Abstract
Second-generation androgen receptor (AR) inhibitors such as enzalutamide are the first-line treatments for castration-resistant prostate cancer (CRPC). Resistance to enzalutamide will greatly increase the difficulty of prostate cancer treatment and reduce the survival time of patients. However, drug-resistant cancer cells seem to be more sensitive to ferroptosis. Therefore, we constructed a biomimetic tumor-targeting magnetic lipid nanoparticle (t-ML) to codeliver dihomo-γ-linolenic acid (DGLA) and 2,4-dienoyl-CoA reductase 1 (DECR1) siRNA (t-ML@DGLA/siDECR1). DGLA is a dietary polyunsaturated fatty acid (PUFA), while DECR1 is overexpressed in prostate cancer and can inhibit the generation of PUFAs. The combination of DGLA and siDECR1 can efficiently induce ferroptosis by peroxidation of PUFAs, which has been verified both in vitro and in vivo. With the assistance of an external magnet, t-ML showed good tumor targeting ability and biocompatibility, and t-ML@DGLA/siDECR1 exhibited significant ferroptosis induction and tumor suppression capabilities. Moreover, in a nude mouse model of prostate cancer fed on a high-fat diet (HFD), there was no distant organ metastasis when the tumor-bearing mice were treated with t-ML@DGLA/siDECR1 and an external magnet, with upregulated PUFAs and downregulated monounsaturated fatty acids (MUFAs). Hence, this study has broadened the way of treating drug-resistant prostate cancer based on ferroptosis induction.
Keywords: Enzalutamide-resistant prostate cancer, Polyunsaturated fatty acids, Ferroptosis, Dihomo-γ-linolenic acid, DECR1 siRNA1
Graphical abstract
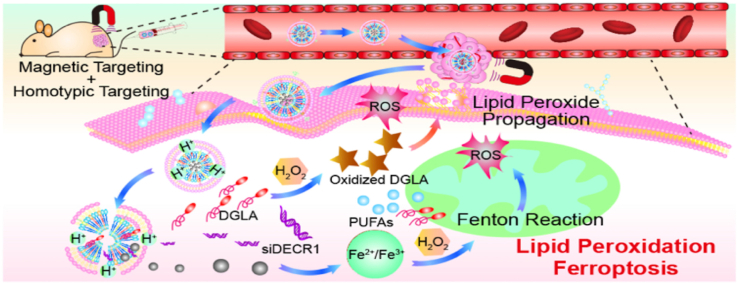
The biomimetic nanocomposite t-ML@DGLA/siDECR1 with an external magnetic field can accurately target the prostate cancer microenvironment accurately based on homotypic-magnetic dual-targeting abilities, thereby regulating lipid metabolism and inducing ferroptosis in enzalutamide-resistant prostate cancer.
1. Introduction
Prostate cancer is the second most prevalent malignant tumor among men worldwide [1]. In the USA, the 5-year relative survival rate for all stages of prostate cancer is 98% due to the development of treatments and diagnosis methods. However, approximately 6–7% of patients will metastasize distantly and progress to metastatic castration-resistant prostate cancer (CRPC), with a sharp decline in 5-year relative survival rate (from 98% to 30%) [2]. To date, first-line drugs for metastatic CRPC, such as second-generation androgen receptor (AR) inhibitors enzalutamide (Enz) and abiraterone acetate have shown considerable effects. However, drug resistance and low responsiveness have limited the application of these drugs [3].
Recently, ferroptosis, a newly witnessed form of programmed cell death, is characterized by the accumulation of iron-dependent lethal lipid peroxides [4]. Fe2+/Fe3+ can react with peroxides and generate reactive oxygen species (ROS), which was called Fenton reaction. The accumulation of ROS leads to ferroptosis in cells. Cancer cells usually have higher iron demands and susceptibility to ferroptosis, and AR inhibitor-resistant prostate cancer cells are even more susceptible to ferroptosis [5]. AR inhibitors can reprogram the metabolic state of prostate cancer, leading to an accumulation of lipids to supply bioenergetic processes and cell proliferation. These increased lipids, especially polyunsaturated fatty acids (PUFAs), can enhance cell membrane fluidity and lipid peroxidation. Thus, AR inhibitor-resistant CRPC cells are highly sensitive to ferroptosis, suggesting that ferroptosis may have significant advantages for AR inhibitor-resistant CRPC therapy [6]. However, ferroptosis is a double-edged sword. Excessive ferroptosis may also lead to damage to body functions, such as exacerbating inflammatory bowel disease and cardiovascular disease [7,8]. Therefore, we need to precisely induce ferroptosis in CRPC tumor sites to avoid the toxic side effects of ferroptosis. It is reported that ingestion of dihomo-γ-linolenic acid (DGLA), a dietary PUFA, can induce ferroptosis in cancer cells [9]. In addition, the mitochondrial enzyme 2,4-dienoyl-CoA reductase 1 (DECR1) is overexpressed in CRPC,which is involved in the degradation of PUFAs. Knockdown or inhibition of DECR1 can increase PUFAs and decrease monounsaturated fatty acids (MUFAs) in prostate cancer cells and induce ferroptosis in prostate cancer by inhibiting GPX4, thereby suppressing the growth of cancer [10].
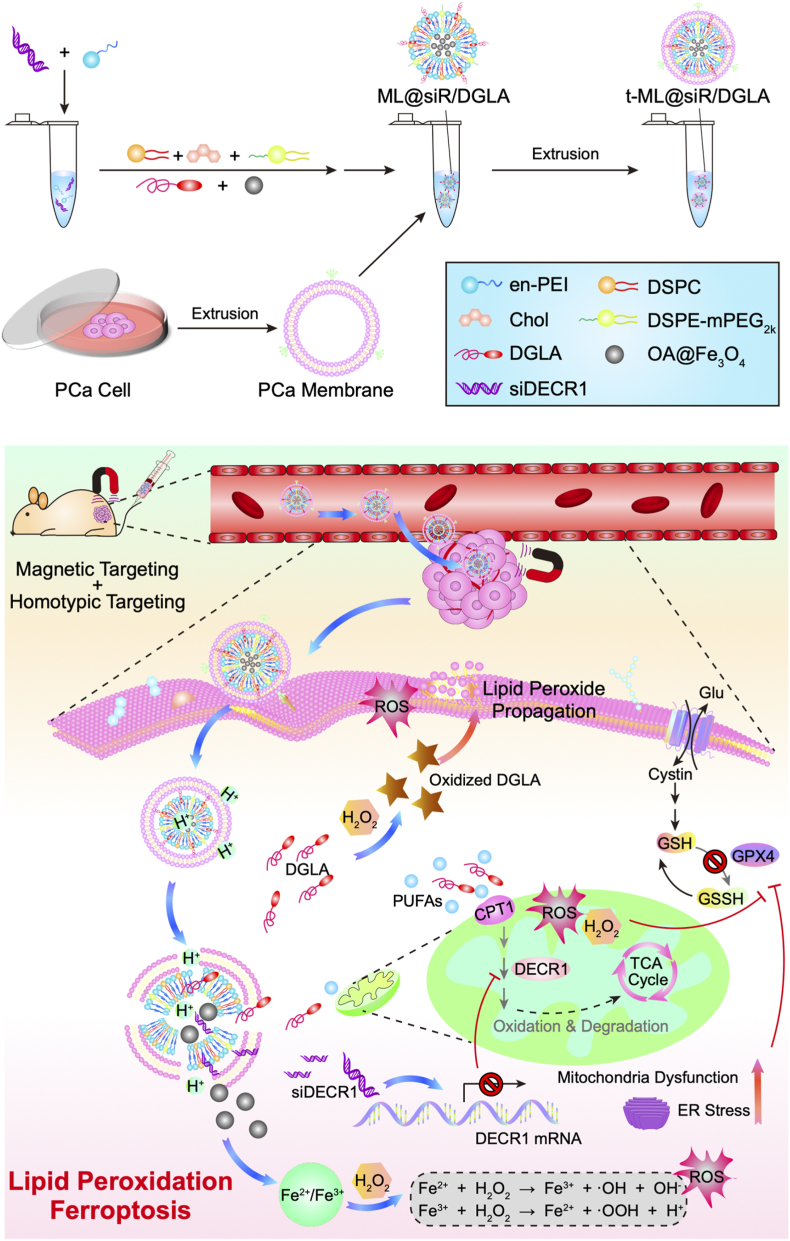
Herein, we report a biomimetic nanocomposite that codeliver DGLA and DECR1 siRNA (siDECR1) for the treatment of enzalutamide-resistant prostate cancer based on the regulation of PUFAs and ferroptosis (Scheme 1). To codeliver DGLA and siDECR1, magnetic lipid nanoparticles (ML) was prepared as a drug-loaded core (ML@DGLA/siDECR1). Lipid nanoparticles, composed of lipidoids, helper lipids, cholesterol and positive lipids, are usually used to deliver gene drugs such as siRNA and mRNA [11]. And obviously, lipid drugs such as DGLA can be easily loaded onto lipid nanoparticles due to the similarity solution principle. Moreover, Fe3O4 superparamagnetic magnetosomes could induce ferroptosis by increasing iron levels in cancer cells, with excellent magnetic targeting ability [12]. Here, the ML was composed of ethylenediamine-capped polyethyleneimine (en-PEI), 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethylene glycol)-2000] (DSPE-mPEG2k), and oleic acid-coated Fe3O4 nanoparticles (OA@Fe3O4), with a molar ratio of 52:10:32:2:4. However, if without a specific modification, lipid nanoparticles are cytotoxic and easily accumulate in the liver [13]. Cell membrane coating technology is an emerging bionic technology, and nanoparticles camouflaged with cancer cell membranes have a unique tumor targeting ability owing to inherent homotypic binding with good biocompatibility [14]. Therefore, to improve the active tumor targeting ability and safety of the nanoparticles, the prostate cancer cell membrane was extracted as a shell to camouflage the ML (t-ML).
Scheme 1.
Schematic illustrations of the establishment and mechanism of t-ML@DGLA/siDECR1.
We demonstrated that the drug loaded biomimetic nanocomposite (t-ML@DGLA/siDECR1) can accurately target the tumor site, thereby inducing ferroptosis in drug-resistant prostate cancer by Fenton reaction and increased PUFAs. Moreover, administration of t-ML@DGLA/siDECR1 with an external magnet could significantly reduce distant organ metastasis in a mouse model of drug-resistant CRPC fed a high fat diet (HFD), showing considerable safety and significantly inhibited tumor growth.
2. Results
2.1. Establishment and characterization of an enzalutamide-resistant prostate cancer cell line
The establishment of drug-resistant cell lines is a continuous and laborious process that usually takes at least 3 months [15], and the established enzalutamide-resistant C4–2B cell line (C4-2BEnz) was characterized after 6 months of coincubation with 10 μM enzalutamide. As shown in Fig. S1A, compared with enzalutamide-sensitive C4–2B cells, C4-2BEnz cells had fewer pseudopodia bulges, smaller cell volumes and denser organelles. C4-2BEnz cells presented a densely adherent growth mode and had a significantly faster growth rate than C4–2B cells. The CCK-8 experiment (Fig. S1B) showed that C4-2BEnz cells exhibited strong drug resistance with an IC50 value of 284.5 μg/mL, which was 178 times that of the C4–2B group. Moreover, the cell clone formation experiment further proved that the proliferation and cloning ability of drug-resistant cells was significantly enhanced (Fig. S1C). The established enzalutamide-resistant C4-2BEnz cell line was applied in subsequent experiments.
2.2. Preparation and characterization of the biomimetic magnetic nanoparticles t-ML
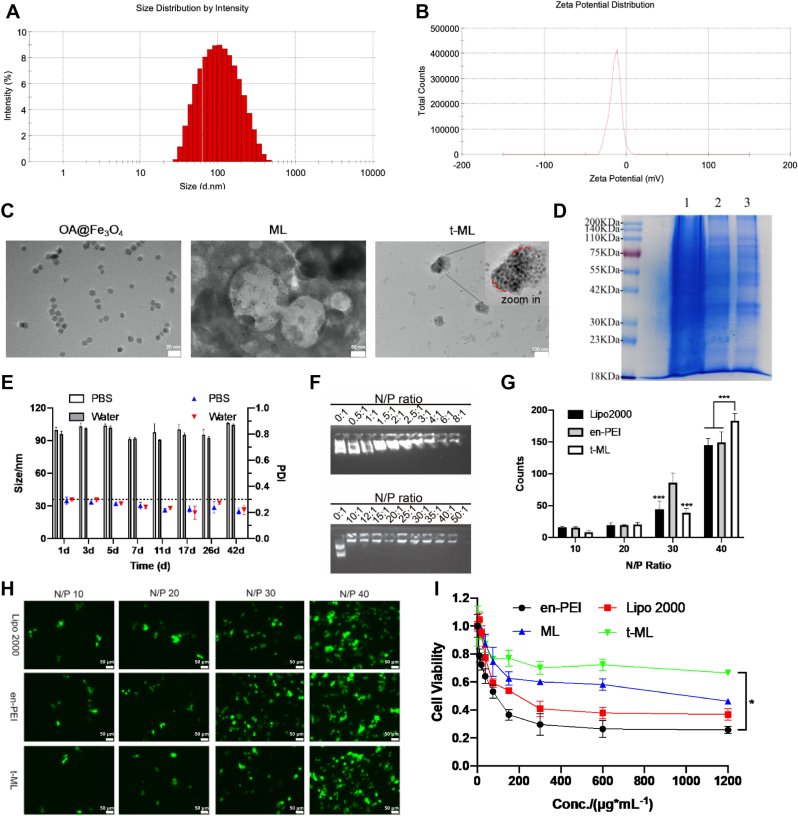
As shown in Fig. 1A and B and Fig. S2A, ML was loaded with OA@Fe3O4 and had a particle size of 92.49 ± 0.24 nm, with a near neutral zeta potential (−0.76 ± 0.46 mV). After camouflaging the prostate cancer cell membrane, the size of t-ML (96.37 ± 2.38 nm) was slightly higher, while the zeta potential of t-ML was −9.90 ± 0.80 mV, which was due to the negative potential of the C4–2B (−19.93 ± 0.50 mV) or C4-2BEnz cell membrane (−24.13 ± 1.12 mV). The morphology of OA@Fe3O4, ML, and t-ML could be observed more intuitively through transmission electron microscopy (TEM) images (Fig. 1C). OA@Fe3O4 nanoparticles were ∼10 nm spheroids or cubes. ML was an ∼100 nm spherical particle, with OA@Fe3O4 nanoparticles scattered in it. Relevently, t-ML was an ∼100 nm irregular spherical particle, and the inner OA@Fe3O4 nanoparticles and the surface prostate cancer cell membrane could be clearly witnessed. Meanwhile, t-ML fully inherited the membrane proteins of prostate cancer cell membranes (Fig. 1D). In addition, the stability of t-ML in PBS buffer, ultrapure water and fetal bovine serum (FBS) was evaluated at 4 °C. As shown in Fig. 1E and Fig. S3, t-ML had good stability, it was stable for 42 days in PBS or ultrapure water and 5 days in FBS. All these results indicated that t-ML was an irregular spherical nanoparticle with negative zeta potential, which was easily taken up by cells (∼100 nm) and had good biocompatibility (∼−10 mV) [16,17].
Fig. 1.
Characterization of t-ML. A,B) Size and zeta potential of t-ML; C) TEM images of OA@Fe3O4 (scale bar = 20 nm), ML (scale bar = 50 nm), t-ML (scale bar = 100 nm); D) SDS-PAGE protein analysis of prostate cancer cells (1), prostate cancer cell membranes (2), and t-ML (3); E) the stability test results of t-ML in PBS or ultrapure water in 42 days (n = 3, mean ± SD); F) Results of agarose gel electrophoresis (pEGFP: 1 μg); G) statistical analysis of the gene transfection assay (n = 3, mean ± SD, multiple t-test, ∗∗∗P < 0.001); H) gene transfection fluorescence images of t-ML at different N/P ratios (bars: 50 μm); I) CCK-8 assay of the toxicity of Lipo 2000, en-PEI, ML and t-ML (n = 3, mean ± SD, one-way ANOVA, ∗P < 0.05).
2.3. Investigation of t-ML's drug loading capacity and gene transfection ability
Owing to the rule of “like dissolves like” [18], t-ML had a good entrapment capacity for lipid drugs like DGLA, and the entrapment efficiency (EE) rate and drug loading (DL) rate of DGLA in t-ML were 87.14% ± 14.94% and 22.73% ± 3.90%, respectively (Fig. S2B). To evaluate the gene loading capacity of t-ML, a plasmid of enhanced green fluorescence protein (pEGFP) was used as a model drug. As shown in Fig. 1F, when the N/P ratio was ≥ 10, pEGFP could be completely compressed in t-ML, showing a strong gene binding ability. As shown in Fig. 1G and H, when the N/P ratio was 40, the gene transfection ability of t-ML was better than that of traditional cationic materials Lipofectamine 2000 (Lipo 2000) and en-PEI (P < 0.001). Therefore, in subsequent evaluation experiments, the N/P ratio of t-ML and gene drugs was set at 50. Moreover, the drug release of siRNA is acid-dependent, and its drug release rate was much faster in pH 5.5 (in endolysosomes) than in pH 7.4 (in blood), which may due to the proton sponge effect (Fig. S4) [19]. Additionally, after coating a film of prostate cancer cell membrane, the cytotoxicity of ML to HEK-293T cells was reduced, and the cell viability of the t-ML group was maintained at more than 70% at 600 μg/mL, which was much higher than that of the Lipo 2000 group and en-PEI group (P < 0.05) (Fig. 1I). Moreover, t-ML also had no toxicity to C4–2B or C4-2BEnz cells, and the cell survival rate was greater than 75% at a concentration of 1200 μg/mL for 24 h (Fig. S5). All these results indicated that t-ML had good safety and biocompatibility.
2.4. In vitro biodistribution assays of t-ML@DGLA/siDECR1
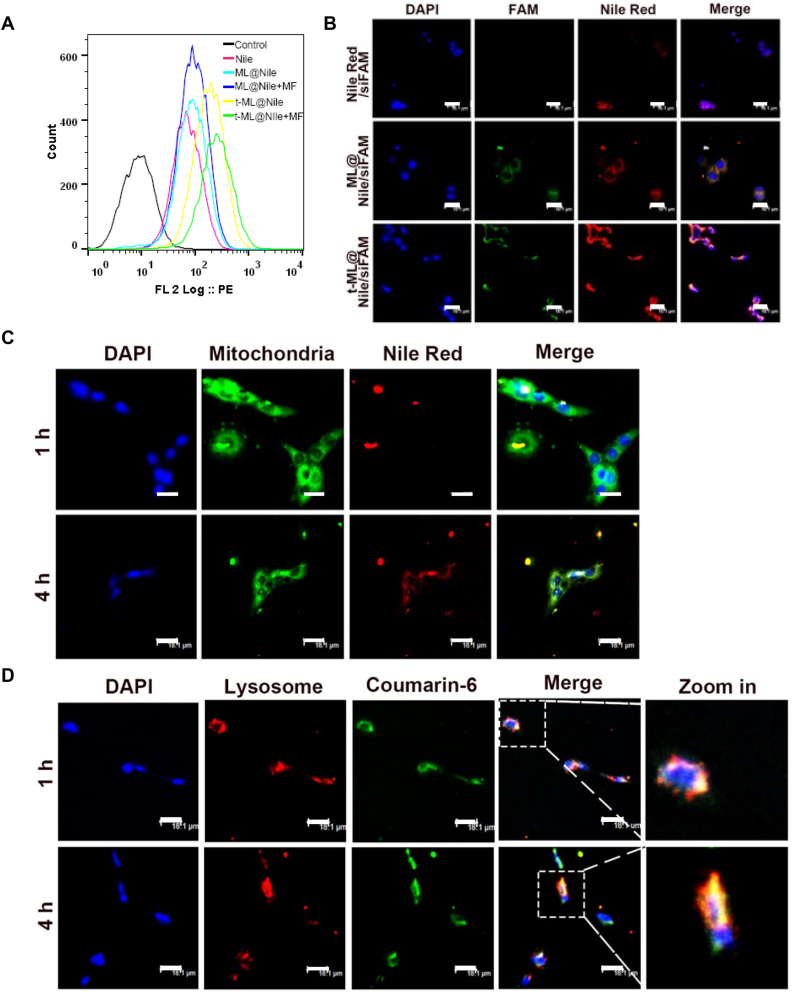
To obtain active tumor targeting ability, t-ML was endowed with superparamagnetic OA@Fe3O4 nanoparticles and homologous cancer cell membranes. Our previous study has demonstrated the homologous targeting capability of the prostate cancer cell membrane [20]. To investigate the tumor targeting ability of t-ML on prostate cancer cells, the fluorescent substance – Nile red (Nile) was used as a model drug. Moreover, 1.5 T external magnets were applied to evaluate the in vitro magnetic targeting ability of t-ML. As shown in Fig. 2A and Fig. S6, after camouflaging the cancer cell membrane with or without external magnets, the rate of positive cells in the t-ML@Nile groups was 2.2 times higher than that of the ML@Nile groups (P < 0.01). Moreover, after applying external magnets to the bottom of the cell plate for 2 h, the rate of positive cells in the t-ML group increased by 10.7% (P < 0.001). Moreover, the intracellular colocalization test was used to verify that t-ML could deliver drugs to the effect site (Fig. 2B). The fluorescence of Nile and siFAM in the t-ML@Nile/siFAM group overlapped around the nucleus. The fluorescence intensity of the t-ML@Nile/siFAM group was much stronger than that of the other groups, following the order of t-ML@Nile/siFAM > ML@Nile/siFAM > Nile/siFAM, which was in line with the results of cellular uptake study. Besides, due to the sensitivity and degradability of naked siRNA to ribozymes [21], there was no obvious FAM fluorescence in the Nile/siFAM group.
Fig. 2.
Cellular uptake and biodistribution. A) Flow cytometry test results of cellular uptake assays (Nile: 250 ng/well); B) intracellular colocalization results of Nile/siFAM, ML@Nile/siFAM, and t-ML@Nile/siFAM, visualized by a CLSM (Nile: 100 ng/mL, siFAM: 0.5 μg/mL, bars: 18.1 μm); C) mitochondria targeting study (Nile: 100 ng/mL, MitoTracker Green: 50 ng/mL, bars: 18.1 μm); D) endolysosomal escape study (coumarin-6: 100 ng/mL, LysoTracker Red: 50 ng/mL, bars: 18.1 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To assess whether t-ML requires energy to enter cells and whether t-ML can deliver drugs into mitochondria for mitochondrial regulatiory effects, t-ML@Nile was coincubated with C4-2BEnz cells for 1–4 h, and the mitochondria were labeled with MitoTracker Green. After 1 h of coincubation, a small amount of Nile entered the mitochondria; while at 4 h, a large amount of Nile had entered the mitochondria, indicating that the entry of t-ML into cells was an active transport process (Fig. 2C). Moreover, endolysosome escape ability is critical for gene drug delivery systems to reduce the degradation of gene drugs in endolysosomes [22]. Therefore, lysosomes were labeled with LysoTracker Red to observe the intracellular biodistribution of t-ML@coumarin-6. As shown in Fig. 2D, after 1 h of coincubation, coumarin-6 had entered the cytoplasm, and its green fluorescence were overlapped well with the red fluorescence of lysosomes, emitting strong orange fluorescence around the nucleus in the merged image. However, after 4 h of administration, the green fluorescence of coumarin-6 was separated from the red fluorescence of lysosomes, suggesting an emancipation of coumarin-6 from lysosomes. All these results demonstrated the in vitro acitive tumor targeting ability and high drug delivery efficiency of t-ML, which could actively target prostate cancer and escape phagocytic degradation by the endolysosome system. In addition, t-ML could be localized in mitochondria for drug release, providing evidence for the regulation of mitochondrial lipid metabolism and ferroptosis.
2.5. In vitro cell proliferation, migration and invasion study
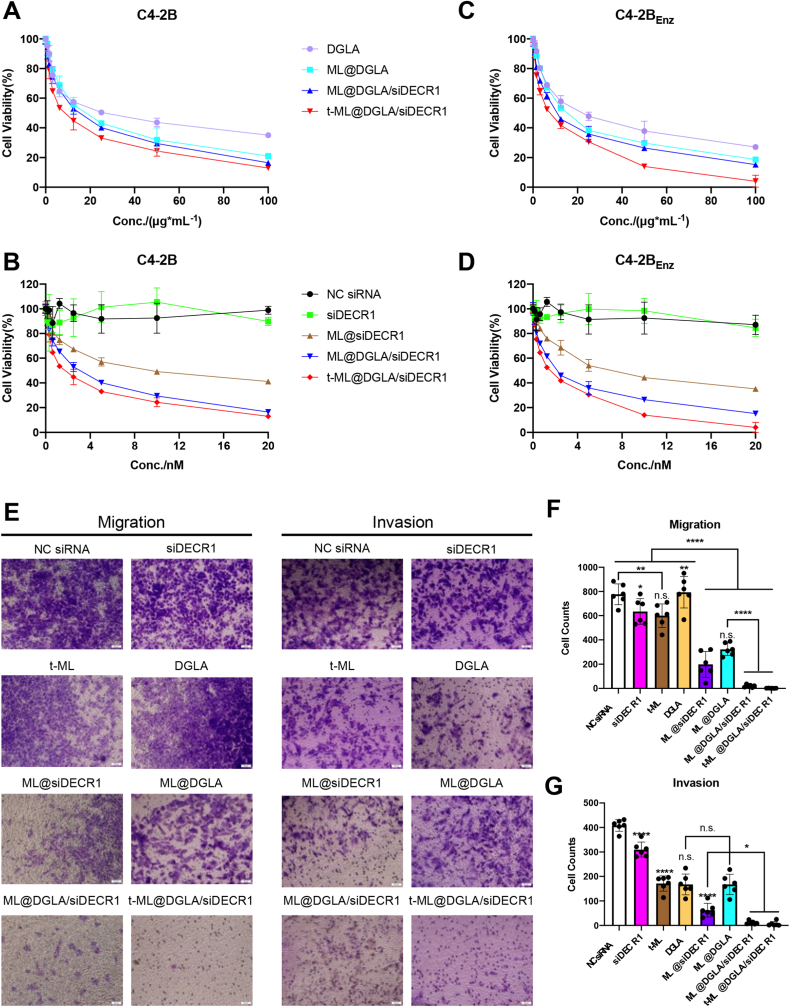
As shown in Fig. 3A and C and Fig. S7A, for the DGLA groups, either in C4–2B or C4-2BEnz cells, the antiproliferation ability sequence from high to low was as follows: t-ML@DGLA/siDECR1, ML@DGLA/siDECR1, ML@DGLA, and DGLA. The half maximal inhibitory concentration (IC50) values of the DGLA group in C4–2B or C4-2BEnz cells were 27.36 μg/mL and 22.76 μg/mL, respectively. After loading in ML, the drug efficiency of DGLA was enhanced, and the IC50 values of ML@DGLA in C4–2B or C4-2BEnz cells were 1.52-fold or 1.45-fold lower than that in the DGLA group, respectively. Moreover, after synergy with siDECR1, the IC50 values were further reduced, and the IC50 values of the t-ML@DGLA/siDECR1 group on C4–2B and C4-2BEnz cells were 3.08-fold and 3.21-fold lower than those of the DGLA group, respectively. Correspondingly, for the siRNA groups, due to the instability of naked siRNA, the free NC siRNA and siDECR1 had almost no cytotoxicity (Fig. 3B,D and Fig. S7B). The IC50 values of the ML@siDECR1 group in C4–2B and C4-2BEnz cells were 9.158 nM and 7.279 nM, respectively. The IC50 values of the ML@DGLA/siDECR1 and t-ML@DGLA/siDECR1 groups were 3.2-fold and 5.1-fold lower than those of the ML@siDECR group, respectively. All these results showed a synergistic effect of DGLA and siDECR1. While DGLA and siDECR1 were coloaded in t-ML, the antiproliferation effect was significantly enhanced, which might be due to the tumor homotypic targeting ability of t-ML. In addition, it is worth noting that enzalutamide-resistant C4-2BEnz cells were more sensitive to DGLA and siDECR1 treatment than C4–2B cells. Thus, with increased iron demands, drug-resistant cancer cells are more sensitive to ferroptosis, and ferroptosis-inducing agents can enhance the effect of AR inhibitors in CRPC [[23], [24], [25], [26]].
Fig. 3.
Antiproliferation, antimigration and anti-invasion assays. A-D) CCK-8 test of the antiproliferation ability of t-ML@DGLA/siDECR1 and other control groups to A,B) C4–2B or C,D) C4-2BEnz cells (n = 3, mean ± SD); E-G) antimigration and anti-invasion test (DGLA: 100 μg/mL, siDECR1: 20 nM, n = 6, mean ± SD, one-way ANOVA, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001).
The results of the antimigration and anti-invasion studies were basically in line with those of the antiproliferation studies. DGLA and siDECR1 showed a synergistic effect in t-ML@DGLA/siDECR1 and exhibited the strongest antimigration and anti-invasion abilities among all the groups (Fig. 3E–G). Consistent with a previous study [27], siDECR1 in vectors (ML, t-ML) showed significant antimigration and anti-invasion abilities (P < 0.0001). When C4-2BEnz cells were treated with DGLA alone, it had no antimigration effect but had a certain degree of anti-invasion ability. This might be due to the rapid growth and migration of drug-resistant cells, free DGLA did not have enough time to be taken up by the cells to exert its antimigration effect. After loading in ML, ML@DGLA exhibited considerable antimigration ability with increased cellular uptake of DGLA (P < 0.0001). Indeed, ML also had some antimetastatic effect, as the inner OA@Fe3O4 nanoparticles induced ferroptosis in the presence of increased iron supplements. Based on these factors, there were almost no migrating or invading cells in the t-ML@DGLA/siDECR1 group, indicating its strong antimigration and anti-invasion abilities.
2.6. In vitro ferroptosis induction study
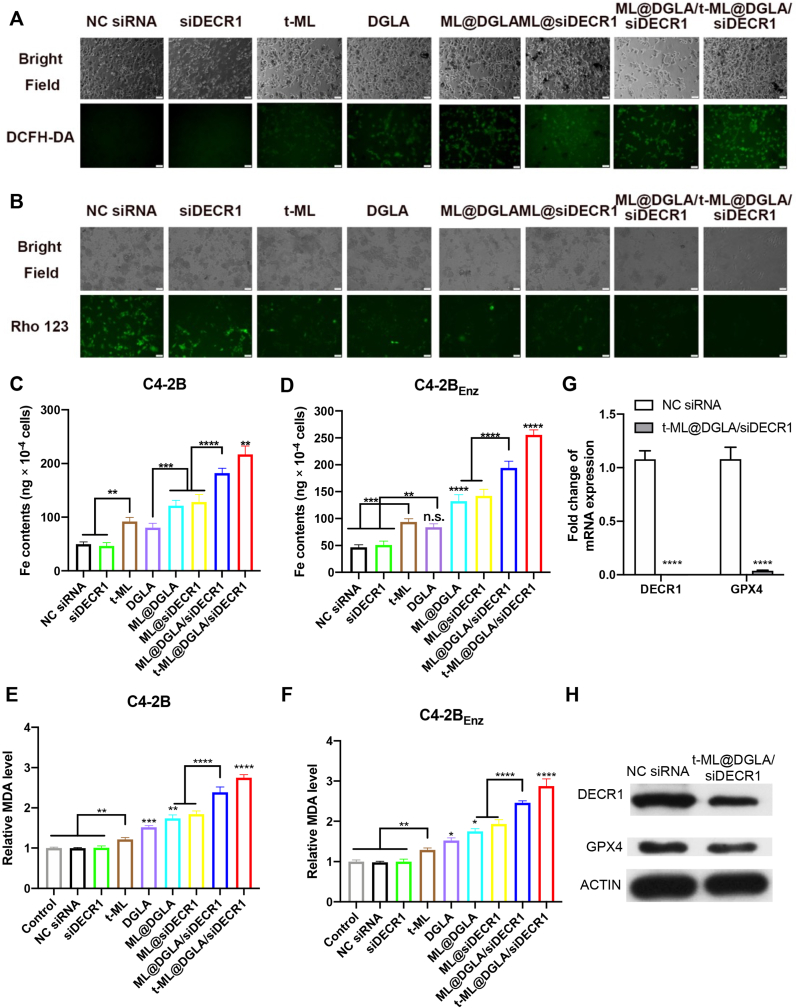
Ferroptosis is characterized by the accumulation of ROS and lipid peroxidation [23]. Fe3O4 nanoparticle-based naniplatforms can release Fe2+/Fe3+ ions in the acidic tumor microenvironment to catalyze the Fenton reaction, leading to an increased level of ROS [28,29]. After 8 h of coincubation with C4-2BEnz cells, the generation of ROS was detected with the green fluorescence probe 2,7-dichlorofluorescin diacetate (DCFH-DA). In line with the antiproliferation study, there was almost no signal in the naked NC siRNA and siDECR1 groups. The free DGLA group exhibited considerable green fluorescence, and the fluorescence intensity was enhanced upon loading of DGLA in ML. Correspondingly, ML@siDECR1 induced a certain amount of ROS under the protection of ML. The fluorescence intensity of ML@DGLA/siDECR1 group was further enhanced, and the fluorescence intensity of t-ML@DGLA/siDECR1 group was the strongest (P < 0.05) (Fig. 4A, Fig. S8A). Moreover, mitochondria play a vital role in ferroptosis, and mitochondrial membrane potential (MMP) hyperpolarization is highly associated with ferroptosis [30]. Rhodamine 123 (Rho 123) is a probe for MMP, and its fluorescence intensity decreases with decreasing MMP, indicating damage to the mitochondrial membrane [31,32]. Compared with the NC siRNA group, the fluorescence intensity of ML@DGLA/siDECR1 and t-ML@DGLA/siDECR1 groups decreased sharply (P < 0.0001), indicating boosted MMP loss and ferroptosis in prostate cancer cells (Fig. 4B, Fig. S8B). Moreover, the intracellular concentration of Fe2+/Fe3+ ions was detected by an intracellular iron colorimetric assay kit. As shown in Fig. 4C,D and Fig. S9, in both C4–2B cells and C4-2BEnz cells, compared with the naked NC siRNA and siDECR1 groups, the Fe content in the t-ML and DGLA groups was slightly increased. The Fe content of the ML@DGLA and ML@siDECR1 groups was further increased due to the OA@Fe3O4 encapsulated by ML itself and its tumor-targeting ability, and the Fe content of the t-ML@DGLA/siDECR1 group was the highest (P < 0.01). Besides, a malondialdehyede (MDA) test showed that lipid peroxidation explosively increased about 3-fold in cells after administration of t-ML@DGLA/siDECR1 compared to control (Fig. 4E and F). Additionally, the mRNA and protein expression levels of DECR1 and GPX4 were significantly attenuated in cells treated with t-ML@DGLA/siDECR1 compared to control (Fig. 4G and H). All these results suggested that ferroptosis was strongly induced in prostate cancer cells, especially in enzalutamide-resistant prostate cancer cells.
Fig. 4.
In vitro ferroptosis evaluation. A) ROS generation assay of C4-2BEnz cells treated with t-ML@DGLA/siDECR1 and other control groups (DGLA: 100 μg/mL, siRNA: 10 nM, bars: 50 μm), 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) was used as a probe; B) mitochondrial potential depolarization assay (DGLA: 100 μg/mL, siRNA: 10 nM, bars: 50 μm), rhodamine 123 (Rho 123) was used as a probe; C,D) Intracellular Fe content of C4–2B or C4-2BEnz cells was detected by an intracellular iron colorimetric assay kit (OA@Fe3O4: 10 μM, DGLA: 25 μg/mL, siRNA: 2.5 nM, n = 3, mean ± SD, one-way ANOVA, n.s.: no significance, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001); E,F) Relative MDA content of C4–2B or C4-2BEnz cells was detected by a Lipid Peroxidation (MDA) Assay kit (OA@Fe3O4: 10 μM, DGLA: 25 μg/mL, siRNA: 2.5 nM, n = 3, mean ± SD, one-way ANOVA, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001); G) QF-PCR analysis of DECR1 and GPX4 mRNA levels (DGLA: 100 μg/mL, siRNA: 10 nM, n = 3, mean ± SD, multiple t-test, ∗∗∗∗P < 0.0001); H) Western blot analysis of DECR1 and GPX4 protein levels (DGLA: 100 μg/mL, siRNA: 10 nM).
2.7. In vivo biodistribution and pharmacodynamics study
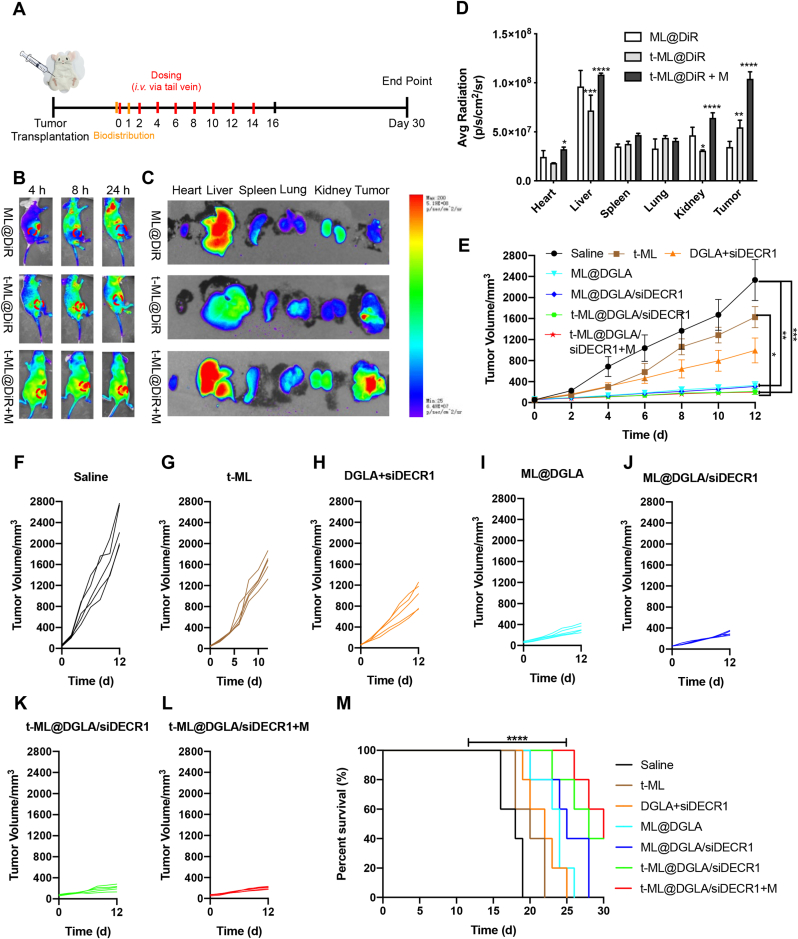
In vivo animal experiments were performed according to Fig. 5A. To investigate the biodistribution behaviors of t-ML, the deep red fluorescent dye 1,1-dioctadecyl-3,3,3,3-tetramethylindotricarbocyaine iodide (DiR) was used as a model drug. As shown in Fig. 5B–D, comparing with the ML@DiR group, prostate cancer membrane camouflaged biomimetic lipid nanoparticles t-ML@DiR showed significant tumor targeting ability (P < 0.01), and less drug accumulation in liver (P < 0.001) and kidney (P < 0.05) sites. Moreover, with the help of an external magnetic field, the t-ML@DiR + M group showed strong tumor targeting ability. The drug was obviously accumulated in the tumor site with 4 h of in vivo injection, and the fluorescence intensity did not decline for 24 h. In contrast, the fluorescence intensity in the tumor site of the t-ML@DiR group was significantly reduced at 24 h. Therefore, the t-ML@DiR + M group exhibited a long-circulating effect, and the tumor targeting ability was significantly better than that of the t-ML@DiR group (P < 0.0001). It is worth mentioning that compared with the t-ML@DiR group, the accumulation of the t-ML@DiR + M group in the liver and kidney also increased correspondingly. This may be due to the fact that when the t-ML@DiR was guided by an external magnetic field to the tumor site, the nanoparticles passed through the liver and kidney, the flux of nanoparticles increased, and the amount of nanoparticles retained by the liver and kidney was relatively increased at the same time.
Fig. 5.
In vivo evaluation of biodistribution and pharmacodynamics. A) Scheme of animal experiments. In vivo biodistribution study (orange line segment): Mice were treated with ML@DiR, t-ML@DiR or t-ML@DiR + M on Day 0 and sacrificed after 1 day to observe the tissue fluorescence distribution. In vivo pharmacodynamics study: red line segment: intravenous injection of t-ML@DGLA/siDECR1 and other groups via tail vein; black line segment: some mice of each group were sacrificed on the 16th day for physiological and biochemical index detection and lipidomics research. The remaining mice were used for survival time investigation, and the 30th day was set as the end point of the experiment; B) In vivo fluorescence imaging of C4-2BEnz tumor-bearing nude mice after intravenous injection of ML@DiR, t-ML@DiR, t-ML@DiR + M (with a 1.5 T external magnet) via tail vein at different time points (DiR: 1 mg/kg); C) Ex vivo imaging and D) corresponding average radiant efficiency of the tumor and major organs 24 h after injection (n = 3, mean ± SD, two-way ANOVA, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001); E) tumor volume growth curves (DGLA: 5 mg/kg, siDECR1: 0.2 mg/kg, n = 5, mean ± SD, one-way ANOVA, ∗P < 0.05, ∗∗∗P < 0.001); F-J) tumor volume growth curves of each group (n = 5); M) survival curves (n = 5, mean ± SD, log-rank analysis, ∗∗∗∗P < 0.0001). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In the pharmacodynamics study, all mice were fed an HFD. Tumors in HFD-fed mice grew rapidly, reaching 100 mm3 in 3–5 days. During the 16-day observation period, comparing with the rapidly increasing tumor volume in the saline group, the tumor growth rate in the other groups was relatively slow (Fig. 5E-L, S10). The slowdown of tumor growth in the t-ML group may be due to the fact that OA@Fe3O4 contained in t-ML has a certain ferroptosis-inducing effect based on the catalytic effect of the Fenton reaction. Although the free drug group (DGLA + siDECR1) showed a significant effect compared with the saline group (P < 0.05), its tumor volume continued to grow, reaching 1632.9 ± 421.5 mm3 on day 16. Although there was no statistical difference between the last three groups, the tumor volume in the t-ML@DGLA/siDECR1 + M group (228.0 ± 23.8 mm3) was 1.4-fold and 1.9-fold smaller than that in the t-ML@DGLA/siDECR1 (311.9 ± 93.9 mm3) and ML@DGLA/siDECR1 (436.9 ± 133.9 mm3) groups on day 16, respectively. Moreover, in the survival observation experiment, the survival time of the t-ML@DGLA/siDECR1 + M group was significantly longer than that of the saline group (P < 0.0001), and the median survival time was extended from 18 days to 30 days (Fig. 5M).
2.8. In vivo evaluation of ferroptosis
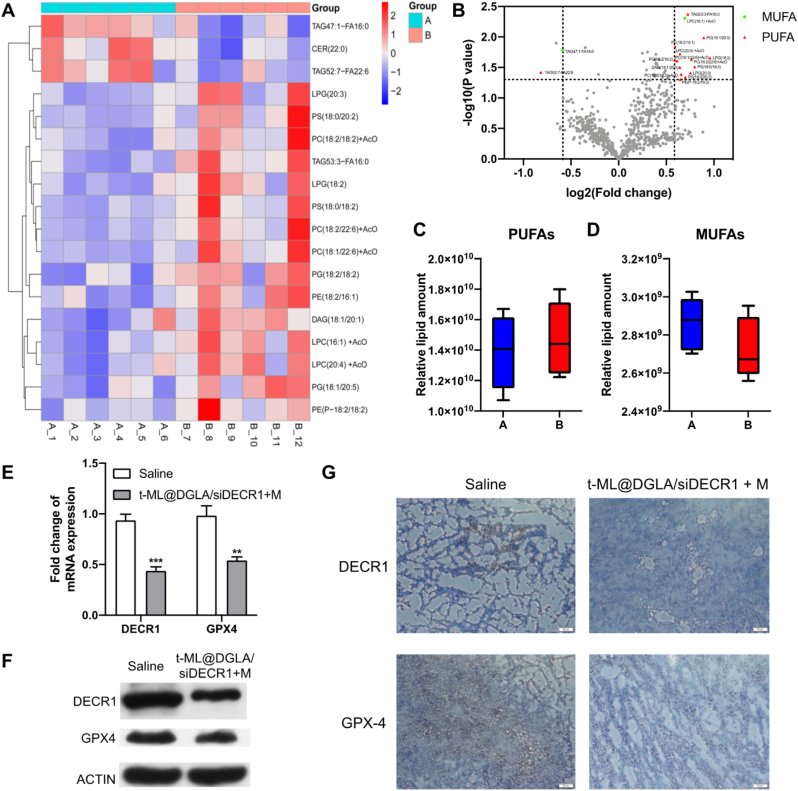
One of the main mechanism of ferroptosis is the peroxidation of unsaturated fatty acids, and interestingly, different types of unsaturated fatty acids have different effects on ferroptosis [23]. PUFAs can be peroxidized by the Fe2+/Fe3+ ion-catalyzed Fenton reaction. Correspondingly, endogenous MUFAs can inhibit ferroptosis through compensatory action, thereby maintaining a balance of PUFAs/MUFAs in the body [4]. However, in tumor tissue, the balance of PUFAs/MUFAs is broken, it has higher iron and lipid demands [4,23]. Therefore, PUFA supplementation and/or MUFA inhibition can inhibit tumor growth and metastasis by inducing ferroptosis. In the targeted lipidomics study, all the targeted lipids were analyzed by UPLC–MS/MS. 423 lipids were quantified in positive ion mode, and 251 lipids were quantified in negative ion mode. As shown in Fig. 6A and B, Fig. S11 and Supplementary Material 2,3, compared with the normal saline group, in the significant difference with |FC| > 1.5 and P < 0.05 as the screening criteria, the upregulated lipids in t-ML@DGLA/siDECR1 + M group were basically PUFAs. In addition, although there was no significant difference among the tested lipids, PUFAs in the tumor tissues of the t-ML@DGLA/siDECR1 + M group showed an upregulated trend, while MUFAs showed a downregulated trend, which was conducive to the process of ferroptosis (Fig. 6C and D and Supplementary Material 4). In line with the in vitro study, the mRNA and protein levels of DECR1 and GPX4 in tumor tissues of t-ML@DGLA/siDECR1+M group were significantly downregulated, suggesting that t-ML@DGLA/siDECR1 with an external magnetic field has efficient DECR1 knockdown and ferroptosis-inducing abilities (Fig. 6E–F).
Fig. 6.
In vivo ferroptosis-inducing study. A) Heatmap (A: saline group: B: t-ML@DGLA/siDECR1 + M group) and B) volcano plots (red: PUFAs with a significant change, green: MUFAs with a significant change, gray: no significant change or saturated lipids) illustrating the levels of significantly regulated lipids in tumor tissues of t-ML@DGLA/siDECR1 + M group when compared to normal saline group (n = 6, |FC| > 1.5, p < 0.05, one-way ANOVA, values were expressed as log (FC)); C,D) Relative lipid amount change of PUFAs and MUFAs in saline (A) and t-ML@DGLA/siDECR1 + M (B) groups (n = 6, multiple t-test); E) QF-PCR analysis of DECR1 and GPX4 mRNA levels in tumors (n = 3, mean ± SD, multiple t-test, ∗∗P < 0.01, ∗∗∗P < 0.001); F) Western blot and G) immunohistochemistry (IHC) analysis of DECR1 and GPX4 protein levels in tumors (scale bars = 50 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.9. In vivo antimetastasis and safety evaluation
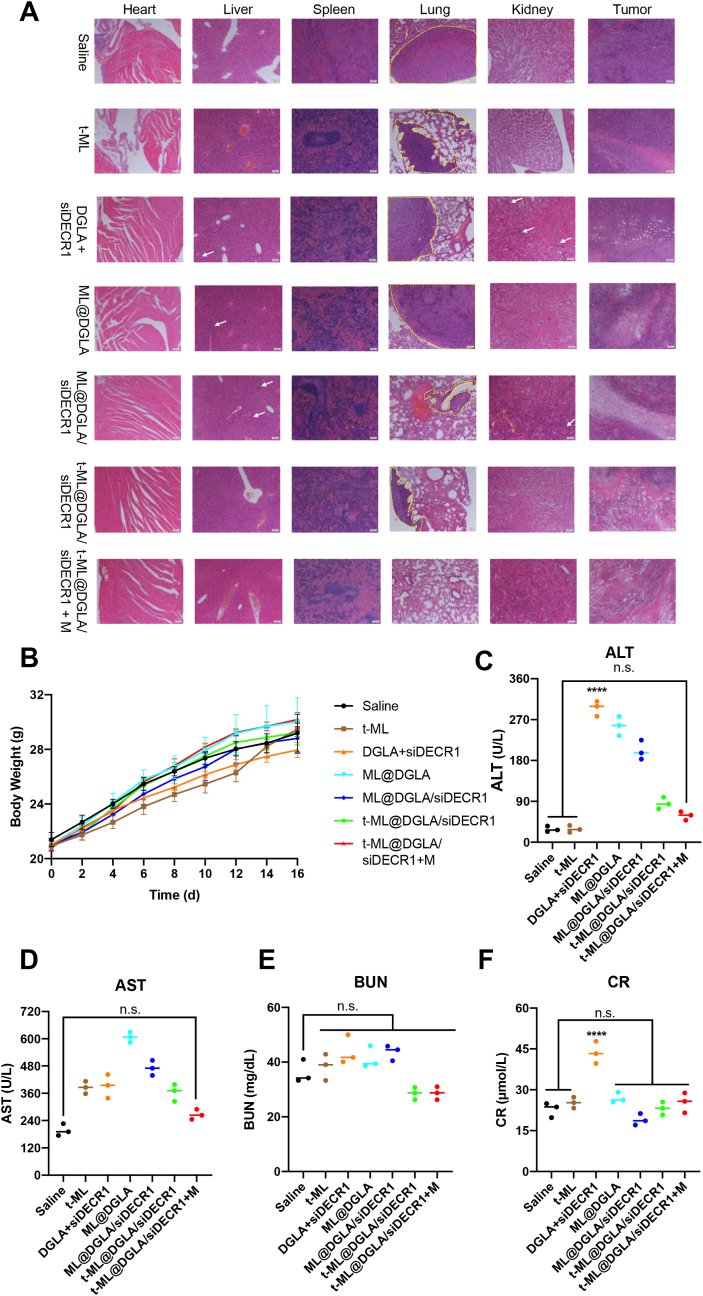
As shown in Fig. 7A, tumors from HFD-fed mice exhibited a high degree of malignancy and metastatic capacity. All groups except t-ML@DGLA/siDECR1 + M had lung metastases. Meanwhile, DGLA alone and ML showed a certain degree of toxicity and inflammatory damage in liver and kidney. After camouflaging with prostate cancer membrane, no inflammatory damage was witnessed in major organs in both t-ML@DGLA/siDECR1 and t-ML@DGLA/siDECR1 + M groups. Correspondingly, from histological observation, the damage of tumor sites in each group was consistent with the results of the drug efficacy experiments. Except for the normal saline control group, the tumor sites in the other groups had a certain degree of ablation. The t-ML@DGLA/siDECR1 + M group had the strongest drug effect, most of the tumor cells were ablated, and there was almost no complete nuclear morphology. Drug treatment had no effect on weight gain of mice in each group, and the weight of mice in each group maintained a steady increase (P > 0.05) (Fig. 7B). Additionally, the free drug group, DGLA + siDECR1, showed an increase in all blood biochemical indiexes, and alanine aminotransferase (ALT) and creatinine (CR) were significantly higher than those in the saline group (P < 0.0001), suggesting its toxicity to the liver and kidneys. Accordingly, there was no significant difference between the saline and t-ML@DGLA/siDECR1 + M groups (Fig. 7C–F). These results indicated that t-ML@DGLA/siDECR1 with external magnets had good biocompatibility and safety, and can significantly inhibit tumor growth and distant organ metastasis.
Fig. 7.
Evaluation of the antimetastatic ability and safety of t-ML@DGLA/siDECR1 with 1.5 T external magnets. A) Representative hematoxylin-eosin (HE) staining slices of major organs and tumors in each group (white arrow: inflammatory injury, yellow dotted line: tumor metastases in distant organs, scale bars = 500 μm); B) body weight growth curves (n = 5, mean ± SD, one-way ANOVA); C–F) plasma levels of C) alanine aminotransferase (ALT); D) aspartate aminotransferase (AST); E) blood urea nitrogen (BUN); F) creatinine (CR) in each group (n = 3, mean ± SD, one-way ANOVA, n.s.: no significance, ∗∗∗∗P < 0.0001). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
Drug resistance and metastasis of prostate cancer present great obstacles to its treatment, and the malignant progression of prostate cancer is characterized by high levels of lipogenesis [33]. Aberrant lipogenesis has become a metabolic hallmark of prostate cancer, and lipid accumulation favors carbon and energy storage in prostate cancer, promoting its progression and metastasis [[34], [35], [36]]. Moreover, it has been reported that a high-fat diet fuels prostate cancer progression by reprogramming the metabolome, which may explain why the prevalence of prostate cancer in Western countries is much higher than that in Eastern countries [37,38]. More importantly, lipid metabolism is closely related to ferroptosis, as it was characterized by the accumulation of iron-dependent lethal lipid peroxides. It is worth noting that lipid free radicals generated by the iron-mediated Fenton reaction promote lipid peroxidation of cell membranes, thereby damaging cell membrannes and inducing ferroptosis [4]. Moreover, not all cell membrane lipids are susceptible to peroxidation. It can be devided into 3 types: saturated fatty acids, MUFAs, and PUFAs. Only PUFAs, especially those in phospholipids, appear to be susceptible to peroxidation. However, peroxidation of PUFAs by lipoxygenase is compenstad by MUFAs to protect cancer cells from ferroptosis [4,39]. Peroxidation of lipids is regulated by the glutathione peroxidase system, which is directly or indirectly regulated by different ferroptosis-related genes [40]. GPX4 is a vital factor in lipid homeostasis and ferroptosis, converts lipid hydroperoxides to lipid alcohols, and prevents the formation and accumulation of toxic lipid ROS in cancer cells [41]. However, the function of GPX4 may be disrupted by up-regulated PUFA levels, which lead to significant ferroptosis. Dietaty intake of DGLA increases levels of PUFAs and lipid peroxidation in prostate cancer and antagonizes the effects of ferroptosis inhibitors such as GPX4, Vitamin E, and ferrostatin. Instead, dietary and endogenous MUFAs act as a compensatory mechanism to suppress ferroptosis [9]. Moreover, deleption of DECR1, a mitochondrial enzyme involved in the degradation of PUFAs, increases PUFA levels and increases the sensitivity of prostate cancer cells to ferroptosis [10]. In addition, knockdown of DECR1 with siRNA can significantly reduce prostate cancer cell migration and invasion [27].
In the current study, we developed a biomimetic nanocomposite to codeliver DGLA and siDECR1 for the treatment of enzalutamide-resistant prostate cancer by regulating PUFAs and ferroptosis in prostate cancer. The biomimetic nanocomposite t-ML@DGLA/siDECR1 was developed and characterized, showing good biocompatibility, stability, dispersibility and excellent gene transfection ability (Fig. 1). After camouflaging the prostate cancer cell membrane, t-ML can be efficiently taken up by prostate cancer cells with the help of an external magnetic field, avoiding the phagocytosis and degradation of lysosomes due to endolysosomal escape, and the drugs can be localized and released in the cytoplasm and mitochondria, which is beneficial to DGLA and siDECR1 induces ferroptosis in prostate cancer cells (Fig. 2). Due to the superparamagnetic and homologous targeting effect of t-ML, t-ML exhibited excellent active tumor targeting under 1.5 T external magnetic field, which can reduce drug accumulation in liver, kidney, and lung regions (Fig. 2, Fig. 5). Moreover, both in vivo and in vitro studies demonstrated that t-ML@DGLA/siDECR1 + M had favorable antitumor effects, provided PUFAs and induced ferroptosis in drug-resistant prostate cancer by knocking down DECR1 and supplementing DGLA. With the upregulation of PUFAs and the downregulation of MUFAs, the sensitivity of prostate cancer to ferroptosis was enhanced, which can inhibit the growth and distant organ metastasis of prostate cancer and prolong the survival time. In addition, it was biocompatible and had no obvious damage to major organs (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7).
4. Conclusion
In conclusion, the established biomimetic nanocomposite t-ML was stable and biocompatible with active tumor targeting and mild ferroptosis-inducing abilities. The drug-loaded t-ML@DGLA/siDECR1 with external magnets showed significant antitumor efficacy. Based on the regulation of PUFAs/MUFAs in HFD-fed drug-resistant prostate cancer-bearing nude mice, t-ML@DGLA/siDECR1 + M had a significant ferroptosis-inducing effect. In this mouse model of rapid tumor growth and metastasis, the tumor gowth of mice in the t-ML@DGLA/siDECR1 + M group was remarkably inhibited without distant organ metastasis. Therefore, this biomimetic nanocomposite provides a ferroptosis-based treatment for drug-resistant prostate cancer.
5. Experimental section
All the materials and methods are provided in Supplementary Material 1.
Credit author statement
Jiyuan Chen: Conception and Design, Investigation, Methodology, Visualization, Writing-Reviewing and Editing; Yujie Wang: Conception and Design, Investigation, Writing-Reviewing and Editing; Lu Han: Conception and Design, Investigation, Methodology, Writing-Reviewing and Editing; Rong Wang: Conception and Design, Investigation, Methodology, Writing-Reviewing and Editing; Chunai Gong: Data Analysis, Supervision; Gang Yang: Data Analysis, Supervision; Ze Li: Data Analysis, Supervision; Shen Gao: Supervision; Yongfang Yuan: Conception and Design, Supervision, Writing-Reviewing and Editing.
The authors declare that all data supporting the findings of this study are available within the article and its supplementary material files, or from the corresponding author on reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC 81973275, 82102902, and 81903141), and Shanghai Sailing Program (No. 20YF1424000 and 21YF1422700). Besides, the authors would like to express their profound gratitude to Shanghai Bioprofile Technology Co., Ltd. for their technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100484.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. Ca - Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Galletti G., Leach B.I., Lam L., Tagawa S.T. Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treat Rev. 2017;57:16–27. doi: 10.1016/j.ctrv.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Li D., Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Targeted Ther. 2020;5:108. doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordini J., Morisi F., Elia A.R., Santambrogio P., Pagani A., Cucchiara V., Ghia P., Bellone M., Briganti A., Camaschella C., Campanella A. Iron induces cell death and strenthens the efficacy of antiandrogen therapy in prostate cancer models. Clin. Cancer Res. 2020;26:6387–6398. doi: 10.1158/1078-0432.CCR-20-3182. [DOI] [PubMed] [Google Scholar]
- 6.Tousignant K.D., Rockstroh A., Poad B.L.J., Talebi A., Young R.S.E., Fard A.T., Gupta R., Zang T., Wang C., Lehman M.L., Swinnen J.V., Blanksby S.J., Nelson C.C., Sadowski M.C. Therapy-induced lipid uptake and remodeling unperpin ferroptosis hypersensitivity in prostate cancer. Cancer Metabol. 2020;8:11. doi: 10.1186/s40170-020-00217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayr L., Grabherr F., Schwärzler J., Reitmeier I., Sommer F., Gehmacher T., Niederreiter L., He G.W., Ruder B., Kunz K.T.R., Tymoszuk P., Hilbe R., Haschka D., Feistritzer C., Gerner R.R., Enrich B., Przysiecki N., Seifert M., Keller M.A., Oberhuber G., Sprung S., Ran Q., Koch R., Effenberger M., Tancevski I., Zoller H., Moschen A.R., Weiss G., Becker C., Rosenstiel P., Kaser A., Tilg H., Adolph T.E. Dietary lipids fuel GPX4-restricted enteritis resembling crohn's disease. Nat. Commun. 2020;11:1775. doi: 10.1038/s41467-020-15646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang X., Ardehali H., Min J., Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat. Rev. Cardiol. 2022 doi: 10.1038/s41569-022-00735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez M.A., Magtanong L., Dixon S.J., Watts J.L. Dietary lipids induce ferroptosis in Caenorhabditis elegans and human cancer cells. Dev. Cell. 2020;54:30498–30506. doi: 10.1016/j.devcel.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomme A., Ford C.A., Mui E., Patel R., Ntala C., Jamieson L.E., Planque M., McGregor G.H., Peixoto P., Hervouet E., Nixon C., Salji M., Gaughan L., Markert E., Repiscak P., Sumpton D., Blanco G.R., Lilla S., Kanphorst J.J., Graham D., et al. 2,4-Dienoyl-CoA reductase regulates lipid homeostasis in treatment-resistant prostate cancer. Nat. Commun. 2020;11:2508. doi: 10.1038/s41467-020-16126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ball R.L., Hajj K.A., Vizelman J., Bajaj P., Whitehead K.A. Lipid nanoparticle formulations for enhanced Co-delivery of siRNA and mRNA. Nano Lett. 2018;18:3814–3822. doi: 10.1021/acs.nanolett.8b01101. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F., Li F., Lu G.H., Nie W., Zhang L., Lv Y., Bao W., Gao X., Wei W., Pu K., Xie H.Y. Engineering magnetosomes for ferroptosis/immunomodulation synergism in cancer. ACS Nano. 2019;13:5662–5673. doi: 10.1021/acsnano.9b00892. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer J.R., Sinegra A.J., Ivancic D., Yeap X.Y., Qiu L., Wang J.J., Zhang Z.J., Wertheim J.A., Mirkin C.A. Structure-dependent biodistribution of liposomal spherical nucleic acids. ACS Nano. 2020;14:1682–1693. doi: 10.1021/acsnano.9b07254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Y., Su J., Ran W., Zhang P., Yin Q., Zhang Z., Yu H., Li Y. Preparation and application of cell membrane-camouflaged nanoparticles for cancer therapy. Theranostics. 2017;7:2575–2592. doi: 10.7150/thno.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xavier C.P.R., Pesic M., Vasconcelos M.H. Understanding cancer drug resistance by developing and studying resistant cell line models. Curr. Cancer Drug Targets. 2016;16:226–237. doi: 10.2174/1568009616666151113120705. [DOI] [PubMed] [Google Scholar]
- 16.Sindhwani S., Syed A.M., Ngai J., Kingston B.R., Maiorino L., Rothschild J., MacMillan P., Zhang Y., Rajesh N.U., Hoang T., Wu J.L.Y., Wilhelm S., Zilman A., Gadde S., Sulaiman A., Ouyang B., Lin Z., Wang L., Egeblad M., Chan W.C.W. The entry of nanoparticles into solid tumours. Nat. Mater. 2020;19:566–575. doi: 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- 17.Nam J., Won N., Bang J., Jin H., Park J., Jung S., Jung S., Park Y., Kim S. Surface engineering of inorganic nanoparticles for imaging and therapy. Adv. Drug Deliv. Rev. 2013;65:622–648. doi: 10.1016/j.addr.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Shruti U., Patel M.M. In: Chapter 2 – Lipid Nanoparticulate Systems: Modern Versatile Drug Carriers. Lipid Nanocarriers for Drug Targeting. Grumezescu A.M., editor. Elsevier; 2018. pp. 49–138. [Google Scholar]
- 19.Wojnilowicz M., Glab A., Bertucci A., Caruso F., Cavalieri F. Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA. ACS Nano. 2019;13:187–202. doi: 10.1021/acsnano.8b05151. [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Wu Z., Ding W., Xiao C., Zhang Y., Gao S., Gao Y., Cai W. SREBP1 siRNA enhance the doxetacel effect based on a bone-cancer dual-targeting biomimetic nanosystem against bone metastatic castration-resistant prostate cancer. Theranostics. 2020;10:1619–1632. doi: 10.7150/thno.40489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subhan M.A., Torchilin V.P. SiRNA based drug Design, quality, delivery and clinical translation. Nanomedicine: NBM (NMR Biomed.) 2020;29 doi: 10.1016/j.nano.2020.102239. [DOI] [PubMed] [Google Scholar]
- 23.Angeli J.P.F., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 24.Hassannia B., Vandenabeele P., Berghe T.V. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y., Yu C., Luo M., Cen C., Qiu J., Zhang S., Hu K. Ferroptosis in cancer treatment: another way to rome. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.571127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghoochani A., Hsu E.C., Aslan M., Rice M.A., Nguyen H.M., Brooks J.D., Corey E., Paulmurugan R., Stoyanova T. Ferroptosis inducers are a novel therapeutic approach for advanced prostate cancer. Cancer Res. 2021;81:1583–1594. doi: 10.1158/0008-5472.CAN-20-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassar Z.D., Mah C.Y., Dehairs J., Burvenich I.J.G., Irani S., Centenera M.M., Helm M., Shrestha R.K., Moldovan M., Don A.S., Holst J., Scott A.M., Horvath L.G., Lynn D.J., Selth L.A., Hoy A.J., Swinnen J.V., Butler L.M. Human DECR1 in an androgen-repressed survival factor that regulates PUFA oxidation to protect prostate tumor cells from ferroptosis. Elife. 2020;9 doi: 10.7554/eLife.54166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Z., Liu T., Li Y., Lau J., Yang Z., Fan W., Zhou Z., Shi C., Ke C., Bregadze V.I., Mandal S., Liu Y., Li Z., Xue T., Zhu G., Munasinghe J., Niu G., Wu A., Chen X. Fenton-reaction-accelerable magnetic nanoparticles for ferroptosis therapy of orthotopic brain tumors. ACS Nano. 2018;12:11355–11365. doi: 10.1021/acsnano.8b06201. [DOI] [PubMed] [Google Scholar]
- 29.Min H., Qi Y., Zhang Y., Han X., Cheng K., Liu Y., Liu H., Hu J., Nie G., Li Y. A graphdiyne oxide-based iron sponge with photothermally enhanced tumor-specific Fenton chemistry. Adv. Mater. 2020;32 doi: 10.1002/adma.202000038. [DOI] [PubMed] [Google Scholar]
- 30.Gao M., Yi J., Zhu J., Minikes A.M., Monian P., Thompson C.B., Jiang X. Role of mitochondria in ferroptosis. Mol. Cell. 2019;73:354–363. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W., Wang X., Zhao J., Liu R., Liu J., Wang Z., Peng J., Wu H., Zhang X., Long Z., Kong D., Li W., Hai C. Stat2-Drp1 mediated mitochondrial mass increase is necessary for pro-inflammatory differentiation of macrophages. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Li Y., Wang K., Chen Y., Shi M., Zhang X., Pan W., Li N., Tang B. GSH-Responsive nanoprodrug to inhibit glycolysis and alleviate immunosuppression for cancer therapy. Nano Lett. 2021;21:7862–7869. doi: 10.1021/acs.nanolett.1c03089. [DOI] [PubMed] [Google Scholar]
- 33.Bader D.A., McGuire S.E. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat. Rev. Urol. 2020;17:214–231. doi: 10.1038/s41585-020-0288-x. [DOI] [PubMed] [Google Scholar]
- 34.Muunir R., Lisec J., Swinnen J.V., Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer. 2019;120:1090–1098. doi: 10.1038/s41416-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J., Thompson C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019;20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galbraith L., Leung H.Y., Ahmad I. Lipid pathway deregulation in advanced prostate cancer. Pharmacol. Res. 2018;131:177–184. doi: 10.1016/j.phrs.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Labbé D.P., Zadra G., Yang M., Reyes J.M., Lin C.Y., Cacciatore S., Ebot E.M., Creech A.L., Giunchi F., Fiorentino M., Elfandy H., Syamala S., Karoly E.D., Alshalalfa M., Erho N., Ross A., Schaeffer E.M., Gibb E.A., Takhar M., Den R.B., et al. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC Program. Nat. Commun. 2019;10:4358. doi: 10.1038/s41467-019-12298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culp M.B., Soerjomataram I., Efstathiou J.A., Bray F., Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur. Urol. 2020;77:38–52. doi: 10.1016/j.eururo.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Cao F., Yin H., Huang Z., Lin Z., Mao N., Sun B., Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forcina G.C., Dixon S.J. GPX4 at the crossroads of lipid homeostasis and ferroptosis. Proteomics. 2019;19 doi: 10.1002/pmic.201800311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.