901) to R.C., and NIH grants HG004659 to Y.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures and 3 tables.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue), W202–208. 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal PA, and Dervan PB. (1991). Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science 251, 1360–1363. 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Black BE, and Cleveland DW (2011). Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144, 471–479. 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD (2016). Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep. 15, 1624–1633. 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Rodríguez MG, Moreno N, and Jansen LE (2012). Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr. Protoc. Cell Biol Chapter 8, Unit8.8. 10.1002/0471143030.cb0808s55. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busan S, and Weeks KM (2018). Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148. 10.1261/rna.061945.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, Bauer DC, Mattick JS, and Bailey TL (2012). Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 22, 1372–1381. 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, and Straight AF (2010). Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol 189, 1143–1155. 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catasti P, Gupta G, Garcia AE, Ratliff R, Hong L, Yau P, Moyzis RK, and Bradbury EM (1994). Unusual structures of the tandem repetitive DNA sequences located at human centromeres. Biochemistry 33, 3819–3830. 10.1021/bi00179a005. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, and Chang HY (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD (2017). Molecular Mechanisms of Spindle Assembly Checkpoint Activation and Silencing. Prog. Mol. Subcell Biol 56, 429–455. 10.1007/978-3-319-58592-5_18. [DOI] [PubMed] [Google Scholar]
- DiChiacchio L, and Mathews DH (2016). Predicting RNA-RNA Interactions Using RNAstructure. Methods Mol. Biol 1490, 51–62. 10.1007/978-1-4939-6433-8_4. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Topp CN, and Dawe RK (2010). DNA Binding of Centromere Protein C (CENPC) Is Stabilized by Single-Stranded RNA. PLoS Genet. 6, e1000835. 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudé C, François JC, Sun JS, Ott G, Sprinzl M, Garestier T, and Hélène C (1993). Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 21, 5547–5553. 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. (2013). A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol 15, 1056–1066. 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, and Cleveland DW (2015). DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev. Cell 33, 314–327. 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. (2015). CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703. 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, and Francastel C (2009). Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37, 5071–5080. 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, and Earnshaw WC (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavís M, Méndez-Lago M, Gabelica V, Whitehead SL, González C, and Villasante A (2015). The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep 5, 13307. 10.1038/srep13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, et al. (2017). Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun 8, 15775. 10.1038/ncomms15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, and Straight AF (2011). In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–358. 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Wang D, Wu S, Li X, Shao C, Zhang P, Chen JY, Lim DH, Fu XD, Chen R, et al. (2020). Active retrotransposons help maintain pericentromeric heterochromatin required for faithful cell division. Genome Res. 30, 1570–1582. 10.1101/gr.256131.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, and Diekmann S (2008). Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 180, 1101–1114. 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Hervé S, Cleveland DW, and Fachinetti D (2016). CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell Rep. 17, 2394–2404. 10.1016/j.celrep.2016.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Emerman AB, Schweidenback CTH, and Blower MD (2014). RNA Stimulates Aurora B Kinase Activity During Mitosis. PLoS One 9, e100748. 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, and Cleveland DW (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol 176, 795–805. 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalwa M, Hänzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, et al. (2016). The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 44, 10631–10643. 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, and Bai Y (2013). A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113. 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, and Musacchio A (2015). CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol 210, 11–22. 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding HM, Butty VL, Torrey L, Haas S, et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, and Gao G (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35(Web Server issue), W345–349. 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noe L, and Touzet H (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Hänzelmann S, Cetin NS, Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt I, et al. (2019). Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 47, e32. 10.1093/nar/gkz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou E, and Heun P (2018). High-resolution mapping of centromeric protein association using APEX-chromatin fibers. Epigenetics Chromatin 11, https://doi.org/68. 10.1186/s13072-018-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, and Almouzni G (2014). Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53, 631–644. 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, and Fu XD (2019). Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat. Rev. Genet 20, 503–519. 10.1038/s41576-019-0135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Syed J, and Sugiyama H (2016). RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol 23, 1325–1333. 10.1016/j.chembiol.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, and Kellis M (2011). PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282. 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YH, and Yuen KWY (2019). Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. U S A 116, 6270–6279. 10.1073/pnas.1821384116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qu Q, Warrington R, Rice A, Cheng N, and Yu H (2015). Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol. Cell 59, 426–436. 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, and Black BE (2019). Human Artificial Chromosomes that Bypass Centromeric DNA. Cell 178, 624–639.e19. 10.1016/j.cell.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, and Cheeseman IM (2015). The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell 60, 886–898. 10.1016/j.molcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, and Cheeseman IM (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol 17, 16–29. 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SM, Sullivan LL, and Sullivan BA (2017). Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Dev. Cell 42, 226–240.e6. 10.1016/j.devcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. (2013). Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10. 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, and Kent WJ (2014). Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24, 697–707. 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al. (2015). MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun 6, 7743. 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, and Okazaki T (1992). Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol 116, 585–596. 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal H, Hori T, Furukawa A, Sugase K, Osakabe A, Kurumizaka H, and Fukagawa T (2015). Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 26, 3768–3776. 10.1091/mbc.E15-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechemia-Arbely Y, Miga KH, Shoshani O, Aslanian A, McMahon MA, Lee AY, Fachinetti D, Yates JI 3rd, Ren B, and Cleveland DW (2019). DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat. Cell Biol 21, 743–754. 10.1038/S41556-019-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, and Sullivan KF (2011). Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 9, e1001082. 10.1371/journal.pbio.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quénet D, and Dalal Y (2014). A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 3, e03254. https://doi.org/e41593. 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, and Dalal Y (2018). Correction: A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 7, 10.7554/eLife.41593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rošić S, Köhler F, and Erhardt S (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol 207, 335–349. 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CK, Hsieh TS, Wang JC, and Hearst JE (1977). Photochemical cross-linking of DNA-RNA helices by psoralen derivatives. J. Mol. Biol 116, 661–679. 10.1016/0022-2836(77)90265-0. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, and Weeks KM (2014). RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, and Weeks KM (2015). Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry 54, 6867–6875. 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, and Weeks KM (2015). Selective 2'-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 10, 1643–1669. 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, and Sullivan BA (2020). Genomic and functional variation of human centromeres. Exp. Cell Res 6, 111896. 10.1016/j.yexcr.2020.111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, and Zhao Y (2013). Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41, e166. 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, and Henikoff S (2018). Transcribing Centromeres: Noncoding RNAs and Kinetochore Assembly. Trends Genet. 34, 587–599. 10.1016/j.tig.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, and Chang HY (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, and Salmon ED (2012). The KMN protein network--chief conductors of the kinetochore orchestra. J. Cell Sci 125, 5927–5936. 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, and Chang HY (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, and Li W (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74. 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Li L, Xue X, Xie H, Shi H, and Hu Y (2020). A lncRNA coordinates with Ezh2 to inhibit HIF-1α transcription and suppress cancer cell adaption to hypoxia. Oncogene 39, 1860–1874. 10.1038/s41388-019-1123-9. [DOI] [PubMed] [Google Scholar]
- Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischböck J, Pentakota S, Keller J, Pesenti ME, Pan D, et al. (2016). Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 537, 249–253. 10.1038/nature19333. [DOI] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. (2007).Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17, 1146–1160. 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, and Chen LL (2017). SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 169, 664–678. 10.1016/j.cell.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Xu SLY, Grullon S, Ge K, and Peng WQ (2014). Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol 1150, 97–111. 10.1007/978-1-4939-0512-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YC, Zhou Y, Wu TB, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. (2009). Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36, 996–1006. 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Yang J, Zhang Z, McLaughlin SH, Chang L, Fasci D, Ehrenhofer-Murray AE, Heck AJR, and Barford D (2019). Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282. 10.1038/S41586-019-1609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, and Gage FH (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol 16, 130–137. 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, and Peng W (2009). A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958. 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, and Khavari PA (2016). irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 13, 489–492. 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Kobert K, Flouri T, and Stamatakis A (2013). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu SS, Yang L, and Chen LL (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers and Probes Used in the Study, Related to Star Methods
Table S3. The Possible CENP-C Binding Regions of All ncRNAs According to irCLIP-seq, Related to Figure 5
Data Availability Statement
The accession number for RIP-seq, AMT-ChIRP-seq, irCLIP-seq, and SHAPE-MaP from HeLa cells is GEO: GSE149534.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.