Summary
Several in vitro models have been developed to recapitulate mouse embryogenesis solely from embryonic stem cells (ESCs). Despite mimicking many aspects of early development, they fail to capture the interactions between embryonic and extraembryonic tissues. To overcome this difficulty, we have developed a mouse ESC-based in vitro model that reconstitutes the pluripotent ESC lineage and the two extra-embryonic lineages of the post-implantation embryo by transcription factor-mediated induction. This unified model recapitulates developmental events from embryonic day 5.5 to 8.5, including gastrulation, and formation of the anterior-posterior axis, brain, a beating heart structure, and the development of extraembryonic tissues, including yolk sac and chorion. Comparing single-cell RNA sequencing from individual structures with time-matched natural embryos identified remarkably similar transcriptional programs across lineages, but also showed when and where the model diverges from the natural program. Our findings demonstrate an extraordinary plasticity of ESCs to self-organise and generate a whole embryo-like structure.
Graphical Abstract
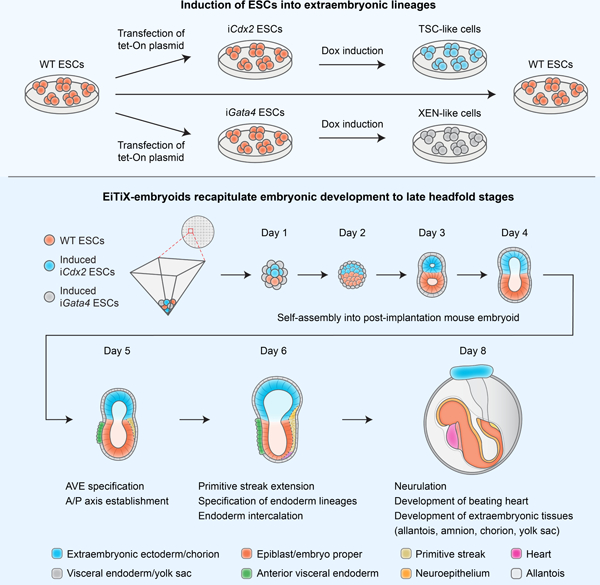
In brief:
Lau and colleagues generated an in vitro mouse embryo model with embryonic and extraembryonic lineages using exclusively embryonic stem cells and showed that the embryoids can undergo advanced development to late headfold stages. Single-cell RNA sequencing demonstrates similarity between embryoids and natural mouse embryos.
Introduction
At the time of implantation, the mouse blastocyst comprises three lineages: the epiblast (EPI), the trophectoderm (TE), and the primitive endoderm (PE) that will give rise to the embryo proper, the placenta, and the yolk sac respectively. By using stem cells derived from these lineages, several in vitro models have been developed to recapitulate various events of post-implantation development. One approach has been to take solely mouse embryonic stem cells (ESCs) and by applying exogenous stimuli induce them to establish anterior-posterior polarity (ten Berge et al., 2008) and mimic basic body axis formation, and aspects of gastrulation, somitogenesis, cardiogenesis and neurulation (Van Den Brink et al., 2014; Turner et al., 2017; Beccari et al., 2018; Rossi et al., 2020; Veenvliet et al., 2020; Xu et al., 2021). Such so-called “gastruloids” are a powerful system and demonstrate the ability of ESCs to be directed into complex developmental programs. However, these systems fail to capture the entire complexity of signalling and morphological events along the complete body axes. This is largely because they fail to recapitulate the spatio-temporal interplay of signalling pathways between embryonic and extraembryonic tissues, which is crucial to pattern the post-implantation mouse embryo. Consequently, they do not represent complete embryonic structures and lack the overall morphological resemblance to natural post-implantation mouse embryos.
We have therefore adopted a second approach to fully model the post-implantation mouse embryo by promoting assembly of mouse ESCs with either extraembryonic trophoblast stem cells (TSCs), to direct formation of a post-implantation egg cylinder showing appropriate posterior development (Harrison et al., 2017), or a mixture of TSCs and extraembryonic endoderm (XEN) stem cells to generate “ETX” embryos that develop anterior and posterior identity and gastrulation movements (Sozen et al., 2019). Subsequently, by replacing XEN cells with ESCs harbouring inducible Gata4 expression (iGata4 ESCs), it proved possible to generate XEN cells at an earlier stage of development which could contribute to iETX embryos that were fully able to complete gastrulation movements (Amadei et al., 2021). One remaining complication of the iETX embryo model is that TSCs and ESCs require different culture media, necessitating the use of undefined culture conditions and increasing the difficulty of developing embryoids in the laboratory. Therefore, we have developed an entirely ESC-based in vitro model that reconstitutes the three fundamental cell lineages of the post-implantation mouse embryo through transcription factor-mediated reprogramming. In addition to replacing XEN cells with induced XEN cells, we now further substitute TSCs with ESCs that transiently overexpress Cdx2 upon doxycycline induction. We show that such induced TSCs could effectively replace TSCs to form embryo-like structures which we termed “EiTiX-embryoids”. These EiTiX-embryoids undergo development from pre-gastrulation stages to neurulation stages, developing headfolds, brain, a beating heart structure, and extraembryonic tissues, including a yolk sac and chorion. In agreement with the similar overall morphology, our single cell, single structure analysis reveals a robust recapitulation of cell states spanning both embryonic and extraembryonic lineages, with strikingly little variation in the overall gene expression program in these states. Yet, our approach also demonstrates that Cdx2-expressing cells can contribute to the chorion but not the ectoplacental cone lineage in the extraembryonic ectoderm compartment.
Results
Cdx2-induced ESCs self-assemble with Gata4-induced ESCs and ESCs into post-implantation-like mouse embryoids
Cdx2 is a key transcription factor driving TE development and its overexpression leads ESCs to transdifferentiate into TSC-like cells (Niwa et al., 2005). To determine whether Cdx2-expressing ESCs could replace TSCs in generating ETiX-embryoids, we generated a transgenic ESC line carrying a doxycycline (Dox)-inducible Cdx2 gene. The resulting clones of iCdx2-ESCs showed a 100- to 200-fold increase in Cdx2 mRNA expression after 6 hours of Dox induction (Figure S1A). From the four clones we tested, we selected the clone with the highest level of Cdx2 overexpression for subsequent experiments. This clone showed a substantial upregulation of both Cdx2 mRNA (Figure S1B) and protein, as detected by qRT-PCR and immunofluorescence respectively, after 6 hours of induction (Figure S1C). To assess the long-term effect of Cdx2 overexpression on cell fate, we compared three different types of cell aggregates: either induced iCdx2 ESCs, uninduced iCdx2 ESCs; or TSCs (Figure 1A). After three days, we observed a significant upregulation of the TSC marker, Eomes, and downregulation of the ESC marker, Oct4, in the aggregates of induced iCdx2 ESCs (Figures 1B, 1C, S1D and S1E). Transcripts of the TSC markers Elf5, Eomes and Gata3 were also upregulated in the induced iCdx2 ESC aggregates (Figures S1F–H). Together, these findings suggest that upon Cdx2 overexpression, iCdx2 ESCs lose their ESC identity and acquire TSC-like cell fate.
Figure 1. Cdx2-induced ESCs self-assemble with Gata4-induced ESCs and ESCs into post-implantation-like mouse embryoids.
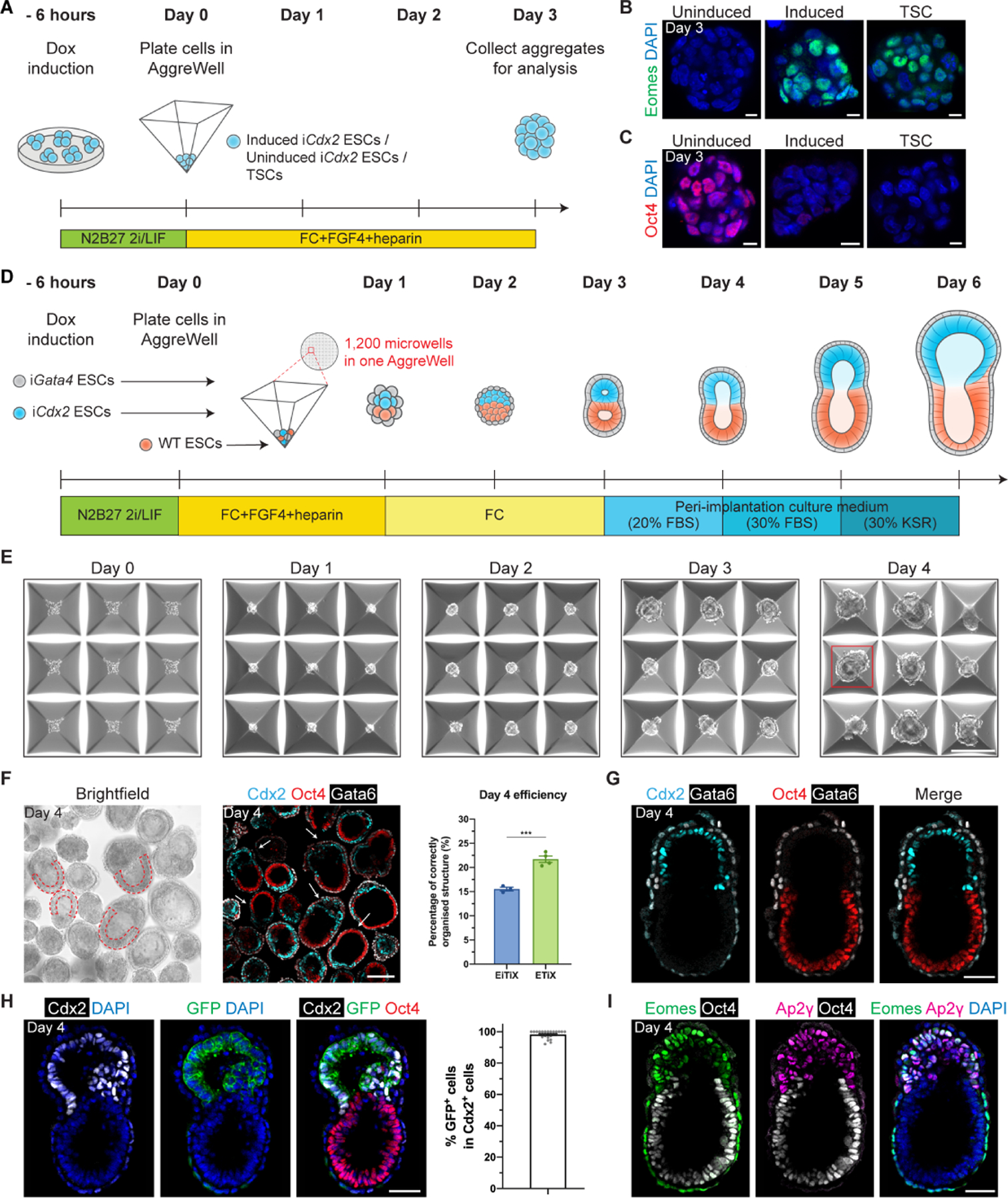
(A) Schematic of the formation of cell aggregates in AggreWells. Day 3 iCdx2 ESC aggregates show elevated Eomes expression (B) and downregulated Oct4 (C) upon induction of Cdx2. (D) Schematic of EiTiX-embryoid generation. FBS: fetal bovine serum, KSR: knockout serum replacement. (E) Representative brightfield images of structures developing in AggreWells from Day 0 to Day 4. A structure resembling the early post-implantation mouse embryo can be seen on Day 4 in the well outlined in red. (F) All structures in the combined microwells from one AggreWell were collected at Day 4 and stained to reveal Cdx2 (cyan), Oct4 (red) and Gata6 (white). Arrows indicate structures considered to exhibit correct organisation. Such cylindrical structures with two cellular compartments and an epithelialised EPI-like cell layer (red dashed outline) were selected under brightfield microscopy. The efficiency of obtaining organised structures in EiTiX-embryoid system and ETiX-embryoid system is shown. Efficiency of ETiX-embryoid is taken from our previous publication (referring to these embryoids as iETX-embryoids, Amadei et al., 2021). (G) Day 4 EiTiX-embryoids stained to reveal Cdx2 (cyan), Oct4 (red) and Gata6 (white). (H) Day 4 EiTiX-embryoid stained to reveal Cdx2 (white), GFP (green) and Oct4 (red). The percentage of the Cdx2-positive cells that are also GFP-positive is shown. n = 49 structures. (I) Day 4 EiTiX-embryoids stained to reveal Eomes (green), Ap2γ (magenta) and Oct4 (white). n = 35/35 structures are positive for both Eomes and Ap2γ. All experiments were performed minimum 3 times. Scale bars: 10μm, (B-C); 150μm, (E-F); 50μm, (G-I). See also Figure S1.
We then asked whether induced iCdx2 ESCs could replace TSCs in generating embryoids when aggregated with wild-type (WT) ESCs and Gata4 induced ESCs. To this end, we adapted our previously described protocol (Amadei et al., 2021) by inducing expression of Cdx2 and Gata4 by treating both iCdx2- and iGata4-ESC lines for 6 hours with doxycycline before combining them with WT ESCs in AggreWell plates (Figure 1D). Over the course of four days, we observed drastic morphological changes of the resulting cell aggregates such that by Day 4 we could observe structures that resembled post-implantation embryos, which naturally comprise EPI and TE-derived extra-embryonic ectoderm (ExE) compartments surrounded by visceral endoderm (VE) (Figure 1E). The random nature of the interactions of the three cell types results in a variety of structures (Figure S1I) but we optimised the efficiency of correct structure formation by adding Fgf4 and heparin during the first 24 hours after plating and by doubling the number of iCdx2 ESCs seeded from 16 to 32 per microwell (Figures 1D and 1F). We showed that the efficiency of correct structure formation on Day 4 was 15.5%, which is slightly lower than ETiX-embryoid system (formerly termed iETX-embryoid) (Amadei et al., 2021), reflecting differences between using iCdx2 ESCs and TSCs (Figure 1F). We note that by setting up several AggreWells that each contain 1200 microwells at the start of the experiment, we would routinely obtain around 350 well-developed embryo-like structures per experiment on Day 4 (Figures 1G, Methods), which exceeded our capacity to transfer all of them to post-implantation culture conditions due to the costs of serum for culture media.
When we examined the expression of the constitutive membrane GFP marker of iCdx2 ESCs, we found that iCdx2 ESCs had given rise to the Cdx2-positive cells that correctly localised within the ExE-like compartment (Figure 1H). We also detected the expression of other ExE markers including Ap2γ and Eomes in the putative ExE compartment, suggesting downstream TSC markers were also upregulated after Cdx2 overexpression (Figure 1I). Finally, we compared the dimensions of EiTiX-embryoids, E5.5 mouse embryos and Day 4 ETiX-embryoids and found that EiTiX-embryoids were most similar to E5.5 embryos (Figures S1J–M). Together, our findings show that iCdx2 ESCs can replace TSCs to generate post-implantation embryo-like structures expressing canonical lineage markers. Since TSCs were replaced by iCdx2 ESC, we termed the structures “EiTiX-embryoids.”
EiTiX-embryoids establish an anterior-posterior axis and undergo gastrulation
We then asked whether EiTiX-embryoids could recapitulate key events of post-implantation development. We first asked whether the critical anterior signalling center can be formed, which breaks mouse embryo symmetry and establishes the anterior-posterior axis (Thomas and Beddington, 1996). This center first appears as the distal visceral endoderm (DVE) at the distal tip of the egg cylinder before migrating to the anterior side of the egg cylinder to become the anterior visceral endoderm (AVE), which is characterised by the expression of Cer1, Lefty1 and Dkk1 (Figure 2A). To this end, we formed EiTiX-embryoids using iGata4 ESCs with a Cer1-GFP reporter and observed the co-expression of GFP with Dkk1 or Lefty1 in EiTiX-embryoids at Day 4 and Day 5 (Figures 2B, S2A and S2B). To follow the development of the Cer1-GFP-positive domain, we determined the extent of AVE anterior migration (see Methods) and binned the measurements into three groups: ‘proximal’, >67% migration; ‘lateral’, 33–67% migration, and ‘distal’, <33% migration. Anterior migration of the AVE was evident from the higher proportion of Day 5 EiTiX-embryoids with proximal Cer1-GFP and Dkk1 expression than in Day 4 EiTiX-embryoids, while the distribution of Lefty1-positive domain remained similar (Figures 2C and S2C).
Figure 2. EiTiX-embryoids establish an anterior-posterior axis and undergo gastrulation.
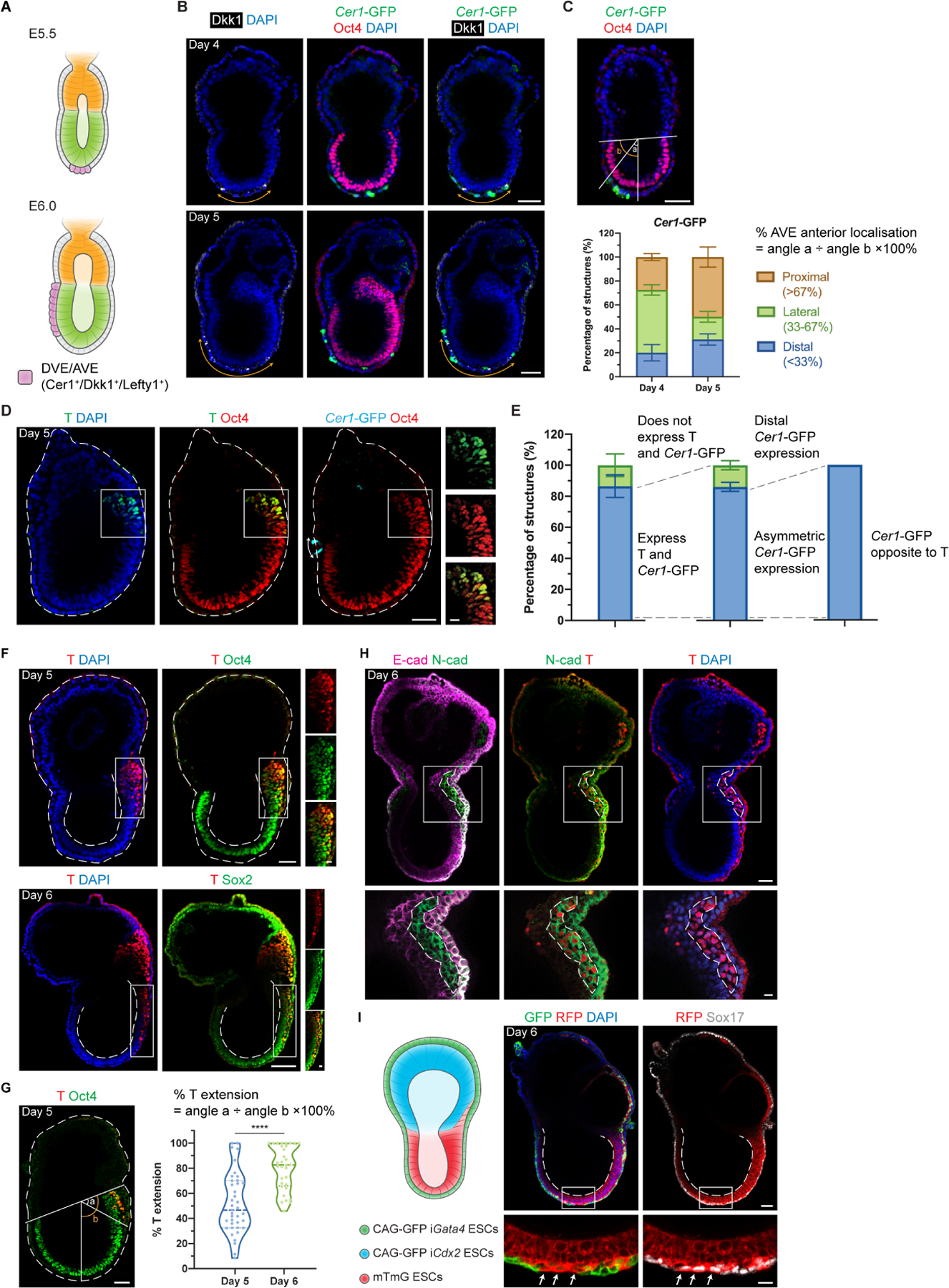
(A) Schematic showing the position of DVE/AVE in E5.5 and E6.5 mouse embryos. (B) Day 4 and Day 5 EiTiX-embryoids stained to reveal Cer1-GFP (green), Oct4 (red) and Dkk1 (white). Orange double-headed arrows indicate Dkk1-positive domains. (C) Localisation of Cer1-GFP in Day 4 and Day 5 EiTiX-embryoids (See Materials and Methods for quantification method). n = 57 Day 4 structures; 45 Day 5 structures. (D) Day 5 EiTiX-embryoids stained to reveal T (green), Oct4 (red) and Cer1-GFP (cyan). Double-headed arrow, Cer1-GFP-expressing domain; white box outline, T- and Oct4-positive domain. (E) Percentages of Day 5 EiTiX-embryoids showing 1) expression of both Cer1-GFP and T in same structure, 2) asymmetric Cer1-GFP expression, and 3) T expression on the opposite side from Cer1-GFP. n = 42 structures. (F) Day 5 and Day 6 EiTiX-embryoids stained to reveal T (red) and Oct4 (green) or Sox2 (green). White boxes enclose T-positive domain while dotted lines outline the structure and the lumen of ES compartment. n = 39/42 Day 5 structures and 32/32 Day 6 structures. (G) Percentage of T extension in Day 5 and Day 6 EiTiX-embryoids. See Materials and Methods for quantification method. n = 39 Day 5 structures and 32 Day 6 structures. ****p < 0.0001. (H) Day 6 EiTiX-embryoid stained to reveal E-cadherin (magenta), N-cadherin (green) and T (red). Dotted line indicates T- and N-cadherin-positive domain. n = 14/21 structures with N-cadherin upregulation and E-cadherin downregulation from 4 experiments. (I) Schematic of EiTiX-embryoids using CAG-GFP iGata4 ESCs (with membrane GFP) and mTmG ESCs (with membrane tdTomato) to construct the VE-like layer and EPI-like compartment, respectively. Day 6 EiTiX-embryoid stained to reveal GFP (green), RFP (red) and Sox17 (white). Dotted line indicates the lumen of ES compartment while arrows mark definitive endoderm-like cells intercalated into the VE-like layer. n = 8/12 structures. All experiments were performed minimum 3 times. Scale bars: 50μm; 15μm (zoomed). See also Figure S2.
In the natural post-implantation embryo, the AVE is critical to restrict primitive streak formation to the posterior EPI through the secretion of Nodal and Wnt inhibitors (Stower and Srinivas, 2017). We therefore asked whether these events could be recapitulated in EiTiX-embryoids and analysed the expression of Brachyury (T), a primitive streak marker, in relation to the Cer1-GFP domain at Day 5. We found that 86.7% of Day 5 EiTiX-embryoids expressed Cer1-GFP and T, and of these, 86% showed opposed Cer1-GFP and T expression (Figures 2D and 2E). Similarly, we found that 94.7% of structures with asymmetric AVE expression of Cer1-, Dkk1-, or Lefty1 showed expression of the primitive streak marker, Eomes, on the opposite side (Figures S2D–F). Thus, EiTiX-embryoids correctly establish both the AVE and primitive streak, recapitulating anterior-posterior patterning as in natural post-implantation embryos.
After establishment of the anterior-posterior axis and onset of gastrulation in the posterior EPI, the primitive streak extends to the distal end of the egg cylinder (Bardot and Hadjantonakis, 2020). Accordingly, as EiTiX-embryoids developed, we could detect T- and Oct4-positive cells at the posterior end of the EPI-like compartment on Day 5, that had extended to the distal-most part of the egg cylinder on Day 6 (Figure 2F). To quantify the percentile extension of this T-positive domain, we measured the angle between the posterior boundary of the Oct4-positive domain and the most anterior T-positive cells (Figure 2G, angle a) as well as the angle subtended by the Oct4-positive domain boundary and the distal tip (angle b), where the percentage of angle a divided by angle b indicates the percentile extension. This showed the degree of extension approached its fullest extent at Day 6 (Figure 2G). As cells egress from the EPI to form the primitive streak, they undergo epithelial-to-mesenchymal transition, downregulating E-cadherin and upregulating N-cadherin (Arnold and Robertson, 2009). We observed a T-positive domain that had robust N-cadherin expression but unlike surrounding cells, did not express E-cadherin in 66.7% of EiTiX-embryoids at Day 6 (Figure 2H).
As development progresses, the primitive streak undergoes further specification to produce a range of cell types, including axial mesendoderm and definitive endoderm (Bardot and Hadjantonakis, 2020; Scheibner et al., 2021). In Day 6 EiTiX-embryoids, we could detect the presence of T- and Foxa2-positive cells identifying axial mesendoderm (93.8%), as well as Foxa2- and Sox17-positive cells identifying definitive endoderm (88.2%) (Figures S2G and S2H). In the natural mouse embryo, the EPI-derived definitive endoderm gradually displaces and intercalates with the VE which covers the egg cylinder (Kwon, Viotti and Hadjantonakis, 2008). To visualise whether such endoderm intercalation takes place in EiTiX-embryoids, we used ESCs expressing membrane tdTomato (mTmG ESCs) to generate the EPI-like compartment and iGata4 ESCs expressing membrane GFP (CAG-GFP iGata4 ESCs) to generate the VE-like layer in the embryoids. We observed a discontinuous GFP-positive cell layer interspersed with RFP-positive cells (Figure 2I, 66.7%). These cells also expressed Sox17 which is a critical factor for endoderm specification and for the egression of definitive endoderm cells into the VE (Viotti, Nowotschin and Hadjantonakis, 2014). Thus, the intercalation of definitive endoderm into the VE is recapitulated in the EiTiX-embryoids.
Finally, we asked whether the ExE-like compartment in EiTiX-embryoid could recapitulate functions of ExE in natural mouse embryos. Bmp4 is a crucial signalling molecule expressed specifically in the ExE, which is important for patterning the embryos through cross-talk with embryonic tissues and specifying mesoderm and primordial germ cells (PGCs) (Winnier et al., 1995; Lawson et al., 1999; Arnold and Robertson, 2009). We assessed Bmp4 expression by in situ hybridisation and could detect robust Bmp4 expression in the ExE-like compartment in both Day 5 and Day 6 EiTiX-embryoids (Figure S2I). Notably, we observed a stronger Bmp4 signal in the posterior ExE-like compartment, in line with the natural mouse embryo where Bmp4 expression is gradually restricted to the posterior (Arnold and Robertson, 2009). Since Bmp4 is necessary for PGC specification, we asked whether PGC progenitors were present in EiTiX-embryoids but we could not reliably detect PGC progenitors in Day 6 EiTiX-embryoids, which might reflect insufficient Bmp signalling.
Day 6 EiTiX-embryoids capture major cell types of gastrulation
After finding that Day 6 EiTiX-embryoids could capture numerous processes of gastrulation, we sought to understand the overall cell type composition of these gastrulating EiTiX-embryoids in comparison to natural embryos. We utilised a recently established time-resolved model of mouse gastrulation consisting of ~68,000 single cells derived from 287 individually processed embryos spanning egg cylinder stage to early somitogenesis (Figure 3A, Mittnenzweig et al., 2021), which: (i) enables a quantitative evaluation of transcriptional states; and (ii) describes the natural flux of embryonic and extraembryonic lineage differentiation, thus allowing analysis of cell state composition within individual structures. We generated Day 6 EiTiX-embryoids by combining iCdx2 ESCs with constitutive membrane GFP-expression (CAG-GFP), unlabelled WT ESCs and iGata4 ESCs carrying the Cer1-GFP reporter. GFP signals confirmed the appearance of the ExE-like compartment and the AVE-like domain in EiTiX-embryoids (Figure 3B). Next, we performed single-cell RNA sequencing (scRNA-seq) on 14 individual EiTiX-embryoids using MARS-seq by index-sorting into barcoded 384-well plate as previously reported (Mittnenzweig et al., 2021).
Figure 3. Day 6 EiTiX-embryoids capture major cell types of gastrulation.
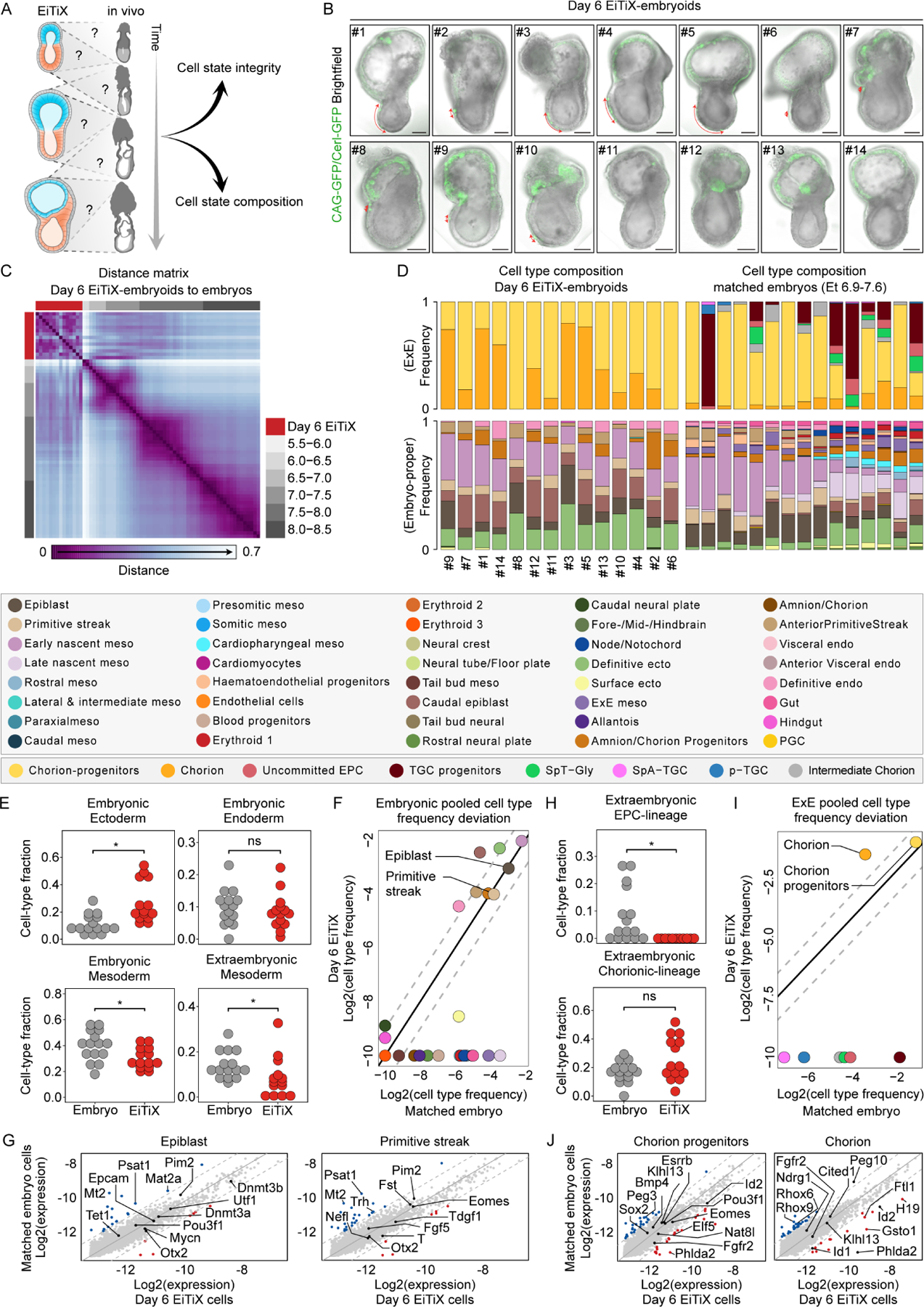
(A) Evaluation of cell state integrity and cell type composition of Day 6 EiTiX-embryoids using our recently established time-resolved model of mouse gastrulation. (B) Brightfield images of Day 6 EiTiX-embryoids (n = 14, annotated as S#1–14) collected for single-structure, single-cell RNA sequencing, overlaid with GFP expression (Cer1-GFP reporter in VE-like layer and membrane CAG-GFP from iCdx2 ESCs in ExE-like compartment). Red arrow indicates Cer1-GFP expression. Scale bar: 100μm. (C) Embryo-embryo cell type composition similarity matrix. Natural embryos are annotated based on embryonic age groups (grey), Day 6 EiTiX-embryoids (red). (D) Cell type composition bars of individual Day 6 EiTiX-embryoids (left) and matched natural embryos (right, annotated according to the key shown below). (E, H) Comparison of major embryonic germ layers frequency (E) and ExE lineages frequency (H) between Day 6 EiTiX-embryoids and matching natural embryos. Medians of frequencies were compared using a Wilcoxon-Mann-Whitney rank sum test after down sampling of cell state specific cells to corresponding number of Day 6 EiTiX cell state specific cells (i.e. for each cell state individually). q Values were calculated from p values according to the Benjamini-Hochberg procedure. ns, not significant; *, q value < 0.05. Major germ layers - Embryonic ectoderm; Forebrain/Midbrain/Hindbrain, Rostral neural plate, Surface ectoderm, Caudal neural plate, Definitive ectoderm. Embryonic endoderm; Definitive endoderm, Gut, Hindgut, Visceral and Anterior Visceral endoderm. Embryonic mesoderm; Tail bud-, Early and Late nascent-, Caudal-, Presomitic-, Somitic-, Paraxial-, Rostral-, Cardioparyngeal- and Lateral & intermediate-mesoderm. ExE Mesoderm; Amnion/Chorion progenitor, Amnion/Chorion, Allantois and ExE mesoderm. EPC-lineage; SpT-Gly, TGC progenitors, uncommitted EPC, pTGC and SpA-TGCs. Chorion-lineage; intermediate ExE, Chorion progenitors and Chorion. (F, I) Pooled embryonic (F) and ExE (I) cell type frequencies comparison between Day 6 EiTiX-embryoids and matched natural embryos. (G, J) Bulk differential gene expression per cell type of Day 6 EiTiX cells against matched embryo cells in embryonic cell types (G, EPI and primitive streak) and ExE cell types (J, chorion progenitors and chorion). Dots represent individual genes. Color annotated dots mark genes with a two-fold change in expression (blue – above two-fold decrease in Day 6 EiTiX cells, red – above two-fold increase in Day 6 EiTiX cells). See also Figure S3.
A strategy for ranking embryos by K-nn similarities among their single-cell profiles identified high similarity between individual Day 6 EiTiX-embryoids. In agreement with the morphological assessment of these embryoids, their overall transcriptional ranking was found to be most similar to E6.5–7.5 gastrulation stages (Figures 3C and S3A). Next, we constructed and annotated a transcriptional manifold of EiTiX-embryoids (see Methods). Remarkably, we found robust mapping to unmodified embryonic and extraembryonic cell states and did not detect any non-coherent transcriptional programs in the metacells of Day 6 EiTiX-embryoids, suggesting they conserve the transcriptional programs of the corresponding cell states in natural embryos (Figure S3B). Focusing first on embryonic cell state compositions, we observed a high degree of similarity among the 14 Day 6 EiTiX-embryoids, despite variable morphologies (Figure 3D). However, when compared to natural embryos from corresponding time bins (Methods), we found deviations from the natural program. First, we noted some of the lineages were not in synchrony. Specifically, while the relatively high proportions of EPI and embryonic nascent mesoderm coincides with the cell state composition of natural early to mid-gastrulating embryos (~E6.5–7.0), the specification of extraembryonic mesoderm derivatives (i.e. chorion/amnion progenitors), caudal EPI and definitive ectoderm frequencies resembles the composition of advanced gastrulating natural embryo stages (~E7.0–7.5, Figures 3D–F). Yet, the transcriptional profile of properly represented cell states was nearly identical to comparable cell states in natural embryos (Figures 3G and S3C–D).
Ectoplacental cone (EPC) lineage were absent from Day 6 EiTiX-embryoids compared to respective natural embryos. In contrast, the chorion lineage, comprising both chorion progenitors and their differentiated progenies, was largely intact (Figures 3D, 3H–I and S3C). Gene expression analysis showed both programs to be overall highly similar to the natural embryos (Figure 3J). It also revealed down-regulation of bonafide chorion genes Rhox6 and Rhox9, together with upregulation of Id2, consistent with a lack of proximal signals emanating from the EPC compartment. Taken together, our analysis identified remarkably similar transcriptional states between Day 6 EiTiX-embryoids and their natural counterparts, but it also revealed pausing in mesoderm differentiation and over-accumulation of posterior cell types, most likely reflecting alterations in synchronicity between the mesoderm and ectoderm lineages. We therefore next asked whether further culture of embryoids would enhance the synchronicity of lineage development.
EiTiX-embryoids develop to late headfold stages with heart and chorion development
To assess the full developmental potential of EiTiX-embryoids, we next transferred them from peri-implantation (Bedzhov and Zernicka-Goetz, 2014) to post-implantation culture medium (Sturm and Tam, 1993; Aguilera-Castrejon et al., 2021) (Figure 4A). We found that EiTiX-embryoids developed to establish headfolds, a beating heart, allantois and chorion structures over the next three days in culture and that they shared highly similar morphologies with E8.5 natural embryos cultured in vitro (Figures 4B, movie S1 and S2). EiTiX-embryoids also developed amnion-like and yolk sac-like membranes which properly enveloped the embryonic structures. Moreover, regions resembling blood islands, both in spatial localisation and stereotypical red pigmentation, could be observed on the yolk sac-like membrane (Figure 4C). The efficiencies of EiTiX-embryoids progressing from Day 5 to 6, Day 6 to 7 and Day 7 to 8 were between 65.4% and 75%, which are comparable to ETiX-embryoids (Figure 4D). Successfully developed Day 8 EiTiX-embryoids had well defined structures resembling headfolds, heart and tail (Figure S4A). The most commonly observed phenotypes of underdeveloped Day 8 EiTiX-embryoids include stunted overall development and impaired axial elongation to generate posterior structures (Figure S4B).
Figure 4. EiTiX-embryoids develop to late headfold stages with heart and chorion development.
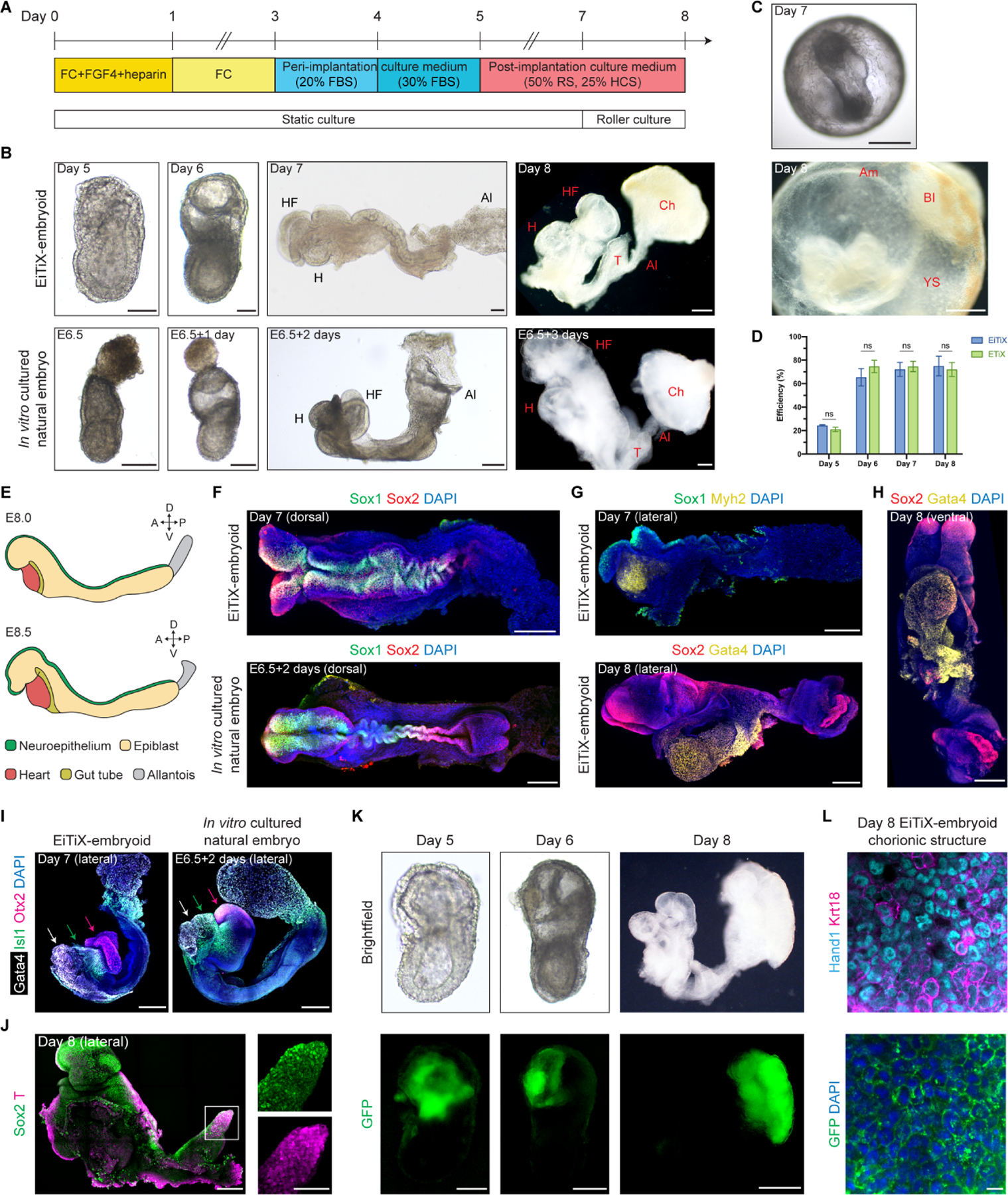
(A) Schematic showing culture conditions of EiTiX-embryoids to Day 8. FBS: fetal bovine serum, RS: rat serum, HCS: human cord serum. (B) Representative brightfield images EiTiX-embryoids cultured from Day 5 to 8 and E6.5 natural embryo cultured in vitro for 3 days. Al: allantois, Ch: chorion, H: heart, HF: headfolds, T: tail. Scale bar, 100μm. (C) Brightfield images of Day 7 and Day 8 EiTiX-embryoids before dissecting yolk sac-like membrane. Am: amnion, BI: blood island, YS: yolk sac. Scale bar, 200μm (Day 7); 500μm (Day 8). (D) Efficiency of EiTiX-embryoid and ETiX-embryoid progression from Day 4 to 5, Day 5 to 6, Day 6 to 7, and Day 7 to 8. Efficiency of ETiX-embryoid is taken from our publication (Amadei et al., 2022). Embryoids were selected on each day for further culture and experiments were performed minimum 3 times. Multiple t-tests. ns, non-significant. (E) Schematic showing major cell types in E8.0 and E8.5 embryos. (F) Dorsal view of Day 7 EiTiX-embryoid and E6.5 natural embryo cultured in vitro for 2 days and stained to reveal Sox1 (green) and Sox2 (red). Scale bar, 200μm. (G) Lateral view of Day 7 and Day 8 EiTiX-embryoids stained to reveal neuroepithelial markers Sox1 (green) or Sox2 (red) and heart markers Myh2 or Gata4 (yellow). Scale bar, 200μm. (H) Ventral view of Day 8 EiTiX-embryoid stained to reveal Sox2 (red) and Gata4 (yellow), resembling the linear heart tube stage. Scale bar, 200μm. (I) Lateral view of Day 7 EiTiX-embryoid and E6.5 natural embryo cultured in vitro for two days stained to reveal heart marker Gata4 (white), pharyngeal mesoderm marker Isl1 (green), and forebrain marker Otx2 (magenta). Scale bar, 200μm. (J) Lateral view of Day 8 EiTiX-embryoids stained to reveal Sox2 (green) and T (red). Magnified panel showing co-expression in tail region (white square). Scale bar, 200μm; 100μm (zoomed). (K) Representative brightfield and GFP fluorescence image of Day 5 to 8 EiTiX-embryoids to track the contribution of CAG-GFP iCdx2 ESCs. Structures show GFP expression in chorion-like region. Scale bar, 50μm (Day 5); 200μm (Day 6); 500μm (Day 8). (L) Dissected chorionic structure from Day 8 EiTiX-embryoid stained to reveal GFP (green), Hand1 (cyan) and Keratin18 (magenta). Scale bar, 100μm. See also Figure S4 and Movie S1–2.
Similar to natural E8.0 embryos and E6.5 embryos cultured in vitro for 2 days, the neuroepithelium markers Sox1 and Sox2 were expressed along the anterior-posterior axis of Day 7 EiTiX-embryoids (Figures 4E and 4F), indicative of neurulation. Interestingly we observed twisting of the neural tube-like region in both Day 7 EiTiX-embryoid and in vitro cultured embryo, suggesting that this could be a defect of in vitro culture. The heart markers Myh2 and Gata4 were expressed below the headfolds (Figure 4G), and a ventral view of the Gata4-expressing heart region revealed a morphology that resembled the linear heart tube (Figure 4H). Importantly, the anterior region of the headfolds showed robust Otx2 expression, indicating development of the forebrain (Figures 4I and S4C). We also detected the expression of Islet1 (Isl1), a pharyngeal mesoderm marker, between the Gata4-expressing heart region and Otx2-expressing forebrain region, recapitulating the expression pattern in the in vitro cultured embryos (Figures 4I and S4C). At the posterior end of the body axis, we observed robust co-expression of Sox2 and T at the region resembling the tail, which identifies the neuromesodermal progenitor population (Figure 4J).
As the neurulating EiTiX-embryoids arise from ESC and two different types of induced extraembryonic ESC types, we wished to determine the extent of extraembryonic tissue development. We detected chorion, an ExE-derived tissue that forms part of the placenta, and chorion progenitors in Day 6 EiTiX-embryoids by scRNA-seq (Figure 3D) and could observe a region that resembled the chorion in Day 8 EiTiX-embryoids (Figure 4K). To ask whether this region was derived from iCdx2 ESCs, we generated Day 8 EiTiX embryos using iCdx2 ESCs constitutively expressing membrane associated GFP and combined these with unlabelled WT and iGata4 ESCs. We observed membrane-associated GFP in the ExE region in Day 5 and Day 6 EiTiX-embryoids and at Day 8, the membrane GFP was exclusively found in the region resembling the chorion (Figure 4K). Further examination of this latter region showed co-expression of the chorion markers, Hand1 and Keratin18 (Figure 4L).
Another crucial extraembryonic cell type is the extraembryonic mesoderm, which contributes to amnion and yolk sac in the natural mouse embryo (Jin et al., 2016). In Day 6 EiTiX-embryoids, we could observe cells expressing both T, a mesoderm marker, and Runx1, a haematopoietic marker that is enriched in extraembryonic mesoderm (Saykali et al., 2019) in the region resembling the proximal EPI (Figure S4D), suggesting the emergence of extraembryonic mesoderm. Runx1 is also a key transcription factor driving definitive haematopoiesis and is expressed in blood islands (Tanaka et al., 2014). As we had observed blood islands on the yolk sac-like membrane in Day 8 EiTiX-embryoids (Figure 4C), we dissected out the yolk sac-like membrane and could detect robust Runx1 expression, indicative of extraembryonic haematopoiesis (Figure S4E).
Taken together, these results indicate that EiTiX-embryoids have the remarkable ability to develop to headfold stages. They not only give rise to advanced embryonic structures such as neuroepithelium, a beating heart and mesodermal populations, but importantly, also develop extraembryonic tissues including yolk sac and chorion. The tracking of membrane GFP-positive iCdx2 ESCs further confirmed iCdx2 ESCs can effectively develop into chorion, an ExE-derived tissue, demonstrating that we could generate an embryoid with embryonic and extraembryonic tissues entirely from ESCs.
Cell state and composition analysis of neurulating embryoids using scRNA-seq
To undertake a comprehensive analysis of cell state integrity and composition in neurulating embryoids, we collected four Day 8 EiTiX-embryoids for scRNA-seq (Figure 5A). Transcriptional similarity analysis showed Day 8 EiTiX-embryoids overall most resemble E8.0–8.5 stages (Figure 5B). Analysis of cell state composition confirmed the high similarity between individual embryoids. In addition, Day 8 embryoids displayed high synchronicity between lineages and exhibited advanced cell states consistent with late head fold stages (Figures 5C, S5A–B). Quantitative analysis of cell-state frequency deviations identified depletion of tail bud cell types and haematoendothelial progenitors. Furthermore, we found an over-representation of cardiomyocytes cell state (Figure 5D), which may correspond to the apparent larger cardiac domain (Figure S4A). We note that these deviations must be viewed cautiously, given the low frequency associated with some of these cell types. For example, although presomitic mesoderm was not present in Day 8 EiTiX-embryoids, a bonafide somitic mesoderm population could be detected, suggesting that this progenitor population was merely not sampled. Finally, we detected similar gene expression patterns in high-frequency cell states compared to controls (Figures 5E and S5C).
Figure 5. Cell state and composition analysis of neurulating embryoids using scRNA-seq.
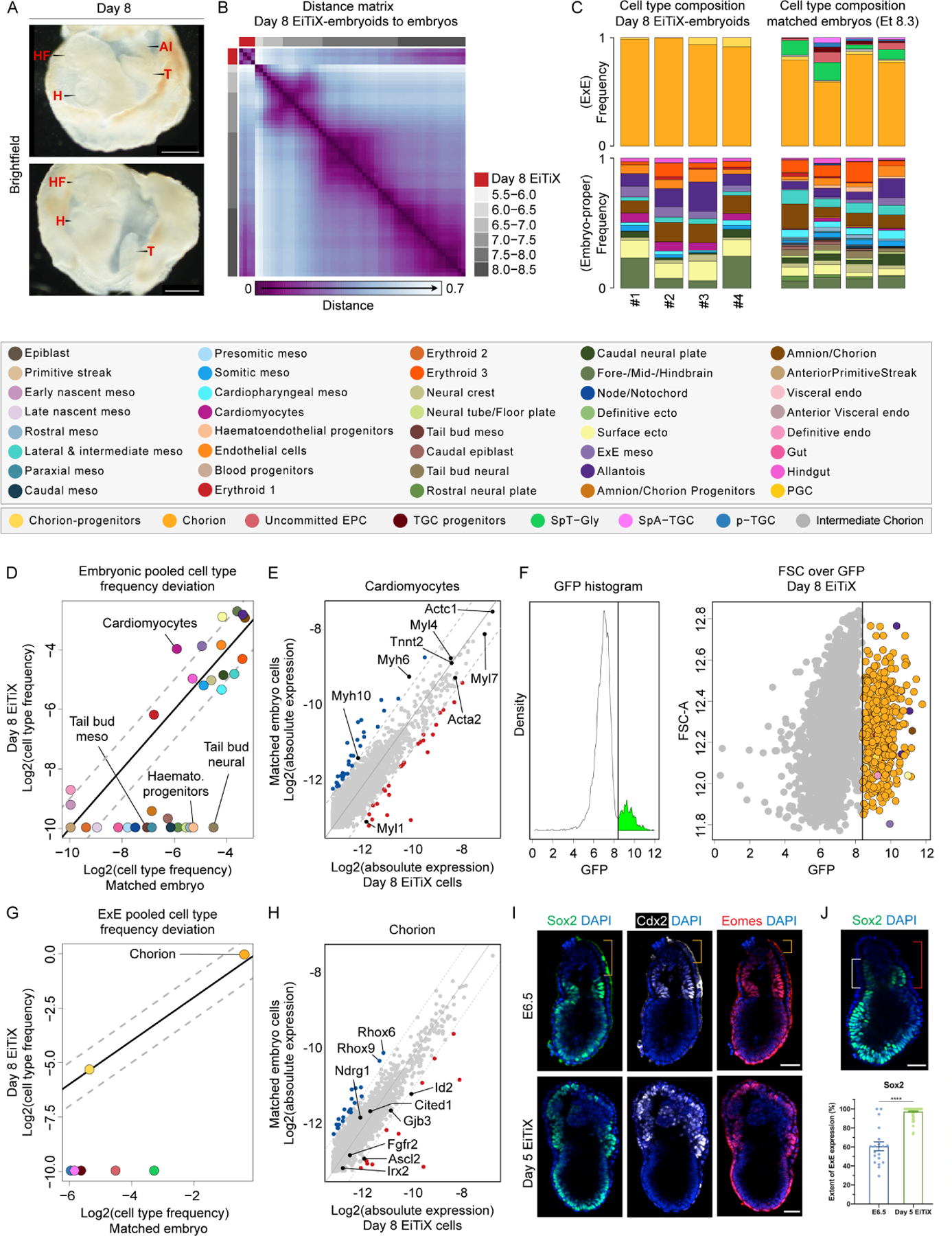
(A) Brightfield images of Day 8 EiTiX-embryoids collected for single-structure, single-cell RNA sequencing. The yolk sac-like membrane was partially opened to reveal embryonic structures. Al: allantois, H: heart, HF: headfolds, T: tail. Scale bar: 500μm. (B) Embryo-embryo cell type composition similarity matrix. Natural embryos are annotated based on embryonic age groups (grey), Day 8 EiTiX-embryoids (red). (C) Cell type composition bars of individual Day 8 EiTiX-embryoids (left) and matched natural embryos (right, annotated according to the legend below). (D, G) Pooled embryonic (D) and ExE (G) cell type frequencies comparison between Day 8 EiTiX-embryoids and matched natural embryos. (E, H) Bulk differential gene expression per cell state of Day 6 EiTiX cells against matched embryo cells in embryonic cell type (E, cardiomyocyte) and ExE cell type (H, chorion). Dots represent individual genes. Colour annotated dots mark genes with a two-fold change in expression (blue – above two-fold decrease in Day 6 EiTiX cells, red – above two-fold increase in Day 6 EiTiX cells). (F) GFP channel bimodal distribution (left) with threshold use to define GFP+ cell population shown as dots, annotated accordingly to cell type (right). (I) Day 5 EiTiX-embryoids and E6.5 natural embryos stained to reveal trophoblast stem markers Sox2 (green), Cdx2 (white) and Eomes (red). Orange brackets show the absence of trophoblast stem markers in the tip of ExE in E6.5 embryos. Scale bar: 50μm. (J) Quantification of the extent of ExE expression of Sox2. It was determined by dividing the height of the expression domain (white bracket) by the height of ExE (red bracket), multiplied by 100%. n = 19 E6.5 embryos from 2 experiments and 78 Day 5 EiTiX-embryoids from 3 experiments; ****p < 0.0001. Scale bar: 50μm. See also Figure S5.
Analysing ExE differentiation showed expected progression in the chorion lineage (i.e. most chorion progenitors fully converting to their differentiated progenies). However, we could not detect any cell types associated with the EPC lineage, including uncommitted EPCs and trophoblast giant cells (TGC) progenitors (Figure 5C and S5A). We could confirm that the vast majority of GFP-positive iCdx2 ESCs (97.26%) gave rise to chorion lineage, although we noted a few embryonic cell types among the GFP positive cells (Figure 5F). Indeed, we occasionally observed GFP-positive cells in the EPI-like compartment in Day 4 EiTiX-embryoids, and we suspected that these cells might have retained ESC fate and would eventually give rise to embryonic lineages. Overall, cells in the chorion lineage appeared with comparable frequencies and exhibited highly similar gene expression signatures when compared to time-matched natural embryos (Figures 5G and 5H). Our inability to detect EPC and TGC subtypes in both Day 6 and Day 8 EiTiX-embryoids suggested iCdx2 ESCs exhibit restricted ExE differentiation potential. In the natural embryo, the ExE can be subdivided into proximal ExE (adjacent to the EPI/ExE boundary) and distal ExE (towards the tip of the embryo). The proximal ExE is characterised by the expression of TSC-like markers such as Sox2, Cdx2 and Eomes, which are downregulated in the distal ExE, where the EPC and TGC subtypes are found (Donnison, Broadhurst and Pfeffer, 2015). We confirmed the absence of TSC-like markers in the distal ExE in E6.5 embryos (orange bracket), whereas in Day 5 EiTiX-embryoids, we observed strong expression of these genes throughout the ExE compartment (Figure 5I), with an extended-expression domain (Figure 5J and S5D).
Discussion
Here we show that ESCs carrying a Cdx2 transgene can adopt a TSC-like cell fate upon doxycycline-induced Cdx2 overexpression. The resulting iCdx2 ESCs have the ability to self-assemble with ESCs induced to overexpress Gata4 (induced XEN cells) and wildtype ESCs to generate an in vitro model of mouse post-implantation development with embryonic and extraembryonic lineages. The embryonic-extraembryonic embryo model we present here, which we term “EiTiX embryoids”, is derived entirely of ESCs and thus circumvents the use of undefined media to culture conventional extraembryonic cell lines. When we culture EiTiX-embryoids in the peri-implantation culture medium in stationary culture (Bedzhov and Zernicka-Goetz, 2014), they specify the DVE and AVE, establish an anterior-posterior axis, and undergo gastrulation. Following transfer into roller-bottle system and enriched post-implantation culture medium (Sturm and Tam, 1993; Aguilera-Castrejon et al., 2021), EiTiX-embryoids undertake neurulation and form headfolds, brain, a beating heart structure, and develop extraembryonic tissues including yolk sac and chorion.
We performed single-structure scRNA-seq of 14 Day 6 and 4 Day 8 EiTiX-embryoids and projected the data on a temporal model describing the parallel differentiation in embryonic and extraembryonic lineages. This enabled direct comparisons with time-matched natural embryos, providing an analytical framework for quantifying the fidelity of intracellular transcriptional programs and overall cell composition within individual structures. We observed overall similarity of Day 6 EiTiX-embryoids with natural embryos of E6.0 to E7.5 stages, whereas Day 8 EiTiX-embryoids were most similar to natural embryos from E8.0 to E8.5. Despite the morphological variability of EiTiX-embryoids, transcriptional states appeared remarkably conserved compared to corresponding ones in natural embryos. We noted two main types of deviation from the natural flow of the embryo proper: (i) First, some differentiated cell types were missing in both Day 6 and Day 8 embryoids, and (ii) synchronicity between lineages was impaired in Day 6 embryos. Nevertheless, adaptations in culture conditions significantly improved lineage synchronicity in Day 8 embryoids, resulting in much comparable cell compositions to that of time-matched embryos. The analytical approach described here can complement future screening aimed at improved culture conditions, by providing a robust quantitative readout on embryoid development.
The EiTiX-embryo is thus able to develop many more tissues than structures derived solely from homogeneous populations of ESCs induced to differentiate by various exogenous molecules, generating a more complete in vitro model with both embryonic and extraembryonic tissues. Hence, our data substantiate the essential role of extraembryonic tissues in driving the self-organisation of mouse embryo-like structures. For example, the role of ESCs induced to express Gata4 is of critical importance in establishing the formation of the AVE, which is required to direct the formation of anterior structures, particularly such as those of the forebrain.
Although the induced extraembryonic structures contribute to the correct development of diverse embryonic cell types and overall structure in EiTiX-embryoids, the development of the ExE lineage is incomplete as reflected by the lack of EPC and TGC cell types. This can be partly because EiTiX-embryoids lack the interactions with the maternal environment that they would have in utero; and partly because transcription factor-mediated induction biases iCdx2 ESCs to differentiate into chorionic cell types. It is possible that there are two types of progenitor cells in the ExE splitting immediately after implantation and iCdx2 ESCs resemble most the chorion lineage progenitors. Unlike the ExE in E6.5 natural embryos, we showed that there is a strong and extended expression of TSC-like markers throughout the ExE-like compartment in Day 5 EiTiX-embryoids. Thus, it might be necessary to introduce the expression of genes promoting ExE differentiation or induce the downregulation of TSC-like genes to generate EPC and TGC subtypes in EiTiX embryo. Moreover, by incorporating TE-derived cell types, our system offers future possibilities for dissecting the precise roles of such cells in the developmental process.
We have also recently reported advanced development of ETiX-embryoids (which use TSCs, unmodified ESCs and Gata4 inducible ESCs) to late headfold stages. Interestingly ETiX-embryoids also lack TGCs and spongiotrophoblasts, which could again be attributed to the lack of maternal interaction (Amadei et al., 2022). Here we show that EiTiX-embryoids and ETiX-embryoids share similar efficiency to progress to late headfold stages and that they undergo similar development of major tissues, including embryonic tissues such as headfolds and heart, and extraembryonic tissues such as allantois, chorion and yolk sac blood island. Nevertheless, we note that ETiX-embryoids capture more cell types than EiTiX-embryoids. For example, PGCs could be detected near the allantois in Day 7 and Day 8 ETiX-embryoids but not in Day 8 EiTiX-embryoids. This could reflect differences in signalling molecules secreted by TSCs compared to iCdx2 ESCs. Another group has also recently reported the generation of embryoids from mouse ESCs adapting our previously described protocol (Amadei et al., 2021) and using a similar induction approach to the one we describe here, although with a much longer induction period (1 to 14 days as compared to 6 hours here) (Tarazi et al., 2022). Similar to our EiTiX-embryoids, their embryoids reached a developmental stage resembling E8.5 embryos and did not develop further. They also noted the absence of some TGC and spongiotrophoblast markers.
Despite not recapitulating the later stages of development of extraembryonic tissues and their lack of some cell types present in ETiX-embryoids, the substitution of TSCs by iCdx2 ESCs in EiTiX-embryoid permits remarkable development of the embryo per se, with the development of a yolk sac and chorion. The reconstitution of the three principal lineages of peri-implantation development exclusively from ESCs ensures simplified, defined and consistent culture conditions to recapitulate the interactions between embryonic and extraembryonic tissues that facilitate development through gastrulation to neurulation-like stages.
Limitations of the study
We have shown that iCdx2 ESCs could replace TSCs to generate embryoids, but the extend of their contribution to ExE lineages in natural embryos was not tested here. However, we note that it has been reported that Cdx2-overexpressing ESCs could contribute to the placenta of E12.5 chimeric embryos (Niwa et al., 2005). Secondly, we found that the level of Cdx2 overexpression is critical to successful generation of EiTiX-embryoids. We have tested the generation of EiTiX-embryoids using the clone of iCdx2 ESCs that had the lowest level of Cdx2 overexpression (Figure S1A, clone B) and found the efficiency of obtaining organised structures on Day 4 was very low (approximately 3%), which could be due to insufficient reprogramming of iCdx2 ESCs into TSC-like cells. Furthermore, EiTiX-embryoids generated using different ESC lines had different developmental potential. In our hands, EiTiX-embryoids generated using a combination of CD1 unmodified ESCs, CD1 Gata4 inducible ESCs and CAG-GFP Cdx2 inducible ESCs could consistently undergo advanced development to late headfold stages. Lastly, we note that the efficiency of EiTiX-embryoids progressing to Day 4 and Day 5 is only 15.5% and 24.4% respectively. However, by setting up several AggreWells at the start of the experiment, we would routinely obtain around 350 well-developed Day 4 EiTiX-embryoids per experiment, which exceeded our capacity to transfer all of them to post-implantation culture conditions due to the costs and availability of serum for culture media.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to Magdalena Zernicka-Goetz (magdaz@caltech.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
Single-cell RNA sequencing data have been deposited at ArrayExpress and are publicly available as of the date of publication. Accession number is listed in the key resources table.
Code used in the study has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-AP-2 gamma | R&D Systems | Cat# AF5059, RRID: AB_2255891 |
| Mouse monoclonal anti-AP-2gamma | Santa Cruz Biotechnology | Cat# sc-12762, RRID: AB_667770 |
| Goat polyclonal anti-Brachyury | R&D Systems | Cat# AF2085, RRID: AB_2200235 |
| Mouse monoclonal anti-Cdx2 | BioGenex | Cat# MU392-UC, RRID: AB_2335627 |
| Rabbit monoclonal anti-Cdx2 | Abcam | Cat# ab76541, RRID: AB_1523334 |
| Rat monoclonal anti-Cerberus 1 | R&D Systems | Cat# MAB1986, RRID: AB_2275974 |
| Goat polyclonal anti-Dkk1 | R&D Systems | Cat# AF1096, RRID: AB_354597 |
| Rat monoclonal anti-E-Cadherin | Thermo Fisher Scientific | Cat# 13–1900, RRID: AB_2533005 |
| Rabbit polyclonal anti-TBR2 / Eomes | Abcam | Cat# ab23345, RRID: AB_778267 |
| Rabbit monoclonal anti-FoxA2 / HNF3 | Cell Signaling Technology | Cat# 8186, RRID: AB_10891055 |
| Rabbit polyclonal anti-GATA-4 | Santa Cruz Biotechnology | Cat# sc-9053, RRID: AB_2247396 |
| Goat polyclonal anti-GATA-6 | R&D Systems | Cat# AF1700, RRID: AB_2108901 |
| Rat monoclonal anti-GFP | Nacalai Tesque | Cat# GF090R, RRID: AB_2314545 |
| Mouse monoclonal anti-eHAND | Santa Cruz Biotechnology | Cat# sc-390376 |
| Rabbit polyclonal anti-Keratin18 | Sigma-Aldrich | Cat# SAB4501665, RRID: AB_10746153 |
| Goat polyclonal anti-Lefty | R&D Systems | Cat# AF746, RRID: AB_355566 |
| Mouse monoclonal anti-Myh2 | R&D Systems | Cat# MAB4470, RRID: AB_1293549 |
| Mouse monoclonal anti-N-Cadherin | BD Biosciences | Cat# 610920, RRID: AB_2077527 |
| Goat polyclonal anti-Nkx2.5 | R&D Systems | Cat# AF2444, RRID: AB_355269 |
| Mouse monoclonal anti-Oct-3/4 | Santa Cruz Biotechnology | Cat# sc-5279, RRID: AB_628051 |
| Goat polyclonal anti-Otx2 | R&D Systems | Cat# AF1979, RRID: AB_2157172 |
| Rabbit polyclonal anti-RFP | Rockland | Cat# 600–401-379, RRID: AB_2209751 |
| Rabbit monoclonal anti-RUNX1 / AML1 | Abcam | Cat# ab92336, RRID: AB_2049267 |
| Rabbit polyclonal anti-Sox1 | Cell Signaling Technology | Cat# 4194, RRID: AB_1904140 |
| Mouse monoclonal anti-Sox2 | Santa Cruz Biotechnology | Cat# sc-365823, RRID: AB_10842165 |
| Rat monoclonal anti-Sox2 | Thermo Fisher Scientific | Cat# 14–9811-82, RRID: AB_11219471 |
| Goat polyclonal anti-Sox17 | R&D Systems | Cat# AF1924, RRID: AB_355060 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_2535792 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11055, RRID: AB_2534102 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10037, RRID: AB_2534013 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10042, RRID: AB_2534017 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11057, RRID: AB_2534104 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571, RRID: AB_162542 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573, RRID: AB_2536183 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21447, RRID: AB_2535864 |
| Donkey Anti-Rat IgG H&L (Alexa Fluor® 647) preadsorbed antibody | Abcam | Cat# ab150155, RRID: AB_2813835 |
| Bacterial and virus strains | ||
| 5-alpha competent E. coli | New England Biolabs | C2987I |
| Chemicals, peptides, and recombinant proteins | ||
| β-estradiol | Sigma-Aldrich | Cat# E8875–1G |
| CHIR99021 | Cambridge Stem Cell Institute | N/A |
| Doxycycline | Sigma-Aldrich | Cat# D9891–5G |
| Fgf2 | Cambridge Stem Cell Institute | N/A |
| Recombinant Mouse FGF-4 (aa 67–202) | R&D Systems | Cat# 7486-F4–025 |
| Heparin | Sigma-Aldrich | Cat# H3149–25KU |
| Insulin-transferrin-selenium-ethanolamine | ThermoFisher Scientific | Cat# 51500–056 |
| Leukaemia inhibitory factor | Cambridge Stem Cell Institute | N/A |
| N-acetyl-L-cysteine | Sigma-Aldrich | Cat# A7250 |
| PD0325901 | Cambridge Stem Cell Institute | N/A |
| Progesterone | Sigma-Aldrich | Cat# P8783–1G |
| Y-27632 | STEMCELL Technologies |
Cat# 72304 |
| Critical commercial assays | ||
| Gateway™ BP Clonase™ II Enzyme mix | Invitrogen | Cat# 11789–100 |
| Gateway™ LR Clonase™ II Enzyme mix | Invitrogen | Cat# 11791–100 |
| Lipofectamine™ 3000 Transfection Reagent | Invitrogen | Cat# L3000001 |
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4368708 |
| Deposited data | ||
| Single-cell RNA sequencing data | This manuscript | ArrayExpress: E-MTAB-12140 |
| Code for analysing single-cell RNA sequencing data | This manuscript | DOI: 10.5281/zenodo.7021607 |
| Experimental models: Cell lines | ||
| Mouse: CAG-GFP ESCs | Rhee et al., 2006 | N/A |
| Mouse: CAG-GFP/tetO-mCherry/tetO-Cdx2 ESCs | This manuscript | N/A |
| Mouse: CAG-GFP/tetO-mCherry/tetO-Gata4 ESCs | Amadei et al., 2021 | N/A |
| Mouse: Cerl-GFP/tetO-Gata4 ESCs | Amadei et al., 2021 | N/A |
| Mouse: CD1 ESCs | Prof Jennifer Nichols (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Mouse: CD1/tetO-Gata4 ESCs | Amadei et al., 2022 | N/A |
| Mouse: Confetti TSCs | Prof Jennifer Nichols (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Mouse: mT/mG mouse ESCs | Muzumdar et al., 2007 | N/A |
| Mouse: Sox2-Venus/Brachyury-mCherry/Oct4-Venus mouse ESCs | Dr Jesse Veenvliet and Prof Bernhard G. Herrmann (Max Planck Institute of Molecular Cell Biology and Genetics) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1 | Charles River Laboratories | N/A |
| Oligonucleotides | ||
| PCR primer: Cdx2-attB forward: GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTACGTGAGCTACCTTCTGGAC | This manuscript | N/A |
| PCR primer: Cdx2-attB reverse: GGGGACCACTTTGTACAAGAAAGCTGGGTTTCACTGGGTGACAGTGGAGTTTAAAAC | This manuscript | N/A |
| qPCR primer: Cdx2 forward: AAACCTGTGCGAGTGGATG | Blij et al., 2012 | N/A |
| qPCR primer: Cdx2 reverse: TCTGTGTACACCACCCGGTA | Blij et al., 2012 | N/A |
| qPCR primer: Elf5 forward: ATTTCTACAGTCCGCTGGTGC | This manuscript | N/A |
| qPCR primer: Elf5 reverse: ACATCACCGTGAAGACAAGTGG | This manuscript | N/A |
| qPCR primer: Gapdh forward: CGTATTGGGCGCCTGGTCAC | Amadei et al., 2021 | N/A |
| qPCR primer: Gapdh reverse: ATGATGACCCTTTTGGCTCC | Amadei et al., 2021 | N/A |
| qPCR primer: Gata3 forward: TGTCTGCGAACACTGAGCTG | This manuscript | N/A |
| qPCR primer: Gata3 reverse: CGATCACCTGAGTAGCAAGGAG | This manuscript | N/A |
| Recombinant DNA | ||
| PB-tetO-hygro | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| pBAse | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| rtTA-zeocyin | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji |
| metacell_0.3.7 | Baran et al., 2018 | https://github.com/tanaylab/metacell |
| Metacells 0.9.0-dev.1 | Ben-Kiki et al., 2022 | https://github.com/tanaylab/metacells#id3 |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| RStudio | RStudio | https://www.rstudio.com/ |
| Smart Denoise | Gurdon Institute | N/A |
| Other | ||
| AggreWell400 | STEMCELL Technologies |
Cat# 34415 |
| Anti-Adherence Rinsing Solution | STEMCELL Technologies |
Cat# 07010 |
| FACSAria III | BD Biosciences | N/A |
| Human cord serum | Cambridge Blood and Stem Cell Biobank | N/A |
| Knockout serum replacement | Gibco | Cat# 10828010 |
| Leica SP5 | Leica Microsystems | N/A |
| Leica SP8 | Leica Microsystems | N/A |
| M-MuLV Reverse Transcriptase | New England Biolabs | Cat# M0253S |
| Non-adherent 6 well multiwell plate | Greiner Bio-One | Cat# 657185 |
| 48-well multi-well plate | Greiner Bio-One | Cat# 677102 |
| Rat serum | Charles River Laboratories | N/A |
| StepOnePlus™ Real-Time PCR System | Applied Biosystems | N/A |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture conditions
We have used the following ESC lines:
CAG-GFP mouse ESCs: ESCs with constitutive membrane GFP expression, derived from CAG-GFP reporter mice (Rhee et al., 2006).
CAG-GFP tetO-Cdx2 mouse ESCs: ESCs overexpressing Cdx2 upon Dox induction, generated in-house using methods described below.
CAG-GFP tetO-Gata4 mouse ESCs: ESCs overexpressing Gata4 upon Dox induction generated as previously reported (Amadei et al., 2021)
Cer1-GFP tetoO-Gata4 mouse ESCs: ESCs with GFP expression under Cer1 promoter which overexpress Gata4 upon Dox induction, generated as previously reported (Amadei et al., 2021)
CD1 mouse ESCs: a generous gift from Jennifer Nichols
CD1 tetO-Gata4 mouse ESCs: ESCs overexpressing Gata4 upon Dox induction generated as previously reported (Amadei et al., 2022)
Confetti TSCs: a generous gift from Jennifer Nichols
mT/mG mouse ESCs: ESCs expressing membrane tdTomato, derived from mT/mG mice (Muzumdar et al., 2007)
Sox2-Venus/Brachyury-mCherry/Oct4-Venus mouse ESCs: a generous gift from Jesse Veenvliet and Bernhard G. Herrmann
All cell lines were cultured and passaged as previously reported (Amadei et al., 2021). Briefly, all cell lines were cultured at 37°C, 5% CO2 and 21% O2. All ESC lines were cultured on gelatin-coated plates in N2B27 consisting of 50% DMEM/F12 (Gibco 21331–020), 50% Neurobasal A (Gibco 10888–022), 1% B27 (Gibco A1895601), 0.5% N2 (made in-house), 0.1mM beta-mercaptoethanol (Gibco 31350–010), 1% penicillin/streptomycin (Gibco 15140–122) and 2mM Glutamax (Thermo Fisher Scientific 35050–038), supplemented with 3μM CHIR99021 (Cambridge Stem Cell Institute), 1μM PD0325901 (Cambridge Stem Cell Institute) and 10ng/ml mouse leukaemia inhibitory factor (Cambridge Stem Cell Institute). TSCs were cultured on mitotically inactivated mouse embryonic fibroblasts (MEFs, Insight Biotechnology, ASF-1201) in FC media supplemented with 1 μg ml−1 heparin (Sigma-Aldrich H3149–25KU), 25 ng ml−1 Fgf4 (R&D Systems 7486-F4–025) and 25 ng ml-1 Fgf2 (Cambridge Stem Cell Institute). FC media is a DMEM-based media (Gibco 41966–029) with 15% heat-inactivated FBS (Gibco 10270–098), 0.1mM beta-mercaptoethanol (Gibco 31350–010), 1% penicillin/streptomycin (Gibco 15140–122), 2mM Glutamax (Thermo Fisher Scientific 35050–038), 1% NEAA (Gibco 11140–035) and 1mM sodium pyruvate (Gibco 11360–039). Cell lines were routinely passaged every 3–4 days by treating with trypsin-EDTA (Gibco 25300054) for 4 minutes, followed by inactivation with double volume of FC media. ESC cell pellet was washed with PBS before resuspending in N2B27 2i/LIF for replating. TSC cell pellet was washed with PBS, resuspended in FC media and added to gelatin-coated well to deplete MEFs for 20 minutes. TSC cell suspension was then collected and replated on to MEFs. Mycoplasma tests were carried out every two weeks to exclude contamination. Genotyping was not performed determine the sex of the cell lines.
Mouse model and recovery of mouse embryos
CD-1 mice were maintained in the University of Cambridge’s University Biomedical Services Combined Animal Facility, adhering to national and international guidelines. Experiments were performed under the regulation of the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 and were reviewed by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). Experiments were also approved by the Home Office.
Natural mating was performed with six-week-old CD-1 females and mouse embryos were recovered at embryonic days E6.5 by dissecting from the deciduae in M2 medium, as we described before (Zernicka-Goetz et al., 1997).
METHOD DETAILS
Plasmids and transfection
Cdx2 DNA flanked by attB sites was PCR-amplified from TSC cDNA using Cdx2-attB primers. Using Gateway technology (Thermo Fisher Scientific), it was then cloned into PB-tetO-hygromycin according to the manufacturer’s instructions and the resulting plasmid, PB-tetO-hygro-Cdx2 was verified by Sanger sequencing.
PB-tetO-hygro-Cdx2 was then transfected into 12,000 CAG-GFP ESCs together with pBAse and rtTA-zeocyin (0.25 μg each) using Lipofectamine 3000 Transfection Reagent (Invitrogen L3000001). Antibiotic selection was performed for 7 days with hygromycin (1:250; Gibco 10687010) and zeocyin (1:1000; InvivoGen ant-zn-1), followed by clonal expansion.
PB-tetO-hygro, pBAse and rtTA-zeocyin were generously gifted by Dr Jose Silva from the Stem Cell Institute (Cambridge, UK).
RNA extraction and qRT-PCR
RNA was extracted from cell pellets using either Trizol Reagent (Invitrogen 15596–026) or RNeasy Mini Kit (Qiagen 74104). It was subsequently reverse transcribed into cDNA with M-MuLV reverse transcriptase (New England Biolabs M0253S). qRT-PCR was carried out using SYBR Green PCR Master Mix (Applied Biosystems 4368708) and StepOnePlus™ Real-Time PCR System (Applied Biosystems). ΔΔCt method was used to calculate fold change using GAPDH as endogenous control. Sequences of qPCR primers for Cdx2 and Gapdh were taken from published papers (Blij et al., 2015; Amadei et al., 2021) while other qPCR primers were designed and validated in this study.
Formation of cell aggregates and EiTiX-embryoids
Cell aggregates and EiTiX-embryoids were generated largely following the previously described method (Amadei et al., 2021) with some modifications.
To generate cell aggregates, 1 mg/ml doxycycline (Sigma-Aldrich D9891–5G) was added to iCdx2 ESCs for 6 hours. Then, each AggreWell (STEMCELL Technologies 34415) was treated with 500μl anti-adherence rinsing solution (STEMCELL Technologies 07010) and the AggreWell plate was centrifuged at 2000g for 5 minutes to remove bubbles in microwells. Anti-adherence rinsing solution was kept in the AggreWell for 20 minutes. ESCs were then dissociated using the above method (Cell lines and culture conditions) and cell density in the cell suspension was determined using a haemocytometer. For cell aggregates, 38,400 6-hour induced iCdx2 ESCs, 19,200 uninduced iCdx2 ESCs, and 19,200 TSCs were needed per AggreWell. After determining the volume of cell suspension needed, cell suspension was centrifuged at 1,000 rpm for 4 minutes before resuspending in 1ml FC media supplemented with 1 μg ml−1 heparin (Sigma-Aldrich H3149–25KU), 25 ng ml−1 Fgf4 (R&D Systems 7486-F4–025) and 7.5nM Y27632 (STEMCELL Technologies 72304). The anti-adherence rinsing solution was aspirated from AggreWell and each AggreWell was washed twice with 1ml PBS before adding 500μl FC media with heparin and Fgf4 (FCF4H media) per well. Cell suspension was then added drop-wise onto AggreWell and the plate was centrifuged at 100g for 3 minutes. In each AggreWell, 1ml of media was replaced with 1ml fresh FCF4H media on Day 1 and Day 2.
To generate EiTiX-embryoids, 1 mg/ml doxycycline (Sigma-Aldrich D9891–5G) was added to both iCdx2 ESCs and iGata4 ESCs for 6 hours. AggreWell was treated same as the above for generating cell aggregates. 6-hour induced iCdx2 ESCs, 6-hour induced iGata4 ESCs and WT ESCs were dissociated and cell density was determined. For each AggreWell, 38,400 6-hour induced iCdx2 ESCs, 6,000 6-hour induced iGata4 ESCs, and 6,000 WT ESCs were needed and the cell suspensions were pooled together to be centrifuged at 1,000 rpm for 4 minutes. The resulting cell pellet was resuspended in 1ml FC media supplemented with 1 μg ml−1 heparin, 25 ng ml−1 Fgf4 and 10nM ROCK inhibitor. The anti-adherence rinsing solution was aspirated from AggreWell and each AggreWell was washed twice with 1ml PBS before adding 500μl FC media with heparin and Fgf4 per well. Cell suspension was then added drop-wise onto AggreWell and the plate was centrifuged at 100g for 3 minutes. On Day 1, 1ml of media was replaced with 1ml fresh FC media and media change was repeated once to ensure removal of Y27632. On Day 2, 1ml of media was replaced with 1ml fresh FC media. On Day 3, 1.2ml media was removed and 1.5ml peri-implantation media (20% FBS) was added per well. Peri-implantation media (also known as IVC media,(Bedzhov and Zernicka-Goetz, 2014)) contained 80% advanced DMEM/F12 (Gibco 12634–010 21331–020) supplemented with 20% FBS, 2mM GlutaMax, 1% penicillin–streptomycin, 1X insulin-transferrin-selenium-ethanolamine (ThermoFisher Scientific 51500–056), 8 nM β-estradiol (Sigma-Aldrich E8875–1G), 200 ng/ml progesterone (Sigma-Aldrich P8783–1G) and 25 mM N-acetyl-L-cysteine (Sigma-Aldrich A7250). On Day 4, structures were selected under dissection microscope (see below for inclusion criteria of EiTiX embryos) and up to 50 structures were transferred to each well of non-adherent 6 well multiwell plate (Greiner Bio-One 657185), which contained 5ml peri-implantation media (with 30% FBS). On Day 5, structures were transferred to peri-implantation media (with 30% knockout serum replacement, Gibco 10828010). Peri-implantation media was equilibrated in the incubator for 20 minutes in advance.
Culture of EiTiX-embryoids in post-implantation culture medium
Post-implantation culture medium was prepared as described in Sturm and Tam (1993) and Aguilera-Castrejon et al., (2021). It comprises 25% DMEM (Gibco 11054 (using HCO3 buffer) or Gibco 11880 (using HEPES)) with 1x Glutamax (Gibco 35050061), 100 units/ml penicillin/streptomycin (ThermoFisher 15140122) and 11 mM HEPES (Gibco 15630056 added to DMEM-Gibco 11880), plus 50% rat serum (Charles River Laboratories) and 25% human cord serum (Cambridge Blood and Stem Cell Biobank). Rat serum and human cord serum were thawed at room temperature and heat-inactivated for 30 minutes at 56°C. After preparation, post-implantation culture medium was filter-sterilised and equilibrated at 37°C for 1 hour. Each of the selected Day 5 EiTiX-embryoids was transferred to one well of a 48-well multi-well plate for suspension culture (Greiner Bio-One 677102), with 250μl post-implantation culture medium per well. On Day 6, 100μl medium was removed and 250μl fresh post-implantation culture medium was added per well. On Day 7, EiTiX-embryoids were transferred to a rotating bottle culture chamber apparatus. Up to 3 EiTiX-embryoids were cultured in the same rotating bottle that contained 2ml post-implantation medium supplemented with 3.0 mg/ml of D-Glucose (Sigma G8644).
Immunofluorescence
Samples were fixed with 4% paraformaldehyde for 20 minutes at room temperature and then washed three times with PBST (0.1% Tween-20 in PBS). Samples were permeabilised in 0.1 M glycine and 0.3% Triton X-100 in PBS for 30 minutes at room temperature. After washing with PBST for three times, primary antibodies diluted in blocking buffer (10% FBS and 0.1% Tween 20 in PBS) were added and the samples incubated overnight at 4°C. Primary antibodies were removed the following day, and samples were washed three times with PBST, before adding secondary antibodies and DAPI. After incubating overnight at 4°C, samples were washed three times with PBST and mounted in a glass-bottom dish for imaging.
Whole mount in situ hybridisation
Whole mount in situ hybridisation was performed following a published protocol (Piette et al., 2008). Briefly, EiTiX-embryoids were fixed with 4% paraformaldehyde at 4°C overnight and were dehydrated with methanol series. After rehydrating with inverted methanol series, Day 5 and Day 6 EiTiX-embryoids were treated with proteinase K for 3 and 4 minutes respectively and were subsequently washed with PBST and refixed in 4% paraformaldehyde / 0.2% glutaraldehyde. Then, EiTiX-embryoids were washed in 50% and 100% hybridisation buffer and denatured Bmp4 probe (a generous gift from Dr Jose Silva from the Stem Cell Institute (Cambridge, UK)) were added for overnight hybridisation at 70°C. Probe was removed by washing in hybridisation buffer, SSC, MA buffer, PBS and PBST. Next, EiTiX-embryoids were incubated in antibody buffer and preblocked antidigoxigenin antibodies were added and incubated overnight at 4°C. EiTiX-embryoids were then washed in PBST / 0.1% BSA, PBST, and AP1 buffer. AP1 buffer with BM purple was added for colour development and the reaction was stopped by washing in STOP solution.
Dissociation of EiTiX-embryoids for MARS-seq
Individual Day 6 EiTiX-embryoids were dissected into four pieces with needles in PBS which were then dissociated with 70μl TrypLE Express Enzyme (Gibco 12604021) at 37°C for 15 minutes, pipetting up and down every 5 minutes. Dissociation was stopped by adding 500μl FC media supplemented with Y27632 (1:2,000) and DAPI (1:2,000). Day 8 samples were dissociated with 200μl TrypLE Express Enzyme and 800μl media was added to stop the dissociation. The cell suspension was subsequently filtered through a 40μm cell strainer (Merck CLS431750) and further diluted with 2ml FC media. It was then sorted by FACSAria III (BD Biosciences) using index sorting into 384-well plates.
MARS-seq library preparation
Single-cell cDNA libraries were prepared as previously described (Cheng et al., 2021; Mittnenzweig et al., 2021) following the MARS-seq protocol. MARS-seq libraries were processed using NextSeq 500 or NovaSeq 6000. The output reads were processed following the MARS-seq2.0 protocol (Keren-Shaul et al., 2019) with the same specifications as previously reported, using STAR aligner for sequence alignment (Dobin et al., 2013). Here, we processed 8832 wells. To analyse Day 8 EiTiX embryoids, we used FACS index sorting to record fluorescence per cell in addition to structure identity per well. We then distinguished GFP positive cells using the green channel bimodal distribution.
QUANTIFICATION AND STATISTICAL ANALYSIS
Inclusion criteria of EiTiX-embryoids
All EiTiX-embryoids were collected from AggreWell on Day 4 and their morphologies were examined under a dissection microscope. Structures were selected for analyses or further culture if 1) there were two distinct cellular compartments enclosed by a thin outer cell layer, and 2) there was a clear epithelialised ES compartment with a central lumen. For Day 5 and Day 6 EiTiX-embryoids, we selected structures that were elongated and had a thick epithelial cell layer in the ES compartment that resembled the EPI in natural mouse embryos. On Day 7, we selected EiTiX-embryoids that had developed headfolds structure within a yolk sac-like membrane.
Atlas projection and cell type annotation of EiTiX-embryoids
To identify marker-genes for metacell construction (Baran et al., 2019; Ben-Kiki et al., 2022), we selected all the genes displaying a minimal variance over mean (T_vm = 0.1) and coverage threshold (Ttot = 50, Ttop3 = 3). These genes were clustered into 137 clusters based on their gene-gene correlation overall the UMI mat. Gene-cluster enriched with stress- and cell-cycle-related genes were manually removed (n = 625) leaving 807 feature genes. The final metacell object (Knn = 100, minimal metacell size = 30 cells) contained 60 metacells comprised of 7076 cells (2184 from Day 6 EiTiX structures and 4892 from Day 8 EiTiX-embryoids) with a 5362 median UMIs per cell. Metacells were annotated with cell types by projection on the gastrulation wildtype atlas, as previously reported (Cheng et al., 2021; Mittnenzweig et al., 2021).
Natural embryo matching
For each EiTiX-embryoid, we inferred a best-matching natural embryo based on the similarity (Euclidean distance) between their cell state compositions. Only natural embryos with at least 161 embryonic cells and 29 extraembryonic ectoderm cells were included (threshold fits the calculated median number of ExE cells per EiTiX-embryoid). For each EiTiX-embryoid, we included the three closest natural embryos in the matching natural cohort, counting each natural embryo only once. Natural embryos were temporally ordered as previously reported (Cheng et al., 2021; Mittnenzweig et al., 2021).
Mean differential expression among transcriptional states from EiTiX-embryoids
For each EiTiX Day, we computed bulk (average) gene expression profiles per cell type and compared them with the corresponding natural embryo expression profiles from the matching natural embryos (log2 absolute expression). For each cell type, the number of included natural embryo cells was down sampled to the corresponding number of cells from EiTiX-embryoids compared. Cell-cycle and stress-related genes were not included in that comparison. Highlighted cell-type-specific genes for each included cell type were defined as being on average at least two-fold enriched in the metacells from this cell type relative to the global average among all natural embryo Metacells and additional known cell type markers were added.
Image acquisition, processing and analysis
Leica SP5 and SP8 confocal microscopes (Leica Microsystems) with either a 40x oil objective or a 25x water objective were used to acquire immunofluorescence images. A 405 nm diode laser, 488 nm argon laser, 543 nm HeNe laser and 633 nm HeNe laser (Alexa Fluor 647) were used to excite the fluorophores. Fiji (Schindelin et al., 2012) and Smart Denoise (Gurdon Institute) were used for image processing and analysis.
Quantification of the extent of AVE anterior localisation
The angle between the distal tip and the most anterior cell expressing AVE marker was termed as angle a (white) while the angle between the distal tip and the boundary of Oct4-positive domain was termed as angle b (orange) (Figure 2D). % AVE migration was obtained by dividing angle a by angle b and multiplying by 100%. It was then classified as proximal (>67%), lateral (33–67%) and distal (<33%).
Quantification of the extent of Brachyury (T) extension
The angle between the posterior boundary of Oct4-positive domain and the most anterior T-positive cell was termed as angle a (white) while the angle between the posterior boundary of Oct4-positive domain and the distal tip The percentage of T extension was obtained by dividing angle a by angle b and multiplying by 100%.
Statistics
Statistical analyses were performed using GraphPad Prism 8 and quantitative data were presented as mean ± S.E.M. or as violin plots with median and quartiles. Student’s t-test was used to determine statistical significance between two samples while one-way ANOVA followed by Bonferroni’s multiple comparisons test was used to determine statistical significance between more than two groups. Sample size and number of experimental replicates were indicated in figure legends.
Supplementary Material
Movie S1-2. Movie showing the beating heart region of Day 8 EiTiX-embryoids, related to Figure 4.
Highlights:
A unified in vitro mouse embryo model derived exclusively from embryonic stem cells
Embryoids undergoes advanced development to late headfold stages
Single-cell RNA sequencing shows similar transcriptional programs across lineages
Acknowledgements
The authors would like to thank David Glover for helpful comments, Yoav Mayshar for his assistance with sample collection for scRNA-seq, Netta Reines for technical support with scRNA-seq processing, Joana Cerveira and Mercedes Cabrera Jarana from the flow cytometry facility from the School of the Biological Sciences, University of Cambridge for their assistance in this work, and Jacob Hanna and Alejandro Aguilera-Castrejon for providing a pressurising chamber that we used for the last day of embryo culture. The grants to M.Z.-G that supported this work are: NIH Pioneer Award (DP1 HD104575–01), the Allen Discovery Center for Lineage Tracing; European Research Council (669198), the Wellcome Trust (207415/Z/17/Z), Open Philanthropy/Silicon Valley Community Foundation and Weston Havens Foundation. The grants to YS that supported this work are: European Research Council (ERC_StG 852865) and Helen and Martin Kimmel Stem Cell Institute. K.Y.C.L. is supported by the Croucher Foundation and the Cambridge Trust. C.W.G. is supported by a Leverhulm Early Career Research Fellowship. Research in the M.Z.-G and Y.S. labs is supported by the Schwartz/Reisman Collaborative Science Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
M.Z.-G. is an advisory board member of Cell Stem Cell. M.Z.-G. and G.A. wish to declare the filing of a patent on May 5th, 2022 by Caltech and the University of Cambridge on the “Generation of synthetic embryos from multiple stem cell types”. The Patent was filed under the following numbers: CIT File No.: CIT-8826-P and Serial Number: 63/344,251
References
- Aguilera-Castrejon A et al. (2021) ‘Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis’, Nature, 593(7857), pp. 119–124. doi: 10.1038/s41586-021-03416-3. [DOI] [PubMed] [Google Scholar]
- Amadei G et al. (2021) ‘Inducible Stem-Cell-Derived Embryos Capture Mouse Morphogenetic Events In Vitro’, Developmental Cell, pp. 1–17. doi: 10.1016/j.devcel.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadei G et al. (2022) ‘Synthetic embryos derived from mouse embryonic and extra-embryonic stem cells recapitulate gastrulation, neurulation and early organogenesis’, Nature doi: 10.1038/s41586-022-05246-3. [DOI] [Google Scholar]
- Arnold SJ and Robertson EJ (2009) ‘Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo’, Nature Reviews Molecular Cell Biology, 10(2), pp. 91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Baran Y et al. (2019) ‘MetaCell: Analysis of single-cell RNA-seq data using K-nn graph partitions’, Genome Biology, 20(1). doi: 10.1186/s13059-019-1812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot ES and Hadjantonakis AK (2020) ‘Mouse gastrulation: Coordination of tissue patterning, specification and diversification of cell fate’, Mechanisms of Development, 163(February), p. 103617. doi: 10.1016/j.mod.2020.103617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari L et al. (2018) ‘Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids’, Nature, 562(7726), pp. 272–276. doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- Bedzhov I and Zernicka-Goetz M (2014) ‘Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation’, Cell, 156(5), pp. 1032–1044. doi: 10.1016/j.cell.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Kiki O et al. (2022) ‘Metacell-2: a divide-and-conquer metacell algorithm for scalable scRNA-seq analysis’, Genome Biology, 23(1), pp. 1–18. doi: 10.1186/s13059-022-02667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D et al. (2008) ‘Wnt Signaling Mediates Self-Organization and Axis Formation in Embryoid Bodies’, Cell Stem Cell, 3(5), pp. 508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blij S et al. (2015) ‘Cdx2 efficiently induces trophoblast stem-like cells in naïve, but not primed, pluripotent stem cells’, Stem Cells and Development, 24(11), pp. 1352–1365. doi: 10.1089/scd.2014.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Brink SC et al. (2014) ‘Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells’, Development (Cambridge), 141(22), pp. 4231–4242. doi: 10.1242/dev.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S et al. (2021) ‘The Intrinsic and Extrinsic Effects of Tet Proteins During Gastrulation’, SSRN Electronic Journal, pp. 1–17. doi: 10.2139/ssrn.3959334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A et al. (2013) ‘STAR: Ultrafast universal RNA-seq aligner’, Bioinformatics, 29(1), pp. 15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnison M, Broadhurst R and Pfeffer PL (2015) ‘Elf5 and Ets2 maintain the mouse extraembryonic ectoderm in a dosage dependent synergistic manner’, Developmental Biology, 397(1), pp. 77–88. doi: 10.1016/j.ydbio.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Harrison SE et al. (2017) ‘Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro’, Science, 356(6334). doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Jin JZ et al. (2016) ‘Analysis of extraembryonic mesodermal structure formation in the absence of morphological primitive streak’, Development Growth and Differentiation, 58(6), pp. 522–529. doi: 10.1111/dgd.12294. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H et al. (2019) ‘MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing’, Nature Protocols, 14(6), pp. 1841–1862. doi: 10.1038/s41596-019-0164-4. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M and Hadjantonakis AK (2008) ‘The Endoderm of the Mouse Embryo Arises by Dynamic Widespread Intercalation of Embryonic and Extraembryonic Lineages’, Developmental Cell, 15(4), pp. 509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA et al. (1999) ‘Bmp4 is required for the generation of primordial germ cells in the mouse embryo’, Genes and Development, 13(4), pp. 424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnenzweig M et al. (2021) ‘A single-embryo, single-cell time-resolved model for mouse gastrulation’, Cell, 184(11), pp. 2825–2842.e22. doi: 10.1016/j.cell.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD et al. (2007) ‘A Global Double-Fluorescent Cre Reporter Mouse’, Genesis, 45, pp. 593–605. doi: 10.1002/dvg. [DOI] [PubMed] [Google Scholar]
- Niwa H et al. (2005) ‘Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation’, Cell, 123(5), pp. 917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Piette D et al. (2008) ‘An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies’, Nature Protocols, 3(7), pp. 1194–1201. doi: 10.1038/nprot.2008.103. [DOI] [PubMed] [Google Scholar]
- Rhee JM et al. (2006) ‘In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice’, Genesis, 44, pp. 202–218. doi: 10.1002/dvg.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G et al. (2020) ‘Capturing Cardiogenesis in Gastruloids’, Cell Stem Cell, 28(2), pp. 230–240.e6. doi: 10.1016/j.stem.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykali B et al. (2019) ‘Distinct mesoderm migration phenotypes in extra-embryonic and embryonic regions of the early mouse embryo’, eLife, 8, pp. 1–27. doi: 10.7554/eLife.42434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibner K et al. (2021) ‘Epithelial cell plasticity drives endoderm formation during gastrulation’, Nature Cell Biology, 23(7), pp. 692–703. doi: 10.1038/s41556-021-00694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J et al. (2012) ‘Fiji: An open-source platform for biological-image analysis’, Nature Methods, 9(7), pp. 676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B et al. (2019) ‘Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid’, Developmental Cell, 51(6), pp. 698–712.e8. doi: 10.1016/j.devcel.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stower MJ and Srinivas S (2017) The Head’s Tale: Anterior–Posterior Axis Formation in the Mouse Embryo. 1st edn, Cell Fate in Mammalian Development. 1st edn. Elsevier Inc doi: 10.1016/bs.ctdb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Sturm K and Tam PPL (1993) ‘Isolation and Culture of Whole Postimplantation Embryos and Germ Layer Derivatives’, Methods in Enzymology, 225, pp. 164–190. [DOI] [PubMed] [Google Scholar]
- Tanaka Y et al. (2014) ‘Circulation-Independent Differentiation Pathway from Extraembryonic Mesoderm toward Hematopoietic Stem Cells via Hemogenic Angioblasts’, Cell Reports, 8(1), pp. 31–39. doi: 10.1016/j.celrep.2014.05.055. [DOI] [PubMed] [Google Scholar]
- Tarazi S et al. (2022) ‘Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs’, Cell, 185, pp. 1–17. doi: 10.1016/j.cell.2022.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P and Beddington R (1996) ‘Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo’, Current Biology, 6(11), pp. 1487–1496. doi: 10.1016/S0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- Turner DA et al. (2017) ‘Anteroposterior polarity and elongation in the absence of extraembryonic tissues and of spatially localised signalling in gastruloids: Mammalian embryonic organoids’, Development (Cambridge), 144(21), pp. 3894–3906. doi: 10.1242/dev.150391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenvliet JV et al. (2020) ‘Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites’, Science, 370(6522). doi: 10.1126/science.aba4937. [DOI] [PubMed] [Google Scholar]
- Viotti M, Nowotschin S and Hadjantonakis AK (2014) ‘SOX17 links gut endoderm morphogenesis and germ layer segregation’, Nature Cell Biology, 16(12), pp. 1146–1156. doi: 10.1038/ncb3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G et al. (1995) ‘Bone morhogenetic protein-4 is required for mesoderm formation and patterning in the mouse’, Genes and Development, pp. 2105–2116. [DOI] [PubMed] [Google Scholar]
- Xu PF et al. (2021) ‘Construction of a mammalian embryo model from stem cells organized by a morphogen signalling centre’, Nature Communications, 12(1), pp. 1–22. doi: 10.1038/s41467-021-23653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M et al. (1997) ‘Following cell fate in the living mouse embryo’, Development, 1137, pp. 1133–1137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1-2. Movie showing the beating heart region of Day 8 EiTiX-embryoids, related to Figure 4.
Data Availability Statement
Single-cell RNA sequencing data have been deposited at ArrayExpress and are publicly available as of the date of publication. Accession number is listed in the key resources table.
Code used in the study has been deposited at Zenodo and is publicly available as of the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-AP-2 gamma | R&D Systems | Cat# AF5059, RRID: AB_2255891 |
| Mouse monoclonal anti-AP-2gamma | Santa Cruz Biotechnology | Cat# sc-12762, RRID: AB_667770 |
| Goat polyclonal anti-Brachyury | R&D Systems | Cat# AF2085, RRID: AB_2200235 |
| Mouse monoclonal anti-Cdx2 | BioGenex | Cat# MU392-UC, RRID: AB_2335627 |
| Rabbit monoclonal anti-Cdx2 | Abcam | Cat# ab76541, RRID: AB_1523334 |
| Rat monoclonal anti-Cerberus 1 | R&D Systems | Cat# MAB1986, RRID: AB_2275974 |
| Goat polyclonal anti-Dkk1 | R&D Systems | Cat# AF1096, RRID: AB_354597 |
| Rat monoclonal anti-E-Cadherin | Thermo Fisher Scientific | Cat# 13–1900, RRID: AB_2533005 |
| Rabbit polyclonal anti-TBR2 / Eomes | Abcam | Cat# ab23345, RRID: AB_778267 |
| Rabbit monoclonal anti-FoxA2 / HNF3 | Cell Signaling Technology | Cat# 8186, RRID: AB_10891055 |
| Rabbit polyclonal anti-GATA-4 | Santa Cruz Biotechnology | Cat# sc-9053, RRID: AB_2247396 |
| Goat polyclonal anti-GATA-6 | R&D Systems | Cat# AF1700, RRID: AB_2108901 |
| Rat monoclonal anti-GFP | Nacalai Tesque | Cat# GF090R, RRID: AB_2314545 |
| Mouse monoclonal anti-eHAND | Santa Cruz Biotechnology | Cat# sc-390376 |
| Rabbit polyclonal anti-Keratin18 | Sigma-Aldrich | Cat# SAB4501665, RRID: AB_10746153 |
| Goat polyclonal anti-Lefty | R&D Systems | Cat# AF746, RRID: AB_355566 |
| Mouse monoclonal anti-Myh2 | R&D Systems | Cat# MAB4470, RRID: AB_1293549 |
| Mouse monoclonal anti-N-Cadherin | BD Biosciences | Cat# 610920, RRID: AB_2077527 |
| Goat polyclonal anti-Nkx2.5 | R&D Systems | Cat# AF2444, RRID: AB_355269 |
| Mouse monoclonal anti-Oct-3/4 | Santa Cruz Biotechnology | Cat# sc-5279, RRID: AB_628051 |
| Goat polyclonal anti-Otx2 | R&D Systems | Cat# AF1979, RRID: AB_2157172 |
| Rabbit polyclonal anti-RFP | Rockland | Cat# 600–401-379, RRID: AB_2209751 |
| Rabbit monoclonal anti-RUNX1 / AML1 | Abcam | Cat# ab92336, RRID: AB_2049267 |
| Rabbit polyclonal anti-Sox1 | Cell Signaling Technology | Cat# 4194, RRID: AB_1904140 |
| Mouse monoclonal anti-Sox2 | Santa Cruz Biotechnology | Cat# sc-365823, RRID: AB_10842165 |
| Rat monoclonal anti-Sox2 | Thermo Fisher Scientific | Cat# 14–9811-82, RRID: AB_11219471 |
| Goat polyclonal anti-Sox17 | R&D Systems | Cat# AF1924, RRID: AB_355060 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_2535792 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11055, RRID: AB_2534102 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10037, RRID: AB_2534013 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10042, RRID: AB_2534017 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11057, RRID: AB_2534104 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571, RRID: AB_162542 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573, RRID: AB_2536183 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21447, RRID: AB_2535864 |
| Donkey Anti-Rat IgG H&L (Alexa Fluor® 647) preadsorbed antibody | Abcam | Cat# ab150155, RRID: AB_2813835 |
| Bacterial and virus strains | ||
| 5-alpha competent E. coli | New England Biolabs | C2987I |
| Chemicals, peptides, and recombinant proteins | ||
| β-estradiol | Sigma-Aldrich | Cat# E8875–1G |
| CHIR99021 | Cambridge Stem Cell Institute | N/A |
| Doxycycline | Sigma-Aldrich | Cat# D9891–5G |
| Fgf2 | Cambridge Stem Cell Institute | N/A |
| Recombinant Mouse FGF-4 (aa 67–202) | R&D Systems | Cat# 7486-F4–025 |
| Heparin | Sigma-Aldrich | Cat# H3149–25KU |
| Insulin-transferrin-selenium-ethanolamine | ThermoFisher Scientific | Cat# 51500–056 |
| Leukaemia inhibitory factor | Cambridge Stem Cell Institute | N/A |
| N-acetyl-L-cysteine | Sigma-Aldrich | Cat# A7250 |
| PD0325901 | Cambridge Stem Cell Institute | N/A |
| Progesterone | Sigma-Aldrich | Cat# P8783–1G |
| Y-27632 | STEMCELL Technologies |
Cat# 72304 |
| Critical commercial assays | ||
| Gateway™ BP Clonase™ II Enzyme mix | Invitrogen | Cat# 11789–100 |
| Gateway™ LR Clonase™ II Enzyme mix | Invitrogen | Cat# 11791–100 |
| Lipofectamine™ 3000 Transfection Reagent | Invitrogen | Cat# L3000001 |
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4368708 |
| Deposited data | ||
| Single-cell RNA sequencing data | This manuscript | ArrayExpress: E-MTAB-12140 |
| Code for analysing single-cell RNA sequencing data | This manuscript | DOI: 10.5281/zenodo.7021607 |
| Experimental models: Cell lines | ||
| Mouse: CAG-GFP ESCs | Rhee et al., 2006 | N/A |
| Mouse: CAG-GFP/tetO-mCherry/tetO-Cdx2 ESCs | This manuscript | N/A |
| Mouse: CAG-GFP/tetO-mCherry/tetO-Gata4 ESCs | Amadei et al., 2021 | N/A |
| Mouse: Cerl-GFP/tetO-Gata4 ESCs | Amadei et al., 2021 | N/A |
| Mouse: CD1 ESCs | Prof Jennifer Nichols (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Mouse: CD1/tetO-Gata4 ESCs | Amadei et al., 2022 | N/A |
| Mouse: Confetti TSCs | Prof Jennifer Nichols (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Mouse: mT/mG mouse ESCs | Muzumdar et al., 2007 | N/A |
| Mouse: Sox2-Venus/Brachyury-mCherry/Oct4-Venus mouse ESCs | Dr Jesse Veenvliet and Prof Bernhard G. Herrmann (Max Planck Institute of Molecular Cell Biology and Genetics) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: CD-1 | Charles River Laboratories | N/A |
| Oligonucleotides | ||
| PCR primer: Cdx2-attB forward: GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTACGTGAGCTACCTTCTGGAC | This manuscript | N/A |
| PCR primer: Cdx2-attB reverse: GGGGACCACTTTGTACAAGAAAGCTGGGTTTCACTGGGTGACAGTGGAGTTTAAAAC | This manuscript | N/A |
| qPCR primer: Cdx2 forward: AAACCTGTGCGAGTGGATG | Blij et al., 2012 | N/A |
| qPCR primer: Cdx2 reverse: TCTGTGTACACCACCCGGTA | Blij et al., 2012 | N/A |
| qPCR primer: Elf5 forward: ATTTCTACAGTCCGCTGGTGC | This manuscript | N/A |
| qPCR primer: Elf5 reverse: ACATCACCGTGAAGACAAGTGG | This manuscript | N/A |
| qPCR primer: Gapdh forward: CGTATTGGGCGCCTGGTCAC | Amadei et al., 2021 | N/A |
| qPCR primer: Gapdh reverse: ATGATGACCCTTTTGGCTCC | Amadei et al., 2021 | N/A |
| qPCR primer: Gata3 forward: TGTCTGCGAACACTGAGCTG | This manuscript | N/A |
| qPCR primer: Gata3 reverse: CGATCACCTGAGTAGCAAGGAG | This manuscript | N/A |
| Recombinant DNA | ||
| PB-tetO-hygro | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| pBAse | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| rtTA-zeocyin | Dr José Silva (Stem Cell Institute, University of Cambridge, UK) | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji |
| metacell_0.3.7 | Baran et al., 2018 | https://github.com/tanaylab/metacell |
| Metacells 0.9.0-dev.1 | Ben-Kiki et al., 2022 | https://github.com/tanaylab/metacells#id3 |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| RStudio | RStudio | https://www.rstudio.com/ |
| Smart Denoise | Gurdon Institute | N/A |
| Other | ||
| AggreWell400 | STEMCELL Technologies |
Cat# 34415 |
| Anti-Adherence Rinsing Solution | STEMCELL Technologies |
Cat# 07010 |
| FACSAria III | BD Biosciences | N/A |
| Human cord serum | Cambridge Blood and Stem Cell Biobank | N/A |
| Knockout serum replacement | Gibco | Cat# 10828010 |
| Leica SP5 | Leica Microsystems | N/A |
| Leica SP8 | Leica Microsystems | N/A |
| M-MuLV Reverse Transcriptase | New England Biolabs | Cat# M0253S |
| Non-adherent 6 well multiwell plate | Greiner Bio-One | Cat# 657185 |
| 48-well multi-well plate | Greiner Bio-One | Cat# 677102 |
| Rat serum | Charles River Laboratories | N/A |
| StepOnePlus™ Real-Time PCR System | Applied Biosystems | N/A |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |


