Abstract
Background
Immunosuppressive M2 macrophages in the tumor microenvironment (TME) can mediate the therapeutic resistance of tumors, and seriously affect the clinical efficacy and prognosis of tumor patients. This study aims to develop a novel drug delivery system for dual-targeting tumor and macrophages to inhibit tumor and induce macrophage polarization.
Methods
The anti-tumor effects of methyltransferase like 14 (METTL14) were investigated both in vitro and in vivo. The underlying mechanisms of METTL14 regulating macrophages were also explored in this study. We further constructed the cyclic (Arg-Gly-Asp) (cRGD) peptide modified macrophage membrane-coated nanovesicles to co-deliver METTL14 and the TLR4 agonist.
Results
We found that METTL14 significantly inhibits the growth of tumor in vitro. METTL14 might downregulate TICAM2 and inhibit the Toll-like receptor 4 (TLR4) pathway of macrophages, meanwhile, the combination of METTL14 and the TLR4 agonist could induce M1 polarization of macrophages. Macrophage membrane-coated nanovesicles are characterized by easy modification, drug loading, and dual-targeting tumor and macrophages, and cRGD modification can further enhance its targeting ability. It showed that the nanovesicles could improve the in vivo stability of METTL14, and dual-target tumor and macrophages to inhibit tumor and induce M1 polarization of macrophages.
Conclusions
This study anticipates achieving the dual purposes of tumor inhibition and macrophage polarization, and providing a new therapeutic strategy for tumors.
Keywords: Cell membrane-coated nanovesicles, Targeted drug delivery system, Tumor microenvironment, Macrophage polarization, Osteosarcoma
Graphical abstract
Highlights
-
•
Methyltransferase like 14 (METTL14) significantly inhibits the growth of tumor in vitro.
-
•
The combination of METTL14 and TLR4 agonist could induce M1 polarization of macrophages.
-
•
We constructed the cyclic (Arg-Gly-Asp) (cRGD) peptide modified macrophage membrane-coated nanovesicles to co-deliver METTL14 and TLR4 agonist.
-
•
This nanovesicles could dual-target tumor and macrophages to inhibit tumor and induce M1 polarization of macrophages.
1. Introduction
Tumor microenvironment (TME) refers to the surrounding microenvironment of tumor cells, including surrounding cells, signaling molecules, and extracellular matrix [1]. A large number of immune cells infiltrate the TME, but tumor cells achieve immune escape and change the surrounding microenvironment. Therefore, it is necessary to reveal the characteristics of TME in tumors, and further develop novel targeted anti-tumor strategies to remodel immunosuppressive TME and improve the efficacy of tumor treatment. It is of great significance to improve the cure and survival rates of tumor patients. So far, there is no effective anti-tumor therapy that can both inhibit tumor and regulate TME synergistically.
Tumor associated macrophages (TAM) serve as an important participant in tumor immunosuppression [2]. TAMs can be divided into two subtypes: M1 type TAMs mainly play an anti-tumor role, while M2 could promote tumor progression. Based on the functional characteristics of TAMs, inducing polarization of M2 to M1 type is an important strategy to remodel TME for tumor therapy [3]. Toll-like receptor (TLR) is the most widely studied pattern recognition receptor in the natural immune system. TLR4 exists on a variety of cell surfaces, such as macrophages, endothelial cells, epithelial cells, and so on, which plays an important role in infiltrating immune cells or tumor cells in TME. The intracellular domain of TLR4 is responsible for the recruitment of downstream adaptor proteins including myeloid differentiation factor 88 (MyD88), β interferon-TIR domain adaptor protein (TRIF), and TRIF-associated adaptor molecules (TRAM/TICAM2). M1 polarization of TAMs could be induced by interferon-γ (IFN-γ) or lipopolysaccharides (LPS) and participate in a series of inflammatory responses [2]. It has been reported that TLR agonists might induce M1 polarization of TAMs to play an anti-tumor role [4]. Paclitaxel inhibits tumor growth by activating the TLR4 signaling pathway to reprogram TAMs into M1 type [5]. Therefore, the TLR4 signaling pathway is involved in regulating M1 polarization of macrophages. However, TLR4 agonists alone have poor therapeutic effects on tumors, so it should be used in combination with other anti-tumor agents.
N6-methyladenosine (m6A) has been proved to be one of the most common internal chemical modifications in mRNA [6]. The abundance and distribution of m6A are regulated by methyltransferases (writers), demethylases (erasers), and RNA binding proteins (readers) [7,8]. Writers mainly consist of METTL3 and METTL14, and erasers include FTO and ALKBH5. Readers include YTHDC1/2, YTHDF1/2/3, IGF2BP1/2/3, and HNRNPC [9]. Nowadays, it is indicated that aberrant expressions of m6A regulators get involved in the progression of tumors [[10], [11], [12], [13]]. Whereas, the expressions of m6A regulators, as well as the immune, infiltrates and macrophage polarization in tumor have not been explored thoroughly. It is reported that the loss of METTL14 in TAMs promotes CD8+ T cell dysfunction and tumor growth [14]. Ablation of METTL14 in myeloid cells might lead to the overactivation of TLR4/NF-κB signaling, and exacerbate macrophage responses to acute bacterial infection in mice [15].
More effective drug delivery systems are in great demand for anti-tumor strategies [16]. Most of conventional nanoparticles (NPs) are easily recognized and cleared by the immune system, and the functional design of NPs is complicated and laborious [17]. In this situation, biomimetic NPs coated with cell membranes have received growing interest [18,19]. Because of the reserved antigens and cell membrane structure, membrane-coated nanovesicles could obtain the following advantages: effective targeting, prolonged circulation, immune escaping, and so on [20,21]. Moreover, the membranes of red blood cells, macrophages, platelets, and even tumor cells, could be utilized for different functional requirements [[22], [23], [24]]. Macrophage membrane-coated nanovesicles show the same characteristic antigenic properties as macrophages, indicating their potential for obtaining homologous targeting ability [25]. Cyclic (Arg-Gly-Asp) (cRGD) peptide as both a tumor-targeting and penetrating peptide, could be modified on the surface of nanovesicles, which might promote drug accumulation in tumor [26,27].
In this study, we found that METTL14 significantly inhibits the growth of tumor in vitro, but METTL14 is easily degraded in vivo and difficult to be used for the treatment of tumor in vivo. METTL14 downregulates TICAM2 and inhibits the TLR4 pathway of macrophages. Accordingly, we developed macrophage membrane-coated nanovesicles to co-deliver METTL14 and the TLR4 agonist. We further investigated that the nanovesicles might improve the in vivo stability of drugs, inhibit tumor, and induce macrophage polarization. This study intends to clarify the therapeutic roles of the nanovesicles in dual-targeting tumor and macrophages and achieve the dual purposes of tumor inhibition and TME remodeling, which provides a new therapeutic strategy for tumors.
2. Materials and methods
2.1. Patient samples
This study was approved by the institutional review board and the medical ethics committee of Wuhan Union Hospital, Huazhong University of Science and Technology. Each participant provided written informed consent. We collected the medical data and samples of three patients with osteosarcoma in our hospital.
2.2. GEPIA and TIMER database analysis
The database GEPIA (http://gepia.cancer-pku.cn/index.html) was used for exploring the expression of m6A regulators in tumor. In the TIMER database (https://cistrome.shinyapps.io/timer/), the correlations between METTL14 and the abundance of immune infiltrates were conducted by “Gene module”. Moreover, the correlations between METTL14 and gene markers were detected by “Correlation module”.
2.3. Cell culture and treatment
Tumor cell line (MNNG/HOS, osteosarcoma cell line) and macrophage (RAW264.7) were obtained from the Cell Bank of the China Academy of Sciences (Shanghai, China). Tumor cells were cultured in α-MEM medium and macrophages were cultured in DMEM medium with high glucose with 10% FBS. The METTL14 sequence was synthesized and subcloned into the pCDNA3.1 vector (GeneChem, Shanghai, China). Cells were transfected with pMETTL14 using Lipofectamine 3000 reagent (Invitrogen, USA). The fluorescence images in cells were recorded by a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
2.4. Cell viability and proliferation assay
Cells were seeded onto 96-well plates with 100 μl medium. Cells were then treated and incubated in 90 μl medium with 10 μl CCK-8 solution (Dojindo Laboratories Co. Ltd, Kumamoto, Japan). After incubation for 4 h, the absorbance was recorded at 450 nm.
Cells (1 × 103 cells/well) were seeded into 6-well plates and cultured for two weeks. Then cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet.
2.5. Single-cell RNA sequencing and bioinformatic analysis
The raw scRNA-seq data of GSE152048 and GSE162454 (osteosarcoma tissues) were utilized for bioinformatic analysis. The identified clusters were visualized on the 2D map with the t-distributed t-SNE or UMAP method. The Louvain approach was used to compute a shared nearest-neighbor graph for further clustering analysis. Differentially expressed genes (DEGs) with strong group-level discrimination were found in Seurat using the standard non-parametric Wilcoxon rank sum test. Based on the DEGs and well-known cellular markers from the literature, the cell clusters were annotated [28].
2.6. Xenograft tumor model
Nude mice (BALB/c-nu, 5–6 weeks, females) were injected subcutaneously with 5 × 106 MNNG/HOS cells. Tumor volume (mm3) = ab2/2, the length (a) and the width (b). Subsequently, mice were divided into different groups randomly and prepared for treatment with 1.5 mg/kg bodyweight pMETTL14, RS09, pMETTL14+RS09, pMETTL14@cRGD-M, and (pMETTL14+RS09)@cRGD-M or the same volume of PBS (control group) via caudal vein when the subcutaneous tumor volume reached 50 mm3. After three weeks, the animals were sacrificed and the tumors were harvested. Hematoxylin-eosin (HE) staining was conducted to stain tumors and organ tissues. Immunohistochemistry (IHC) staining for Ki67 was conducted to stain proliferative cells and TUNEL staining was conducted to stain apoptotic cells. Immunofluorescence (IF) staining of tumor tissues for METTL14, CD163, and iNOS was performed. All antibodies were purchased from Proteintech (Wuhan, China).
2.7. Preparation of (pMETTL14+RS09)@cRGD-M
The METTL14 sequence was synthesized and subcloned into the pCDNA3.1 vector (GeneChem, Shanghai, China), named pMETTL14. The drugs RS09 and DSPE-PEG2000-cRGD were purchased from MedChemExpress (MCE, Shanghai, China). The macrophage cell membrane was extracted by the process of hypotonic lysis, mechanical membrane disruption, and differential centrifugation. Then, pMETTL14 and RS09 were coated into cell membrane nanovesicles by coextruding through a 200 nm polycarbonate membrane to form (pMETTL14+RS09)@M. Subsequently, cRGD micelle was formed by incubating 1 mg phospholipid (DSPE)-PEG2000-cRGDyK with 1 ml ddH2O at 60 °C for 20 min and then mixed the cRGD micelle with obtained (pMETTL14+RS09)@M membrane nanovesicles. The mixture was stirred at 40 °C for 2 h until the cRGD micelles were inserted into the membranes of nanovesicles to form (pMETTL14+RS09)@cRGD-M.
2.8. Characterization of (pMETTL14+RS09)@cRGD-M
Transmission electron microscopy (TEM) was performed by JEM-2000EXII TEM (JEOL, Tokyo, Japan) to evaluate the morphological structures. Dynamic light scattering was conducted by Zetasizer Nano ZS (Malvern Zetasizer, Malvern, UK) to test the size of the nanovesicles. Zeta potential was conducted to evaluate the stability of colloidal dispersions.
2.9. Drug loading and release study
A solution containing pMETTL14 or RS09 (0.01–0.2 mg) and 0.1 mg of cRGD-M was prepared in 1 ml PBS (pH 7.4) and stirred at 4 °C for 8 h and sonicated for 0.5 h in dark. pMETTL14@cRGD-M or RS09@cRGD-M were collected by ultra-centrifugation and washed several times with PBS until the supernatant became color free. The amount of unbound pMETTL14 or RS09 in the solution was determined by measuring the absorbance using a UV–Vis spectrophotometer (Beckman Coulter, Indianapolis, IN, USA), respectively. The drug loading efficiency (%) and the drug loading content (mg/mg) were calculated as described before [29]:
| Loading Efficiency (%) = MEncapsulated / MUsed × 100% |
where MEncapsulated is the net weight (mg) of encapsulated agent inside the nanoparticles, and MUsed represents the total weight (mg) of agents used.
| Loading Content (mg/mg) = MAgent / MTotal |
where MAgent is the weight (mg) of pMETTL14 or RS09 encapsulated inside the nanoparticles, while the MTotal represents the total weight (mg) of agent-laden nanocarriers, including both the encapsulated agents and the empty cRGD-M nanovesicles.
The drug release behavior was investigated with pH = 7.4 or pH = 5.6. The amount of pMETTL14 or RS09 released into the solution was determined by measuring the absorbance, respectively. The cumulative release percentage (%) was calculated with the formulas:
| Cumulative Release Percentage (%) = MReleased / MLoaded × 100% |
where MReleased represents the total weight (mg) of the released drug and MLoaded is the total weight (mg) of the initial loaded drug.
2.10. In vitro cellular uptake
pMETTL14 was labeled with FITC (green), the membrane was labeled with DiD (red), and the nucleus was stained with DAPI (blue). Tumor cells and macrophages were incubated with nanovesicles for 12 h. The images were obtained via a fluorescence microscope.
2.11. In vivo imaging and biodistribution analysis
When the tumor volumes of the xenograft tumor model reached 100–200 mm3, the mice were injected with nanovesicles via the tail vein. After injection for 24 h, the in vivo imaging system (IVIS, CRI Maestro, USA) was performed to investigate the accumulation site of nanovesicles.
2.12. Statistical analysis
All experiments were repeated at least three times. The results were presented as mean ± standard deviation (SD) via GraphPad 6.0 software. P values were calculated using one-way analysis of variance. A p-value of <0.05 was considered as a statistically significant result (*p < 0.05 and **p < 0.01).
3. Results
3.1. The anti-tumor effects of METTL14 in vitro and in vivo
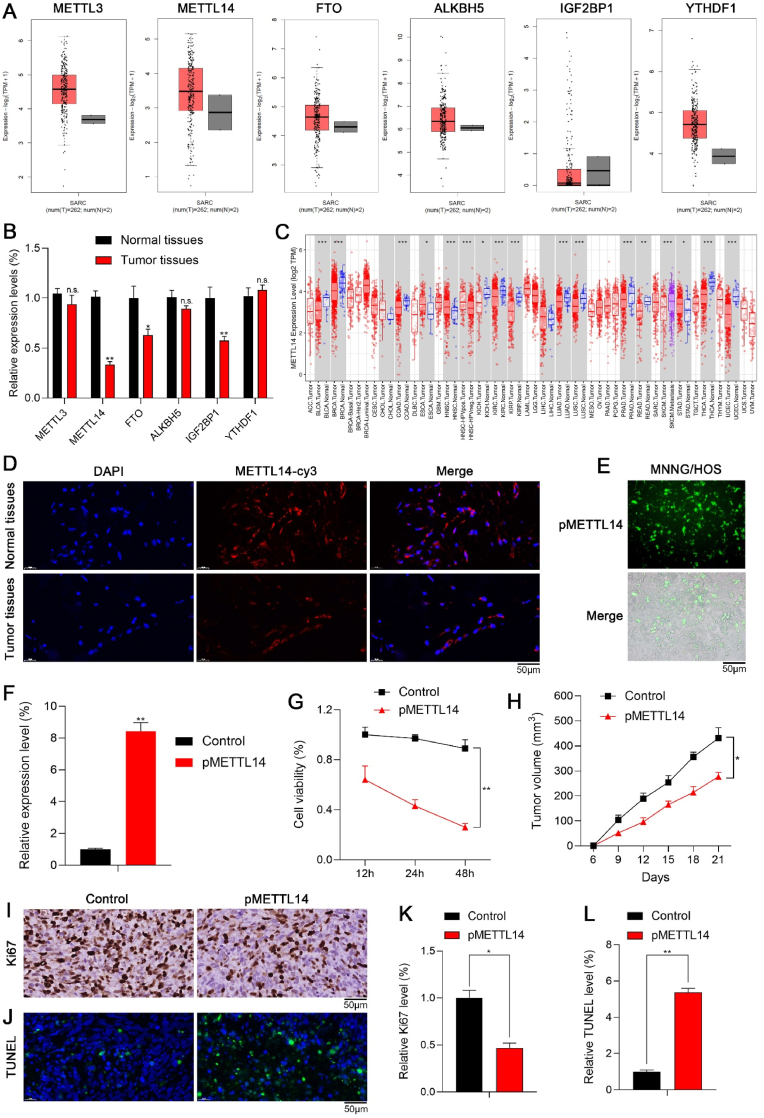
We firstly investigated the dysregulated expression levels of m6A regulators in tumors. Fig. 1A showed the expression levels of m6A regulators in sarcoma via the GEPIA database. Compared with normal tissues, METTL14 was one of the most significantly decreased m6A regulators in tumor tissues (Fig. 1B), which might play a vital role in tumor progression. Furthermore, METTL14 was remarkably downregulated in most types of tumors via the TIMER database (Fig. 1C). Immunofluorescence (IF) staining of METTL14 in tumor tissues was consistent with the above results (Fig. 1D). Therefore, we aimed to reveal the effects of dysregulated METTL14 in tumor in the following experiments.
Fig. 1.
The anti-tumor effects of METTL14 in vitro and in vivo. (A) The expression levels of m6A regulators in sarcoma via GEPIA database. (B) qPCR showed the expression levels of m6A regulators in normal tissues and tumor tissues (n.s.: no significance; **p < 0.01; *p < 0.05). (C) METTL14 was remarkably downregulated in most types of tumors via TIMER database (***p < 0.001; **p < 0.01; *p < 0.05). (D) IF staining of METTL14 in normal tissues and tumor tissues. (E) The fluorescence analysis indicated that pMETTL14 was successfully transfected in tumor cells. (F) qPCR showed that METTL14 was significantly upregulated in tumor cells of pMETTL14 group (**p < 0.01). (G) The CCK-8 assay indicated that the cell viability of tumor cells reduced significantly in pMETTL14 group (**p < 0.01). (H) The tumor growth pMETTL14 group was remarkably inhibited compared with control group (*p < 0.05). (I, K) The number of Ki67 positive cells via IHC staining in pMETTL14 group (scale bar: 50 μm; *p < 0.05). (J, L) The number of TUNEL positive cells in pMETTL14 group (scale bar: 50 μm; **p < 0.01).
METTL14 was overexpressed by transfecting tumor cells with pCDNA-METTL14 (pMETTL14). The fluorescence analysis indicated that pMETTL14 was successfully transfected in tumor cells (Fig. 1E), and METTL14 was significantly upregulated in tumor cells of pMETTL14 group (Fig. 1F). Then we detected the anti-tumor effects of METTL14. In the CCK-8 assay, the cell viability of tumor cells reduced significantly in pMETTL14 group (Fig. 1G). In the xenograft tumor model, the tumor growth of pMETTL14 group was significantly inhibited compared with control group (Fig. 1H). Furthermore, the number of Ki67 positive cells via immunohistochemistry (IHC) staining was remarkably decreased (Fig. 1I and 1K), and the number of TUNEL positive cells was remarkably increased (Fig. 1J and 1L) in pMETTL14 group. However, we observed that METTL14 significantly inhibits the growth of tumor in vitro, but the anti-tumor effect of METTL14 in vivo was unsatisfactory. The underlying mechanism was not yet clear.
3.2. The characteristics of TME and METTL14 might regulate immunosuppressive states of TME
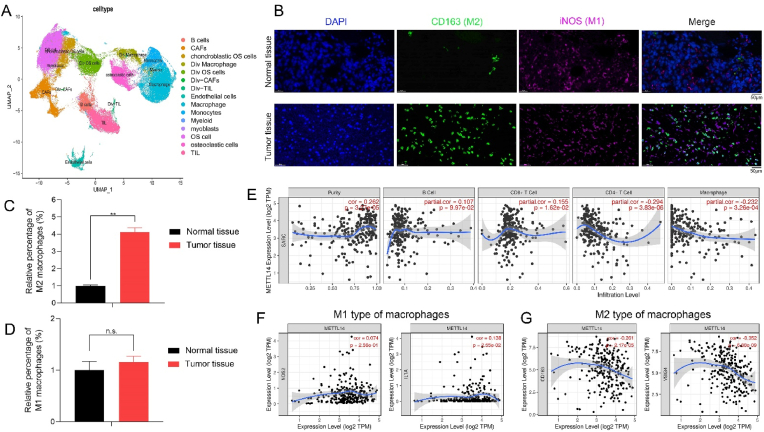
To reveal the characteristics of TME, we conducted single-cell RNA sequencing and bioinformatic analysis to investigate the cellular constitution of tumor lesions by using GSE152048 and GSE162454 (data sets of osteosarcoma patients) according to our previous study [28]. After quality control, the cells of GSE152048 and GSE162454 were clustered into 14 major clusters via the T-distributed stochastic neighbor embedding (t-SNE) method (Fig. 2A). To further confirm the above results, we utilized IF staining and a significantly increased number of M2 macrophages (CD163) was observed in tumor tissues when compared with normal tissues (Fig. 2B and 2C). No significant change in the number of M1 macrophages in these tissues was shown (Fig. 2D). It suggested that immunosuppressive M2 macrophages in TME might contribute to the unsatisfactory anti-tumor effects of METTL14 in vivo.
Fig. 2.
The characteristics of TME and METTL14 might regulate immunosuppressive states in TME. (A) The t-SNE plot of 14 identified cell types in tumor lesions. (B, C, D) The relative proportion of macrophages in tumor tissues and normal tissues was shown by IF staining (CD163: M2 type of macrophages; iNOS: M1 type of macrophages; scale bars: 50 μm; **p < 0.01; n.s.: no significance). (E) METTL14 showed significant negative associations with infiltrating levels of CD4+ T cells (r = −0.294, p = 3.83e-06) and macrophages (r = −0.232, p = 3.26e-04) in sarcoma via the TIMER database. (F, G) METTL14 had a significantly positive association with IL1A (r = 0.138, p = 2.55e-02) of M1 type and negative correlations with CD163 (r = −0.261, p = 2.17e-05) and VSIG4 (r = −0.352, p = 6.88e-09) of M2 type of macrophages.
Moreover, METTL14 had significant negative associations with infiltrating levels of CD4+ T cells (r = −0.294, p = 3.83e-06) and macrophages (r = −0.232, p = 3.26e-04) in sarcoma via the TIMER database (Fig. 2E). METTL14 showed a significantly positive association with IL1A (r = 0.138, p = 2.55e-02) of M1 type and negative correlations with CD163 (r = −0.261, p = 2.17e-05) and VSIG4 (r = −0.352, p = 6.88e-09) of M2 type of macrophages (Fig. 2F and 2G). Accordingly, METTL14 was correlated with immune infiltration levels and immunosuppressive states of TEM in tumor, which might be a novel therapeutic target for tumor.
3.3. METTL14 combined with TLR4 agonist inhibit tumor and regulate macrophage polarization
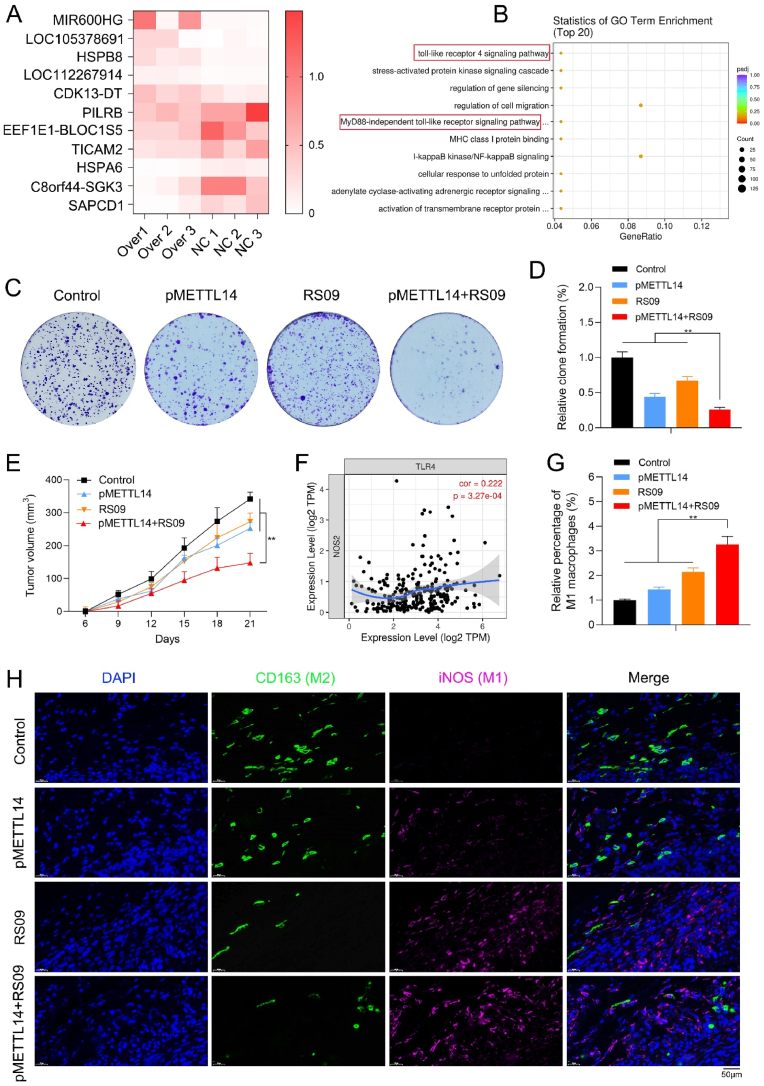
To further investigate the role of METTL14 in macrophage polarization, we conducted the cluster analysis of differentially expressed genes in pMETTL14 and normal groups of macrophages (Fig. 3A) and found that METTL14 significantly downregulated TICAM2 expression of macrophages. The GO enrichment analysis showed that METTL14 was remarkably associated with the Toll-like receptor 4 (TLR4) signaling pathway of macrophages (Fig. 3B). Therefore, METTL14 might downregulate TICAM2 expression and inhibit the TLR4 pathway of macrophages.
Fig. 3.
METTL14 combined with TLR4 agonist inhibit tumor and regulate macrophage polarization. (A) The cluster analysis of differentially expressed genes in pMETTL14 and normal groups of macrophages. (B) The GO enrichment analysis showed that METTL14 was remarkably correlated with the TLR4 signaling pathway of macrophages. (C, D) RS09 is one of the most common TLR4 agonists. The clone formation assay indicated that pMETTL14+RS09 group remarkably decreased tumor cell proliferation than other groups (**p < 0.01). (E) pMETTL14+RS09 significantly reduced the tumor volume when compared with other groups (**p < 0.01). (F) TLR4 showed a remarkably positive association with NOS2/iNOS (r = 0.222, p = 3.27e-04) of M1 type of macrophages via TIMER database. (G, H) The relative proportion of M1 (iNOS) and M2 macrophages (CD163) were observed in tumor tissues of control, pMETTL14, RS09, and pMETTL14+RS09 groups via IF staining (scale bar: 50 μm; **p < 0.01).
We further confirmed whether METTL14 could inhibit tumor and regulate macrophage polarization via the TLR4 pathway. RS09 is one of the most common TLR4 agonists. The clone formation assay showed that pMETTL14+RS09 group remarkably decreased tumor cell proliferation than other groups (Fig. 3C and 3D). It suggested that METTL14 combined with TLR4 agonist exerted high anti-tumor efficacy in vitro. The anti-tumor effects of pMETTL14+RS09 in vivo were further explored. pMETTL14+RS09 significantly reduced the tumor volume when compared with other groups (Fig. 3E). By TIMER database, TLR4 showed a remarkably positive association with NOS2/iNOS (r = 0.222, p = 3.27e-04) of M1 type of macrophages (Fig. 3F). To further confirm the above results, a significantly increased number of M1 macrophages (iNOS) and a decreased number of M2 macrophages (CD163) were observed in tumor tissues of pMETTL14+RS09 group via IF staining (Fig. 3G and 3H). It suggested that METTL14 combined with TLR4 agonist might inhibit tumor and induce macrophage polarization.
3.4. Fabrication and characteristics of (pMETTL14+RS09)@cRGD-M nanovesicles
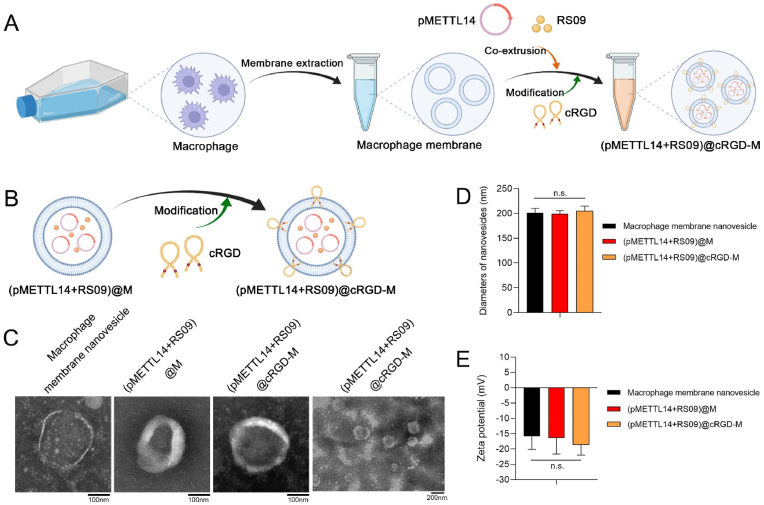
The fabrication of (pMETTL14+RS09)@cRGD-M mainly consists of three steps (Fig. 4A and 4B): (a) extracting macrophage cell membranes; (b) coating pMETTL14 and RS09 into cell membrane nanovesicles by co-extrusion to form (pMETTL14+RS09)@M; (c) forming cRGD modified cell membrane nanovesicles, which were named (pMETTL14+RS09)@cRGD-M. The morphology of the nanovesicles was conducted by TEM, respectively (Fig. 4C). The (pMETTL14+RS09)@cRGD-M nanovesicle had a diameter of around 200 nm (Fig. 4D). Because of the biomimetic cell membrane, the Zeta potential of (pMETTL14+RS09)@cRGD-M was only about −18 mV (Fig. 4E).
Fig. 4.
Fabrication and characteristics of nanovesicles. (A, B) Fabrication procedure of (pMETTL14+RS09)@cRGD-M nanovesicles. (C) The morphology of macrophage membrane nanovesicle, (pMETTL14+RS09)@M, and (pMETTL14+RS09)@cRGD-M was characterized by TEM, respectively (scale bar: 100 nm and 200 nm). (D) The three types of nanovesicles had an approximate diameter of 200 nm (n.s.: no significance). (E) The Zeta potential of three types of nanovesicles (n.s.: no significance).
The drug loading efficiency and the drug loading content of (pMETTL14+RS09)@cRGD-M nanovesicle were further investigated [29]. Reacting 0.08 mg RS09 with 0.1 mg cRGD-M (RS09/cRGD-M = 0.8) was deemed to be the best condition for preparing RS09@cRGD-M, considering both loading efficiency and loading content of the drug, with the former being 32.0% and the latter being 0.21 mg/mg. Similarly, pMETTL14/cRGD-M = 0.8 was determined to be the best condition for preparing pMETTL14@cRGD-M with 46.2% loading efficiency and 0.27 mg/mg loading content. It showed that cRGD-M nanovesicle is an effective nanovesicle for the loading and delivery of RS09 and pMETTL14 to finally prepare (pMETTL14+RS09)@cRGD-M (Supplementary Fig. 1).
Furthermore, the drug release behaviors of (pMETTL14+RS09)@cRGD-M nanovesicles were investigated with pH = 7.4 or pH = 5.6. With pH = 7.4, the nanoparticles exhibited less than 35% release of RS09 and pMETTL14 within 100 h. In contrast, RS09 and pMETTL14 exhibited a staggered release within 24 h with pH = 5.6 (Supplementary Fig. 2).
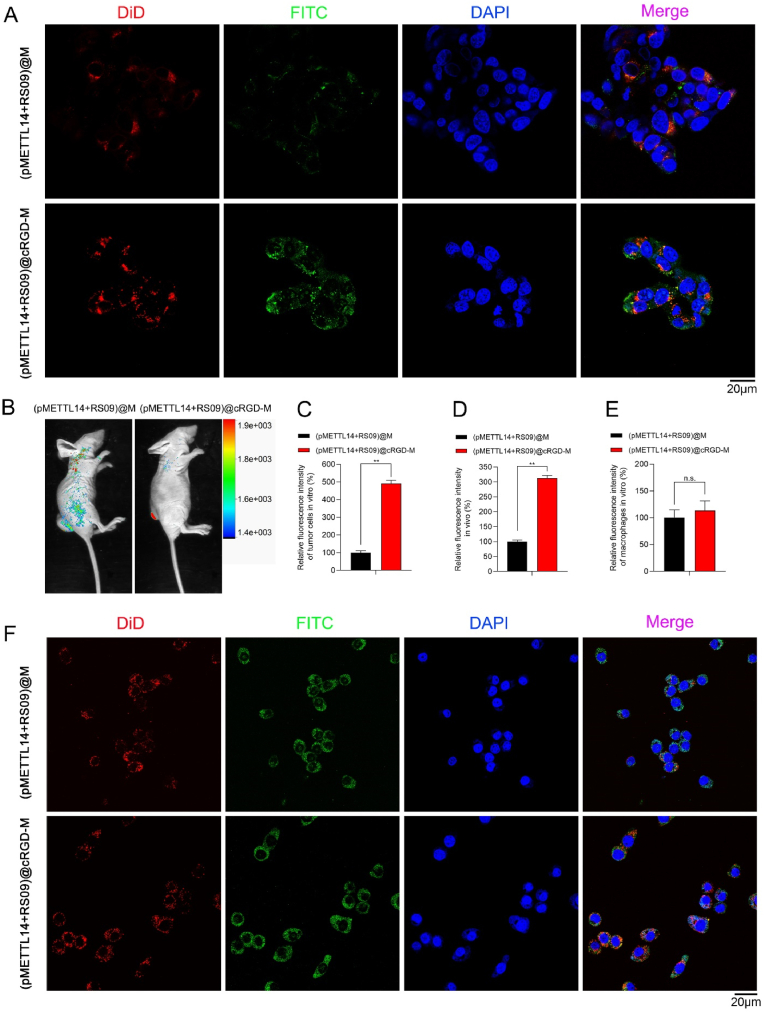
We further explored the dual-targeting effects of (pMETTL14+RS09)@cRGD-M. pMETTL14 was labeled with FITC (green) and the membrane was labeled with DiD (red). After incubated with (pMETTL14+RS09)@M and (pMETTL14+RS09)@cRGD-M, we detected remarkably stronger fluorescence intensity (both green and red) in tumor cells incubated with (pMETTL14+RS09)@cRGD-M than that with (pMETTL14+RS09)@M group (Fig. 5A and 5C). The results indicated that cRGD modification significantly enhanced the tumor-targeting ability of the nanovesicles in vitro. In the xenograft tumor model, (pMETTL14+RS09)@M and (pMETTL14+RS09)@cRGD-M were intravenously injected into nude mice. The fluorescence signal in the tumor sites of (pMETTL14+RS09)@cRGD-M group was remarkably higher than (pMETTL14+RS09)@M group (Fig. 5B and 5D). No organ accumulation of nanovesicles (pMETTL14+RS09)@cRGD-M in vivo was observed (Supplementary Fig. 3). (pMETTL14+RS09)@cRGD-M showed effective tumor-targeting ability both in vitro and in vivo. Moreover, (pMETTL14+RS09)@M and (pMETTL14+RS09)@cRGD-M could also target macrophages in vitro, and no significant difference in the fluorescence signal between these two groups was observed (Fig. 5E and 5F). To sum up, (pMETTL14+RS09)@cRGD-M nanovesicle had the ability of dual-targeting tumor and macrophages. Additionally, cRGD could significantly enhance the tumor-targeting ability of macrophage membrane-coated nanovesicles but have no influence on their macrophage-targeting ability. Macrophage membrane-coated nanovesicles showed homologous targeting ability.
Fig. 5.
The dual-targeting effects of (pMETTL14+RS09)@cRGD-M. (A, C) pMETTL14 was labeled with FITC (green) and the membrane was labeled with DiD (red). Tumor cell-targeting ability of (pMETTL14+RS09)@M and (pMETTL14+RS09)@cRGD-M was investigated in vitro (scale bar: 20 μm; **p < 0.01). (B, D) The tumor accumulation ability of (pMETTL14+RS09)@cRGD-M was investigated in vivo (**p < 0.01). (E, F) Macrophage-targeting ability of (pMETTL14+RS09)@M and (pMETTL14+RS09)@cRGD-M was investigated in vitro (scale bar: 20 μm; n.s.: no significance).
In the xenograft tumor model, pMETTL14 was labeled with FITC (green), and pMETTL14 and pMETTL14@cRGD-M were intravenously injected into nude mice. The fluorescence signal decreased rapidly over time in the pMETTL14 group. In contrast, pMETTL14@cRGD-M gradually cumulated in tumor sites and no significant change in the fluorescence signal was observed over time. It showed that the nanovesicles could improve the in vivo stability of METTL14 (Supplementary Fig. 4).
HE staining of the control, pMETTL14, cRGD-M, and (pMETTL14+RS09)@cRGD-M groups did not show obvious cardiac damage, pulmonary toxicity, inflammatory infiltrates in the spleen, or liver and kidney injury (Supplementary Fig. 5). Accordingly, (pMETTL14+RS09)@cRGD-M could function as a promising drug delivery system for biomedical applications because of its low organ toxicity.
3.5. Effects of (pMETTL14+RS09)@cRGD-M on tumor inhibition and macrophage polarization
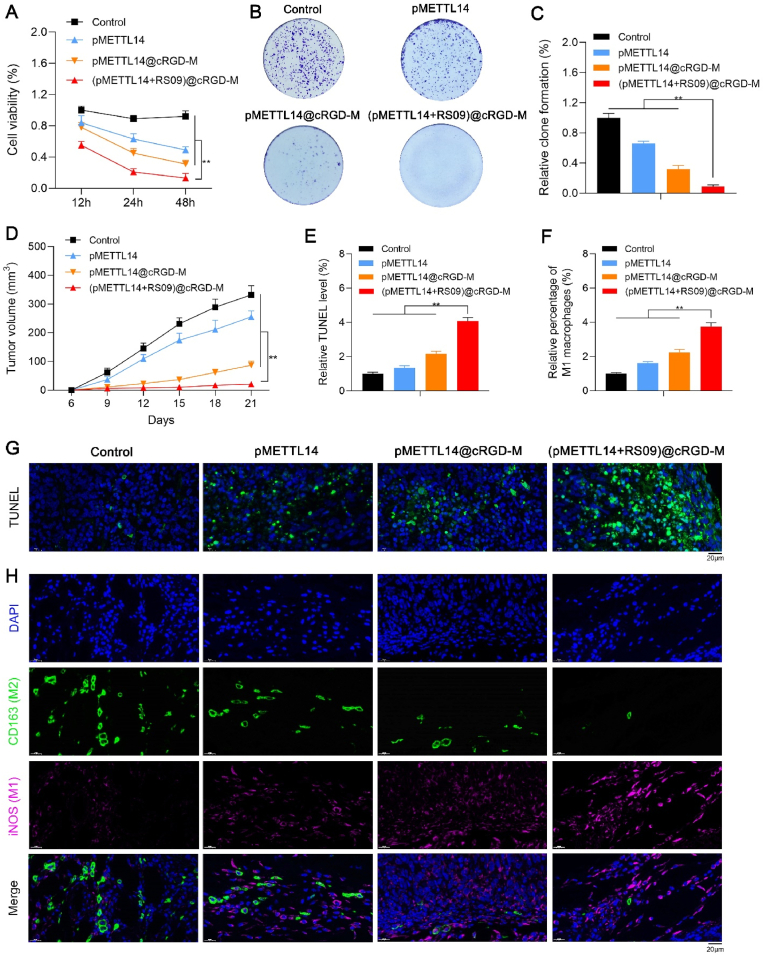
We explored the anti-tumor effects of (pMETTL14+RS09)@cRGD-M in vitro. Tumor cells were incubated with control, pMETTL14, pMETTL14@cRGD-M, and (pMETTL14+RS09)@cRGD-M. The CCK-8 assay showed that (pMETTL14+RS09)@cRGD-M remarkably decreased tumor cell viability than other groups (Fig. 6A). The clone formation assay showed that (pMETTL14+RS09)@cRGD-M remarkably decreased cell proliferation when compared with other groups (Fig. 6B and 6C). It suggested that (pMETTL14+RS09)@cRGD-M exerted high anti-tumor efficacy in vitro.
Fig. 6.
Effects of (pMETTL14+RS09)@cRGD-M on tumor inhibition and macrophage polarization. (A) The CCK-8 assay indicated the role of (pMETTL14+RS09)@cRGD-M in tumor cell validity (**p < 0.01). (B, C) The clone formation assay indicated the role of (pMETTL14+RS09)@cRGD-M in tumor cell proliferation (**p < 0.01). (D) Nude mice were divided into four groups of control, pMETTL14, pMETTL14@cRGD-M, and (pMETTL14+RS09)@cRGD-M, and the tumor volumes were calculated (**p < 0.01). (E, G) TUNEL staining indicated cell apoptosis in tumor tissues (scale bar: 20 μm; **p < 0.01). (F, H) The numbers of M1 (iNOS) and M2 macrophages (CD163) were observed in tumor tissues of control, pMETTL14, pMETTL14@cRGD-M, and (pMETTL14+RS09)@cRGD-M groups via IF staining (scale bar: 20 μm; **p < 0.01).
The anti-tumor effects of (pMETTL14+RS09)@cRGD-M in vivo were also explored. Mice were treated with control, pMETTL14, pMETTL14@cRGD-M, and (pMETTL14+RS09)@cRGD-M for 21 days. Compared with other groups, (pMETTL14+RS09)@cRGD-M significantly reduced the tumor volume (Fig. 6D). The number of TUNEL positive cells was remarkably increased (Fig. 6E and 6G) in (pMETTL14+RS09)@cRGD-M group. It indicated that (pMETTL14+RS09)@cRGD-M had stronger anti-tumor effects in vivo.
More importantly, a significantly increased number of M1 macrophages (iNOS) and a decreased number of M2 macrophages (CD163) were observed in tumor tissues of (pMETTL14+RS09)@cRGD-M group (Fig. 6F and 6H). With the help of macrophage membrane-coated nanovesicles, (pMETTL14+RS09)@cRGD-M could dual-target tumor and macrophages, and further release pMETTL14 and RS09 in TME. METTL14 inhibited tumor and regulate macrophage polarization via the TLR4 pathway. Accordingly, METTL14 combined with the TLR4 agonist (RS09) could induce M1 polarization of macrophages in TME, thereby remodeling immunosuppressive TME. To sum up, (pMETTL14+RS09)@cRGD-M could efficiently inhibit tumor and induce macrophage polarization.
4. Discussion
By investigating the dysregulated expressions of m6A regulators in tumors, this study revealed that METTL14 was one of the most significantly decreased m6A regulators in tumor tissues, which might play a vital role in tumor progression. Therefore, we aimed to reveal the effects of dysregulated METTL14 in tumor in the following experiments. We observed that METTL14 significantly inhibits the growth of tumor in vitro, but the anti-tumor effect of METTL14 in vivo was not ideal. The underlying mechanism is not yet clear. To sum up, the unsatisfied anti-tumor effect of METTL14 in vivo might due to the following two aspects: (a) METTL14 was easily degraded in vivo and difficult to be used for the treatment of tumor in vivo; (b) immunosuppressive states of TME could mediate the therapeutic resistance of tumor. Accordingly, we aimed to develop new strategies to (a) improve the in vivo stability of METTL14, and (b) achieve the dual purposes of tumor inhibition and TME remodeling.
Inducing polarization of M2 type to M1 type is an effective strategy to remodel TME, which might be a key target for tumor therapy. In this study, the GO enrichment analysis indicated that METTL14 was significantly related with the TLR4 signaling pathway of macrophages. METTL14 might downregulate TICAM2 and inhibit the TLR4 pathway of macrophages, which reduced the anti-tumor effects of METTL14 in vivo. Accordingly, it is necessary to combine the TLR4 agonist to induce M1 polarization of macrophages and remodel the TME. In our study, we further confirmed that METTL14 combined with the TLR4 agonist (RS09) could induce M1 polarization of macrophages and enhance the anti-tumor effects in vivo.
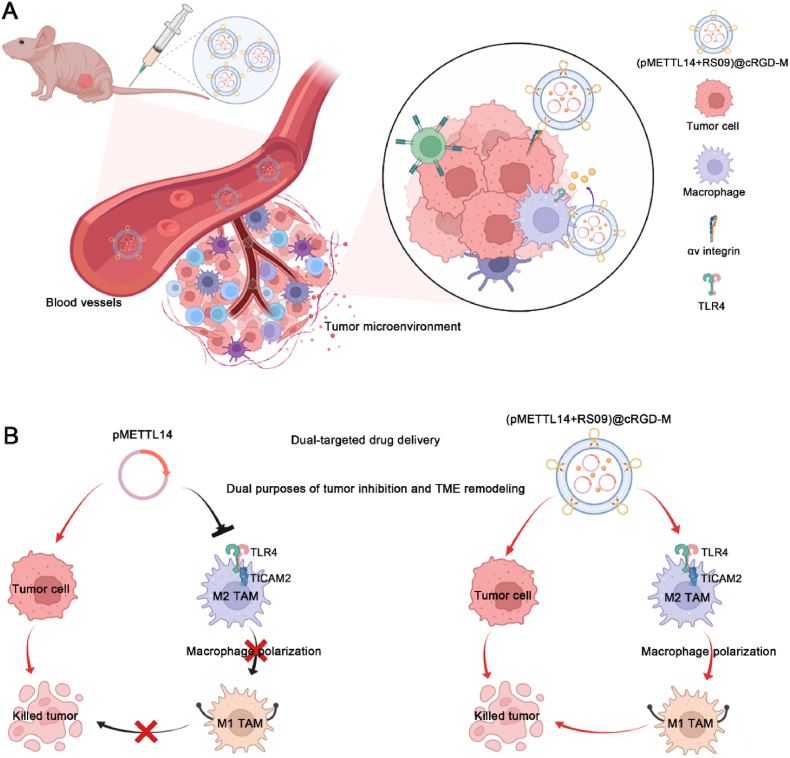
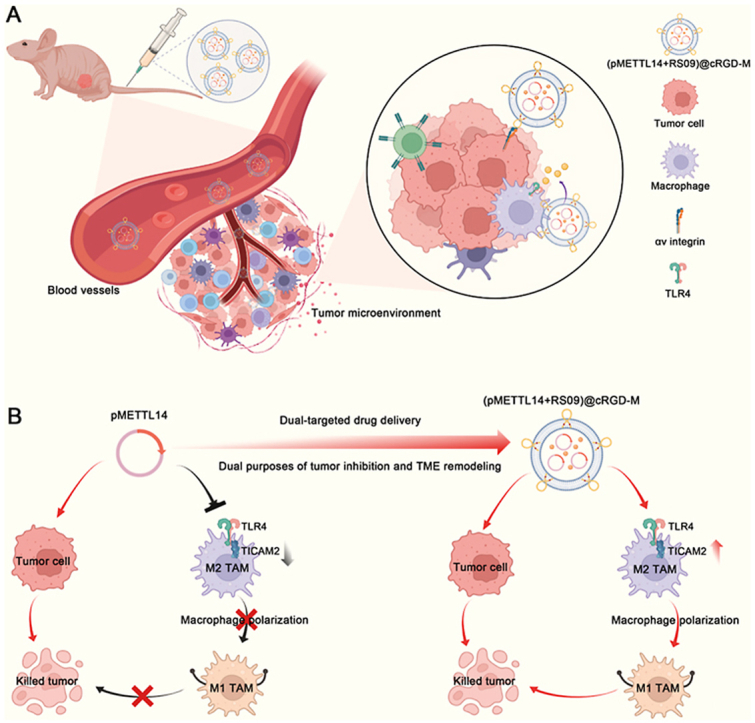
Macrophage membranes carry adhesion molecules, therefore, coating nanoparticles with macrophage membrane could actively target tumor tissues and avoid immune recognition [30]. Macrophage membrane-coated nanovesicles are characterized by easy modification, drug loading, and dual-targeting tumor and macrophage, and cRGD modification can further enhance its targeting ability. Accordingly, our study further constructed cRGD modified macrophage membrane-coated nanovesicles to co-deliver METTL14 and the TLR4 agonist. The results confirmed that (pMETTL14+RS09)@cRGD-M nanovesicles had the ability of dual-targeting tumor and macrophages (Fig. 7A). This study further showed that (pMETTL14+RS09)@cRGD-M nanovesicles could improve the in vivo stability of METTL14, target to inhibit tumor and induce M1 polarization of macrophages (Fig. 7B). Therefore, this study achieved the dual purposes of tumor inhibition and macrophage polarization via macrophage membrane-coated nanovesicles.
Fig. 7.
Overview of (pMETTL14+RS09)@cRGD-M in dual-targeted tumor therapy. (A) (pMETTL14+RS09)@cRGD-M nanovesicles had the ability of dual-targeting tumor and macrophages. (B) (pMETTL14+RS09)@cRGD-M nanovesicles could target to inhibit tumor and induce M1 polarization of macrophages, thereby achieving the dual purposes of tumor inhibition and macrophage polarization.
Whereas, these limitations are warranted to be solved before membrane-coated nanovesicles could be used in the clinic. Firstly, from an economic perspective, the membrane preparation needs to be simplified and optimized thoroughly. At the same time, the standardized process should be designed to control the quality and enable the repeatability of membrane preparation [31]. Moreover, protein loss during the process of cell membrane extraction might result in decreased tumor targeting ability of membrane-coated nanovesicles [32], which might influence its anti-tumor effects. Finally, owing to the unsatisfied tumor targeting ability, the membrane structure of the nanovesicle is changed and its biocompatibility is reduced during the modification of targeting ligands. Therefore, membrane hybridization has been regarded as a novel approach to resolving the above obstacles [18]. Hybrid membranes might combine the membrane properties and functions of different cells. The most important biological activities of hybrid membranes include (a) enhanced targeting ability and (b) inherent properties conferred by the membranes of the source cells. For example, the membranes of RAW264.7 cells show a high expression level of integrin α4β1. Therefore, a hybrid membrane composed of macrophages and tumor cells might have the ability to target homologous tumor cells and improve the potential of targeting metastases [33].
5. Conclusions
In this study, we developed macrophage membrane-coated nanovesicles to co-deliver METTL14 and the TLR4 agonist for dual purposes of tumor inhibition and macrophage polarization. It showed that the nanovesicles could improve the in vivo stability of drugs, inhibit tumor, and induce M1 polarization of macrophages. This study intends to clarify the therapeutic roles of the nanovesicles in dual-targeting tumor and macrophages and achieve the dual purposes of tumor inhibition and TME remodeling, which provides a novel therapeutic strategy for tumors.
Ethics approval and consent to participate
This study was approved by the institutional review board and the medical ethics committee of Wuhan Union Hospital, Huazhong University of Science and Technology. Each participant provided written informed consent. We collected the medical data and samples of three patients with osteosarcoma in our hospital.
Consent for publication
All authors have read and approved this version of the article for publication.
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author.
Funding
This study is supported by the National Natural Science Foundation of China (No. 82203059) and the China Postdoctoral Science Foundation (2021M701335).
CRediT authorship contribution statement
Xin Huang: Formal analysis, Methodology, Investigation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Lutong Wang: Formal analysis, Data curation, Methodology, Software. Haoyu Guo: Formal analysis, Investigation, Methodology, Software. Weiyue Zhang: Formal analysis, Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
No conflict of interest exits in the submission of this manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.09.027.
Contributor Information
Xin Huang, Email: xin_huang@hust.edu.cn.
Weiyue Zhang, Email: zhangweiyuehust@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hinshaw D.C., Shevde L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly B., O'Neill L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson N.R., Minutolo N.G., Gill S., Klichinsky M. Macrophage-Based approaches for cancer immunotherapy. Cancer Res. 2021;81:1201–1208. doi: 10.1158/0008-5472.CAN-20-2990. [DOI] [PubMed] [Google Scholar]
- 4.Luo Z.W., Liu P.P., Wang Z.X., Chen C.Y., Xie H. Macrophages in osteosarcoma immune microenvironment: implications for immunotherapy. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.586580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanderley C.W., Colón D.F., Luiz J.P.M., Oliveira F.F., Viacava P.R., Leite C.A., Pereira J.A., Silva C.M., Silva C.R., Silva R.L., Speck-Hernandez C.A., Mota J.M., Alves-Filho J.C., Lima-Junior R.C., Cunha T.M., Cunha F.Q. Paclitaxel reduces tumor growth by reprogramming tumor-associated macrophages to an M1 profile in a TLR4-dependent manner. Cancer Res. 2018;78:5891–5900. doi: 10.1158/0008-5472.CAN-17-3480. [DOI] [PubMed] [Google Scholar]
- 6.Du K., Zhang L., Lee T., Sun T. m(6)A RNA methylation controls neural development and is involved in human diseases. Mol. Neurobiol. 2019;56:1596–1606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 7.Lan Q., Liu P.Y., Haase J., Bell J.L., Hüttelmaier S., Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 8.Patil D.P., Pickering B.F., Jaffrey S.R. Reading m(6)A in the transcriptome: m(6)a-binding proteins. Trends Cell Biol. 2018;28:113–127. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai F., Wu Y., Lu Y., An C., Zheng X., Dai L., Guo Y., Zhang L., Li H., Xu W., Gao W. Crosstalk between RNA m(6)A modification and non-coding RNA contributes to cancer growth and progression, molecular therapy. Nucleic acids. 2020;22:62–71. doi: 10.1016/j.omtn.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi Y.C., Chen X.Y., Zhang J., Zhu J.S. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng C., Huang W., Li Y., Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X., Guo H., Wang L., Yang L., Shao Z., Zhang W. Recent advances in crosstalk between N6-methyladenosine (m6A) modification and circular RNAs in cancer, Molecular therapy. Nucleic acids. 2022;27:947–955. doi: 10.1016/j.omtn.2022.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L., Chen C., Zhang Y., Guo P., Wang Z., Li J., Liu Y., Liu J., Chang R., Li Y., Liang G., Lai W., Sun M., Dougherty U., Bissonnette M.B., Wang H., Shen L., Xu M.M., Han D. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39:945–957. doi: 10.1016/j.ccell.2021.04.016. e910. [DOI] [PubMed] [Google Scholar]
- 15.Du J., Liao W., Liu W., Deb D.K., He L., Hsu P.J., Nguyen T., Zhang L., Bissonnette M., He C., Li Y.C. N(6)-Adenosine methylation of Socs1 mRNA is required to sustain the negative feedback control of macrophage activation. Dev. Cell. 2020;55:737–753. doi: 10.1016/j.devcel.2020.10.023. e737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patra J.K., Das G., Fraceto L.F., Campos E.V.R., Rodriguez-Torres M.D.P., Acosta-Torres L.S., Diaz-Torres L.A., Grillo R., Swamy M.K., Sharma S., Habtemariam S., Shin H.S. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018;16:71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy S.A., Davies G.L., Gun'ko Y.K. Preparation of multifunctional nanoparticles and their assemblies. Nat. Protoc. 2012;7:1677–1693. doi: 10.1038/nprot.2012.082. [DOI] [PubMed] [Google Scholar]
- 18.Chen H.Y., Deng J., Wang Y., Wu C.Q., Li X., Dai H.W. Hybrid cell membrane-coated nanoparticles: a multifunctional biomimetic platform for cancer diagnosis and therapy. Acta Biomater. 2020;112:1–13. doi: 10.1016/j.actbio.2020.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W., Huang X. Stem cell membrane-camouflaged targeted delivery system in tumor, Materials today. Bio. 2022;16 doi: 10.1016/j.mtbio.2022.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain Z., Rahim M.A., Jan N., Shah H., Rawas-Qalaji M., Khan S., Sohail M., Thu H.E., Ramli N.A., Sarfraz R.M., Abourehab M.A.S. Cell membrane cloaked nanomedicines for bio-imaging and immunotherapy of cancer: improved pharmacokinetics, cell internalization and anticancer efficacy. J. Contr. Release : official journal of the Controlled Release Society. 2021;335:130–157. doi: 10.1016/j.jconrel.2021.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Oroojalian F., Beygi M., Baradaran B., Mokhtarzadeh A., Shahbazi M.A. Immune cell membrane-coated biomimetic nanoparticles for targeted cancer therapy. Small (Weinheim an der Bergstrasse, Germany) 2021;17 doi: 10.1002/smll.202006484. [DOI] [PubMed] [Google Scholar]
- 22.Fang R.H., Hu C.M., Luk B.T., Gao W., Copp J.A., Tai Y., O'Connor D.E., Zhang L. Cancer cell membrane-coated nanoparticles for anticancer vaccination and drug delivery. Nano Lett. 2014;14:2181–2188. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y., He G., Liu Z., Wang J., Li M., Zhang Z., Bao Q., Wen J., Zhu X., Zhang C., Zhang W. DNA base pairing-inspired supramolecular nanodrug camouflaged by cancer-cell membrane for osteosarcoma treatment. Small (Weinheim an der Bergstrasse, Germany) 2022;18 doi: 10.1002/smll.202202337. [DOI] [PubMed] [Google Scholar]
- 24.Fu L., Zhang W., Zhou X., Fu J., He C. Tumor cell membrane-camouflaged responsive nanoparticles enable MRI-guided immuno-chemodynamic therapy of orthotopic osteosarcoma. Bioact. Mater. 2022;17:221–233. doi: 10.1016/j.bioactmat.2022.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. Ying, Y. Zhu, J. Wan, C. Zhan, Y. Wang, B. Xie, P. Xu, H. Pan, H. Wang, Macrophage membrane-biomimetic adhesive polycaprolactone nanocamptothecin for improving cancer-targeting efficiency and impairing metastasis, Bioact. Mater., 20 (2023) 449-462. [DOI] [PMC free article] [PubMed]
- 26.Huang X., Wu W., Jing D., Yang L., Guo H., Wang L., Zhang W., Pu F., Shao Z. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J. Contr. Release : official journal of the Controlled Release Society. 2022;343:107–117. doi: 10.1016/j.jconrel.2022.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Sugahara K.N., Teesalu T., Karmali P.P., Kotamraju V.R., Agemy L., Greenwald D.R., Ruoslahti E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science (New York, N.Y.) 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X., Wang L., Guo H., Zhang W., Shao Z. Single-cell transcriptomics reveals the regulative roles of cancer associated fibroblasts in tumor immune microenvironment of recurrent osteosarcoma. Theranostics. 2022;12:5877–5887. doi: 10.7150/thno.73714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Agarwal P., Zhao S., Yu J., Lu X., He X. Combined cancer therapy with hyaluronan-decorated fullerene-silica multifunctional nanoparticles to target cancer stem-like cells. Biomaterials. 2016;97:62–73. doi: 10.1016/j.biomaterials.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Si J., Shao S., Shen Y., Wang K. Macrophages as active nanocarriers for targeted early and adjuvant cancer chemotherapy. Small (Weinheim an der Bergstrasse, Germany) 2016;12:5108–5119. doi: 10.1002/smll.201601282. [DOI] [PubMed] [Google Scholar]
- 31.Lei W., Yang C., Wu Y., Ru G., He X., Tong X., Wang S. Nanocarriers surface engineered with cell membranes for cancer targeted chemotherapy. J. Nanobiotechnol. 2022;20:45. doi: 10.1186/s12951-022-01251-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Y., Rao L., Yao H., Wang Z., Ning P., Chen X. Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater. 2020;32 doi: 10.1002/adma.202002054. [DOI] [PubMed] [Google Scholar]
- 33.Gong C., Yu X., You B., Wu Y., Wang R., Han L., Wang Y., Gao S., Yuan Y. Macrophage-cancer hybrid membrane-coated nanoparticles for targeting lung metastasis in breast cancer therapy. J. Nanobiotechnol. 2020;18:92. doi: 10.1186/s12951-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding author.