Loss of the dNTPase and DNA repair enzyme SAMHD1 is associated with cancer and causes systemic autoimmunity. We show transformation-promoting spontaneous DNA damage and MDA5-driven, but cGAS/STING-dependent, chronic type I interferon production in SAMHD1-deficient mice.
Abstract
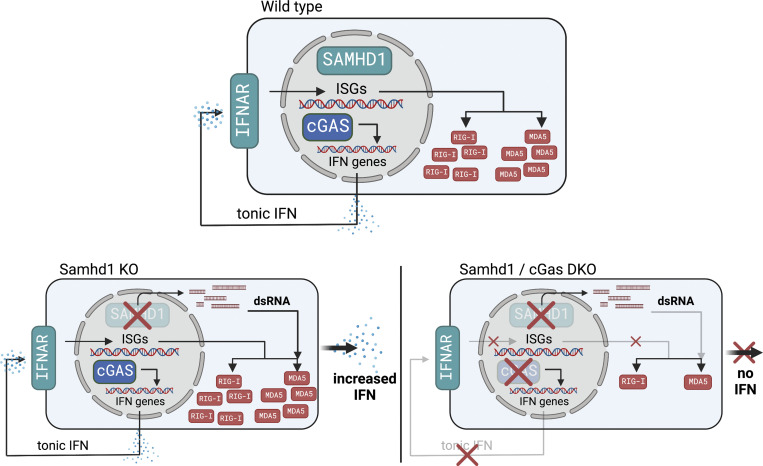
Defects in nucleic acid metabolizing enzymes can lead to spontaneous but selective activation of either cGAS/STING or RIG-like receptor (RLR) signaling, causing type I interferon–driven inflammatory diseases. In these pathophysiological conditions, activation of the DNA sensor cGAS and IFN production are linked to spontaneous DNA damage. Physiological, or tonic, IFN signaling on the other hand is essential to functionally prime nucleic acid sensing pathways. Here, we show that low-level chronic DNA damage in mice lacking the Aicardi-Goutières syndrome gene SAMHD1 reduced tumor-free survival when crossed to a p53-deficient, but not to a DNA mismatch repair-deficient background. Increased DNA damage did not result in higher levels of type I interferon. Instead, we found that the chronic interferon response in SAMHD1-deficient mice was driven by the MDA5/MAVS pathway but required functional priming through the cGAS/STING pathway. Our work positions cGAS/STING upstream of tonic IFN signaling in Samhd1-deficient mice and highlights an important role of the pathway in physiological and pathophysiological innate immune priming.
Graphical Abstract
Introduction
Intracellular recognition of nucleic acids is essential in antiviral and in antitumor immunity, but uncontrolled activation of this machinery in the context of severe viral infections and tissue damage can cause detrimental inflammation. The same potent and therefore dangerous response is initiated when cells fail to control the emergence of endogenous nucleic acids in amounts that exceed normal physiological levels or that lack secondary modifications, which would prevent autoactivation of nucleic acid receptors such as the double-stranded DNA (dsDNA) sensor cyclic GMP-AMP synthase (cGAS) or the retinoic acid-inducible gene (RIG)–like dsRNA receptors (RLRs) RIG-I, MDA5, and LGP2 (Ablasser and Hur, 2020). RIG-I senses blunt-end dsRNA based on the presence or absence of 5′-modifications, while MDA5 requires dsRNA stem structures of so far unknown minimal length (Ablasser and Hur, 2020). The enzyme cGAS senses nucleosome-free dsDNA inside of cells and produces the second messenger 2′3′cGAMP, a direct ligand for the cyclic-dinucleotide sensor stimulator of IFN genes (STING; de Oliveira Mann and Hopfner, 2021). Nucleosome-free dsDNA accumulates in cells after DNA damage, and therefore DNA damage has been identified as a primary pathogenic event in an increasing number of sterile inflammatory conditions that are driven by cGAS/STING-dependent cytokine production (Crow and Stetson, 2022). Both activated STING and mitochondrial antiviral signaling (MAVS) recruit the kinase TBK1 leading to downstream activation of type I IFN and NF-κB responses. IFN in turn acts through autocrine and paracrine signaling to stimulate expression of primarily antiviral genes via the type I IFN receptor (IFNAR). In many, but not in all cell types, expression of several pattern recognition receptors, including cGAS and the RLRs, is regulated via this positive-feedback loop and depends on tonic IFN signaling, which leads to severely reduced pattern recognition receptor (PRR) levels in IFNAR-deficient cells compared with IFNAR-competent cells (Behrendt et al., 2013; Schaupp et al., 2020). This results in a broad antiviral immune-defect in mice (Cervantes-Barragán et al., 2009; Schaupp et al., 2020) and humans with inborn errors of the IFN system (Zhang et al., 2020). How exactly tonic IFN signaling is established and how this impacts physiological and pathophysiological immune priming is an emerging topic.
Aicardi-Goutières syndrome (AGS) is a monogenic systemic autoimmune disease that is associated with high levels of IFN in peripheral blood and cerebrospinal fluid (Rodero and Crow, 2016). Mutations in AGS genes lead to spontaneous but selective activation of either RLR or cGAS/STING signaling (Crow and Stetson, 2022). The latter pathway has been implicated in the pathogenesis of AGS with underlying defects in the gene SAMHD1 (AGS5; Rice et al., 2009; Maelfait et al., 2016; Daddacha et al., 2017; Coquel et al., 2018). SAMHD1 has at least two different functions: It is an enzyme with deoxynucleoside triphosphate triphosphohydrolase (dNTPase) activity (Goldstone et al., 2011). Through this activity, SAMHD1 limits the availability of dNTPs in resting cells, which hinders replication of pathogens like retroviruses that depend on cellular dNTP supply (Hrecka et al., 2011; Laguette et al., 2011). Furthermore, increased levels of SAMHD1 in relapsed hematopoietic tumors have been shown to degrade nucleotide analogs, thereby diminishing the efficacy of chemotherapy (Schneider et al., 2017; Herold et al., 2017). Moreover, SAMHD1-deficient tumor cells can be selectively killed by targeting the nucleotide metabolism, (Davenne et al., 2020) making it an attractive anticancer drug target. A second function of SAMHD1 is reflected by the recruitment of the enzyme to sites of DNA double-strand breaks (DSB) and stalled replication forks. There, it interacts with the endonuclease CtIP and with the MRE11–RAD50–NBS1 (MRN) complex to facilitate end resection in preparation for DSB repair by homologous recombination and to enable fork-restart (Daddacha et al., 2017; Coquel et al., 2018). The latter function does not require dNTPase activity and is mediated by the C-terminal region in SAMHD1. Failure in recruiting the DNA repair machinery by SAMHD1 results in spontaneous DNA damage and in the release of self-DNA that has been suggested to activate cGAS (Daddacha et al., 2017; Coquel et al., 2018). In agreement with its functions in DNA repair, SAMHD1-deficient patient fibroblasts showed a spontaneous transcriptional signature of IFN-stimulated genes (ISGs) and increased numbers of DNA double-strand breaks (Kretschmer et al., 2015). The latter caused a chronic activation of the p53 pathway and senescence (Kretschmer et al., 2015). Impaired DNA repair predisposes to malignant transformation, and consequently, mutations in SAMHD1 have been identified in many different tumors (Clifford et al., 2014; Rentoft et al., 2016). However, mutations found in cancer cells scatter across the whole SAMHD1 gene (reviewed by Mauney and Hollis, 2018) and do not allow for a general conclusion about a definitive mechanism that could explain how the protein prevents malignant transformation. Here, two scenarios seem plausible: loss of SAMHD1 dNTPase activity could affect the composition of cellular dNTP pools, which has a direct effect on the fidelity of replicative polymerases and could cause a mutator phenotype. This has been widely studied in cancers originating from deregulation of the ribonucleotide reductase complex (Aye et al., 2015). On the other hand, loss of SAMHD1-mediated DNA repair activity could cause increased numbers of DSBs and delay their repair, which might promote the selection of cell clones that inactivated cell cycle checkpoints to overcome this block. However, none of these scenarios have been experimentally addressed using in vivo models.
In addition to these established functions of SAMHD1, one group reported an exoribonuclease activity of the protein (Choi et al., 2015; Ryoo et al., 2014; Ryoo et al., 2016). However, RNAse activity of SAMHD1 was not reproduced by other studies (Seamon et al., 2015; Antonucci et al., 2016; Bloch et al., 2017; Yu et al., 2021), including our own (Wittmann et al., 2015). Therefore, if and how SAMHD1 regulates RNA metabolism in cells remains to be fully elucidated.
In contrast to patients, loss of SAMHD1 in mice caused a mild activation of the type I IFN system but not systemic autoimmunity (Behrendt et al., 2013; Rehwinkel et al., 2013; Thientosapol et al., 2018). The IFN response was shown to be meditated via the cGAS/STING pathway (Maelfait et al., 2016). Furthermore, as opposed to studies in human cells lacking SAMHD1, no spontaneous DNA damage and no increased frequency of spontaneous tumors have been described in three independently generated SAMHD1-deficient mouse strains (Behrendt et al., 2013; Rehwinkel et al., 2013; Thientosapol et al., 2018). The lack of detectable DNA damage but the presence of a spontaneous IFN response in these mutant mice remained an unresolved incoherence with the current understanding of how IFN is induced in SAMHD1-deficient cells.
Here, we show low-level chronic DNA damage in SAMHD1-deficient mice that is detected by the p53 pathway. We found that inactivation of SAMHD1 in p53-deficient mice, but not in mice with defective DNA mismatch repair, reduced tumor-free survival. Surprisingly, increased DNA damage did not amplify the spontaneous IFN response in SAMHD1-deficient mice. In contrast, we found that IFN is induced via the RNA sensor MDA5 but not via RIG-I. Using SAMHD1-deficient mice as a model, we show that innate immune sensing of endogenous RNA through the RLR pathway requires functional priming via the cGAS/STING pathway.
Results
Low-level chronic DNA damage in SAMHD1-deficient mice
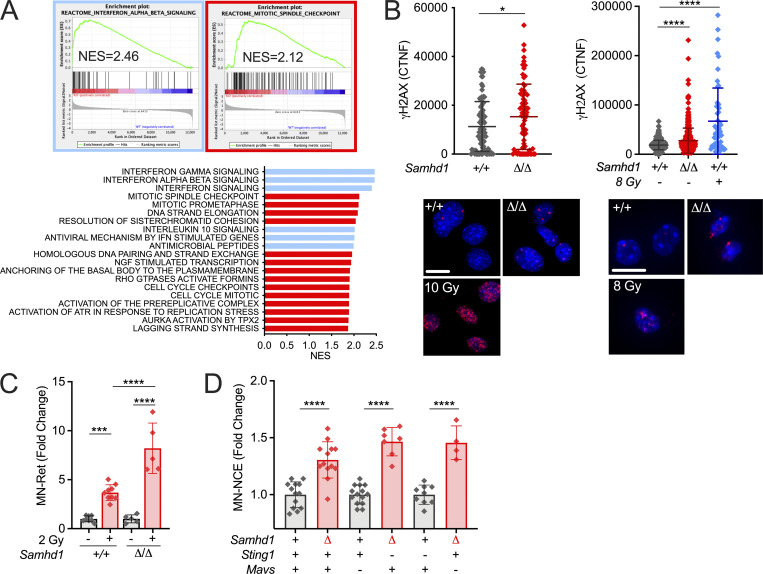
We and others previously reported a mild spontaneous IFN response in Samhd1 KO mice, which was dependent on the cGAS/STING pathway, suggesting that it was triggered by endogenous DNA (Behrendt et al., 2013; Maelfait et al., 2016). So far, however, there were no reports about spontaneous DNA damage in SAMHD1-deficient mice, which led us to ask if IFN in these mice is induced by an alternative mechanism to the human or if evidence of spontaneous DNA damage has been overlooked. Indeed, gene set enrichment analysis (GSEA) of whole transcriptome data from peritoneal macrophages revealed that only two types of pathways reflecting an ongoing inflammatory response, including type I IFN, and replication stress were enriched in SAMHD1-deficient macrophages over control macrophages (Fig. 1 A). This is in line with previous reports about human cells and suggests that also mouse SAMHD1 acts on stalled replication forks and in DNA repair (Daddacha et al., 2017; Coquel et al., 2018). Furthermore, nuclei of primary SAMHD1-deficient mouse embryonic fibroblasts (MEFs) and peritoneal macrophages showed slightly elevated levels of γH2AX, a genuine marker for DNA strand breaks, when compared with littermate control cells (Fig. 1 B). Of note, in MEFs, this difference equilibrated after passage four and was no longer detectable in clones that overcame replicative senescence (not shown). DNA damage in erythroid precursors results in DNA DSBs and the rapid emergence of micronucleated reticulocytes followed by an increase of micronucleated erythrocytes, which can act as a short- and long-term memory of genotoxic insults, respectively. After sublethal whole-body irradiation, frequencies of micronucleated reticulocytes were increased by fourfold in irradiated vs. nonirradiated Samhd1+/+ mice (Fig. 1 C). This increase was doubled in Samhd1Δ/Δ mice, indicating higher susceptibility of SAMHD1-deficient mice to genotoxic stress (Fig. 1 C). Next, we compared the steady-state frequencies of micronucleated erythrocytes in the peripheral blood of several SAMHD1-deficient mouse strains in our colony. Compared with the respective SAMHD1-proficient control of the same mutant strain, animals that lacked SAMHD1 consistently showed higher frequencies of micronucleated erythrocytes in peripheral blood, indicative of low-level chronic spontaneous DNA damage in these mice (Fig. 1 D). Taken together, our results suggest that in SAMHD1-deficient mice, genome replication and DNA repair are impaired resulting in low levels of chronic DNA damage.
Figure 1.
Low-level chronic DNA damage in SAMHD1-deficient mice. (A) GSEA against the Reactome gene set collection (MSigDB) showing that exclusively gene sets of immune pathways (blue) and DNA replication (red) are enriched in Samhd1Δ/Δ vs. Samhd1+/+ peritoneal macrophages. NES, normalized enrichment score. (B) CTNF of the γH2AX signal in pre-senescent primary MEFs and CD11b+F4/80HI peritoneal macrophages of the indicated genotypes (Student’s t test) and representative immunofluorescence pictures. Scale bars = 10 µM; 10 and 8 Gy = positive controls. (C) Change in micronucleated reticulocytes (MN-Ret) before (−) and 48 h after (+) whole body γ-irradiation with a dose of 2 Gy in Samhd1+/+ (n = 8) and Samhd1Δ/Δ (n = 5) mice. Fold change compared with the mean of Samhd1+/+ before irradiation is shown (one-way ANOVA followed by Tukey’s multiple comparison test). (D) Relative change in MN-NCEs from peripheral blood of mice with the indicated genotypes. Fold change was calculated for each genetic background between Samhd1+/+ (+) and Samhd1Δ/Δ (Δ). For Mavs and Sting1: + = WT/WT, − = KO/KO, n ≥ 4 for each group (Student’s t test). * = P < 0.05, *** = P < 0.001, **** = P < 0.0001.
Loss of SAMHD1 reduces tumor-free survival of mice lacking p53, but not of mice with defective DNA mismatch repair
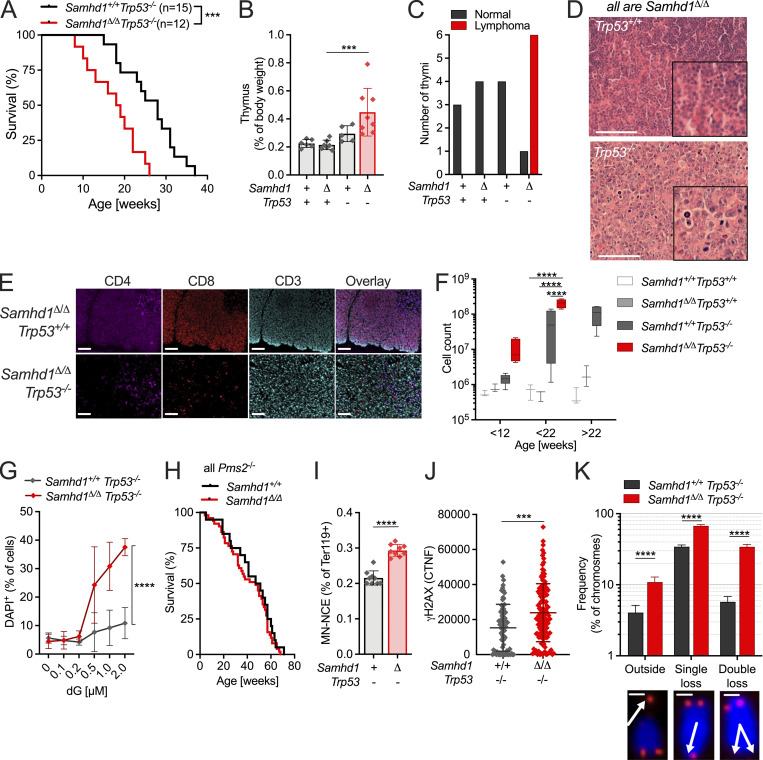
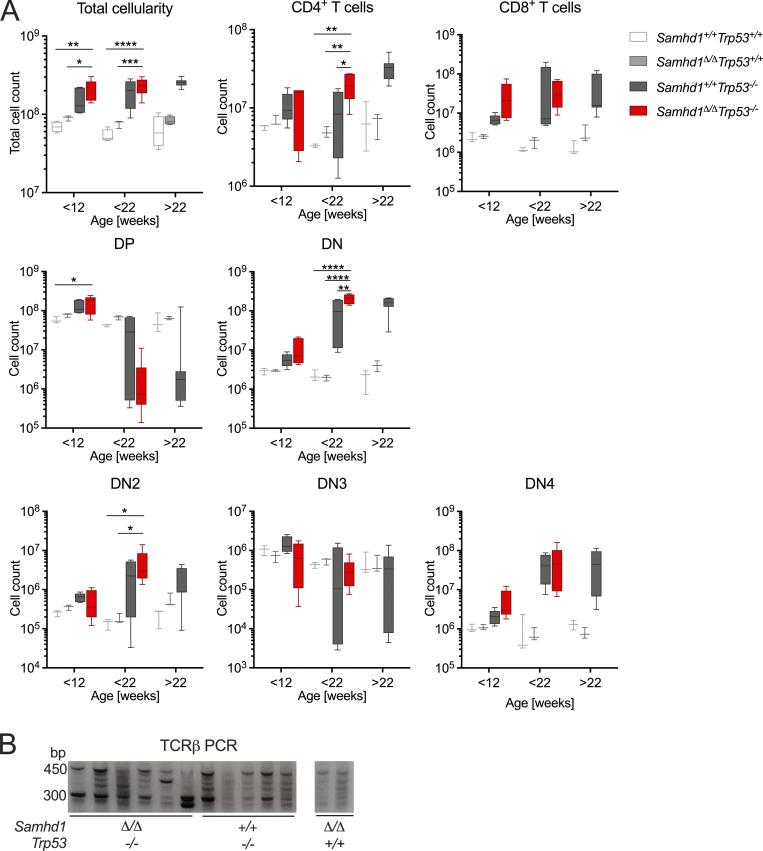
Next, we asked why SAMHD1-deficient mice did not show an increased frequency of spontaneous tumor formation in spite of spontaneous DNA damage in various cell types. Patient fibroblasts lacking SAMHD1 activate the p53 pathway in response to DNA damage (Kretschmer et al., 2015). We reasoned that the low-level DNA damage in Samhd1Δ/Δ mice can be kept in check by p53-mediated damage responses and that inactivation of the p53 pathway might reveal how loss of SAMHD1 impacts genome stability in vivo. To address this question, we crossed SAMHD1-deficient mice to Trp53−/− mice, which predominantly develop spontaneous thymic lymphoma (Jacks et al., 1994). In our colony, Trp53−/− mice showed a mean tumor-free survival of 28 wk, which was reduced to 18 wk in Samhd1Δ/ΔTrp53−/− mice (Fig. 2 A). In a cohort of Trp53−/− mice sacrificed at 12 wk of age, we found slightly enlarged thymi compared with control mice. At the same age, Samhd1Δ/ΔTrp53−/− thymi were already significantly larger than thymi of the Trp53−/− group (Fig. 2 B). Histopathologic examination of thymic sections from this cohort revealed that in five out of six Samhd1Δ/ΔTrp53−/− mice, the disease had already progressed to thymic lymphoma, while at that time point no lymphoma cells were identified in thymus sections from Trp53−/− mice (Fig. 2, C and D). Subsequent immunophenotypic analysis by multicolor immunohistochemistry demonstrated that CD4−CD8− double negative CD3+ T cells were the dominant population in thymi of Samhd1Δ/ΔTrp53−/− (Fig. 2 E) mice, and this population emerged in Samhd1Δ/ΔTrp53−/− mice before 12 wk of age, most evident in the CD25− T cell subsets DN1 (Fig. 2 F) and DN4 (Fig. S1 A). Longitudinal quantification of T cell development further supported that the disease of Trp53−/− mice develops faster but not qualitatively different in the absence of SAMHD1 (Fig. S1 A). PCR on total thymus DNA amplifying specific recombination events in the TCRβ genes (Martins et al., 2014) demonstrated T cell bi- or oligoclonality indicative of thymic T cell lymphoma (Fig. S1 B). Our data thus show that additional loss of SAMHD1 accelerated malignant transformation in Trp53−/− mice most likely by enhancing DNA damage.
Figure 2.
SAMHD1 prevents spontaneous DNA DSBs and accelerated transformation in p53-deficient mice. (A) Tumor-free survival of Samhd1+/+Trp53−/− (n = 15) and Samhd1Δ/ΔTrp53−/− (n = 12) mice (log-rank test). (B and C) + = WT/WT, − = KO/KO, Δ = Δ/Δ. (B) Relative thymus weight of mice with the indicated genotypes at 12 wk of age, n ≥ 5 per group (one-way ANOVA followed by Sidak’s multiple comparison test). (C) Thymi of 12-wk-old mice with the indicated genotypes were examined for lymphoma formation by a trained histopathologist. Numbers of analyzed thymi in each group are shown and categorized according to the disease state. (D) Representative sections of a normal Samhd1Δ/ΔTrp53+/+ (upper) and a Samhd1Δ/ΔTrp53−/− lymphoma-bearing thymus. Sections were stained with H&E. Scale bar = 100 µm. Insets were created with digital zooming. (E) Representative multicolor immunohistochemistry staining for T cell lineage markers of thymic sections from mice with the indicated genotypes. Scale bar = 100 µm. (F) Cell counts of CD4−CD8−CD44+CD25− DN1 immature T cells over time in the thymus of mice with the indicated genotypes. Complete dataset of T cell development in Fig. S2, n ≥ 3 for each group and time point (two-way ANOVA followed by Tukey’s multiple comparison test). (G) Survival of Samhd1Δ/ΔTrp53−/− (n = 3) and of Samhd1+/+Trp53−/− (n = 3) immortalized thymic fibroblasts after treatment for 48 h with dG at the indicated concentrations. Representative of two independent experiments is shown (two-way ANOVA). (H) Tumor-free survival of Samhd1Δ/ΔPms2−/− (n = 51) and of Samhd1+/+Pms2−/− mice (n = 20). (I) Frequency of MN-NCEs in peripheral blood of Samhd1Δ/ΔTrp53−/− (n = 9) and of Samhd1+/+Trp53−/− (n = 9; Student’s t test). (J) CTNF of the γH2AX signal in presenescent primary MEFs from Samhd1Δ/ΔTrp53−/− and Samhd1+/+Trp53−/− mice. Representative result of two independent experiments is shown (Student’s t test). (K) Telomer integrity was quantified by FISH in 20 metaphases of immortalized thymic fibroblasts from Samhd1Δ/ΔTrp53−/− and from Samhd1+/+Trp53−/− mice (Student’s t test). Scale bar = 5 µm. *** = P < 0.001, **** = P < 0.0001.
Figure S1.
Aberrant T cell development in Samhd1Δ/ΔTrp53−/− and in Samhd1+/+Trp53−/− mice. Related to Fig. 2. (A) Thymus parameters recorded by flow cytometry. Cells were gated based on scatter (exclude debris) and DAPI− for living cells, before gating on the respective markers. DP = CD4+CD8+, DN = CD4−CD8−, DN1 = CD4−CD8−CD44+CD25− (shown in Fig. 2 F), DN2 = CD4−CD8−CD44+CD25+, DN3 = CD4−CD8−CD44−CD25+, DN4 = CD4−CD8−CD44−CD25− (two-way ANOVA followed by Tukey’s multiple comparison test). (B) DNA was extracted from total thymus of mice with the indicated genotypes. TCRβ loci were amplified by PCR using a combination of 22 primers binding in a V segment combined with one primer binding in J1.7. Similar results were obtained with primer J2.7 (not shown). Strategy according to Martins et al. (2014). * = P < 0.05, ** = P < 0.001, *** = P < 0.001, **** = P < 0.0001. Source data are available for this figure: SourceData FS1.
We then asked if the altered nucleotide metabolism might contribute to malignant transformation in Samhd1Δ/ΔTrp53−/− mice. Due to altered dNTP levels, tumor cells lacking SAMHD1 can be selectively killed by 2′-deoxy-guanosin (dG; Davenne et al., 2020). Indeed, we observed that immortalized Samhd1Δ/ΔTrp53−/−, but not Trp53−/−, thymic fibroblasts were hypersensitive to dG treatment, confirming that aberrant nucleotide metabolism caused by deficiency for SAMHD1 in cancer cells is an attractive drug target (Fig. 2 G). Imbalanced dNTP levels decrease the fidelity of replicative polymerases and increase numbers of DNA mismatch mutations (Aye et al., 2015). In such a scenario, loss of SAMHD1 would be expected to reduce the tumor-free survival of DNA mismatch repair (MMR)–deficient mice. To test this hypothesis, we crossed Samhd1Δ/Δ mice to Pms2−/− mice, which lack functional MMR and develop spontaneous lymphoma (Baker et al., 1995). Surprisingly, and in contrast to our observations in Trp53−/− mice, loss of SAMHD1 did not significantly reduce the tumor-free survival of Pms2−/− mice (Fig 2 H; 50% mean survival Samhd1Δ/ΔPms2−/− 47 wk, Pms2−/− 49 wk, log-rank P = 0.4052). This suggested that loss of SAMHD1 in mice is not associated with a strong mutator phenotype and that accelerated transformation seen in Trp53−/− mice lacking SAMHD1 is mainly driven by other forms of DNA damage.
To better understand the molecular events leading to reduced survival of Samhd1Δ/ΔTrp53−/− mice, we quantified the frequency of micronucleated erythrocytes in peripheral blood and found higher frequencies in Samhd1Δ/ΔTrp53−/− mice compared with Trp53−/− mice (Fig. 2 I). In line with these observations, we detected a higher γH2AX signal in Samhd1Δ/ΔTrp53−/− versus Trp53−/− primary MEFs, further supporting overall increased spontaneous DNA damage inflicted by additional loss of SAMHD1 in Trp53−/− mice (Fig. 2 J). Independently of its role as a dNTPase, SAMHD1 recruits the MRN complex to sites of DSBs to promote homologous recombination and to restart stalled replication forks. Telomeres consist of repetitive DNA sequences and form R-loop structures, both of which can lead to replication fork stalling. In cells with a defective shelterin complex, which protects telomeres from being recognized by the DNA repair machinery, SAMHD1 has been shown to prevent telomere breakage and the formation of extrachromosomal (“outsider”) telomere signals (Majerska et al., 2018). Quantification of telomere integrity in transformed Samhd1Δ/ΔTrp53−/− versus Trp53−/− thymic fibroblasts by telomere fluorescence in situ hybridization (FISH) revealed higher frequencies of chromosomes displaying single or double telomere loss and the characteristic outsider telomere signals (Fig. 2 K). Our data suggest that loss of SAMHD1 in mice inflicts spontaneous DNA damage that is counteracted by a p53 response, and in the absence of p53 accelerates tumor development. Accelerated transformation is more likely to be the result of increased numbers of DSBs in “difficult-to-replicate” regions, like telomeres, rather than being caused by a pronounced mutator phenotype.
Inhibition of ataxia telangiectasia mutated (ATM) kinase but not loss of p53 or DNA mismatch repair amplified the spontaneous IFN response in SAMHD1-deficient cells
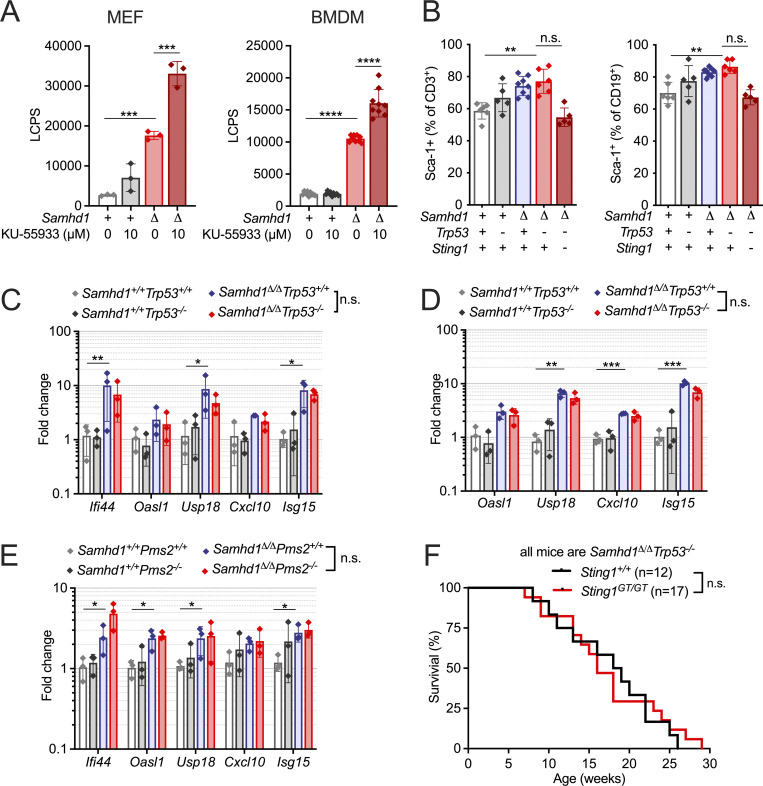
In eukaryotes, ATM and ataxia telangiectasia and Rad3-related protein (ATR) are the apex kinases of DNA damage signaling responding to DSBs and replication stress, respectively. Prolonged replications stress can promote the generation of DSBs, therefore activation of ATR and ATM can coincide. Spontaneous activation of ATR was implicated in limiting the IFN response in SAMHD1-deficient human cells (Coquel et al., 2018), but the role of ATM has not been addressed so far. We could not detect spontaneous phosphorylation of ATM in SAMHD1-deficient MEFs or bone marrow–derived macrophages (BMDMs; not shown). However, treatment with the ATM inhibitor KU-55933 blocked ATM phosphorylation after irradiation of Samhd1Δ/Δ MEFs (Fig. S2) and increased the IFN levels in the supernatant of SAMHD1-deficient cells but not of WT control cells (Fig. 3 A), suggesting that ATM limits the IFN response in cells lacking SAMHD1. ATM activation is followed by activation of p53 to halt the cell cycle and allow for DNA repair. Hence, we expected that loss of p53 might increase IFN response via cGAS/STING activation in response to DNA damage as previously observed by others and us in RNASEH2-deficient cells (Hiller et al., 2018; Giordano et al., 2022). While we measured increased levels of the surface ISG Sca-1 on peripheral blood lymphocytes (Fig. 3 B) and increased transcription of several ISGs in peripheral blood of Samhd1Δ/Δ vs. Samhd1+/+ control mice (Fig. 3 C), this response was not further increased by additional loss of p53 (Fig. 3, B and C). We made similar observations in BMDMs from Samhd1Δ/ΔTrp53−/− and control mice (Fig. 3 D). Of note, like in the p53 model, ISG transcription was similar between Samhd1Δ/Δ and Samhd1Δ/Δ Pms2−/− mice (Fig. 3 E). Cancer cell–intrinsic activation of the cGAS/STING pathway restricts tumor growth. This can be achieved by exogenous stimulation using synthetic ligands (reviewed in Demaria et al., 2019) or by promoting unphysiological accumulation of endogenous nucleic acids (Vanpouille-Box et al., 2019; Ishizuka et al., 2019). To investigate the role of endogenous DNA sensing in controlling tumor development in Samhd1Δ/ΔTrp53−/− mice, we crossed this mouseline to a STING-deficient background (Goldenticket mouse, Sting1GT/GT). While loss of STING abrogated the IFN response induced by loss of SAMHD1 (Fig. 3 B, last column), it had no effect on tumor-free survival of Samhd1Δ/ΔTrp53−/− mice (Fig. 3 F), suggesting that STING signaling is not crucial in controlling the growth of Samhd1Δ/ΔTrp53−/−-deficient tumors and further corroborating the absence of significant cGAS/STING activation in our model.
Figure S2.
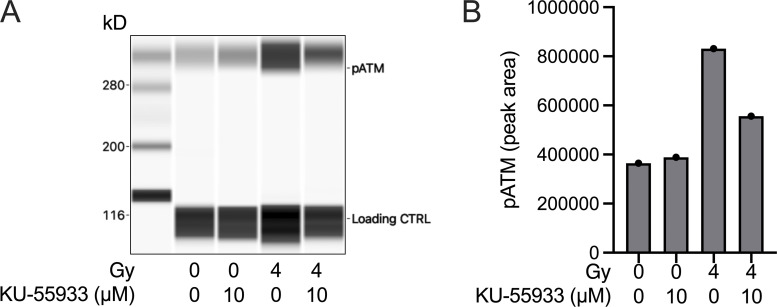
KU-55933 reduces ATM phosphorylation in Samhd1Δ/Δ MEFs after irradiation. Related to Fig. 3. Samhd1Δ/Δ MEFs were incubated with 10 µM KU-55933 or DMSO for 1 h and then irradiated with a dose of 4 Gy or left untreated. 1 h after irradiation cells were collected in RIPA buffer and subjected to capillary western analysis using a WES system (Protein Simple). (A) Virtual blot of the signals obtained in WES. The antibody produced an unspecific signal at 115 kD that was used as a loading control. (B) Quantification of the signal area for the pATM signal at 300 kD. Source data are available for this figure: SourceData FS2.
Figure 3.
Differential roles of ATM, p53, and STING in controlling tumor growth and ISG transcription in Samhd1Δ/Δ mice. (A) MEFs (n = 3 per group) and BMDMs (n = 3 per group, three independent measurements, pooled data shown) from Samhd1Δ/Δ and control mice were incubated with DMSO or the ATM inhibitor KU-55933 for 48 h. IFN bioactivity in the supernatant was quantified using LL171 ISG-LUC reporter cells. LCPS, light counts per second. Two-way ANOVA followed by Tukey’s multiple comparison test. (B) Frequency of DAPI−Sca-1+CD3+ T cells (left) and DAPI−Sca-1+CD19+ B cells (right) in peripheral blood of mice with the indicated genotypes (one-way ANOVA followed by Tukey’s multiple comparison test). (C–E) Relative transcript levels of ISGs in peripheral blood (C and E) and BMDMs (D) of mice with the indicated genotypes. Fold change compared to the mean of Samhd1+/+Trp53+/+ (C and D) or Samhd1+/+Pms2+/+ (E) are shown, n = 3 for each group in each experiment (multiple t tests were performed). (F) Tumor-free survival of Samhd1Δ/ΔTrp53−/− mice on STING-deficient (Sting1GT/GT) and STING-proficient (Sting1+/+) genetic backgrounds (log-rank test). * = P < 0.05, ** = P < 0.01, *** = P < 0.001,**** = P < 0.0001.
Although we observed that inhibition of ATM increased the spontaneous IFN production in SAMHD1-deficient cells, concomitant inactivation of Samhd1 and Trp53 in mice did not increase the ISG transcription above levels that were induced by the absence of SAMHD1 alone. In this context, STING signaling was irrelevant for the control of tumor growth. We conclude that the p53-dependent DNA damage response does not function to prevent the generation of IFN-inducing endogenous nucleic acids in SAMHD1-deficient mice.
MDA5 but not RIG-I drives IFN production in Samhd1Δ/Δ mice
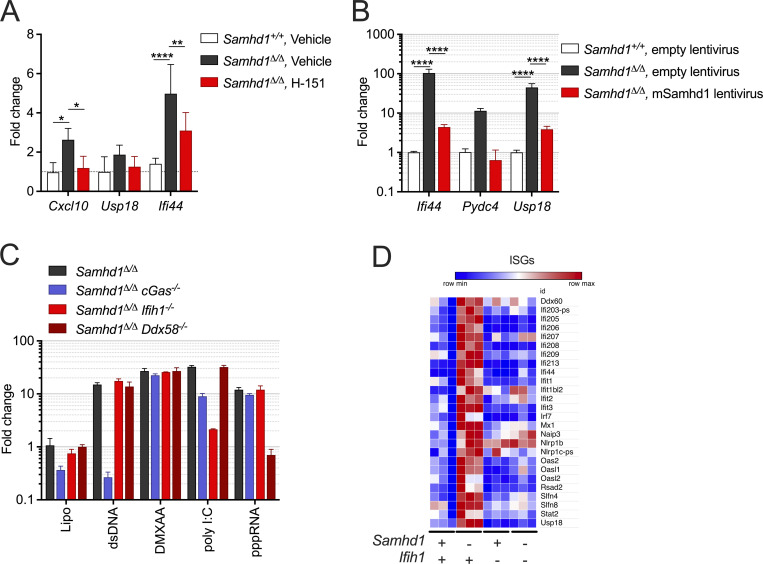
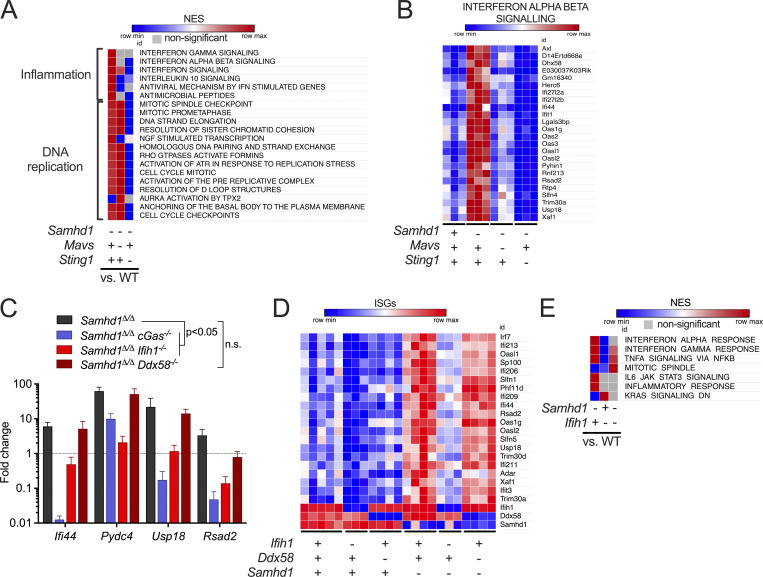
Our observation that increased DNA damage did not boost the IFN response in SAMHD1-deficient mice led us to investigate the exact role of cGAS/STING signaling in this model. To address whether STING signaling is important in SAMHD1-deficient mice, we treated Samhd1Δ/Δ mice for 2 wk with 10 mg/kg of the STING antagonist H-151 (Haag et al., 2018). Pharmacologic inhibition of STING was able to reduce the transcription of ISGs in peripheral blood, demonstrating that STING is required for spontaneous IFN production in SAMHD1-deficient mice and that it represents a valuable therapeutic target to treat inflammatory conditions ensuing from defects in SAMHD1 (Fig. S3 A). Until now, spontaneous IFN production caused by biallelic mutations in any of the AGS-related genes could be explained by activation of either the cGAS/STING pathway or the RLR pathway. To confirm that loss of SAMHD1 selectively activated the cGAS/STING pathway, we turned to a genetic approach to directly compare the relevance of intracellular DNA and RNA sensing in SAMHD1-deficient mice. As expected from our data with the STING inhibitor, KO of Sting1 completely blunted the ISG response in Samhd1Δ/Δ mice (Fig. 4, A and B). To our surprise, also loss of MAVS abrogated the ISG response in SAMHD1-deficient peritoneal macrophages. This suggested that in contrast to mutations in other AGS enzymes, both intact STING and MAVS signaling are required for the spontaneous IFN production in SAMHD1-deficient mice (Fig. 4, A and B). Next, we asked if the nucleic acid sensors upstream of MAVS and STING were chronically activated in SAMHD1-deficient cells or if there was a direct crosstalk between the two pathways at the level of STING and MAVS. To this end, we used post-replicative senescence Samhd1Δ/Δ MEFs that retained a spontaneous ISG response, which could be rescued by lentiviral expression of murine SAMHD1 (Fig. S3 B). In these MEFs, we inactivated cGAS, RIG-I (Ddx58), and MDA5 (Ifih1) using CRISPR/Cas9 (Fig. S3 C). As observed before (Maelfait et al., 2016), KO of cGas completely blunted the transcription of several ISGs in SAMHD1-deficient cells (Fig. 4 C). For the RIG-like receptors, only loss of MDA5, but not of RIG-I, was able to reduce the mRNA levels of the ISGs tested (Fig. 4 C). To further substantiate our finding that the spontaneous ISG response is indeed MDA5-dependent, we crossed Samhd1Δ/Δ mice to Ifih1−/− mice and to Ddx58−/− mice and analyzed the transcriptome of peritoneal macrophages. Like in previous experiments, Samhd1Δ/Δ peritoneal macrophages displayed elevated spontaneous transcription of ISGs, which was abrogated in peritoneal macrophages from Samhd1Δ/Δ Ifih1−/− mice but not in macrophages of Samhd1Δ/Δ Ddx58−/− mice (Fig. 4 D). The essential role of MDA5 in driving the spontaneous inflammatory response in Samhd1Δ/Δ mice was confirmed in another independent transcriptome analysis of peritoneal macrophages. Pathways related to IFN signaling, inflammation, and DNA replication stress were enriched in peritoneal macrophages from Samhd1Δ/Δ mice compared with control mice (Fig. 4 E and Fig. S3 D). In contrast, in peritoneal macrophages from Samhd1Δ/Δ Ifih1−/− mice, only pathways related to DNA replication and cell cycle progression remained enriched when compared with macrophages from littermate control mice (Fig. 4 E). The absence of upregulated inflammatory pathways in Samhd1Δ/Δ Ifih1−/− mice indicated selective chronic activation of MDA5 but not of RIG-I in SAMHD1-deficient mice and suggested the presence of endogenous immune-stimulatory dsRNA structures.
Figure S3.
MDA5 drives spontaneous IFN production in Samhd1Δ/Δ mice. Related to Fig. 4. (A) Samhd1+/+ and Samhd1Δ/Δ mice were treated i.p. with 10 mg/kg/d H-151 or vehicle for 14 d. Transcript levels of the indicated ISGs were determined in the spleen. Fold change compared with the WT-vehicle group is shown, n = 4 in each group (two-way ANOVA followed by Tukey’s multiple comparison test). (B) Post-replicative senescence Samhd1Δ/Δ and Samhd1+/+ MEFs were transduced with empty lentivirus or a lentivirus, which expresses the cDNA of murine Samhd1 isoform1 as well as EYFP. Transduced cells were enriched by FACS for EYFP and transcript levels of the indicated ISGs were determined by qRT-PCR. Data of two independent measurements are displayed as fold change compared with the mean of Samhd1+/+ MEFs transduced with empty lentivirus (two-way ANOVA followed by Tukey’s multiple comparison test). * = P < 0.05, ** = P < 0.01,**** = P < 0.0001. (C) Relative transcript levels of the indicated ISGs measured by qRT-PCR in post-replicative senescence Samhd1Δ/Δ MEFs with additional CRISPR-mediated inactivation of the genes cGas (n = 4), Ifih1 (n = 3), and Ddx58 (n = 2) after lipofection with 1 µg/ml plasmid DNA (dsDNA), 100 ng/ml poly I:C, 100 ng/ml 3´-triphosphate RNA (pppRNA), or incubation with 10 μg/ml 5,6-dimethylxanthenone-4-acetic acid (DMXAA) for 16 h. Fold change compared to Lipo-treated Samhd1+/+ MEFs is shown. (D) Normalized read counts for the indicated ISG transcripts from the experiment shown in Fig. 4 E.
Figure 4.
MDA5 drives spontaneous IFN production in Samhd1Δ/Δ mice. For the whole figure − = homozygous null, + = homozygous WT. (A) Enrichment of reactome gene sets (MSigDB) in the transcriptome of peritoneal macrophages from mutant mice compared with littermate WT controls of Samhd1Δ/Δ mice. (B) Normalized read counts for the indicated ISG transcripts from the analysis shown in A. (C) Relative transcript levels of the indicated ISGs measured by qRT-PCR in post-replicative senescence Samhd1Δ/Δ MEFs with additional CRISPR-mediated inactivation of the genes cGas (n = 4), Ifih1 (n = 3), and Ddx58 (n = 2). Data from two independent experiments were pooled and displayed as fold change compared to the mean of Samhd1+/+ MEFs (multiple t tests, summary of results is shown with P < 0.05 as lowest significance level). (D) Differentially expressed (padj<0.1) ISG in transcriptomes of peritoneal macrophages from mutant mice compared with littermate WT controls of Samhd1Δ/Δ mice. (E) Enrichment of reactome gene sets (MSigDB) of another independently generated transcriptome using peritoneal macrophages from Samhd1Δ/ΔIfih1+/+, Samhd1+/+Ifih1−/−, and Samhd1Δ/ΔIfih1−/− compared with littermate Samhd1+/+Ifih1+/+control mice. NES, normalized enrichment score.
Spontaneous activation of MDA5 in Samhd1Δ/Δ mice depends on cGAS/STING signaling
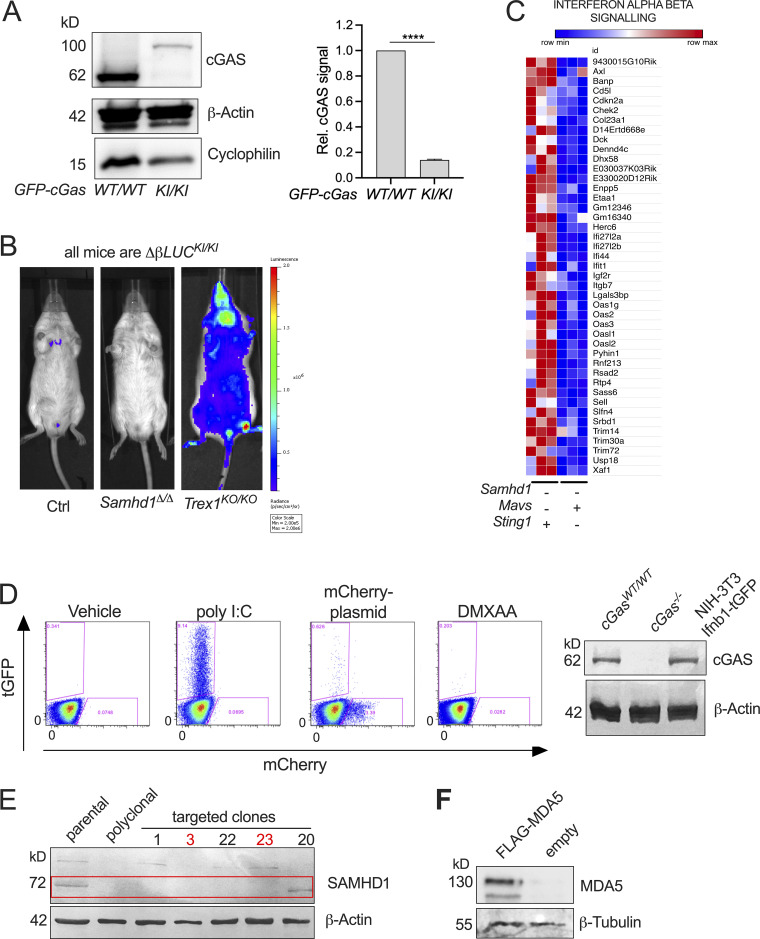
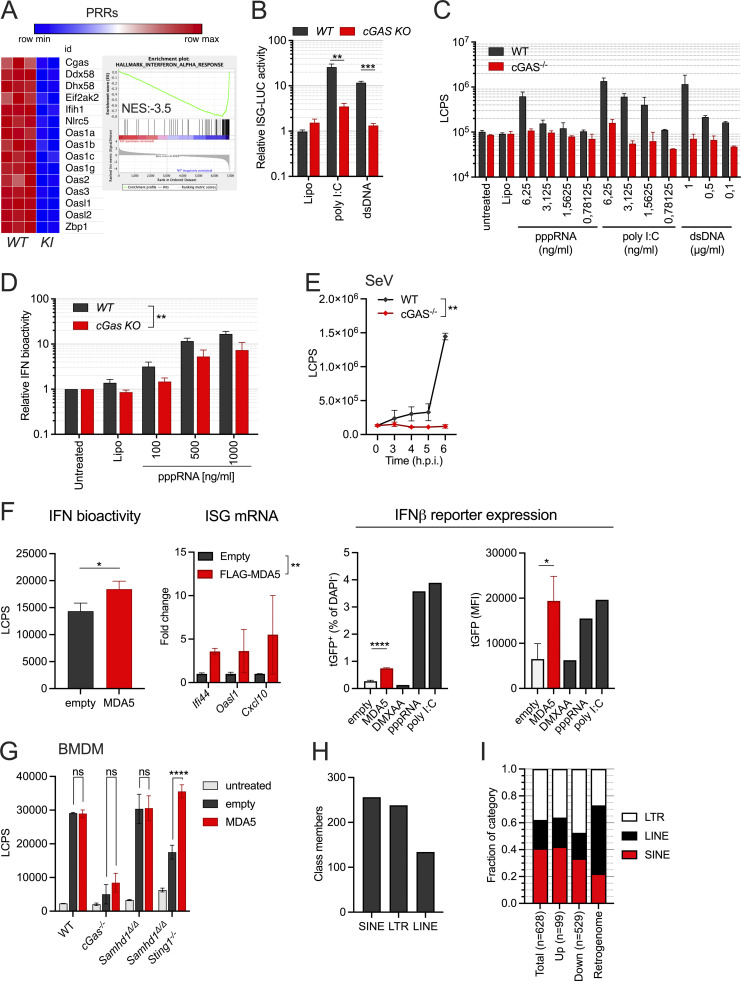
We were still puzzled by the requirement for cGAS in this system that was independently observed by us and by other groups (Maelfait et al., 2016; Coquel et al., 2018). Our transcriptomes of STING-deficient peritoneal macrophages (Fig. 4 B) and as well quantitative PCR (qPCR) results of cGAS-deficient MEFs (Fig. 4 C) showed that ISG expression in the mutant cells is below the levels found in cGAS/STING-competent control cells. This suggested that cGAS primes the antiviral immune system most likely by its sporadic activation in response to endogenous DNA. To address the relevance of cGAS-mediated immune priming in the absence of SAMHD1, we generated SAMHD1-deficient mice that express GFP-tagged cGAS (Gentili et al., 2019) from a hypomorphic allele reducing the cGAS protein level to around 20% compared to that in WT mice (Fig. S4 A). Whole transcriptome sequencing of BMDMs from Samhd1Δ/ΔGFP-cGasKI/KI and control mice demonstrated reduced expression of ISGs, including several pattern recognition receptors, among these RIG-I (Ddx58) and MDA5 (Ifih1; Fig. 5 A). Haploinsufficiency for cGas rescues mice lacking the DNase Trex1 from lethal autoimmunity (Gao et al., 2015; Gray et al., 2015), suggesting that even in terminally sick Trex1−/− mice, the amount of cGAS ligands is only slightly above the threshold of tolerance. In contrast to Trex1−/− mice, the spontaneous IFN response in SAMHD1-deficient mice is very weak, as illustrated by the lack of a spontaneous signal in Samhd1Δ/Δ Ifnβ-LUC reporter mice (Fig. S4 B), rendering spontaneous IFN production even more sensitive to fluctuations of cGAS protein levels. As reported for reduced levels of cGAS, reduced levels of RIG-I and MDA5 might desensitize the intracellular RNA sensing pathways. To test if the sensitivity of the RLR pathway is controlled by cGAS, we knocked out cGas in LL171 ISG-luciferase reporter cells (Uzé et al., 1994). As expected, no increased luciferase activity was detected after transfection of plasmid DNA. However, also the response to transfected poly I:C was almost completely blunted (Fig. 5 B). Next, we isolated MEFs from cGas−/− and littermate control mice and transfected them with increasing amounts of 5′triphosphate dsRNA or with high molecular weight poly I:C to stimulate a RIG-I or MDA5 response, respectively. As observed in cGAS-deficient LL171 cells, the same amounts of these ligands induced lower IFN levels in the supernatant of cGas−/− MEFs compared with control MEFs (Fig. 5 C). Similar results were obtained for BMDMs (Fig. 5 D). Likewise, cGAS-deficient, but not cGAS-proficient, MEFs failed to secrete IFN into the cell culture supernatant upon infection with Sendai Virus (Fig. 5 E). Our results suggest that cGAS keeps expression of PRRs at functional levels, which enables fast and efficient sensing of even low amounts of cytoplasmic dsRNA by RLRs, and that cGAS/STING signaling controls the expression of MDA5 in Samhd1Δ/Δ cells through tonic IFN signaling.
Figure S4.
Spontaneous activation of MDA5 in Samhd1Δ/Δ mice depends on cGAS/STING signaling. Related to Fig. 5. (A) Representative Western blot for cGAS in GFP-cGasKI/KI and GFP-cGasWT/WT control mice (left). Data from two independent experiments for densitometric quantification of cGAS signal relative to the signal for β-actin (right, Student’s t test). cGAS = 62 kD, GFP-cGAS around 92 kD. (B) Spontaneous in vivo Ifnb1-luciferase signal in Samhd1+/Δ (Ctrl), Samhd1Δ/Δ, and Trex1KO/KO mice. All mice were homozygous for the luciferase knock in (ΔβLUCKI/KI). (C) Normalized read counts of genes belonging to the Reactome Interferon_Alpha_Beta_Signalling gene set showing lower expression of ISGs in Samhd1Δ/ΔSting1GT/GT vs. between Samhd1Δ/ΔMavs−/− peritoneal macrophages. The heatmap was generated with data presented in Fig. 4, A and B. (D) Left: Transfection of NIH-3T3-Ifnb1-tGFP reporter cells with 1 µg/ml poly I:C, 3 µg/ml mCherry-expression plasmid, or incubation with 20 μg/ml DMXAA. Frequency of tGFP+ cells was determined by flow cytometry 16 h later. One of three experiments with identical results is shown. Note the lack of a response to plasmid DNA and DMXAA consistent with a lack of STING in these cells. The amounts of poly I:C used here are 10- to 1,000-fold above the amounts used in Fig. 5, B and C. Right: Western blot confirming cGAS protein in Ifnb1-tGFP reporter cells. (E) Confirmation of SAMHD1 KO (deletion of exon 4, not shown) in Ifnb1-tGFP. Numbers above designate single clones. Clone 20 shows truncation and was not used. Studies in the manuscript show results from clone 3 and were confirmed in clone 23; both are marked in red. (F) Western blot 72 h after transfection with of Ifnb1-tGFP with 5 μg/ml of either empty plasmid or FLAG-MDA5 expression plasmid. Detection was done using FLAG-antibody. **** = P < 0.0001. Source data are available for this figure: SourceData FS4.
Figure 5.
Spontaneous activation of MDA5 in Samhd1Δ/Δ mice depends on cGAS/STING signaling. (A) Normalized read counts for transcripts of PRRs in BMDMs from Samhd1Δ/ΔGFP-cGasKI/KI (n = 2) vs. Samhd1+/+ (n = 3) control mice and the enrichment plot for the gene set INTERFERON_ALPHA_RESPONSE of the Hallmark gene set (MSigDB). (B) Relative ISG-luciferase reporter activity in cGAS-competent (WT) and cGAS-deficient (cGas KO) LL171 cells 16 h after lipofection with 100 ng/ml poly I:C and 1 µg/ml plasmid DNA (3 kb). Luciferase activity was normalized to the mean of Lipo-treated WT LL171 cells (Student’s t test). (C) cGAS−/− and control MEFs were lipofected with the indicated concentrations of ligands for pppRNA, high molecular weight poly I:C and plasmid DNA (dsDNA) for 6 h and washed. 18 h later, IFN bioactivity was quantified in the cell culture supernatant using LL171 ISG-LUC reporter cells. LCPS, light counts per second. (D) Similar assay as in C but with BMDMs isolated from cGas+/+ (WT) and cGas−/− (cGas KO) mice lipofected the indicated amounts of pppRNA for 6 h. Two-way ANOVA. (E) cGAS−/− and control MEFs were infected with Sendai Virus (MOI = 1.5) for 2 h and washed. At the indicated time points IFN bioactivity in the supernatant was determined as in C. (F) SAMHD1 and STING-deficient Ifnb1-tGFP reporter cells were transfected with an empty plasmid or a plasmid expressing Flag-tagged murine MDA5 for 16 h before washing. After 72 h post washing IFN bioactivity as in C, ISG transcription by qRT-PCR and Ifnb1-tGFP reporter gene expression by FACS were quantified (T test). (G) Similar experiment as in H but in BMDMs from mice with the indicated genotypes. IFN bioactivity was quantified as described in C 72 h after washing the cells (two-way ANOVA). (H) Numbers of significantly differentially expressed retroelements in transcriptomes of Samhd1Δ/Δ vs. control peritoneal macrophages. Numbers for each class as annotated in TEtranscript are shown. (I) Relative abundance of the classes shown in H within each of the categories (total, upregulated, downregulated, retrogenome). Retrogenome refers to the genomic frequency of retroelements in the respective class among all mouse retroelements. LINE = 19.2%, SINE = 8.2%, LTR = 10%, all together = 37.4% of the mouse genome (Kassiotis and Stoye, 2016); relative abundance among genomic retroelement sequences for LINE = 51.4%; SINE = 21.9%, LTR = 26.7%. * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001.
To investigate if the abrogation of IFN production in Samhd1Δ/Δ cells lacking cGAS/STING can be attributed to a failure in priming MDA5 expression, we sought to overcome the need for tonic IFN signaling by ectopic expression of MDA5. To prevent induction of IFN by transfection of the MDA5-expression plasmid, we inactivated SAMHD1 by CRISPR/Cas9 in NIH-3T3-Ifnb1-tGFP reporter cells (from here on “Ifnb1-tGFP” [Rand et al., 2012]), which are deficient for STING1 (Fig. S4, D and E). Transfection of the MDA5 expression plasmid into SAMHD1-deficient Ifnb1-tGFP reporter cells (Fig. S4 F) increased IFN bioactivity in the cell culture supernatant, transcription of ISGs and Ifnb1-tGFP reporter expression compared with empty plasmid transfected cells (Fig. 5 F). Similar results were obtained when transfecting these plasmids into BMDMs of Samhd1Δ/Δ STING1GT/GT and control mice (Fig. 5 G). Our data suggest that in SAMHD1-deficient cells, spontaneous IFN-production via MDA5 requires priming through the cGAS/STING pathway and suggests endogenous dsRNA as the primary IFN-inducing nucleic acid.
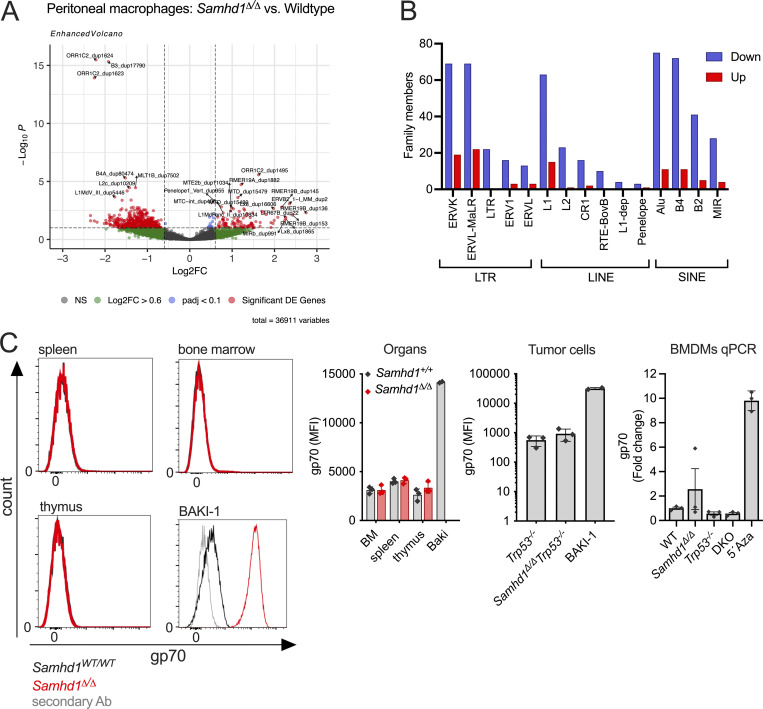
SAMHD1 has been implicated in controlling the activity of retroviruses and endogenous retroelements (ERE; Behrendt et al., 2013; Rehwinkel et al., 2013; Zhao et al., 2013; Hu et al., 2015; Herrmann et al., 2018). Replication intermediates of such elements have recurrently been suggested to activate nucleic acid sensors, including MDA5. To investigate if transcription of ERE is altered in SAMHD1-deficient mice, we quantified retroelement transcripts in one of the whole transcriptome datasets obtained from peritoneal macrophages. Reads originating from ERE were identified using a reference file from TEtranscripts (mm39_rmsk_TE.gtf, 2021-04-07), which allowed us to detect expression of 36911 transposable elements (TEs). Among these, we identified 628 differentially expressed (padj < 0.1, Log2FC > 0.6) retroelements of the LTR, long interspersed nuclear elements (LINE), and short interspersed nuclear elements (SINE) classes between Samhd1Δ/Δ and control peritoneal macrophages (Fig. 5 H and Fig. S5 A). The vast majority (81%, n = 529) of these ERE transcripts were downregulated in Samhd1Δ/Δ peritoneal macrophages and about 19% (n = 99) were derepressed compared with Samhd1-proficient cells (Fig. S5 B). Interestingly, compared with their abundance in the mouse retrogenome, SINE elements (22% of the retrogenome vs. 42% among upregulated ERE), and to a lesser extent LTR elements (27 vs. 36%), were overrepresented among all upregulated ERE by roughly a factor of two (Fig. 5 I). In contrast, LINE elements were 2.5-fold underrepresented compared with their frequency in the retrogenome (51 vs. 21%). Although around 50 endogenous retrovirus (ERV) family members were significantly upregulated in Samhd1Δ/Δ vs. control peritoneal macrophages (Fig. S5 B), we failed to detect increased expression of gp70 env on the cell surface of various cell types and tumor cells lacking SAMHD1 arguing against a strong retro-viremia in mice lacking SAMHD1 (Fig. S5 C).
Figure S5.
Analysis of endogenous retrolements (ERE) in Samhd1Δ/Δ mice. Related to Fig. 5. (A) Volcano plot of ERE differentially expressed in peritoneal macrophages of Samhd1Δ/Δ vs. control peritoneal macrophages as identified by a TEtranscript reference (see main text). (B) Numbers of differentially expressed ERE for each family in the three ERE classes (all red dots from A). Down (blue) and Up (red) indicate the expression level in Samhd1Δ/Δ vs. control cells. B1 elements were designated as “Alu” in the mouse genome TEtranscript reference file, although the term was originally reserved for primate SINEs of this family. (C) Single-cell suspensions of the indicated organs from Samhd1Δ/Δ (red, n = 3) vs. control mice (black, n = 3) were stained for the surface expression of MLV gp70 and analyzed by flow cytometry. Histogram overlays are shown for DAPI− living cells (left) and next to it the quantification. Bar graph in the middle shows the same analysis on immortalized thymic fibroblasts from tumor bearing mice with the indicated genotypes (n = 3 per genotype). BAKI-1 cells express high levels of the recombinant MLV from the EMV2 locus were used as a positive control (Yu et al., 2012). Graph on the right shows qPCR for MLV gp70 in BMDMs from mice of the indicated genotypes (n = 3). BMDMs were differentiated for 7 d in RPMI with 30% L929 supernatant and then seeded for 72 h in RPMI +15% L929. Samhd1Δ/Δ BMDMs cultured in the presence of 500 nM 5-Azacytidin were used as a positive control. Fold change in gp70 mRNA abundance is shown in comparison to WT. MFI = mean fluorecence intensity.
Collectively, we show that priming of MDA5-expression via the cGAS/STING pathway enables sensing of an endogenous MDA5 ligand in SAMHD1-deficient cells. Quantification of deregulated retroelement transcripts hints toward an increased abundance of SINE-derived RNA in cells lacking SAMHD1.
Discussion
We showed that loss of SAMHD1 in mice leads to a DNA replication defect, which causes DNA damage that is counteracted by activation of a p53 response. Loss of SAMHD1 in tumor cells exacerbated DNA damage in difficult-to-replicate regions like telomeres, which might contribute to accelerated malignant transformation when DNA repair pathways are impaired. Our observations of increased telomere damage in tumor cells lacking SAMHD1 are in line with previous reports of SAMHD1 being part of the telomere proteome (Majerska et al., 2018; Lin et al., 2021) and with increased frequencies of R-loop structures in SAMHD1-deficient cells (Park et al., 2021). Contrary to our observations in the p53-deficient tumor model, loss of SAMHD1 in MMR-deficient mice had no effect on tumor-free survival. This was unexpected as earlier reports demonstrated that even Samhd1+/− mice had elevated dNTP levels, and mutations in the ribonucleotide reductase complex cause a cancerogenic mutator phenotype as a result of altered dNTP levels (Rentoft et al., 2016; Aye et al., 2015). Samhd1Δ/Δ mice lack dNTPase and DNA repair activity, suggesting that loss of neither activity leads to significant DNA damage that would be detected by the MMR machinery in vivo. Hence, our data suggest that loss of SAMHD1 in mice does not cause a strong mutator phenotype.
We recently found that the IFN response in Trex1−/− mice is linked to DNA replication (Schubert et al., 2022) and similar findings have been reported for SAMHD1−/− cells (Coquel et al., 2018). In both models, loss of p53 did not amplify the IFN response, while such p53-dependent amplification was observed in cells lacking RNaseH2 in which chromatin fragments activate cGAS (Hiller et al., 2018; Mackenzie et al., 2017; Aditi et al., 2021; Giordano et al., 2022). Similar to the loss of p53, inhibition of ATM increased IFN production in cells deficient for RNASEH2 (Aditi et al., 2021; Giordano et al., 2022). Although we found no evidence for increased ISG transcription in Samhd1Δ/ΔTrp53−/− mice (Fig. 3, B–D), we observed that inhibition of ATM in cells isolated from SAMHD1-deficient mice increased the spontaneous IFN response, further suggesting the presence of DSB (Fig. 3 A). Our data confirm observations in SAMHD1-depleted U2OS cells (Park et al., 2021) and add a new link between ATM and IFN signaling in SAMHD1-deficient cells. This might point to a differential involvement of the p53 pathway in the generation of immune stimulatory DNA as a result of DNA replication in mice lacking TREX1 or SAMHD1 compared with postreplicative DNA damage found in RNaseH2-deficient cells. However, it should also be noted that p53 represents only one of many ATM targets, and as opposed to suppressing cGAS/STING-dependent IFN induction, direct regulation of ISG transcription through ATM has been observed previously (Purbey et al., 2017). Therefore, it remains to be established if and how spontaneous DNA damage in SAMHD1-deficient mice translates into an aberrant immune response. Accumulation of ssDNA species and concomitant activation of cGAS has been reported in SAMHD1-deficient cells, but it still remains unclear whether these oligonucleotides represent direct ligands for the DNA sensor, which, under physiological conditions, is known to nucleate only in the presence of unprotected long dsDNA (Andreeva et al., 2017; Du and Chen, 2018). Thus, it remains possible that in SAMHD1-deficient cells cGAS activation had a different culprit.
The lack of increased IFN might explain why STING had no role in regulating the tumor-free survival of Samhd1Δ/ΔTrp53−/− mice, suggesting that endogenous DNA damage in p53-deficient tumors only weakly, if at all, activates STING (Fig. 3, B–D). The relevance of our observation is illustrated by the fact that every other tumor in humans carries homozygous inactivation of the p53 gene (Baugh et al., 2018). However, this is in sharp contrast to targeted activation of the STING pathway in tumor tissue, which is currently being explored as a potent strategy to boost antitumor immunity (Demaria et al., 2019).
Our results challenge a role of pathogenic DNA sensing in SAMHD1-deficient mice as inactivation of RLR sensing in SAMHD1-deficient, but cGAS/STING-competent, cells was sufficient to blunt the spontaneous IFN response. This cannot be explained by a lack of RLR-mediated immune priming because ISGs levels in MAVS-deficient mice were similar to that of WT mice, while in the absence of STING they were below levels found in WT mice (Fig. 4 B). ISG transcription was also lower in Samhd1Δ/ΔSting1GT/GT when compared with Samhd1Δ/ΔMavs−/− mice (Fig. S4 C). We previously observed that plasmacytoid dendritic cells are the main producers of tonic IFN in mice (Peschke et al., 2016), which was later shown to be induced in response to commensal bacteria activating TLR and MAVS signaling pathways (Schaupp et al., 2020). In the skin, microbiota induced derepression of endogenous retroelements and a cGAS/STING-dependent IFN response, but in this study, MAVS signaling was not investigated (Lima-Junior et al., 2021). Recently, an alternative scenario was observed in which gut microbiota-derived membrane vesicles transfer DNA into the cytoplasm of host cells to induce cGAS/STING-driven tonic IFN signaling (Erttmann et al., 2022). Interestingly, in human macrophages phagocytosed gut commensal bacteria evoked an IFN response, which was codependent on STING and MAVS expression (Gutierrez-Merino et al., 2020), suggesting that innate immune priming in response to low-level chronic stimuli can be driven by innate sensing of endogenous DNA and RNA. Our transcriptome data indicated that the cGAS/STING pathway establishes tonic IFN signaling and baseline expression of antiviral genes, including RLRs. This places cGAS/STING signaling upstream of RLR sensing in the analyzed cell types because the absence of this pathway leads to impaired cytoplasmic RNA sensing (Fig. 5, B–E). Similar findings have been reported in the context of RNA virus infections (Schoggins et al., 2014; Parker et al., 2018 Preprint; Lima-Junior et al., 2021; Erttmann et al., 2022). As the IFN response in SAMHD1-deficient mice is weak, we propose that loss of cGAS/STING signaling in cells lacking SAMHD1 desensitizes the RLR pathways and increases tolerance against RNA ligands, thereby preventing spontaneous induction of IFN despite the presence of an endogenous MDA5 ligand. Consistently, we showed that ectopic expression of MDA5 in cells lacking SAMHD1 and STING1 reinstates increased IFN production (Fig. 5, F and G). To this end, dsRNA originating from endogenous retroelements has been shown to activate MDA5 in cells lacking the AGS gene ADAR1 (Ahmad et al., 2018) and after DNA damage induced by chemotherapy (Clapes et al., 2021). Derepression of endogenous retroelements is not only a physiological response (Lima-Junior et al., 2021; Young et al., 2012; Yu et al., 2012) but is also a general stress response (Simon et al., 2019; De Cecco et al., 2019), and SAMHD1-deficient cells display signs of replication stress including spontaneous DNA damage as shown here and previously by other groups (Daddacha et al., 2017; Coquel et al., 2018). Quantification of retroelement transcripts in bulk RNA sequencing data revealed an overrepresentation of SINE transcripts compared with their abundance in the mouse retrogenome (Fig. 5 I), suggesting that their derepression might not simply reflect homogenous global chromatin remodeling in Samhd1Δ/Δ mice. Interestingly, in the human system, Alu-repeats (belonging to the SINE class) have been implicated as potent endogenous MDA5 ligands (Chung et al., 2018; Ahmad et al., 2018; Mehdipour et al., 2020). Therefore, it is tempting to speculate that the stress response alone is sufficient to promote aberrant transcription and processing of endogenous RNA from, but not limited to, the vast numbers of retroelement loci, which might lead to autorecognition by RNA sensors in SAMHD1-deficient cells.
Taken together, our work suggests that in SAMHD1-deficient cells, endogenous dsRNA represents the primary nucleic acid ligand that drives IFN production and implicates an important role of the cGAS/STING pathway in physiological and pathophysiological innate immune priming.
Material and methods
Mice
Samhd1∆/∆ (Behrendt et al., 2013), cGas−/− (Schoggins et al., 2014), Mavs−/− (Michallet et al., 2008), Sting1GT/GT (Sauer et al., 2011), Ifih1−/− (Gitlin et al., 2006), Trp53−/− (Jacks et al., 1994), Pms2−/− (Baker et al., 1995), Trex1−/− (Morita et al., 2004), ΔβLUCKI/KI (Lienenklaus et al., 2009), and GFP-cGasKI/KI (Gentili et al., 2019) mice were described previously. Trp53−/− (#002101) and Pms2−/− (#010945) were purchased from The Jackson Laboratory. Ddx58−/− mice on a C57BL/6 background were generated by Stefan Bauer (unpublished). Mice were housed under specific pathogen–free conditions at the Experimental Center of the University of Technology Dresden. All animal experiments were done according to institutional guidelines on animal welfare and were approved by the Landesdirektion Sachsen (11-1/2010-33, 24-1/2013-12, 24/2017; 88/2017).
MEFs
MEFs were generated by standard procedures. In brief, E11.5 mouse embryos were dissected and decapitated. After removal of internal organs, tissue was cut into small pieces, digested with 1× trypsin (0.25%; Invitrogen) for 30 min at 37°C, and disaggregated by pipetting. The cell suspension was cultured in DMEM (Gibco) supplemented with 10% heat-inactivated FCS, 100 U/ml Penicillin, 100 mg/ml Streptomycin, 1× non-nonessential amino acids (all Biochrom), and 100 µM β−mercaptoethanol (Gibco). After 24 h, nondigested tissue aggregates were removed and the cells were kept cultivated in complete DMEM medium at 37°C and 5% CO2 under atmospheric oxygen.
Thymic fibroblasts
Thymi were homogenized and passed through a 40-μm cell filter. The single cell suspension was cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, 1 mM sodium pyruvate, and 2 mM L-alanyl L-glutamine (all Biochrom). Surviving cells were kept cultivated in complete RPMI 1640 medium at 37°C and 5% CO2 under atmospheric oxygen.
In vitro differentiation of BMDMs
Bone marrow cells were cultured overnight in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, 1 mM sodium pyruvate, and 2 mM L-alanyl L-glutamine (all Biochrom). The next day, nonadherent cells were transferred to new dishes and differentiated for 6 d in RPMI medium (supplemented as described) containing 30% L929 supernatant. An equal amount of fresh differentiation medium was added after 3 d. 4 d later, the attached cells were harvested, counted, and seeded in RPMI + 15% L929 supernatant to perform the experiments. Cells were cultured at 37°C and 5% CO2 under atmospheric oxygen.
Micronucleus flow assay
Retrobulbar blood sample was mixed with heparin/PBS (250 U/ml; Biochrom) and fixed by adding 20 μl of the mixture to 2 ml ice-cold methanol, inverted, and stored at −80°C. Quantification of micronucleated erythrocytes was performed as described previously (Balmus et al., 2015). Briefly, fixed blood cells were washed in bicarbonate buffer and stained using antibodies against CD71 (1:200, #1720-02; Southern Biotech) and Ter119 (1:500, #48-5921-82; eBioscience) in the presence of 1 µg/μl RNase A. After washing and addition of 1 µg/ml propidium iodide (PI), cells were analyzed by flow cytometry and gated for single Ter119+ CD71+ PI+ micronucleated reticulocytes and Ter119+ CD71− PI+ micronucleated normochromatic erythrocytes (MN-NCE).
Transcriptomics of peritoneal macrophages
Peritoneal macrophages (DAPI−, CD11bhi, F4/80hi) were isolated by FACS using antibodies against CD11b (1:1,600, #11-0112; eBioscience) and F4/80 (1:200, #123114; Biolegend), and total RNA was extracted using RNeasy Plus Mini Kit (Qiagen). For each experiment, equal amounts of total RNA were used for poly-dT enrichment before library preparation and sequencing was performed as described before (Schubert et al., 2022).
Differential gene expression: Reads were mapped to mouse genome GRCm39 followed by normalization, exploratory, and differential expression analysis using DESeq2 (Love et al., 2014). Unless otherwise stated, differentially expressed gene lists were generated by comparing all mutant mouse lines to the respective WT group in each experiment and sorted according to ascending padj. All transcripts with padj < 0.05 were subjected to GSEA (Subramanian et al., 2005). To generate heatmaps, all transcripts with padj < 0.05 in the comparison Samhd1∆/∆ vs. Samhd1+/+ were extracted from lists containing normalized read counts of all genotypes in the respective experiment, and the resulting sublist was displayed as a heatmap. Differential gene expression in Fig. 4 D: The processing of the whole transcript RNA sequencing data was carried out using the nf-core RNA-seq pipeline v3.8.1 (Ewels et al., 2020). Mapping of the raw reads to the reference genome (GRCm38.94) and quantification of aligned reads were done by STAR (Dobin et al., 2013) and Salmon (Patro et al., 2017), respectively. The statistical was performed in R environment v4.1.2 with the Bioconductor R-package DESeq2 (Love et al., 2014). The Benjamini–Hochberg method was used to calculate multiple testing adjusted P values. Downstream analysis was performed on the genes with a minimum expression level of 10 in at least three samples.
Quantification of retroelements transcripts: RNA sequencing reads were preprocessed using FASTQC (https://github.com/s-andrews/FastQC) and cutadapt (Martin, 2011) and aligned to the Genome Reference Consortium Mouse Build 39 using STAR (Dobin et al., 2013), including multimapping reads. BAM files were indexed using Samtool (Danecek et al., 2021). Mapped reads were annotated to transposable elements using a GTF file (mm39_rmsk_TE.gtf, 2021-04-07) obtained from TEtranscripts (Jin et al., 2015) and quantified via HT-Seq (Putri et al., 2022). Differential expression analysis was performed with DESeq2 and visualizations were prepared using the EnhancedVolcano R package (https://github.com/kevinblighe/EnhancedVolcano).
Heatmaps were generated using the mighty Morpheus (https://software.broadinstitute.org/morpheus).
Gene expression datasets are available at GEO database: Fig. 1 A: GSE45358; Fig. 4, A and B, and Fig. S3 C: GSE211209; Fig. 4, D and E: GSE214713; Fig. 5 A: GSE211208, Fig. 4, A and B, and Fig. 5, H and I: GSE41879.
Quantification of phosphorylated histone H2AX
25,000 MEFs or 20,000 peritoneal macrophages (sorted as CD11b+ F4/80HI) were plated onto 8-well chamber slides. Cells were left at 37°C overnight to attach to the slide. As positive control, slides were γ-irradiated with the indicated dose 1 h before fixation. After incubation, cells were washed in PBS, fixed in ice-cold methanol, and washed again. For quantification of phosphorylated histone H2AX (γH2AX), slides were blocked at room temperature for 1 h in 1× blocking reagent (Roche) and incubated at 4°C overnight with a phospho-histone H2AX (pSer139) antibody (1:50, #2577; Cell Signaling Technology). After washing with PBS, slides were incubated with a goat anti-rabbit-AF488 antibody (1:500, #A-11034; Thermo Fisher Scientific) for 1 h at room temperature in the dark. Slides were washed and nuclei were counterstained with 10 µg/ml DAPI in the mounting solution. Imaging was done on a Keyence fluorescence microscope or on Leica Thunder Imaging System. Nuclei were segmented with the DAPI channel recording using Cellpose (Stringer et al., 2021) and extracted as masks. These masks were loaded as an overlay on the corresponding recoding of the γH2AX channel to quantify integrated density, mean intensity, and area using ImageJ software (National Institutes of Health [NIH]). The mean of four background (outside of nuclei masks and cells) measurements per picture was used to calculate the corrected total nuclear fluorescence (CTNF) of γH2AX signal using the formula CTNF = integrated density − (area * mean of the background).
Histology
Thymi were formalin-fixed, paraffin-embedded, and cut into 3-µm sections. For H&E staining, sections were dyed with Mayer’s hemalum solution for 2 min, followed by staining with eosin and rinsed with water for 30 s. The preparations were dehydrated again in an ascending alcohol series and washed in xylene. H&E sections were evaluated by a board-certified pathologist on a Zeiss Axioskop 2 microscope and photographs were made with an Axiocam 503 color camera using ZEN 2.5 (blue edition) software (Zeiss).
Multiplex immunohistochemical staining against CD3, CD4, and CD8 to assess T cell composition in thymi was performed on a Ventana Discovery Ultra Instrument. Briefly, antigen retrieval using cell conditioning 1 solution (Ventana Medical Systems) was performed at 95°C for 32 min, followed by incubation with the primary antibody against CD8 (1:100, #14-0195-82; eBioscience) at 36°C for 32 min, the HRP-coupled secondary anti-rat OmniMap antibody (Ventana Medical Systems) for 12 min, and finally Opal 520 fluorophore (1:100; Akoya Biosciences) at room temperature for 8 min. Primary and secondary antibodies were removed by denaturation at 100°C for 24 min in cell conditioning 2 buffer (Ventana Medical Systems). The above-described steps were repeated for CD4 (1:500, #ab183685; Abcam) with OmniMap anti-rabbit-HRP and Opal 570 fluorophore (1:1,000; Akoya Biosciences), and lastly, CD3 (1:50, #ab16669; Abcam) with OmniMap anti-rabbit-HRP and Opal 690 fluorophore (1:50; Akoya Biosciences). Finally, sections were counterstained with DAPI (Merck) and mounted with Fluoromount G mounting media (Southern Biotech). Sections were scanned at 100× magnification, regions of interest were defined using Phenochart software (Akoya Biosciences), and multispectral images were acquired at ×200 magnification using the Ventra 3.0 Automated Imaging System (Akoya Biosciences). Upon spectral unmixing using inForm Software (Akoya Biosciences), images were exported and processed in ImageJ (NIH).
Flow cytometry
Thymi were homogenized and passed through a 70-μm cell filter. Following washing with ice-cold FACS buffer (PBS/2%FCS/2 mM EDTA), cells were filtered again through a 70-μm cell filter. On samples from peripheral blood, erythrocytes were lysed if leucocytes were analyzed. Cells were incubated with anti-CD16/CD32 (1:200, #101302; Biolegend) at room temperature for 10 min to block Fc receptors and stained with the following antibodies in FACS buffer at 4°C for 30 min: CD3e (# 11-0031, 1:200 or # 17-0031, 1:100; eBioscience), CD4 (1:200, #53-0041; eBioscience), CD8a (1:600, #25-0081; eBioscience), CD11b (1:1,600, #11-0112; eBioscience), CD19 (1:200, #25-0193; eBioscience), CD25 (1:800, #12-0251; eBioscience), CD44 (1:200, # 48-0441; eBioscience), CD45R (B220; 1:100, # 47-0452; eBioscience), and Ly-6A/E (Sca-1; 1:200, #17-5981; eBioscience). After incubation, cells were washed and resuspended in FACS buffer. For dead cell exclusion, 1 μg/ml of PI was added to the cell suspension shortly before the analysis. Cells were analyzed using the FACSAria III (BD Bioscience) and evaluated with FlowJo Version 10 (Tree Star).
Peripheral blood was stained for Sca-1+ within the CD3+ and CD19+ populations.
Telomere integrity
Quantification of telomere integrity was done with metaphase telomere FISH. Metaphase spreads were performed as previously published (Poon and Lansdorp, 2001). Briefly, the cells were cultured in 10-cm Petri dishes and grown to 60% confluency. The cells were treated with 0.2 µg/ml Colcemid (#10 295 892 001; Merck) for 3 h and incubated with hypotonic solution (75 mM KCl). Swollen cells were washed with fixative solution (methanol:glacial acetic acid 3:1) and dropped on superfrost microscopic slides. Telomeres were stained with TelC-Alexa488 labeled PNA probe (#F1004; Panagene), as previously published (Awad et al., 2020). The slides were mounted with Fluoroshield mounting media containing DAPI (#F6057-20ML; Sigma-Aldrich) to stain the chromosomes. Images were acquired using a ZEISS Axio Observer microscope. The obtained images were analyzed by evaluating the average telomere integrity per metaphase. Depending on the signal, the telomere phenotypes were categorized as fragile, outside, apposition, and fusion.
qRT-PCR
Total RNA was isolated using NucleoSpin RNA Kit (Macherey-Nagel) and reverse transcribed into cDNA using PrimeScript RT Reagent Kit (Takara) following the manufacturer’s instructions. qRT-PCR using Luna Universal qPCR Master Mix (New England BioLabs) was performed with the following cycling conditions on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad): 10 min 95°C, 40 cycles of 95°C for 20 s, and 60°C for 30 s. The used qRT-PCR primers are listed in Table S1. Transcript levels were normalized to the housekeeping gene Tbp1. All samples were run in technical triplicates.
CRISPR/Cas9 gene targeting
Cells were transfected with pSpCas9(BB)-2A-GFP (px458; Addgene) containing guide RNAs targeting genes cGas, Samhd1, Ifih1, or Ddx58. Target sequences are given in Table S1. Cells were selected with 3 µg/ml puromycin for 72 h and single-cell clones were isolated in a 96-well format. Genotyping was performed by amplicon deep sequencing on a MiSeq using a protocol, described by Lange et al. (2014), which was adapted to the target loci. KO of the target genes was determined genetically using the Outknocker tool (Schmid-Burgk et al., 2014) and functionally by the lack of response to specific ligands (Fig. S2). KO of Samhd1 in Ifnb-1-tGFP cells was confined by PCR and Western blot (Fig. S3).
LL171 luciferase reporter assay
ISRE luciferase reporter expressing LL171 cells were cultured in DMEM (Gibco) supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, 1× non-nonessential amino acids (all Biochrom), and 600 µg/ml G418. To analyze luciferase activity in cell supernatant, LL171 cells were seeded in supplemented DMEM without G418 in 96-well plates. Once cells are attached, DMEM was removed and IFN-containing cell supernatant was added overnight to the cells. Luciferase assay was performed using the SpectraMax Glo Steady-Luc Reporter Assay Kit (Molecular Devices) according to the manufacturer’s instructions, and relative luciferase activity was measured at the LUMIstar Omega (BMG Labtech) microplate reader.
Western blot
Cell pellets from GFP-cGasKI/KI and control BMDMs were lysed in 2× Laemmli buffer and incubated at 95°C for 5 min. Proteins were separated on a 12% denaturing acrylamide gel and subsequently transferred onto a nitrocellulose membrane (Amersham Hybond-ECL, GE Healthcare). Membrane was blocked using 1× RotiBlock (Carl Roth) for 1 h and incubated overnight at 4°C with the primary antibodies against cGAS (1:1,000, #31659; Cell Signaling Technology), β-actin (1:10,000, #4970; Cell Signaling Technology), and Cyclophilin B (1:20,000, #43603; Cell Signaling Technology) diluted in 1× RotiBlock. Following washing with TBS/0.1% Tween, the membrane was incubated for 1 h at room temperature with peroxidase-conjugated goat anti-rabbit secondary antibody (1:1,000, #7074; Cell Signaling Technology) and washed again. The Amersham ECL Prime Western blotting Detection Reagent (GE Healthcare) was used for protein visualization using the Fusion FX (Vilber Lourmat) and Fusion FX7 Advanced imaging software. Signals were densitometrically analyzed using ImageJ software (NIH).
Statistical analysis
Data are shown as means ± SD. Statistical analysis was performed using GraphPad Prism 9. To compare the mean of two groups, Student’s t test (unpaired t test, two-tailed, 95% confidence intervals) was used. For the comparison of more groups, one-way ANOVA or two-way ANOVA followed by either Tukey’s or Sidak’s multiple comparison test was used. Log-rank test was used to compare survival data. Significance levels in each figure are stated as follows: *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Online supplemental material
Fig. S1 is related to Fig. 2 and shows flow cytometric data and TCR recombination data of malignant thymi. Fig. S2 is related to Fig. 3 A and shows inhibition of ATM phosphorylation by KU-55933. Fig. S3 is related to Fig. 4 and shows control experiments and additional data on the in vivo role of MDA5 in SAMHD1-deficient mice. Fig. S4 is related to Fig. 5 and shows important controls. Fig. S5 is related to Fig. 5, H and I, and shows additional data on the activation of endogenous retroelements in Samhd1-deficient mice. Table S1 lists oligonucleotide sequences used in this study.
Supplementary Material
lists oligonucleotides.
contains original blots for Fig. S1.
contains original blots for Fig. S2.
contains original blots for Fig. S4.
Acknowledgments
We thank the Dresden Concept Genome Center, Christa Haase, and Livia Schulze for excellent technical support. We would like to thank Alesja Dernst for her assistance with the WES analysis. Graphical abstract was created using BioRender.com.
R. Behrendt, A. Roers, K. Paeschke, S. Bauer, T. Zillinger, H. Kato, and G. Hartmann were supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project-ID 369799452-TRR237. R. Behrendt was additionally supported by the DFG (BE-5877/2-1) and by an AGS Research Award of the Aicardi-Goutières Syndrome Advocacy Association. T. Gramberg was supported by DFG 401821119/GRK 2504. A. Gerbaulet was supported by the Fritz-Thyssen-Stiftung (Az. 10.19.1.013MN).
Author contributions: Conceptualization—R. Behrendt; Writing—Original Draft—R. Behrendt, T. Schumann, and N. Schubert; Writing—Review & Editing—R. Behrendt, T. Schumann, and N. Schubert; Methodology—R. Behrendt, S.C. Ramon, T. Schumann; A. Gerbaulet, K.I. Maser, L. Muhandes, Validation. Verification—L. Muhandes, M. Haase, and R. Behrendt; Formal Analysis—R. Behrendt, A. Gerbaulet, M.A. Mayo, M. Haase, K.I. Maser, Y. Ge, S.-R. Ada, R.O., N. Schubert, Lu. M., L. Muhandes, M. Schmitz, F. Shakeri, A. Buness, and S. Lienenklaus; Investigation—R. Behrendt, T. Schumann, S.C. Roman, M.A. Mayo, Y. Ge, S.-R. Ada, R.O., L. Sydow, N. Schubert, B. Utess, Lu.M., L. Muhandes, S. Lienenklaus, M. Schmitz, A. Gerbaulet, and P.Müller; Resources—G. Hartmann, H. Kato, M. Schmitz, S. Bauer, T. Zillinger, N. Manel, and B. Rupf; Data Curation—Y. Ge, F. Shakeri, and A. Buness; Visualization—R. Behrendt, N. Schubert, M. Hega, and L. Muhandes; Supervision—R. Behrendt, A. Roers, K. Paeschke, and A. Gerbaulet; Funding Acquisition—R. Behrendt, A. Roers, K. Paeschke, S. Bauer, G. Hartmann, and H. Kato.
References
- Ablasser, A., and Hur S.. 2020. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 21:17–29. 10.1038/s41590-019-0556-1 [DOI] [PubMed] [Google Scholar]
- Aditi, S.M., Downing S.M., Schreiner P.A., Kwak Y.D., Li Y., Shaw T.I., Russell H.R., and McKinnon P.J.. 2021. Genome instability independent of type I interferon signaling drives neuropathology caused by impaired ribonucleotide excision repair. Neuron. 109:3962–3979.e6. 10.1016/j.neuron.2021.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, S., Mu X., Yang F., Greenwald E., Park J.W., Jacob E., Zhang C.-Z., and Hur S.. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell. 172:797–810.e13. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva, L., Hiller B., Kostrewa D., Lässig C., de Oliveira Mann C.C., Jan Drexler D., Maiser A., Gaidt M., Leonhardt H., Hornung V., and Hopfner K.-P.. 2017. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 549:394–398. 10.1038/nature23890 [DOI] [PubMed] [Google Scholar]
- Antonucci, J.M., St Gelais C., de Silva S., Yount J.S., Tang C., Ji X., Shepard C., Xiong Y., Kim B., and Wu L.. 2016. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat. Med. 22:1072–1074. 10.1038/nm.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad, A., Glousker G., Lamm N., Tawil S., Hourvitz N., Smoom R., Revy P., and Tzfati Y.. 2020. Full length RTEL1 is required for the elongation of the single-stranded telomeric overhang by telomerase. Nucleic Acids Res. 48:7239–7251. 10.1093/nar/gkaa503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye, Y., Li M., Long M.J.C., and Weiss R.S.. 2015. Ribonucleotide reductase and cancer: Biological mechanisms and targeted therapies. Oncogene. 34:2011–2021. 10.1038/onc.2014.155 [DOI] [PubMed] [Google Scholar]
- Baker, S.M., Bronner C.E., Zhang L., Plug A.W., Robatzek M., Warren G., Elliott E.A., Yu J., Ashley T., Arnheim N., et al. 1995. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 82:309–319. 10.1016/0092-8674(95)90318-6 [DOI] [PubMed] [Google Scholar]
- Balmus, G., Karp N.A., Ng B.L., Jackson S.P., Adams D.J., and McIntyre R.E.. 2015. A high-throughput in vivo micronucleus assay for genome instability screening in mice. Nat. Protoc. 10:205–215. 10.1038/nprot.2015.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh, E.H., Ke H., Levine A.J., Bonneau R.A., and Chan C.S.. 2018. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 25:154–160. 10.1038/cdd.2017.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt, R., Schumann T., Gerbaulet A., Nguyen L.A., Schubert N., Alexopoulou D., Berka U., Lienenklaus S., Peschke K., Gibbert K., et al. 2013. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 4:689–696. 10.1016/j.celrep.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, N., Gläsker S., Sitaram P., Hofmann H., Shepard C.N., Schultz M.L., Kim B., and Landau N.R.. 2017. A highly active isoform of lentivirus restriction factor SAMHD1 in mouse. J. Biol. Chem. 292:1068–1080. 10.1074/jbc.M116.743740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragán, L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., and Ludewig B.. 2009. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 182:1099–1106. 10.4049/jimmunol.182.2.1099 [DOI] [PubMed] [Google Scholar]
- Choi, J., Ryoo J., Oh C., Hwang S., and Ahn K.. 2015. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology. 12:46. 10.1186/s12977-015-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H., Calis J.J.A., Wu X., Sun T., Yu Y., Sarbanes S.L., Dao Thi V.L., Shilvock A.R., Hoffmann H.-H., Rosenberg B.R., and Rice C.M.. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 172:811–824.e14. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapes, T., Polyzou A., Prater P., SagarMorales-Hernández A., Ferrarini M.G., Ferrarini M.G., Kehrer N., Lefkopoulos S., Bergo V., Hummel B., et al. 2021. Chemotherapy-induced transposable elements activate MDA5 to enhance haematopoietic regeneration. Nat. Cell Biol. 23:704–717. 10.1038/s41556-021-00707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, R., Louis T., Robbe P., Ackroyd S., Burns A., Timbs A.T., Wright Colopy G., Dreau H., Sigaux F., Judde J.G., et al. 2014. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 123:1021–1031. 10.1182/blood-2013-04-490847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel, F., Silva M.-J., Técher H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A.-L., et al. 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 557:57–61. 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- Crow, Y.J., and Stetson D.B.. 2022. The type I interferonopathies: 10 years on. Nat. Rev. Immunol. 22:471–483. 10.1038/s41577-021-00633-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddacha, W., Koyen A.E., Bastien A.J., Head P.E., Dhere V.R., Nabeta G.N., Connolly E.C., Werner E., Madden M.Z., Daly M.B., et al. 2017. SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 20:1921–1935. 10.1016/j.celrep.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., and Li H.. 2021. Twelve years of SAMtools and BCFtools. GigaScience. 10:giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenne, T., Klintman J., Sharma S., Rigby R.E., Blest H.T.W., Cursi C., Bridgeman A., Dadonaite B., De Keersmaecker K., Hillmen P., et al. 2020. SAMHD1 limits the efficacy of forodesine in leukemia by protecting cells against the cytotoxicity of dGTP. Cell Rep. 31:107640. 10.1016/j.celrep.2020.107640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco, M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D., et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 566:73–78. 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Mann, C.C., and Hopfner K.-P.. 2021. Nuclear cGAS: Guard or prisoner? EMBO J. 40:e108293. 10.15252/embj.2021108293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria, O., Cornen S., Daëron M., Morel Y., Medzhitov R., and Vivier E.. 2019. Harnessing innate immunity in cancer therapy. Nature. 574:45–56. 10.1038/s41586-019-1593-5 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, M., and Chen Z.J.. 2018. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 361:704–709. 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erttmann, S.F., Swacha P., Aung K.M., Brindefalk B., Jiang H., Härtlova A., Uhlin B.E., Wai S.N., and Gekara N.O.. 2022. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity. 55:847–861.e10. 10.1016/j.immuni.2022.04.006 [DOI] [PubMed] [Google Scholar]
- Ewels, P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., and Nahnsen S.. 2020. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38:276–278. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- Gao, D., Li T., Li X.-D., Chen X., Li Q.-Z., Wight-Carter M., and Chen Z.J.. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA. 112:E5699–E5705. 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili, M., Lahaye X., Nadalin F., Nader G.P.F., Lombardi E.P., Herve S., De Silva N.S., Rookhuizen D.C., Zueva E., Goudot C., et al. 2019. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26:3798. 10.1016/j.celrep.2019.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, A.M.S., Luciani M., Gatto F., Abou Alezz M., Beghè C., Della Volpe L., Migliara A., Valsoni S., Genua M., Dzieciatkowska M., et al. 2022. DNA damage contributes to neurotoxic inflammation in Aicardi-Goutières syndrome astrocytes. J. Exp. Med. 219:e20211121. 10.1084/jem.20211121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin, L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R.A., Diamond M.S., and Colonna M.. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 103:8459–8464. 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone, D.C., Ennis-Adeniran V., Hedden J.J., Groom H.C.T., Rice G.I., Christodoulou E., Walker P.A., Kelly G., Haire L.F., Yap M.W., et al. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 480:379–382. 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- Gray, E.E., Treuting P.M., Woodward J.J., and Stetson D.B.. 2015. Cutting edge: cGAS is required for lethal autoimmune disease in the trex1-deficient mouse model of aicardi-goutières syndrome. J. Immunol. 195:1939–1943. 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Merino, J., Isla B., Combes T., Martinez-Estrada F., and Maluquer De Motes C.. 2020. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microb. 11:771–788. 10.1080/19490976.2019.1707015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., and Ablasser A.. 2018. Targeting STING with covalent small-molecule inhibitors. Nature. 559:269–273. 10.1038/s41586-018-0287-8 [DOI] [PubMed] [Google Scholar]
- Herold, N., Rudd S.G., Ljungblad L., Sanjiv K., Myrberg I.H., Paulin C.B.J., Heshmati Y., Hagenkort A., Kutzner J., Page B.D.G., et al. 2017. Targeting SAMHD1 with the Vpx protein to improve cytarabine therapy for hematological malignancies. Nat. Med. 23:256–263. 10.1038/nm.4265 [DOI] [PubMed] [Google Scholar]
- Herrmann, A., Wittmann S., Thomas D., Shepard C.N., Kim B., Ferreirós N., and Gramberg T.. 2018. The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mobile DNA. 9:11. 10.1186/s13100-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, B., Hoppe A., Haase C., Hiller C., Schubert N., Müller W., Reijns M.A.M., Jackson A.P., Kunkel T.A., Wenzel J., et al. 2018. Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res. 78:5917–5926. 10.1158/0008-5472.CAN-18-1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka, K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M.P., and Skowronski J.. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 474:658–661. 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S., Li J., Xu F., Mei S., Le Duff Y., Yin L., Pang X., Cen S., Jin Q., Liang C., and Guo F.. 2015. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet. 11:e1005367. 10.1371/journal.pgen.1005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka, J.J., Manguso R.T., Cheruiyot C.K., Bi K., Panda A., Iracheta-Vellve A., Miller B.C., Du P.P., Yates K.B., Dubrot J., et al. 2019. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 565:43–48. 10.1038/s41586-018-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., and Weinberg R.A.. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1–7. 10.1016/s0960-9822(00)00002-6 [DOI] [PubMed] [Google Scholar]
- Jin, Y., Tam O.H., Paniagua E., and Hammell M.. 2015. TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 31:3593–3599. 10.1093/bioinformatics/btv422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis, G., and Stoye J.P.. 2016. Immune responses to endogenous retroelements: Taking the bad with the good. Nat. Rev. Immunol. 16:207–219. 10.1038/nri.2016.27 [DOI] [PubMed] [Google Scholar]
- Kretschmer, S., Wolf C., König N., Staroske W., Guck J., Häusler M., Luksch H., Nguyen L.A., Kim B., Alexopoulou D., et al. 2015. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 74:e17. 10.1136/annrheumdis-2013-204845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette, N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M.. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 474:654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, V., Böhme I., Hofmann J., Lang K., Sauter J., Schöne B., Paul P., Albrecht V., Andreas J.M., Baier D.M., et al. 2014. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics. 15:63. 10.1186/1471-2164-15-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienenklaus, S., Cornitescu M., Zietara N., Łyszkiewicz M., Gekara N., Jabłónska J., Edenhofer F., Rajewsky K., Bruder D., Hafner M., et al. 2009. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J. Immunol. 183:3229–3236. 10.4049/jimmunol.0804277 [DOI] [PubMed] [Google Scholar]
- Lima-Junior, D.S., Krishnamurthy S.R., Bouladoux N., Collins N., Han S.-J., Chen E.Y., Constantinides M.G., Link V.M., Lim A.I., Enamorado M., et al. 2021. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell. 184:3794–3811.e19. 10.1016/j.cell.2021.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.-Y.G., Näger A.C., Lunardi T., Vančevska A., Lossaint G., and Lingner J.. 2021. The human telomeric proteome during telomere replication. Nucleic Acids Res. 49:12119–12135. 10.1093/nar/gkab1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, K.J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548:461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait, J., Bridgeman A., Benlahrech A., Cursi C., and Rehwinkel J.. 2016. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 16:1492–1501. 10.1016/j.celrep.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerska, J., Feretzaki M., Glousker G., and Lingner J.. 2018. Transformation-induced stress at telomeres is counteracted through changes in the telomeric proteome including SAMHD1. Life Sci. Alliance. 1:e201800121. 10.26508/lsa.201800121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martins, V.C., Busch K., Juraeva D., Blum C., Ludwig C., Rasche V., Lasitschka F., Mastitsky S.E., Brors B., Hielscher T., et al. 2014. Cell competition is a tumour suppressor mechanism in the thymus. Nature. 509:465–470. 10.1038/nature13317 [DOI] [PubMed] [Google Scholar]
- Mauney, C.H., and Hollis T.. 2018. SAMHD1: Recurring roles in cell cycle, viral restriction, cancer, and innate immunity. Autoimmunity. 51:96–110. 10.1080/08916934.2018.1454912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdipour, P., Marhon S.A., Ettayebi I., Chakravarthy A., Hosseini A., Wang Y., de Castro F.A., Loo Yau H., Ishak C., Abelson S., et al. 2020. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature. 588:169–173. 10.1038/s41586-020-2844-1 [DOI] [PubMed] [Google Scholar]
- Michallet, M.-C., Meylan E., Ermolaeva M.A., Vazquez J., Rebsamen M., Curran J., Poeck H., Bscheider M., Hartmann G., König M., et al. 2008. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 28:651–661. 10.1016/j.immuni.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Morita, M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., and Barnes D.E.. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 24:6719–6727. 10.1128/MCB.24.15.6719-6727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K., Ryoo J., Jeong H., Kim M., Lee S., Hwang S.-Y., Ahn J., Kim D., Moon H.C., Baek D., et al. 2021. Aicardi-Goutières syndrome-associated gene SAMHD1 preserves genome integrity by preventing R-loop formation at transcription–replication conflict regions. PLoS Genet. 17:e1009523. 10.1371/journal.pgen.1009523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M.T., Gopinath S., Perez C.E., Linehan M.M., Crawford J.M., Iwasaki A., and Lindenbach B.D.. 2018. Innate immune priming by cGAS as a preparatory countermeasure against RNA virus infection. bioRxiv:434027. (Preprint posted October 03, 2018). 10.1101/434027 [DOI] [Google Scholar]
- Patro, R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke, K., Achleitner M., Frenzel K., Gerbaulet A., Ada S.R., Zeller N., Lienenklaus S., Lesche M., Poulet C., Naumann R., et al. 2016. Loss of Trex1 in dendritic cells is sufficient to trigger systemic autoimmunity. J. Immunol. 197:2157–2166. 10.4049/jimmunol.1600722 [DOI] [PubMed] [Google Scholar]
- Poon, S.S., and Lansdorp P.M.. 2001. Measurements of telomere length on individual chromosomes by image cytometry. Methods Cell Biol. 64:69–96. 10.1016/s0091-679x(01)64007-x [DOI] [PubMed] [Google Scholar]
- Purbey, P.K., Scumpia P.O., Kim P.J., Tong A.-J., Iwamoto K.S., McBride W.H., and Smale S.T.. 2017. Defined sensing mechanisms and signaling pathways contribute to the global inflammatory gene expression output elicited by ionizing radiation. Immunity. 47:421–434.e3. 10.1016/j.immuni.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri, G.H., Anders S., Pyl P.T., Pimanda J.E., and Zanini F.. 2022. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 38:2943–2945. 10.1093/bioinformatics/btac166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, U., Rinas M., Schwerk J., Nöhren G., Linnes M., Kröger A., Flossdorf M., Kály-Kullai K., Hauser H., Höfer T., and Köster M.. 2012. Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol. Syst. Biol. 8:584. 10.1038/msb.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J., Maelfait J., Bridgeman A., Rigby R., Hayward B., Liberatore R.A., Bieniasz P.D., Towers G.J., Moita L.F., Crow Y.J., et al. 2013. e SousaSAMHD1-dependent retroviral control and escape in mice. EMBO J. 32:2454–2462. 10.1038/emboj.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentoft, M., Lindell K., Tran P., Chabes A.L., Buckland R.J., Watt D.L., Marjavaara L., Nilsson A.K., Melin B., Trygg J., et al. 2016. Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc. Natl. Acad. Sci. USA. 113:4723–4728. 10.1073/pnas.1519128113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M., Fuller J.C., Jackson R.M., Lamb T., Briggs T.A., et al. 2009. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41:829–832. 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero, M.P., and Crow Y.J.. 2016. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med. 213:2527–2538. 10.1084/jem.20161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, J., Choi J., Oh C., Kim S., Seo M., Kim S.-Y., Seo D., Kim J., White T.E., Brandariz-Nuñez A., et al. 2014. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med. 20:936–941. 10.1038/nm.3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo, J., Hwang S.-Y., Choi J., Oh C., and Ahn K.. 2016. Reply to SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat. Med. 22:1074–1075. 10.1038/nm.4164 [DOI] [PubMed] [Google Scholar]
- Sauer, J.-D., Sotelo-Troha K., von Moltke J., Monroe K.M., Rae C.S., Brubaker S.W., Hyodo M., Hayakawa Y., Woodward J.J., Portnoy D.A., and Vance R.E.. 2011. The N-ethyl-N-Nitrosourea-Induced goldenticket mouse mutant reveals an essential function of sting in the in vivo interferon response to listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79:688–694. 10.1128/IAI.00999-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp, L., Muth S., Rogell L., Kofoed-Branzk M., Melchior F., Lienenklaus S., Ganal-Vonarburg S.C., Klein M., Guendel F., Hain T., et al. 2020. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell. 181:1080–1096.e19. 10.1016/j.cell.2020.04.022 [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk, J.L., Schmidt T., Gaidt M.M., Pelka K., Latz E., Ebert T.S., and Hornung V.. 2014. OutKnocker: A web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 24:1719–1723. 10.1101/gr.176701.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Oellerich T., Baldauf H.-M., Schwarz S.-M., Thomas D., Flick R., Bohnenberger H., Kaderali L., Stegmann L., Cremer A., et al. 2017. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med. 23:250–255. 10.1038/nm.4255 [DOI] [PubMed] [Google Scholar]
- Schoggins, J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., et al. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 505:691–695. 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, N., Schumann T., Daum E., Flade K., Ge Y., Hagedorn L., Edelmann W., Müller L., Schmitz M., Kuut G., et al. 2022. Genome replication is associated with release of immunogenic DNA waste. Front. Immunol. 13:880413. 10.3389/fimmu.2022.880413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon, K.J., Sun Z., Shlyakhtenko L.S., Lyubchenko Y.L., and Stivers J.T.. 2015. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 43:6486–6499. 10.1093/nar/gkv633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M., Meter M.V., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., Cecco M.D., Leonova K.I., Kogan V., Helfand S.L., et al. 2019. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29:871–885.e5. 10.1016/j.cmet.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, C., Wang T., Michaelos M., and Pachitariu M.. 2021. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods. 18:100–106. 10.1038/s41592-020-01018-x [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thientosapol, E.S., Bosnjak D., Durack T., Stevanovski I., van Geldermalsen M., Holst J., Jahan Z., Shepard C., Weninger W., Kim B., et al. 2018. SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc. Natl. Acad. Sci. USA. 115:4921–4926. 10.1073/pnas.1719771115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzé, G., Di Marco S., Mouchel-Vielh E., Monneron D., Bandu M.-T., Horisberger M.A., Dorques A., Lutfalla G., and Mogensen K.E.. 1994. Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 243:245–257. 10.1006/jmbi.1994.1651 [DOI] [PubMed] [Google Scholar]
- Vanpouille-Box, C., Hoffmann J.A., and Galluzzi L.. 2019. Pharmacological modulation of nucleic acid sensors: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Dis. 18:845–867. 10.1038/s41573-019-0043-2 [DOI] [PubMed] [Google Scholar]
- Wittmann, S., Behrendt R., Eissmann K., Volkmann B., Thomas D., Ebert T., Cribier A., Benkirane M., Hornung V., Bouzas N.F., and Gramberg T.. 2015. Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology. 12:103. 10.1186/s12977-015-0229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, G.R., Eksmond U., Salcedo R., Alexopoulou L., Stoye J.P., and Kassiotis G.. 2012. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 491:774–778. 10.1038/nature11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, P., Lübben W., Slomka H., Gebler J., Konert M., Cai C., Neubrandt L., Prazeres da Costa O., Paul S., Dehnert S., et al. 2012. Nucleic acid-sensing toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 37:867–879. 10.1016/j.immuni.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Yu, C.H., Bhattacharya A., Persaud M., Taylor A.B., Wang Z., Bulnes-Ramos A., Xu J., Selyutina A., Martinez-Lopez A., Cano K., et al. 2021. Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification. Nat. Commun. 12:731. 10.1038/s41467-021-21023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 370:eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., Du J., Han X., Goodier J.L., Li P., Zhou X., Wei W., Evans S.L., Li L., Zhang W., et al. 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by aicardi-goutières syndrome-related SAMHD1. Cell Rep. 4:1108–1115. 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists oligonucleotides.
contains original blots for Fig. S1.
contains original blots for Fig. S2.
contains original blots for Fig. S4.