Abstract
As enteric pathogens, Salmonella spp. are resistant to the actions of bile. Salmonella typhimurium and Salmonella typhi strains were examined to better define the bile resistance phenotype. The MICs of bile for wild-type S. typhimurium and S. typhi were 18 and 12%, respectively, and pretreatment of log-phase S. typhimurium with 15% bile dramatically increased bile resistance. Mutant strains of S. typhimurium and S. typhi lacking the virulence regulator PhoP-PhoQ were killed at significantly lower bile concentrations than wild-type strains, while strains with constitutively active PhoP were able to survive prolonged incubation with bile at concentrations of >60%. PhoP-PhoQ was shown to mediate resistance specifically to the bile components deoxycholate and conjugated forms of chenodeoxycholate, and the protective effect was not generalized to other membrane-active agents. Growth of both S. typhimurium and S. typhi in bile and in deoxycholate resulted in the induction or repression of a number of proteins, many of which appeared identical to PhoP-PhoQ-activated or -repressed products. The PhoP-PhoQ regulon was not induced by bile, nor did any of the 21 PhoP-activated or -repressed genes tested play a role in bile resistance. However, of the PhoP-activated or -repressed genes tested, two (prgC and prgH) were transcriptionally repressed by bile in the medium independent of PhoP-PhoQ. These data suggest that salmonellae can sense and respond to bile to increase resistance and that this response likely includes proteins that are members of the PhoP regulon. These bile- and PhoP-PhoQ-regulated products may play an important role in the survival of Salmonella spp. in the intestine or gallbladder.
Mounting evidence has suggested that the success or failure of a bacterial pathogen during infection of a host relies upon its ability to sense and respond to its immediate environment. Salmonella spp. are a prime example of this concept, as these organisms encounter numerous different environments upon infection of a host. Many of these environments are potentially lethal to the bacterium; therefore, the requisite survival response often includes mechanisms of resistance to these lethal factors. In the human host, harsh environments encountered by Salmonella spp. include the acid environment of the stomach, the bloodstream, epithelial and phagocytic cell intercompartments, and the intestinal lumen.
Within the intestine, Salmonella spp. encounter and must be able to resist the action of bile salts. Bile salts are detergents made by the liver and secreted into and stored in high concentrations in the gallbladder. Bile containing these salts is released into the intestine to aid in the dispersion and degradation of fats. Enteric bacteria, including Salmonella spp., are resistant to the effects of bile, a finding that has been used clinically in the selective enrichment of these organisms (e.g., MacConkey agar).
A percentage (1 to 3%) of individuals infected with Salmonella typhi become chronic carriers, and the prime location of the persistent infection is the gallbladder. In the carrier state, organisms are continuously released into the intestine and shed in the feces. It is thought that bile duct or gallbladder abnormalities (including gallstones) play a role in the development of the carrier state (24). Therefore, while all Salmonella spp. infecting a human host through oral means encounter bile in the intestine, organisms in the carrier state likely encounter and must resist the action of even higher concentrations of bile salts.
Although bile resistance in enteric organisms has been known for some time, relatively little is known about the molecular mechanisms responsible for the resistance. The outer membrane of gram-negative bacteria is thought to be the main barrier to bile salts. Changes in lipopolysaccharide and membrane proteins (including porins) have been shown to affect bile salt tolerance (29, 34). However, bile salts are known to enter the periplasm and cytoplasm of Escherichia coli. Recently, Thanassi et al. (36) demonstrated that the E. coli acr and emr loci encode efflux pumps that actively transport bile salts. Several studies of enteric organisms, such as Enterococcus faecalis and Enterobacter cloacae, have been conducted to examine and compare the responses to bile salts and other detergents, such as sodium dodecyl sulfate (SDS) (7, 28). While it had been thought that the responses to these two detergents would be similar, recent studies have demonstrated this notion to be untrue, as both elicit the production or increased production of 34 to 45 proteins with limited overlap.
The PhoP-PhoQ two-component regulatory system is necessary for the virulence of Salmonella spp. (6, 26). PhoQ is a membrane-bound kinase (13) that, upon sensing specific environmental cues (such as Mg2+ concentration [8]), initiates a phosphorylation cascade to activate PhoP, a transcriptional regulator. A number of genes are both transcriptionally activated and transcriptionally repressed by PhoP-PhoQ (27). Activation of the PhoP regulon occurs in vivo when the organism is within the macrophage phagosome, an environment within which Salmonella must survive to cause disease (1). Therefore, it is thought that PhoP-activated genes are necessary for intramacrophage survival. Because homologs of PhoP, PhoQ, and both PhoP-activated (pag) and -repressed (prg) products exist in a variety of bacterial organisms (10–12), including intestinal pathogens that do not survive within macrophage phagosomes, other important functions of the PhoP regulon likely exist.
This study was designed to examine in depth the resistance and response of Salmonella typhimurium and S. typhi to bile and bile salts. We report that both serovars are highly resistant to the effects of bile and significantly alter protein expression in response to bile or bile components in the growth medium. Furthermore, high-level resistance of both S. typhimurium and S. typhi to bile requires the pleotropic virulence regulator PhoP-PhoQ.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and reagents.
Bacterial strains used in this study include S. typhimurium 14028s (American Type Culture Collection), a constitutively active PhoP (PhoPc) strain (pho24; CS022), and a PhoP− strain (phoP::Tn10d-cam; CS015), previously described by Miller et al. (26). S. typhi strains include Ty2, provided by Carolyn Hardegree at the U.S. Food and Drug Administration; a PhoPc strain (pho24) provided by Renato Morona, University of Adelaide, Adelaide, South Australia, Australia (2); and a PhoP− deletion derivative of Ty2 (20). Cultures were grown overnight at 37°C with aeration in Luria-Bertani (LB) broth or in microtiter plates as described below. When necessary, the medium was supplemented with chloramphenicol (25 μg/ml), ampicillin (50 μg/ml), or kanamycin (45 μg/ml).
Conjugated and unconjugated bile salts were purchased from Sigma Chemical Co. (St. Louis, Mo.). The bile used in this work was labeled “sodium choleate,” but is a crude ox bile extract which contains salts of taurocholic, glycocholic, deoxycholic, and cholic acids.
Protein gel electrophoresis.
Whole-cell bacterial extracts and membrane protein extracts were prepared from stationary-phase or log-phase cultures grown at 37°C with aeration in LB broth with or without bile or deoxycholic acid. Whole-cell samples were collected by centrifugation and lysed by boiling for 10 min in 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (0.125 M Tris [pH 6.8], 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.1% bromophenol blue). Membrane protein samples were prepared as previously described (13). Proteins were separated by SDS-PAGE (10%) and stained with Coomassie brilliant blue.
Two-dimensional (2-D) gel analysis with silver staining was performed by Kendrick Laboratories, Inc. (Madison, Wis.). Overnight cultures grown with or without 3% bile were pelleted by centrifugation, washed once with LB broth, and flash frozen. 2-D electrophoresis was performed by the method of O’Farrell (31) as follows. Isoelectric focusing was carried out with glass tubes having an inner diameter of 2.0 mm and with 2.0% pH 4 to 8 ampholines (Hoefer Scientific Instruments, San Francisco, Calif.) for 9,600 V-h. Fifty nanograms of an isoelectric focusing internal standard, tropomyosin protein, with a molecular weight of 33,000 and a pI of 5.2, was added to the samples.
After equilibration for 10 min in buffer O (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, 0.0625 M Tris [pH 6.8]), the tube gel was sealed to the top of a stacking gel, which was on top of a 10% acrylamide slab gel (0.75 mm thick). SDS slab gel electrophoresis was then carried out for about 4 h at 12.5 mA/gel. The slab gels were fixed in a solution of 10% acetic acid–50% methanol overnight. The following proteins (Sigma) were added as molecular weight standards to the agarose which sealed the tube gel to the slab gel: myosin (220,000), phosphorylase a (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000), and lysozyme (14,000). These standards appeared as horizontal lines on the silver-stained 10% acrylamide slab gel. The gels were dried between sheets of cellophane paper with the acid edge to the left.
Standard MIC and MBC assays of bile resistance.
Bacterial cells in either the log or the stationary phase were challenged with bile, unconjugated bile acids (cholic and deoxycholic), or conjugated bile acids (glycocholic, taurocholic, and glychochenodeoxycholic) and assayed for MICs under both aerated and nonaerated conditions. Polypropylene microtiter plates (Costar Corp., Cambridge, Mass.) were used for nonaerated conditions, and 1-ml cultures were grown on a roller drum for aerated conditions. Stationary- and log (optical density, 0.8)-phase cultures were diluted such that 2 × 103 to 5 × 103 CFU/ml was subjected to various concentrations of bile and bile acids. All assay mixtures were incubated overnight at 37°C and visually analyzed for MICs. MBCs were determined by plating well or tube cultures exhibiting no apparent growth in the MIC assays.
Assays of adaptation to bile.
Adaptation experiments were conducted by either an MIC challenge assay or a time-kill assay. For the MIC challenge assay, cells were grown to the log or stationary phase in the absence or presence of bile. Cultures were washed twice and diluted such that 2 × 103 to 5 × 103 CFU/ml was challenged with a range of bile salt concentrations in microtiter plates. Time-kill assays were accomplished in an identical manner, except that after washing and dilution, cells were challenged with a single bile concentration (most frequently, 24%). Aliquots of 100 μl were washed and plated at 2-h intervals for up to 10 h. Percent survival was determined by comparing the number of colonies on the time point plates to that present before the addition of the challenge concentration of bile.
Transcription assays.
Strains carrying chromosomal pag and prg MudJ (β-galactosidase) or TnphoA (alkaline phosphatase) transposon-generated gene fusions were grown to the log or stationary phase with 3 or 15% bile or without bile. Cells were recovered by centrifugation, washed twice with LB broth, and assayed for β-galactosidase or alkaline phosphatase activity as previously described (15). Activity was expressed in units determined by the method of Miller (25). Firefly luciferase assays were accomplished as previously described (15).
Construction of acr and mar fusions and assays of PhoP-PhoQ regulation.
DNA internal to the marAB genes and the acrB gene was amplified by PCR with primers JB134-JG135 and JG136-JG137, respectively. Primers were constructed to contain a KpnI or EcoRI site at the 5′ end. PCR fragments were cloned into the firefly luciferase reporter-suicide vector pGPL01 (15). Recombination on the chromosome accomplished both a gene fusion and a gene knockout. PhoP− and PhoPc strains carrying the fusion were assayed for luciferase activity after overnight growth and in the log phase.
RESULTS
Salmonella tolerance of bile and bile salts.
Although it has been known for some time that Salmonella spp. are resistant to bile salts, the MIC and MBC of bile and bile salts for Salmonella have not been clearly defined. To determine the MIC and MBC of bile for S. typhimurium and S. typhi, stationary-phase cultures were diluted and incubated with various concentrations of bile. After overnight microaerophilic incubation in 96-well plates or aerated incubation in tubes, cultures were assessed for growth. As shown in Table 1, the MIC of bile for S. typhimurium was 18%, while the MIC for S. typhi was slightly lower at 12%. To confirm that this phenomenon was not limited to the S. typhimurium (14028s) and S. typhi (Ty2) laboratory strains analyzed but was generalizable to other isolates of these serovars, several S. typhimurium and S. typhi isolates were examined. The results showed MICs identical to those obtained for 14028s and Ty2 (Table 1). The MICs reported in Table 1 are from microaerophilic assays (96-well plates) but were identical for cultures examined under aerated experimental growth conditions (data not shown). In addition, the MICs for cultures assayed in the log phase instead of the stationary phase were similar.
TABLE 1.
MICs and MBCs of bile for S. typhimurium and S. typhi
| Organism | Strain | MIC (% bile) | MBC (% bile) |
|---|---|---|---|
| S. typhimurium | 14028s | 18 | >60a |
| LT2 | 18 | >60a | |
| SR-11 | 18 | >60a | |
| S. typhi | Ty2 | 12 | 18 |
| Clinical isolate | 12 | 18 | |
| Clinical isolate | 12 | 18 |
Low numbers of survivors (∼1%) were detectable at this concentration of bile, although the major reduction in survival occurred at a bile concentration of 42%.
To determine what concentration of bile was bactericidal for S. typhimurium and S. typhi, MBCs were determined by plating well or tube cultures exhibiting no apparent growth in the MIC assays (Table 1). Surprisingly, the MBC of bile for S. typhimurium (>60%) was dramatically higher (>3-fold) than the MIC. Although a significant reduction in survival was seen at 42%, bacteria were recovered at 60%. It became technically difficult to examine bile concentrations exceeding 60%, as this percentage of bile appears near the verge of solubility, and solution viscosity above 60% too severely reduced pipetting accuracy. Contrary to what was found with S. typhimurium, the MBC of bile for S. typhi (18%) was only slightly higher (1.5-fold) than the MIC. The MBCs for other isolates of each serovar were identical to the MBCs of 14028s and Ty2 (data not shown). Therefore, while wild-type strains of both S. typhimurium and S. typhi were able to survive at bile concentrations exceeding the MIC, S. typhimurium was better able to survive in high concentrations of bile.
PhoP-PhoQ is necessary for enhanced resistance to bile.
The PhoP-PhoQ virulence regulator both activates and represses the production of a number of membrane or secreted proteins (5, 27). In addition, activation of this two-component system results in structural and charge modifications of lipopolysaccharide (16). Both types of gram-negative bacterial cell surface alterations have been implicated in resistance to bile. To determine if PhoP-activated or -repressed gene products played a role in bile resistance, MIC assays were conducted with S. typhimurium and S. typhi wild-type, PhoP−, and PhoPc strains. PhoP− strains, in which phoP is absent or is inactivated by a transposon, mimic the environmental repression of the PhoP-PhoQ regulon, while PhoPc strains show activation of the regulon to high levels even in the absence of environmental signals. As shown in Table 2, an S. typhimurium PhoP− strain was fourfold more sensitive to bile than a PhoPc strain and threefold more sensitive than a wild-type strain. The bile-sensitive phenotype of an S. typhi PhoP− strain was also evident but was slightly less pronounced (2.5-fold less than that of a PhoPc strain and 2-fold less than that of a wild-type strain) than that in the S. typhimurium strain.
TABLE 2.
| Organism | Strain | Bile
|
Deoxycholate MIC | Glycochenodeoxy-cholate MIC | Taurocholate MIC | Glycocholate MIC | SDS MIC | Triton X-100 MIC | |
|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | ||||||||
| S. typhimurium | Wild type | 18 | >60 | 5 | 20 | 20 | 20 | 14.5 | 14.5 |
| PhoP− | 6 | 12 | 0.3 | 0.3 | 10 | 10 | 14.5 | 14.5 | |
| PhoPc | 24 | >60 | 5 | 20 | 20 | 20 | 14.5 | 14.5 | |
| S. typhi | Wild type | 12 | 18 | 20 | 10 | 20 | 20 | 7.3 | 14.5 |
| PhoP− | 6 | 12 | 0.3 | 0.3 | 5 | 5 | 7.3 | 14.5 | |
| PhoPc | 15 | >60 | 20 | 10 | 20 | 20 | 7.3 | 14.5 | |
Presented as percentage of bile, bile acid, or detergent.
Presented as percentage of bile.
In addition to the MIC analysis of PhoP− and PhoPc strains, the MBCs were also determined. The results for S. typhimurium, as shown in Table 2, indicate that the MBC for the PhoPc strain was extremely high (>60%) and that the MBC for the PhoP− strain was >5-fold lower. The observed MBC for the S. typhimurium PhoP− strain was nearly identical to the MIC, suggesting that for this strain, the bile concentrations necessary for the cessation of growth and for bactericidal activity are roughly equivalent. Although the MBCs for both wild-type and PhoPc S. typhimurium strains were >60%, the number of surviving cells for the PhoPc strain was often 10- to 100-fold higher than that for the wild-type strain at concentrations of up to and including 60% bile (data not shown).
As in S. typhimurium, the MBC for the S. typhi PhoP− strain was nearly identical to the MIC (Table 2). However, the S. typhi PhoPc strain was recovered in significant numbers at bile concentrations of up to and including 60%. Therefore, induction of the PhoP regulon by a constitutive mutation (PhoPc) in both S. typhimurium and S. typhi resulted in increased survival at extremely high concentrations of bile.
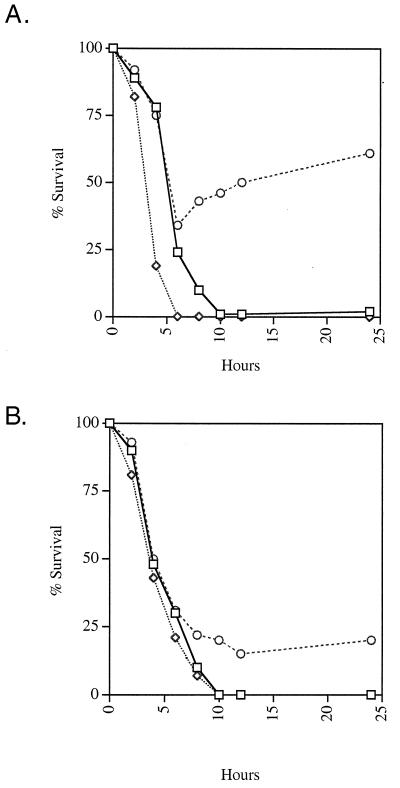
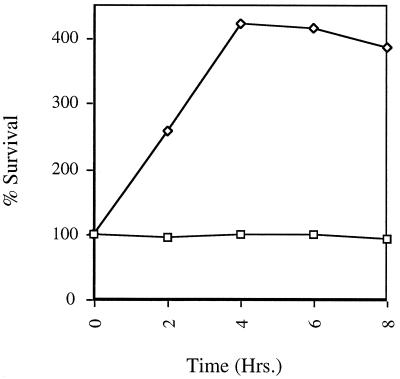
In order to determine the kinetics of bile action on Salmonella spp. and to further explore the role of PhoP-PhoQ in bile resistance, the sensitivity of S. typhimurium and S. typhi cultures to a high concentration (30%) of bile was measured over time. As shown in Fig. 1, bile was not rapidly bactericidal for these organisms. This result is in sharp contrast to those for some other enteric organisms that are killed within several minutes at bile concentrations higher than the MIC (7, 28). The PhoP− strain of S. typhimurium shows a slow decline in cell numbers culminating in complete killing in approximately 6 h. The wild-type and PhoP− S. typhi strains, which are also completely killed by this concentration of bile, survive until approximately 10 h. Besides providing information about the kinetics of bile action, these data corroborate the MBC results. While this bile concentration is bactericidal for PhoP− and wild-type strains of S. typhi and a PhoP− strain of S. typhimurium, a wild-type strain of S. typhimurium survives in low numbers and PhoPc mutants of both serovars survive an initial decrease in cell numbers followed by a slow recovery. The results presented above (MIC, MBC, and kinetic experiments) were accomplished with stationary-phase organisms but were similar to those for cells assayed in the log phase (data not shown).
FIG. 1.
Bile survival assay of S. typhimurium (A) and S. typhi (B). Strains grown to the stationary phase in LB broth were diluted and incubated in the presence of 30% bile. Aliquots were removed at various times, diluted or washed, and plated to determine the number of surviving cells. Symbols: □, wild type; ◊, PhoP−; ○, PhoPc.
PhoP-PhoQ plays a more significant role in resistance of Salmonella spp. to deoxycholic and chenodeoxycholic acids than in resistance to other bile acids or detergents.
To test the effect of several bile salts individually on Salmonella growth, wild-type, PhoP−, and PhoPc strains were tested with deoxycholic acid and conjugated forms of cholic and chenodeoxycholic acids in MIC assays. Cholic and chenodeoxycholic acids are conjugated with glycine or taurine after biosynthesis, and these conjugated bile acids, along with deoxycholic acid, constitute ∼90% of the bile acids found in the gallbladder or small intestine (19). The PhoP− strain of both serovars was dramatically more sensitive to deoxycholic acid and a conjugated form of chenodeoxycholic acid than the PhoPc strain (17- and 67-fold for S. typhimurium and 67- and 33-fold for S. typhi for deoxycholate and glycochenodeoxycholate, respectively) (Table 2), while only 2- to 4-fold differences in the sensitivity of S. typhimurium or S. typhi to glycocholic and taurocholic salts were observed for PhoP− versus wild-type or PhoPc strains.
To determine if the observed resistance mediated by PhoP-PhoQ was specific for bile or common to other membrane-active agents, MIC assays of S. typhimurium and S. typhi were conducted with a nonionic detergent (Triton X-100) and an ionic detergent (SDS). No differences were observed in the MICs for wild-type, PhoP−, or PhoPc strains of either serovar with Triton X-100 (14.5%) or SDS (14.5% for S. typhimurium and 7.3% for S. typhi) (Table 2), demonstrating that the resistance mediated by PhoP-PhoQ was not generalizable to other detergents and likely was specific for bile.
Individual PhoP-activated and -repressed loci examined play no role in PhoP-PhoQ-mediated bile resistance.
To attempt to identify the PhoP-activated gene product(s) necessary for resistance to bile, stationary- and log-phase cultures carrying PhoP-activated gene mutants (pagA-pagP) in a PhoPc background were examined for a reduction in the MIC of bile. None of the pag PhoPc double mutant strains exhibited a reduction in the MIC (data not shown). Because the PhoP-PhoQ-regulated factors affecting the bile phenotype may not be induced genes causing increased resistance but rather repressed genes causing increased susceptibility, MICs for prg mutants (prgA, prgB, prgC, prgE, and prgH) in a PhoP− background were determined. A PhoP− background was used because prg will be maximally induced, and if Prg results in susceptibility to bile, the loss of the gene encoding it should result in a dramatic increase in bile resistance. However, none of the prg PhoP− mutants showed increased resistance to bile (data not shown).
In addition to the individual prg and pag mutants tested, a strain containing mutations in seven pag genes (pagCDKMJNP) and pmrA also was tested; there was no reduction in the MIC of bile. These results are significant, as the genes encoding the PmrA-PmrB regulatory system (which are regulated by PhoP-PhoQ [15, 33]) as well as pagP produce products that mediate charge or structural alterations of LPS (16–18, 37), and pagCDKMJ and pagN encode membrane or secreted proteins (12). These types of surface alterations (LPS and membrane protein) have been previously shown to affect bile resistance in other organisms and were likely candidates as mediators of the PhoP-PhoQ bile phenotype. Therefore, the gene product(s) that is regulated by PhoP-PhoQ and that is responsible for resistance to bile remains to be identified.
It has been shown that acr and mar loci affect bile resistance in E. coli and S. typhimurium (30, 35, 36). To determine if these loci are regulated by PhoP-PhoQ and therefore are responsible for the bile resistance phenotype, chromosomal fusions of the marA and acrB genes with the firefly luciferase gene (luc) were constructed. The insertion of the luc gene both inactivated the marA and acrB genes and generated a transcriptional luc fusion. Upon transduction of this fusion into PhoP− and PhoPc backgrounds, both bile MIC and luciferase assays were conducted. These experiments confirmed the roles of these loci in Salmonella bile resistance (>5-fold reduction); however, neither was regulated by PhoP-PhoQ (data not shown).
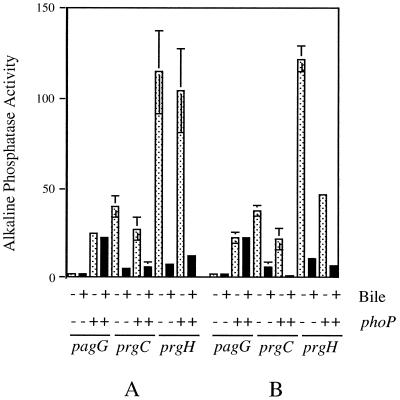
prgH and prgC transcription is repressed by bile.
To test whether the PhoP-PhoQ regulon was induced in the presence of bile, a collection of 16 transposon-generated pag-reporter gene fusions and 5 prg-reporter gene fusions was examined for altered transcription when strains were grown with or without bile in the medium. Of the 21 fusions examined, 19 showed no significant alteration in expression due to the presence of bile, suggesting that the PhoP-PhoQ regulon does not sense and respond to bile. However, two PhoP-repressed fusions, prgH and prgC, were repressed by bile 12.5- and 9.4-fold, respectively (Fig. 2). Furthermore, these fusions were still repressed by bile to nearly the same levels when assayed in a PhoP− background. In addition, the bile effect on these loci was growth phase independent, as similar levels of repression were observed with both stationary-phase and log-phase organisms. Therefore, although prgH and prgC are not individually responsible for the PhoP-PhoQ-mediated bile resistance phenotype, their transcription is affected by bile independent of PhoP-PhoQ.
FIG. 2.
PhoP-PhoQ-independent effect of bile on prgC and prgH transcription. Strains grown to the stationary (A) or logarithmic (B) phase in the presence or absence of bile (3% for PhoP− and 15% for PhoP+) were assayed for alkaline phosphatase activity. The pagG results are representative of PhoP-PhoQ-regulated fusions unaffected by bile. Stippled bars show results from cultures grown without bile, and black bars show those with bile. The means of three independent experiments with associated standard errors are shown.
Alteration of Salmonella protein expression in response to bile.
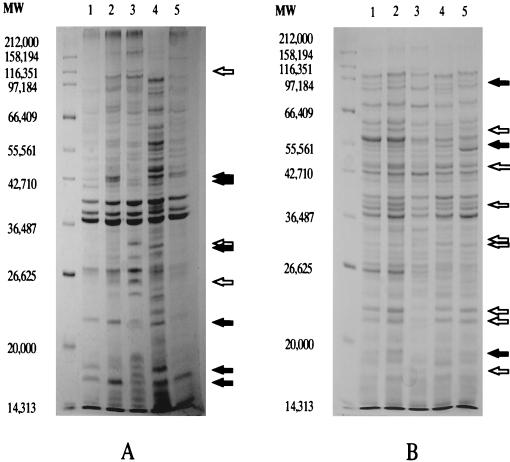
Protein profiles of organisms grown in the presence of bile or bile salts were examined by SDS-PAGE for potential alterations. Whole-cell and membrane protein preparations of S. typhimurium or S. typhi cultures grown in LB broth, LB broth with 3% bile, or LB broth with 1% deoxycholate were electrophoretically separated and visualized by Coomassie brilliant blue staining. As shown in Fig. 3, the growth of S. typhimurium in the presence of bile or deoxycholate affected numerous proteins, with bile resulting in ∼15 easily observable changes (14 increased protein species and 1 decreased protein species) and deoxycholate resulting in ∼14 easily observable changes (7 increased protein species and 7 decreased protein species). Of these bile- and deoxycholate-induced protein alterations, only five protein alterations (e.g., ∼106-kDa species; Fig. 3A) appeared common to both. PhoPc and PhoP− lysates (whole cell or membrane) were also examined, and of the proteins affected by bile or deoxycholate, 13 appeared similar in size to PhoP-activated or -repressed products. The proteins which appeared to be affected by bile or deoxycholate and PhoP-PhoQ were still induced in a PhoP− strain grown in the presence of bile, further demonstrating, as had been shown with the repression of prgC and prgH transcription, that the effect of bile on pag or prg loci is independent of PhoP-PhoQ (data not shown).
FIG. 3.
Comparison of S. typhimurium proteins affected by bile or deoxycholate in the growth medium and proteins regulated by PhoP-PhoQ. Membrane extracts (A) and whole-cell extracts (B) were separated by SDS–10% PAGE. Lanes 1 to 5 show protein profiles of the following strains, respectively: wild type, wild type plus 3% bile, wild type plus 1% deoxycholate, PhoPc, and PhoP−. Molecular weight (MW) markers are indicated to the left of the panels. Open arrows denote proteins affected by bile or deoxycholate but not by PhoP-PhoQ. Closed arrows denote proteins affected by bile or deoxycholate and by PhoP-PhoQ. Only those protein species most obviously affected are noted.
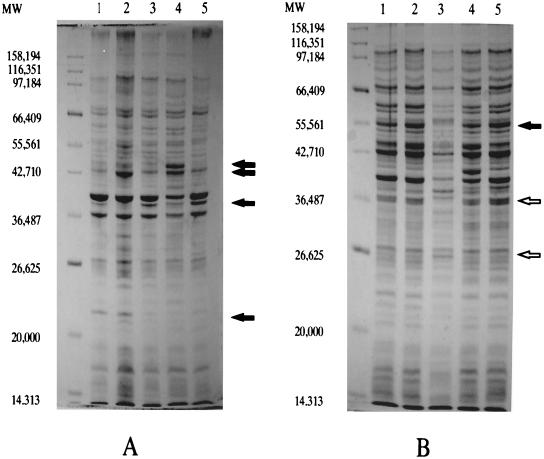
Upon examination of S. typhi outer membrane or whole-cell protein profiles, a smaller number of alterations was observed than for S. typhimurium grown in the presence of bile or deoxycholate (Fig. 4). Two alterations were observed in the presence of bile (one increased protein species and one decreased protein species), while six changes were observed in the presence of deoxycholate (two increased protein species and four decreased protein species). None of the individual bile- or deoxycholate-induced protein alterations overlapped. When examined against S. typhi PhoPc or PhoP− lysates, three of the bile- or deoxycholate-induced alterations appeared similar to those induced by PhoP-PhoQ.
FIG. 4.
Comparison of S. typhi proteins affected by bile or deoxycholate in the growth medium and proteins regulated by PhoP-PhoQ. Membrane extracts (A) and whole-cell extracts (B) were separated by SDS–10% PAGE. Lanes 1 to 5 show protein profiles of the following strains, respectively: wild type, wild type plus 3% bile, wild type plus 1% deoxycholate, PhoPc, and PhoP−. Molecular weight (MW) markers are indicated to the left of the panels. Open arrows denote proteins affected by bile or deoxycholate but not by PhoP-PhoQ. Closed arrows denote proteins affected by bile or deoxycholate and by PhoP-PhoQ. Only those protein species most obviously affected are noted.
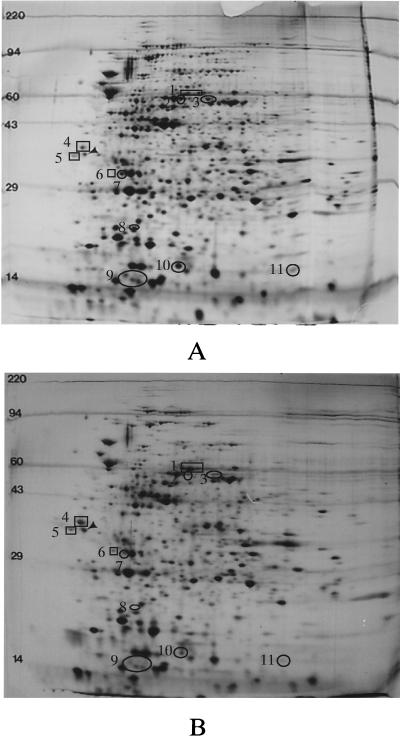
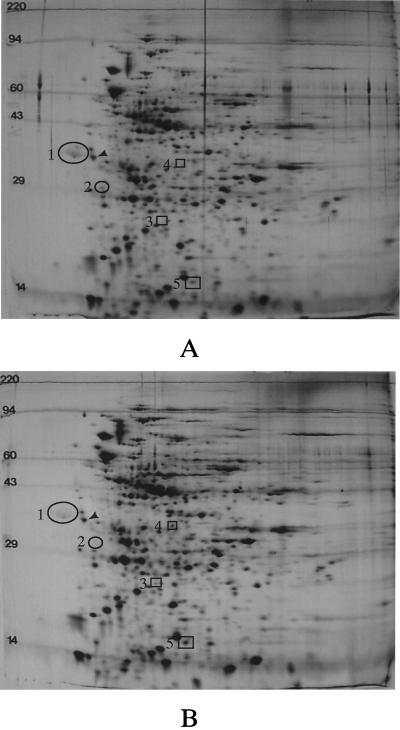
To further confirm protein variations seen in cultures grown in the presence of bile, 2-D gel electrophoresis was performed on S. typhimurium whole-cell protein extracts (Fig. 5). Numerous (∼15) protein species were easily observed as altered by growth in the presence of bile; both increased protein production and decreased protein production were observed. Several protein species were observed at molecular masses corresponding to those seen in the 1-D gel analysis (e.g., ∼16 kDa and ∼32 kDa). In addition, when protein spots altered by the presence of bile were compared to those altered by PhoP-PhoQ activation (27), several overlaps were observed, including protein spots 1, 4, and 5 (Fig. 5). Therefore, as in the 1-D SDS-PAGE analyses and the prgC and prgH transcription assays, significant overlap exists between proteins affected by bile and those affected by PhoP-PhoQ. 2-D gel electrophoresis was also performed on S. typhi; about eight easily observed protein alterations (including both repressed and activated species) were seen (Fig. 6). Surprisingly, little overlap was apparent between proteins affected by bile in S. typhi and in S. typhimurium.
FIG. 5.
Examination by 2-D gel electrophoresis of S. typhimurium proteins affected by bile. (A) Wild-type cells grown in LB broth. (B) Cells grown in LB broth plus 3% bile. Protein species in circles denote those repressed by bile, and protein species in squares denote those activated by bile. Only those proteins most obviously affected by bile are noted. The arrowhead indicates the isoelectric focusing standard (molecular mass, 33 kDa; pI 5.2). Molecular mass standards (in kilodaltons) are noted to the left of each panel.
FIG. 6.
Examination by 2-D gel electrophoresis of S. typhi proteins affected by bile. (A) Wild-type cells grown in LB broth. (B) Cells grown in LB broth plus 3% bile. Protein species in circles denote those repressed by bile, and protein species in squares denote those activated by bile. Only those proteins most obviously affected by bile are noted. The arrowhead indicates the isoelectric focusing standard (molecular mass, 33 kDa; pI 5.2). Molecular mass standards (in kilodaltons) are noted to the left of each panel.
Adaptation of S. typhimurium to bile.
Several enteric organisms have been shown to adapt to lethal concentrations of detergents (including bile) by a short exposure to a sublethal detergent concentration (7). To determine if S. typhimurium possessed the ability to increase resistance to bile or bile salts by pretreatment, cultures were grown to the stationary or log phase in concentrations of bile or deoxycholate lower than the MIC (1 to 3%). Upon challenge of these cells with bile or deoxycholate in a standard MIC assay or in a time-kill assay, surprisingly no significant increase in resistance was observed for pretreated cells (data not shown). With increases in the concentrations of bile pretreatment and challenge (15 and 24%, respectively), still no effect was observed with stationary-phase cultures; however, log-phase cultures demonstrated a significant increase in resistance to bile (Fig. 7). This experiment could not be performed with PhoP− organisms to determine the role of PhoP-PhoQ in adaptation, as they were killed in 15% bile before reaching the log phase.
FIG. 7.
S. typhimurium adaptation to bile. Wild-type S. typhimurium grown logarithmically with no bile or with 15% bile was washed, diluted, and incubated with 24% bile. The percentage of survival was determined at various times. While the results presented are from a single experiment, the experiment was repeated three times, with nearly identical outcomes. Symbols: □, no bile; ◊, 15% bile.
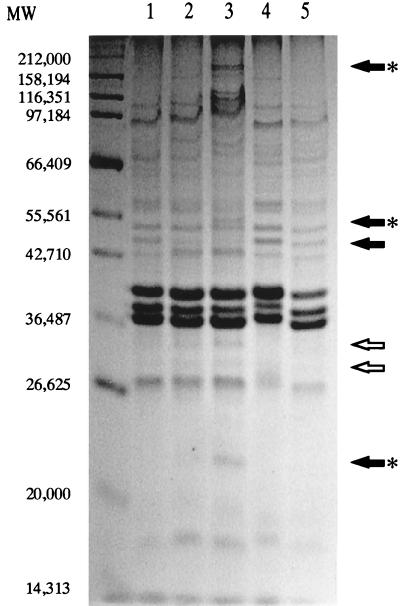
Because a 15% bile concentration was required for adaptation of wild-type S. typhimurium but the protein expression experiments were performed with a 3% bile concentration, we compared the responses of log-phase S. typhimurium to both 3 and 15% bile (Fig. 8). The results showed protein species that were affected similarly and differently by 3 and 15% bile. Of those affected differently, some protein species appeared in 15% bile but not in 3% bile (Fig. 8), and these proteins are candidates as mediators of the adaptation effect.
FIG. 8.
Comparison of logarithmic-phase S. typhimurium proteins affected by 3 and 15% bile. Membrane extracts were separated by SDS–10% PAGE. Lanes 1 to 5 show protein profiles of the following strains, respectively: wild type, wild type plus 3% bile, wild type plus 15% bile, PhoPc, and PhoP−. Molecular weight (MW) markers are indicated to the left of the panels. Open arrows denote obvious proteins affected by both 3 and 15% bile. Closed arrows denote obvious proteins affected by only 15% bile or 3% bile. The asterisks denote examples of proteins that are visible in lane 3 (15% bile) but not lane 2 (3% bile) and that may represent mediators of the adaptation effect seen in organisms grown with 15% bile.
DISCUSSION
Salmonella spp. infecting vertebrate hosts interact with bile in the intestine and, if involved in the carrier state, may interact with bile in the gallbladder or bile duct. Because bile salts are such a potent detergent, Salmonella must possess mechanisms of bile resistance in order to survive. In this work, we demonstrate that Salmonella dramatically alters protein expression in response to bile and that the PhoP-PhoQ two-component regulatory system is a major part of the bile resistance mechanism of Salmonella.
The PhoP-PhoQ two-component regulatory system is required for the virulence of S. typhi in humans (21) and S. typhimurium in mice (6, 26). Genes activated by PhoP-PhoQ include those necessary for the in vivo modification of LPS (16) and survival against the action of cationic antimicrobial peptides (14, 15, 17). This regulatory system has been shown to be induced within macrophage phagosomes (1) and to both activate and repress the transcription of a number of genes (26, 27). Inducing factors in vitro and/or in vivo are pH (1, 4) and the divalent cations Mg2+ and Ca2+, which have been shown to bind to the periplasmic domain of PhoQ (38). Current data suggest that the activation of PhoP-PhoQ within macrophages results in the survival of Salmonella within these cells—a trait highly correlated with the ability of Salmonella to cause disease (3, 6, 26). When not within professional phagocytes, for example, when in the proximity of the intestinal epithelium, PhoP-PhoQ is thought to be uninduced, leading to the expression of PhoP-repressed gene products. PhoP-repressed gene products have been shown to be involved in eukaryotic cell invasion and, more specifically, to be required for the secretion of proteins by a type III mechanism (22, 32). Genes homologous to pag, including phoP and phoQ, have been identified in a wide variety of bacteria other than Salmonella (10–12). This fact has led to the speculation that PhoP-PhoQ, although necessary for Salmonella intramacrophage survival, likely activates the expression of genes whose products may be important for survival in other environments.
S. typhi and S. typhimurium wild-type strains can survive in concentrations of bile greatly exceeding the MIC, while PhoPc strains of each serovar can survive in concentrations of bile exceeding 60%. PhoP− organisms are killed at bile concentrations approximating the MIC (6 to 12%). No individual PhoP-activated or -repressed gene mutant tested showed a reduction in bile resistance, and the S. typhimurium acr and mar loci which are involved in the production of efflux pumps and which are known to be involved in bile resistance (30, 35), were not regulated by PhoP-PhoQ. In addition, combinations of pag mutants, including those in which outer membrane proteins or LPS modifications are known to be affected, showed no effect on bile resistance. Therefore, the PhoP-regulated loci responsible for the bile resistance phenotype are not known.
When individual bile salts were tested against Salmonella wild-type, PhoPc, and PhoP− strains, the resistance mediated by PhoP-PhoQ was specific for deoxycholate and glycochenodeoxycholate but not glycocholate or taurocholate. In addition, PhoP-PhoQ played no role in the resistance of Salmonella to ionic (SDS) or nonionic (Triton X-100) detergents. Primary bile (found in the gallbladder and small intestine) is mainly composed of glyco- or taurocholic acid, glyco- or taurochenodeoxycholic acid, and deoxycholic acid (19). Salmonella spp., which invade primarily in the distal ileum, are likely to encounter only these types of bile salts during infection. Deoxycholate and chenodeoxycholate (for which PhoP-PhoQ plays the largest role in resistance) are dihydroxy bile salts with similar hydrophilic-hydrophobic balances, while the trihydroxy cholate differs considerably in structure and amphipathicity. Dihydroxy bile salts have been shown to penetrate biological membranes better than trihydroxy bile salts (9). It is unclear if the dihydroxy salts have more of an effect on PhoP− strains due to the increased permeability of these mutants or if there exists a more specific, PhoP-mediated mechanism of resistance to these bile acids based on structural or hydrophobic balance differences.
The adaptive response of an organism to pretreatment with a deleterious agent suggests a sensory response that often involves numerous protein species. Enteric organisms, such as E. faecalis, rapidly respond to bile (<30 s) to be able to survive the action of higher bile concentrations (7). S. typhimurium and S. typhi were also able to mount an adaptive response, but only when log-phase organisms were adapted with a high concentration (15%) of bile. Resistance to bile was not increased in stationary- or log-phase organisms grown in 3% bile or stationary-phase organisms grown in 15% bile. Therefore, only actively growing S. typhimurium in the presence of a high concentration of bile is able to adapt to growth in even higher bile concentrations. This bacterial growth phase and the bile environment likely mimic most closely those occurring in the vertebrate host.
The levels of numerous protein species were seen to increase or decrease in both S. typhimurium and S. typhi with the addition of bile or deoxycholate into the medium. Minimal overlap was observed in the proteins affected by bile and by deoxycholate for each serovar, suggesting that the regulatory factors or the regulated target genes differ greatly between S. typhimurium and S. typhi. As determined from the examination of numerous pag and prg reporter gene fusions from cells in both the log and the stationary growth phases, PhoP-PhoQ does not sense and respond to bile in the growth medium. However, a subset of bile- or deoxycholate-activated or -repressed proteins was shown by 1-D or 2-D gel analysis to be identical in pI and/or molecular weight to PhoP-activated and PhoP-repressed proteins, suggesting an overlap in the responses initiated by bile or PhoP-PhoQ sensing.
Data corroborating the above-mentioned protein expression results demonstrated that two PhoP-repressed fusions (prgC and prgH) were dramatically regulated by bile. This regulation was independent of PhoP-PhoQ, as PhoP-PhoQ does not respond to bile and repression of the prgC and prgH fusions was still observed in a PhoP− background. The regulation of the prgH locus is complex, as several regulatory factors appear to be important in prgH expression (23). Therefore, it is possible that a regulatory factor other than PhoP-PhoQ is responsive to bile. As the products of the prgH locus are components of the type III secretion apparatus, a reduction in the expression of these components could lead to the loss of the ability of Salmonella to secrete proteins known to be necessary for epithelial cell invasion. If so, it is possible that bile is an environmental signal for controlling type III secretion, such that type III secretion is shut off within the gallbladder or intestinal lumen but, upon penetration of the mucus layer and arrival of the bacterium in close proximity to the intestinal wall, where the apparent bile concentration is lower, type III secretion and cell invasion are initiated.
Several enteric organisms are rapidly killed by bile; e.g., E. faecalis is nearly completely killed within 30 min by 0.08% bile (7). The kinetics of bile action on Salmonella are not rapid, as complete killing of even PhoP− organisms takes ∼6 h in concentrations of bile that are fivefold higher than the MIC. In addition, even without the high level of bile resistance mediated by PhoP-PhoQ (i.e., PhoPc strains), wild-type S. typhimurium and S. typhi strains are able to survive in higher bile concentrations than are other enteric bacterial pathogens. For example, the MIC for S. typhi and S. typhimurium is >2-fold higher than that for E. coli, Shigella flexneri, Vibrio cholerae, and Aeromonas hydrophila; unlike the MBC for Salmonella, the MBC for these other organisms is identical to the MIC (37a). It is intriguing to speculate that Salmonella is uniquely able to enter into a carrier state associated with the gallbladder because it has developed mechanisms of increased bile resistance, including protein products that are regulated by PhoP-PhoQ.
ACKNOWLEDGMENTS
We are grateful to Karl Klose for helpful suggestions.
This work was supported by the Department of Microbiology at the University of Texas Health Science Center at San Antonio and by an award to the University of Texas Health Science Center at San Antonio for the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute (to J.S.G.).
REFERENCES
- 1.Alpuche-Aranda C M, Swanson J A, Loomis W P, Miller S I. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S J, Daniels C, Morona R. PhoP/Q regulated genes in Salmonella typhi: identification of melittin sensitive mutants. Microb Pathog. 1997;22:165–179. doi: 10.1006/mpat.1996.0099. [DOI] [PubMed] [Google Scholar]
- 3.Barrows P A, Huggins M B, Lovell M A. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun. 1994;62:4602–4610. doi: 10.1128/iai.62.10.4602-4610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearson B L, Wilson L, Foster J W. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belden W J, Miller S I. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flahaut S, Frere J, Boutibonnes P, Auffray Y. Comparison of the bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl Environ Microbiol. 1996;62:2416–2420. doi: 10.1128/aem.62.7.2416-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Vescovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 9.Gordon G S, Moses C, Silver R D, Flier J S, Carey M C. Nasal adsorption of insulin: enhancement by hydrophobic bile salts. Proc Natl Acad Sci USA. 1985;82:7419–7423. doi: 10.1073/pnas.82.21.7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groisman E A, Chiao E, Lipps C J, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groisman E A, Saier M H, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 1992;11:1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn J S, Belden W J, Miller S I. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb Pathog. 1998;25:77–90. doi: 10.1006/mpat.1998.0217. [DOI] [PubMed] [Google Scholar]
- 13.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 15.Gunn J S, Miller S I. PhoP/PhoQ activates transcription of pmrA/B, encoding a two-component system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, Lim K, Gunn J S, Bainbridge B, Darveau R, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 17.Guo, L., K. B. Lim, C. M. Poduje, M. Danial, J. S. Gunn, M. Hackett, and S. I. Miller. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell, in press. [DOI] [PubMed]
- 18.Helander I M, Kato Y, Kilpelainen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem. 1996;237:272–278. doi: 10.1111/j.1432-1033.1996.0272n.x. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann A F. Bile secretion and the enterohepatic circulation of bile acids. In: Feldman M, Scharschmidt B F, Sleisenger M H, editors. Sleisenger and Fordtran’s gastrointestinal and liver disease. 6th ed. Philadelphia, Pa: The W. B. Saunders, Co.; 1998. pp. 937–948. [Google Scholar]
- 20.Hohmann E L, Oletta C A, Killeen K P, Miller S I. phoP/phoQ-deleted Salmonella typhi (TY800) is a safe and immunogenic single dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 21.Hohmann E L, Oletta C A, Miller S I. Evaluation of phoP/phoQ deleted aroA-deleted live oral S. typhi vaccine in humans. Vaccine. 1995;14:19–24. doi: 10.1016/0264-410x(95)00173-x. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 23.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 24.Lai C W, Chan R C Y, Cheng A F B, Sung J Y, Leung J W C. Common bile duct stones: a cause of chronic salmonellosis. Am J Gastroenterol. 1992;87:1198–1199. [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickerson K W, Aspedon A. Detergent-shock response in enteric bacteria. Mol Microbiol. 1992;6:957–961. doi: 10.1111/j.1365-2958.1992.tb02161.x. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido H. Outer membrane of Salmonella typhimurium: transmembrane diffusion of some hydrophobic substances. Biochim Biophys Acta. 1976;433:118–132. doi: 10.1016/0005-2736(76)90182-6. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Basina M, Nguyen V, Rosenberg E Y. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 32.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 33.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukupolvi S, Vaara M. Salmonella typhimurium and Escherichia coli mutants with increased outer membrane permeability to hydrophobic compounds. Biochim Biophys Acta. 1989;988:377–387. doi: 10.1016/0304-4157(89)90011-7. [DOI] [PubMed] [Google Scholar]
- 35.Sulavik M C, Dazer M, Miller P F. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaara M. Increased outer membrane resistance to ethylenediaminetetraacetate and cations in novel lipid A mutants. J Bacteriol. 1981;148:426–434. doi: 10.1128/jb.148.2.426-434.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Van Velkinburgh, J. C., and J. S. Gunn. Unpublished data.
- 38.Waldburger C D, Sauer R T. Signal detection by the PhoQ sensor-transmitter. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]