Abstract
The gene encoding a protective protein antigen of the gram-positive bacterium Erysipelothrix rhusiopathiae, an important veterinary pathogen responsible for erysipelas in swine and a variety of diseases in animals, was cloned and sequenced. The gene encodes a polypeptide of 597 amino acids plus a putative signal sequence of 29 amino acids, resulting in a mature protein with a molecular mass of 69,017 Da. Sequence analysis of the gene product revealed a C-terminal region composed of nine tandem repeats of 20 amino acids and a total sequence that is nearly identical to that of the 64-kDa cell surface protein (SpaA) of the bacterium. Because of this similarity, the protein was designated SpaA.1. In this study, we examined whether the SpaA.1 protein could induce protective antibodies and whether we could identify the region involved in protective immunity. Both the mature SpaA.1 protein and its C-terminal repeat region, but not the N-terminal segment, were expressed in Escherichia coli and purified as a histidine-tagged fusion recombinant protein. Rabbit antiserum raised against the mature SpaA.1 protein passively protected mice from lethal challenge with a virulent homologous strain, Fujisawa-SmR, suggesting that protection is mediated by humoral antibodies. To determine which domain of the SpaA.1 protein is responsible for the observed protection, mice were actively immunized with either the mature SpaA.1 protein or the C-terminal repeat region and then challenged with Fujisawa-SmR. The result showed that mice immunized with the mature SpaA.1 protein, but not the C-terminal repeat region, were protected, suggesting that the protection-eliciting epitope(s) is located within the N-terminal two-thirds of the SpaA.1 molecule. This was confirmed by passive immunization experiments in which the protective activity of rabbit antiserum, raised against mature SpaA.1 protein, was not abolished by absorption with the purified recombinant C-terminal repeat region. In addition, antibodies specific for the C-terminal repeat region were unable to protect mice from lethal challenge. These results show that the N-terminal two-thirds of the SpaA.1 molecule may constitute a good vaccine candidate against erysipelas.
The gram-positive bacterium Erysipelothrix rhusiopathiae is the causative agent of erysipelas in animals and erysipeloid in humans (17, 18). The organism causes great economic losses to the swine and turkey industries (17, 18). In swine, the organism may cause acute septicemia or chronic disease typically characterized by endocarditis and polyarthritis (18). Live attenuated vaccines or bacterins have been used for the control of swine erysipelas for many years. However, it has been suggested that the currently available vaccines do not prevent the chronic form of the disease and that vaccination may cause an increase in arthritis lesions (18). Thus, there is a clear need for the development of a more effective and safer vaccine.
In erysipelas, antibodies against E. rhusiopathiae have been known to play an important role in protection (17–19), suggesting that antibodies against a cell surface component(s) of the organism are important in protective immunity. However, studies on the immunogenicity of antigens in these organism have been few (4), and those antigens involved in protection have not been fully characterized. Recently, we showed that the capsule is the major virulence determinant of E. rhusiopathiae, protecting the organism from phagocytosis by polymorphonuclear leukocytes and intracellular killing by macrophages (12, 13). However, the capsule of E. rhusiopathiae is poorly immunogenic, and mice immunized with purified capsular antigen were not protected from subsequent lethal challenge with a virulent homologous strain (13a), suggesting that a molecule(s) on the cell surface other than capsular antigen may be important in inducing protective antibodies.
It has been reported that a 200-kDa glycolipoprotein complex in the culture supernatant is a protective antigen of E. rhusiopathiae (15, 16). This antigen was able to adsorb passively protective antibodies from rabbit antiserum produced by immunization with whole culture, suggesting that this molecule is a major protective antigen of the organism. It has also been reported that a 66- to 64-kDa protein in a Triton X-100 extract of cell surface antigens is also a protective molecule (1). However, mice immunized with the recombinant 66- to 64-kDa protein showed incomplete protection (1). Recently, the cloning of a gene encoding the cell surface protein of E. rhusiopathiae (SpaA) was described (6). In that study, however, protection experiments with the isolated SpaA protein were not conducted. Thus, the identification of antigens in E. rhusiopathiae which can induce protective antibodies has been inconclusive, and it is unknown whether a single or multiple antigens are necessary to provide complete protection from infection.
To address this question, we examine whether SpaA.1 can induce antibodies that protect against lethal challenge and further determine the region responsible for protection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. rhusiopathiae strains used were Fujisawa (serovar 1a), Fujisawa-SmR (serovar 1a) (12), 422/1E1 (serovar 1b), ATCC 19414T (serovar 2), and SE-9 (serovar 2). These strains were grown in brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) containing 0.1% Tween 80 (pH 7.6) (BHI-T80). Escherichia coli strains were grown in Luria-Bertani medium. When appropriate, medium was supplemented with tetracycline (25 μg/ml), ampicillin (100 μg/ml), kanamycin (50 μg/ml), or isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM).
Construction of genomic libraries of E. rhusiopathiae.
Genomic DNA from E. rhusiopathiae Fujisawa was prepared as previously described (1). The genomic DNA was partially digested with Sau3AI and fractionated on a sucrose gradient to yield 3- to 5-kb fragments. The DNA fragments were ligated to BamHI-predigested ZAP Express vector (Stratagene, La Jolla, Calif.), and in vitro packaging was conducted by using Gigapack III Gold packaging extract (Stratagene) according to the manufacturer’s instructions.
Cloning of spaA.1 gene.
The genomic library was plated to give approximately 200 to 400 plaques per plate. The plates were overlaid with nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) previously soaked in 10 mM IPTG and incubated at 37°C overnight. The membranes were incubated for 2 h at room temperature with convalescent serum from a pig which was experimentally inoculated with the wild-type E. rhusiopathiae strain Fujisawa. After washing, the membranes were incubated for 2 h at room temperature with peroxidase-labeled goat anti-pig immunoglobulin G (IgG) (Rockland, Inc., Gilbertsville, Pa.). Detection of the peroxidase-labeled goat anti-pig IgG was performed with 0.03% 3,3′-diaminobenzidine tetrahydrochloride dihydrate and 0.003% hydrogen peroxide in phosphate-buffered saline (PBS). Immunoreactive plaques were isolated and used for further experiments.
For protection experiments, phage lysates from the positive clones that reacted with the pig antiserum were prepared for immunization as described previously (1). Briefly, groups of 13 BALB/c mice were immunized subcutaneously (s.c.) with the phage lysates emulsified with complete Freund adjuvant and then boosted s.c. with the same antigen preparation 2 and 3 weeks after the first immunization. The mice were challenged s.c. with about 10 50% lethal doses (LD50) (approximately 1.6 × 102 CFU) of Fujisawa-SmR strain 7 days after final boosting, and deaths were recorded for 14 days. Two recombinant phage clones which showed protective activities were subjected to in vivo excision to form recombinant phagemid pBK-CMV by using ExAssist helper phage and the E. coli XLOLR system (Stratagene).
DNA sequencing and data analysis.
Nucleotide sequencing of both strands of the cloned DNA in pBK-CMV was performed by the dideoxy chain termination method (7) with fluorescent dye terminators and cycle sequencing reactions. Samples were electrophoresed, detected, and analyzed on an Applied Biosystems DNA sequencer. Initial sequencing reactions were performed with pBK-CMV vector primers T3 (5′-AATTAACCCTCACTAAAGGG-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′). Subsequent primers were synthesized on the basis of sequence data previously obtained with vector primers. The sequences were analyzed with the GENETYX-MAC program, version 7.3 (SDC, Tokyo, Japan). Sequence similarity searches were performed with GenBank sequences by using the BLAST network service.
Construction and purification of recombinant fusion proteins.
Construction and purification of histidine (His)-tagged fusion protein were performed with the QIAexpress kit (Qiagen Inc., Santa Clarita, Calif.). Insert DNA was amplified by PCR from subclone plasmids of pERc6 (Fig. 1), with primers constituting an in-frame BamHI restriction site at the 5′ end and T7 (5′-GTAATACGACTCACTATAGGGC-3′) primers. Primers ORF-1 (5′-CATTGGATCCAGTCTTATGTCGTGCTTACT-3′) and T7 were used to amplify spaA.1 gene regions ranging from bp 34 to 1878, 34 to 1329, and 34 to 587 from subclones pERc6.1, pERc6.3, and pERc6.4, respectively; ORF-1.1 (5′-CTCAGGATCCTAGTTCCTCTAAGAGATAGA-3′) and T7 were used for the region ranging from bp 569 to 1329 from subclone pERc6.3; ORF-1.2 (5′-CATTGGATCCTCAAAAGAAGGGTGGATTA-3′) and T7 were used for the region ranging from bp 1333 to 1878 from subclone pERc6.1; and WG1 (5′-CCCCGGATCCTACAACAAAATGACTGATGC-3′) and T7 were used for the region ranging from bp 178 to 1329 from subclone pERc6.3. The PCR, consisting of denaturation at 94°C for 1 min, annealing at 50°C for 30 s, and extension at 72°C for 1 min, was performed as previously described (10). The PCR products were digested with BamHI and KpnI and then cloned in frame into one of the pQE series expression vectors to generate plasmids pQEr1 to pQEr6 (Fig. 2).
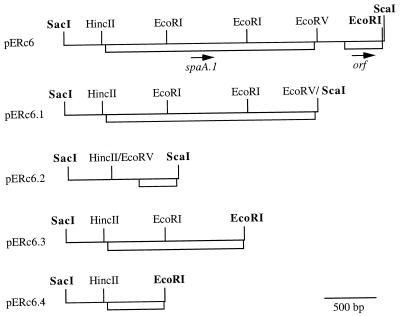
FIG. 1.
Partial restriction map of the insert of pERc6 and its deletion derivatives. The positions of the ORFs and the incomplete ORFs are indicated by open boxes; arrows indicate the direction of transcription. Sites of restriction enzymes presented in boldface are located within the multicloning site of the vector pBK-CMV.
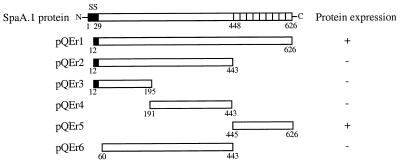
FIG. 2.
Schematic representation of the expression plasmids and the expression of fusion proteins. The expression of the fusion proteins is indicated on the right side. The signal sequence (SS) is indicated as black boxes. Numbers indicate positions of amino acids in the SpaA.1 protein.
Rabbit immunization.
Purified recombinant SpaA.1 protein (approximately 4.5 μg of total protein per lane) was run on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel by the method of Laemmli (5). The gel regions where the recombinant SpaA.1 protein band migrated were excised from the gel, ground, and mixed with distilled water for preparation of antigen. Ten gel slices were combined, yielding approximately 450 μg of total protein based on the bicinchoninic acid protein estimation method (Pierce, Rockford, Ill.). New Zealand White rabbits (Charles River) were immunized s.c. at multiple sites with the gel fragments containing protein. The rabbits were boosted once with the same gel preparation 30 days later and then bled 15 days after the boosting. Sera were filter sterilized and stored at −70°C until use. The sera had an IgG titer of 1:102,400 (with an optical density endpoint of 1.0) as determined by an enzyme-linked immunosorbent assay that used the recombinant SpaA.1 protein as the antigen.
Absorption of anti-SpaA.1 rabbit serum with the C-terminal SpaA.1 truncate.
Approximately 3.5 mg of purified recombinant C-terminal region of SpaA.1 protein was coupled with 100 mg of beads (3M Emphaze Biosupport medium; Pierce) according to the manufacturer’s instructions. As a control, the beads coupled with 3.5 mg of bovine serum albumin (BSA) were used. The anti-SpaA.1 rabbit serum (1.0 ml) was incubated under rotation with the protein-coupled beads at 4°C overnight. After separation of the beads from the antiserum by centrifugation, the beads were washed three times with 10 ml of PBS (pH 7.6) and then treated with 2 ml of 100 mM glycine-HCl buffer (pH 2.2) for elution of bound antibody specific for the C-terminal repeat region of SpaA.1 protein. The antibody solution was neutralized immediately after elution with 1 M Trizma base. The beads were then washed three times with PBS and used for further absorption of the antiserum. These absorption procedures were repeated three times with the same 1.0-ml antiserum sample, and the absorbed antiserum was used for Western immunoblot analysis and passive immunization experiments. The eluted antibody solution was pooled, dialyzed against PBS (pH 7.6), and concentrated to 1.0 ml, and BSA was added to a final concentration of 1%. This final preparation was used for Western immunoblot analysis and passive immunization experiments.
Immunization experiments.
Nine- to twelve-week-old female BALB/c mice (Charles River) were used. For passive immunization experiments, mice were injected intraperitoneally (i.p.) with either 0.1 ml of rabbit antiserum or 0.1 ml of eluted C-terminal region-specific antibodies (approximately 60 μg of protein). Eight hours after injection of the antiserum or the antibody, mice were challenged with Fujisawa-SmR and were observed for clinical symptoms and death for 14 days. For active immunization experiments, mice were immunized i.p. with approximately 50 μg of purified recombinant protein emulsified with complete Freund adjuvant and then boosted s.c. with approximately 37 μg of the same protein emulsified with incomplete Freund adjuvant 3 weeks later. Seven days after boosting, mice were challenged s.c. with Fujisawa-SmR and were observed for clinical symptoms and death for 14 days.
Challenge inoculation.
The strain Fujisawa-SmR was grown in BHI-T80 at 37°C for 14 h and diluted with BHI-T80. Mice were inoculated with 0.1 ml of appropriate dilutions of the bacterial suspension.
Cell surface extract and supernatant preparation.
Bacterial strains were grown in 10 ml of BHI-T80 for 14 h at 37°C. The cultures were centrifuged, and the supernatants were filtered (0.22-μm pore size; Millipore Corp., Bedford, Mass.) and used for Western immunoblot analysis. For cell surface antigen preparation, the pelleted cells were treated with Triton X-100 as previously described (1, 4, 11). Briefly, the cells were washed once with 20 mM Tris-HCl (pH 7.6) and suspended in 0.5 ml of 20 mM Tris (pH 7.6) containing 0.5% Triton X-100. The cells were incubated at 37°C for 1 h with rotation. Cells were removed by centrifugation, and the supernatants were used for Western immunoblot analysis.
Western immunoblotting.
For localization of SpaA.1, a 7.5-μl volume of the supernatant fluids or the Triton X-100-solubilized surface antigen extracts was electrophoresed in an SDS–12% polyacrylamide gel by the method of Laemmli (5) and transferred to a nitrocellulose membrane (Schleicher & Schuell). The nitrocellulose membranes were incubated for 1 h in blocking solution, which contained 5% skim milk in TTBS (20 mM Tris, 500 mM NaCl, 0.05% Tween 20 [pH 7.5]), and then incubated for 1 h with the anti-SpaA.1 rabbit serum diluted 1:500 in TTBS containing 5% skim milk. The membranes were washed in TTBS and then incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Bio-Rad Laboratories, Richmond, Calif.) diluted 1:2,000 in TTBS containing 5% skim milk. Bound antibodies were detected with chemiluminescence (ECL; Amersham).
For confirmation of absorption of anti-SpaA.1 rabbit serum with the C-terminal repeat region of SpaA.1 protein, nitrocellulose strips containing the C-terminal region fragment were incubated with various dilutions of the absorbed antiserum or the eluted antibody for 1 h at room temperature. The bound antibodies were detected by alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma Chemical Co., St. Louis, Mo.).
Southern hybridization.
Genomic DNAs from E. rhusiopathiae strains were digested with the restriction enzyme EcoRV and transferred to nitrocellulose membranes (Schleicher & Schuell). The PCR product used for construction of recombinant mature SpaA.1 protein was used for labeling as a probe. Subsequent hybridization and detection of the probe were performed with ECL direct nucleic acid labeling and detection systems (Amersham) according to the manufacturer’s instructions.
Statistical methods.
Statistical analyses were performed by the Fisher exact test.
Nucleotide sequence accession number.
The nucleotide sequence of spaA.1 has been deposited in the DDBJ-EMBL-GenBank databases under accession no. AB017447.
RESULTS
Cloning of spaA.1 gene.
The genomic DNA library of E. rhusiopathiae Fujisawa was screened with E. rhusiopathiae convalescent pig serum, and 22 positive clones were detected from a total of 5,000 plaques. Of 22 positive clones, 5 which showed a strong positive signal were chosen and further analyzed for their protective activities. Groups of 13 mice immunized s.c. with phage lysates of each clone were challenged s.c. with 10 LD50 of virulent strain Fujisawa-SmR. Two clones (clone 6 and clone 20) which showed protective activities (85 and 92% survival, respectively) were subsequently subjected to in vivo excision to generate phagemids pERc6 and pERc20, respectively. Restriction analysis of the insert in these phagemids showed the same restriction pattern, suggesting that inserts of the two clones are identical. Therefore, one clone (pERc6) was subjected to sequence analysis.
The DNA sequence of the insert of pERc6 revealed that it contains one complete and one incomplete open reading frame (ORF) (Fig. 1). To determine which ORF product is responsible for protection, deletion clones of pERc6 were constructed. The subclones pERc6.1 and pERc6.2, which contain each ORF, respectively, were obtained by deletion of the EcoRV-ScaI and HincII-EcoRV fragments, respectively (Fig. 1). Lysates of E. coli DH5α containing each subclone were examined by immunoblotting with the convalescent pig serum, and a positive reaction was detected in subclone pERc6.1 (data not shown).
Analysis of the spaA.1 gene sequence.
The deduced amino acid sequence of the cloned gene revealed a protein of 626 amino acids with a potential signal peptidase cleaving site, predicted by the method of von Heijne (14), between amino acids 29 and 30, resulting in a mature protein of 597 amino acids with a deduced molecular mass of 69,017 Da. Comparison of the deduced amino acid sequence of the gene product with known protein sequences deposited in databases showed that its sequence is nearly identical to that of SpaA (6), which was recently reported. Sequence differences were observed at residues 426 and 435 and in the C-terminal region, in which our molecule contains an additional 20-amino-acid repeat, resulting in a size difference between these molecules. Because of the similarities with the SpaA molecule, the gene was designated spaA.1.
Construction of His-tagged fusion proteins.
To produce the recombinant SpaA.1 protein, the DNA fragment encoding the mature SpaA.1 was amplified from pERc6.1 and cloned into pQE30 expression vector. To produce SpaA.1 truncates, the subclones pERc6.3 and pERc6.4, which were constructed from pERc6 by deleting EcoRI fragments (Fig. 1), and the DNA fragments which encode different regions were amplified from these subclones and cloned into pQE expression vectors. The recombinant pQE plasmids were transformed into E. coli M15, and transformants were checked for the expression of fusion proteins by purification with chromatography on Ni-nitrilotriacetic acid resins followed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western immunoblot analyses.
The mature SpaA.1 protein and its C-terminal repeat region were efficiently expressed (Fig. 2 and 3). However, regions other than the C-terminal repeats could not be expressed in several different experiments with various conditions. We found that lysates of E. coli DH5α containing either plasmid pERc6.3 or pERc6.4 were not reactive with convalescent pig serum.
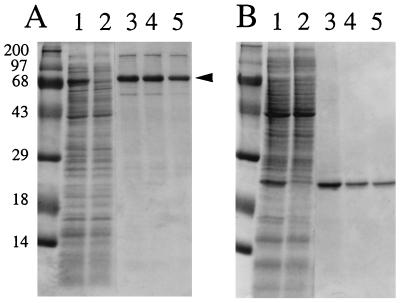
FIG. 3.
Purification of recombinant E. rhusiopathiae SpaA.1 proteins. Mature SpaA.1 and its C-terminal repeat region were expressed as His-tagged fusion proteins, purified on a Ni-nitrilotriacetic acid resin, and separated by SDS-PAGE. The gels were stained for protein with Coomassie brilliant blue. (A) Expression of the mature SpaA.1 protein with pQEr1. The 69-kDa band (arrowhead) was excised from the gel and used for immunization of rabbits. (B) Expression of the C-terminal repeat region of SpaA.1 protein with pQEr5. Lanes: 1, lysates of IPTG-induced culture; 2, uninduced culture; 3 to 5, affinity-purified recombinant proteins. Molecular size markers (kilodaltons) are shown on the left.
The purified mature SpaA.1 protein gave a prominent 69-kDa band with a larger band at approximately 140 kDa and smaller minor bands on SDS-PAGE gels (Fig. 3A). The gel regions where the prominent 69-kDa protein migrated were excised and injected into rabbits for the preparation of anti-SpaA.1 serum. When this antiserum was used in Western immunoblots with recombinant SpaA.1, it was reactive with not only the 69-kDa protein but also the 140-kDa protein and smaller bands similar to that seen in Fig. 3A (data not shown). Thus, we conclude that the larger and smaller bands are likely dimer and degradation products or processed forms of SpaA.1, respectively. The purified C-terminal region of SpaA.1 gave a prominent 21-kDa band (Fig. 3B).
Passive immunization experiments.
In E. rhusiopathiae infection, humoral as well as cellular immunity is involved in protection (11). To determine which type of immunity is responsible for the protection induced by SpaA.1, passive immunization experiments were performed with the anti-SpaA.1 rabbit serum. Since the LD50 of E. rhusiopathiae Fujisawa-SmR varies according to the route of the challenge inoculation (s.c. LD50, 101.2; i.p. LD50, 103.5) (11, 12), protection experiments were conducted by both s.c. and i.p. inoculations. Eight hours after injection of 0.1 ml of the antiserum, mice were challenged with Fujisawa-SmR and observed for clinical symptoms and death for 14 days. As shown in Table 1, when challenged with 5 LD50, all the antiserum-treated mice survived without any clinical symptoms. When challenged with 100 LD50, half of the antiserum-treated mice showed clinical symptoms, such as depression; one mouse challenged s.c. died on each of days 4, 8, and 11, and one mouse challenged i.p. died on each of days 8 and 14. All control mice died within 4 days after challenge. Thus, the anti-SpaA.1 serum passively protected mice, and the route of challenge inoculation did not affect the protective activity of the antiserum.
TABLE 1.
Passive immunization of mice against virulent E. rhusiopathiae strain by anti-SpaA.1 rabbit seruma
| Challenge route | Challenge dose (LD50) | No. of survivors/total no.
|
|
|---|---|---|---|
| Control | Anti-SpaA.1 serum | ||
| s.c. | 5 | 0/5 | 5/5* |
| 100 | 7/10** | ||
| i.p. | 5 | 0/5 | 5/5* |
| 100 | 8/10* | ||
Eight hours after i.p. injection of 0.1 ml of the anti-SpaA.1 rabbit serum, mice were challenged (s.c. or i.p.) with virulent E. rhusiopathiae Fujisawa-SmR. Control mice were injected with 0.1 ml of preimmune serum. Asterisks indicate differences (*, P < 0.01; **, P < 0.05) compared to control by the Fisher exact test.
Determination of the protective domain of the SpaA.1 protein.
To examine whether the C-terminal repeat region of SpaA.1 protein alone can induce complete protection, mice were actively immunized with either the purified mature protein or the C-terminal repeat region and then challenged s.c. with 10 LD50 of the Fujisawa-SmR strain (Table 2). All the mice immunized with the mature SpaA.1 protein survived without any clinical symptoms, whereas all the control mice and four of the five mice immunized with the C-terminal repeat region protein died within 4 days after challenge. This result strongly suggests that the C-terminal repeat region of SpaA.1 does not play a major role in protection and that the protection-eliciting epitope(s) of SpaA.1 is located within the N-terminal two-thirds of the protein.
TABLE 2.
Active immunization of mice against virulent E. rhusiopathiae strain by truncated SpaA.1 proteins
Mice were immunized with recombinant protein with adjuvant and boosted 3 weeks later. Control mice were treated with PBS with adjuvant.
Mice were challenged s.c. with 10 LD50 of the virulent E. rhusiopathiae strain Fujisawa-SmR. Asterisk indicates difference (P < 0.05) compared to control by the Fisher exact test.
The result suggesting that the C-terminal repeat region of the SpaA.1 protein does not play a major role in protection was further confirmed by passive immunization experiments. The protective anti-SpaA.1 rabbit serum was absorbed with the purified recombinant C-terminal repeat region of SpaA.1, and C-terminal region-specific antibodies were obtained. In Western immunoblot analysis, a 10−4 dilution of the absorbed serum was very weakly reactive with the recombinant C-terminal region of the SpaA.1 protein; however, the eluted C-terminal region-specific antibody was strongly reactive at a 10−5 dilution (data not shown). These results suggest that the anti-SpaA.1 rabbit serum was significantly reduced in C-terminal region-specific antibodies and that a high titer of the C-terminal region-specific antibodies was obtained by eluting the bound antibody from the antigen-coupled beads. Mice were then treated with either the absorbed antiserum or the eluted antibody and then challenged s.c. with 10 LD50 of Fujisawa-SmR (Table 3). In the control group, all the mice died within 4 days after challenge. In the antiserum-treated groups, only one mouse treated with C-terminal region-absorbed antiserum died on day 4, and two mice treated with BSA-absorbed antiserum died (days 7 and 9). All the survivors did not show any clinical symptoms. Thus, mice treated with C-terminal region-absorbed antiserum were protected from lethal challenge, showing that absorption of anti-SpaA.1 serum with the C-terminal repeat region had little to no effect on protection. Furthermore, all the mice treated with the C-terminal region specific-antibody were not protected; four mice died within 4 days after challenge, and the last mouse died on day 6. These results, taken together with those of the active immunization experiments, strongly suggest that the C-terminal repeat region of SpaA.1 protein does not play a major role in protection and that antibodies directed against the N-terminal region are responsible for protection.
TABLE 3.
Passive immunization of mice against virulent E. rhusiopathiae strain by absorbed serum or C-terminal region-specific antibodya
| Treatment | No. of survivors/total no. |
|---|---|
| Control | 0/4 |
| Anti-SpaA.1 serum absorbed with BSA | 3/5 |
| Anti-SpaA.1 serum absorbed with C-terminal region | 4/5* |
| C-terminal region-specific antibody | 0/5 |
Eight hours after injection of 0.1 ml of the absorbed serum or antibody, mice were challenged s.c. with 10 LD50 of the virulent E. rhusiopathiae strain Fujisawa-SmR. Control mice were injected with 0.1 ml of PBS containing 1% BSA. Asterisk indicates difference (P < 0.05) compared to control by Fisher exact tests.
Presence of SpaA.1 in different strains of E. rhusiopathiae.
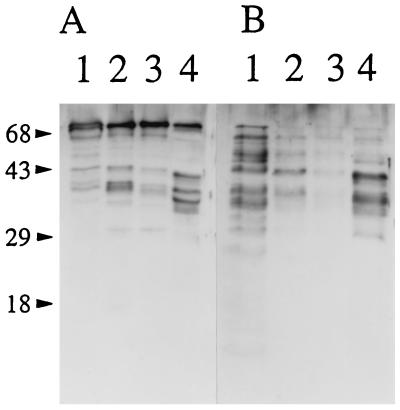
To determine the molecular weight and the localization of the native SpaA.1 protein in different strains of E. rhusiopathiae, Western immunoblotting was performed with the anti-SpaA.1 rabbit serum. Cell surface antigen extracts prepared by treatment with Triton X-100 and unconcentrated culture supernatants from different strains were analyzed for the presence of the antigen. The antiserum detected a prominent 69-kDa band and multiple minor bands ranging in size from 32 to 66 kDa in the cell surface extracts from all the strains (Fig. 4A), suggesting that SpaA.1 is expressed in these different strains of E. rhusiopathiae. The antiserum also detected multiple bands in the unconcentrated culture supernatants from the strains (Fig. 4B). We found that an increase of the smaller bands in the cell surface extracts and the culture supernatant corresponded to a reduction of the 69-kDa band (data not shown), suggesting that these smaller bands may be degradation products or normally processed forms of the 69-kDa protein.
FIG. 4.
Detection of E. rhusiopathiae SpaA.1 protein by Western immunoblotting with anti-SpaA.1 rabbit serum in cell surface antigen preparations (A) and unconcentrated culture supernatants (B). Lanes: 1, SE-9; 2, ATCC 19414T; 3, 422/1E1; 4, Fujisawa-SmR. Molecular size markers (kilodaltons) are shown on the left. Preimmune rabbit serum was not reactive with these bands (data not shown).
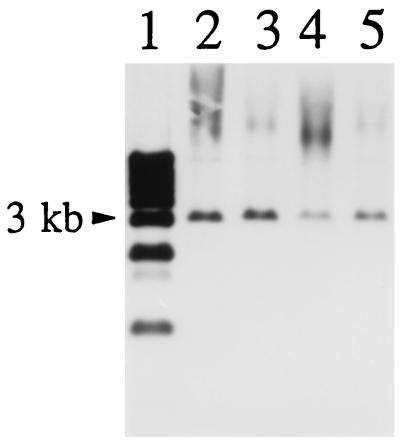
The presence of a spaA.1 gene in these strains was confirmed by Southern hybridization. The results showed that the probe hybridized with an approximately 3.2-kb EcoRV fragment in the genomic DNAs from all the strains examined (Fig. 5).
FIG. 5.
Detection of spaA.1 gene from E. rhusiopathiae strains by Southern hybridization with spaA.1 gene as a probe. Genomic DNAs of E. rhusiopathiae strains were digested with EcoRV and transferred onto nitrocellulose membranes. Lanes: 1, 1-kb ladder (GIBCO-BRL); 2, Fujisawa-SmR; 3, 422/1E1; 4, ATCC 19414T; 5, SE-9.
DISCUSSION
In this study, we demonstrate that SpaA.1, the 69-kDa protective antigen of E. rhusiopathiae, can elicit protective antibodies in animals and that the protective epitope(s) is located within the N-terminal two-thirds of the SpaA.1 molecule.
Although humoral immunity has been known to play an important role in protection against E. rhusiopathiae infection (17–19), the antigen(s) responsible for inducing protective antibodies has not been identified. Galán and Timoney (1) reported that the 66- to 64-kDa cell surface protein antigen in Triton X-100 extracts is a protective antigen of E. rhusiopathiae. In that study, mice immunized with a recombinant 66- to 64-kDa protein, which was fused with β-galactosidase, were poorly protected. The authors assumed that the incomplete protection might be derived from a less efficient protective immune response by the form of the protein fused with β-galactosidase. Although subsequent studies on the protective antigens of the organism have drawn attention to the 66- to 64-kDa cell surface protein (2, 3, 8), the identification of a protective antigen which is involved in humoral immunity against E. rhusiopathiae has been inconclusive.
Recently, a cell surface protein gene (spaA) of E. rhusiopathiae was cloned by Makino et al. (6). In that study, mice were inoculated with various recombinant E. coli strains carrying the spaA gene or its deletion derivatives and challenged with a virulent E. rhusiopathiae strain only 7 days after inoculation with E. coli. The authors reported that only mice inoculated with the recombinant E. coli carrying the intact spaA gene were protected from lethal challenge. Although they concluded that the whole molecule, particularly the C-terminal repeat region, is essential for the observed protection, neither purified protein nor experiments designed to implicate the C-terminal region in this protection were reported in their publication.
In our experiments, the protective activity of SpaA.1 was examined by passive immunization with an anti-SpaA.1 rabbit serum prepared against purified SpaA.1. Mice treated with this antiserum were protected from lethal challenge with 10 LD50 of the virulent strain, suggesting that SpaA.1 alone was sufficient for protection against infection with low doses of challenge organisms. However, when the mice were challenged with a high dose (100 LD50), half of the mice which were treated with anti-SpaA.1 rabbit serum showed clinical symptoms and 20 to 30% died. These results suggest that protective antibodies induced by immunization of a single antigen may not be sufficient for protection against infection with a high dose of organisms. Either several different protective antigens or higher antibody titers may be required to induce complete protection with this dose of bacteria.
The protection-eliciting domain of the SpaA.1 protein was determined by active and passive immunization. In active immunization experiments, mice immunized with recombinant mature SpaA.1 protein, but not the C-terminal repeat region, were protected from lethal challenge with the virulent strain Fujisawa-SmR, showing that the C-terminal repeat region does not play a major role in protection. This was confirmed by passive immunization experiments in which the protective activity of the rabbit antiserum against the mature SpaA.1 protein could not be abolished by absorption with the recombinant C-terminal region and the specific antibody against this region was not protective. Taken together, these results strongly indicate that antibodies to the N-terminal two-thirds of the SpaA.1 molecule are responsible for protection.
It has been reported that the protective antigen of E. rhusiopathiae is also present in the culture supernatant of the organism and that antiserum against the culture supernatant fluids is protective (8, 9, 15, 16). Although we found that SpaA.1 is also present in the culture supernatant, we do not yet know the relationship between the bound SpaA.1 protein and the protective antigens found in the culture supernatant reported so far (8, 9, 15, 16).
In Western and Southern blotting analysis, the spaA.1 gene and its product are found to be present in all strains tested, suggesting that the protein is well conserved among E. rhusiopathiae bacteria. In erysipelas, the protective antigen has been known to be species specific (9). If SpaA.1 is in fact responsible for the observed species-specific protection, this protective antigen, particularly the N-terminal segment, will be a good candidate for a new vaccine against erysipelas.
ACKNOWLEDGMENTS
We thank T. Sekizaki and Y. Yokomizo for valuable suggestions in the experiments.
Y.S. is a recipient of a fellowship from the Science and Technology Agency of Japan. The work was supported in part by USPHS grant AI11822 to V.A.F.
ADDENDUM
When E. coli clones containing pQEr2, pQEr3, pQEr4, or pQEr6 were grown at 28°C, we found that only one clone containing the pQEr3 plasmid (Fig. 2) (coding amino acid residues 12 to 195 of the SpaA.1 protein) produced a small amount of recombinant protein. This protein was purified as described for SpaA.1, and mice were immunized with the same quantity and immunization schedule as described in the text for SpaA.1. Control mice were immunized with either whole SpaA.1 or PBS. All mice were challenged s.c. with approximately 70 LD50 cells of Fujisawa-SmR and observed for clinical symptoms and death for 14 days. The result showed that all five mice immunized with either the 12-195 fragment or the whole SpaA.1 protein survived without any clinical symptoms (P < 0.01), whereas all five mice immunized with PBS died within 5 days after challenge. The arithmetic mean of ELISA IgG titers (against peptide 12-195) of sera taken before challenge from individual mice was 76,800 for peptide 12-195-immunized mice, 66,560 for animals immunized with whole SpaA.1, and <100 for PBS-immunized control mice. These results support the results presented in the paper and further localize the protective epitope to the N-terminal 30% of the SpaA.1 protein.
REFERENCES
- 1.Galán J E, Timoney J F. Cloning and expression in Escherichia coli of a protective antigen of Erysipelothrix rhusiopathiae. Infect Immun. 1990;58:3116–3121. doi: 10.1128/iai.58.9.3116-3121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groschup M H, Cussler K, Weiss R, Timoney J F. Characterization of a protective protein antigen of Erysipelothrix rhusiopathiae. Epidemiol Infect. 1991;107:637–649. doi: 10.1017/s0950268800049335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitajima T, Oishi E, Amimoto K, Ui S, Nakamura H, Okada N, Sasaki O, Yasuhara H. Protective effect of NaOH-extracted Erysipelothrix rhusiopathiae vaccine in pigs. J Vet Med Sci. 1998;60:9–14. doi: 10.1292/jvms.60.9. [DOI] [PubMed] [Google Scholar]
- 4.Lachmann P G, Deicher H. Solubilization and characterization of surface antigenic components of Erysipelothrix rhusiopathiae T28. Infect Immun. 1986;52:818–822. doi: 10.1128/iai.52.3.818-822.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 6.Makino S, Yamamoto K, Murakami S, Shirahata T, Uemura K, Sawada T, Wakamoto H, Morita Y. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb Pathog. 1998;25:101–109. doi: 10.1006/mpat.1998.0216. [DOI] [PubMed] [Google Scholar]
- 7.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato H, Hirose K, Saito H. Protective activity and antigenic analysis of fractions of culture filtrates of Erysipelothrix rhusiopathiae. Vet Microbiol. 1995;43:173–182. doi: 10.1016/0378-1135(95)92533-h. [DOI] [PubMed] [Google Scholar]
- 9.Sawada T, Takahashi T. Cross protection of mice and swine inoculated with culture filtrate of attenuated Erysipelothrix rhusiopathiae and challenge exposed to strains of various serovars. Am J Vet Res. 1987;48:239–242. [PubMed] [Google Scholar]
- 10.Shimoji Y, Mori Y, Hyakutake K, Sekizaki T, Yokomizo Y. Use of an enrichment broth cultivation-PCR combination assay for rapid diagnosis of swine erysipelas. J Clin Microbiol. 1998;36:86–89. doi: 10.1128/jcm.36.1.86-89.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimoji Y, Mori Y, Sekizaki T, Shibahara T, Yokomizo Y. Construction and vaccine potential of acapsular mutants of Erysipelothrix rhusiopathiae: use of excision of Tn916 to inactivate a target gene. Infect Immun. 1998;66:3250–3254. doi: 10.1128/iai.66.7.3250-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimoji Y, Yokomizo Y, Sekizaki T, Mori Y, Kubo M. Presence of a capsule in Erysipelothrix rhusiopathiae and its relationship to virulence for mice. Infect Immun. 1994;62:2806–2810. doi: 10.1128/iai.62.7.2806-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimoji Y, Yokomizo Y, Mori Y. Intracellular survival and replication of Erysipelothrix rhusiopathiae within murine macrophages: failure of induction of the oxidative burst of macrophages. Infect Immun. 1996;64:1789–1793. doi: 10.1128/iai.64.5.1789-1793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Shimoji, Y., et al. Unpublished data.
- 14.von Heijne G. Patterns of amino acids near signal-sequence cleaving sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 15.White R R, Verwey W F. Isolation and characterization of a protective antigen-containing particle from culture supernatant fluids of Erysipelothrix rhusiopathiae. Infect Immun. 1970;1:380–386. doi: 10.1128/iai.1.4.380-386.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White R R, Verwey W F. Solubilization and characterization of a protective antigen of Erysipelothrix rhusiopathiae. Infect Immun. 1970;1:387–393. doi: 10.1128/iai.1.4.387-393.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood R L. Swine erysipelas—a review of prevalence and research. J Am Vet Med Assoc. 1984;184:944–949. [PubMed] [Google Scholar]
- 18.Wood R L. Erysipelas. In: Leman A D, et al., editors. Diseases of swine. Ames, Iowa: Iowa State University Press; 1992. pp. 475–486. [Google Scholar]
- 19.Yokomizo Y, Isayama Y. Antibody activity of IgM and IgG fractions from rabbit anti-Erysipelothrix rhusiopathiae sera. Res Vet Sci. 1972;13:294–296. [PubMed] [Google Scholar]