Abstract
Streptococcus pyogenes produces several extracellular proteins, including streptococcal erythrogenic toxin B (SPE B), also known as streptococcal pyrogenic exotoxin B and streptococcal proteinase. Several reports suggest that SPE B contributes to the virulence associated with S. pyogenes; however, little is known about its regulation. Nucleotide sequence data revealed the presence, upstream of the speB gene, of a gene, designated rgg, that was predicted to encode a polypeptide similar to previously described positive regulatory factors. The putative Rgg polypeptide of S. pyogenes NZ131 consisted of 280 amino acids and had a predicted molecular weight of 33,246. To assess the potential role of Rgg in the production of SPE B, the rgg gene was insertionally inactivated in S. pyogenes NZ131, which resulted in markedly decreased SPE B production, as determined both by immunoblotting and caseinolytic activity on agar plates. However, the production of other extracellular products, including streptolysin O, streptokinase, and DNase, was not affected. Complementation of the rgg mutant with an intact rgg gene copy in S. pyogenes NZ131 could restore SPE B production and confirmed that the rgg gene product is involved in the production of SPE B.
Streptococcus pyogenes produces a variety of extracellular proteins including streptococcal erythrogenic toxin B (SPE B), also known as streptococcal pyrogenic exotoxin B and streptococcal proteinase (7, 17, 19, 33). The speB gene encodes a polypeptide consisting of 371 amino acids with a predicted molecular weight of 40,000 (19). Following secretion, the SPE B zymogen is thought to be activated by proteolysis and reduction to form a 28-kDa sulfhydryl proteinase (15, 16, 27). In contrast to the genes encoding streptococcal erythrogenic toxins A and C, which are bacteriophage encoded, the speB gene is present in the chromosome of all strains of S. pyogenes (55), although certain strains, including some isolates from invasive infections, do not produce the protein in vitro (8, 14, 15, 22, 32, 38, 42, 43, 47). Although it is not known if those isolates which did not produce SPE B in vitro were capable of SPE B production in vivo, recent results showed that in a murine model of necrotizing fasciitis, speB is not required for soft-tissue necrosis (1). Nonetheless, other studies indicate that SPE B does contribute to the virulence of S. pyogenes in mice (25, 29).
The proteolytic activity of SPE B has been suggested to enable S. pyogenes to alter its interaction with the human host during the course of infection. Specifically, SPE B has been shown to cleave the streptococcal adhesin M1 protein from the bacterial surface (4) and to degrade host cell extracellular matrix factors that also participate in adherence, including fibronectin and vitronectin (2, 24, 51). This strategy could facilitate dissemination of S. pyogenes after colonization has been established. In addition, the proteolytic activity of SPE B has been shown to activate a variety of host cell molecules including kininogens, which results in the release of kinins (20), interleukin-1β (23), and a human matrix metalloprotease (5). Such activation may be relevant to the clinical manifestations associated with streptococcal infection.
Despite the potential importance of SPE B in streptococcal pathogenesis, relatively little is known about the molecular mechanisms involved in the regulation of its production. Previous research has identified culture conditions that influence SPE B production, including an acidic pH and the presence of neopeptone (12). In addition, SPE B was detected in sterile culture supernatant fluid primarily during the transition from the exponential to the stationary phase of growth when strain NZ131 was grown in dialyzed Todd-Hewitt (TH) medium (9), which confirmed an earlier report by Lo et al. (28). However, SPE B production in NZ131 did not strictly correlate with the growth phase of the culture, based on the observation that the addition of catabolic compounds during the exponential phase of growth, including glucose and Casamino Acids, inhibited SPE B production even as the culture entered the stationary phase (9). Production of SPE B also did not strictly correlate with nutrient depletion, based on the finding that exponential phase cultures resuspended in conditioned medium did not secrete SPE B (9). Not surprisingly, the molecular mechanisms involved in the regulation of SPE B expression are also proving to be complex. Podbielski et al. (37) reported that SPE B is expressed as part of the multiple gene activator (Mga) regulon, which consists of several coregulated virulence-associated genes (6, 10, 34). In addition, inactivation of components of both the oligopeptide and dipeptide transport systems diminished speB mRNA levels, suggesting that a regulatory link exists between peptide transport and the expression of the extracellular proteinase (35, 36). Thus, several previous studies suggest that SPE B production is regulated, at least in part, by the metabolic state of the culture; however, the molecular mechanisms responsible for this regulation are not well understood.
The purpose of this study was to assess the potential role of the streptococcal rgg gene, which was identified upstream of the speB locus, in the production of SPE B. Inactivation of the rgg gene in S. pyogenes NZ131 resulted in a marked decrease in SPE B production, as determined by immunoblotting and an abrogation of caseinolytic activity on agar plates. Extracellular SPE B production could be restored by complementation of the rgg mutant with an intact rgg gene copy, confirming that the rgg gene is involved in the production of SPE B.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are described in Table 1. S. pyogenes NZ131 was grown in TH medium (Becton Dickinson, Cockeysville, Md.) containing 0.2% yeast extract (Difco Laboratories, Detroit, Mich.). Dialyzed TH medium was prepared by suspending 30 g of TH medium (Becton Dickinson) in 100 ml of water and dialyzing (in dialysis tubing with an approximate molecular weight cutoff of 3,500; Spectrum Medical Industries, Inc., Los Angeles, Calif.) against 1 liter of water overnight at 4°C. The dialyzed material was filter sterilized by using a 0.2-μm-pore-size filter sterilization unit (Nalge Nunc International, Rochester, N.Y.). When appropriate, erythromycin and chloramphenicol were added to TH medium at final concentrations of 3.0 and 10.0 μg/ml, respectively. Escherichia coli was grown in Luria-Bertani (LB) broth or LB agar plates containing, when appropriate, erythromycin (10 μg/ml) or chloramphenicol (80 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or referenceb |
|---|---|---|
| Strains | ||
| S. pyogenes NZ131 | D. R. Martin, Porirua, New Zealand | |
| E. coli | ||
| DB11 | Erythromycin sensitive | 13 |
| JM109 | JM107 recAJ | |
| DH10B | recA1 | BRL |
| Plasmids | ||
| pCRII | Apr; cloning vector | Invitrogen |
| pUC18::ermAM | Emr | 44 |
| pOU118 | 1,500-bp I-PCR product cloned into pCRII; independently isolated colony | This study |
| pOU424 | 1,500-bp I-PCR product cloned into pCRII; independently isolated colony | This study |
| pVA891-2 | Emr | H. Malke, Jena, Germany 31 |
| pOU500 | EcoRI fragment from pOU118 consisting of nucleotides 25–599 of the rgg gene cloned into pVA891-2; Emr | This study |
| pCRII::rgg | 1.185-kb PCR product (Rgg-F, Rgg-R) cloned into pCRII | This study |
| pAM401 | Shuttle vector; Cmr | B. Jett, University of Oklahoma HSC, (53) |
| pAM401::rgg | EcoRV-BamHI 1.18-kb fragment of pCRII::rgg cloned into pAM401 | This study |
Apr, Emr, and Cmr, resistance to ampicillin, erythromycin, and chloramphenicol, respectively.
BRL, Gibco-Bethesda Research Laboratories Inc., Gaithersburg, Md.; HSC, Health Sciences Center.
PCR.
Southern hybridization analysis using a radiolabeled probe specific to the 5′ portion of the speB gene indicated the presence of ClaI restriction endonucleases sites within and upstream of the speB gene that resulted in a fragment of suitable size for inverse PCR (I-PCR). Based on these results, chromosomal DNA was isolated from S. pyogenes NZ131 and digested with ClaI overnight at 37°C. Following digestion, total DNA was purified by using a Qiaex DNA purification kit (Qiagen, Chatsworth, Calif.). The DNA was eluted in water, and T4 DNA ligase (New England Biolabs Inc., Beverly, Mass.) was used to ligate the DNA under conditions that favored intramolecular ligation. A 1-μl aliquot of the ligation mix was used as template DNA, and the genomic DNA upstream of the speB gene was amplified by using oligonucleotide primers purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). The first primer, designated SpeB-1 (5′-ACGAGCAAAGTTTTGATC-3′), corresponded to bp 99 to 82 of the speB gene (19). The second primer, designated SpeB-2 (5′-AAAGTAGGCGGACATGCC-3′), was complementary to nucleotides 1006 to 1023 of the speB gene. Thirty cycles of amplification were carried out in a Perkin-Elmer (Branchburg, N.J.) DNA thermocycler with strand denaturation (1 min at 94°C), annealing (1 min at 48°C), and elongation (3 min at 72°C). The total volume of the PCR mixture was 50 μl and consisted of 1 μl of template DNA, 0.4 μM each primer, 20 μM deoxynucleoside triphosphates, 4 mM Mg2+, and 2.5 U of Taq polymerase (Promega Corp., Madison, Wis.). Following amplification, the amplicon was cloned into the TA cloning vector pCRII (Invitrogen, San Diego, Calif.) as described by the manufacturer. Two independently isolated clones were selected for further analysis and designated pOU118 and pOU424.
Southern hybridization.
Streptococcal chromosomal DNA was isolated as previously described (7). DNA was digested with various restriction endonucleases obtained from New England Biolabs, and the fragments were separated by agarose gel electrophoresis. Following electrophoresis, DNA was transferred to Nytran (Schleicher & Schuell, Keene, N.H.) by the method of Southern (46) as described elsewhere (40). Probes were labeled with [α-32P]dATP (New England Nuclear, Dupont Corp.) using a random-primer DNA labeling kit (U.S. Biochemical Corp. Cleveland, Ohio) as instructed by the manufacturer. The rgg-specific probe consisted of nucleotides 25 to 559 of the rgg gene and was isolated from an agarose gel by using a Qiaex purification kit (Qiagen) following digestion of pOU118 with EcoRI. The ermAM-specific probe was prepared by digesting pUC18::ermAM (obtained from D. Simon) (44) with EcoRI and BamHI. The approximately 1.1-kb ermAM fragment was then purified following agarose electrophoresis using a Qiaex purification kit (Qiagen). Hybridization was done under stringent conditions (65°C, 0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) essentially as previously described (40).
Cloning of the rgg gene from NZ131.
Using the nucleotide sequence data obtained from sequencing of pOU118 and pOU424 and data from the unannotated streptococcal genome sequencing project (39), oligonucleotides were designed to amplify a 1.185-kb fragment containing the complete rgg gene from NZ131. The oligonucleotides, Rgg-F (5′-TTATGGCTATATCATAGCTGC-3′) and Rgg-R (5′-ATCGCCCTGGAGCTGTTGAG-3′), were synthesized by Genemed Biotechnologies, Inc. (San Francisco, Calif.). Rgg-F corresponded to the nucleotide sequence beginning 247 bases upstream of the predicted translational start codon of rgg, and thus the amplicon may have included a functional promoter. Thirty cycles were carried out in a Perkin-Elmer 9600 DNA thermocycler with strand denaturation (15 s at 94°C), annealing (30 s at 47°C), and elongation (1.5 min at 70°C). The total volume of the PCR mixture was 50 μl and consisted of 0.1 μg of genomic DNA isolated from strain NZ131, 0.4 μM each primer, 20 μM deoxynucleoside triphosphates, 1.5 mM Mg2+, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). Following amplification, the amplicon was cloned into the TA cloning vector pCRII (Invitrogen) as described by the manufacturer. Restriction enzyme fragment analysis and PCR confirmed that rgg had been cloned into pCRII. The recombinant plasmid was designated pCRII::rgg.
Nucleotide sequence determination.
Plasmid DNA was isolated from pOU118 and pOU424 by using a Wizard MiniPrep plasmid isolation kit (Promega) according to the manufacturer’s instructions. The nucleotide sequences of both strands of the insert DNA were determined by using Sequenase as described by the manufacturer (U.S. Biochemical Corp.) and custom-designed oligonucleotides purchased from Integrated DNA Technologies. The complete rgg gene of NZ131 was amplified from purified genomic DNA in a Perkin-Elmer 9600 thermocycler with High Fidelity polymerase (Boehringer Mannheim), using primers Rgg-F and Rgg-R. The approximately 1.1-kb PCR product was purified by using a Centricon-100 column (Amicon Inc., Beverly, Mass). Subsequently, the nucleotide sequences of both strands were determined by using custom-designed oligonucleotide primers (Genemed Synthesis) and a Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.). The sequencing reactions were analyzed on an Applied Biosystems 373 DNA sequencer (PE Applied Biosystems). The sequencing data were analyzed with Sequencer version 3.0 software (Gene Codes Corp., Ann Arbor, Mich.).
Inactivation of the rgg gene in S. pyogenes NZ131 and complementation of the mutant with an intact gene.
The chromosomal rgg gene of NZ131 was insertionally inactivated following electrotransformation (45) of S. pyogenes NZ131 with the suicide plasmid pOU500. Erythromycin-resistant (Emr) transformants were selected on agar plates containing 3.0 μg of erythromycin per ml. Three Emr transformants were selected for further study.
To complement the rgg mutant, the recombinant plasmid pAM401::rgg was constructed by subcloning the complete rgg gene from pCRII::rgg into pAM401 and transforming E. coli DH10B as previously described (11). The purified plasmid was then used to electrotransform the rgg mutant, and transformants were selected on agar plates containing chloramphenicol. Cell lysates from selected transformants were prepared by boiling bacterial cell suspension in an alkaline solution as previously described (18). PCR was performed with cell lysates as template DNA and oligonucleotide primers specific to the rgg gene and to the gene conferring chloramphenicol resistance. The results from the PCR analysis confirmed that transformants possessed the recombinant plasmid pAM401::rgg.
Caseinolytic assay.
The proteolytic activity of SPE B was assessed by the casein agar plate assay as described by Hynes and Tagg (22). Briefly, the strain to be tested was stab inoculated into TH agar plates containing 10% skim milk (Difco). The plates were then incubated anaerobically at 37°C for approximately 18 h in an anaerobic GasPak (Becton Dickinson). Caseinolytic activity resulted in a zone of translucence surrounding the stab site.
Streptokinase and DNase assays.
A plate assay was used to assess streptokinase activity in broth culture supernatant fluid as previously described (21). DNase activity was assessed on agar plates containing methyl green (Becton Dickinson).
Supernatant fluid preparation, SDS-PAGE, and immunoblot analysis.
Culture supernatant fluid was prepared following growth in dialyzed TH medium at 37°C. The A600 of each culture was adjusted to 0.5 by dilution with sterile medium. Typically 1 ml of the suspension was centrifuged to pellet the cells, and the supernatant fluid was removed and passed through a 0.2-μm-pore-size filter unit (Millipore Corp., Bedford, Mass.). Between 2 and 5 μl of culture supernatant fluid was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using a 12.5% acrylamide resolving gel (26). Following electrophoresis, proteins were transferred to Immobilon-NC nitrocellulose membranes (Millipore Corp.), using a Bio-Rad transfer apparatus and Towbin’s buffer (50). After transfer, the nitrocellulose membrane was blocked with 5% skim milk in phosphate-buffered saline containing 0.02% Tween 20 (PBST) for 45 min. The membrane was washed with PBST, rabbit antiserum raised against purified SPE B (diluted 1:1,000 in PBST) was added, and the blot was incubated for 1 h. Next, protein A-horseradish peroxidase conjugate (Amersham) was added at a 1:2,000 dilution in PBST for 1 h. The antibody-antigen complexes were visualized by using the Amersham enhanced chemiluminescence Western blotting detection system as described by the manufacturer.
Nucleotide sequence accession number.
Nucleotide sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession no. AF091252.
RESULTS
I-PCR and nucleotide sequence analysis of rgg.
To identify potential regulatory elements involved in the production of SPE B, the genomic region upstream of the speB gene was amplified by I-PCR. The PCR product was cloned, and the nucleotide sequences of both strands of the amplicon were determined from two independently isolated clones (designated pOU118 and pOU424). One terminus of the I-PCR product included 99 bp that were identical to the first 99 bp of the speB structural gene, including the sequence used to design SpeB-2, one of the I-PCR oligonucleotide primers. Adjacent to the structural gene were 160 bp that corresponded to the region upstream of the speB translational start site. The other terminus of the amplicon contained 30 bp that corresponded to nucleotides 1006 to 1036 of the speB structural gene (19) and included the sequence used to design SpeB-1, the second I-PCR primer. Thus, the nucleotide sequence at the termini of the I-PCR product indicated that the intervening 1,500 bp of sequenced DNA represented the region of the streptococcal chromosome that is upstream of speB in strain NZ131.
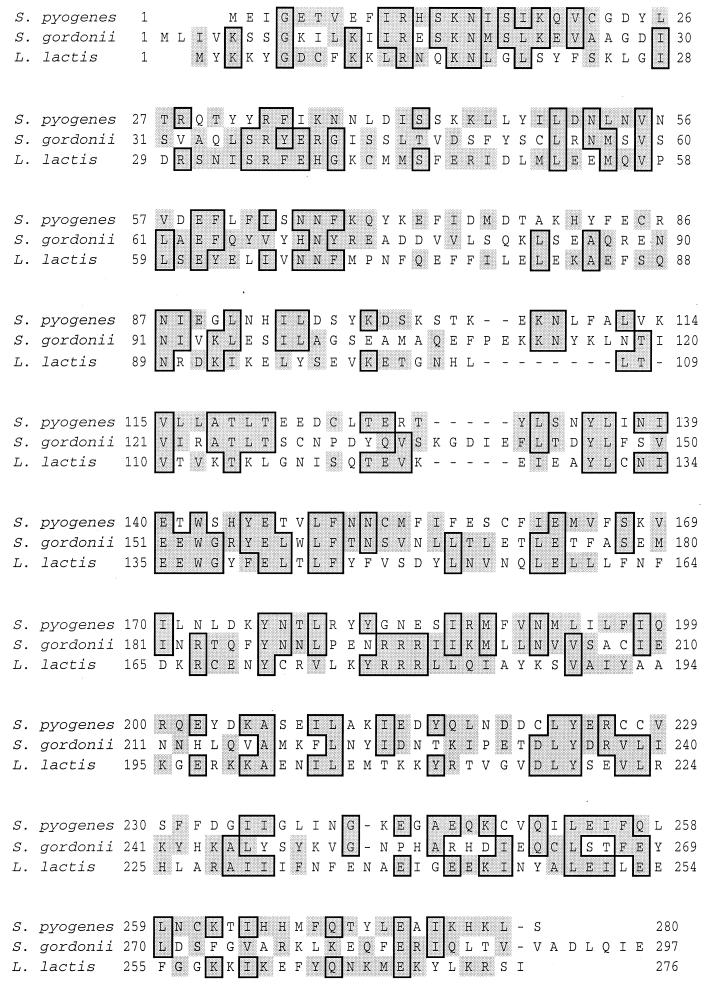
A single incomplete open reading frame (ORF) was identified upstream of the speB gene. The translational start site of the ORF is 941 bp upstream of the translational start site of speB. The ORF is encoded on the strand opposite that encoding SPE B. A putative ribosome-binding site (GGAAGG) was identified 8 bp upstream of the predicted ATG translational start site. The ORF did not include a termination codon within the I-PCR-amplified region, and thus the entire gene was not amplified. Using nucleotide sequence data from the streptococcal genome sequencing project, we amplified the complete ORF from NZ131 genomic DNA by PCR and determined the nucleotide sequence of both strands. Subsequently, the nucleotide sequence of the region upstream of speB in strain CS101 was submitted to GenBank (54). The nucleotide sequence from strain NZ131 had five single base pair differences in the noncoding region of NZ131 compared to the nucleotide sequence determined from strain CS101; both NZ131 and CS101 are M49 serotypes. In addition, there were several single base pair differences in the noncoding region compared to strain SF370, the strain used in the S. pyogenes genome sequencing project (data not shown). The complete ORF identified in strain NZ131 was predicted to encode a polypeptide composed of 280 amino acids that is 22% identical and 34% similar to the Rgg polypeptide of S. gordonii (49). The polypeptide is also 22% identical and 35% similar to the GadR polypeptide of Lactobacillus lactis (41). An alignment of these polypeptides is shown in Fig. 1. Based on the similarity of ORF to Rgg of S. gordonii, we have designated the gene rgg and the corresponding polypeptide Rgg.
FIG. 1.
Alignment of the deduced amino acid sequence of the Rgg polypeptide from S. pyogenes NZ131 with amino acid sequences of the Rgg polypeptide from S. gordonii (49) and the GadR polypeptide of L. lactis (41). Boxed and shaded regions indicate identical and similar amino acids, respectively.
Inactivation of the rgg gene in S. pyogenes NZ131 and complementation of the mutant with an intact gene copy.
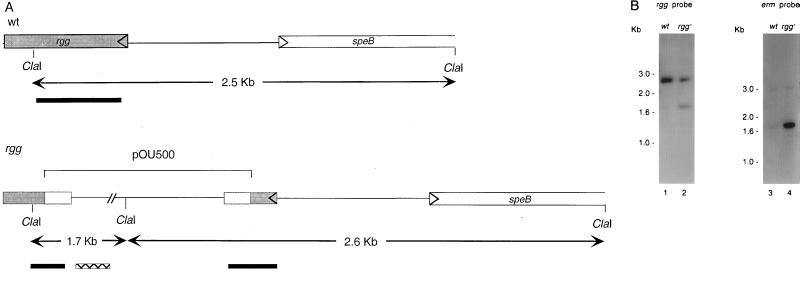
To assess the potential role of the rgg gene in SPE B production, isogenic strains of NZ131 that differed with respect to the presence of an intact rgg gene were created by directed mutagenesis. To inactivate rgg, we constructed a recombinant plasmid, designated pOU500, that contained 536 bp of the rgg gene to facilitate homologous recombination with the genomic rgg locus. The plasmid also contained a marker that conferred erythromycin resistance but not an origin of replication that is functional in S. pyogenes. Following electrotransformation of NZ131 with pOU500, transformants were selected on agar plates containing erythromycin. To confirm that Emr transformants resulted following integration of pOU500 into the genomic rgg locus, Southern hybridization was performed with chromosomal DNA isolated from the parental S. pyogenes NZ131 strain (NZ131 wild type [wt]) and from three independently isolated Emr transformants. A schematic representation of the Southern analysis is shown in Fig. 2A. As expected, ClaI restriction of chromosomal DNA isolated from NZ131 wt resulted in a single fragment of approximately 2.5 kb that hybridized with the rgg-specific probe (Fig. 2B). In contrast, ClaI restriction of chromosomal DNA isolated from three Emr transformants resulted in two fragments that hybridized to the rgg-specific probe; the results obtained with a representative transformant are shown in Fig. 2B. Only chromosomal DNA isolated from the Emr transformants hybridized to the erythromycin-specific probe (Fig. 2B). These results were confirmed by Southern hybridization following PstI restriction of chromosomal DNA isolated from the wild-type NZ131 strain and from the three Emr transformants (results not shown). Together, these results demonstrated that pOU500 had integrated into the rgg locus of the streptococcal chromosome.
FIG. 2.
Southern blot analysis of chromosomal DNA isolated from NZ131 wt and an NZ131 rgg mutant. DNA isolated from NZ131 wt and an rgg mutant strain was digested with ClaI and analyzed by Southern blot hybridization. (A) Schematic presentation of the rgg locus showing relative positions of the ClaI restriction sites, approximate sizes of the ClaI fragments, and locations of the rgg (solid bar) and ermAM (hatched bar) probes. (B) DNA from NZ131 wt (lane 1) and the NZ131 rgg mutant (lane 2) was hybridized with an rgg-specific probe. DNA from NZ131 wt (3) and the NZ131 rgg mutant (4) was probed with a probe specific to ermAM.
To control for potential polar affects associated with the insertion of pOU500 into the streptococcal chromosome, the complete rgg gene was cloned into the shuttle plasmid pAM401, and the recombinant plasmid was designated pAM401::rgg. Plasmid pAM401 replicates episomally in S. pyogenes (10) and confers chloramphenicol resistance (3). The recombinant plasmid (pAM401::rgg) was used to electrotransform the rgg mutant, and the resulting transformants (designated NZ131 rgg/pAM401::rgg+) were selected on agar plates containing chloramphenicol. The presence of the intact rgg gene and the gene which conferred chloramphenicol resistance was confirmed by PCR (data not shown). As a control, NZ131 wt was similarly electrotransformed with pAM401::rgg; however, no transformants were obtained. In contrast, both NZ131 wt and the rgg mutant were readily transformed with the vector (pAM401) alone.
SPE B caseinolytic activity in NZ131 requires an intact rgg gene.
The casein agar assay was used as an initial step to determine if inactivation of the rgg gene affected the caseinolytic activity previously associated with SPE B production (7). The caseinolytic activity associated with SPE B creates a zone of translucence surrounding either a colony or stab site following growth on opaque, casein-containing TH agar medium due to the proteolytic activity of SPE B (Fig. 3) (22). As shown in Fig. 3, a zone of translucence formed around the wild-type stab site, indicative of SPE B production. In contrast, an NZ131 speB mutant, created by insertional duplication mutagenesis, showed markedly reduced caseinolytic activity; however, as previously reported (7), a slight zone of clearing was still evident surrounding the stab site (Fig. 3). Inactivation of the rgg gene ablated caseinolytic activity (Fig. 3). Two phenotypes were observed on skim milk-containing plates among transformants obtained following electroporation of the rgg mutant with pAM401::rgg. The majority of the chloramphenicol-resistant (Cmr) colonies showed a low level of caseinolytic activity, reminiscent of that observed with a speB mutant (NZ131 rgg/pAM401::rgg+-2) (Fig. 3). The second phenotype (designated NZ131 rgg/pAM401::rgg+-1) was observed in 1 of 20 Cmr transformants and showed a significantly greater level of caseinolytic activity compared to the parental strain (Fig. 3).
FIG. 3.
Casein agar assay for SPE B proteolytic activity. Genetic derivatives of NZ131 were stab inoculated into agar plates containing skim milk and incubated for 18 h anaerobically. Proteinase activity is manifest as a zone of translucence surrounding the stab sites of NZ131 wt and NZ131 rgg/pAM::rgg+-1.
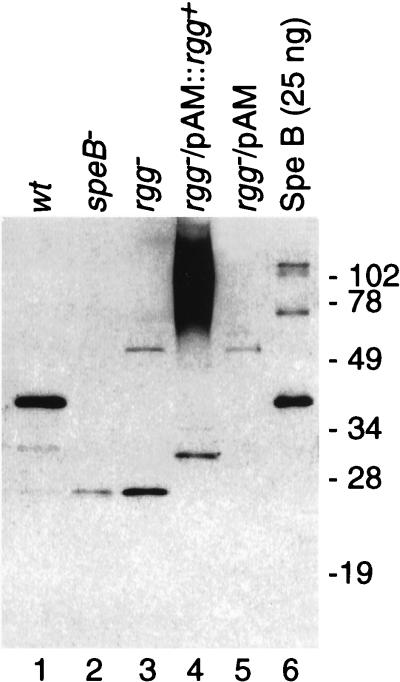
To confirm that inactivation of the rgg gene diminished SPE B production, culture supernatant fluid was prepared from cultures with an equal A600 (following approximately 18 h of growth in dialyzed TH broth medium) from strains NZ131, NZ131 speB, NZ131 rgg, NZ131 rgg/pAM401::rgg+-1, and NZ131 rgg/pAM401. Sterile culture supernatant fluid was prepared and analyzed by immunoblotting using antiserum prepared against purified SPE B. As expected, culture supernatant fluid prepared from the parental NZ131 strain (Fig. 4, lane 1) contained a significant amount of protein that migrated similarly to purified SPE B (lane 6) and reacted with antiserum to SPE B. Also detected were several faster-migrating species (lane 1) which likely represent modified forms of the SPE B zymogen. As a control, supernatant fluid was similarly prepared from the speB mutant, and relatively little protein that reacted with antiserum to SPE B was present (lane 2). Similarly, the amount of anti-SPE B-reactive protein in supernatant fluid prepared from both the rgg mutant (lane 3) and the rgg mutant transformed with pAM401 (lane 5) was less than for the wild-type strain. In supernatant fluid prepared from the complemented rgg mutant (NZ131 rgg/pAM401::rgg+-1), anti-SPE B-reactive protein that migrated as a high-molecular-weight smear was detected (lane 4). The smear did not resolve into distinct bands following either serial dilution of the sample or treatment with various reducing agents (results not shown). In contrast, no protein was detected with antiserum to SPE B in supernatant fluids prepared from the complemented rgg mutants (NZ131 rgg/pAM401::rgg+-2) that showed only a minor level of caseinolytic activity (not shown). Thus, the results obtained from immunoblots of culture supernatant fluid and the casein agar assay showed that the rgg gene positively influences the production of SPE B in S. pyogenes NZ131.
FIG. 4.
Immunoblot analysis of culture supernatant fluid from isogenic derivatives of strain NZ131. Sterile supernatant fluid was prepared from derivatives of strain NZ131. Proteins in the supernatant fluid were separated by SDS-PAGE and transferred to a nitrocellulose membrane. SPE B was then detected by using rabbit antiserum raised against purified SPE B. Lanes 1 to 5, 2 μl of supernatant fluid prepared from the strains indicated at the top; lane 6, 25 ng of purified SPE B protein. Sizes are indicated in kilodaltons.
Inactivation of the rgg gene does not affect extracellular hemolytic, streptokinase, or DNase activity.
Despite the similarity of Rgg to previously described transcriptional regulatory factors, it remained possible that inactivation of the rgg gene had a general negative effect on the secretion apparatus of NZ131. In this regard, NZ131 rgg mutants were beta-hemolytic on blood agar plates, indicating that the production of the extracellular streptolysin S and O was not affected by inactivation of the rgg gene (data not shown). In addition, the rgg mutant showed normal production of both extracellular streptokinase and DNase activities (data not shown). Taken together, these results suggest that the lack of SPE B production in rgg mutants is unlikely to be related to secretion defects. In addition, these results showed that rgg is not required for the production of all extracellular products.
DISCUSSION
The rgg gene product of S. pyogenes is similar in amino acid sequence to the rgg gene product of S. gordonii (49). In S. gordonii, the rgg gene is present upstream and in the same transcriptional orientation as the gtfG gene, which encodes an extracellular glucosyltransferase (52). Sulavik and Clewell recently showed that Rgg is a positive transcriptional regulator of gtfG expression (48). In addition, the rgg genes of S. gordonii and S. pyogenes are also similar to the gadR gene of L. lactis, which encodes an activator of the gadCB operon (41). The location of the rgg gene upstream of speB and the similarity of the protein it encodes to other positive regulatory proteins prompted this investigation to determine whether the Rgg protein is involved in the regulation of extracellular proteins in S. pyogenes. Toward this end, inactivation of the rgg gene in strain NZ131 resulted in an ablation of caseinolytic activity and a decrease in the secretion of SPE B, as determined by immunoblots of sterile culture supernatant fluid. However, the rgg mutant did possess hemolytic, streptokinase, and DNase activity, indicating that production of the exoproteins responsible for those activities was not affected by rgg inactivation. Complementation of the rgg mutant with an intact rgg gene could restore SPE B production, confirming that the gene is involved in the production of SPE B in strain NZ131.
As previously reported (7), a speB mutant constructed in NZ131 by insertional duplication mutagenesis had markedly reduced caseinolytic activity when grown on skim milk-containing agar plates compared to the parental strain. Nonetheless, some caseinolytic activity remained (Fig. 3). Interestingly, the rgg mutant showed no detectable caseinolytic activity under these conditions (Fig. 3). The results from immunoblotting of sterile culture supernatant fluid prepared from both the speB and rgg mutant showed a similar, albeit minor, amount of an approximately 28-kDa protein that reacted with SPE B antiserum. Therefore, the amount of SPE B detected in supernatant fluid from these mutants did not correlate with the level of caseinolytic activity observed following growth on solid medium. This result suggested that the difference in caseinolytic activity observed between speB and rgg mutants was not due to low level SPE B production. The disparity in the levels of caseinolytic activity between the mutants on solid medium may be related to the relative amount of acid produced by these strains. S. pyogenes generates ATP by fermentation and produces primarily lactic acid as the fermentation end product, which could potentially hydrolyze casein to result in a small zone of translucence on casein-containing medium. However, no difference in the pH of the medium during or following growth of the mutants in broth medium was apparent (6a). Nonetheless, it is unclear if the production of lactic acid or the buffering capacity of the medium was different following growth in broth medium compared to growth on solid medium containing skim milk. An additional explanation is that rgg inactivation may have affected the production of a previously uncharacterized protein (i.e., a protein other than SPE B) with caseinolytic activity. The rgg mutant was hemolytic on blood agar plates and produced levels of streptokinase and DNase activity similar to those produced by the wild-type strain, which indicated that the rgg mutation did not hinder exoprotein transport in general, nor was it required for the production of all extracellular products. Further experiments are necessary to determine if the production of other proteins, in addition to SPE B, is influenced by rgg.
Complementation of the rgg mutant with an intact gene resulted in partial restoration of caseinolytic activity on skim milk-containing agar plates compared to both the mutant and the mutant transformed with the plasmid only (Fig. 3). This result confirmed that the rgg gene positively influenced SPE B production. In addition, these results indicated that the rgg gene product can function in trans to influence production. The majority of the anti-SPE B-reactive material in the supernatant fluid prepared from the complemented strain (NZ131 rgg/pAM401::rgg+-1) migrated in the gel as a high-molecular-weight smear. To complement the mutant, an intact rgg gene was introduced into the mutant on a multicopy plasmid. Thus, it seems likely, based on the Western blotting results (Fig. 4), that SPE B was overproduced due to the presence of multiple copies of the rgg gene. Overproduction of SPE B may have resulted in the formation of aggregates of SPE B with other polypeptides. Thus, the smear likely represents a heterologous aggregate of SPE B-associated proteins, since it did not resolve into distinct bands upon dilution of the sample as would be expected if the smear was composed entirely of SPE B. In this regard, we noted that purified SPE B formed homoaggregates that migrated as distinct bands (Fig. 4, lane 6).
Two phenotypes were observed following growth of complemented rgg mutants on skim milk-containing agar. The majority of the Cmr colonies showed a low level of caseinolytic activity, reminiscent of that observed with the speB mutant (Fig. 3). However, approximately 1 of 20 Cmr transformants showed significantly greater caseinolytic activity (approximately 70% of that detected in the parental strain [Fig. 3]). Results from PCR analysis of colonies displaying both phenotypes showed the presence of a full-length rgg gene; however, we cannot rule out the possibility that point mutations were present in the plasmid-borne rgg gene. This seems unlikely, however, since the caseinolytic activities of all Cmr colonies were significantly greater than those of both the mutant and the mutant transformed with the vector alone, both of which did not produce any detectable caseinolytic activity (Fig. 3). Thus, all of the Cmr colonies analyzed appeared to be at least partially complemented for caseinolytic activity. The recombinant plasmid was unstable and NZ131 rgg/pAM401::rgg+-1 failed to grow after several passages on selective medium, suggesting that there was a toxic effect associated with the presence of rgg on a multicopy plasmid. Consistent with this interpretation was the lack of transformants when wild-type NZ131 was electrotransformed with pAM401::rgg. Thus, it seems reasonable to speculate that the colonies that showed a low level of caseinolytic activity following complementation may have had additional genotypic or phenotypic changes that were selected for during growth on media containing chloramphenicol. However, the molecular basis for the different phenotypes remains unknown. Thus, in general the results following complementation of the rgg mutant indicated that an intact rgg gene could restore SPE B production and caseinolytic activity to an rgg mutant, but production was not restored to the levels observed in the parental strain.
The similarity of the rgg gene product to previously described positive transcriptional regulators suggested that the basis for the lack of SPE B production in an rgg mutant was likely to be at the level of transcription. Recently Lyon et al. (30) showed that inactivation of the rgg gene (ropB) in strain HSC5 was associated with an absence of speB transcription. Together, the data indicate that Rgg is a positive transcriptional activator required for the expression of speB; however, it remains to be determined if Rgg acts directly or indirectly to regulate speB expression.
The rgg gene product is homologous to the gadR gene product of L. lactis throughout the primary structure of the polypeptide. This information, coupled with results of the present study, suggests that the regulation of SPE B production may be related to pH homeostasis in S. pyogenes. In L. lactis, GadR positively regulates the expression of gadC, which encodes a putative glutamate–γ-aminobutyrate (GABA) antiporter, and gadB, which encodes a glutamate decarboxylase. Together, these proteins contribute to pH homeostasis by facilitating the exchange of an acidic extracellular glutamate residue for a less acidic intracellular derivative (GABA). In addition, an intracellular proton is consumed during the decarboxylation of glutamate to form GABA. The expression of gadCB begins in the late exponential phase of growth and is maximal during the stationary phase of growth (41). Thus, one possible function of SPE B may be to degrade host cell proteins, such as fibronectin and vitronectin, to form peptides and amino acids that could potentially be important in pH homeostasis. To date, however, homologues of GadC and GadB have not been identified in S. pyogenes. In addition, peptides formed by the proteolytic activity of SPE B could potentially serve as a nitrogen source, and we note that the expression of speB has been previously shown to be linked to both the oligopeptide (36) and dipeptide transport (35) systems of S. pyogenes, as well as nutrient starvation (7). Clearly the regulation of SPE B production is complex, and additional information is required to understand how the various regulons involved in SPE B production interact. Understanding the conditions of SPE B production coupled with the knowledge of the molecular basis for SPE B regulation should enhance our understanding of how SPE B potentially affects the interaction between S. pyogenes and its human host.
ACKNOWLEDGMENTS
We thank Dieter Gerlach for providing purified SPE B and SPE B antiserum, Jim Bono, Joe Hinnebusch, Tom Schwan, and Jos van Putten for critical review of the manuscript; and Bob Evans and Gary Hettrick for help in preparing the figures.
Sequencing data were provided by the members of the streptococcal genome sequencing project including B. A. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, R. McLaughlin, and M. McShan and funded by USPHS/NIH grant AI38406.
REFERENCES
- 1.Ashbaugh C D, Warren H B, Carey V J, Wessels M R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Investig. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beachey E H, Giampapa C S, Abraham S M. Adhesin receptor mediated attachment of pathogenic bacteria to mucosal surfaces. Am Rev Respir Dis. 1988;138:S45–S48. doi: 10.1164/ajrccm/138.6_Pt_2.S45. [DOI] [PubMed] [Google Scholar]
- 3.Behnke D, Gilmore M S. Location of antibiotic resistance determinants, copy control and replication functions on the double-selective-streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184:115–120. doi: 10.1007/BF00271206. [DOI] [PubMed] [Google Scholar]
- 4.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 5.Burns E H, Marciel A M, Musser J M. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun. 1996;64:4744–4750. doi: 10.1128/iai.64.11.4744-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Chaussee, M. Unpublished observation.
- 7.Chaussee M S, Gerlach D, Yu C-E, Ferretti J J. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect Immun. 1993;61:3719–3723. doi: 10.1128/iai.61.9.3719-3723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee M S, Liu J, Stevens D L, Ferretti J J. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J Infect Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee M S, Phillips E R, Ferretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 11.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J O. Effect of culture medium composition and pH on the production of M protein and proteinase by group A streptococci. J Bacteriol. 1969;99:737–744. doi: 10.1128/jb.99.3.737-744.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta N, Hedges R W, Becker D, Davies J. Plasmid-determined fusidic acid resistance in the Enterobacteriaceae. J Gen Microbiol. 1974;83:191–196. doi: 10.1099/00221287-83-1-191. [DOI] [PubMed] [Google Scholar]
- 14.Deibel R H. Hydrolysis of proteins and nucleic acids by Lancefield group A and other streptococci. J Bacteriol. 1963;86:1270–1274. doi: 10.1128/jb.86.6.1270-1274.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott S D. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J Exp Med. 1945;81:191–196. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott S D, Dole V P. An inactive precursor of streptococcal proteinase. J Exp Med. 1947;85:305–320. doi: 10.1084/jem.85.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach D, Knöll H, Köhler W, Ozegowski J H, Hríbalova V. Isolation and characterization of erythrogenic toxins, V. Communication: identify of erythrogenic toxin type B and streptococcal proteinase precursor. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1983;255:221–233. [PubMed] [Google Scholar]
- 18.Hartas J, Hibble M, Sriprakash K S. Simplification of a locus-specific DNA typing method (Vir typing) for Streptococcus pyogenes. J Clin Microbiol. 1998;36:1428–1429. doi: 10.1128/jcm.36.5.1428-1429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4542. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herwald H, Collin M, Müller-Esterl W, Björck L. Streptococcal cysteine proteinase releases kinins: a novel virulence mechanism. J Exp Med. 1996;184:665–673. doi: 10.1084/jem.184.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang T-T, Malke H, Ferretti J J. Heterogeneity of the streptokinase gene in group A streptococci. Infect Immun. 1989;57:502–506. doi: 10.1128/iai.57.2.502-506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes W L, Tagg J R. A simple plate assay for detection of group A streptococcus proteinase. J Microbiol Methods. 1985;4:25–31. [Google Scholar]
- 23.Kapur V, Majesky M W, Li L-L, Black R A, Musser J M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapur V, Topouzis S, Jajesky M W, Li L-L, Hamrick M R, Patti R J, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 25.Kuo C-F, Wu J-J, Lin K-Y, Tsai P-J, Lee S-C, Jin Y-T, Lei H-Y, Lin Y-S. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect Immun. 1998;66:3931–3935. doi: 10.1128/iai.66.8.3931-3935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu T-Y, Elliott S D. Activation of streptococcal proteinase and its zymogen by bacterial cell walls. Nature (London) 1965;206:33–34. doi: 10.1038/206033a0. [DOI] [PubMed] [Google Scholar]
- 28.Lo S-S, Liang S-M, Liu T-Y. Intracellular form of streptococcal proteinase: a clue to a novel mechanism of secretion. Anal Biochem. 1984;136:89–92. doi: 10.1016/0003-2697(84)90309-9. [DOI] [PubMed] [Google Scholar]
- 29.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Investig. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion, and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malke H, Mechold U, Gase K, Gerlach D. Inactivation of the streptokinase gene prevents Streptococcus equisimilis H46A from acquiring cell-associated plasmin activity in the presence of plasminogen. FEMS Microbiol Lett. 1994;116:107–112. doi: 10.1111/j.1574-6968.1994.tb06683.x. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima K, Ichiyama S, Iinuma Y, Hasegawa Y, Ohta M, Ooe K, Shimizu Y, Igarashi H, Murai T, Shimokata K. A clinical and bacteriologic investigation of invasive streptococcal infections in Japan on the basis of serotypes, toxin production, and genomic DNA fingerprints. Clin Infect Dis. 1997;25:260–266. doi: 10.1086/514543. [DOI] [PubMed] [Google Scholar]
- 33.Ohara-Nemoto Y, Sasaki M, Kaneko M, Nemoto T, Ota M. Cysteine protease activity of streptococcal pyrogenic exotoxin B. Can J Microbiol. 1994;40:930–936. doi: 10.1139/m94-149. [DOI] [PubMed] [Google Scholar]
- 34.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podbielski A, Leonard B A B. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol. 1998;28:1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 36.Podbielski A, Pohl B, Woischnik M, Körner C, Schmidt K-H, Rozdzinski E, Leonard B A B. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (Opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- 37.Podbielski A, Woischnik M, Pohl B, Schmidt K H. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 38.Reichardt W, Müller-Alouf H, Alouf J F, Köhler W. Erythrogenic toxins A, B, and C: occurrence of the genes and exotoxin formation from clinical Streptococcus pyogenes strains associated with streptococcal toxic shock-like syndrome. FEMS Microbiol Lett. 1992;100:313–322. doi: 10.1111/j.1574-6968.1992.tb14058.x. [DOI] [PubMed] [Google Scholar]
- 39.Roe, B. A., S. P. Linn, L. Song, X. Yuan, S. Clifton, M. McShan, and J. J. Ferretti. 18 December 1998, revision date. Streptococcal genome sequencing project. [Online.] http://www.genome.ou.edu/strep.html. [17 February 1998, last date accessed.]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sanders J W, Leenhouts K, Burghoorn J, Brands J R, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz B, Facklam R, Breiman R. Changing epidemiology of group A streptococcal infection in the USA. Lancet. 1990;336:1167–1171. doi: 10.1016/0140-6736(90)92777-f. [DOI] [PubMed] [Google Scholar]
- 43.Sherwood N P, Paretsky D, Nachtigall A, McLain A R, Truffelli G T. Studies on streptococci. V. A study of streptococcal proteinases. J Infect Dis. 1954;95:1–12. doi: 10.1093/infdis/95.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 45.Simon D, Ferretti J J. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;82:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 46.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 47.Stevens D L, Tanner M H, Winship J, Swarts R, Ries K M, Schlievert P M, Kaplan E. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;312:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- 48.Sulavik M C, Clewell D B. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J Bacteriol. 1996;178:5826–5830. doi: 10.1128/jb.178.19.5826-5830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sulavik M C, Tardif G, Clewell D B. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J Bacteriol. 1992;174:3577–3586. doi: 10.1128/jb.174.11.3577-3586.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valentin-Weigand P, Grulich-Henn J, Chhatwal G S, Müller-Berghaus G, Blobel H, Preissner K T. Mediation of adherence of streptococci to human endothelial cells by complement S protein (vitronectin) Infect Immun. 1988;56:2851–2855. doi: 10.1128/iai.56.11.2851-2855.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vickerman M M, Sulavik M C, Nowak J D, Gardner N M, Jones G W, Clewell D B. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene gtfG. DNA Seq. 1997;7:83–95. doi: 10.3109/10425179709020155. [DOI] [PubMed] [Google Scholar]
- 53.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woischnik, M., and A. Podbielski. 1996. Direct submission to GenBank.
- 55.Yu C-E, Ferretti J J. Frequency of the erythrogenic toxin B and C genes (speB and speC) among clinical isolates of group A streptococci. Infect Immun. 1991;59:211–215. doi: 10.1128/iai.59.1.211-215.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]