Abstract
In Escherichia coli, extracytoplasmic stress is partially controlled by the alternative sigma factor, RpoE (ςE). In response to environmental stress or alteration in the protein content of the cell envelope, ςE upregulates the expression of a number of genes, including htrA. It has been shown that htrA is required for intramacrophage survival and virulence in Salmonella typhimurium. To investigate whether ςE-regulated genes other than htrA are involved in salmonella virulence, we inactivated the rpoE gene of S. typhimurium SL1344 by allelic exchange and compared the phenotype of the mutant (GVB311) in vitro and in vivo with its parent and an isogenic htrA mutant (BRD915). Unlike E. coli, ςE is not required for the growth and survival of S. typhimurium at high temperatures. However, GVB311 did display a defect in its ability to utilize carbon sources other than glucose. GVB311 was more sensitive to hydrogen peroxide, superoxide, and antimicrobial peptides than SL1344 and BRD915. Although able to invade both macrophage and epithelial cell lines normally, the rpoE mutant was defective in its ability to survive and proliferate in both cell lines. The effect of the rpoE mutation on the intracellular behavior of S. typhimurium was greater than that of the htrA mutation. Both GVB311 and BRD915 were highly attenuated in mice. Neither strain was able to kill mice via the oral route, and the 50% lethal dose (LD50) for both strains via the intravenous (i.v.) route was very high. The i.v. LD50s for SL1344, BRD915, and GVB311 were <10, 5.5 × 105, and 1.24 × 107 CFU, respectively. Growth in murine tissues after oral and i.v. inoculation was impaired for both the htrA and rpoE mutant, with the latter mutant being more severely affected. Neither mutant was able to translocate successfully from the Peyer’s patches to other organs after oral infection or to proliferate in the liver and spleen after i.v. inoculation. However, the htrA mutant efficiently colonized the livers and spleens of mice infected i.v., but the rpoE mutant did not. Previous studies have shown that salmonella htrA mutants are excellent live vaccines. In contrast, oral immunization of mice with GVB311 was unable to protect any of the mice from oral challenge with SL1344. Furthermore, i.v. immunization with a large dose (∼106 CFU) of GVB311 protected less than half of the orally challenged mice. Thus, our results indicate that genes in the ςE regulon other than htrA play a critical role in the virulence and immunogenicity of S. typhimurium.
Salmonella species can infect both warm- and cold-blooded hosts and cause a spectrum of diseases ranging from mild enteritis to severe systemic infections. This range reflects the ability of salmonellae to adapt to a range of different environments, including the interior of macrophages, in the vertebrate host. Although a number of virulence genes have been characterized and there is a basic understanding of how salmonellae cause infection, a number of questions remain unanswered about how the bacterium adapts and survives in different environments in vivo.
A clue to this adaptation is given by the reduced virulence of salmonella strains harboring mutations in the htrA gene (6, 29). HtrA (also called DegP) is a stress-induced serine protease. In E. coli, where it was first identified, HtrA is required for survival at high temperatures (>42°C). E. coli htrA accumulate abnormal periplasmic proteins, indicating that HtrA was active in the periplasm, where it is thought to assist in the degradation of denatured or damaged proteins which may result from enteric bacteria encountering a toxic environment (50). Unlike their E. coli counterparts, S. typhimurium htrA strains are not temperature sensitive; they are, however, more sensitive to the oxidizing agents H2O2 and menadione (a superoxide radical generator) than are wild-type strains (29). S. typhimurium htrA strains are also less able to survive within macrophages and exhibit a profound reduction in virulence in mice (2, 6). Thus, HtrA is required as part of the salmonella adaptive response to the host environment, in particular the oxidative stress present in the interior of macrophages (2, 29).
Homologues of htrA have been identified in a number of different pathogenic and nonpathogenic bacteria, including Brucella abortus and Yersinia enterocolitica (31, 44, 47, 54). A strain of Y. enterocolitica harboring a mutation in the htrA gene showed a reduced ability to colonize the livers and spleens of BALB/c mice and an increased sensitivity to oxidative killing (31). Several studies indicate that HtrA expression is upregulated in the interior of eucaryotic cells. LacZ expression was induced in a salmonella strain harboring a lacZ reporter gene under the control of the htrA promoter when it entered macrophages or epithelial cells (15). A similar finding was obtained by using the promoter of gsrA, the htrA homologue from Y. enterocolitica (55).
E. coli htrA mutants are temperature sensitive, indicating that HtrA is part of the heat shock response. However, expression of htrA is independent of sigma 32 (ς32), the classical heat shock sigma factor, but is controlled instead by a novel sigma factor (ςE) encoded by the rpoE gene (13, 33). Under extreme stress (50°C or 10% ethanol), ςE is also required in E. coli for expression of ς32 (26). Sequence analysis of RpoE reveals a similarity with a family of sigma factors classified as extracytoplasmic sigma factors which control the expression of gene products required in the extracellular compartments (34). The accumulation of misfolded proteins in the periplasm or cytoplasmic membrane is thought to induce ςE to activate expression of htrA.
A ςE homologue, AlgU, was isolated from Pseudomonas aeruginosa prior to its identification in E. coli (35). P. aeruginosa algU mutants show increased sensitivity to chemically or enzymatically produced halogenated reactive oxygen intermediates and increased sensitivity to phagocytic killing, indicating a similar function for the AlgU and ςE regulons in protecting bacteria from environmental stress (56). Strangely, an algU mutant had a slightly lower 50% lethal dose (LD50) in normal inbred mice and killed neutropenic mice faster than wild-type P. aeruginosa (56). Both P. aeruginosa AlgU and E. coli ςE negatively, as well as positively, affect the expression of a number of polypeptides (45, 56).
We were interested in investigating the involvement of ςE and ςE-regulated genes in salmonella virulence. In particular, we wished to determine whether the involvement of the rpoE regulon was confined to htrA or if it involved other genes. To this end, we constructed an rpoE mutant of S. typhimurium and compared its phenotype in vitro and in vivo with that of its parent or an isogenic htrA mutant. Compared to its wild-type parent and an isogenic htrA mutant, the S. typhimurium rpoE mutant was more sensitive to oxidizing agents and antimicrobial peptides, survived less well in eucaryotic cells, and was highly attenuated in mice, thus implicating other genes in the ςE regulon in S. typhimurium virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used or constructed in this study are listed in Table 1. All strains were maintained on Luria-Bertani (LB) or M9 minimal media that was prepared as described earlier (49). Where required, media were supplemented with 1.5% agar; 100 μg of ampicillin (Ap), 50 μg of kanamycin (Km), 100 μg of streptomycin (Sm), or 50 μg of chloramphenicol (Cm) per ml; 0.4% glucose, or 0.4% succinate.
TABLE 1.
Bacterial strains and plasmids used or constructed in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | S. typhimurium his mutant, mouse virulent strain | 28 |
| BRD509 | SL1344 aroA aroD mutant | 51 |
| BRD915 | SL1344 ΔhtrA | 15 |
| GVB311 | SL1344 rpoE::Kmr | This study |
| SM10λpir | E. coli thi-1 thr1 leuB6 tonA21 lacY1 supE44 recA::RP4-2-Tc::MuKmRλpir | 30 |
| Plasmids | ||
| pCR II | PCR cloning vector | Invitrogen |
| pRDH10 | λpir-based suicide vector: Tetr Cmr SacB | R. Haigh (Leicester University, Leicester, United Kingdom) |
| pUC4K | Kmr cassette | 43 |
| pSH101 | rpoE in pCR II | This study |
| pSH102 | pCR II rpoE::Kmr | This study |
| pWSK29 | Apr, low copy number vector | 52a |
| pSH103 | pRDH10 rpoE::Kmr | This study |
| pSH117 | rpoE in pWSK29 | This study |
PCR and template preparation.
All PCR reactions were carried out with Taq polymerase (Gibco BRL) with the manufacturer’s buffer and deoxynucleoside triphosphate mix. The S. typhimurium rpoE gene was isolated by PCR with the primers 181 (5′-GTCTACAACATGACAAACAAAAACAAATGC) and 182 (5′-CCTTTTCCAGTATCCCGCTATCGTCAACGC) from an S. typhimurium BRD509 colony template (25). The amplified DNA fragment was cloned into the vector pCR II (Invitrogen) to create plasmid pSH101.
Recombinant DNA manipulations.
Standard methods were used for the preparation of plasmid or chromosomal DNA, for restriction analysis, and for ligation (49). DNA hybridization was carried out by using the Gene Images labelling and detection kit (Amersham Life Sciences). DNA for sequencing was isolated by using Qiagen plasmid preparation columns. Sequencing reactions were carried out by using the Thermo Sequenase kit (Amersham) and run on an LI-COR 4000L automated DNA sequencer. Standard methods were used for the transformation and conjugation of plasmid DNA from E. coli to S. typhimurium.
Construction of an rpoE mutant.
The rpoE gene isolated by PCR was mutated by insertion of a kanamycin antibiotic resistance cassette. The kanamycin antibiotic resistance cassette was isolated from pUC4K by digestion with HincII and inserted into a unique StuI site within the coding sequence of the rpoE gene to form pSH102. The mutated copy of the rpoE gene (rpoE::Kmr) was isolated from pSH102 by digestion with BamHI and SalI and ligated into similarly digested pRDH10 to form pSH103. The suicide vector pRDH10 (25a) requires the pir product for replication and possesses sacB, from Bacillus subtilus, which allows positive selection of allelic exchange (11). pSH103 was introduced into S. typhimurium SL1344 from E. coli SM10λpir by conjugation. Merodiploids were isolated by selection with Km, Cm, and Sm. Chromosomal DNA was isolated from a number of putative merodiploids (Kmr, Cmr, and Smr) for Southern analysis. Southern hybridization with a DNA probe consisting of the last 528 bp of the rpoE gene revealed that the rpoE locus had been altered, indicating that the plasmid had inserted into the correct region of the chromosome (data not shown). The merodiploids were resolved by growing the bacteria in the presence of 6% sucrose. Kmr Cms colonies were isolated, and mutation of the rpoE gene was confirmed by Southern blotting and PCR with primers 181 and 182. Both techniques revealed that in the mutant the size of the rpoE gene had increased by ∼1.2 kb, which could be attributed to the presence of the Kmr gene (data not shown).
Complementation.
The rpoE gene, along with its natural P2 promoter, was excised from pSH101 by digestion with XbaI and HindIII and ligated into the low-copy-number vector pWSK29 (52a), which had been similarly digested. The resulting plasmid, pSH117, was used in complementation studies.
Analysis of bacterial growth.
Strains were grown overnight in the appropriate medium. To analyze the growth curve of the strains, the overnight cultures were diluted 100-fold into 50-ml portions of fresh media. Bacteria were incubated at 30, 37, or 42°C with aeration, 1-ml samples were removed at intervals, and the absorbance at 600 nm was recorded. The number of CFU per milliliter was determined by plating out serial dilutions of bacterial samples.
Disk diffusion assay.
Bacteria cultured overnight in M9-glucose (M9-G) media were diluted to an optical density at 600 nm of 0.25 in fresh M9-G and grown for 2 h at 30 or 37°C. Then, 100 μl was used to inoculate 3 ml of top agar to form a lawn on M9-G plates. Next, 6-mm filter paper disks were soaked in 10 μl of the toxic agent (3% H2O2 or 2% paraquat [methyl viologen]; both from Sigma). These disks or disks containing 300 U of polymyxin B (Oxoid) were added to the agar surface. Plates were incubated aerobically overnight at 37 or 30°C. The diameter of the zones of inhibition were measured.
Invasion and persistence of S. typhimurium strains in phagocytic and nonphagocytic cells.
The ability of the different S. typhimurium strains to invade and survive in phagocytic and nonphagocytic cells was assessed by using the macrophage-like cell line RAW264.7 and the epithelial cell line HEp-2, respectively. Cells were routinely cultured in Dulbecco modified Eagle medium (DMEM; Gibco BRL) supplemented with 4 mM l-glutamine, 10% (vol/vol) fetal bovine serum and 1× Antibiotic-Antimycotic mix (Gibco-BRL). For invasion assays, cells were seeded into 24-well tissue culture plates (Costar) at 2 × 105 cells per well and incubated overnight at 37°C with 5% CO2. Prior to infection, the monolayers were washed twice in antibiotic-free DMEM. Bacteria from overnight culture in LB-glucose (LB-G) broth were diluted in DMEM to give 2 × 105 CFU/ml. Then, 1 ml of bacterial suspension was added to each well to give an approximate 1:1 multiplicity of infection. Bacterial invasion was synchronized by centrifugation at 1,000 rpm for 10 min followed by incubation for 1 to 2 h as described above. The monolayers were then washed twice with sterile phosphate-buffered saline (PBS) and overlaid with 1 ml of DMEM containing 100 μg of polymyxin B per ml and incubated for 1 h as described above. The monolayers were washed twice with PBS, and either the cells were lysed with sterile water or they were overlaid with 1 ml of DMEM containing 10 μg of polymyxin B per ml and incubated for a further 21 h before being washed and lysed as described above. After lysis of the cells, the number of viable bacteria released from the cells was determined by plating serial dilutions on LB-agar (LA) plates.
Analysis of salmonella virulence, in vivo growth, and immunity in mice.
Virulence and immunity studies were carried out as previously described (6, 50). S. typhimurium strains grown statically overnight in LB-G were recovered by centrifugation and resuspended in sterile PBS (pH 7.2) to approximately 1 × 1010 to 1010 CFU/ml. The actual number of bacteria present was determined by a viable counting. Female BALB/c mice (6 to 8 weeks old; Charles River, Margate, United Kingdom) were challenged either orally with a gavage tube or intravenously (i.v.) by injection into the tail vein as described previously (6, 50). For the virulence studies, mice were challenged with different numbers of organisms and were then closely observed, and deaths were recorded for 28 days. The LD50s of the strains were determined by the method of Reed and Muench (46). To study the in vivo growth and survival of S. typhimurium strains, groups of mice were inoculated by the oral or i.v. route as described above. On various days after infection, groups of four mice were sacrificed and the spleen, liver, mesenteric lymph nodes, and Peyer’s patches were removed from each animal. Then, 10 ml of sterile water was added to each organ in a separate sterile bag, and the organs were homogenized with a Stomacher 80 (Steward Lab System). To determine the number of viable organisms present, the homogenates were serially diluted and plated onto LA-Sm plates. To examine the immunogenicity of the mutant strain, mice were immunized either orally or i.v. with different doses of this strain. The mice were challenged orally 28 days later with 2 × 108 CFU of SL1344 and deaths were recorded for 6 weeks.
RESULTS
Construction of a S. typhimurium rpoE mutant.
The complete S. typhimurium rpoE gene was isolated by PCR. The sequence was found to be 100% identical to the published S. typhimurium rpoE gene (36). The rpoE gene was insertionally inactivated in vitro by using a Km resistance cassette, and this construct was used to produce an S. typhimurium SL1344 rpoE mutant (GVB311) by allelic exchange as described above. The disruption of the rpoE gene in GVB311 was confirmed by PCR and Southern blotting as described above.
The S. typhimurium rpoE mutant (GVB311) exhibits aberrant growth under certain conditions.
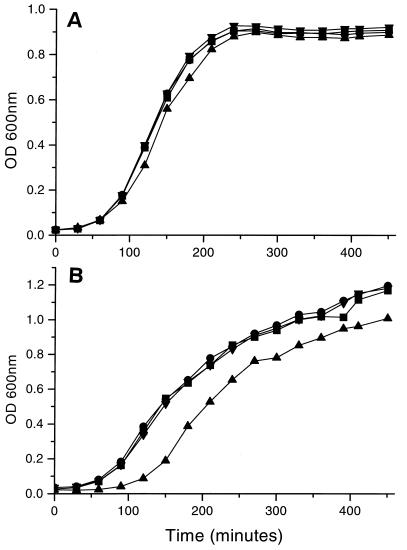
Inactivation of the E. coli rpoE gene confers a temperature-sensitive phenotype such that the bacteria begin to lyse at growth temperatures above 43°C (26). Therefore, we analyzed GVB311 for any temperature-dependent effects on growth. The mutant formed normal-sized colonies on LA after overnight growth at 30, 37, or 42°C (data not shown). When the bacteria were grown in LB at 30, 37 (data not shown), or 42°C (Fig. 1B), GVB311 exhibited a longer lag phase than SL1344 or an SL1344 htrA mutant (BRD915). The growth rate during the log phase for GVB311 was the same as for SL1344 and BRD915; however, GVB311 did not reach the same final optical density after overnight growth as the other two strains. This reflected fewer CFU of GVB311 per milliliter in the overnight culture than with SL1344 and BRD915 (data not shown). Introduction of the wild-type rpoE gene into GVB311 on a low-copy-number plasmid complemented the growth defect in LB (Fig. 1B).
FIG. 1.
Effect of media and temperature on the growth of S. typhimurium rpoE mutant. Overnight cultures of the S. typhimurium strains SL1344 (■), BRD915 (●), GVB311 (▴), and GVB311(pSH117) (▾) were used to inoculate fresh media, which was then incubated aerobically. Growth was followed spectrophotometrically. (A) Growth in LB-G at 42°C. (B) Growth in LB at 42°C.
In contrast to the growth in LB, GVB311 grew normally in M9-G medium (data not shown). However, when glucose was replaced with succinate as the carbon source, the lag period of GVB311 was greatly protracted (4 to 6 h [data not shown]). If LB medium was supplemented with glucose, then the growth curve of GVB311 was normal (Fig. 1A [note that our LB medium does not normally contain glucose]). These results suggest that GVB311 is defective in its ability to utilize carbon sources other than glucose.
GVB311 shows increased sensitivity to H2O2 and paraquat compared to SL1344.
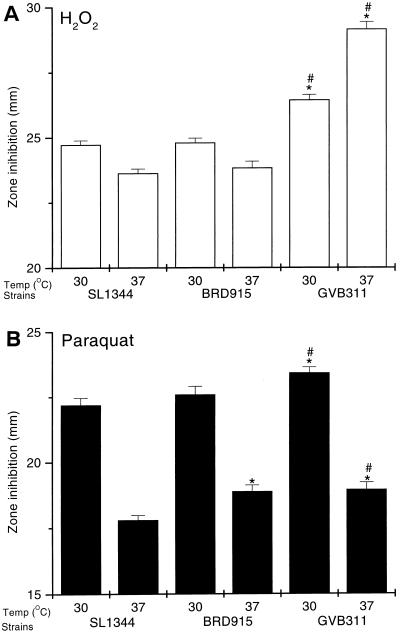
It was previously reported that an S. typhimurium htrA mutant was more sensitive to H2O2 and menadione (a superoxide generator) than its wild-type parent, indicating a role for HtrA in the defense against oxidative stress (29). If, as well as htrA, S. typhimurium rpoE regulates other genes required to repair the damage caused by oxidative stress, then the rpoE mutant should be at least as sensitive as, if not more sensitive than, an htrA mutant to oxidative agents. The sensitivity of GVB311 and other Salmonella strains to H2O2 and paraquat (a superoxide anion generator) was assessed by a disk diffusion assay. The assays were performed at 30 and 37°C to analyze whether temperature affected the ability of the bacteria to adapt to oxidative stress. The rationale for this is that at high temperatures both the ς32 and ςE regulons will be upregulated (13, 14); therefore, the bacteria may be better adapted to survive oxidative stress. The results are shown in Fig. 2. BRD915 was not more sensitive to H2O2 than SL1344 at either temperature. However, BRD915 and SL1344 were both significantly (P < 0.05) more sensitive to H2O2 at 30°C than at 37°C. GVB311 was significantly more sensitive to H2O2 than SL1344 and BRD915 at both temperatures and, unlike the other two strains, GVB311 is more sensitive to the effect of H2O2 at 37 than at 30°C.
FIG. 2.
Effect of the rpoE and htrA mutations on the sensitivity of S. typhimurium to oxidizing agents. The salmonella strains were tested for sensitivity to 3% H2O2 (A) or 2% paraquat (B) by disk diffusion assay. The plates were incubated overnight at 30 or 37°C as indicated in the figure below each lane. Each bar represents the mean diameter of the zone of inhibition, and the error bar shows the standard deviation (SD) of 30 replica assays. The asterisk indicates that the mean diameter of the zones is statistically different (P < 0.05) from that of SL1344 incubated at the same temperature; the number sign (#) indicates that the mean is statistically different from BRD915 grown at the same temperature (one-way analysis of variance [ANOVA]).
All three strains were more sensitive to paraquat at 30°C than at 37°C. At 37°C, there was no significant difference in the paraquat sensitivities of BRD915 and GVB311, but both were significantly more sensitive to paraquat than SL1344. At 30°C, GVB311 (but not BRD915) was again more sensitive to paraquat than SL1344. The effect of the rpoE and htrA mutations on the sensitivity of S. typhimurium to reactive nitrogen intermediates was assayed by disk diffusion assay by using SIN-1. SIN-1 spontaneously generates peroxynitrate radicals under aerobic conditions. There was no significant difference in the sensitivities of the three strains to SIN-1 (500 mM [Sigma]; data not shown).
GVB311 is more sensitive to the antimicrobial peptide polymyxin B than SL1344 and BRD915.
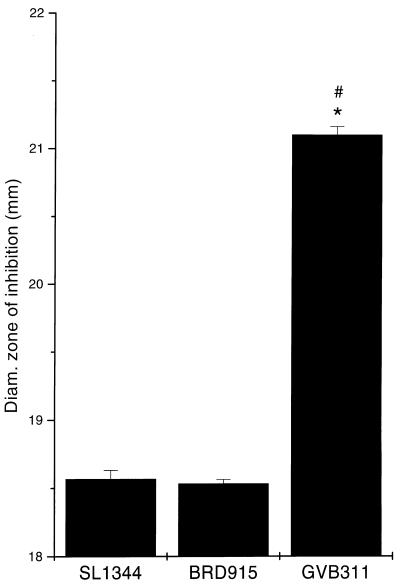
The sensitivity of GVB311 to polymyxin B was analyzed by disk diffusion assay. GVB311 was significantly more sensitive to killing by polymyxin B than were SL1344 and BRD915, as shown in Fig. 3. This peptide binds to the lipid A core of lipopolysaccharide (LPS). Analysis of the LPS of GVB311 and SL1344 on silver-stained gels revealed no obvious differences between the LPS profiles (data not shown).
FIG. 3.
Effect of the rpoE and htrA mutations on the sensitivity of S. typhimurium to polymyxin B. SL1344, BRD915, and GVB311 were tested for sensitivity to polymyxin B by disk diffusion assay. Disks containing 300 U of polymyxin B (Oxoid) were placed on the plates and incubated overnight at 37°C. The diameter of the zone of inhibition was then measured in millimeters. Each bar represents the mean diameter of the zone of inhibition, and the error bar shows the SD of 30 replica assays. The asterisk indicates that the mean diameter of the zones is statistically different (P < 0.05) from that of SL1344, and the number sign (#) indicates that the mean is statistically different from BRD915 (one-way ANOVA).
The sensitivity of the rpoE and htrA mutants to other membrane-damaging agents was also assessed. The ability of the strains to grow in the presence of detergents was assessed by growing the organisms on MacConkey and desoxycholate-citrate agar containing 0.5% bile salts and 0.5% sodium deoxycholate, respectively. There was no difference in the plating efficiencies or in the sizes of the colonies of the three strains on these two media. The sensitivity of the strains to killing by complement was assessed by exposing the strains to 10% normal human serum. There was no difference in the sensitivities of the three strains to complement killing (data not shown). These studies indicate that the rpoE and htrA mutations do not cause a generalized defect in outer-membrane integrity.
RpoE is involved in intracellular survival within macrophages and nonphagocytic cells.
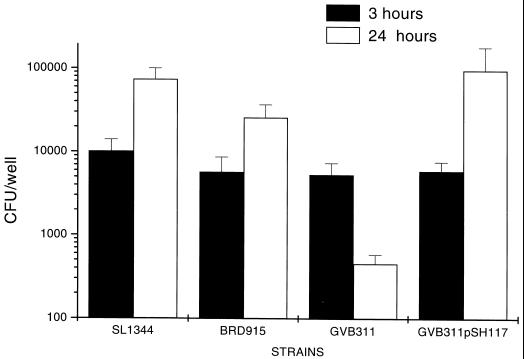
Previous studies with S. typhimurium showed that an htrA mutant strain survived less well in macrophages than did a wild-type bacteria. It is therefore likely that a strain that lacks rpoE would also survive less well in macrophages (2). Furthermore, if ςE-regulated genes, in addition to htrA, are involved in intramacrophage survival, then an rpoE mutant may be more attenuated in macrophages than an htrA strain. In order to address this SL1344, BRD915, GVB311, and GVB311(pSH117) were assayed to assess their abilities to invade and survive in the murine macrophage cell line RAW264.7. We also examined the capacity of the strains to invade and survive in nonphagocytic HEp-2 cells. GVB311 invaded macrophages as well as had SL1344 and BRD915 (Fig. 4). However, after 24 h the number of GVB311 inside RAW264.7 cells had decreased ∼6-fold, whereas the numbers of BRD915, SL1344, and GVB311(pSH117) had increased ca. 6- to 40-fold (Fig. 4). This indicates that rpoE-regulated genes other than htrA do participate in intramacrophage survival. GVB311 was also able to invade HEp-2 normally. However, by between 3 and 24 h the number of GVB311 organisms present intracellularly had decreased 5-fold, whereas the numbers of SL1344 and BRD915 had increased 10- and 5-fold, respectively (data not shown).
FIG. 4.
Effect of the rpoE and htrA mutations on the ability of S. typhimurium to invade and survive in macrophages. The ability of the different S. typhimurium strains to invade and survive in macrophages was examined by using the murine macrophage-like cell line RAW264.7. RAW264.7 cells in the wells of tissue culture plates were infected with bacteria at a multiplicity of infection of ∼1:1. The assay was performed as described in the text. The graph shows the number of viable bacteria inside macrophages at 3 and 24 h after infection. Each bar represents the mean of CFU from triplicate experiments, and the error bars indicate the SD.
Recently, it has been reported that Salmonella spp. can induce apoptosis in infected macrophages (7, 32, 42). We compared the cytotoxicity of the three strains for RAW264.7 cells by using a commercial assay (Cytotox; Promega). No difference was found between the cytotoxicity induced in RAW264.7 cells by these three strains (data not shown). This suggests that the rpoE and htrA mutations are not affecting the different effector mechanisms that are reported to be responsible for the induction of apoptosis.
RpoE is critical for the virulence and immunogenicity of S. typhimurium.
The virulence of GVB311 in mice was compared with SL1344 and BRD915 after oral or parenteral (i.v.) challenge. As shown in Table 2, GVB311 and BRD915 were unable to kill mice after oral challenge, even at a dose of ca. 1010 CFU. Whereas SL1344 had an LD50 at least 4 logs lower, the LD50 of GVB311 after i.v. inoculation was approximately 20-fold higher than that of the htrA mutant and 106 times greater than the LD50 of the wild-type strain.
TABLE 2.
Effect of the rpoE mutation on S. typhimurium virulencea
| Strain | Genotype | LD50
|
|
|---|---|---|---|
| Oral | i.v. | ||
| SL1344 | Wild type | 1.20 × 106 | <10 |
| BRD915 | htrA | >1010 | 5.5 × 105 |
| GVB311 | rpoE | >1010 | 1.24 × 107 |
Groups of five mice were challenged with different doses of each strain of S. typhimurium by the route indicated, and the deaths were recorded for 6 weeks.
Although highly attenuated, S. typhimurium htrA mutants are excellent live vaccines that can induce solid immunity to lethal challenge with wild-type S. typhimurium (5). To determine whether inactivation of rpoE affected the ability of S. typhimurium to induce a protective immune response, we immunized BALB/c mice with different doses of GVB311 by the oral or i.v. routes and 28 days later challenged the mice with 100× LD50s (∼2 × 108 CFU) of SL1344; the results are shown in Table 3. Even at the highest dose of ∼1010 CFU, none of the mice immunized orally were protected. The i.v. immunization usually induces a stronger immunity than an oral immunization. Nevertheless, an i.v. immunization with a dose of 106 CFU of GVB311 protected fewer than half of the mice. This inactivation of rpoE severely compromised the immunogenicity of S. typhimurium.
TABLE 3.
Effect of the rpoE mutation on S. typhimurium immunogenicitya
| Immunization route | Dose (CFU) | Protection (no. of survivors/no. challenged) |
|---|---|---|
| Oral | 8.75 × 109 | 0/5 |
| 9.63 × 107 | 0/5 | |
| 1.0 × 106 | 0/5 | |
| i.v. | 1.11 × 106 | 2/5 |
| 1.0 × 104 | 2/5 |
Mice were immunized as indicated; 28 days later they were challenged orally with 2 × 108 CFU of SL1344. Mice were observed for 6 weeks, and the number of deaths were recorded.
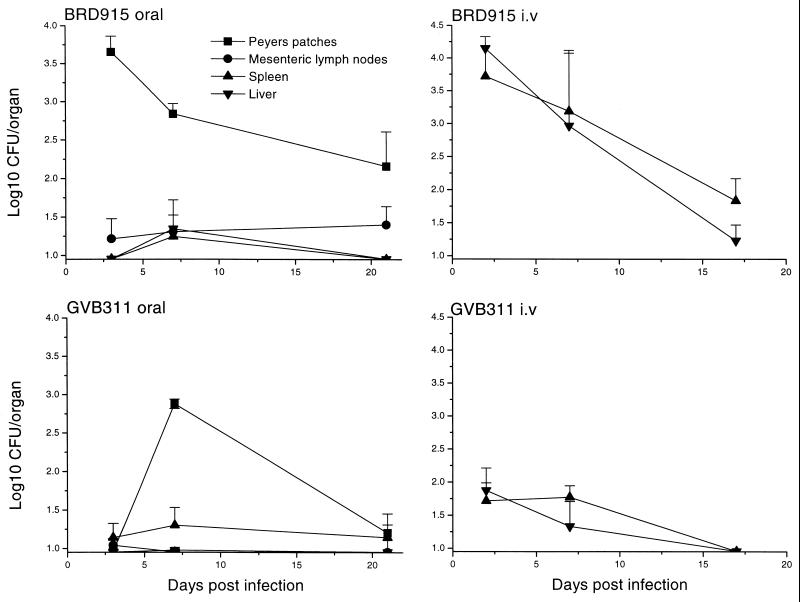
In order to understand further the reduced virulence of GVB311, the ability of the organism to colonize, survive, and replicate in different murine tissues was compared with BRD915 after oral or i.v. inoculation (Fig. 5). After oral inoculation, BRD915 was able to colonize Peyer’s patches (PPs) efficiently in high numbers but was slowly eliminated from this organ after day 3; however, hundreds of bacteria were still detected in the PPs at day 21. GVB311 could not be detected in the PPs on day 3, although levels comparable to BRD915 were present on day 7. Thereafter, GVB311 was cleared rapidly from the PPs and was detectable in only one of four mice on day 21. Both BRD915 and GVB311 translocated very poorly from the PPs to deeper tissues.
FIG. 5.
Colonization, growth, and survival of BRD915 and GVB311 in murine tissues after oral or i.v. inoculation. Groups of mice were infected with BRD915 or GVB311 orally (1010 CFU) or i.v. (106 CFU). At the indicated periods after infection, organs were removed from groups of four mice and homogenized and the number of viable organisms present was determined. Symbols: ■, PPs; ●, mesenteric lymph nodes; ▴, spleen; and ▾, liver. Each point represents the mean CFU/organ for four mice, and the error bars indicate one standard error of the mean.
After i.v. infection, strain BRD915 colonized both livers and spleens well, but it did not demonstrate an increase in numbers over the course of the study; instead it slowly cleared from both organs as previously described (6). GVB311 was present in the liver and spleen on day 2 but in much lower numbers than BRD915. For example, there were ca. 100 CFU in the livers and spleens of GVB311-infected mice but ca. 15,000 CFU in the corresponding organs of BRD915-infected mice. On day 17 only one of the four mice sampled had bacteria in the liver, as was also the case on day 21, by which point the organism was undetectable in the spleen.
DISCUSSION
E. coli ςE is one of the factors that control the response of the bacterium to extracytoplasmic stress (26, 45, 48). Our results indicate that ςE plays a similar role in S. typhimurium and also that it is intimately involved in salmonella virulence. However, S. typhimurium ςE is not required for survival at high temperatures as it is in E. coli (26). GVB311 appeared to grow normally at different temperatures on solid medium. However, aberrant growth was noted in liquid media. In general, the growth rate of GVB311 was very similar to SL1344 and BRD915, but in media lacking glucose the lag period was extended and the final cell yields were lower. Growth was normal in M9 containing glucose as the sole carbon source, but if glucose was replaced with the nonfermentable carbon source succinate it took up to 7 h for GVB311 to begin growing (data not shown). Supplementing LB with glucose abolished the extended lag period of GVB311. These results suggest that rpoE mutants may be defective in their ability to utilize nonfermentable carbon sources or carbon sources other than glucose.
A clue to understanding this phenomena may come from studies with E. coli mutants that are hypersensitive to redox-cycling drugs, such as paraquat, because they lack cytosolic superoxide dismutase (20). Such mutants can only utilize fermentable carbon sources because O2− generated normally during aerobic respiration inactivates enzymes involved in the tricarboxylic acid cycle (5). The rpoE strain is more sensitive to O2− than wild-type S. typhimurium, and so it may grow poorly in the presence of nonfermentable carbon sources because it cannot detoxify the endogenously produced O2−. Alternatively, the products of one or more ςE-regulated genes other than htrA may be involved in the correct folding or integrity of one or more polypeptides involved in the transport of certain carbon sources across the outer and/or inner membranes into the cell.
GVB311 is more sensitive than SL1344 and BRD915 to oxidative stress imposed by both H2O2 and paraquat. Although the differences were not large, they were statistically significant. This indicates that genes in the ςE regulon other than htrA are required for defense against H2O2 and paraquat. Both SL1344 and BRD915 are more resistant to H2O2 at 37 than at 30°C but, interestingly, the reverse was true for GVB311. We hypothesized that strains may be more resistant to oxidants at the higher temperatures because of the increased activity of ςE and RpoH (ς32). Lower sensitivity of the strains to the oxidant at the higher temperature would indicate that there is involvement of the intracellular stress regulon (controlled by ς32) and/or the extracellular stress regulon (controlled by ςE). The rpoE mutation also had a bigger effect on the sensitivity of GVB311 to H2O2 than on its sensitivity to paraquat. This suggests that ςE is more important for the defense against H2O2 than for the defense against paraquat. This is as expected because paraquat causes the generation of superoxide in the cytoplasm where it exerts its effect, whereas H2O2 readily crosses biological membranes and is likely to cause damage in both the cytoplasmic and extracytoplasmic compartments (37). It may be that ςE-regulated genes are required to combat damage caused by H2O2 in the cell envelope. Also, ςE may be involved in controlling ς32-mediated responses to H2O2 damage in the cytoplasm. H2O2 is known to induce a subset of the classical (ς32-dependent) heat shock genes. ςE can activate the transcription of rpoH from the rpoH3 promoter under particular conditions, such as higher temperature (>42°C) and 20% ethanol (14). It may be that exposure of salmonellae to H2O2 also stimulates ςE-dependent rpoH transcription and, because this response would be absent in GVB311, this may account for the results we obtained. Unlike an earlier study, we did not find here that htrA was required for resistance to H2O2 (29). The previous work used an S. typhimurium C5 htrA mutant produced by TnphoA mutagenesis, and it may be that variation in the strain background (SL1344 versus C5), the nature of the mutations, or the assay methodology may account for the different results. Salmonella has a number of genes that are involved in the defense against oxidizing agents, including the kat genes, the sod genes, oxyR, rpoS, soxS, slyA, htrA, and now rpoE (3, 4, 9, 16–18, 29, 37).
Unlike the other genes listed above, some of which are also transcriptional regulators, rpoE also contributes towards the ability of salmonella to resist the activity of antimicrobial peptides. GVB311 was more sensitive to the antibacterial activity of the peptide antibiotic polymyxin B, and preliminary experiments also indicate that GVB311 is more sensitive to cecropin P1 and a peptide (P2) derived from the bactericidal-permeability increasing protein (reference 53 and data not shown). These peptides bind to the negatively charged phosphoryl groups on the lipid A core of LPS. After this binding, the inner and outer membranes are permeabilized, resulting in cell death (21, 22). A number of genes have been reported to influence the sensitivity of salmonella to antibacterial peptides, including polymyxin B, which is probably the most well studied and is regulated by the two-component regulators PhoPQ and PmrAB (23, 24). PhoPQ activates or represses a total of 40 genes, and the regulon is essential for virulence (38, 39). As regards its involvement in peptide resistance, PhoPQ activates the pmrAB genes and, in turn, PmrAB positively regulates the expression of two genes, pmrE and pmrF, that mediate antimicrobial peptide resistance. The function of the product of these genes is to add an aminoarabinose residue onto the lipid A core of LPS, reducing the anionic charge and thereby lowering its ability to be bound by cationic peptides (23).
GVB311 does not have a generalized defect in membrane integrity because it is no more sensitive to detergents or serum than SL1344 or BRD915 and there were no apparent defects in its LPS profile. How ςE affects peptide resistance is unknown, but our results indicate that it is not via activation of htrA. Interestingly, it has been suggested that proteases in the bacterial envelope may also mediate peptide resistance (21). It may be that ςE controls the expression of an extracytoplasmic protease other than HtrA that mediates peptide resistance.
The ability of salmonella strains to invade and survive in macrophages in vitro usually correlates with the virulence of the strain in vivo (2, 19). Many genes are involved in salmonella intramacrophage survival and growth (1). Our results indicate that ςE-regulated genes other than htrA are also involved in this process because GVB311 is less able to survive in macrophages than is BRD915. One candidate ςE-regulated gene is fkpA. FkpA is a peptidyl-proly-cis-trans isomerase involved in protein folding in the periplasm of E. coli (40). Expression of fkpA in E. coli is positively regulated by ςE, making it the second such gene, along with htrA, that is involved in extracytoplasmic protein folding or stability (8). FkpA shows homology to Mip proteins, which were identified as important for intracellular survival in Legionella pneumophilia and Chlamydia trachomatis (27). Recently, an S. typhimurium fkpA mutant was shown to survive less well in macrophages and epithelial cells (27). GVB311 showed reduced survival in HEp-2 cells compared to wild type, indicating a similar phenotype as the fkpA mutant. The affect of the fkpA mutation on salmonella virulence in vivo was not investigated. Whether HtrA and FkpA can account for all of the intracellular survival (and virulence) functions regulated by ςE will require the construction and analysis of S. typhimurium htrA fkpA mutants.
The rpoE mutant strain was able to invade eucaryotic cells as well as did the wild-type strain. This indicates that ςE is not involved in either regulating the genes required for invasion or the correct assembly or translocation of the invasion factors. The same applies to the ability of salmonellae to kill macrophages, since we found no difference in ability of the three strains to mediate macrophage lysis.
The rpoE mutation greatly reduced the ability of S. typhimurium to cause disease in BALB/c mice by either the natural or parenteral routes of infection. As expected, the htrA mutant BRD915 was also highly attenuated compared with SL1344. It was not possible to differentiate between the virulence levels of GVB311 and BRD915 when administered via the oral route because all mice that received the largest dose survived. However, the LD50 of GVB311 was ca. 20-fold higher than that of BRD915 delivered via the i.v. route. The number of CFU of GVB311 that need to be given to kill mice is very high, and it is likely that some of the mice are dying directly from endotoxinemia rather than from salmonella infection per se.
Analysis of the growth and survival of BRD915 and GVB311 in murine tissues after oral or parenteral challenge confirmed that both strains are defective in survival and/or replication in vivo and that the rpoE mutation has a greater effect than the htrA mutation. Both strains were able to enter PPs after oral inoculation. BRD915 was able to persist at high levels in this tissue throughout the course of the experiment but appears unable to translocate efficiently to deeper tissues as has been previously reported (12). GVB311 was only found in high numbers in the PPs at day 7 postinfection. This result is not due to large numbers of organisms present in a single mouse because the PPs of the four mice sampled contained a similar number of organisms. The reason that only low numbers of GVB311 were found at day 3 is not known. It may be that higher numbers were present but that the organisms were fragile and were damaged during homogenization or that they required prolonged incubation in vitro for the colonies to appear.
The htrA mutant was able to colonize the liver and spleen efficiently after inoculation directly into the bloodstream, but thereafter the numbers of organisms slowly decreased, indicating that the organism cannot replicate in these tissues or that bacterial killing exceeds replication as previously reported (6). In contrast only a tiny percentage of the GVB311 inoculated i.v. remained 2 days after challenge. This may be because the organism is unable to enter the liver and spleen or, more likely, because the organisms are rapidly killed, as suggested from the studies in macrophages. Our results suggest that genes regulated by ςE other than htrA are required for full virulence of salmonella (at least in mice) after oral and systemic infection. The low levels of GVB311 in murine tissues following oral or i.v. immunization probably accounts for the poor immunogenicity of this strain.
We cannot conclude definitively why the rpoE strain is so attenuated at present. This is probably due to a combination of the phenotypes that it exhibits in vitro. As mentioned previously, the ability of strains to survive and grow within macrophages correlates with virulence. GVB311 is defective in this regard, but whether this is because of (i) its increased sensitivity to oxidizing agents, (ii) antimicrobial peptides, (iii) the reduced ability of GVB311 to grow when carbon sources other than glucose are used, (iv) a combination of these effects, or (v) some other defect(s) is not known. The sensitivity of salmonella strains to oxidizing agents in vitro does not necessarily correlate with the loss of virulence. For example, mutations in soxS and kat render S. typhimurium more sensitive to paraquat and H2O2, respectively, but does not affect the ability of the organism to survive in macrophages or cause infection in mice (4, 16).
The structural genes so far known to be positively regulated by ςE, htrA and fkpA, both affect intramacrophage survival and are involved with protein folding and degradation outside of the cytoplasm. Presumably, the inability to degrade, fold, or refold extracytoplasmic proteins that have been damaged in vivo is a major reason why rpoE strains are attenuated.
It will be interesting to learn which signals lead to activation of ςE in vivo. The ςE-dependent htrA promoter is activated upon the entry of salmonellae into epithelial and macrophage cell lines (5). However, the cue(s) from the eucaryotic cell that trigger this response are unknown. A strong local and serum antibody response to the tetanus toxin fragment C antigen is seen in mice orally immunized with attenuated salmonellae expressing fragment C from the htrA promoter. These data suggest that ςE and ςE-regulated genes are highly upregulated within salmonellae in vivo.
In E. coli, ςE activity is regulated positively at the transcriptional level and negatively at the posttranslational levels. Negative regulation of ςE is mediated by the anti-sigma factors RseA and RseB (and possibly RseC) (41). RseA is situated in the cytoplasmic membrane and may have periplasmic and cytoplasmic domains. It is thought that the cytoplasmic domain binds ςE and prevents it from interacting with RNA polymerase, whereas the periplasmic domain interacts with RseB. RseB is thought to sense changes in protein content in the cell envelope that are brought about by environmental stress. This is transmitted to RseA, causing a conformational change that releases ςE, which can then bind to specific recognition sequences, allowing transcription of htrA and other genes in its regulon (10, 41). We are currently analyzing how Rse regulation of ςE affects the ability of salmonellae to interact with their host.
ACKNOWLEDGMENTS
This work was supported by a grant 17/P05639 from the BBSRC.
We thank D. O’Connor, Southampton University, Southampton, United Kingdom, for the gift of the BPI P2 peptide.
REFERENCES
- 1.Baumler A J, Heffron F. Microbial resistance to macrophage effector functions: strategies for evading microbial mechanisms and scavenging nutrients within mononuclear phagocytes. In: Roth J A, Bolin C A, Brogden K A, Minion F C, Wannemuehler M J, editors. Virulence mechanisms of bacterial pathogens. Washington, D.C: ASM Press; 1995. pp. 115–132. [Google Scholar]
- 2.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier N A, Bossie S, Chen C-Y, Fang F C, Guiney D G, Libby S J. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect Immun. 1997;65:3725–3730. doi: 10.1128/iai.65.9.3725-3730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier N A, Libby S J, Xu Y S, Loewen P C, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for salmonella virulence in mice. J Clin Invest. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlioz A, Touati D. Isolation of superoxide dismutase in Escherichia coli: is superoxide dismutase essential for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatfield S N, Strahan K, Pickard D, Charles I, Hormaeche C, Dougan G. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb Pathog. 1992;12:145–151. doi: 10.1016/0882-4010(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen L M, Kaniga K, Galen J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 8.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 9.DeGroote M A, Ochsner U A, Shiloh M U, Nathan C, Mccord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y S, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DelasPenas A, Connolly L, Gross C A. The ςE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of ςE. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Kaper J J. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunstan S J, Simmons C P, Strugnell R A. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect Immun. 1998;66:732–740. doi: 10.1128/iai.66.2.732-740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson J W, Gross C A. Identification of the γE subunit of Escherichia coli RNA polymerase: a second alternative γ factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 14.Erickson J W, Vaughn V, Walter W A, Neidhart F C, Gross C A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987;1:419–432. doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- 15.Everest P H, Frankel G, Li J, Lund P, Chatfield S N, Dougan G. Expression of LacZ from the htrA, nirB and groE promoters in a Salmonella vaccine strain: influence of growth in mammalian cells. FEMS Microbiol Lett. 1994;126:97–102. doi: 10.1111/j.1574-6968.1995.tb07398.x. [DOI] [PubMed] [Google Scholar]
- 16.Fang F C, Vazquez-Torres A, Xu Y. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect Immun. 1997;65:5371–5375. doi: 10.1128/iai.65.12.5371-5375.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 19.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gort A S, Inzuka M. Balance between endogenous superoxide stress and antioxidant defenses. J Bacteriol. 1998;180:1402–1410. doi: 10.1128/jb.180.6.1402-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groisman E A. How bacteria resist killing by host-defense peptides. Trends Microbiol. 1994;2:444–449. doi: 10.1016/0966-842x(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 22.Groisman E A. Bacterial responses to host-defense peptides. Trends Microbiol. 1996;4:127–128. doi: 10.1016/0966-842x(96)30013-9. [DOI] [PubMed] [Google Scholar]
- 23.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 24.Gunn J S, Miller S I. phoP-phoQ activates transcription of pmrAB, encoding a 2-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Güssow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res. 1989;17:4000. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Haigh, R. Personal communication.
- 26.Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes ςE, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horne S M, Kottom T J, Nolan L K, Young K D. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect Immun. 1997;65:806–810. doi: 10.1128/iai.65.2.806-810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosieth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 29.Johnson K, Charles I, Dougan G, Pickard D, Ogaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 30.Kolter R, Inzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 31.Li S R, Dorrell N, Everest P H, Dougan G, Wren B W. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement htrA isogenic mutant. Infect Immun. 1996;64:2088–2094. doi: 10.1128/iai.64.6.2088-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindgren S W, Stojiljkovic I, Heffron F. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:4197–4201. doi: 10.1073/pnas.93.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli—a sigma-32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D W, Holloway B W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa algU shows sequence similarities with a Bacillus sigma-factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma-factor algU regulating conversion to mucoidy in Pseudomonas aeruginosa—relationship to ςE and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller R A, Britigan B E. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival in macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Missiakas D, Betton J-M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 41.Missiakas D, Mayer M P, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli ςE RpoE heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 42.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oka A, Sugisaki M, Takanmai M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 44.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 45.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigmaE (sigma 24) Heat-shock sigma-factor of Escherichia coli. EMBO J. 1995;14:1043–1051. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- 47.Roop R M, Fletcher T W, Sriranganathan N M, Boyle S M, Schurig G G. Identification of an immunoreactive Brucella abortus htrA stress-response protein homolog. Infect Immun. 1994;62:1000–1007. doi: 10.1128/iai.62.3.1000-1007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouviere P E, Penas A L, Mecsas J, Lu C Z, Rudd K E, Gross C A. RpoE, the gene encoding the 2nd heat-shock sigma-factor, sigmaE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Strauch K L, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell-envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strugnell R A, Dougan G, Chatfield S N, Charles I, Fairweather N, Tite J, Li J, Beesley J, Roberts M. Characterization of a Salmonella typhimurium aro vaccine strain expressing the P.69 antigen of Bordetella pertussis. Infect Immun. 1992;60:3994–4002. doi: 10.1128/iai.60.10.3994-4002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tacket C O, Sztein M B, Losonsky G A, Wasserman S S, Nataro J P, Edelman R, Pickard D, Dougan G, Chatfield S N, Levine M M. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect Immun. 1997;65:452–456. doi: 10.1128/iai.65.2.452-456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 53.Weiss J, Elsbach P, Olsson I, Ogata S. Purification and characterization of a potent bactericidal and membrane active protein from the granules of human polymorphonuclear leukocytes. J Biol Chem. 1978;253:2664–2672. [PubMed] [Google Scholar]
- 54.Yamamoto T, Hanawa T, Ogata S, Kamiya S. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress-induced by macrophage phagocytosis and to extracellular environmental stress. Infect Immun. 1996;64:2980–2987. doi: 10.1128/iai.64.8.2980-2987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto T, Hanawa T, Ogata S, Kamiya S. The Yersinia enterocolitica gsrA stress protein, involved in intracellular survival, is induced by macrophage phagocytosis. Infect Immun. 1997;65:2190–2196. doi: 10.1128/iai.65.6.2190-2196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu H, Boucher J C, Hibler N S, Deretic V. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma-factor algU (SigmaE) Infect Immun. 1996;64:2774–2781. doi: 10.1128/iai.64.7.2774-2781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]