Abstract
Surface-associated M protein is a major virulence factor in Streptococcus pyogenes which confers bacterial resistance to phagocytosis. However, many S. pyogenes strains also express additional structurally related so-called M-like proteins. The strain studied here is of the clinically important M1 serotype and expresses two structurally related surface proteins, the M1 protein and protein H. Mutants were generated that expressed only one or none of these proteins at the bacterial surface. For survival in human blood either protein H or M1 protein was sufficient, whereas the double mutant was rapidly killed. The protein-binding properties of protein H, M1 protein, and the mutants suggest that bacterial binding of immunoglobulin G and factor H or factor H-like protein 1, which are regulatory proteins in the complement system, contribute to the antiphagocytic property.
Streptococcus pyogenes (group A streptococcus) is an important human pathogen, with the ability to cause a variety of infections. Primary disease manifestations include pharyngitis, impetigo, and erysipelas, which may lead to serious sequelae like rheumatic fever and glomerulonephritis (7). Since the late 1980s there has been an increase in severe invasive infections of skin and soft tissues (8), associated with streptococcal toxic shock syndrome, with high mortality (54). This type of severe infection has mainly been reported to be associated with the serotypes M1 and M3 (22, 38, 41).
Clinical isolates of S. pyogenes grow rapidly in human blood, a property associated with resistance against phagocytosis (33). This antiphagocytic activity has been ascribed to the expression of surface-associated M protein (for references, see reference 15). There are more than 80 different antigenically distinguishable M proteins, and immunity against S. pyogenes is known to be M type specific. Each strain was originally believed to express a single antiphagocytic M protein. However, the M proteins have been shown to be members of a larger family of structurally related proteins, the M-like proteins, that includes three subtypes of proteins known as Mrp, Emm, and Enn (29), and many S. pyogenes strains express more than one M-like protein. The expression of these proteins is under the control of the trans-acting regulator Mga (11, 30, 39, 47). Many M proteins with known antiphagocytic activities belong to the Emm subfamily (14, 44, 50). Recently, Mrp proteins also were shown to contribute to S. pyogenes resistance against phagocytosis (48, 56). However, not all M proteins have antiphagocytic activity, as demonstrated for Arp4, which belongs to the Emm subfamily (25).
The M and M-like proteins have affinity for several human plasma proteins and often display different ligand-binding properties. For instance, many of the proteins have affinity for factor H or C4b-binding protein (C4BP) (24, 55), which are inhibitors of complement. Both of these proteins have been suggested to contribute to the resistance to phagocytosis (23, 56). Moreover, many Emm and Mrp proteins bind fibrinogen, another human plasma protein that has been implicated in the resistance to phagocytosis (14, 59). Several M and M-like proteins also bind human immunoglobulin G (IgG) and/or IgA through the constant Fc region (19, 20, 36, 53), but the importance of the nonimmune interaction with Ig-Fc for the virulence of S. pyogenes is unclear (12). However protein H, an IgG-Fc-binding M-like protein, was shown to inhibit complement activation at the bacterial surface, suggesting that Ig binding could contribute to resistance to phagocytosis (6).
The S. pyogenes strain used in this study, AP1, is of the clinically important M1 serotype. An insertional inactivation of Mga in AP1 rendered this strain sensitive to phagocytosis. The transcriptional regulator Mga in AP1 was shown to coregulate the expression of the M1 protein, protein H, protein SIC, and the C5a peptidase (30). The proteins M1 and H are structurally related and belong to the Emm subfamily, but these proteins have different ligand-binding properties. Protein SIC has been shown to interfere with complement-mediated cell lysis (4), and the C5a peptidase is a cell wall-anchored enzyme with the ability to inactivate C5a, a chemotactic factor of complement (58). Here we evaluate the importance of the M1 protein and protein H for the resistance to phagocytosis. This was done by generating mutants that were selectively affected in the expression of one or both of these proteins, followed by analyzing the properties of the mutants.
MATERIALS AND METHODS
Bacteria, proteins, and plasmids.
S. pyogenes AP1 is strain 40/58 of the M1 serotype from the World Health Organization Streptococcal Reference Laboratory in Prague and has a nonmucoid morphology. S. pyogenes was cultured in Todd-Hewitt (TH) medium supplemented with 0.2% yeast extract (THY medium) at 37°C. Luria-Bertani medium (51) was used for the culturing of Escherichia coli. Unless otherwise indicated, antibiotics were used at the following concentrations: tetracycline at 5 μg/ml, streptomycin at 1,000 μg/ml, kanamycin at 25 μg/ml for E. coli and 500 μg/ml for S. pyogenes, and erythromycin at 300 μg/ml for E. coli and 1 μg/ml for S. pyogenes.
Human fibrinogen and human IgG3κ were from Sigma (St. Louis, Mo.). Purified human factor H was kindly provided by U. Sjöbring, and a rabbit antiserum against human factor H was a generous gift from A. Sjöholm. Proteins were labeled with 125I by using the Bolton and Hunter reagent (Amersham, Little Chalfont, England) or the chloramine-T method.
Plasmid pMC10 (13) is a recombinant vector consisting of a 960-bp internal PCR fragment from emm1, the gene encoding the M1 protein, ligated into the vector pFW13 (49). To generate the truncated emm1 sequence the oligonucleotide primers 5′-ATAGAAGATCTAGAAGCAAACAAT-3′ and 5′-TCAGCTTTTTCTAGATCTGTTAATTTCTTG-3′ were used in a PCR experiment with chromosomal DNA from AP1 as the template. After XbaI digestion of the PCR-generated emm1 product, this fragment was ligated into the SpeI site in the multiple cloning site II of plasmid pFW13. pBK37 was generated by the insertion of an 803-bp internal PCR fragment from sph, the gene encoding protein H, into pJRS233 (46), a derivative of the temperature-sensitive plasmid pG+host4 (37). A detailed description of pBK37 has been published previously (6). pBH was generated by cloning the complete nucleotide sequence for sph, including promoter and termination sequences, into the EcoRI site in pLZ12-21K (45). To generate the sph sequence the primers hybridizing to the following sequences were used: 5′-GCTATCACTTTGTAATACTGAGTG-3′ and 5′-GTGACCTCTCCTTAACCTCATTC-3′.
Generation of S. pyogenes mutants.
Transposon mutants of S. pyogenes AP1 were generated by Tn916 mutagenesis. Tn916 was transferred by conjugation essentially as described previously (10), with some modifications as described previously (30). Transconjugants potentially lacking surface-associated M1 protein or protein H were identified by colony blotting, as previously described (30), by their reduced binding of fibrinogen and IgG3, respectively. Chromosomal DNA from the transconjugants was digested with HindIII and hybridized with a Tn916-specific probe to analyze the number of transposons that had been integrated.
Insertional inactivation of sph in AP1, generating BM27.6, has been described previously (6). Briefly, S. pyogenes was transformed with the temperature-sensitive recombinant plasmid pBK37, which in BM27.6 was shown to be integrated into sph by a single homologous recombination event (6). The insertional inactivation of sph in BMJ11, generating BM22.1, was performed in the same way.
The insertional inactivation of emm1 in AP1, generating MC25, was performed as described previously (13). Briefly, the emm1 mutant was generated by a single homologous crossover recombination event with plasmid pMC10. Recombinant clones were selected by plating on TH agar containing 150 μg of kanamycin per ml.
The bacterial strains used, the bacterial mutants generated, and the plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Description | Reference or source |
|---|---|---|---|
| S. pyogenes | |||
| AP1 | Wild type | Strain 40/58 of the M1 serotype | World Health Organization Streptococcal Reference Laboratory, Prague |
| MC25 | emm1::Km | Deletion of the 3′ end of emm1 gene in AP1 by homologous recombination with plasmid pMC10 | 13 |
| BM27.6 | sph::Erm | Deletion of the 3′ end of sph gene in AP1 by homologous recombination with plasmid pBK37 | 6 |
| BMJ11 | Tn916::xyz | Tn916 insertional inactivation in AP1 | This study |
| BM22.1 | Tn916::xyz sph::Erm | Deletion of the 3′ end of sph in BMJ11 by homologous recombination with plasmid pBK37 | This study |
| Plasmids | |||
| pMC10 | 960-bp internal fragment of emm1 inserted into plasmid pFW13 (47) | 13 | |
| pBK37 | 803-bp internal fragment of sph inserted into plasmid pJRS233 (44) | 6 | |
| pBH | Complete sph gene cloned into the EcoRI site in pLZ12-21K (43) | This study |
DNA preparation and other DNA techniques.
Chromosomal DNA was prepared from S. pyogenes as previously described (9), with the addition that the cells were treated with 500 U of mutanolysin (Sigma) per ml at 37°C for 2 h and then lysed with 1% sodium dodecyl sulfate (SDS) and 0.2% Tween 20 on ice for 2 h.
Plasmid DNA preparations and other standard recombinant DNA techniques were performed as described previously (51). PCR (26) were performed with Taq DNA polymerase (Promega, Madison, Wis.) in the presence of 1.5 mM MgCl2 and 0.1 μM concentrations of each primer (25 cycles were performed at an annealing temperature of 55°C and 1 to 3 min of extension at 72°C, depending on the length of the product). E. coli JM109 was used to propagate plasmids and was made competent for transformation according to the procedure of Nishimura et al. (42). S. pyogenes was transformed by electroporation (45).
Binding of human plasma proteins and analysis of protein expression in S. pyogenes.
Bacteria to be analyzed in a direct binding assay were grown overnight, washed twice in PBST (10 mM sodium phosphate buffer, pH 7.2, containing 0.12 M NaCl and 0.05% Tween 20), and resuspended in PBST to a concentration of 2 × 109 bacteria/ml. A serial dilution of bacteria (200 μl) was mixed with 25 μl of 125I-labeled protein (104 cpm) and incubated at room temperature for 1 h. Cells were spun down, and the radioactivity associated with the pellet was determined in a gamma counter. All samples were run in duplicate. The absorption of human plasma proteins to S. pyogenes was performed by incubating the bacteria in human heparinized plasma at 37°C as described previously (1). Plasma proteins bound to the bacteria were eluted with 0.1 M glycine-HCl buffer, pH 2.0, and then neutralized.
CNBr treatment was used to release surface-associated proteins M1 and H, as previously described (43). M1 protein released into the medium was precipitated with 70% (wt/vol) (NH4)2SO4, and this material was further purified on a fibrinogen-Sepharose CL-4B column as described previously (1). Protein SIC was isolated from the growth medium of S. pyogenes by precipitation with 30% (wt/vol) (NH4)2SO4, followed by gel filtration as previously described (4). C5a peptidase was solubilized from the bacterial surface with streptococcal cysteine proteinase as described previously (5).
SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) were performed according to standard methods (32, 57). Gels were stained with Coomassie blue.
Bactericidal assay.
The ability of S. pyogenes strains to survive in blood was tested essentially as described previously (46). Cells grown to early mid-log phase (A620 = 0.15) were serially diluted in TH medium, and 100 μl of the bacterial solution was mixed with 1 ml of heparin-treated blood from a donor lacking type-specific antibodies and rotated end over end for 3 h at 37°C. In some experiments purified M1 protein was added to a concentration of 50 μg/ml. Samples of 100 μl were withdrawn at indicated time points, added to 2.5 ml of TH medium with 0.5% agar, spread on TH agar plates, and incubated at 37°C overnight. For strains complemented with plasmid pBH, the blood and the plates were supplemented with kanamycin at a concentration of 500 μg/ml.
RESULTS
Generation and characterization of emm and sph mutants.
Transposon mutagenesis of AP1 was used to generate mutants affected in the expression of proteins H and M1. Two types of mutants were generated with the Tn916 conjugative transposon. The first type was deficient in the expression of both proteins, and a detailed description and characterization of this mutant have been published (30). The second type was deficient in M1 protein, whereas the expression of protein H was unaffected. One of the mutants, BMJ11, was selected for further studies. Despite extensive screening, a mutant that had selectively lost its ability to express surface-associated protein H could not be detected. Such a protein H mutant, BM27.6, was therefore generated from AP1 by homologous recombination (6). The same technique was used to isolate the double mutant BM22.1 from BMJ11, which lacks both surface-expressed M1 protein and protein H.
Further analysis of BMJ11 demonstrated that only a single transposon was present in this mutant. However, the transposon in BMJ11 was not inserted either into or close to the M1 protein gene (emm1) (data not shown). Another M1 protein mutant, named MC25, was therefore generated from AP1 by deleting the 3′ end of the emm1 gene by homologous recombination (13), and the MC25 strain is the M1 protein-negative mutant used in this study.
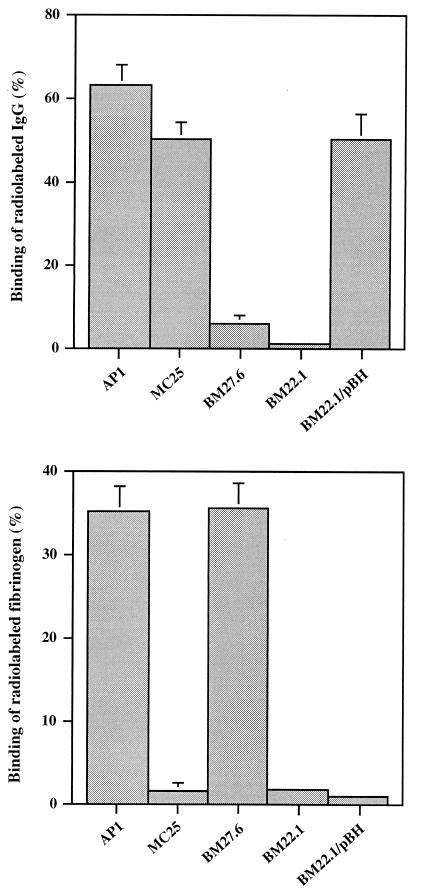
The M1 protein expressed by AP1 has been shown to bind fibrinogen, and it also has affinity for human IgG. Compared to protein H, which does not bind fibrinogen, the affinity for IgG is lower, especially for the IgG3 subclass (2, 3). Therefore, the binding of human fibrinogen and IgG3 to intact AP1 bacteria was used as a marker for surface-expressed M1 protein and protein H, respectively. A direct binding assay demonstrated that MC25 had no affinity for fibrinogen, whereas only a slight reduction was seen in the affinity for IgG3 (Fig. 1). The sph mutant BM27.6 bound fibrinogen, but the binding of IgG3 was significantly reduced (Fig. 1). The reduction of IgG3 binding seen with MC25, as well as the residual IgG3 binding exhibited by BM27.6, is explained by the affinity between M1 protein and IgG (see above). The double-mutant strain BM22.1 had no affinity for either IgG3 or fibrinogen (Fig. 1). This shows that MC25 lacks surface-associated M1 protein, whereas BM27.6 does not express protein H. BM22.1, finally, does not express M1 protein or protein H at its surface. When BM27.6 was complemented with pBH, a plasmid containing the sph gene, including promoter and termination sequences, it regained its ability to bind IgG3 (data not shown). BM22.1 trans-complemented with pBH bound IgG3 almost as well as MC25 (Fig. 1). This shows that the IgG3-binding property is predominately associated with the expression of protein H.
FIG. 1.
Binding of human fibrinogen or IgG3 to S. pyogenes bacteria. 125I-labeled proteins were incubated with indicated strains of bacteria at a concentration of 2 × 109 bacteria/ml. Data are means + standard deviations of at least three independent binding experiments.
Since the trans-acting regulator Mga in AP1 is known to coregulate the expression of M1 protein, protein H, protein SIC, and the C5a peptidase (30), the expression of the two latter proteins in the mutants was also studied. Protein SIC is an extracellular protein and can be purified from the growth medium (4), whereas C5a peptidase can be released from the streptococcal surface by a cysteine proteinase produced by S. pyogenes (5). The same amounts of protein SIC and C5a peptidase could be purified from AP1 and the mutants (data not shown), demonstrating that the expression of M1 protein and protein H was selectively affected in the mutants studied here.
Characterization of protein expression in the mutants.
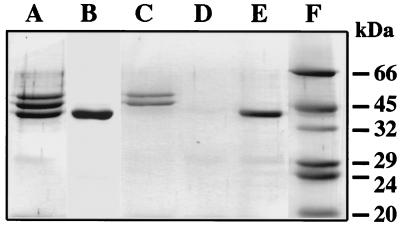
To investigate whether the binding data described above correlate with the expression of proteins M1 and H, we investigated the presence of surface-associated proteins by cyanogen bromide (CNBr) treatment. This treatment releases three protein fragments of 54, 49, and 44 kDa from wild-type AP1 bacteria (Fig. 2, lane A). The 54- and 49-kDa fragments originate from the M1 protein, and the 44-kDa band represents a protein H fragment (30). CNBr treatment of MC25 solubilized only the 44-kDa protein H-related fragment (Fig. 2, lane B). CNBr released the 54- and 49-kDa M1 fragments from BM27.6, whereas the 44-kDa band was missing (Fig. 2, lane C), which is consistent with an insertional mutation of sph in this strain. CNBr treatment of BM27.6 trans-complemented with pBH released the same three protein bands seen in extracts from wild-type AP1 bacteria (data not shown). No protein band was released by CNBr treatment of the double mutant BM22.1, whereas the 44-kDa fragment was seen after trans complementation of this strain with pBH (Fig. 2, lanes D and E). These results confirmed the binding data described above.
FIG. 2.
Analysis of surface-anchored M1 protein and protein H. AP1 (A), MC25 (B), BM27.6 (C), BM22.1 (D), or BM22.1/pBH (E) bacteria were treated with CNBr. Molecular mass markers are shown in lane F. Solubilized proteins were separated on SDS–10% PAGE gels followed by Coomassie blue staining.
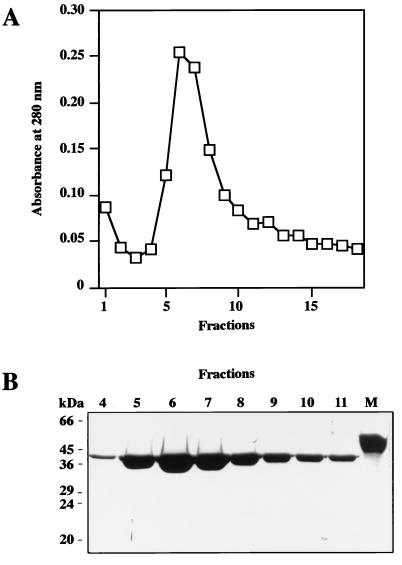
In the MC25 strain the mutant M1 protein was expected to be exported into the growth medium due to the absence of the COOH-terminal cell wall-anchoring LPXTGE amino acid motif. Therefore, growth medium from MC25 was purified on a Sepharose CL-4B column coupled with human fibrinogen. The eluted material contained a single protein component with a molecular mass of approximately 42 kDa (Fig. 3). The molecular masses of M proteins are overestimated by SDS-PAGE (21), and the value of 42 kDa is therefore compatible with the mass of 38 kDa predicted from the sequence of the truncated M1 protein.
FIG. 3.
Analysis of M1 protein released into the growth medium by MC25 bacteria. Culture supernatant from MC25 was affinity purified on Sepharose CL-4B coupled with human fibrinogen. (A) Absorbance at 280 nm of eluted fractions. (B) Eluted fractions 4 to 11 were separated on SDS–10% PAGE gels and stained with Coomassie blue. Lane M, purified recombinant M1 protein. Molecular mass markers are indicated to the left.
Surface-expressed protein H is sufficient for the survival of AP1 in human blood.
The antiphagocytic property of S. pyogenes is associated with the type-specific M protein (for references, see references 15 and 29). However, M-like proteins have also been shown to contribute to the antiphagocytic property (48, 56). The MC25 mutant expressed protein H but not M protein, and by using the classical bactericidal test (34) it was found that MC25 survived in human blood (Table 1), although the rate of multiplication was reduced compared to that of the wild-type AP1 strain. This demonstrates that surface-associated protein H is sufficient for the antiphagocytic property. MC25 secretes a fragment of the M1 protein into the growth medium (Fig. 3). To investigate whether the soluble M1 protein fragment contributes to the resistance to phagocytosis, an AP1 mutant carrying a transposon in mga was tested in the bactericidal assay. This mutant expresses no M or M-like proteins (30), and it was rapidly killed in human blood also in the presence of added purified M protein (data not shown), further emphasizing that protein H alone is sufficient for the antiphagocytic property. When BM27.6 was tested in the bactericidal test it was shown that it survived and multiplied rapidly, whereas BM22.1 was killed (Table 2). Also BM22.1/pBH survived in human blood (Table 2) but multiplied less well than the parent strain BMJ11. Thus, it can be concluded that M1 protein and protein H both contribute to resistance against phagocytosis.
TABLE 2.
Survival of S. pyogenes in human blood
| Strain | Relevant pheno-type | CFU/mla after time (h)
|
MF (3 h)b | |||
|---|---|---|---|---|---|---|
| 0 | 1.5 | 3 | 4.5 | |||
| AP1 (wild type) | M+ H+ | 380 | 7.6 × 103 | 8.9 × 104 | ndc | 234 ± 68 |
| MC25 | M− H+ | 340 | 2.1 × 103 | 2.2 × 104 | nd | 64 ± 17 |
| BM27.6 | M+ H− | 220 | 2.0 × 103 | 2.8 × 104 | nd | 127 ± 35 |
| BM22.1 | M− H− | 300 | 0 | 0 | 0 | 0 |
| BM22.1/pBH | M− H+ | 200 | 520 | 1.6 × 103 | 7.8 × 103 | 8 ± 2 |
The data represent the means of at least two independent bactericidal assays.
The increases in titers (multiplication factor [MF]) after 3 h of incubation in human blood are given as means ± standard deviations.
nd, not determined.
Analysis of the binding of human factor H to the M1 protein and protein H mutants.
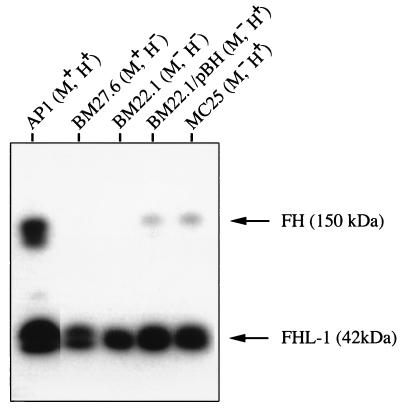
The resistance of S. pyogenes to phagocytosis has been linked to the expression of M protein and the ability of this protein to bind fibrinogen and/or factor H (24, 59). MC25 has selectively lost its fibrinogen-binding activity. However, soluble protein H, as well as protein M1, has been shown to bind purified factor H (30), which could explain the antiphagocytic property of MC25. Moreover, it is known that wild-type AP1 bacteria absorb albumin, fibrinogen, and IgG from human plasma (3). To investigate whether factor H could bind to AP1 and the mutants also in plasma environment, the different strains were separately incubated with human plasma. Following incubation, bacteria were washed, and proteins bound to the surface were eluted and subjected to SDS-PAGE and Western blot analysis with antibodies against factor H (Fig. 4). The most efficient absorption of factor H and its naturally occurring splice variant, factor H-like protein 1 (FHL-1), was seen with wild-type AP1 bacteria. All mutants also showed affinity for FHL-1, whereas factor H was clearly detected in the eluates from mutant bacteria expressing protein H (BM22.1/pBH and MC25). The data suggest that protein H and M1 protein both contribute to the binding of factor H-FHL-1 to the surface of AP1 bacteria, but other surface components also show affinity. Finally, AP1 mutants that survive in fresh human blood bind both factor H-FHL-1 and IgG (Table 3).
FIG. 4.
Absorption of factor H (FH) and FHL-1 from human plasma by wild-type AP1 bacteria and various AP1 mutants. Bacteria were incubated with human plasma, and the proteins absorbed were eluted and subjected to SDS–10% PAGE, followed by electrotransfer to a polyvinylidene difluoride membrane. The blot was incubated with antibodies against factor H, and the antibodies were visualized with peroxidase-labeled protein A.
TABLE 3.
Summary of properties of the S. pyogenes strains studied
| Strain | Genotype | In trans | Phenotype | Result of binding to:
|
Survival in blood | ||
|---|---|---|---|---|---|---|---|
| Fibrinogen | IgG3 | Factor H-FHL-1 | |||||
| AP1 | Wild type | M+ H+ | + | + | + | + | |
| MC25 | emm1::Km | M− H+ | − | + | + | + | |
| BM27.6 | sph::Erm | M+ H− | + | − | + | + | |
| BM22.1 | Tn916::xyz sph::Erm | M− H− | − | − | + | − | |
| BM22.1/pBH | Tn916::xyz sph::Erm | psph | M− H+ | − | + | + | + |
DISCUSSION
We have previously shown that the insertional inactivation of the regulator gene mga in the AP1 strain of the M1 serotype resulted in the loss of its antiphagocytic property (30). Most opacity factor-negative S. pyogenes strains express a single Emm protein (29), whereas AP1 bacteria express two structurally related Emm proteins, the M1 protein and protein H, with different ligand-binding properties (3, 17, 18). The present work demonstrates that these proteins both contribute to the phagocytic resistance. Mutants expressing either the M1 protein or protein H multiplied rapidly, whereas the double mutant lacking both proteins was killed in human blood. The double mutant trans-complemented with protein H on a plasmid (BM22.1/pBH) regained the ability to survive in human blood. However, BM22.1/pBH multiplied less well than the other mutants. This could be due to the insertion of several extra DNA segments. Both double-mutant bacteria and BM22.1/pBH grew somewhat slower in THY media than the other strains studied. The fact that BM22.1/pBH, but not the double mutant, is resistant to phagocytosis also indicates that proteins M1 and H are the only factors expressed by AP1 that are directly involved in the resistance to phagocytosis in human blood.
The M and M-like proteins are structurally related and are coexpressed but often display different ligand-binding properties. Our results, in line with the results of previous studies (48, 56), demonstrate that apart from the type-specific M protein, other M-like proteins also can be involved in the resistance to phagocytosis. Opacity factor-positive S. pyogenes strains express M-like proteins known as the Mrp proteins (29). The Mrp protein, as well as the coexpressed M protein, has been shown to be antiphagocytic in several strains (48, 56). However, the abilities of the individual proteins to confer resistance against phagocytosis seem to vary. Furthermore, not all M-like proteins are antiphagocytic. The IgA-binding protein Arp, an M-like protein expressed by an M4 strain (35), does not have this property (25). Attempts to complement heterologous nonresistant S. pyogenes JRS145, used by Husmann et al. (25), with pBH did not succeed. JRS145 could be transformed with pBH, but the recombinant strain expressed very small amounts of protein H, and this transformed strain was killed by human phagocytes (30a).
It has been demonstrated that surface-associated protein H can block C3 deposition on the bacterial surface, due to the inhibition of the classical complement pathway. This inhibitory activity of protein H was dependent on its ability to bind IgG (6). A significantly higher amount of C3 deposition was measured on the mutant of AP1 that expresses the M1 protein but lacks protein H (BM27.6) than on wild-type AP1 (6). However, despite the observed C3 deposition, BM27.6 was found to be resistant against phagocytosis in this study. Protein H has also been shown to bind C4BP (28), an inhibitor of the classical complement pathway, and purified protein H and M1 protein both bind factor H (30), another inhibitor of complement. Moreover, it was recently demonstrated that FHL-1, a splice variant of factor H (40), interacts with M5 and M6 proteins (31) by binding to their hypervariable NH2-terminal regions, whereas the factor H-binding site is separate (27) and located further towards the COOH-terminal end (16, 27, 52). In the case of M5 protein, fibrinogen was found to inhibit the binding of factor H (27). The binding of fibrinogen to M1 protein could therefore explain the low degree of absorption of factor H from plasma by the protein H-negative mutant expressing M1 protein. Among the tested strains, wild-type AP1 bacteria absorbed both factor H and FHL-1 most efficiently. However, the fact that the mutants, including the double mutant, still had FHL-1-binding activity demonstrates that AP1 bacteria have surface structures apart from protein H and M1 protein that also mediate FHL-1 binding. Thus, the relative importance of the binding of factor H, FHL-1, C4BP, and IgG for phagocytosis resistance remains unclear.
There is no doubt that the molecular basis for the antiphagocytic property of S. pyogenes is highly complex. Nevertheless, some conclusions can be made from the present observations, at least concerning the M1 strain used in this study. Firstly, proteins H and M1 are both antiphagocytic, and the presence of one of them at the bacterial surface is sufficient for the survival of bacteria in human blood. Secondly, the binding of fibrinogen to the bacterial surface is not required. Thus, the M1 protein mutant (MC25) devoid of fibrinogen-binding activity still survived. Similar observations were made for a strain of the M22 serotype (56). In contrast, fibrinogen binding was found to be necessary for the antiphagocytic activity of M proteins of some other serotypes (14). Thirdly, the surviving mutants bind both factor H-FHL-1 and IgG, suggesting that the presence of these host proteins at the surface of the M1 strain used here is crucial for its antiphagocytic property. As mentioned above, it has been demonstrated that the binding of IgG to the streptococcal surface through interaction with the Fc region of IgG inhibits the activation of the classical pathway (6), whereas factor H binding interferes with the activation of the alternative pathway of complement (24). Therefore, the present data suggest that the antiphagocytic property of S. pyogenes could be based on the interference with both complement pathways.
ACKNOWLEDGMENTS
The excellent technical assistance of Ingbritt Gustafsson and Ulla Johannesson is acknowledged.
This work was supported by grants from the Swedish Medical Research Council (projects 7480 and 13062), the Foundations of Crafoord, Kock, Lundberg, Nilson, and Österlund, the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine, ACTINOVA Ltd., and the Medical Faculty, Lund University.
REFERENCES
- 1.Åkerström B, Lindqvist A, Vander Maelen V, Grubb A, Lindahl G, Vaerman J-P. Interaction between streptococcal protein Arp and different molecular forms of human immunoglobulin A. Mol Immunol. 1994;5:393–400. doi: 10.1016/0161-5890(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 2.Åkesson P, Cooney J, Kishimoto F, Björck L. Protein H-A novel IgG binding bacterial protein. Mol Immunol. 1990;27:523–531. doi: 10.1016/0161-5890(90)90071-7. [DOI] [PubMed] [Google Scholar]
- 3.Åkesson P, Schmidt K-H, Cooney J, Björck L. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994;300:877–886. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Åkesson P, Sjöholm A G, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 5.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 6.Berge A, Kihlberg B-M, Sjöholm A, Björck L. Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J Biol Chem. 1997;272:20774–20781. doi: 10.1074/jbc.272.33.20774. [DOI] [PubMed] [Google Scholar]
- 7.Bisno A L. Group A streptococcal infections and acute rheumatic fever. New Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 8.Bisno A L, Stevens D L. Streptococcal infections of skin and soft tissues. N Engl J Med. 1996;334:240–245. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- 9.Björck L, Kastern W, Lindahl G, Widebäck K. Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Mol Immunol. 1987;24:1113–1122. doi: 10.1016/0161-5890(87)90080-0. [DOI] [PubMed] [Google Scholar]
- 10.Caparon M G, Scott J R. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 11.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleary P, Retnoningrum D. Group A streptococcal immunoglobulin-binding proteins: adhesins, molecular mimicry or sensory proteins? Trends Microbiol. 1994;2:131–136. doi: 10.1016/0966-842x(94)90600-9. [DOI] [PubMed] [Google Scholar]
- 13.Collin, M., and A. Olsén. 1998. Unpublished data.
- 14.Courtney H S, Liu S, Dale J B, Hasty D L. Conversion of M serotype 24 of Streptococcus pyogenes to M serotype 5 and 18: effect on resistance to phagocytosis and adhesion to host cells. Infect Immun. 1997;65:2472–2474. doi: 10.1128/iai.65.6.2472-2474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischetti V A, Horstmann R D, Pancholi V. Location of the complement factor H binding site on streptococcal M6 protein. Infect Immun. 1995;63:149–153. doi: 10.1128/iai.63.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frick I-M, Crossin K L, Edelman G M, Björck L. Protein H—a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J. 1995;14:1674–1679. doi: 10.1002/j.1460-2075.1995.tb07156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frick I-M, Åkesson P, Cooney J, Sjöbring U, Schmidt K-H, Gomi H, Hattori S, Tagawa C, Kishimoto F, Björck L. Protein H—a surface protein of Streptococcus pyogenes with separate binding sites for IgG and albumin. Mol Microbiol. 1994;12:143–151. doi: 10.1111/j.1365-2958.1994.tb01003.x. [DOI] [PubMed] [Google Scholar]
- 19.Gomi H, Hozumi T, Hattori S, Tagawa C, Kishimoto F, Björck L. The gene sequence and some properties of protein H—a novel IgG binding protein. J Immunol. 1990;144:4046–4052. [PubMed] [Google Scholar]
- 20.Heath D G, Cleary P P. Fc-receptors and M-protein genes of group A streptococci are products of gene duplication. Proc Natl Acad Sci USA. 1989;86:4741–4745. doi: 10.1073/pnas.86.12.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingshead S K, Fischetti V A, Scott J R. Complete nucleotide sequence of the type 6 M protein of the group A streptococcus. Repetitive structure and membrane anchor. J Biol Chem. 1986;261:1677–1686. [PubMed] [Google Scholar]
- 22.Holm S E, Norrby A, Bergholm A-M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 23.Hong K, Kinoshita T, Takeda J, Kozono H, Pramoonjago P. Inhibition of the alternative C3 convertase and classical C5 convertase of complement by group A streptococcal M protein. Infect Immun. 1990;58:2535–2541. doi: 10.1128/iai.58.8.2535-2541.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horstmann R D, Sievertsen H J, Knobloch J, Fischetti V A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husmann L, Scott J, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols; a guide to methods and applications. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 27.Johnsson E, Berggård K, Kotarsky H, Hellwage J, Zipfel P F, Sjöbring U, Lindahl G. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161:4894–4901. [PubMed] [Google Scholar]
- 28.Johnsson E, Thern A, Dahlbäck B, Hedén L-O, Wikström M, Lindahl G. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J Immunol. 1996;157:3021–3029. [PubMed] [Google Scholar]
- 29.Kehoe M A. Cell-wall-associated proteins in Gram-positive bacteria. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier; 1994. pp. 217–261. [Google Scholar]
- 30.Kihlberg B-M, Cooney J, Caparon M G, Olsén A, Björck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 30a.Kihlberg, B.-M., and L. Björck. Unpublished data.
- 31.Kotarsky H, Hellwage J, Johnsson E, Skerka C, Svensson H G, Lindahl G, Sjöbring U, Zipfel P F. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J Immunol. 1998;160:3349–3354. [PubMed] [Google Scholar]
- 32.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 34.Lancefield R C. Differentiation of group A streptococci with a common R antigen into three serological types with special reference to the bactericidal test. J Exp Med. 1948;106:525–544. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindahl G. Cell surface proteins of a group A streptococcus type M4: the IgA receptor and a receptor related to M proteins are coded for by closely linked genes. Mol Gen Genet. 1989;216:372–379. doi: 10.1007/BF00334378. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl G, Stenberg L. Binding of IgA and/or IgG is a common property among clinical isolates of group A streptococci. Epidemiol Infect. 1990;105:87–93. doi: 10.1017/s0950268800047683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin D R, Single L A. Molecular epidemiology of group A streptococcus M type 1 infections. J Infect Dis. 1993;167:1112–1117. doi: 10.1093/infdis/167.5.1112. [DOI] [PubMed] [Google Scholar]
- 39.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol Lett. 1995;128:45–52. doi: 10.1111/j.1574-6968.1995.tb07498.x. [DOI] [PubMed] [Google Scholar]
- 40.Misasi R, Huemer H P, Schwaeble W, Solder E, Larcher C, Dierich M P. Human complement factor H: an additional gene product of 43 kDa isolated from human plasma shows cofactor activity for the cleavage of the third component of complement. Eur J Immunol. 1989;19:1765–1768. doi: 10.1002/eji.1830190936. [DOI] [PubMed] [Google Scholar]
- 41.Musser J M, Kapur V, Kanjilal S, Shah U, Musher D M, Barg N L, Johnston K H, Schlievert P M, Henrichsen J, Gerlach D, Rakita R M, Tannna A, Cookson B D, Huang J C. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin) J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura A, Morita M, Nishimura Y, Sugino Y. A rapid and highly efficient method for preparing of competent Escherichia coli cells. Nucleic Acids Res. 1990;18:6169. doi: 10.1093/nar/18.20.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otten R, Raeder R, Heath D G, Lottenberg R, Cleary P P, Boyle M D P. Identification of two type IIa IgG-binding proteins expressed by a single group A streptococcus. J Immunol. 1992;148:3174–3182. [PubMed] [Google Scholar]
- 44.Perez-Casal J, Caparon M G, Scott J R. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res Microbiol. 1992;143:549–558. doi: 10.1016/0923-2508(92)90112-2. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Casal J, Price J A, Maguin E, Scott J R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequence to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 47.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Podbielski A, Schnitzler N, Beyhs P, Boyle M D P. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 49.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 50.Poirier T P, Kehoe M A, Whitnack E, Dockter M E, Beachey E H. Fibrinogen binding and resistance to phagocytosis of Streptococcus sanguis expressing cloned M protein of Streptococcus pyogenes. Infect Immun. 1989;57:29–35. doi: 10.1128/iai.57.1.29-35.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 52.Sharma A K, Pangburn M K. Localization by site-directed mutagenesis of the site in human complement factor H that binds to Streptococcus pyogenes M protein. Infect Immun. 1997;65:484–487. doi: 10.1128/iai.65.2.484-487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stenberg L, O’Toole P, Lindahl G. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol Microbiol. 1992;6:1185–1194. doi: 10.1111/j.1365-2958.1992.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 54.Stevens D L. Streptococcal toxic-shock syndrome: spectrum of disease, pathogenesis, and new concepts in treatment. Emerg Infect Dis. 1995;1:69–78. doi: 10.3201/eid0103.950301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thern A, Stenberg L, Dahlbäck B, Lindahl G. Ig-binding surface proteins of Streptococcus pyogenes also bind C4B-binding protein (C4BP), a regulatory component of the complement system. J Immunol. 1995;154:375–386. [PubMed] [Google Scholar]
- 56.Thern A, Wästfelt M, Lindahl G. Expression of two different antiphagocytic M proteins by Streptococcus pyogenes of the OF+ lineage. J Immunol. 1998;160:860–869. [PubMed] [Google Scholar]
- 57.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wexler D E, Chenoweth D E, Cleary P P. Mechanism of action of the group A streptococcal C5a inactivator. Proc Natl Acad Sci USA. 1985;82:8144–8148. doi: 10.1073/pnas.82.23.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitnack E, Beachey E H. Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J Clin Investig. 1982;69:1042–1045. doi: 10.1172/JCI110508. [DOI] [PMC free article] [PubMed] [Google Scholar]