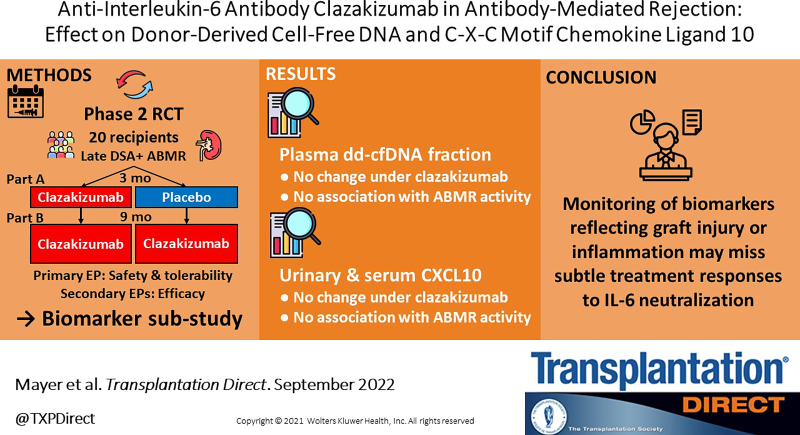
Background.
Targeting interleukin-6 (IL-6) was shown to counteract donor-specific antibody production and antibody-mediated rejection (AMR) activity. It is not known whether, or to what extent, IL-6 antagonism modulates biomarkers indicative of tissue damage (donor-derived cell-free DNA [dd-cfDNA]) and parenchymal inflammation (C-X-C motif chemokine ligand [CXCL] 10).
Methods.
We report a secondary endpoint analysis of a phase 2 trial of anti-IL-6 antibody clazakizumab in late AMR (ClinicalTrials.gov, NCT03444103). Twenty kidney transplant recipients were randomized to treatment with clazakizumab or placebo over 12 wk (part A), followed by an extension in which all recipients received clazakizumab through week 52 (part B). Biomarkers were evaluated at day 0 and after 12 and 52 wk, respectively.
Results.
Fractional dd-cfDNA (dd-cfDNA[%]) did not significantly change under clazakizumab, with no differences between study arms (clazakizumab versus placebo) at week 12 (1.65% [median; interquartile range: 0.91%–2.78%] versus 0.97% [0.56%–2.30%]; P = 0.25) and no significant decrease from weeks 12 to 52 (1.15% [0.70%–2.38%] versus 1.0% [0.61%–1.70%]; P = 0.25). Similarly, urine CXCL10 was not different between groups at week 12 (55.7 [41.0–91.4] versus 60.2 [48.8–208.7.0] pg/mg creatinine; P = 0.44) and did not change over part B (CXCL10 [pg/mg creatinine]: from 58 [46.3–93.1] to 67.4 [41.5–132.0] pg/mL creatinine; P = 0.95). Similar results were obtained for serum CXCL10. There was no association between biomarker levels and resolution of molecular and morphologic AMR activity.
Conclusions.
Our results suggest that IL-6 blockade does not significantly affect levels of dd-cfDNA[%] and CXCL10. Subtle responses to this therapeutic principle may be overlooked by early biomarker surveillance.
INTRODUCTION
Antibody-mediated rejection (AMR) is a leading cause of kidney transplant dysfunction and failure.1,2 Its treatment, particularly that of chronic AMR, remains a major challenge.3,4 Recent studies have suggested therapeutic efficacy of interleukin-6 (IL-6) or IL-6 receptor (IL-6R) blockade to counteract late AMR, presumably via interference with B cell-driven alloimmunity and promotion of regulatory T cells.5-7 Two therapeutic antibodies are now under investigation in this specific context, the anti-IL-6R antibody tocilizumab and clazakizumab, which directly neutralizes IL-6.8 We have recently published the results of a randomized, controlled phase 2 study including 20 kidney transplant recipients diagnosed with late chronic active or active AMR.6 Consistent with other uncontrolled studies,5,7 our trial revealed that IL-6 antagonism was associated with a reduction in donor-specific antibody (DSA) levels and after 9–12 mo of treatment with a decrease in AMR activity. In addition, a preliminary analysis of renal function suggested a possible less pronounced decline in estimated glomerular filtration rate (eGFR).6
Renal biopsies are the current gold standard in the diagnosis of rejection, and apart from widely used surrogates of treatment responsiveness, such as levels of DSA in serum and the early course of renal function, several interventional trials in late/chronic AMR have relied on follow-up biopsies to monitor new treatment strategies.6,9,10 However, the use of serial protocol biopsies to detect treatment effects has its limitations, most importantly due to the invasiveness of the procedure and the risk of bleeding complications. In this context, the implementation of serial noninvasive biomarker monitoring for the early detection of cellular injury and ongoing inflammation in the graft could be of clinical value.11,12
In recent years, there has been a growing interest in quantifying donor-derived cell-free DNA (dd-cfDNA) to detect ongoing rejection processes.13,14 Several studies have shown a pronounced release of dd-cfDNA into the circulation during rejection, with particularly high levels in patients with AMR.15-18 At the same time, however, it has become evident that this molecular biomarker may not be entirely specific for allograft rejection but may also increase in other processes that trigger active graft injury, such as ischemia-reperfusion injury, polyomavirus allograft nephropathy, or glomerulonephritis.17,19,20 Because of a considerable diagnostic and predictive value for rejection, as has been demonstrated in large multicenter cohorts,18,21 measuring fractions of dd-cfDNA (dd-cfDNA[%]) is now increasingly used in transplantation routine, for example‚ in an effort to avoid unnecessary biopsies. There is preliminary evidence from cohort studies that dd-cfDNA[%] could also play a role in monitoring response to rejection treatment,22-24 but systematic studies evaluating this biomarker in the context of a randomized controlled interventional trial are still lacking.
Another promising biomarker is the detection of C-X-C motif chemokine ligand (CXCL) 10, also known as interferon-γ-inducible protein-10. This chemokine, produced by a variety of different cell types, including endothelial cells in the tubulointerstitial area, plays a critical role in leukocyte trafficking, recruitment‚ and activation.25 Observational studies have shown that urine and serum/plasma CXCL10 levels are elevated in T cell-mediated rejection (TCMR) and AMR and may even anticipate increases in serum creatinine or biopsy-based diagnosis of rejection.26-34 However, some of these studies have shown that chemokine levels are also affected by other processes, such as polyomavirus nephropathy or urinary tract infection.27-29,32,34 A recent study has shown that an integrative approach including different confounding variables improves the prediction of rejection.35 A large multicenter trial is currently underway using chemokine monitoring to guide biopsy management and treatment.36 Although observational studies have suggested utility of CXCL10 monitoring to assess responsiveness to (primarily T cell-mediated) rejection treatment,34 there has been no prospective evaluation of this marker in a randomized controlled trial design.
The primary objective of this secondary endpoint analysis of a recently reported randomized controlled interventional trial6 was to assess whether clazakizumab has an effect on (i) dd-cfDNA[%] as an estimate of allograft injury and (ii) urinary/serum CXCL10 reflecting renal parenchymal inflammation. In addition, since only a subset of patients showed resolution of AMR, we were interested in whether biomarker levels are able to detect reversal of molecular and morphological rejection activity. We analyzed samples collected at predefined time intervals to decipher associations of biomarker levels with efficacy endpoints.
MATERIALS AND METHODS
Study Design and Patients
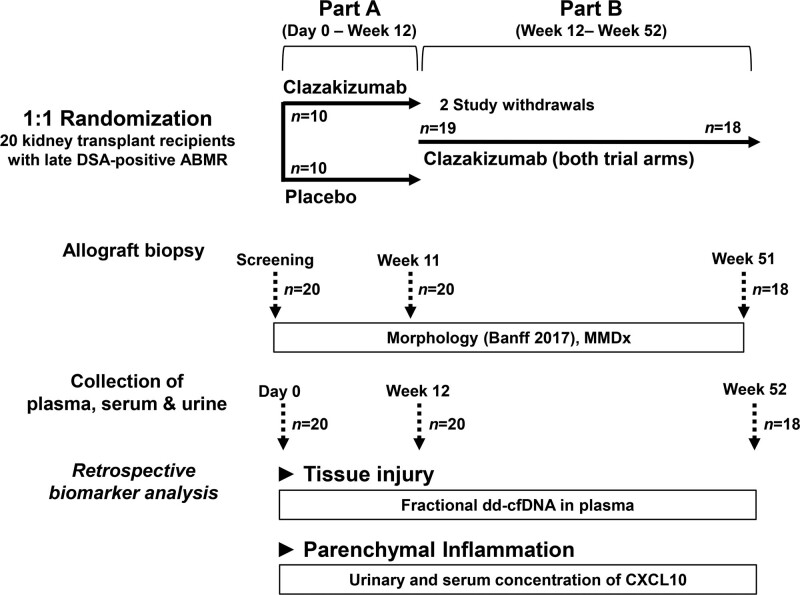
This substudy of biomarkers was performed as part of a randomized, double-blind, placebo-controlled phase 2 trial designed to evaluate the safety (primary endpoint), and efficacy of the anti-IL-6 antibody clazakizumab (Vitaeris Inc., Vancouver, Canada) in late AMR (ClinicalTrials.gov, NCT03444103).6,37 The study was conducted between January 2018 and April 2020 at 2 locations (Medical University of Vienna, Austria; Charité Universitätsmedizin Berlin, Germany). The protocol and eligibility criteria, as well as key safety and efficacy results are described in previous publications.6,37 Briefly, the trial included 20 renal allograft recipients with HLA classes I and II DSA-positive active or chronic active AMR ≥365 d after transplantation. Key patient characteristics are listed in Table 1. As illustrated in Figure 1, the study consisted of a 12-wk randomized placebo-controlled phase (1:1 permuted block randomization) to decipher the short-term effects of treatment (part A), followed by a 40-wk open-label extension in which all subjects received clazakizumab (part B). As previously reported, 11 (55%) subjects did not receive all 13 scheduled clazakizumab injections (12 doses: n = 3; 11 doses: n = 2; 10 doses: n = 2; 9 doses: n = 1; 8 doses: n = 1; 4 doses: n = 1; 2 doses: n = 1), due to adverse events (n = 10) or personal reasons (n = 1). All patients gave written informed consent before study inclusion. The study was approved by the institutional review boards of the Medical University of Vienna and the Berlin State Ethics Committee and conducted in accordance with the principles of the Declaration of Helsinki 2008, and the Declaration of Istanbul.
TABLE 1.
Patient characteristics, serologic data, and biopsy results at baselinea
| Variables | All patients (n = 20) | Clazakizumab (n = 10) | Placebo (n = 10) |
|---|---|---|---|
| Variables recorded at transplantation | |||
| Female sex, n (%) | 10 (50) | 3 (30) | 7 (70) |
| Recipient age (y), median (IQR) | 34.2 (24.6–47.6) | 37.4 (27.1–57.9) | 31.4 (22.3–42.3) |
| Living donor, n (%) | 6 (30) | 3 (30) | 3 (30) |
| Prior kidney transplant, n (%) | 7 (35) | 4 (40) | 3 (30) |
| Preformed anti-HLA DSA, n (%)b | 5 (45.5) | 3 (42.9) | 2 (50) |
| Donor age (y), median (IQR)c | 49.0 (21.8–57.3) | 51.0 (21.8–57.3) | 44.0 (23.3–66.0) |
| HLA mismatch (A, B, DR), median (IQR)d | 3 (2–3) | 3 (3–3) | 3 (2–3) |
| Variables recorded at trial inclusion | |||
| Year to inclusion in the trial | 10.6 (4.4–16.2) | 9.7 (4.1–16.7) | 11.4 (5.9–16.1) |
| eGFR (mL/min per 1.73 m2), median (IQR) | 39.3 (33.6–49.7) | 40.5 (33.3–49.8) | 39.2 (32.9–51.7) |
| Protein/creatinine ratio (mg/g), median (IQR) | 962 (310–1863) | 727 (197–1311) | 1387 (532–3575) |
| Immunosuppressive therapy | |||
| Triple immunosuppression, n (%) | 18 (90) | 9 (90) | 9 (90) |
| Dual immunosuppression without steroids, n (%) | 2 (10) | 1 (10) | 1 (10) |
| Tacrolimus, n (%) | 13 (65) | 6 (60) | 7 (70) |
| Cyclosporin A, n (%) | 6 (30) | 4 (40) | 2 (20) |
| Everolimus, n (%) | 1 (5) | 0 | 1 (10) |
| Mycophenolic acid, n (%) | 20 (100) | 10 (100) | 10 (100) |
| HLA antibody results | |||
| MFI of the peak DSA, median (IQR) | 11 708 (1947–17 709) | 10 789 (3092–15 437) | 14 207 (1252–19 144) |
| Anti-DQ DSA, n (%) | 15 (75) | 7 (70) | 8 (80) |
| AMR morphology (Banff 2017) | |||
| Active AMR, n (%) | 2 (10) | 2 (20) | 0 |
| Chronic/active AMR, n (%) | 18 (90) | 8 (80) | 10 (100) |
| Peritubular capillary C4d staining, n (%) | 7 (35) | 4 (40) | 3 (30) |
| MVI (g+ptc) score, median (IQR)e | 3 (2–4) | 2 (2–3) | 3 (2–4) |
| Transplant glomerulopathy (cg score), median (IQR)e | 3 (2–3) | 3 (1–3) | 3 (3–3) |
| Molecular biopsy results (MMDx) | |||
| AMR score, median (IQR) | 0.65 (0.35–0.81) | 0.70 (0.48–0.81) | 0.44 (0.29–0.82) |
| Acute kidney injury score, median (IQR) | 0.40 (0.18–0.65) | 0.18 (–0.03 to 0.39) | 0.58 (0.40–0.83) |
| Atrophy/fibrosis score, median (IQR) | 0.68 (0.35–0.84) | 0.39 (0.24–0.78) | 0.79 (0.67–0.87) |
aDifferences between clazakizumab and placebo with respect to baseline variables were not significant (P value >0.05).
bPretransplant DSA data were available for 7 recipients in the clazakizumab arm and 4 in the placebo arm (solid-phase HLA antibody screening on the waitlist was implemented at the Vienna transplant unit in July 2009).
cDonor age was not recorded for 2 recipients in the placebo arm.
dHLA mismatch was not recorded for 1 recipient in the placebo arm.
eThe g+ptc could not be calculated in 2 recipients allocated to the placebo arm (no sufficient material for glomerulitis scoring).
AMR, antibody-mediated rejection; DSA, donor-specific antibody; eGFR, estimated glomerular filtration rate; IQR, interquartile range; MFI, mean fluorescence intensity; MMDx, Molecular Microscope Diagnostic System; MVI, microvascular inflammation.
FIGURE 1.
Trial protocol. Twenty kidney allograft recipients with late DSA-positive AMR were randomized 1:1 to receive clazakizumab or placebo for 12 wk (part A). After 12 wk, all patients were scheduled to receive clazakizumab (part B). Two patients allocated to receive clazakizumab were withdrawn from the study because of serious adverse events (diverticular disease complications). Both completed part A. One was withdrawn shortly before, the other after the first dose of clazakizumab in part B. At day 0 and after 12 and 52 wk, biologic material was collected and stored for biomarker evaluation (secondary endpoint analysis). Tested biomarkers included plasma dd-cfDNA and CXCL10 in urine and serum. AMR, antibody-mediated rejection; CXCL, C-X-C motif chemokine ligand; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System.
Measurement of Biomarkers
According to the trial protocol (Figure 1), biologic material (plasma, serum, urine) for biomarker testing was collected at day 0 (n = 20), week 12 (n = 20), and week 52 (n = 18); 2 patients had been withdrawn from the trial, respectively. Samples were stored in aliquots at –80°C until the bioanalysis.
dd-cfDNA
For the quantification of dd-cfDNA fractions (dd-cfDNA[%]), we used biobanked ethylenediaminetetraacetic acid (EDTA)-plasma samples. Following a previously published protocol,17 cfDNA was extracted from ≥0.5 mL plasma using Qiagen’s QIAamp Circulating Nucleic Acid Kit (Qiagen, Venlo, The Netherlands), and a double AMPure clean-up step was included to remove contaminating cell-derived DNA. Levels of dd-cfDNA[%] were measured using the AlloSeq cfDNA assay (CareDx, Fremantle, WA, Australia), a next-generation targeted sequencing assay that uses biallelic single nucleotide polymorphisms to quantify dd-cfDNA without separate recipient or donor genotyping. Sequencing run results generated as FASTQ files were automatically analyzed using AlloSeq cfDNA Software (CareDx). As previously described, the test threshold for dd-cfDNA[%] was 0.15%.17 For 2 of the 58 tested plasma samples sequencing failed to yield a valid dd-cfDNA result (1 of these samples had been collected at day 0 [placebo in part A], a second at week 52 from another patient [placebo in part A]).
Chemokines
Serum and urine CXCL9 and CXCL10 were quantified using Human ProcartaPlex Simplex Immunoassays (Thermo Fisher Scientific, Waltham, MA, United States) according to a previously described protocol.31 Measurements were performed in duplicate on a Luminex 200 instrument (Luminex Corp., Austin, Tx, United States). Urinary chemokine levels were normalized to urinary creatinine (Urinary CXCL10/Cr; pg [biomarker]/mg [creatinine]).
HLA Antibody Detection
As detailed elsewhere,6,37 we used LABscreen Single Antigen assays (One Lambda, Thermo Fisher Scientific, Canoga Park, CA, United States) for HLA antibody detection. Serum samples were pretreated with EDTA (10 mM) to counteract complement interference.38 DSA (mean fluorescence intensity [MFI] threshold >1000) were analyzed according to the results of serological and low- or high-resolution donor/recipient HLA typing (HLA-A, -B, -Cw, -DR, -DQ, and DP).
Transplant Biopsies
According to the trial protocol (Figure 1), renal biopsies were performed at the time of screening, and after 11 and 51 wk, respectively.6,37 Formalin-fixed paraffin-embedded sections were used for standard-of-care histomorphology and C4d staining. Biopsies were evaluated locally (Medical University of Vienna, Charité Universitätsmedizin Berlin) in a blinded fashion according to the rules of the Banff 2017 scheme.39 A sum score of glomerulitis (g) and peritubular capillaritis (ptc) was used to describe the extent of microvascular inflammation (MVI). In addition, biopsy specimens were analyzed at the Alberta Transplant Applied Genomics Centre (ATAGC, University of Alberta, Edmonton, AB, Canada) using the Molecular Microscope Diagnostic System (MMDx).40,41 Molecular scores based on lesion-based classifiers related to rejection (AMR, all rejection, acute [acute kidney injury score], or chronic injury [atrophy/fibrosis score]) were generated using a reference set of 1529 biopsies.6 Biopsy-based responses to treatment at the end of the trial were defined as resolution of molecular (7 patients showed a molecular AMR score <0.2) or morphologic AMR activity (4 patients were diagnosed with C4d-negative chronic inactive AMR according to the Banff 2017 scheme; 3 of them had a molecular AMR score <0.2; 1 had a score of 0.23).
Statistics
Continuous data are presented as median and interquartile range (IQR), and categorical variables as absolute and relative frequencies. For group comparisons, we used Fisher’s exact, Mann-Whitney U, or Wilcoxon tests as appropriate. For nonparametric correlation analysis, we used the Spearman rank test. A 2-sided P value <0.05 was considered statistically significant. IBM SPSS Statistics version 24 (IBM Corporation, Armonk, NY) was used for statistical analysis.
RESULTS
Patient Characteristics and Disposition
Baseline characteristics, DSA data, standard-of-care histomorphology, and MMDx results are provided in Table 1. The study population consisted of 20 patients with late AMR diagnosed a median of 10.6 y after transplantation. Ten (50%) subjects were female, and 6 (30%) were living donor transplant recipients. The majority of the patients were on triple immunosuppressive therapy (90%) and tacrolimus-based immunosuppression (65%). Median levels of eGFR and protein/creatinine ratio were 39.3 mL/min per 1.73 m2 and 962 mg/g, respectively. All participants were (as predefined in the protocol) DSA-positive (75% with anti-DQ DSA) at the time of biopsy, with median MFI of 11 708. Index biopsies showed chronic active AMR in 90%, and positive C4d staining in 35% of the recipients. The median MVI (g+ptc) score was 3. Median molecular AMR, all rejection, acute kidney injury, and atrophy/fibrosis scores were 0.65, 0.69, 0.40, and 0.68, respectively (Table 1).
As shown in Figure 1, study participants were randomized to receive clazakizumab versus placebo for a period of 12 wk. As shown in Table 1, baseline variables were well balanced with some exceptions. Differences in the proportion of female recipients and levels of protein/creatinine ratio, however, were not significant. Thereafter, all patients were scheduled to receive clazakizumab until the end of the trial. Two subjects were withdrawn from the trial, 1 at the end of part A, and 1 after a single clazakizumab injection in part B. Due to adverse events (n = 10) or personal reasons (n = 1), 11 patients did not receive all 13 scheduled clazakizumab injections.6 None of the patients underwent indication biopsies outside of the protocol or received additional antirejection therapy. As described in a previous publication6 and illustrated in Figure S1 (SDC, http://links.lww.com/TXD/A473), treatment with clazakizumab led to an early decline in DSA-MFI with a significant difference to placebo at 3 mo. Between week 12 and week 52, in which all subjects received clazakizumab, we observed a further decrease in DSA along with a reduction in molecular AMR and all rejection classifier scores in second follow-up biopsies (during the initial controlled period, clazakizumab, however, did not consistently decrease molecular rejection activity). The extent of MVI and molecular scores of acute and chronic injury did not change significantly (Figures S1 and S2, SDC, http://links.lww.com/TXD/A473). There was also no change in cg score reflecting the extent of transplant glomerulopathy. Thus, whether the drug has a sustained effect on AMR activity is not entirely clear.
Biomarker Results
Biologic material collected at day 0, week 12, and week 52 (in total 58 plasma, serum, and urine samples) was retrospectively tested for dd-cfDNA[%] and CXCL10. Two samples obtained in 2 different patients (both in the placebo arm; 1 at day 0, the other at week 52) were not adequate for dd-cfDNA analysis. The results of biomarker testing in relation to treatment allocation are presented in Table 2.
TABLE 2.
Biomarker levels in relation to treatment allocation
| Biomarkersa,b | All patients(n = 20) | Results (n) | Clazakizumab(n = 10) | Results(n) | Placebo(n = 10) | Results(n) |
|---|---|---|---|---|---|---|
| Day 0 | ||||||
| dd-cfDNA[%]c | 1.50 (0.79–2.80) | 19 | 2.10 (0.76–3.35) | 10 | 1.10 (0.68–2.10) | 9 |
| Urinary CXCL10/Cr (pg/mg creatinine) | 81.0 (49.1–113.8) | 20 | 69.3 (52.6–103.8) | 10 | 96.6 (45.4–152.9) | 10 |
| Serum CXCL10 | 91.8 (69.1–158.0) | 20 | 94.2 (60.2–167.6) | 10 | 91.8 (75.2–144.0) | 10 |
| Week 12 | ||||||
| dd-cfDNAc | 1.15 (0.70–2.38) | 20 | 1.65 (0.91–2.78) | 10 | 0.97 (0.56–2.30) | 10 |
| Urinary CXCL10/Cr (pg/mg creatinine) | 58.0 (46.3–93.1) | 20 | 55.7 (41.0–91.4) | 10 | 60.2 (48.8–208.7) | 10 |
| Serum CXCL10 | 98.0 (50.3–126.0) | 20 | 98.0 (77.1–305.3) | 10 | 80.7 (47.4–125.4) | 10 |
| Week 52 d | ||||||
| dd-cfDNAc | 1.0 (0.61–1.70) | 17 | 1.35 (0.68–2.85) | 9 | 0.97 (0.60–1.35) | 8 |
| Urinary CXCL10/Cr (pg/mg creatinine) | 67.4 (41.5–132) | 18 | 71.0 (30.5–164.7) | 9 | 65.4 (50.9–132) | 9 |
| Serum CXCL10 | 119.0 (58.6–135.4) | 18 | 102.3 (56.1–136.2) | 9 | 127.1 (58.4–141.9) | 9 |
| Percent baseline—week 12 | ||||||
| dd-cfDNAc | 82.1 (59.1–120.3) | 19 | 100.2 (70.0–123.5) | 10 | 77.3 (56.8–122.1) | 9 |
| Urinary CXCL10/Cr (pg/mg creatinine) | 92.4 (58.6–143.8) | 20 | 103.2 (48.1–147.9) | 10 | 90.5 (58.6–144.3) | 10 |
| Serum CXCL10 | 126.4 (65.4–147) | 20 | 138.4 (120.9–264.6) | 10 | 85.4 (54.8–144) | 10 |
| Percent baseline—week 52 d | ||||||
| dd-cfDNAc | 85.8 (51.1–129.5) | 16 | 84.6 (69.0–135.3) | 8 | 104 (35.4–130.0) | 8 |
| Urinary CXCL10/Cr (pg/mg creatinine) | 80.9 (58.4–149.5) | 18 | 90.1 (38.0–190.3) | 9 | 80.9 (58.4–123.3) | 9 |
| Serum CXCL10 | 124.6 (71.0–163.2) | 18 | 115.0 (84.3–178.5) | 9 | 129.4 (45.4–163.2) | 9 |
aResults are provided as median and interquartile range.
bIntergroup differences with respect to biomarker results (clazakizumab vs placebo; Mann-Whitney U test) and changes from week 12 to week 52 (overall study cohort; paired analysis applying Wilcoxon test) were nonsignificant (P > 0.05).
cFor 2 of the 58 tested plasma samples sequencing did not yield valid dd-cfDNA results (1 of these samples had been collected at day 0, a second from another patient at week 52 [both patients allocated to receive placebo in part A]).
dTwo patients were withdrawn from the study and therefore no biomarker results were available for week 52.
Cr, creatinine; CXCL, C-X-C motif chemokine ligand; dd-cfDNA, donor-derived cell-free DNA.
Fractional dd-cfDNA
Among the 19 recipients with valid dd-cfDNA results at baseline (in 1 patient allocated to placebo, sequencing did not yield valid results), dd-cfDNA[%] was 1.5% (median; IQR: 0.79%–2.8%). Twelve of 19 recipients (63.2%) had fractions ≥1.0%. Although median dd-cfDNA[%] at baseline was numerically higher in patients assigned to the clazakizumab arm (2.1% [0.76% versus 3.35%] versus 1.1% [0.7%–2.1%] in placebo patients), the proportions of recipients with dd-cfDNA[%] ≥1.0% were similar between groups (7 of 10, 70% versus 5 of 9, 55.6%). Baseline dd-cfDNA[%] did not correlate with DSA levels, a g+ptc sum score reflecting the extent of MVI or C4d staining (Figure S3, SDC, http://links.lww.com/TXD/A473), transplant glomerulopathy (cg score), or clinical variables, such as recipient sex, eGFR, or protein excretion. There were also no correlations with molecular AMR and all rejection scores, or classifiers of acute or chronic kidney injury, respectively (Figure S4, SDC, http://links.lww.com/TXD/A473).
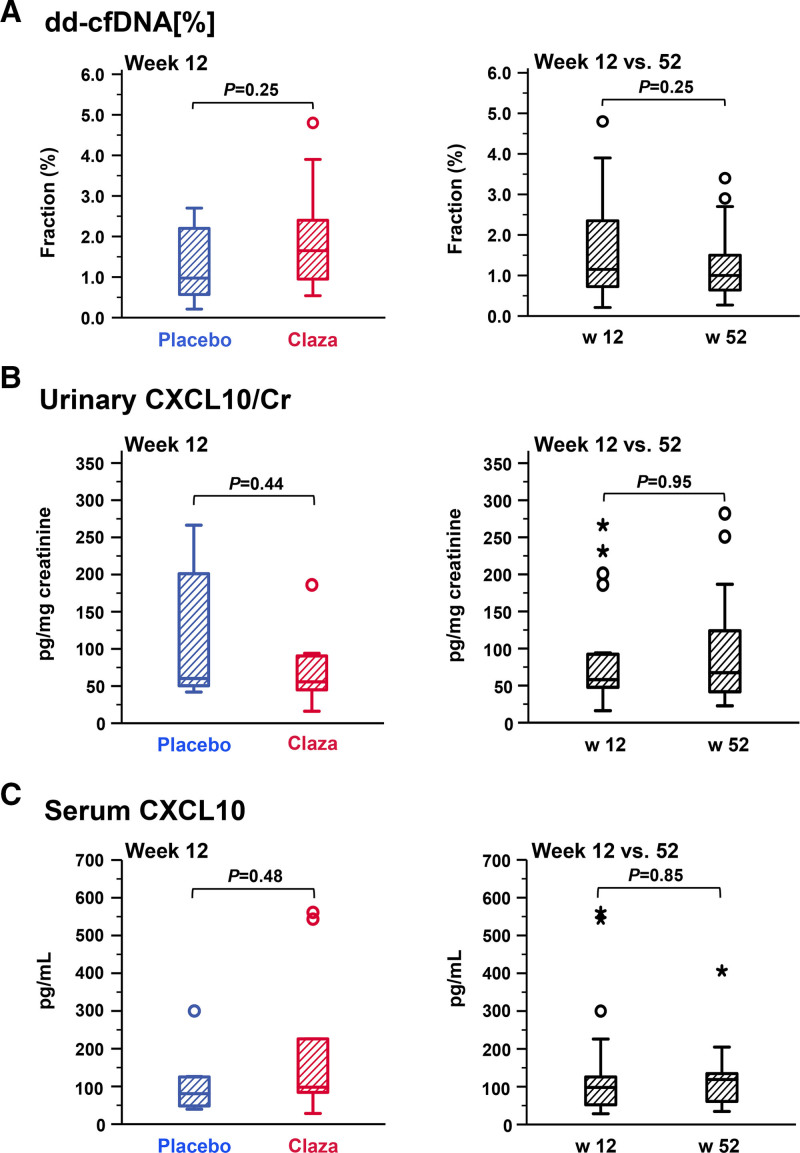
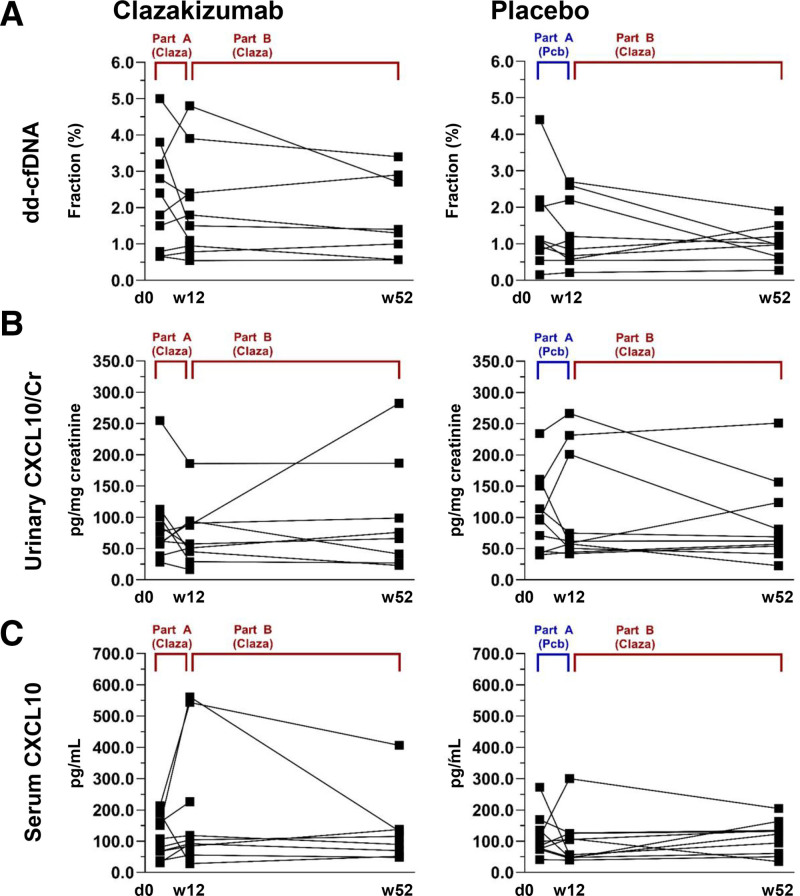
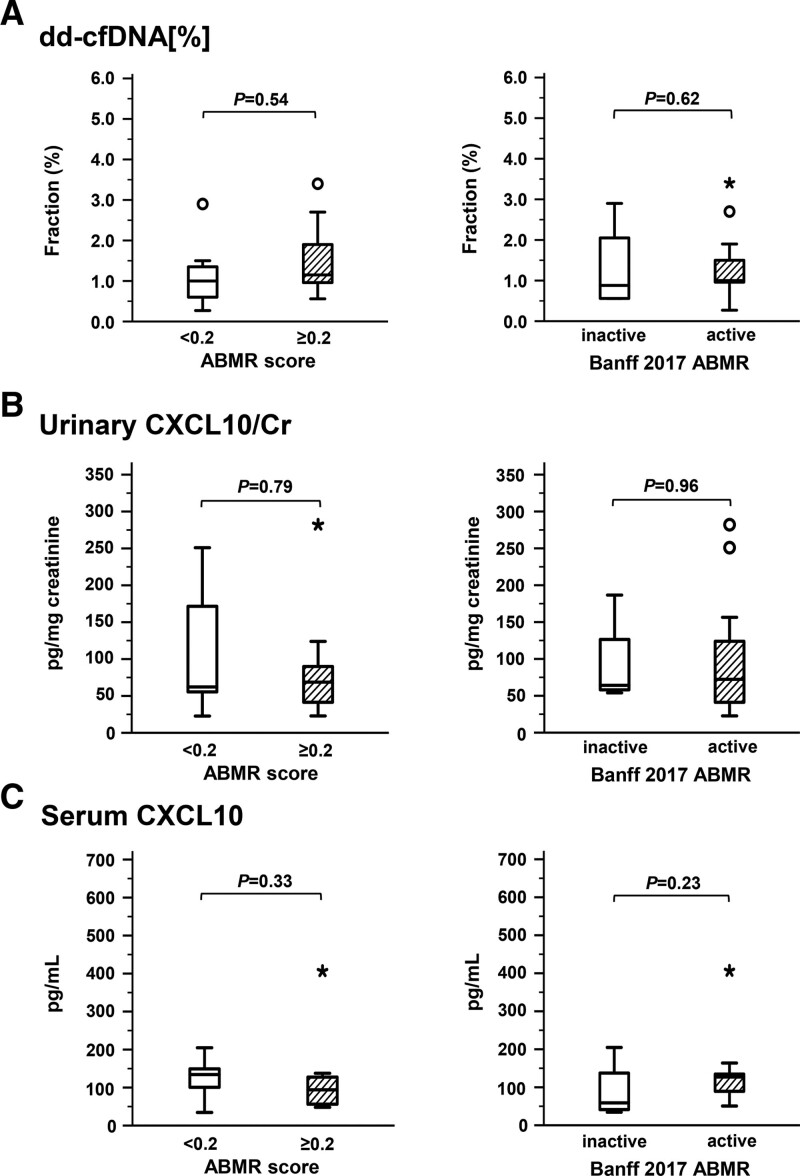
At 12 wk, at the end of part A, we found no significant difference between clazakizumab and placebo groups with respect to dd-cfDNA[%] (1.65% [0.91%–2.78%] versus 1.0% [0.6%–2.3%]; P = 0.25) (Table 2; Figure 2). When evaluating the study cohort over part B (from week 12 to week 52), we found no significant change of dd-cfDNA[%] (median dd-cfDNA[%] from 1.15% [0.70%–2.38%] to 1.0% [0.61%–1.70%], P = 0.25) (Table 2; Figure 2). Individual courses of dd-cfDNA[%] in relation to treatment allocation in parts A and B are illustrated in Figure 3. At the end of the trial (week 52), 10 of 17 patients with dd-cfDNA results still had fractions ≥1% (2 patients had been withdrawn from the trial; in a third patient allocated to placebo in part A, stored plasma did not yield valid test results). Patients with a molecular response to treatment at the end of the study (AMR score <0.2: n = 7) did not differ from patients with persistent activity (AMR score ≥0.2) with respect to dd-cfDNA[%] (Figure 4). Similar results were obtained for the 4 patients who showed resolution of morphologic AMR activity (3 of them showed a molecular AMR score <0.2) (Figure 4).
FIGURE 2.
Biomarkers in relation to clazakizumab treatment. Fractions of dd-cfDNA[%] (A) and concentrations of CXCL10 in urine (B) and serum (C) are shown after 12 wk (end of part A; clazakizumab [Claza] vs placebo) and from week 12 to week 52 in the open-label extension (part B; all patients on clazakizumab), respectively. Box plots indicate the median, interquartile range, and the minimum and maximum of the measures (outliers are indicated by circles and extreme outliers by asterisks). The unpaired Mann-Whitney U test was used for comparisons between study arms and the paired Wilcoxon test to test for differences over time in the overall cohort. Cr, creatinine; CXCL, C-X-C motif chemokine ligand; dd-cfDNA, donor-derived cell-free DNA.
FIGURE 3.
Individual course of biomarker levels in relation to treatment allocation. Biomarkers included (A) fractions of % and concentrations of CXCL10 in urine (B) and serum (C). Two patients in the Claza arm were withdrawn from the trial, and for week 52 no biomarker results are available. Two samples did not yield valid dd-cfDNA results (1 collected at day 0, the other at week 52). Claza, clazakizumab; Cr, creatinine; CXCL, C-X-C motif chemokine ligand; dd-cfDNA, donor-derived cell-free DNA; Pcb, placebo.
FIGURE 4.
Biomarkers in relation to persistent molecular or morphologic AMR persistence. Fractions of dd-cfDNA[%] (A) and concentrations of CXCL10 in urine (B) and serum (C) are shown at the end of the trial, in relation to molecular (AMR score <0.2 [n = 7] vs AMR score ≥0.2 [n = 11]) or morphologic AMR activity according to the Banff 2017 scheme (no activity: n = 4; activity: n = 14). Box plots indicate the median, interquartile range, and the minimum and maximum of the measures (outliers are indicated by circles and extreme outliers by asterisks). For group comparisons, the Mann-Whitney U test was used. AMR, antibody-mediated rejection; Cr, creatinine; CXCL, C-X-C motif chemokine ligand; dd-cfDNA, donor-derived cell-free DNA.
Urinary CXCL10/Cr
In the overall cohort, baseline levels of urinary CXCL10/Cr were 81.0 (median; IQR: 49.1–113.8) pg/mg creatinine, respectively. Urine CXCL10/Cr at baseline was found to correlate with the MFI of the immunodominant DSA (ρ = 0.68, P = 0.001), but not with C4d staining, MVI‚ or molecular rejection scores (Figures S3 and S4, SDC, http://links.lww.com/TXD/A473). In addition, there were no correlations with transplant glomerulopathy (cg score), clinical variables such as recipient sex, eGFR and protein excretion, or with dd-cfDNA[%] (data not shown).
At the end of part A of the study (week 12), we found no significant difference between clazakizumab and placebo in terms of CXCL10/Cr (55.7 [median; IQR: 41.0–91.4] versus 60.2 [48.8–208.7.0] pg/mg creatinine; P = 0.44) (Table 2; Figure 2). Overall, there was no change in median CXCL10/Cr between week 12 and week 52 (from 57.6 [46.3–93.1] to 67.4 [41.5–132.0] pg/mg creatinine; P = 0.95) (Table 2; Figure 2). Figure 3 illustrates individual courses of CXCL10 levels. At the end of the trial, molecular responders (AMR score <0.2) or recipients with morphologic resolution of AMR activity did not differ from recipients with persistent rejection features with respect to CXCL10/Cr (Figure 4). In addition, using logistic regression analysis, we found no significant effect of combined dd-cfDNA and urinary CXCL10 in relation to AMR activity (Table S1, SDC, http://links.lww.com/TXD/A473).
Serum CXCL10
Unlike urinary CXCL10, serum levels of this chemokine were not associated with any serologic, morphologic‚ or molecular parameter at baseline (Figures S3 and S4, SDC, http://links.lww.com/TXD/A473). There was also no correlation with cg or clinical baseline variables (not shown). Furthermore, there was no difference at week 12, at the end of part A (clazakizumab versus placebo: 98.0 [median; IQR: 77.1–305.3] versus 80.7 [47.4–125.4] pg/mL; P = 0.48), and levels did not significantly change from week 12 to the end of the study (from 98.0 [50.3–126.0] pg/mL to 119.0 [58.6–135.4] pg/mL; P = 0.85) (Table 2; Figure 2). Finally, subjects with molecular and morphologic resolution of AMR activity did not differ from those with persistent rejection activity (Figure 4). Logistic regression analysis with interaction terms did not show a significant effect of combined dd-cfDNA and serum CXCL10 in relation to AMR activity at the end of the trial (Table S1, SDC, http://links.lww.com/TXD/A473).
Events Unrelated to Immunologic Rejection
A total of 10 episodes of lower urinary tract infection (in 5 subjects) were reported during the study period, 3 in part A and 7 in part B (data not shown). In addition, 1 patient developed pyelonephritis. Only 1 patient (placebo; diagnosis 1 wk after entry into part B of the trial) developed urinary tract infection at about the time of a study visit. This patient showed urine and serum CXCL10 levels of 71.1 pg/mg creatinine and 75.5 pg/mL at baseline, respectively. There were no apparent increases in urinary levels at week 12 (61.9 pg/mL creatinine), but, at the same time, we observed an approximately 40% increase of serum levels (108.2 pg/mL). At week 52, levels of 62.1 pg/mg creatinine and 34.5 pg/mL were recorded. Fractions of dd-cfDNA were stable over time (0.54%, 0.54%, and 0.56% at 0, 12, and at 52 wk). All other episodes of urinary tract infection and the case of pyelonephritis occurred beyond plus/minus 4 wk of 12- or 52-wk study visits. None of the patients developed polyomavirus infection. There was 1 patient who developed diarrhea-associated acute renal failure 13 d after the first visit, which resolved within a week and was therefore considered irrelevant to biomarker results obtained 2 mo later (data not shown).
DISCUSSION
The main finding of our present study was that treatment of late AMR with the anti-IL-6 monoclonal antibody did not significantly modulate the levels of 2 distinct biomarkers, namely plasma dd-cfDNA[%] as an estimate of allograft injury and CXCL10 in serum or urine reflecting parenchymal inflammation. There was no difference between clazakizumab and placebo at week 12, and, under clazakizumab, no decrease in median urinary CXCL10 or dd-cfDNA[%] until the end of the trial. This was in some contrast to the decline in DSA-MFI, which was observed as early as 3 mo after the start of treatment, as well as changes in a molecular classifier of AMR, which became negative in 7 recipients after 9–12 mo of treatment.
To date, there is no noninvasive rejection biomarker that has proven useful to timely detect treatment responses and thus guide the intensity of immunosuppressive treatment.11 However, there are several observational studies that have examined the course of CXCL10 or dd-cfDNA[%] after rejection treatment. For example, a recently published observational study has shown a decline in levels of urinary CXCL10 within 1–2 mo in 31 recipients undergoing treatment for biopsy-confirmed rejection.34 However, only a few cases of AMR were included, and the authors noted that the number of cases was not sufficient to definitively report on urinary CXCL10/Cr changes in this specific context. Significant changes after rejection treatment were also found for dd-cfDNA[%], although in a recent cohort study changes after treatment of AMR were found to be less pronounced than those in TCMR.23 In 11 pediatric patients with AMR and 5 with mixed rejection, dd-cfDNA[%] after treatment with rituximab/IVIG did not reach baseline levels and fractions remained in a range above 1%. Persistent dd-cfDNA release reflected ongoing rejection processes as also suggested by the results of follow-up biopsies.23 In a retrospective analysis of the DART study, dd-cfDNA[%] showed a numerical decrease in 9 AMR patients receiving a variety of different treatments, but, in contrast to TCMR, levels again remained >1%. In some cases, fractions even continued to rise.22 Whether this was related to ongoing rejection, remained unclear, as no posttreatment biopsies were available. Finally, Gupta et al24 reported 26 kidney transplant recipients with acute rejection (mostly TCMR) or chronic active AMR who were followed via dd-cfDNA[%] monitoring and a posttreatment biopsy approximately 6–8 wk after completion of rejection therapy. Patients with resolution of rejection on MMDx as well as histopathology (n = 8; 50% TCMR) showed a decline in levels (from 0.94% to 0.20%; P = 0.015), whereas 18 nonresponders, most of them (89%) diagnosed with AMR, did not.24
It can be argued that the differences in the behavior of TCMR and AMR observed in the studies mentioned earlier could be due to potentially inadequate treatment of AMR unable to fully resolve ongoing inflammation and injury. It is well known that AMR treatment, especially in late cases, is a great challenge and many cases do not respond to currently used therapeutic approaches.3,4 However, as previously described in detail,6 molecular rejection activity (in some contrast to morphologic MVI) was significantly reduced in our trial, and, after 9–12 mo of IL-6 blockade, 7 recipients presented with a negative AMR score on MMDx. At the same time, DSA-MFI, were in median reduced to 79%. These results were suggestive of a therapeutic effect, at least in some of the patients. Unexpectedly, however, molecular resolution of AMR was not associated with a significant decline in dd-cfDNA[%] or CXCL10. Even for the 4 patients who showed resolution of morphologic AMR activity (3 patients also showed a negative molecular AMR score) no significant effect was observed. When interpreting these results, the limitations of a small sample size must be taken into account, but it may also be of interest that gene expression patterns associated with acute or chronic injury did not change under 9–12 mo of clazakizumab treatment. Persistent acute graft injury, which may not be visible in conventional histomorphology, may at least partly explain the behavior of dd-cfDNA, which from a physiological point of view can be regarded as an indicator of tissue injury. The ongoing release of dd-cfDNA and persistently elevated chemokine levels may suggest incomplete responsiveness to treatment, as also suggested by a continuing decline in eGFR (even though our preliminary data suggested some amelioration of its course) and persistence of morphological MVI in the majority of the patients (4 recipients showed resolution of morphologic AMR activity). One may also argue that treatment was too short to achieve visible changes of all efficacy endpoints and in parallel a decrease in tested biomarkers. However, at the end, our data clearly indicate that apparently in some contrast to DSA monitoring and molecular assessment of follow-up biopsies, they may not be helpful in detecting subtle changes in AMR activity in the first year after initiation of treatment.
At baseline, chemokine levels and dd-cfDNA[%] showed a considerable level of variation, with a significant proportion of patients (n = 7 of 19) showing dd-cfDNA[%] below 1%, but this seemed not to correlate with features reflecting different phenotypic presentations of AMR. Notably, the 2 biomarkers did not correlate with each other, but in this context, a different pathophysiologic background can be assumed. Allograft immune activation indicated by increased chemokine levels may not necessarily be associated with graft damage-induced dd-cfDNA release, and vice versa. Moreover, there was no correlation between biomarker levels and biopsy features indicating rejection activity, such as MVI or scores of molecular rejection classifiers. Our results are in some contrast with the results in the Trifecta study, wherein in a cohort of 289 kidney transplant recipients dd-cfDNA[%] showed a high correlation with different molecular scores from rejection-associated transcript sets and classifiers, including a molecular AMR score (and less tightly with other classifiers including those reflecting acute kidney injury).18 Discrepancies to this large study, which analyzed a heterogeneous case mix (30% of biopsies with AMR), however, could, apart from a limited sample size, be due to a preselection of patients with AMR in our trial. One may argue that associations with the molecular AMR classifier are nonlinear, with increased levels even in residual mild AMR.
Aside from immunologic rejection, there can be various triggers for the release of dd-cfDNA and chemokines, such as polyomavirus nephropathy, urinary tract infection, and acute renal failure.20,35,42 None of the study participants had BK or JC viremia, and only 1 developed moderate acute renal failure due to diarrhea, but this event was not associated with any of the time points, wherein biomarker testing was performed. During the trial, 10 episodes of lower urinary tract infection were recorded, and, in addition, 1 had pyelonephritis. Only 1 subject was diagnosed with urinary tract infection at about the time of biomarker assessment (diagnosis 1 wk after the 12-wk visit). In this patient, we observed an increase of CXCL10 in serum (but not of urinary chemokine levels or plasma dd-cfDNA), which we cannot exclude as being related to infection.
Although the prospective randomized controlled design of our study, which included predefined intervals of serial biomarker monitoring in connection with systematic follow-up biopsies, is a strength, it has several inherent limitations that need to be considered. A major limitation is the small sample size which, given the considerable variation in biomarker levels, may have made it more difficult to detect subtle changes in disease activity and subsequent graft injury. Despite stratified randomization, some baseline variables, such as recipient sex or protein/creatinine ratio, were not well balanced, although differences were not significant. However, this may be of limited relevance as there was no association of these parameters with biomarker levels. Another limitation may be that the trial design included only a short 3-mo phase of a double-blind randomized design. The trial then entered an open-label phase and all patients, including those randomized to placebo, received clazakizumab. This precluded a head-to-head comparison of clazakizumab versus placebo beyond 3 mo. In fact, the only efficacy endpoint that showed a significant intergroup difference after the first 12 wk was the DSA level, whereas no such effect was visible for different morphologic and molecular biopsy features. Given the latter result, our failure to detect early differences between groups regarding the course of biomarkers was not unexpected. Although the validity of 1-y efficacy results may be limited due to the open-label design of the extension phase, significant therapeutic efficacy can be inferred from our study results, particularly given the observed changes of gene expression patterns detected in 12-mo follow-up biopsies and the further reduction of DSA levels. These effects, however, were not complete and one may speculate that treatment was too short and that, as now studied in a large phase 3 trial, an extension of treatment would have significantly increased the observed effects on secondary outcomes. On the other hand, one would expect an ideal biomarker to be able to detect or predict very subtle treatment effects at an early stage, which our study failed to show. When discussing the results on efficacy endpoints and biomarker levels, it is important to consider that 11 of the included patients did not receive all 13 scheduled doses of clazakizumab, primarily due to adverse events. It can be argued that this may have influenced study outcomes to some extent. Finally, a possible limitation could be that data on absolute levels of dd-cfDNA, suggested by some authors to add diagnostic accuracy, were not available for our study.43,44
In conclusion, the results of our study suggest that IL-6 blockade in late AMR may not trigger a significant decline in dd-cfDNA[%] and CXCL10 levels, at least over a 9–12 mo treatment period. These 2 noninvasive biomarkers may not be sensitive enough to timely detect subtle treatment effects of IL-6 neutralization in patients with late AMR.
ACKNOWLEDGMENTS
We wish to thank Sabine Schranz, Sahra Ely, and Dr. Anke Steiner for their valuable assistance.
Supplementary Material
Footnotes
Clinical trial notation: www.clinicaltrials.org, NCT02502903
K.A.M., P.F.H., K.B., B.J., and G.A.B. participated in the research design, performance of the research, data analysis, interpretation of results, and writing of the article. K.D., S.H., J.M., and F.F. participated in performance of research, data analysis, and writing of the article. S.C. and T.V. participated in performance of research and data analysis.
T.V. and S.C. are employed by CareDx Inc., Brisbane, San Francisco, CA. G.A.B. is member of the steering committee for an ongoing pivotal phase 3 trial evaluating clazakizumab in chronic active antibody-mediated rejection (ClinicalTrials.gov number, NCT03744910; sponsored by CSL Behring). The other authors declare no conflicts of interest.
The study was funded by investigator-initiated unrestricted grants from Vitaeris Inc., Vancouver, Canada (a subsidiary of CSL Behring, King of Prussia, PA) (to G.A. Böhmig and B. Jilma) and CareDx Inc., Brisbane, South San Francisco, CA (to K.A. Mayer).
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantationdirect.com).
REFERENCES
- 1.Loupy A, Lefaucheur C. Antibody-mediated rejection of solid-organ allografts. N Engl J Med. 2018;379:1150–1160. [DOI] [PubMed] [Google Scholar]
- 2.Mayrdorfer M, Liefeldt L, Wu K, et al. Exploring the complexity of death-censored kidney allograft failure. J Am Soc Nephrol. 2021;32:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böhmig GA, Eskandary F, Doberer K, et al. The therapeutic challenge of late antibody-mediated kidney allograft rejection. Transpl Int. 2019;32:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schinstock CA, Mannon RB, Budde K, et al. Recommended treatment for antibody-mediated rejection after kidney transplantation: The 2019 Expert Consensus from the Transplantion Society Working Group. Transplantation. 2020;104:911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J, Aubert O, Vo A, et al. Assessment of tocilizumab (anti-interleukin-6 receptor monoclonal) as a potential treatment for chronic antibody-mediated rejection and transplant glomerulopathy in HLA-sensitized renal allograft recipients. Am J Transplant. 2017;17:2381–2389. [DOI] [PubMed] [Google Scholar]
- 6.Doberer K, Duerr M, Halloran PF, et al. A randomized clinical trial of anti-IL-6 antibody clazakizumab in late antibody-mediated kidney transplant rejection. J Am Soc Nephrol. 2021;32:708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan SC, Ammerman N, Choi J, et al. Evaluation of clazakizumab (anti-Interleukin-6) in patients with treatment-resistant chronic active antibody-mediated rejection of kidney allografts. Kidney Int Rep. 2022;7:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer KA, Budde K, Jilma B, et al. Emerging drugs for antibody-mediated rejection after kidney transplantation: a focus on phase II & III trials. Expert Opin Emerg Drugs. 2022;27:151–167. [DOI] [PubMed] [Google Scholar]
- 9.Eskandary F, Regele H, Baumann L, et al. A randomized trial of bortezomib in late antibody-mediated rejection (BORTEJECT). J Am Soc Nephrol. 2018;29:591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreso F, Crespo M, Ruiz JC, et al. Treatment of chronic antibody mediated rejection with intravenous immunoglobulins and rituximab: a multicenter, prospective, randomized, double-blind clinical trial. Am J Transplant. 2018;18:927–935. [DOI] [PubMed] [Google Scholar]
- 11.Brunet M, Shipkova M, van Gelder T, et al. Barcelona consensus on biomarker-based immunosuppressive drugs management in solid organ transplantation. Ther Drug Monit. 2016;38(Suppl 1):S1–S20. [DOI] [PubMed] [Google Scholar]
- 12.Naesens M, Anglicheau D. Precision transplant medicine: biomarkers to the rescue. J Am Soc Nephrol. 2018;29:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oellerich M, Sherwood K, Keown P, et al. Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury. Nat Rev Nephrol. 2021;17:591–603. [DOI] [PubMed] [Google Scholar]
- 14.Kant S, Brennan DC. Donor derived cell free DNA in kidney transplantation: the Circa 2020-2021 update. Transpl Int. 2022;35:10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018;4:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer KA, Doberer K, Tillgren A, et al. Diagnostic value of donor-derived cell-free DNA to predict antibody-mediated rejection in donor-specific antibody-positive renal allograft recipients. Transpl Int. 2021;34:1689–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The Trifecta Study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: results from a prospective observational study. Am J Transplant. 2019;19:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kant S, Bromberg J, Haas M, et al. Donor-derived cell-free DNA and the prediction of BK virus-associated nephropathy. Transplant Direct. 2020;6:e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu L, Gupta G, Pai A, et al. Clinical outcomes from the Assessing Donor-derived Cell-free DNA Monitoring Insights of Kidney Allo Grafts with Longitudinal Surveillance (ADMIRAL) study. Kidney Int. 2022;101:793–803. [DOI] [PubMed] [Google Scholar]
- 22.Wolf-Doty TK, Mannon RB, Poggio ED, et al. Dynamic response of donor-derived cell-free DNA following treatment of acute rejection in kidney allografts. Kidney360. 2021;2:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steggerda JA, Pizzo H, Garrison J, et al. Use of a donor-derived cell-free DNA assay to monitor treatment response in pediatric renal transplant recipients with allograft rejection. Pediatr Transplant. 2022;26:e14258. [DOI] [PubMed] [Google Scholar]
- 24.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 25.Panzer U, Steinmetz OM, Reinking RR, et al. Compartment-specific expression and function of the chemokine IP-10/CXCL10 in a model of renal endothelial microvascular injury. J Am Soc Nephrol. 2006;17:454–464. [DOI] [PubMed] [Google Scholar]
- 26.Schaub S, Nickerson P, Rush D, et al. Urinary CXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am J Transplant. 2009;9:1347–1353. [DOI] [PubMed] [Google Scholar]
- 27.Jackson JA, Kim EJ, Begley B, et al. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am J Transplant. 2011;11:2228–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabant M, Amrouche L, Lebreton X, et al. Urinary C-X-C motif chemokine 10 independently improves the noninvasive diagnosis of antibody-mediated kidney allograft rejection. J Am Soc Nephrol. 2015;26:2840–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho J, Schaub S, Wiebe C, et al. Urinary CXCL10 chemokine is associated with alloimmune and virus compartment-specific renal allograft inflammation. Transplantation. 2018;102:521–529. [DOI] [PubMed] [Google Scholar]
- 30.Millan O, Budde K, Sommerer C, et al. Urinary miR-155-5p and CXCL10 as prognostic and predictive biomarkers of rejection, graft outcome and treatment response in kidney transplantation. Br J Clin Pharmacol. 2017;83:2636–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mühlbacher J, Doberer K, Kozakowski N, et al. Non-invasive chemokine detection: improved prediction of antibody-mediated rejection in donor-specific antibody-positive renal allograft recipients. Front Med (Lausanne). 2020;7:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weseslindtner L, Hedman L, Wang Y, et al. Longitudinal assessment of the CXCL10 blood and urine concentration in kidney transplant recipients with BK polyomavirus replication-a retrospective study. Transpl Int. 2020;33:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millan O, Rovira J, Guirado L, et al. Advantages of plasmatic CXCL-10 as a prognostic and diagnostic biomarker for the risk of rejection and subclinical rejection in kidney transplantation. Clin Immunol. 2021;229:108792. [DOI] [PubMed] [Google Scholar]
- 34.Blydt-Hansen TD, Sharma A, Gibson IW, et al. Validity and utility of urinary CXCL10/Cr immune monitoring in pediatric kidney transplant recipients. Am J Transplant. 2021;21:1545–1555. [DOI] [PubMed] [Google Scholar]
- 35.Tinel C, Devresse A, Vermorel A, et al. Development and validation of an optimized integrative model using urinary chemokines for noninvasive diagnosis of acute allograft rejection. Am J Transplant. 2020;20:3462–3476. [DOI] [PubMed] [Google Scholar]
- 36.Ho J, Sharma A, Kroeker K, et al. Multicentre randomised controlled trial protocol of urine CXCL10 monitoring strategy in kidney transplant recipients. BMJ Open. 2019;9:e024908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eskandary F, Dürr M, Budde K, et al. Clazakizumab in late antibody-mediated rejection: study protocol of a randomized controlled pilot trial. Trials. 2019;20:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwaiger E, Wahrmann M, Bond G, et al. Complement component C3 activation: the leading cause of the prozone phenomenon affecting HLA antibody detection on single-antigen beads. Transplantation. 2014;97:1279–1285. [DOI] [PubMed] [Google Scholar]
- 39.Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant. 2017;17:2851–2862. [DOI] [PubMed] [Google Scholar]
- 41.Reeve J, Böhmig GA, Eskandary F, et al. Precision molecular phenotyping of kidney transplant biopsies using archetypal analysis. JCI Insight. 2017;2:e94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer KA, Omic H, Weseslindtner L, et al. Levels of donor-derived cell-free DNA and chemokines in BK polyomavirus-associated nephropathy. Clin Transplant. 2022:e14785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halloran PF, Reeve J, Madill-Thomsen KS, et al. Combining donor-derived cell-free DNA fraction and quantity to detect kidney transplant rejection using molecular diagnoses and histology as confirmation. Transplantation. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osmanodja B, Akifova A, Budde K, et al. Absolute or relative quantification of donor-derived cell-free DNA in kidney transplant recipients: case series. Transplant Direct. 2021;7:e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.