Abstract
Aims
Coronary vasculature formation is a critical event during cardiac development, essential for heart function throughout perinatal and adult life. However, current understanding of coronary vascular development has largely been derived from transgenic mouse models. The aim of this study was to characterize the transcriptome of the human foetal cardiac endothelium using single-cell RNA sequencing (scRNA-seq) to provide critical new insights into the cellular heterogeneity and transcriptional dynamics that underpin endothelial specification within the vasculature of the developing heart.
Methods and results
We acquired scRNA-seq data of over 10 000 foetal cardiac endothelial cells (ECs), revealing divergent EC subtypes including endocardial, capillary, venous, arterial, and lymphatic populations. Gene regulatory network analyses predicted roles for SMAD1 and MECOM in determining the identity of capillary and arterial populations, respectively. Trajectory inference analysis suggested an endocardial contribution to the coronary vasculature and subsequent arterialization of capillary endothelium accompanied by increasing MECOM expression. Comparative analysis of equivalent data from murine cardiac development demonstrated that transcriptional signatures defining endothelial subpopulations are largely conserved between human and mouse. Comprehensive characterization of the transcriptional response to MECOM knockdown in human embryonic stem cell-derived EC (hESC-EC) demonstrated an increase in the expression of non-arterial markers, including those enriched in venous EC.
Conclusions
scRNA-seq of the human foetal cardiac endothelium identified distinct EC populations. A predicted endocardial contribution to the developing coronary vasculature was identified, as well as subsequent arterial specification of capillary EC. Loss of MECOM in hESC-EC increased expression of non-arterial markers, suggesting a role in maintaining arterial EC identity.
Keywords: Human cardiac development, Single-cell RNA sequencing, Endothelial heterogeneity, Coronary vasculature formation, MECOM, Vascular regeneration
See the editorial comment for this article ‘A new resource for human coronary vessel development’, by Ragini Phansalkar and Kristy Red-Horse, https://doi.org/10.1093/cvr/cvac094.
1. Introduction
While the formation and homeostasis of the coronary vasculature is essential for heart muscle function, the molecular mechanisms underlying coronary vascular development remain incompletely understood. Previous studies using lineage-tracing tools in mouse have provided much needed insight into these mechanisms, including identifying the endocardium and sinus venosus (SV) as the two major sources of coronary vascular endothelium during cardiac development.1,2 A third source, the proepicardium, was previously proposed to contribute a minor population of coronary endothelial cells (ECs),3,4 although this notion has recently been challenged.5 Following the formation of the primitive coronary vascular plexus from these sources and onset of blood flow, subsequent remodelling occurs, giving rise to the distinct EC populations present in the mature vascular bed of the fully developed heart.6,7 Recent studies have elegantly mapped the remodelling of the immature coronary EC plexus in mouse cardiac development, including identification of a role for the transcription factor (TF) Dach1 in potentiating developmental arterial remodelling.7–9 However, given that these advances in our understanding of coronary vascular development primarily originate from murine lineage-tracing studies, the relevance of these findings for human cardiac development remains largely unknown.
Advances in single-cell RNA sequencing (scRNA-seq) have been instrumental in enhancing our understanding of embryonic development, permitting the objective mapping of underlying transcriptional changes at single-cell resolution. In addition, improvements in high-throughput scRNA-seq platforms have facilitated the characterization of tens of thousands of cells in parallel, thus allowing for ‘atlas’ studies to map the gene expression profile of entire organs during embryogenesis.10 In recent years, such scRNA-seq studies have mapped the transcriptional profile of both murine and human heart development.11–19 In the study by Cui et al.,13 scRNA-seq was conducted using cells isolated from specific regions of 18 human foetal hearts, ranging from 5 to 24 weeks gestation. Subsequent dimensionality reduction and clustering analysis revealed an EC cluster of 595 cells characterized by expression of endothelial markers, such as PECAM1.13 Similarly, a clear EC population was identified in a study from Suryawanshi et al.12 in which cells isolated from three healthy human foetal hearts (19–22 weeks) were processed using scRNA-seq. Both studies mapped the expression of known EC marker subtypes to allow annotation of clusters corresponding to endocardium, coronary vascular EC, and valvular EC. Nevertheless, the relatively low numbers of EC in these datasets prevented further characterization of cardiac EC subtypes, including the identification of distinct arterial, venous, capillary, and lymphatic populations. In addition, these low EC numbers also prevented the application of methods to infer the dynamic cellular changes accompanying cardiac EC development.
While scRNA-seq studies of the developing mouse heart yielded large numbers of EC in their datasets,11,16–18 their analysis focused on other cell types, such as the cardiac conduction system, with minimal interpretation of EC heterogeneity and potential function. These included a study from Goodyer et al.,15 which analysed distinct vascular EC and endocardial cell populations from E16.5 mouse hearts.
In this study, we used scRNA-seq to comprehensively map the transcriptional signature of over 10 000 human foetal cardiac ECs isolated by fluorescence activated cell sorting (FACS) from two human foetal hearts at 13- and 14-weeks’ gestation. Unsupervised clustering, gene regulatory analysis, and trajectory inference methods revealed the transcriptional profile of heterogeneous EC populations and predicted dynamic cellular changes including arterial EC specification. In addition, we functionally validated MECOM as a regulator of arterial EC identity, thereby demonstrating the suitability of our novel scRNA-seq dataset to make in-silico predictions, capable of informing future strategies to guide endothelial identity. Collectively, findings from this study complement and expand upon knowledge previously obtained from murine development, bringing insights into human EC heterogeneity and pathways determining specification of subpopulations that are essential for understanding human coronary vascular formation.
2. Methods
2.1 Tissue collection and study approval
Human foetal cardiac tissue was acquired following elective termination of pregnancy. Informed written parental consent was obtained from all participants. Tissue was not collected in cases where termination of pregnancy was conducted due to an identified foetal or pregnancy abnormality. Ethical approval for the collection of foetal tissue was performed in accordance with all relevant guidelines and following study approval from the Lothian Research Ethics Committee (Study code: 08/1101/1) and the Research and Development Office (Study code: 2007/R/RM/10). This study was performed in accordance with the Declaration of Helsinki.
2.2 Isolation of foetal cardiac ECs
Cardiac ECs were isolated from the ventricular tissue of freshly collected human foetal hearts using a method adapted from van Beijnum et al.20 Digestion was performed at 37°C using a digestion solution containing 9 mL 0.1% collagenase II and 1 mL of 2.5 U/mL dispase. Then, 75 µL of 1 mg/mL DNaseI was added following 20 min incubation prior to a further 15 min incubation at 37°C. Digestion was quenched by the addition of 10 mL cold RPMI with 10% FCS and undigested clumps of tissue removed using a 100 µM cell strainer. Red blood cell lysis was performed by incubating cells for 2 min in red blood cell lysis buffer at room temperature prior to neutralizing with RPMI + 0.1% BSA. Cells were stained on ice for 45 min with APC anti-human CD31 and PE anti-human CD45 (Supplementary material online, Table S1). CD31+ CD45− ECs were isolated by FACS with DAPI staining being used to allow exclusion of dead cells.
2.3 scRNA-seq of foetal cardiac ECs
Sorted CD31+ CD45− ECs were counted manually using a haemocytometer with trypan blue staining used to identify non-viable cells. Viability exceeded 85% for both samples. A total of 8000 cells were loaded onto the 10X Chromium controller and library construction conducted using the Single Cell 3’ Reagent Kit (V3.1) in accordance with the manufacturer’s instructions. Libraries were sequenced using the Illumina NovaSeq 6000 platform.
2.4 scRNA-seq data analysis
Raw-sequencing data was processed using the 10X CellRanger pipeline (Version 3.1.0.) aligning reads to the GRCh38-3.0.0 genome reference. Barcodes corresponding to cells were distinguished from those corresponding to empty droplets using both the DropUtils package21 and the default cell calling method applied within the CellRanger pipeline. Cells with a total UMI count exceeding three median absolute deviations (MADs) from the median value were removed from downstream analysis using the R Scater package.22 Similarly, cells with a high proportion of counts from mitochondrial genes (>3 MADs) or with a low total gene count (<2 MADs) were also excluded. Data normalization was performed using the MultiBatchNormalisation method23 prior to merging datasets. Normalized count data were then scaled, and principal component analysis (PCA) applied using genes with the most variable expression across the combined dataset.24 Following batch correction using Harmony,25 non-supervised clustering was performed, and data visualized using Uniform Manifold Approximation and Projection (UMAP).24 A small cluster (155 cells) characterized by increased expression of fibroblast/smooth muscle cell markers (ACTA2 and MYH11) and reduced EC marker expression (PECAM1 and CDH5) was removed from the dataset prior to rerunning data normalization, PCA, and data visualization. Significantly differentially expressed genes (DEGs) within individual clusters were identified using the Wilcoxon signed-rank test (Bonferroni corrected P-value <0.05) and a minimum loge(fold change) threshold of 0.3.24 Additionally, only DEGs expressed in more than 30% of cells within their corresponding cluster were retained for further analysis.
Enriched metagene signatures were identified using the R package SCRAT v1.0.0.26 Gene regulatory analysis was performed using the standard R Single-Cell rEgulatory Network Inference and Clustering (SCENIC) workflow.27 RNA velocity analysis was conducted using the python package scVelo28 with the stochastic model being applied to predict the direction and magnitude of cellular dynamics. Trajectory inference tool Slingshot was performed using the standard workflow.29 Genes significantly differentially expressed over pseudotime were identified using the TradeSeq package30 with the top 2000 most variably expressed genes in the dataset being used to fit the negative binomial generalized additive model.
2.5 Human embryonic stem cell-derived EC differentiation and siRNA-mediated MECOM knockdown
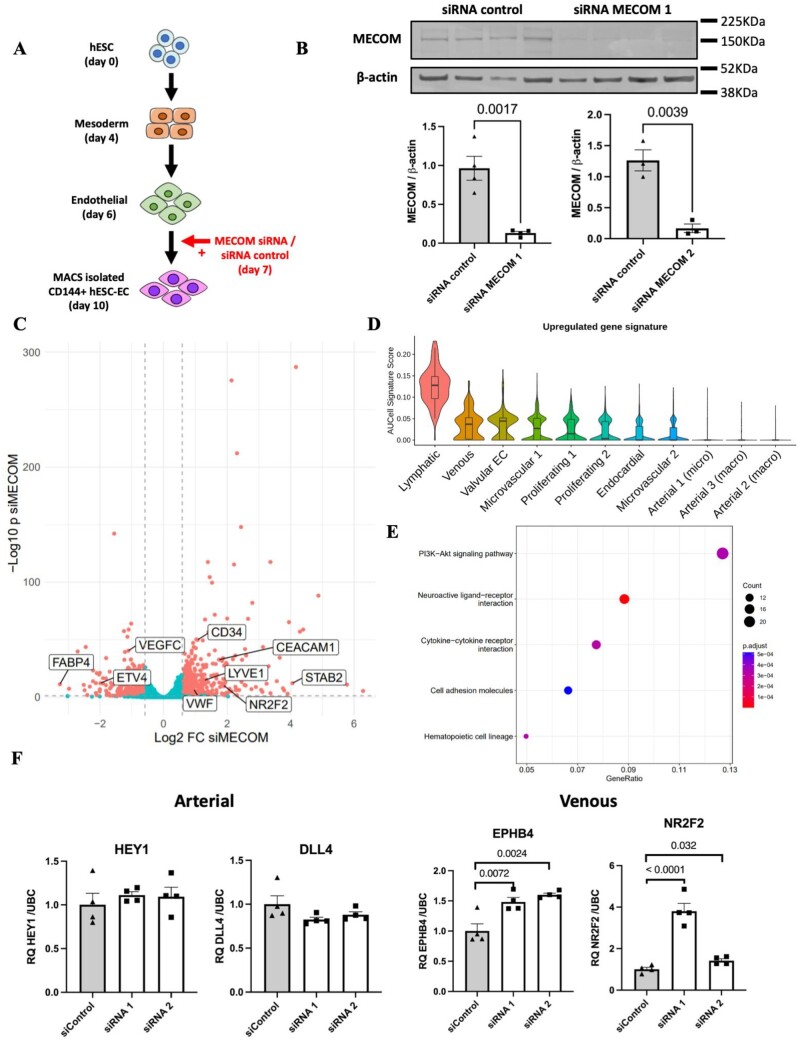
Human ESC lines were used in accordance with the UK Stem Cell Bank Steering Committee guidelines (Project Approvals SCS11-51 and SCSC17-26). H9 hESC were differentiated to human embryonic stem cell-derived EC (hESC-EC) as previously described.31,32 Small interfering RNA (siRNA)-mediated knockdown of MECOM was performed using Day 7 hESC-EC using predesigned siRNA at a final concentration of 5 nM (Supplementary material online, Table S2). After 6 h, transfection media was replaced with EGM-2 media supplemented with 1% human AB serum and 50 ng/mL VEGF-A. At Day 10, CD144+ hESC-EC were isolated by magnetic activated cell sorting and cell pellets stored at −80°C for subsequent isolation of RNA and protein.
2.6 Bulk RNA-sequencing analysis
RNA was isolated from Day 10 CD144+ hESC-EC previously subjected to either transfection with siRNA targeting MECOM (siRNA 1) or control siRNA (n = 4 biological replicates). Illumina strand-specific RNA sequencing libraries with PolyA selection were prepared by GeneWiz (New Jersey, USA) and sequenced using the Illumina NovaSeq sequencer to achieve a read depth of 20 million paired end reads per sample.
Reads from each sample were mapped and quantified using RSEM33 (v1.3.0, –bowtie2) and the GENCODE v38 primary assembly transcriptome. Genes with an average FPKM >1 in one or the other experimental group were considered to be expressed. To identify DEGs, tximport34 (v1.22.0) was used to supply DESeq235 (v1.34.0) with the isoform read counts from RSEM before using the default DESeq2 method (Wald test) to obtain gene-level P-values and fold changes between experimental conditions. Those genes with an absolute fold change value >1.5 (absolute Log2FC value >0.584) and adjusted P-value of <0.05 were considered differentially expressed. Over-represented KEGG terms amongst siMECOM-up-regulated genes were identified using clusterProfiler36 (v4.2.0) and Benjamini–Hochberg multiple hypothesis correction (P < 0.05).
All further experimental and analysis details are included in the Supplementary material online, Methods.
3. Results
3.1 Identification of distinct cardiac endothelial populations
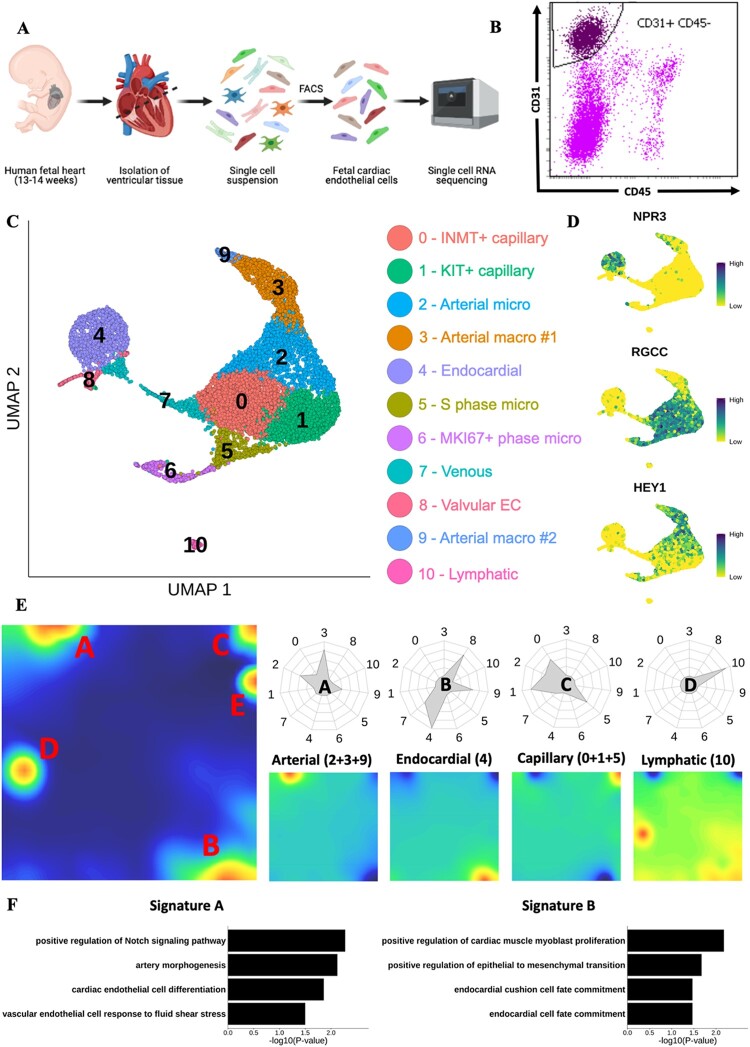
scRNA-seq was performed on CD31+ CD45− cardiac ECs isolated by FACS from ventricular tissue obtained from two human foetuses at 13 and 14 weeks of gestation (Figure 1A and B). At this developmental stage all major structures in the heart have formed, including the coronary vasculature. However, studies from equivalent timepoints in murine development (E15.5–E17.5) have revealed extensive remodelling occurring within the established coronary vasculature producing a mature vascular bed containing heterogeneous EC populations.6,9,37 Following quality control, unsupervised clustering, and UMAP visualization of transcriptomic data from 10 267 cells, 11 distinct clusters (numbered 0–10) were revealed, each with expression of typical pan-EC markers (Figure 1C and Supplementary material online, Figure S1A). Leukocyte marker PTPRC and fibroblast/smooth muscle cell markers ACTA2 and MYH11 demonstrated negligible expression across all clusters (Supplementary material online, Figure S1A). Annotating cells by sample demonstrated successful integration of datasets, with each cluster containing cells from both samples (Supplementary material online, Figure S1B).
Figure 1.
Mapping the human foetal heart endothelium using scRNA-seq. (A) Schematic of experimental design for mapping the human foetal heart endothelium using 10× scRNA-seq. (B) Representative FACS gating strategy used to isolate viable CD31+ CD45− ECs. (C) UMAP visualization of clusters identified in scRNA-seq data from cardiac ECs isolated from human foetal heart (n =2). (D) Feature plots showing expression of key marker genes defining distinct endothelial populations. (E) Metagene analysis of foetal heart scRNA-seq data visualized in self-organized maps for total dataset (left) and subpopulations of EC (right). Radar plots show enrichment of each metagene signature in individual clusters. (F) GO term enrichment analysis conducted using genes from metagenes’ signatures A (left) and B (right).
Expression of NPR3, a known endocardial marker, was localized to cluster 4 (Figure 1D and Supplementary material online, Figure S1C).38 Clusters 0, 1, 2, 5, and 6 were defined by expression of capillary marker RGCC,39 whilst arterial marker, HEY1,40 was expressed predominantly in clusters 2, 3, and 9 (Figure 1D and Supplementary material online, Figure S1C). Metagene analysis revealed five major signatures (A–E) indicating five key populations within the data (Figure 1E and Supplementary material online, File S1). Signature A was enriched in clusters 2, 3, and 9, and included genes involved in arterial EC function, such as JAG1, DLL4, and HEY1 (Figure 1E). Furthermore, Gene Ontology (GO) term enrichment analysis using signature A genes identified ‘artery morphogenesis’ and ‘positive regulation of Notch signalling pathway’ as significantly enriched terms (Figure 1F). Notch signalling is known to be required for arterial EC specification.41
Signature B was predominantly enriched in Cluster 4 and contained known endocardial markers CDH11 and NPR3 (Figure 1E and F).38,42 Lower levels of signature B enrichment were also observed in Clusters 7, 8, and 9 (Figure 1E). Clusters 0, 1, and 5 were enriched for signature C, which included capillary EC marker genes, such as CA4 and RGCC (Figure 1E and Supplementary material online, Figure S1D).39,43 Cluster 10 was enriched for signature D, for which GO term analysis returned terms relating to lymphatic EC (LEC) (Figure 1E and Supplementary material online, Figure S1D). LEC markers,44LYVE1, FLT4, PROX1, and PDPN, were differentially expressed in Cluster 10 (Supplementary material online, Figure S1G). Signature E was selective for Cluster 6 with GO term analysis identifying enriched terms relating to proliferation (Supplementary material online, Figure S1E). In line with this, categorizing cells according to their predicted cell cycle phase revealed that 77% of cells in Cluster 6 were predicted to be in the G2M phase of rapid growth, whereas 95% of cells in cluster 5 were in S phase (Supplementary material online, Figure S1F). The remaining cells in the dataset were predominantly in the G1 phase (79%), with only a small proportion predicted to be in G2M (4%) and S phases (17%) (Supplementary material online, Figure S1F). For clusters not associated with a metagene signature, analysis of DEGs revealed enrichment of NR2F2 and ACKR1 in Cluster 7, suggesting a venous/venular EC identity (Figure 2A and Supplementary material online, Figure S1G).45,46 A valvular identity of Cluster 8 was supported by its differential expression of NFATC1 and BMP4, both with known roles in valvulogenesis (Supplementary material online, Figure S1G).47,48 Collectively, these analyses demonstrate that each major subtype of EC within the heart (endocardial, venous, lymphatic, capillary, arterial, proliferating, and valvular EC) is represented by one or more of the identified 10 clusters.
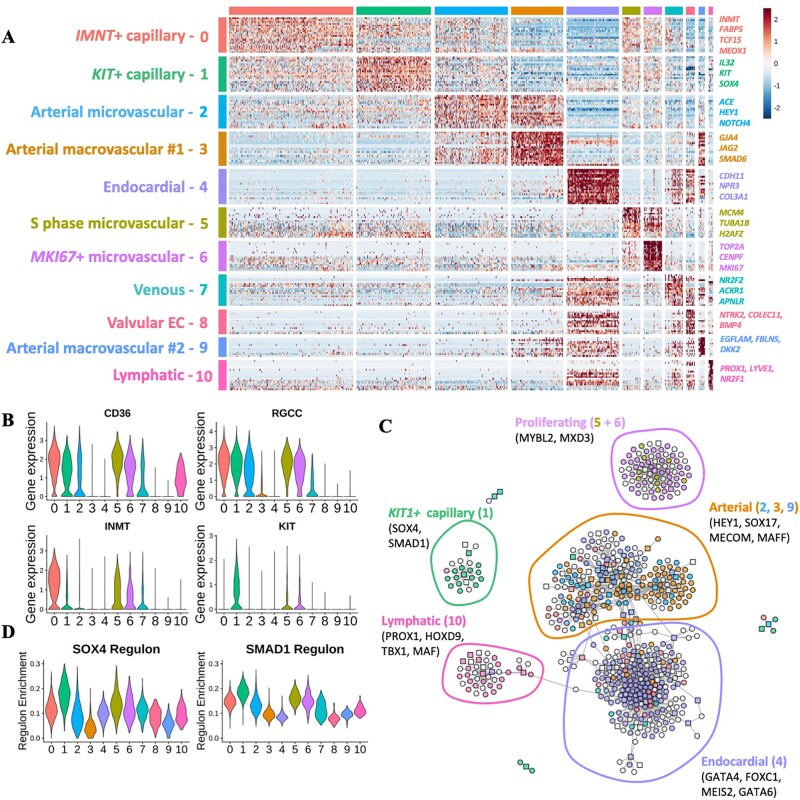
Figure 2.
GRN analysis of human foetal heart endothelium. (A) Heatmap of differentially expressed cluster genes: expression of top 20 DEGs for each cluster identified in the complete dataset. Genes were grouped according to the cluster in which they were differentially expressed. (B) Violin plots: expression of CD36, RGCC, INMT, and KIT across clusters identified in foetal heart EC dataset. (C) GRN constructed using SCENIC analysis. TFs and target genes shown as squares or circles, respectively. Genes are coloured based on the cluster in which they were differentially expressed. White nodes represent gene targets that were not differentially expressed. (D) Violin plots showing enrichment/AUC score of SOX4 and SMAD1 regulons across identified clusters.
3.2 Gene regulatory network analysis of foetal cardiac ECs
Global differential gene expression analysis revealed markers for subpopulations of arterial and capillary cardiac EC (Figure 2A and Supplementary material online, File S2). Notably, Cluster 2, an arterial EC population, is more closely correlated with capillary EC clusters (0 and 1) than with the other two minor arterial clusters (3 and 9) (Supplementary material online, Figure S2A). Together with the co-expression of both arterial and capillary markers, this suggested that Cluster 2 represents an arterial microvascular population. In contrast, arterial clusters 3 and 9 DEGs associated with ECM organization (FBLN5, ELN, and FBN1) and shear stress (KLF4) suggesting a macrovascular identity (Supplementary material online, Figure S2B and C).
Expression of fatty acid translocase encoding CD36 was absent from macrovascular and endocardial populations, corresponding with a previous report of microvasculature-restricted expression (Figure 2A and B).49 Differential gene expression analysis revealed heterogeneity between the two major capillary clusters, 0 and 1. Cluster 0 was defined by differential expression of amine methyltransferase encoding gene, INMT, whilst KIT expression was enriched in Cluster 1 (Figure 2A and B). Selective expression of KIT (also known as C-KIT) in Cluster 1 was accompanied by up-regulated expression of the TF SMAD1 (Supplementary material online, Figure S2D and E).
Gene regulatory network (GRN) analysis was applied to identify gene modules, known as regulons, predicted to be controlled by an individual TF, giving insight into the likely transcriptional regulators of EC heterogeneity. Visualization of differentially expressed TFs and their predicted targets within the GRN largely recapitulated the data structure observed following unsupervised clustering (Figure 2C). Genes differentially expressed in LEC (Cluster 10) localized together in the GRN and included TFs, such as PROX1, a known master regulator of LEC identity,50 as well as HOXD9 and TBX1 (Figure 2C). A set of differentially expressed endocardial TFs (Cluster 4) was evident, including GATA6, MEIS2, and FOXC1, as well as GATA4, known to be implicated in endocardial cushion development.51,52MECOM and MAFF were located amongst known regulators of arterial EC specification, such as HEY1 and SOX17 (Figure 2C).40,53 A distinct cluster of genes differentially expressed in KIT1+ capillary EC (Cluster 1) included SOX4 and SMAD1 (Figure 2C). Enrichment of SOX4 and SMAD1 regulons was also observed in KIT1+ capillary EC (Figure 2D).
3.3 Trajectory analysis predicts an endocardial contribution to the developing coronary vasculature and potential regulators of subsequent arterial specification
Several recent studies using murine models of coronary vascular development have provided insight into the origin of the coronary endothelium and the dynamic changes that occur during its subsequent remodelling.6,54 Endocardial-derived vessels vascularize the heart from the inside-out contributing to vessels of the interventricular septum and inner myocardial wall.1,55,56 Conversely, SV-derived vessels populate the outer ventricular free walls of the heart from the outside-in.38,55,56 Following the formation of the primitive coronary vascular plexus, EC undergo further remodelling to form a functional network of veins, arteries, and capillaries.9
We used trajectory inference methods to determine whether these processes could be identified during human cardiac development and to characterize their accompanying transcriptional changes. Given that these dynamic changes are known to originate from microvascular EC, we excluded the two previously identified arterial macrovascular clusters (Clusters 3 and 9) from the dataset and performed secondary clustering of the remaining cells (Figure 3A). The distinct LEC cluster was also excluded prior to re-clustering. The same marker genes used for annotating the complete dataset were used for the annotation of re-clustered data (Supplementary material online, Figure S3A).
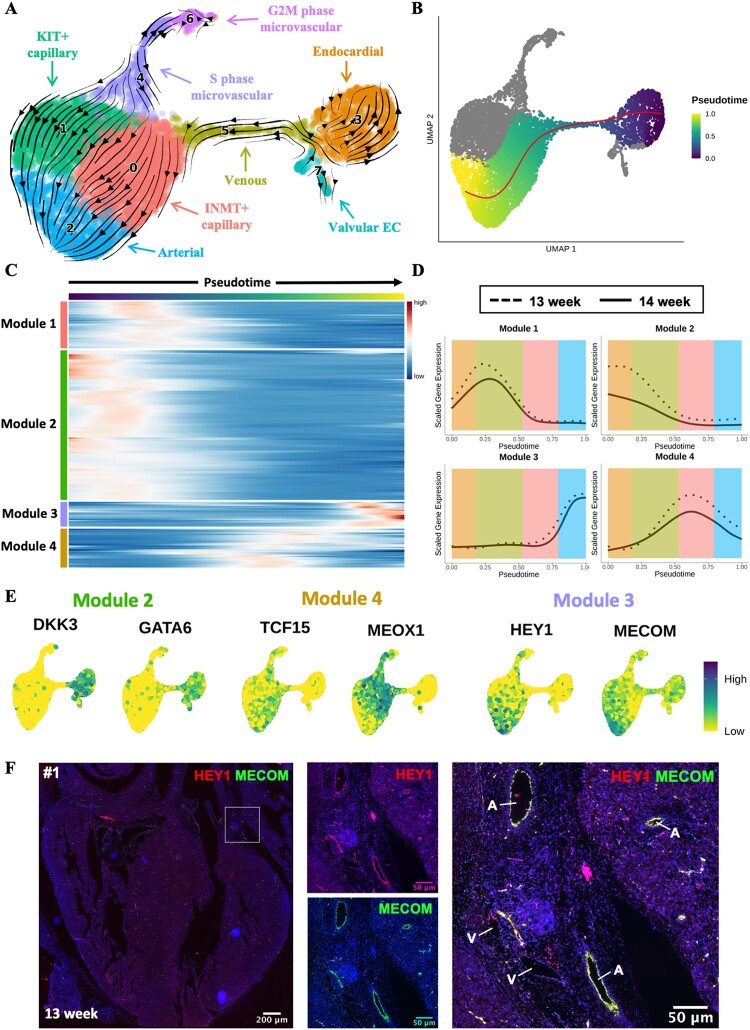
Figure 3.
Trajectory inference analysis of developing cardiac endothelium. (A) RNA velocity analysis of microvascular cardiac endothelium. The RNA velocity field shown superimposed onto a UMAP visualization of microvascular cardiac ECs. (B) Slingshot trajectory demonstrating pseudotemporal cellular dynamics. (C) Heatmap of 200 genes found to be most differentially expressed across pseudotime of trajectory from (B). Genes grouped into modules by k-means clustering (k = 4). (D) Smoothing spline curves show average sample scaled gene expression for genes within modules identified in (A). (E) Feature plots showing expression of selected genes from Modules 2 (DKK3 and GATA6), 3 (MECOM and HEY1), or 4 (TCF15 and MEOX1). (F) ISH validation of co-expression of MECOM (green) with HEY1 (red) in arterial EC of a 13-week human foetal heart (sample #1). See Supplementary material online, Figure S5A for samples #2, #3, and #4. A, artery; V, vein.
RNA velocity analysis,28 which utilizes the ratio of spliced to unspliced transcripts to infer the direction and magnitude of cellular transitions, was first used to gain an overview of the pseudotemporal dynamics of the foetal cardiac endothelium (Figure 3A and Supplementary material online, Figure S3B). We identified a proportion of the endocardial cluster with velocity vectors indicating a probable transition towards a venous identity (Figure 3A and Supplementary material online, Figure S3B). Evidence for this transition was further supported by venous EC-associated genes, such as PLVAP and NR2F2, having positive residuals/velocities in endocardial cells (Supplementary material online, Figure S3C). In turn, venous EC were subsequently predicted to transition to INMT+ capillary EC. This predicted transition of endocardium to coronary vascular EC concurs with studies that identified the endocardium as a significant source of EC for the coronary vasculature. To further substantiate this finding, cells belonging to endocardial, venous, and INMT+ capillary clusters were isolated in silico and re-clustered. UMAP visualization of re-clustered data revealed a comparable result to previous analysis with endocardial and INMT+ capillary populations connected by a ACKR1+ venous population (Supplementary material online, Figure S3D). Additionally, the omission of cell cycle-related genes from clustering and visualization calculations generated a comparable finding, thus confirming localization of identified clusters was not confounded by cell cycle-related effects (Supplementary material online, Figure S3E). Velocity analysis also predicted a likely transition of both capillary EC populations towards an arterial EC fate (Figure 3A and Supplementary material online, Figure S3B). This is an agreement with previous reports of developmental arterial remodelling in mouse.7–9
In addition to the RNA velocity analysis, we also independently applied the trajectory inference tool Slingshot,29 which yielded a comparable interpretation (Figure 3B). Identification of the top 200 genes with most variable expression over pseudotime revealed four temporal patterns of expression, arranged in Modules 1–4 (Figure 3C and D). Average sample module gene expression was visualized over pseudotime to ensure concordant expression dynamics between individual samples. Module 2 genes were expressed early in pseudotime with their reduction in expression occurring in conjunction with the loss of endocardial identity (Figure 3D). These included known markers of endocardium, such as CDH11 and NPR3 as well as the TFs DKK3 and GATA6 (Figure 3E and Supplementary material online, Figure S4A). TFs CEBPD and FOS were identified in Module 1 along with NR2F2, a known regulator of venous EC specification45 (Supplementary material online, Figure S4A). Interestingly, despite not being identified within Module 1, expression of BMP2 was found to increase within the pseudotime range corresponding to the predicted transitioning venous population (Supplementary material online, Figure S4B). As well as demonstrating enriched expression in venous EC in zebrafish,57 BMP2 has also recently been identified as positive regulator of endocardial to coronary vascular EC transition during murine cardiac development.58 Module 4 genes demonstrated peak expression within IMNT+ capillary EC and included TFs TCF15 and MEOX1 (Figure 3E). Expression of DACH1 was found to peak within the IMNT+ capillary cluster before gradually decreasing again within the arterial population (Supplementary material online, Figure S4C). Previous studies have identified Dach1 as a driver of developmental arterial remodelling in murine cardiac development.8,9
The predicted transition of capillary EC to arterial EC was defined by increased expression of Module 3 genes (Figure 3C and D). This included HEY1, known to mediate arterial EC specification.40 Interestingly, Module 3 also contained the TF MECOM, earlier predicted by GRN analysis to underlie arterial EC identity (Figures 2C and 3C–E and Supplementary material online, Figure S4A). Subsequent in situ hybridization (ISH) validation conducted across four independent foetal hearts (aged 13–14 weeks) demonstrated clear co-expression of MECOM with the arterial EC enriched TF HEY1 within arterial vessels (Figure 3F and Supplementary material online, Figure S5A and B). Notably, a lack of MECOM expression was observed in vessels with venous morphology validating its arterial EC specificity (Figure 3F and Supplementary material online, Figure S5A and B). In addition, reanalysis of publicly available scRNA-seq data from healthy human foetal heart data from Suryawanshi et al.12 revealed MECOM expression to be enriched within a subset of the endocardial/endothelial population with minimal expression in other identified cell types (Supplementary material online, Figure S6A–C).
3.4 Comparison with murine coronary developmental gene expression reveals conserved markers of cardiac EC populations
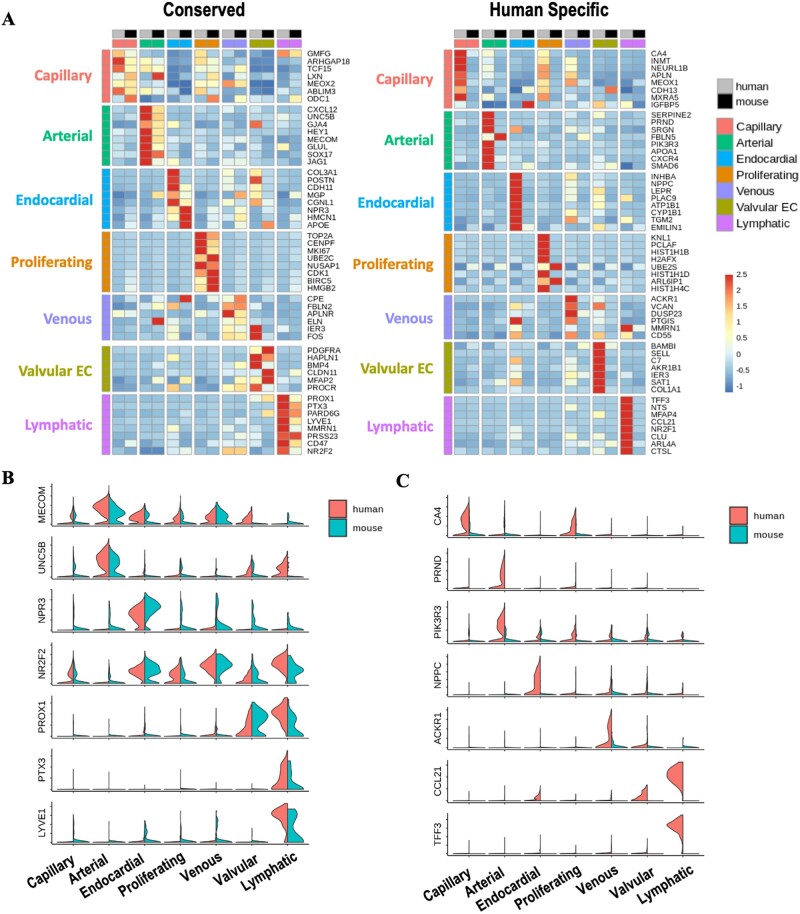
Our current understanding of cardiac vascular development is derived predominantly from murine models. Consequently, we next compared the transcriptional profiles of developing foetal human and embryonic mouse cardiac EC. For this comparison, a publicly available mouse embryonic heart scRNA-seq dataset15 was used due to its good representation of cardiac EC and because its embryonic stage (E16.5) corresponded with the later developmental stage of our human foetal heart data (13–14 weeks).37
Dimensionality reduction revealed successful integration of mouse and human cardiac EC (Supplementary material online, Figure S7A). Unlike our observation in the human heart, no distinct populations of KIT+ or INMT+ capillary populations were observed in the mouse data (Supplementary material online, Figure S7B). Clusters were therefore merged to represent the major subtypes of EC within the heart (endocardial, venous, lymphatic, capillary, arterial, proliferating, and valvular EC). Genes found to be amongst the most significantly differentially expressed in the same population in both human and mouse were classified as conserved markers (Figure 4A). MECOM and UNC5B were among genes with enriched arterial EC expression in both species (Figure 4A and B). In agreement with previous findings in mouse,38NPR3 expression was highly specific to the endocardial population with minimal expression in valvular EC.38 A lack of clear conserved venous EC markers was observed, with partial overlap of DEG in some lymphatic, valvular, and endocardial populations (Figure 4A and B). Interestingly, whilst the known LEC TF, PROX1, was expressed in both valvular EC and LEC in both species, PTX3 and a LEC marker, LYVE1, were found to be highly LEC specific (Figure 4A and B). Species-specific markers were also identified for each EC dataset (Figure 4A and Supplementary material online, Figure S7C). A human endocardial marker, NPCC, described elsewhere as specifically defining human foetal cardiac endocardium,12 was not enriched in the corresponding mouse population (Figure 4C).
Figure 4.
Comparison of the transcriptional profiles of human and mouse foetal cardiac EC populations. (A) Heatmaps showing expression of either conserved (left) or human-specific (right) markers for each subpopulation of foetal cardiac endothelium. (B) Expression of selected conserved markers in EC populations in human and mouse. (C) Expression of markers identified as being human-specific.
3.5 MECOM is required in arterial-like hESC-EC to suppress non-arterial gene expression
Given the in-silico predictions of a role for MECOM in arterial fate and enriched MECOM expression in arterial EC for both human and mouse, suggesting an evolutionarily conserved role in arterial EC, we sought to validate its role in determining human arterial EC identity. Our previous scRNA-seq-based characterization of our 8 day hESC-EC differentiation protocol demonstrated its suitability as an in vitro model of human EC development.32 Additionally, we determined that after acquisition of an early EC identity by Day 6, hESC-EC assume a clear arterial-like EC transcriptional signature by Day 8, characterized by expression of arterial markers, such as SOX17 and DLL4 (Supplementary material online, Figure S8A). Expression of venous (NR2F2 and EPHB4) and lymphatic markers (PROX1) in hESC-EC was low by Day 8 of the differentiation (Supplementary material online, Figure S8A). Notably, in agreement with its arterial EC specificity, MECOM was specifically expressed in hESC-EC at Days 6 and 8 of differentiation (Supplementary material online, Figure S8A).
Using hESC-EC as a developmental model for arterial EC specification, we next determined whether siRNA-mediated MECOM knockdown in hESC-EC resulted in changes to their arteriovenous identity (Figure 5A). Significant knockdown (>50%) of MECOM was observed at the RNA and protein level in hESC-EC 72 h after siRNA transfection (Figure 5B and Supplementary material online, Figure S8B and C). Bulk RNA-sequencing analysis revealed a distinct transcriptional profile for hESC-EC following MECOM knockdown compared to that of control hESC-EC (Supplementary material online, Figure S8D). Differential gene expression analysis demonstrated a reduction of MECOM resulted in a global increase in expression of non-arterial markers, including NR2F245 (Log2FC = 1.89) and VWF59 (Log2FC = 0.95) known to be enriched in venous EC (Figure 5C and Supplementary material online, File S3). Notably, known arterial markers including HEY1 and DLL4 were found not to be significantly down-regulated in response to MECOM knockdown. In addition to the up-regulation of known venous markers, several genes with previously reported differential expression in LEC including LYVE144 (Log2FC = 1.29), STAB260 (Log2FC = 4.05), and CEACAM161 (Log2FC = 1.74) were also found to be significantly up-regulated (Figure 5C). However, expression of key LEC TF PROX1 was not detected in either condition.
Figure 5.
Knockdown of MECOM in hESC-EC. (A) Experimental design for siRNA-mediated knockdown of MECOM in hESC-EC. (B) Quantification of MECOM protein abundance following siRNA knockdown using siRNA MECOM 1 (n = 4 biological replicates) and siRNA MECOM 2 (n = 3 biological replicates). P-values were obtained using an unpaired t-test. (C) Volcano plot showing differential gene expression following MECOM siRNA-mediated knockdown in hESC-EC (n = 4 biological replicates). P-values calculated using the Wald test. (D) Up-regulated gene signature score applied across identified clusters in foetal heart EC scRNA-seq dataset. Up-regulated gene signature constructed using top 20 genes found to be significantly up-regulated following MECOM knockdown in hESC-EC. (E) KEGG pathway enrichment analysis conducted using significantly up-regulated genes following MECOM knockdown. (F) qRT–PCR quantification of known markers of arterial (DLL4 and HEY1) and venous (EPHB4 and NR2F2) EC after MECOM siRNA knockdown (n = 4 biological replicates). P-values were calculated using a one-way ANOVA followed by Dunnett’s post-hoc multiple comparison test. Graphs in (B) and (F) correspond to mean ± standard error of the mean.
Application of an up-regulated gene expression signature (constructed using the top 20 up-regulated genes following MECOM KD in hESC-EC) to our foetal cardiac EC scRNA-seq dataset revealed the lowest level of signature enrichment in arterial populations, with highest levels of enrichment observed in lymphatic and venous clusters (Figure 5D). KEGG pathway enrichment analysis conducted using genes significantly up-regulated following MECOM KD identified enrichment of genes belonging to the PI3K-AKT signalling pathway, reported to play a role in venous EC specification62,63 (Figure 5E). qRT–PCR validation aligned with bulk RNA-seq findings demonstrating knockdown of MECOM resulted in significant up-regulation of venous EC markers (NR2F2 and EPHB4) whilst arterial (HEY1, DLL4, JAG1, and JAG2) markers remained unchanged (Figure 5F and Supplementary material online, Figure S8E). Importantly, reduction of MECOM did not result in altered expression of the pan-endothelial marker, CDH5, suggesting that the changes observed in arteriovenous marker expression are not due to a loss of general EC identity (Supplementary material online, Figure S8E).
4. Discussion
In this study, we comprehensively mapped the transcriptional landscape of the developing human foetal heart endothelium using scRNA-seq. Isolation of foetal cardiac EC by FACS prior to performing high-throughput scRNA-seq empowered this study to identify the full extent of EC heterogeneity. This included identifying distinct endocardial, valvular, venous, capillary, and arterial EC populations each expressing a separate transcriptional signature.
GRN analysis identified the TFs most likely responsible for establishing the observed EC heterogeneity. Application of trajectory inference methods to microvascular ECs was used to map the cellular dynamics accompanying coronary vascular EC development. This revealed a small proportion of endocardial cells that appeared to transition to a vascular EC identity via a venous EC population. In addition, capillary EC was predicted to be undergoing specification to assume an arterial EC identity, defined by increasing expression of the TF, MECOM. Comparison of our human foetal heart EC data with E16.5 murine cardiac EC scRNA-seq data demonstrated the existence of several conserved, as well as species-specific, markers for each of the major cardiac EC populations. This included identifying NPR3 and MECOM as conserved markers of endocardial and arterial populations, respectively. Finally, we demonstrated that loss of MECOM in arterial-like hESC-EC resulted in a global increased expression of non-arterial markers, suggesting a function to maintain identity in arterial EC.
In contrast to the capillary EC cluster defined by differential expression of methyltransferase-encoding gene, INMT, GRN analysis revealed several regulons enriched within the KIT+ capillary population. This included the SMAD1 regulon. BMP/SMAD1 signalling has been demonstrated to promote angiogenesis whilst KIT/C-KIT has been shown to mediate neovascularization in retinal microvascular ECs in response to hypoxia.64,65 This suggests that SMAD1 may mediate angiogenesis within hypoxic regions in the developing heart wall, although this will require further investigation to verify. The existence of two capillary populations with distinct transcriptional signatures, including the differential expression of KIT and INMT, was recently confirmed in an independent study from Phansalkar et al.,19 which performed low-throughput scRNA-seq on EC isolated from 11-, 14-, and 22-week human foetal hearts.
Our in-silico findings suggested a transition of endocardium to coronary vascular endothelium. This is consistent with previous findings from murine lineage-tracing studies, in which a proportion of the endocardium gives rise to coronary vascular EC via angiogenic sprouting.1 A second method of endocardial-derived coronary vessel formation during was also proposed to occur at the murine perinatal stage and involve the formation of new coronary vessels by the segregation of endocardial trabeculae protruding into the myocardium during compaction.66 However, this model has recently been challenged by Lu et al.,67 which concluded that formation of new coronary vessels during the perinatal stage is instead due to angiogenic expansion of the pre-existing coronary plexus.
Although our trajectory analysis indicated the transition of endocardium to coronary vasculature occurs via a venous EC population, the arteriovenous identity of cells undergoing this process has not previously been explored. Whilst studies in mouse have demonstrated a significant proportion of coronary vascular EC to be derived from venous cells of the SV, this is thought to occur much earlier in cardiac development than the comparative gestational age of the human foetal samples used in this analysis.2,38 However, the observed enrichment of BMP2 expression within the identified venous cluster aligns closely with recent scRNA-seq evidence from D’Amato et al.58 identifying Bmp2 as a marker of the transitioning endocardial population in E12 mouse embryos. Additionally, enriched venous expression of bmp2 has previously been described in zebrafish,57 thus, further indicating the identified venous cluster may represent a transitioning endocardial-derived population.
Trajectory inference analysis also revealed subsequent arterial specification of capillary EC. This predicted cellular transition in the human foetal heart was also recently identified by Phansalkar et al.,19 thus, collectively providing human relevance to current understanding of coronary artery development derived from murine studies.7–9 However, in addition to confirming the up-regulation of known mediators of arterial specification, such as HEY140 and SOX17,53MECOM was also identified as having a role in the establishment of an arterial EC identity. Furthermore, enriched arterial expression of MECOM was also observed in coronary EC from E16.5 mouse hearts, suggesting an evolutionarily conserved function.
The localization of MECOM in the developing human heart was validated using ISH methods and its function in arterial EC identity demonstrated by siRNA-mediated knockdown in arterial-like hESC-EC. Previous work from Li et al.68 demonstrated that MECOM acts upstream of Notch signalling during zebrafish nephrogenesis. Given the importance of Notch signalling in arterial EC specification, this suggested that MECOM may alter arteriovenous identity by regulating Notch signalling. Whilst reduction in MECOM expression in hESC-EC did not alter the expression of arterial EC markers, including those belonging to the Notch pathway, a global increase in the expression of non-arterial-enriched genes was observed. This included the TF, NR2F2, which is known to establish venous identity, in part via repression of Notch signalling.45 Although a subset of genes up-regulated following MECOM knockdown have been reported to be differentially expressed in LEC, the absence of increased PROX1 expression indicated the reduction in MECOM does not specify EC towards a lymphatic identity.
Collectively, these findings suggest that MECOM may be required to supress non-arterial gene expression during arterial EC specification. Goyama et al.69 previously demonstrated that loss of MECOM within Tie2+ cells results in severe vascular abnormalities leading to embryonic lethality in mouse between E13.5 and E16.5. However, considering our described findings, further investigation is required to characterize MECOM expression across the murine embryonic and adult coronary vascular endothelium, as well as to evaluate the resultant effect of EC-specific loss of MECOM on arteriovenous identity.
Previous studies from mouse have hypothesized that a venous identity is the default state for EC, with venous identity needing to be repressed via Notch signalling during arterial specification.70,71 Our finding that MECOM knockdown altered venous marker (NR2F2 and EPHB4) expression, without changes to expression of Notch signalling genes, suggests that additional factors are required to supress venous identity during human arterial EC specification. However, further studies simultaneously targeting the expression of characterized arterial EC regulators is required to determine the position of MECOM within the hierarchal network of arteriovenous regulators. Previous findings in mouse demonstrated overexpression of arterial EC regulator Dach1 resulted in an increase in perfused arteries following myocardial infarction.8 Our finding from human data suggesting MECOM may function to maintain the transcriptional identity of arterial EC highlights it as a prime therapeutic candidate to drive arterialization in cardiovascular disease.
Although this study is the most comprehensive of its type to date, due to limited sample availability its data provides only a snapshot of a narrow developmental window (13–14 weeks). This limitation prevented the comparison of gene expression and cluster proportion between different gestational ages. Careful batch correction and visualization of gene expression dynamics across pseudotime for individual samples ensured findings from trajectory inference analysis were not biased by unequal representation of individual clusters. Whilst trajectory inference methods permit the dynamical changes to be characterized within individual datasets, inclusion of foetal samples from a wider range of gestational ages would provide a more comprehensive understanding of human coronary vascular development, especially at earlier stages.
In summary, we have used a high-throughput scRNA-seq platform to comprehensively map the transcriptional landscape of the human foetal heart endothelium at 13–14 weeks. This study complements studies using murine models of cardiovascular development by providing novel insight into EC heterogeneity within the developing human heart, as well as the dynamical changes accompanying coronary vasculature formation. In addition to helping understand the mechanisms giving rise to congenital coronary vascular abnormalities, this information may prove valuable in future strategies to guide coronary vascular formation for the treatment of coronary vascular disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
I.R.M., J.C.M., N.S., M.B., and A.H.B. were involved in the design of the described study. I.R.M. and R.D. carried out foetal tissue collection and sample processing. Bioinformatic analysis was performed by I.R.M. and M.B. In-vitro experiments, qRT–PCR, and western blotting analysis were conducted by I.R.M., R.P., and A.B. A.H.B., C.P.P., and M.B. supervised the research. A.H.B. secured research funding. I.R.M., M.B., N.C.H., J.C.M., P.R.R., C.P.P., N.S., M.B., and A.H.B. were involved in interpreting bioinformatics data. I.R.M. and A.H.B. wrote the manuscript with input from all authors. All authors discussed the data and edited the manuscript.
Supplementary Material
Acknowledgements
The authors thank Andrea Corsinotti for his assistance in generating 10X scRNA-seq libraries and Kathryn Newton for her technical support. FACS was performed with support from both the CRM Flow and Genomic Cytometry Core Facility and the QMRI Flow Cytometry and cell sorting facility, University of Edinburgh. For in-situ hybridization validations, human embryonic and foetal material was provided by the Joint MRC/Wellcome Trust (Grant # MR/006237/1) Human Developmental Biology Resource (http://www.hdbr.org) and performed by the HDBR in house gene expression service.
Funding
This work was supported by Medical Research Council [MRC Precision Medicine Doctoral Training Programme to I.R.M. and MRC program: Computational and Disease Genomics (MC_UU_00007/15) to C.P.P.], Wellcome Trust [Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 219542/Z/19/Z) to N.C.H.], British Heart Foundation [Personal Chair Award (#CH/11/1/28798) to P.R.R., Intermediate Basic Science Research Fellowship (FS/16/4/31831) to M.B., BHF Chair (CH/11/2/28733), BHF Programme grant (RG/20/5/34796), and Centre for Regenerative Medicine (CRMR/21/290009) to A.H.B.], Chief Scientists Office [CSO grant (CGA/19/18) to A.H.B.], and the ERC [Advanced Grant VASCMIR (338991) to A.H.B.].
Contributor Information
Ian R McCracken, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK; Department of Physiology, Anatomy, and Genetics, University of Oxford, Oxford OX1 3PT, UK.
Ross Dobie, Centre for Inflammation Research, University of Edinburgh, Edinburgh EH16 4TJ, UK.
Matthew Bennett, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK.
Rainha Passi, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK.
Abdelaziz Beqqali, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK.
Neil C Henderson, Centre for Inflammation Research, University of Edinburgh, Edinburgh EH16 4TJ, UK; MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK.
Joanne C Mountford, Scottish National Blood Transfusion Service, Edinburgh EH14 4BE, UK.
Paul R Riley, Department of Physiology, Anatomy, and Genetics, University of Oxford, Oxford OX1 3PT, UK.
Chris P Ponting, MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH4 2XU, UK.
Nicola Smart, Department of Physiology, Anatomy, and Genetics, University of Oxford, Oxford OX1 3PT, UK.
Mairi Brittan, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK.
Andrew H Baker, Centre for Cardiovascular Science, University of Edinburgh, Edinburgh EH16 4TJ, UK; Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center, 6229 HX Maastricht, The Netherlands.
This manuscript was handled by Professor Thomas Thum.
Data availability
RNA-sequencing data used in this study are accessible from the Gene Expression Omnibus (accession number: GSE195911).
Translational perspective.
Endogenous blood vessel formation in the adult heart following myocardial infarction is insufficient to support adequate survival of the remaining myocardium, often ultimately leading to heart failure. Improved understanding of the mechanisms regulating human coronary vessel formation is required to inform therapeutic strategies to reactivate developmental pathways promoting therapeutic angiogenesis in patients. We applied scRNA-seq to map the transcriptome of the endothelium of the developing human heart. We identified novel transcriptional signatures underlying the cellular heterogeneity and dynamic changes occurring within the developing cardiac endothelium. This included identifying and validating MECOM as a novel regulator of arterial EC identity, which may serve as a target for therapeutic neovascularization.
References
- 1. Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O'Rourke BP, Sharp DJ, Zheng D, Lenz J, Baldwin HS, Chang C-P, Zhou B. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 2012;151:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010;464:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai C-L, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008;454:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 2012;22:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lupu IE, Redpath AN, Smart N. Spatiotemporal analysis reveals overlap of key proepicardial markers in the developing murine heart. Stem Cell Rep 2020;14:770–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma B, Chang A, Red-Horse K. Coronary artery development: progenitor cells and differentiation pathways. Annu Rev Physiol 2017;79:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su T, Stanley G, Sinha R, D'Amato G, Das S, Rhee S, Chang AH, Poduri A, Raftrey B, Dinh TT, Roper WA, Li G, Quinn KE, Caron KM, Wu S, Miquerol L, Butcher EC, Weissman I, Quake S, Red-Horse K. Single-cell analysis of early progenitor cells that build coronary arteries. Nature 2018;559:356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raftrey B, Williams I, Rios Coronado PE, Fan X, Chang AH, Zhao M, Roth R, Trimm E, Racelis R, D'Amato G, Phansalkar R, Nguyen A, Chai T, Gonzalez KM, Zhang Y, Ang LT, Loh KM, Bernstein D, Red-Horse K. Dach1 extends artery networks and protects against cardiac injury. Circ Res 2021;129:702–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang AH, Raftrey BC, D'Amato G, Surya VN, Poduri A, Chen HI, Goldstone AB, Woo J, Fuller GG, Dunn AR, Red-Horse K. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev 2017;31:1308–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 2018;14:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG, Seidman CE. Single-cell resolution of temporal gene expression during heart development. Dev Cell 2016;39:480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suryawanshi H, Clancy R, Morozov P, Halushka MK, Buyon JP, Tuschl T. Cell atlas of the foetal human heart and implications for autoimmune-mediated congenital heart block. Cardiovasc Res 2020;116:1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep 2019;26:1934–1950.e1935. [DOI] [PubMed] [Google Scholar]
- 14. Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, Österholm C, Ståhl PL, Sundström E, Åkesson E, Bergmann O, Bienko M, Månsson-Broberg A, Nilsson M, Sylvén C, Lundeberg J. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 2019;179:1647–1660.e1619. [DOI] [PubMed] [Google Scholar]
- 15. Goodyer WR, Beyersdorf BM, Paik DT, Tian L, Li G, Buikema JW, Chirikian O, Choi S, Venkatraman S, Adams EL, Tessier-Lavigne M, Wu JC, Wu SM. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ Res 2019;125:379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li G, Tian L, Goodyer W, Kort EJ, Buikema JW, Xu A, Wu JC, Jovinge S, Wu SM. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development 2019;146:dev173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, Wu SM. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev Cell 2016;39:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Soysa TY, Ranade SS, Okawa S, Ravichandran S, Huang Y, Salunga HT, Schricker A, Del Sol A, Gifford CA, Srivastava D. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 2019;572:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phansalkar R, Krieger J, Zhao M, Kolluru SS, Jones RC, Quake SR, Weissman I, Bernstein D, Winn VD, D'Amato G, Red-Horse K. Coronary blood vessels from distinct origins converge to equivalent states during mouse and human development. Elife 2021;10:e70246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Beijnum JR, Rousch M, Castermans K, van der Linden E, Griffioen AW. Isolation of endothelial cells from fresh tissues. Nat Protoc 2008;3:1085–1091. [DOI] [PubMed] [Google Scholar]
- 21. Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, Marioni JC, participants in the 1st Human Cell Atlas Jamboree . EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol 2019;20:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017;33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 2019;16:1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, Belicova L, Bickle M, Barsacchi R, Okuda R, Yoshizawa E, Kimura M, Ayabe H, Taniguchi H, Takebe T, Treutlein B. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017;546:533–538. [DOI] [PubMed] [Google Scholar]
- 27. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408–1414. [DOI] [PubMed] [Google Scholar]
- 29. Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 2020;11:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. MacAskill MG, Saif J, Condie A, Jansen MA, MacGillivray TJ, Tavares AAS, Fleisinger L, Spencer HL, Besnier M, Martin E, Biglino G, Newby DE, Hadoke PWF, Mountford JC, Emanueli C, Baker AH. Robust revascularization in models of limb ischemia using a clinically translatable human stem cell-derived endothelial cell product. Mol Ther 2018;26:1669–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCracken IR, Taylor RS, Kok FO, de la Cuesta F, Dobie R, Henderson BEP, Mountford JC, Caudrillier A, Henderson NC, Ponting CP, Baker AH. Transcriptional dynamics of pluripotent stem cell-derived endothelial cell differentiation revealed by single-cell RNA sequencing. Eur Heart J 2020;41:1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krishnan A, Samtani R, Dhanantwari P, Lee E, Yamada S, Shiota K, Donofrio MT, Leatherbury L, Lo CW. A detailed comparison of mouse and human cardiac development. Pediatr Res 2014;76:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, Zhou B. Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ Res 2016;118:1880–1893. [DOI] [PubMed] [Google Scholar]
- 39. Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen L-A, Veys K, García-Caballero M, Khan S, Geldhof V, Sokol L, Chen R, Treps L, Borri M, de Zeeuw P, Dubois C, Karakach TK, Falkenberg KD, Parys M, Yin X, Vinckier S, Du Y, Fenton RA, Schoonjans L, Dewerchin M, Eelen G, Thienpont B, Lin L, Bolund L, Li X, Luo Y, Carmeliet P. Single-cell transcriptome atlas of murine endothelial cells. Cell 2020;180:764–779.e720. [DOI] [PubMed] [Google Scholar]
- 40. Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev 2004;18:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kume T. Specification of arterial, venous, and lymphatic endothelial cells during embryonic development. Histol Histopathol 2010;25:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou J, Bowen C, Lu G, Knapp Iii C, Recknagel A, Norris RA, Butcher JT. Cadherin-11 expression patterns in heart valves associate with key functions during embryonic cushion formation, valve maturation and calcification. Cells Tissues Organs 2013;198:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, Zhou B, Feinstein JA, Krasnow MA, Metzger RJ. Capillary cell-type specialization in the alveolus. Nature 2020;586:785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliver G, Kipnis J, Randolph GJ, Harvey NL. The lymphatic vasculature in the 21(st) century: novel functional roles in homeostasis and disease. Cell 2020;182:270–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. You L-R, Lin F-J, Lee CT, DeMayo FJ, Tsai M-J, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 2005;435:98–104. [DOI] [PubMed] [Google Scholar]
- 46. Thiriot A, Perdomo C, Cheng G, Novitzky-Basso I, McArdle S, Kishimoto JK, Barreiro O, Mazo I, Triboulet R, Ley K, Rot A, von Andrian UH. Differential DARC/ACKR1 expression distinguishes venular from non-venular endothelial cells in murine tissues. BMC Biol 2017;15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu B, Wang Y, Lui W, Langworthy M, Tompkins KL, Hatzopoulos AK, Baldwin HS, Zhou B. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ Res 2011;109:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCulley DJ, Kang JO, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn 2008;237:3200–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Son NH, Basu D, Samovski D, Pietka TA, Peche VS, Willecke F, Fang X, Yu SQ, Scerbo D, Chang HR, Sun F, Bagdasarov S, Drosatos K, Yeh ST, Mullick AE, Shoghi KI, Gumaste N, Kim K, Huggins LA, Lhakhang T, Abumrad NA, Goldberg IJ. Endothelial cell CD36 optimizes tissue fatty acid uptake. J Clin Invest 2018;128:4329–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wigle JT, Harvey N, Detmar M, Lagutina I, Grosveld G, Gunn MD, Jackson DG, Oliver G. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J 2002;21:1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol 2006;296:421–436. [DOI] [PubMed] [Google Scholar]
- 52. Laforest B, Nemer M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev Biol 2011;358:368–378. [DOI] [PubMed] [Google Scholar]
- 53. Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, Adams RH, Dejana E. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun 2013;4:2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lupu I-E, De Val S, Smart N. Coronary vessel formation in development and disease: mechanisms and insights for therapy. Nat Rev Cardiol 2020;17:790–806. [DOI] [PubMed] [Google Scholar]
- 55. Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivelä R, Saharinen P, Aghajanian H, McKay AS, Bogard PE, Chang AH, Jacobs AH, Epstein JA, Stankunas K, Alitalo K, Red-Horse K. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development 2014;141:4500–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sharma B, Ho L, Ford GH, Chen HI, Goldstone AB, Woo YJ, Quertermous T, Reversade B, Red-Horse K. Alternative progenitor cells compensate to rebuild the coronary vasculature in Elabela- and Apj-deficient hearts. Dev Cell 2017;42:655–666.e653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Neal A, Nornes S, Payne S, Wallace MD, Fritzsche M, Louphrasitthiphol P, Wilkinson RN, Chouliaras KM, Liu K, Plant K, Sholapurkar R, Ratnayaka I, Herzog W, Bond G, Chico T, Bou-Gharios G, De Val S. Venous identity requires BMP signalling through ALK3. Nat Commun 2019;10:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. D’Amato G, Phansalkar R, Naftaly JA, Rios Coronado PE, Cowley DO, Quinn KE, Sharma B, Caron KM, Vigilante A, Red-Horse K. Endocardium-to-coronary artery differentiation during heart development and regeneration involves sequential roles of Bmp2 and Cxcl12/Cxcr4. bioRxiv2021:2021.2010.2025.465773. [DOI] [PMC free article] [PubMed]
- 59. Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018;554:475–480. [DOI] [PubMed] [Google Scholar]
- 60. Park SM, Angel CE, McIntosh JD, Mansell CJ, Mansell CM, Chen C-JJ, Cebon J, Dunbar PR. Mapping the distinctive populations of lymphatic endothelial cells in different zones of human lymph nodes. PLoS One 2014;9:e94781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kilic N, Oliveira-Ferrer L, Neshat-Vahid S, Irmak S, Obst-Pernberg K, Wurmbach JH, Loges S, Kilic E, Weil J, Lauke H, Tilki D, Singer BB, Ergün S. Lymphatic reprogramming of microvascular endothelial cells by CEA-related cell adhesion molecule-1 via interaction with VEGFR-3 and Prox1. Blood 2007;110:4223–4233. [DOI] [PubMed] [Google Scholar]
- 62. Deng Y, Larrivée B, Zhuang ZW, Atri D, Moraes F, Prahst C, Eichmann A, Simons M. Endothelial RAF1/ERK activation regulates arterial morphogenesis. Blood 2013;121:3988–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hong CC, Peterson QP, Hong J-Y, Peterson RT. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr Biol 2006;16:1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim KL, Seo S, Kim JT, Kim J, Kim W, Yeo Y, Sung JH, Park SG, Suh WS. SCF (stem cell factor) and cKIT modulate pathological ocular neovascularization. Arterioscler Thromb Vasc Biol 2019;39:2120–2131. [DOI] [PubMed] [Google Scholar]
- 65. Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, Cheng HW, Raghuram S, Arany Z, Liao R, Croce K, MacRae C, Feinberg MW. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res 2013;113:1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Yan Y, Yang X, Zhong TP, Pu WT, Zhou B. De novo formation of a distinct coronary vascular population in neonatal heart. Science 2014;345:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lu P, Wang Y, Liu Y, Wang Y, Wu B, Zheng D, Harvey RP, Zhou B. Perinatal angiogenesis from pre-existing coronary vessels via DLL4-NOTCH1 signalling. Nat Cell Biol 2021;23:967–977. [DOI] [PubMed] [Google Scholar]
- 68. Li Y, Cheng CN, Verdun VA, Wingert RA. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev Biol 2014;386:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, Chiba S, Kurokawa M. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell 2008;3:207–220. [DOI] [PubMed] [Google Scholar]
- 70. Swift MR, Pham VN, Castranova D, Bell K, Poole RJ, Weinstein BM. SoxF factors and Notch regulate nr2f2 gene expression during venous differentiation in zebrafish. Dev Biol 2014;390:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wolf K, Hu H, Isaji T, Dardik A. Molecular identity of arteries, veins, and lymphatics. J Vasc Surg 2019;69:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data used in this study are accessible from the Gene Expression Omnibus (accession number: GSE195911).
Translational perspective.
Endogenous blood vessel formation in the adult heart following myocardial infarction is insufficient to support adequate survival of the remaining myocardium, often ultimately leading to heart failure. Improved understanding of the mechanisms regulating human coronary vessel formation is required to inform therapeutic strategies to reactivate developmental pathways promoting therapeutic angiogenesis in patients. We applied scRNA-seq to map the transcriptome of the endothelium of the developing human heart. We identified novel transcriptional signatures underlying the cellular heterogeneity and dynamic changes occurring within the developing cardiac endothelium. This included identifying and validating MECOM as a novel regulator of arterial EC identity, which may serve as a target for therapeutic neovascularization.