Abstract
The prevalence and genetic mechanism of antibiotic heteroresistance (HR) have attracted significant research attention recently. However, non-genetic mechanism of HR has not been adequately explored. The present study aimed to evaluate the role of quorum sensing (QS), an important mechanism of behavioral coordination in different subpopulations and consequent heteroresistance. First, the prevalence of HR to 7 antibiotics was investigated in 170 clinical isolates of P. aeruginosa using population analysis profiles. The results showed that P. aeruginosa was significantly heteroresistant to meropenem (MEM), amikacin (AMK), ciprofloxacin (CIP), and ceftazidime (CAZ). The observed HR was correlated with down-regulation of QS associated genes lasI and rhlI. Further, loss-of-function analysis results showed that reduced expression of lasI and rhlI enhanced HR of P. aeruginosa to MEM, AMK, CIP, and CAZ. Conversely, overexpression of these genes or treatment with 3-oxo-C12-HSL/C4-HSL lowered HR of P. aeruginosa to the four antibiotics. Additionally, although downregulation of oprD and upregulation of efflux-associated genes was evident in heteroresistant subpopulations, their expression was not regulated by LasI and RhlI. Moreover, fitness cost measurements disclosed higher growth rates of PAO1ΔlasI and PAO1ΔrhlI in the presence of sub-MIC antibiotic as compared with that of wild-type PAO1. Our data suggest that under temporary antibiotic pressure, downregulation of QS might result in less fitness cost and promote HR of P. aeruginosa.
Keywords: quorum sensing, heteroresistance, Pseudomonas aeruginosa, oprD, efflux pumps gene, fitness cost
Introduction
Heteroresistance (HR) is a phenomenon whereby subpopulations of seemingly isogenic bacteria exhibit different susceptibilities to a particular antibiotic (El-Halfawy and Valvano, 2015). Previous studies have demonstrated the prevalence of heteroresistance in clinical pathogens such as Klebsiella pneumonia (Nimmo et al., 2020), Streptococcus pneumonia (Engel et al., 2014), Mycobacterium tuberculosis (Nimmo et al., 2020), Staphylococcus aureus (Nurjadi et al., 2021), Acinetobacter baumannii (Charretier et al., 2018; Rodriguez et al., 2019), Pseudomonas aeruginosa (Lin et al., 2019; Choby et al., 2021), and Helicobacter pylori (Sun et al., 2018). A heteroresistant subpopulation of bacteria can rapidly replicate in the presence of an antibiotic where other susceptible bacteria cells are killed (Band and Weiss, 2019). This ultimately leads to failure of antibiotic treatment and hence induce bacterial resistance to drugs.
Currently, research on the mechanism of bacterial HR mainly focuses on point mutations. Amplification of chromatin fragments in Gram-negative bacteria has been found to be the prevalent mechanism for balancing resistance and the fitness costs of resistance mutation in the presence of antibiotics by spontaneous tandem amplifications rather than by heritable mutations (Nicoloff et al., 2019). Besides, bacteria can generate subpopulations at a high fitness cost of resistance through point mutations under the selection pressure of antibiotic exposure or compensatory secondary mutations in the absence of antibiotic. Therefore, this reduces the fitness cost and possibly restores the resistance of the native populations (Paradise et al., 1998; Lee et al., 2011). HR may be a strategy for bacteria to adapt to the environment under the antibiotic selection pressure before resistance is acquired. However, it is not yet clear whether a non-genetic mechanism could generate less fitness cost and consequently induce heteroresistance during the short-term evolutionary course. This study evaluated the quorum sensing (QS) system, which is the most common mechanism for coordination of activities in bacterial cells and population-density changes in behavior, such as HR.
Pseudomonas aeruginosa is one of the main causes of nosocomial infections. It is associated with extremely high mortality and morbidity in patients with cystic fibrosis and immunocompromised individuals (Sadikot et al., 2005; Chen et al., 2019). Furthermore, P. aeruginosa, a model bacterium for studying QS systems, has two main QS systems, las and rhl, which are responsible for synthesis of the N-acyl homoserine lactone signal molecules, N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL), and N-butanoyl-L-homoserine lactone (C4-HSL), respectively (Smith and Iglewski, 2003; Chen et al., 2019b). The 3-oxo-C12-HSL and C4-HSL bind to and activate cognate transcription factors, LasR or RhlR, respectively, and induce formation of biofilm and expression of various virulence factors (Moura-Alves et al., 2019). However, resistant P. aeruginosa strains have evolved due to the abuse of antibiotics such as aminoglycosides, quinolones, and β-lactams (De Oliveira et al., 2020). Recently, studies have shown HR in P. aeruginosa to carbapenems (Mei et al., 2015; He et al., 2018), cefepime (Jia et al., 2020), and colistin (Lin et al., 2019). HR may be correlated with upregulation of efflux-associated genes and downregulation of the porin-encoding gene oprD in resistant subpopulations (Ikonomidis et al., 2008). However, the prevalence and mechanism underlying HR are still not clear. Therefore, the current study extensively investigated the mechanism of HR to provide new insights into clinical treatment efficacy for bacterial infections and P. aeruginosa resistance.
The present study focused on the role of QS system in HR of P. aeruginosa. The prevalence of HR to several antibiotics in clinical P. aeruginosa isolates was characterized, which showed that downregulation of LasI/RhlI was associated with increased HR. Further, it was evident that restoration of LasI/RhlI transcription significantly suppressed HR in P. aeruginosa. Notably, the current study showed that fitness cost, but not oprD and efflux-associated genes, was involved in LasI/RhlI-inhibited HR. These findings may provide new and significant insights into regulatory networks of HR, and offer some novel targets for prevention of antibiotic resistance.
Materials and methods
Bacterial strains
A total of 170 non-duplicate clinical P. aeruginosa isolates were obtained from two tertiary hospitals in Guangzhou between 2017 and 2018. The isolates were stored at −80°C and cultured on blood agar plate before the experiments were conducted. All isolates were reconfirmed using matrix-assisted laser desorption ionization-time of flight mass spectrometry. All bacterial strains and plasmids used in the current study are listed in Supplementary Table 1. The PAO1ΔlasI and PAO1ΔrhlI strains had been constructed in our previous study (Zeng et al., 2016; Lu et al., 2017). Briefly, the sacB-based suicide vector system was adapted for knockout of lasI or rhlI in PAO1 bacterial strains.
Plasmid construction
The pROp200 was used to construct plasmid that expressed LasI or RhlI. The coding sequences of LasI or RhlI were cloned into the EcoRΙ site of pROp200 with a Ready-to-Use Seamless Cloning Kit (BBI, United States, Code NO. B632219) to generate expression vectors named pROp200-lasI or pROp200-rhlI.
Antimicrobial susceptibility testing
Meropenem (APE-BIO, United States), Amikacin (BBI Life Science, United States), Ciprofloxacin (BBI Life Science, United States), Ceftazidime (Sigma-Aldrich, United States), Fosfomycin (APE-BIO, United States), polymyxin B (BBI Life Science, United States), and Gentamicin (BBI Life Science, United States) were freshly prepared for each experiment and filter sterilized using a 0.22 μm filter. Mueller-Hinton broth (Oxoid, United Kingdom) supplemented with calcium and magnesium (25.0 mg/l Ca2+ and 12.5 mg/l Mg2+) and Mueller-Hinton II agar (Oxoid, United Kingdom) were used for susceptibility testing in vitro. The antibiotic breakpoints for MEM, AMK, CIP, and CAZ were defined by CLSI-M100-S26. Quality control in the experiments was monitored with P. aeruginosa strain ATCC 27853 and pandrug-resistant 132A5.
Fitness cost measurements
Growth rates were analyzed using the biotek (synergy H1) as previously described (Nicoloff et al., 2019). Four colonies of each test strain were used to inoculate four biological replicates in 3 ml of Mueller-Hinton broth. Overnight cultures were diluted 1:1000 with 3 ml of fresh Mueller-Hinton broth, and then allowed to grow for 24 or 48 h at 37°C. Absorbance was measured at 600 nm (OD600) every 2 h, with cultures shaken between measures.
Population analysis profiles (PAPs)
Heteroresistance of clinical P. aeruginosa isolates and PAO1 was determined using PAPs tests according to a previously published protocol with several modifications (Band et al., 2016). Three colonies were used to start three independent overnight cultures in 3 ml Mueller-Hinton broth. Twenty microliters of gradient dilutions of an overnight culture (101, 102, 103, 104, 105, 106, and 107 CFU/ml) were plated on Mueller-Hinton agar plates supplemented with or without increasing amounts (2-fold increments) of Amikacin (AMK, 1–64 mg/l), Meropenem (MEM, 0.5–16 mg/l), Ciprofloxacin (CIP, 0.25–8 mg/l), Ceftazidime (CAZ, 1–32 mg/l), Fosfomyci (FB, 0.125–8 mg/l), PolymyxinB (PB, 0.125–8 mg/l) or Gentamacin (GM, 0.125–8 mg/l). Plates were incubated overnight at 37°C and colonies were eventually counted and recorded. Heteroresistance was defined as the presence of a subpopulation of cells capable of growing at antibiotic concentrations at least eight-fold higher than the highest concentration that does not affect the replication of the dominant population. Quality control was monitored using P. aeruginosa strain ATCC 27853 and 132A5 (clinical pandrug-resistant strain isolated in our laboratory).
Stability of antibiotic resistance and heteroresistance
Two clones isolated from plates with the highest drug concentration were supplemented with 3 ml of Mueller-Hinton broth and subjected to the same selection pressure overnight. The minimal inhibitory concentrations (MICs) were determined using the broth microdilution method. The cultures were then grown for two more nights. Their MICs after 40 generations in the absence of selective pressure were determined and the resistance was deemed unstable if the MIC clearly decreased or reverted to that of the original parental isolate in at least one of the cultures.
Genes expression analysis
Cultures of original parental strains and heteroresistance subpopulations were grown in CAMHB medium with or without selection pressure at 37°C with shaking to an OD600 of 0.5. Total RNA of each strain was extracted by RNAiso Plus reagent (TaKaRa, Dalian, Liaoning, China). For qPCR analysis, 20 ng of total RNA was subjected to DNase I digestion PrimeScript RT reagent kit (TaKaRa, Dalian, Liaoning, China) at 37°C for 30 min and then to heat inactivation of DNase I at 65°C for 10 min. The cDNA was subjected to quantitative PCR (qPCR) on a ViiATM 7 Dx system (Applied Biosystems, Foster, CA, United States) using the SYBR green qPCR master mixes (TaKaRa, Dalian, Liaoning, China). The temperature cycle profile for the qPCR reactions was 95°C for 30s, and then 40 cycles of 95°C for 5 s, 60°C for 20s, and 72°C for 10s. To verify the specificity of PCR product, melting curve analysis was performed immediately after amplification, as follows: heating to 95°C for 20s, cooling to 60°C for 20s, followed by a temperature increase to 95°C with a transition rate of 0.11°C/s while continuously collecting the fluorescent signal. The transcript levels of the target genes were normalized to the expression of an internal control gene (rpoD) using the 2-ΔΔCt method. All reactions were run in triplicate. The cycle threshold (Ct) values should not differ more than 0.5 among triplicates. Sequences of the primers used in the current study are as listed in Supplementary Table 2.
Pyocyanin production assay
The detection of pyocyanin, rhamnolipid and biofilm formation was performed as described in our previous study (Lu et al., 2019). Three clones isolated from PAPs test plates with 0, 2, 4, 8 or 16 mg/l of AMK were supplemented with 3 ml of Mueller-Hinton broth and subjected to the same selection pressure overnight. Overnight cultures were diluted 1:100 with 10 ml of fresh Mueller-Hinton broth and subjected to the same selection pressure, then allowed to grow for 24 h at 37°C. Pyocyanin in 5 ml of P. aeruginosa culture supernatant was extracted with 3 ml of chloroform and 1 ml of 0.2 N HCl, and then the absorbance of the extract was measured at 520 and 600 nm. The concentration of pyocyanin was determined using the following formula: (A520/A600 × 17.072) = μg/ml (Essar et al., 1990). All of the tests were performed independently and at least in triplicate.
Rhamnolipid assay
Three clones isolated from PAPs test plates with 0, 2, 4, 8 or 16 mg/l of AMK were supplemented with 3 ml of Mueller-Hinton broth and subjected to the same selection pressure overnight. Overnight cultures were diluted 1:100 with 10 ml of fresh M9 (containing 0.4% glucose, 2 mM MgSO4 and 100 μM CaCl2) and subjected to the same selection pressure, then allowed to grow for 20 h at 37°C. Next, 1 ml of culture supernatant was acidified (pH 2.5 ± 0.2) using 1 N HCl, rhamnolipid was extracted with 4 ml chloroform. Then 3 ml of chloroform extract was allowed to react with 100 μl of 1 g/l of methylene blue and 2 ml of distilled water, the OD638 was measured (Pinzon and Ju, 2009). All results were analyzed of three independent experiments.
Biofilm formation assay
Three clones isolated from PAPs test plates with 0, 2, 4, 8 or 16 mg/l of AMK were supplemented with 3 ml of Mueller-Hinton broth and subjected to the same selection pressure overnight. Overnight cultures were diluted 1:100 with 10 ml of fresh Mueller-Hinton broth, 20 ml of the dilutions were plated into 96-well culture and subjected to the same selection pressure, then allowed to grow for 24 h at 37°C. Biofilm cells attached to plates were stained with 1% crystal violet. Then, 2 ml of 95% ethanol was used to solubilize crystal violet-stained biofilm cells and measured at OD600 (Carloni et al., 2017). All analyses included three independent experiments.
Statistical analysis
Data are expressed as the means ± standard errors of the means (SEMs) from at least three independent experiments. The differences between groups were analyzed using the Student’s t-test when two groups were compared or a one-way analysis of variance (ANOVA) when more than two groups were compared. All analyses were performed using GraphPad Prism, version 5 (GraphPad Software, Inc., San Diego, CA, United States). All statistical tests were two-sided; p values of <0.05 were considered statistically significant.
Results
Prevalence of HR in Clinical Pseudomonas aeruginosa isolates
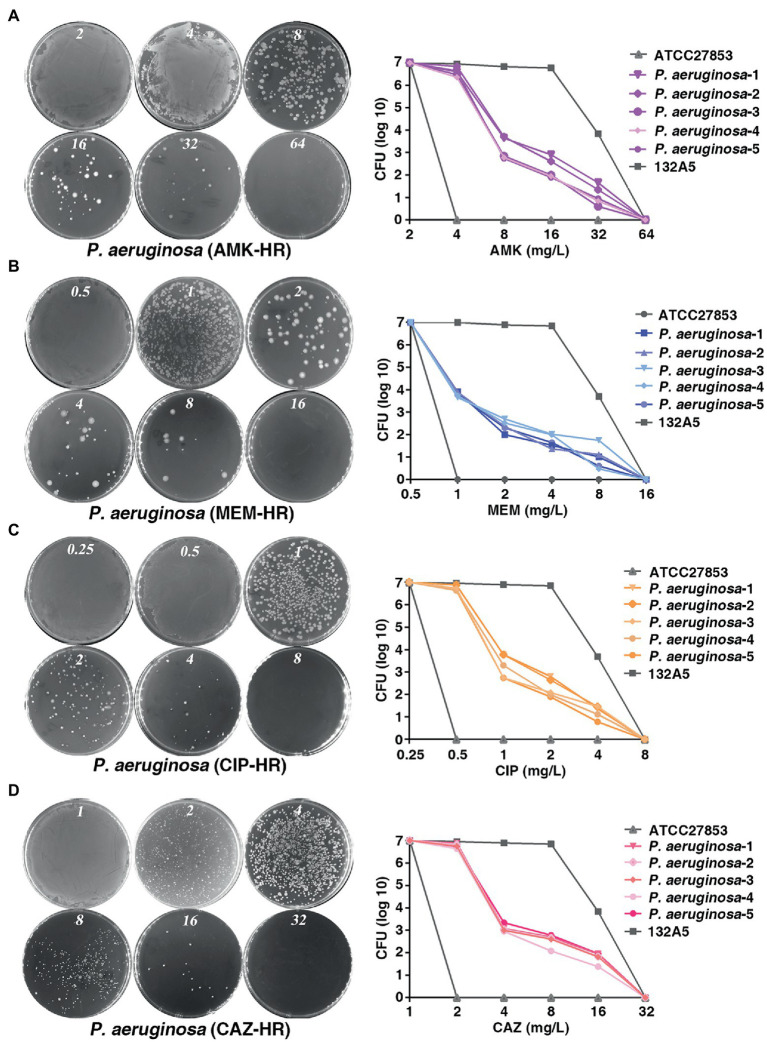
To investigate the prevalence of HR to different antibiotics covering major classes in the clinical P. aeruginosa isolates, a total of 170 strains were randomly selected and PAPs was performed based on the HR definition by Herve and Karin (Nicoloff et al., 2019). They defined HR as the presence of a subpopulation of cells capable of growing at antibiotic concentrations at least eight-fold higher than the highest concentration that does not affect the replication of the dominant population and resistant subpopulations at frequencies of 1 × 10−7 or higher. Of the 170 isolates, 113 (66.5%), 99 (58.2%), 87 (51.2%) and 69 (34.1%) were heteroresistant to amikacin (AMK; Figure 1A), meropenem (MEM; Figure 1B), ciprofloxacin (CIP; Figure 1C) and to ceftazidime (CAZ; Figure 1D), respectively. Notably, 55 isolates (32.4%) showed a cross-HR to all the four antibiotics (AMK, MEM, CIP, and CAZ). In conclusion, a high frequency of HR to AMK, MEM, CIP, and CAZ was observed in different clinical isolates of P. aeruginosa (Table 1). However, HR with fosfomyci (FB), PolymyxinB (PB) and Gentamacin (GM) was not observed in the present investigation. In addition, the current study also assayed the heteroresistance of PAO1, a strain most commonly used in laboratory research. Results of the present study showed that PAO1 was also heteroresistant to MEM, AMK, CIP, and CAZ (Supplementary Figure 1).
Figure 1.
Prevalence of heteroresistance in clinical Pseudomonas aeruginosa isolates. (A–D) P. aeruginosa was heteroresistant to AMK, MEM, CIP and CAZ, respectively. PAPs were performed in five clones of P. aeruginosa isolate. Both the sensitive (ATCC 27853) and resistant strains (132A5) of P. aeruginosa were used as control. AMK, amikacin; MEM, meropenem; CIP, ciprofloxacin; CAZ, ceftazidime.
Table 1.
Heteroresistance (HR) of clinical Pseudomonas aeruginosa isolates(n = 170).
| Antibiotics | Resistance | Resistance Ratio(%) | HR | HR Ratio(%) |
|---|---|---|---|---|
| AMK | 37 | 21.8 | 113 | 66.5 |
| MEM | 50 | 29.4 | 99 | 58.2 |
| CIP | 44 | 25.9 | 87 | 51.2 |
| CAZ | 15 | 8.8 | 58 | 34.1 |
| FOS | 10 | 5.8 | 0 | 0 |
| PB | 15 | 8.8 | 0 | 0 |
| GM | 12 | 7.0 | 0 | 0 |
Expression of LasI and RhlI is negatively correlated with HR of Pseudomonas aeruginosa
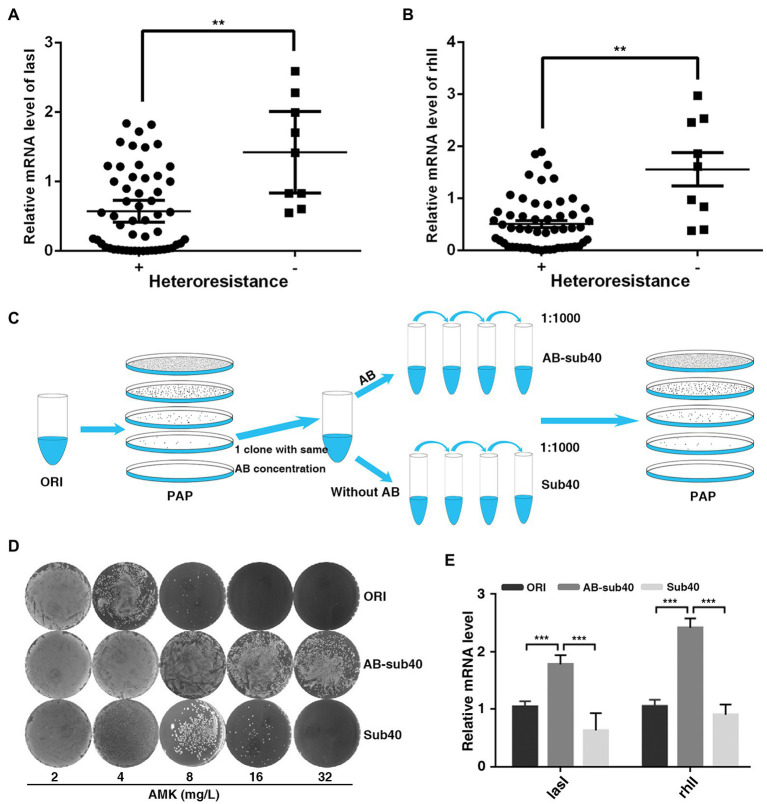
To determine the biological significance of QS system in regulation of heteroresistant, the current study analyzed the correlation between LasI/RhlI levels and heteroresistant features. In total, 55 isolates displayed HR to all four antibiotics (MEM, AMK, CIP, and CAZ) were defined as heteroresistant positive, whereas 9 isolates that showed no heteroresistance to any of these antibiotics were defined as heteroresistant negative. The expression of LasI and RhlI and QS-regulated biofilm formation potential in heteroresistant positive group of isolates was lower than that in the negative group (Figures 2A,B; Supplementary Figure 2A). Furthermore, compared with original, expression of LasI and RhlI and QS-regulated rhamnolipid, pyocyanin synthesis and biofilm formation in resistant subpopulations was also down-regulated (Supplementary Figures 2B–E). These results indicated that QS system may be negatively correlated with heteroresistance in P. aeruginosa.
Figure 2.
Expression of LasI and RhlI is negatively correlated with the HR of P. aeruginosa. (A,B) Expression of LasI and RhlI was lower in the heteroresistance-positive group compared with in the heteroresistance-negative group. (C) A schematic outline of the methods used to assess the stability of HR. A resistant colony from the PAPs test plates was re-streaked and grown at the same concentration of antibiotics (Enrich), followed by serial passages with or without antibiotic selection for 40 generations (1, 1,000 daily dilution). (D) After serial passages in antibiotic-free medium, resistant subpopulations reverted to the heterogeneous resistance phenotype showed by the original population. (E) The expression of LasI and RhlI in the original PAO1 (ORI), resistant subpopulations after 40 generations with or without exposure to antibiotics (AB-sub40 and sub40, respectively). **, p < 0.01; ***, p < 0.001.
Typically, after serial passages in antibiotic-free medium, some highly resistant subpopulations of P. aeruginosa revert to the heterogeneous resistance phenotype shown by the original population (Nicoloff et al., 2019). Therefore, the phenotypic stability and expression of LasI and RhlI was assessed in PAO1 in the present study. PAPs analysis was simultaneously performed on the original PAO1 (ORI) and resistant subpopulations (Enrich), after 40 generations with or without exposure to AMK or CIP (AB-sub40 and Sub40 respectively, Figure 2C). Results showed that ORI and Sub40 were heteroresistant positive, whereas the AB-sub40 became to the resistant strain (Figure 2D; Supplementary Figure 3A). This was an indication that resistance of subpopulations (Enrich) would be effectively enhanced by exposure to long-term antibiotics pressure. It was also demonstrated that the resistance of subpopulations (Enrich) was almost restored to the level of native populations after 40 generations without exposure to antibiotics. Furthermore, it was found that the expression of LasI and RhlI in AB-sub40 group was higher than that in the ORI and Sub40 groups (Figure 2E; Supplementary Figure 3B). These results showed that QS system may be negatively correlated with heteroresistance in P. aeruginosa.
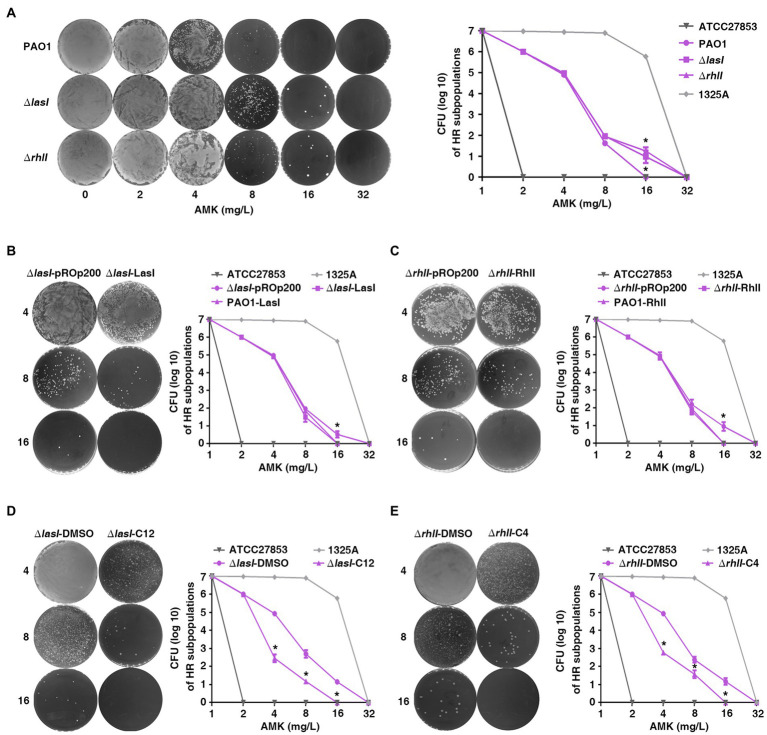
LasI and RhlI inhibit HR in Pseudomonas aeruginosa
To explore the effect of QS on heteroresistance to antibiotics in P. aeruginosa, PAPs was also performed with PAO1 and lasI or the rhlI single gene-deficient strains (PAO1ΔlasI and PAO1ΔrhlI). Subpopulations of PAO1ΔlasI and PAO1ΔrhlI showed an enhanced potential to grow in high concentration of AMK (Figure 3A) compared with the wild-type. This indicated that the deficiency of lasI or rhlI significantly enhanced heteroresistance of P. aeruginosa to AMK. A similar phenomenon was also observed with MEM, CIP, and CAZ antibiotics (Supplementary Figure 4).
Figure 3.
The quorum sensing system of P. aeruginosa inhibits heteroresistance. (A) Deficiency of lasI or rhlI in PAO1 enhanced heteroresistance. *, p < 0.05, PAO1ΔlasI or PAO1ΔrhlI compared with PAO1; (B,C) Over expression of lasI or rhlI inhibited heteroresistance. *, p < 0.05, ΔlasI-LasI compared with ΔlasI-pROp200; ΔrhlI-RhlI compared with ΔrhlI-pROp200. (D,E) Exogenous 3-oxo-C12-HSL (C12) or C4-HSL (C4) inhibited heteroresistance. The HR tests (PAPs) were carried out on PAO1ΔlasI and PAO1ΔrhlI treated with 40 μM of 3-oxo-C12-HSL and C4-HSL, respectively, *, p < 0.05, ΔlasI-C12 compared with ΔlasI-DMSO; ΔrhlI-C12 compared with ΔrhlI-DMSO. ΔlasI-pROp200: PAO1ΔlasI strain transformed with pROp200 plasmid; ΔlasI-LasI: PAO1ΔlasI strain transformed with pROp200 plasmid that expressed LasI; ΔlasI-DMSO: PAO1ΔlasI strain treated with DMSO; ΔlasI-C12: PAO1ΔlasI strain treated with 3-oxo-C12-HSL; ΔrhlI-DMSO: PAO1ΔrhlI strain treated with DMSO; ΔrhlI-C4: PAO1ΔrhlI strain treated with C4-HSL.
To confirm the results of loss-of-function analysis, we performed a gain-of-function analysis using lasI-overexpression plasmid or extracellular signal N-(3-oxododecanoyl)-L-HSL (3-oxo-C12-HSL) for PAO1ΔlasI and rhlI-overexpression plasmid or N-butanoyl-L-homoserine lactone (C4-HSL) for PAO1ΔrhlI. Transformation of lasI-overexpression plasmid (ΔlasI-LasI) or treatment with 3-oxo-C12-HSL (ΔlasI-C12) caused significant decrease in subpopulations at 8 and 16 mg/l of AMK for the PAO1ΔlasI strain as compared with the control group (ΔlasI-pROp200 or ΔlasI-DMSO; Figures 3B,D). Further, the overexpression of RhlI or treatment with C4-HSL also impaired the viability of subpopulations for PAO1ΔrhlI strain in AMK (Figures 3C,E). In general, the data obtained in the current study showed that QS system may negatively regulate the heteroresistance of P. aeruginosa.
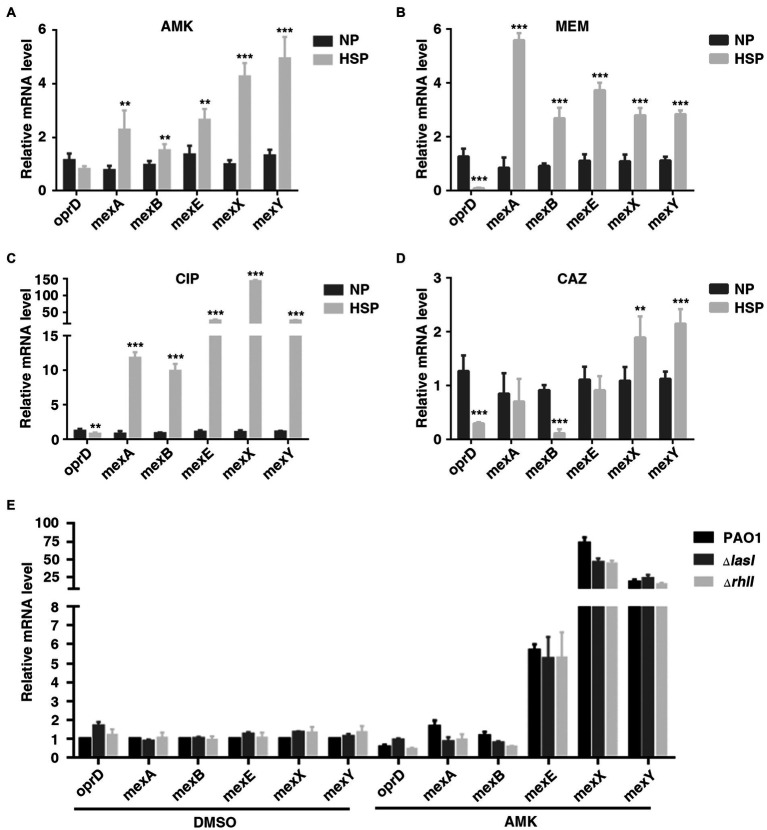
OprD and efflux-associated genes are not involved in QS-inhibited HR in Pseudomonas aeruginosa
A previous study showed that reduced expression of porin gene oprD and increased expression of efflux-associated genes were associated with carbapenem heteroresistance in P. aeruginosa (He et al., 2018). The transcription of these genes in PAO1 HR subpopulations (HSP) and native populations (NP) was also tested in different antibiotics. Consistently, the expression of oprD was lower in HSP compared with NP for antibiotics AMK, MEM, and CAZ. On the other hand, it was found that the transcription of efflux-associated genes, including mexA, mexB, mexE, mexX, and mexY increased in HSP for antibiotics AMK, MEM, and CIP (Figures 4A–D). Therefore, the results of the current study demonstrated that both reduced transcription of porin gene oprD and increased transcription of efflux-associated genes conferred the subpopulations antibiotic resistance.
Figure 4.
OprD and efflux-associated genes were not involved in QS-inhibited HR. (A–D) Expression of OprD and efflux-associated genes in native population (NP) and resistant subpopulation (HSP) of AMK, MEM, CIP, or CAZ. A resistant colony growing on PAPs test plates with highest antibiotic concentration was re-streaked with the same concentration of the indicated antibiotics, followed by real-time PCR analysis. (E) The expression of oprD and efflux-associated genes was not regulated by LasI and RhlI. PAO1, PAO1ΔlasI, and PAO1ΔrhlI were cultured in LB for 6 h before qPCR analysis. The rpoD gene was used as an internal control. **, p < 0.01; ***, p < 0.001.
The present study also confirmed the involvement of oprD and efflux-associated genes in QS system-regulated heteroresistance. It was found that treatment with 2 mg/l of AMK caused downregulation of oprD and up-regulation of MexX/MexY. However, compared with wild-type, PAO1ΔlasI or PAO1ΔrhlI showed no difference in the expression of oprD and the efflux-associated genes (Figure 4E). Generally, the obtained data showed that the inhibitory effect of QS system on heteroresistance of P. aeruginosa was not mediated by oprD and efflux-associated genes.
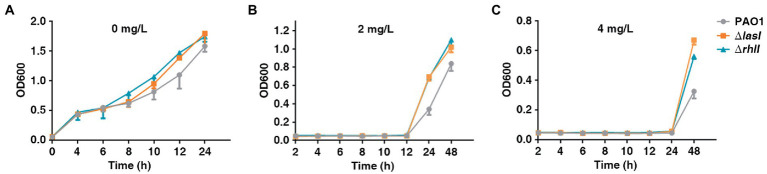
Deficiency of lasI or rhlI favors the survival of PAO1 under sub-MIC concentration of antibiotics
The present study also investigated the growth rate of PAO1 wild type, PAO1ΔlasI, and PAO1ΔrhlI because the fitness cost was found to be significant in reversing HR. PAO1ΔlasI and PAO1ΔrhlI grew at a higher rate in the absence of antibiotics compared with PAO1 wide-type (Figure 5A). Further, PAO1ΔlasI and PAO1ΔrhlI also showed better relative growth rates upon exposure to AMK. Sub-MIC (2 or 4 mg/l) of AMK was used because only a few resistant subpopulations could grow under 2 or 4 mg/l of AMK. Results of this investigation showed that deficiency of lasI or rhlI favored the survival of PAO1 when exposed to 2 or 4 mg/l of AMK (Figures 5B,C). Moreover, previous transcriptome sequencing data (Pu et al., 2022) had found that deficiency of lasI or rhlI observably depressed expression of several genes, especially the virulence-related genes and impaired virulence such as pyocyanin, rhamnolipid synthesis and biofilm formation (Supplementary Table 3; Supplementary Figure 5). Therefore, it was inferred that lasI and rhlI deficiency-mediated downregulation of the genes resulted in lower consumption, which could be one of the strategies for bacteria to adapt to the Sub-MIC of antibiotic pressure before resistance is acquired.
Figure 5.
The growth rate of PAO1, PAO1ΔlasI, and PAO1ΔrhlI in the presence or absence of antibiotic selection. (A–C) PAO1, PAO1ΔlasI, and PAO1ΔrhlI were treated with 0, 2 or 4 mg/l of AMK.
Discussion
Heteroresistance is a common resistance phenotype among clinical isolates that is the least investigated among the standard estimates of MICs. P. aeruginosa shows a high prevalence of HR, which affects the efficacy of clinical treatment courses based antibiotics (Band and Weiss, 2019). HR of P. aeruginosa to carbapenems, colistin, and cefepime has been previously demonstrated. However, there is need for a comprehensive study on the HR of P. aeruginosa. The current study aimed to first detect HR in P. aeruginosa to several antibiotics that are mainly used in clinical treatment. Results of the present study showed a high prevalence of P. aeruginosa-HR to carbapenems (MEM) and β-lactam (CAZ) as well as to quinolones (CIP) and aminoglycosides (AMK) antibiotics (Figure 1). To the best of our knowledge, the HR of P. aeruginosa to CIP, AMK, and CAZ shown in present study has not been previously reported. Therefore, we concluded that heteroresistance phenomenon may significantly contribute to the failure of treatment based on MEM, AMK, CIP, and CAZ antibiotics.
Recent studies have focused on the genetic mechanism-underlying heteroresistance. However, the present study identified QS system as a non-genetic mechanism for heteroresistance. In clinical isolated strains and hereditary stability model, HR was inversely associated with the level of LasI and RhlI (Figure 2A). In addition, gain-of-function and loss-of-function studies showed that QS repressed HR (Figure 3). Similarly, it has been found that indoles, which is a chemical signal produced by the resistant subpopulations of E. coli, can also help sensitive subpopulations adapt to antibiotics (Lee et al., 2010). In addition, resistant subpopulations of Burkholderia cenocepacia were shown to secrete putrescine and YceI proteins to improve the resistance of subpopulations that were sensitive to polymyxin B (Loutet et al., 2011). These studies show that chemical signals involved in the interaction between bacteria may be an important mechanism regulating HR.
The hallmarks of HR are the increase in the frequency of resistant subpopulations when exposed to antibiotic drugs and a decrease after removal of the antibiotic effect (Nicoloff et al., 2019). Previous studies have shown that increased resistance of the subpopulations resulted from spontaneous tandem amplifications, typically including known resistance genes. However, the procedure is costly and unstable, hence it may promote reversion to susceptibility in the absence of antibiotic selection (Nicoloff et al., 2019). In the current study, after 40 passages through antibiotic-free medium, the highly resistant subpopulations reverted to the heterogeneous resistant phenotype exhibited by the original population (Figure 2B). Furthermore, the course was accompanied by the change in expression of LasI, RhlI and QS-regulated virulence such as rhamnolipid, pyocyanin synthesis and biofilm formation (Supplementary Figures 2B–E). Accordingly, lower hamnolipid, pyocyanin synthesis and biofilm formation (Supplementary Figure 5), and better relative growth rates in strains deficient of lasI or rhlI upon exposure to antibiotics (Figure 5). In addition, the clinical isolates of P. aeruginosa naturally evolves lasR mutations during chronic infection. This forms the “cheater” populations that effectively stops the secretion of protease in support of utilizing public goods, thereby establishing a fitness advantage against antibiotics. Therefore, it was speculated that in the temporary presence of selective pressure, downregulation of QS genes may result in less fitness cost, which could be an important reason for the acquisition of heteroresistance against antibiotics.
OprD porin and efflux systems were introduced into mechanism research in the present study. Lower expression of oprD was observed in MEM and CAZ heteroresistant subpopulation compared with the dominant population (Figures 4B,D). While the genes associated with efflux increased in carbapenems, quinolones, and aminoglycosides heteroresistant subpopulation (Figures 4A,B,D). These results evidently confirmed the previous reports that OprD porin and efflux systems could have contributed to HR in P. aeruginosa on different antibiotics (Lin et al., 2019; Jia et al., 2020). Moreover, a little downregulation of oprD and obvious up-regulation of MexX/MexY was observed in PAO1 wild-types, PAO1ΔlasI and PAO1ΔrhlI treated with 2 mg/l of AMK (Figure 4E). These results could be related to the mechanism of P. aeruginosa resistance to antibiotics (Mesaros et al., 2007; Moradali et al., 2017; Xu et al., 2020). However, the expression of OprD and efflux systems showed no significant difference between wild-type and lasI or rhlI deficient strain (Figure 4E). The findings of the current study indicate that the inhibitory effect of QS system on the heteroresistance of P. aeruginosa is not mediated by oprD and efflux-associated genes.
In conclusion, the present study highlighted the regulatory role of the QS system in heteroresistance of P. aeruginosa, expanding the present understanding of the mechanism of antibiotic resistance in P. aeruginosa.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the corresponding authors.
Author contributions
YuL, YL, and CZ performed the experiments. YaL, YiL, QX, and DL described and discussed the results of the study. BH, CC, and YL were involved in data analysis and organization as well as preparation of the draft manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81871703), the Guangzhou Science Technology and Innovation Commission (Grant No. 202102010193), and Guangdong Provincial Hospital of Traditional Chinese Medicine (Grant numbers YN2018QJ01, YN2019QJ03, and YN2019MJ01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1017707/full#supplementary-material
References
- Band V. I., Crispell E. K., Napier B. A., Herrera C. M., Tharp G. K., Vavikolanu K., et al. (2016). Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 1:16053. doi: 10.1038/nmicrobiol.2016.53, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band V. I., Weiss D. S. (2019). Heteroresistance: a cause of unexplained antibiotic treatment failure? PLoS Pathog. 15:e1007726. doi: 10.1371/journal.ppat.1007726, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carloni S., Macchi R., Sattin S., Ferrara S., Bertoni G. (2017). The small RNA ReaL: a novel regulatory element embedded in the Pseudomonas aeruginosa quorum sensing networks. Environ. Microbiol. 19, 4220–4237. doi: 10.1111/1462-2920.13886, PMID: [DOI] [PubMed] [Google Scholar]
- Charretier Y., Diene S. M., Baud D., Chatellier S., Santiago-Allexant E., van Belkum A., et al. (2018). Colistin Heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob. Agents Chemother. 62:18. doi: 10.1128/AAC.00788-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Déziel E., Groleau M.-C., Schaefer A. L., Greenberg E. P. (2019). Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc. Natl. Acad. Sci. U. S. A. 116, 7021–7026. doi: 10.1073/pnas.1819801116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choby J. E., Ozturk T., Satola S. W., Jacob J. T., Weiss D. S. (2021). Widespread cefiderocol heteroresistance in carbapenem-resistant gram-negative pathogens. Lancet Infect. Dis. 21, 597–598. doi: 10.1016/S1473-3099(21)00194-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira D. M. P., Forde B. M., Kidd T. J., Harris P. N. A., Schembri M. A., Beatson S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, 247–255. doi: 10.1128/CMR.00181-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Halfawy O. M., Valvano M. A. (2015). Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. doi: 10.1128/CMR.00058-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H., Mika M., Denapaite D., Hakenbeck R., Mühlemann K., Heller M., et al. (2014). A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 3934–3941. doi: 10.1128/AAC.02547-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Hadero A., Crawford I. P. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172, 884–900. doi: 10.1128/jb.172.2.884-900.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Jia X., Yang S., Xu X., Sun K., Li C., et al. (2018). Heteroresistance to carbapenems in invasive Pseudomonas aeruginosa infections. Int. J. Antimicrob. Agents 51, 413–421. doi: 10.1016/j.ijantimicag.2017.10.014, PMID: [DOI] [PubMed] [Google Scholar]
- Ikonomidis A., Tsakris A., Kantzanou M., Spanakis N., Maniatis A. N., Pournaras S. (2008). Efflux system overexpression and decreased OprD contribute to the carbapenem heterogeneity in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 279, 36–39. doi: 10.1111/j.1574-6968.2007.00997.x, PMID: [DOI] [PubMed] [Google Scholar]
- Jia X., Ma W., He J., Tian X., Liu H., Zou H., et al. (2020). Heteroresistance to cefepime in Pseudomonas aeruginosa bacteraemia. Int. J. Antimicrob. Agents 55:105832. doi: 10.1016/j.ijantimicag.2019.10.013, PMID: [DOI] [PubMed] [Google Scholar]
- Lee H.-Y., Chen C.-L., Wang S.-B., Su L.-H., Chen S.-H., Liu S.-Y., et al. (2011). Imipenem heteroresistance induced by imipenem in multidrug-resistant Acinetobacter baumannii: mechanism and clinical implications. Int. J. Antimicrob. Agents 37, 302–308. doi: 10.1016/j.ijantimicag.2010.12.015, PMID: [DOI] [PubMed] [Google Scholar]
- Lee H. H., Molla M. N., Cantor C. R., Collins J. J. (2010). Bacterial charity work leads to population-wide resistance. Nature 467, 82–85. doi: 10.1038/nature09354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Xu C., Fang R., Cao J., Zhang X., Zhao Y., et al. (2019). Resistance and Heteroresistance to Colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob. Agents Chemother. 63:19. doi: 10.1128/AAC.00556-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet S. A., Di Lorenzo F., Clarke C., Molinaro A., Valvano M. A. (2011). Transcriptional responses of Burkholderia cenocepacia to polymyxin B in isogenic strains with diverse polymyxin B resistance phenotypes. BMC Genomics 12:472. doi: 10.1186/1471-2164-12-472, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Li H., Pu J., Xiao Q., Zhao C., Cai Y., et al. (2019). Identification of a novel RhlI/R-PrrH-LasI/Phzc/PhzD signalling cascade and its implication in P. aeruginosa virulence. Emerg. Microbes Infect. 8, 1658–1667. doi: 10.1080/22221751.2019.1687262, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zeng J., Wu B., E S., Wang L., Cai R., et al. (2017). Quorum sensing N-acyl Homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell. Infect. Microbiol. 7:7. doi: 10.3389/fcimb.2017.00007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S., Gao Y., Zhu C., Dong C., Chen Y. (2015). Research of the heteroresistance of Pseudomonas aeruginosa to imipenem. Int. J. Clin. Exp. Med. 8, 6129–6132. PMID: [PMC free article] [PubMed] [Google Scholar]
- Mesaros N., Nordmann P., Plésiat P., Roussel-Delvallez M., Van Eldere J., Glupczynski Y., et al. (2007). Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 13, 560–578. doi: 10.1111/j.1469-0691.2007.01681.x, PMID: [DOI] [PubMed] [Google Scholar]
- Moradali M. F., Ghods S., Rehm B. H. A. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-Alves P., Puyskens A., Stinn A., Klemm M., Guhlich-Bornhof U., Dorhoi A., et al. (2019). Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 366, –a012427. doi: 10.1126/science.aaw1629, PMID: [DOI] [PubMed] [Google Scholar]
- Nicoloff H., Hjort K., Levin B. R., Andersson D. I. (2019). The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514. doi: 10.1038/s41564-018-0342-0, PMID: [DOI] [PubMed] [Google Scholar]
- Nimmo C., Brien K., Millard J., Grant A. D., Padayatchi N., Pym A. S., et al. (2020). Dynamics of within-host mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMedicine 55:102747. doi: 10.1016/j.ebiom.2020.102747, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurjadi D., Chanthalangsy Q., Zizmann E., Stuermer V., Moll M., Klein S., et al. (2021). Phenotypic detection of Hemin-inducible trimethoprim-Sulfamethoxazole Heteroresistance in Staphylococcus aureus. Microbiol. Spectr. 9:e0151021. doi: 10.1128/Spectrum.01510-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradise M. R., Cook G., Poole R. K., Rather P. N. (1998). Mutations in aarE, the ubiA homolog of Providencia stuartii, result in high-level aminoglycoside resistance and reduced expression of the chromosomal aminoglycoside 2’-N-acetyltransferase. Antimicrob. Agents Chemother. 42, 959–962. doi: 10.1128/AAC.42.4.959, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon N. M., Ju L.-K. (2009). Analysis of rhamnolipid biosurfactants by methylene blue complexation. Appl. Microbiol. Biotechnol. 82, 975–981. doi: 10.1007/s00253-009-1896-9, PMID: [DOI] [PubMed] [Google Scholar]
- Pu J., Zhang S., He X., Zeng J., Shen C., Luo Y., et al. (2022). The small RNA AmiL regulates quorum sensing-mediated virulence in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 10:e0221121. doi: 10.1128/spectrum.02211-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C. H., Traglia G., Bastias N., Pandolfo C., Bruni G., Nastro M., et al. (2019). Discrepancies in susceptibility testing to colistin in Acinetobacter baumannii: the influence of slow growth and heteroresistance. Int. J. Antimicrob. Agents 54, 587–591. doi: 10.1016/j.ijantimicag.2019.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. doi: 10.1164/rccm.200408-1044SO, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. S., Iglewski B. H. (2003). P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6, 56–60. doi: 10.1016/s1369-5274(03)00008-0 [DOI] [PubMed] [Google Scholar]
- Sun L., Talarico S., Yao L., He L., Self S., You Y., et al. (2018). Droplet digital PCR-based detection of clarithromycin resistance in helicobacter pylori isolates reveals frequent Heteroresistance. J. Clin. Microbiol. 56:019. doi: 10.1128/JCM.00019-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang D., Zhang X., Liu H., Zhu G., Wang T., et al. (2020). Mechanisms for rapid evolution of Carbapenem resistance in a clinical isolate of Pseudomonas aeruginosa. Front. Microbiol. 11:1390. doi: 10.3389/fmicb.2020.01390, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Zhang N., Huang B., Cai R., Wu B., E S., et al. (2016). Mechanism of azithromycin inhibition of HSL synthesis in Pseudomonas aeruginosa. Sci. Rep. 6:24299. doi: 10.1038/srep24299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the corresponding authors.