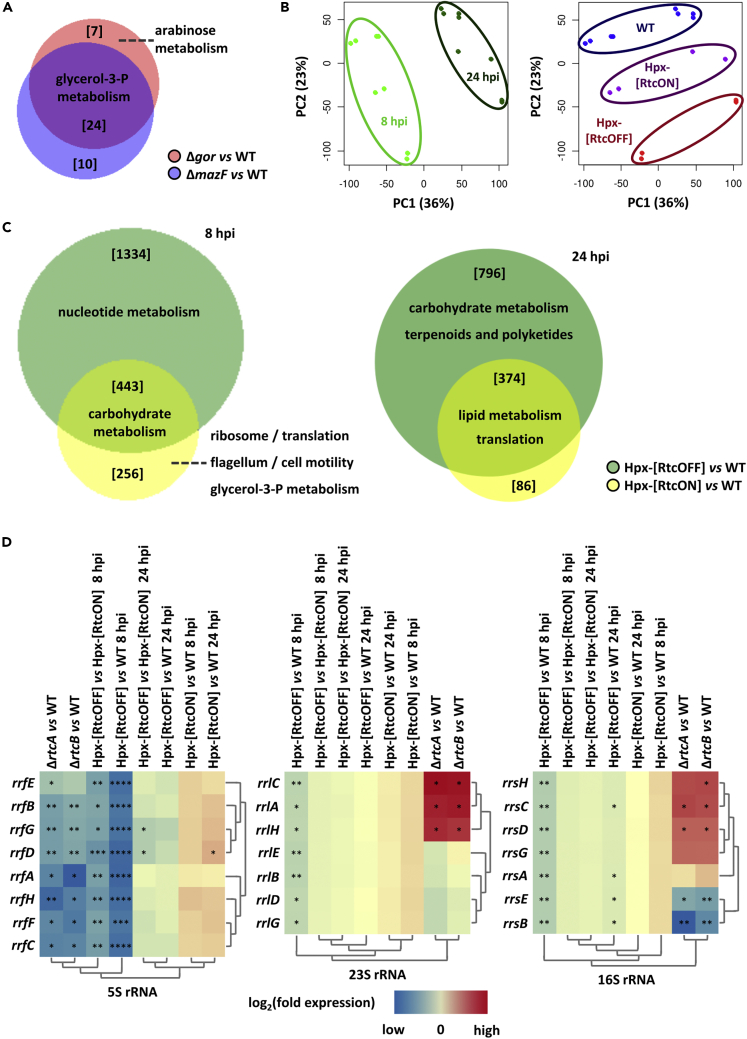
Figure 5.
Transcriptome profiling of E. coli cells under Rtc-inducing conditions
Transcriptome profiling of E. coli wild-type cells, cells lacking gor, mazF or srmB, and Hpx-[RtcON] and Hpx-[RtcOFF] cells was performed by NGS and followed by differential gene expression analysis. Transcriptome profiling of E. coli cells lacking rtcA or rtcB was described previously by Engl et al. (2016).15
(A) Venn diagram of the genes differentially expressed in cells lacking gor or mazF compared to the wild-type and associated GO terms (Tables S7 and S8).
(B) Principal component analysis of the transcriptional profiling of the wild-type, Hpx-[RtcON] and Hpx-[RtcOFF] strains.
(C) Venn diagram of the genes differentially expressed in Hpx-[RtcON[ and Hpx-[RtcOFF] as compared to wild-type cells at 8 and 24 hpi, and associated GO terms and KEGG pathways.
(D) Heat maps of 5S (left; log2fold expression ±5), 23S (left; log2fold expression ±7) and 16S (left; log2fold expression ±6) ribosomal (r) RNA abundance levels in a range of mutants, as shown by NGS; rows and columns have been grouped based on a hierarchical clustering algorithm. Limma/DESeq2 ∗ adjusted p-value < 0.05; ∗∗ adjusted p-value < 0.01; ∗∗∗∗ adjusted p-value < 0.0001.