Abstract
The basic motif-leucine zipper (bZIP) transcription factor neural retina leucine zipper (NRL) determines rod photoreceptor cell fate during retinal development, and its loss leads to cone-only retina in mice. NRL works synergistically with homeodomain protein Cone-Rod Homeobox and other regulatory factors to control the transcription of most genes associated with rod morphogenesis and functional maturation, which span over a period of several weeks in the mammalian retina. We predicted that NRL gradually establishes rod cell identity and function by temporal and dynamic regulation of stage-specific transcriptional targets. Therefore, we mapped the genomic occupancy of NRL at four stages of mouse photoreceptor differentiation by CUT&RUN analysis. Dynamics of NRL binding revealed concordance with the corresponding changes in transcriptome of the developing rods. Notably, we identified c-Jun proto-oncogene as one of the targets of NRL, which could bind to specific cis-elements in the c-Jun promoter and modulate its activity in HEK293 cells. Coimmunoprecipitation studies showed the association of NRL with c-Jun, also a bZIP protein, in transfected cells as well as in developing mouse retina. Additionally, shRNA-mediated knockdown of c-Jun in the mouse retina in vivo resulted in altered expression of almost 1000 genes, with reduced expression of phototransduction genes and many direct targets of NRL in rod photoreceptors. We propose that c-Jun-NRL heterodimers prime the NRL-directed transcriptional program in neonatal rod photoreceptors before high NRL expression suppresses c-Jun at later stages. Our study highlights a broader cooperation among cell-type restricted and widely expressed bZIP proteins, such as c-Jun, in specific spatiotemporal contexts during cellular differentiation.
Introduction
Transcriptional regulation is a critical aspect of cellular control mechanisms that impact a wide spectrum of biological processes, including specification of cell fate, response to intra- and extra-cellular signals, tissue homeostasis and regeneration (1,2). Gene transcription is initiated by transcription factors (TFs) which interact with diverse cis-regulatory elements including enhancers, promoters and silencers in a sequence-specific manner and recruit RNA polymerases to the transcriptional start site (TSS) (3,4). Regulatory function of TFs can be modulated by synergistic or antagonistic activities of co-regulators, repressors, chromatin remodelers and non-coding RNAs (5–8). Combinatorial actions of these multiple regulatory components endow intricate spatiotemporal gene expression profiles that establish cell- and tissue-specific phenotypes and function (9–11). Disruptions in transcriptional control mechanisms can lead to cell-type restricted or syndromic disease phenotypes (12–16).
The mammalian retina has served as an excellent in vivo system to investigate intricacies of development and disease pathogenesis pertaining to the nervous system because of easy accessibility for biological manipulations and clinical phenotyping. Transcriptional regulation has been extensively investigated in the retinal photoreceptors, especially during development (17–19). The two types of photoreceptor cells in the retina (rods and cones) are sensory neurons that detect photons and initiate the visual cascade (20). Rods express the visual pigment rhodopsin (Rho) and transduce visual signals primarily under dim light conditions, whereas cones express distinct opsin pigments to mediate high acuity and color vision under brighter light conditions. Lineage specification and terminal differentiation of photoreceptors require combinatorial interactions of multiple transcriptional regulatory proteins. Homeobox protein OTX2 and PR domain zinc finger protein 1 (PRDM1) promote photoreceptor cell fate and repress sister cell fate choice in developing retina (21–24). Within photoreceptor lineage, the Cone-Rod Homeobox (CRX) protein is critical for the regulation of most genes associated with morphogenesis and maturation (25–27). The basic motif leucine zipper (bZIP) protein of the large-Maf subfamily, NRL, is critical for rod fate specification with high expression levels restricted to rods even in the mature retina (17). The loss of Nrl in mice results in S-cone-like photoreceptors instead of rods whereas its ectopic expression in developing cones leads to rod photoreceptors (28–30). Notably, NRL and the thyroid hormone receptor TRβ2 can together generate the three photoreceptor subtypes (rods, S-cones and M-cones) in developing mouse retina (31). The orphan nuclear receptor NR2E3 is a direct downstream target of NRL and contributes to stabilize rod fate by suppressing the transcriptional program of S-cones (32–35). Mutations in NRL and NR2E3 are associated with human retinopathies and enhanced S-cone syndrome (36–39).
The differentiation and maturation of photoreceptors progresses through a long temporal window during mammalian development. In mice, photoreceptor lineage is established as early as embryonic day (E)12; however, almost all cones are produced prenatally whereas a vast majority of rods are generated after birth between postnatal day (P)0 and P2 (40,41). For both cones and rods, morphogenic differentiation (including outer segment and synapse formation) and functional maturation begins at or after P6 and is complete only around P21 (42–44). NRL controls the expression of a majority of rod-expressed genes as indicated by RNA-seq analysis of Nrl−/− retina (45) and functions synergistically with other regulatory factors, with CRX being a key NRL interactor (46) and modulator of photoreceptor-specific genes (27,47–49). Temporal expression of rhodopsin and other rod genes involved in cilia and presynapse formation is concordant with the rod developmental program; nevertheless, our understanding of how NRL regulates dynamics of gene expression pattern in rods, with a major transition in transcriptome landscape between P6 and P10 (45) is less than clear. Integrated analysis of NRL-ChIP-seq data from adult mouse retina (50) with the transcriptome profiles of Nrl−/− photoreceptors suggested a framework of NRL-directed regulatory network that included four secondary hubs (BHLHE41, ESRRB, FOS and NR2E3) (45).
We noted that the expression of NRL increases dramatically during rod maturation (at and after P10) and predicted that targets of NRL in the early stages of rod development might modulate its activity within specific temporal windows. To test this hypothesis, we performed Cleavage Under Targets and Release Using Nuclease (CUT&RUN) analysis (51,52) to map genomic occupancy of NRL at four stages in mouse retinal development and integrated NRL-binding sites with corresponding gene expression profile in rods (45). Interestingly, we identified c-Jun, a prototype bZIP transcription factor of AP-1 (activator protein 1) complex involved in the control of cell proliferation and apoptosis (53,54), as a direct transcriptional target of NRL in post-mitotic rods. Given that the purified c-Jun and NRL proteins can heterodimerize in vitro (55), we wanted to evaluate its possible role in regulating NRL-mediated transcriptional network. Co-immunoprecipitation and shRNA-mediated knockdown in mouse retina in vivo demonstrated that c-Jun cooperates with NRL to prime the rod transcriptional program during the early stages of differentiation. These studies suggest a previously unrecognized role of broadly expressed bZIP proteins, especially of the AP-1 complex, in modulating transcriptional regulation in cooperation with NRL during photoreceptor development.
Results
Genomic occupancy of NRL during rod photoreceptor development
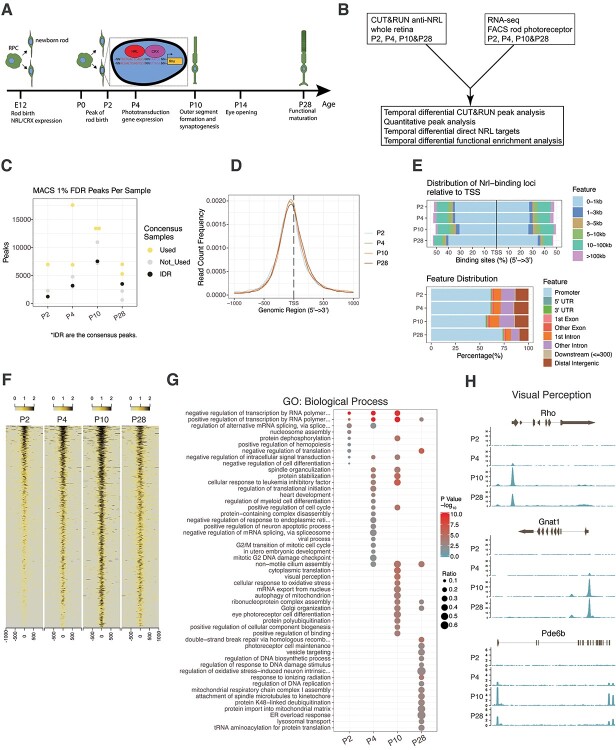
To expand on an early study of NRL occupancy in adult mouse retinas by ChIP-seq (50), we used CUT&RUN, which can overcome the issues related to low NRL protein in developing mouse retina because of the higher sensitivity of the method (51). CUT&RUN analysis was performed with a custom antibody that specifically recognizes NRL protein with multiple isoforms (Supplementary Material, Fig. S1A and B) using mouse retinas at P2, P4, P10 and P28, the time points corresponding to key events in rod development (Fig. 1A and B) (n = 4 for all time points, except P2 where replicate 2 was eliminated from analysis). Supplementary Material, Figure S2A shows the workflow for determining consensus NRL occupancy peaks. We first compared the P28 CUT&RUN data with our previously reported adult retina NRL ChIP-seq data performed with a different antibody and analytical method (50). Irreproducible Discovery Rate (IDR) analysis identified 3494 consensus NRL CUT&RUN peaks (false discovery rate [FDR] ≤ 0.01) in comparison to 2786 NRL ChIP-seq peaks, with 825 common peaks between the two methods (Supplementary Material, Fig. S2B). The use of replicates for analysis and employment of IDR for robust determination of consensus peaks eliminated many MACS-called peaks that were identified in only one replicate (Fig. 1C). CUT&RUN analysis also displayed a greater robustness over ChIP-seq in uncovering genome-wide NRL occupancy (Supplementary Material, Fig. S2C).
Figure 1.
Experimental design and NRL CUT&RUN quality control. (A) Timeline indicating major events in the development of the rod photoreceptor. Insert highlights consensus binding sequences for NRL and CRX in the promoter of rhodopsin (Rho). (B) Experimental paradigm and overview of integrated analysis of CUT&RUN data with RNA-seq data. (C) The number of MACS peaks passing a 1% FDR for each replicate per timepoint. Timepoint consensus peak number determined by IDR analysis are indicated in black. Biological replicates used for determining consensus peaks in IDR analysis are indicated in yellow, whereas replicates not used in IDR are indicated in gray. Further explanation of the IDR process is provided in the Methods section. (D) Enrichment of NRL-bound peak loci within 1000 bp flanking of gene TSS. (E) Upper panel: Percentage of NRL bound IDR peak loci relative to distance from TSS. Lower panel: Percentage of NRL bound IDR peak loci to gene feature distribution. (F) NRL bound loci signal enrichment per timepoint for all identified IDR peaks. (G) Functional gene enrichment of NRL-bound loci at each timepoint for gene sets in GO Biological Process. Top 20 processes per timepoint shown. (H) NRL bound reads in the near genomic region (±2 kb) at each timepoint for selected genes of the visual perception pathway enriched in (G).
We next analyzed the genomic occupancy of NRL during development and noted that the number of NRL peaks increased between P2 and P10 but then declined at P28 (1235, 3188, 7542 and 3493 peaks at P2, P4, P10 and P28, respectively) (Fig. 1C). Mapping of the genomic distribution of these peaks revealed that most peaks were located within 1 kb of gene TSS, and the proportion of NRL peaks within this region increased in the mature rods (62%, 62%, 57.5% and 74.4% peaks near TSS at P2, P4, P10 and P28, respectively) (Fig. 1D and E). A vast majority (>80%) of NRL CUT&RUN peaks were detected within a promoter or gene body, and over 50% of the peaks mapped to the promoter region at all developmental time points with a notable increase after maturation (Fig. 1E). Heatmap visualization presented the developmental dynamics of NRL genomic occupancy, showing the greatest robustness in differentiating rod photoreceptors at P10 (Fig. 1F). These results are consistent with a major change observed in rod transcriptome between P6 and P10 (45) and suggest potential rewiring of NRL-directed gene regulatory network during the transition from early state to the maturation of rod photoreceptors.
To explore whether the dynamics of NRL binding correlates with rod differentiation, we annotated the genes near NRL peaks and identified 1155, 2842, 5816 and 3207 genes at P2, P4, P10 and P28, respectively. The decrease in the gene number at P28 was in line with that of the peak number and potentially reflected a reduced size of NRL targetome in mature rods. Functional gene enrichment analysis using Gene Ontology (GO) uncovered a stepwise enrichment of new Biological Process terms of NRL-bound gene sets with the progression of rod development (Fig. 1G, Supplementary Material, Table S1). Biological processes enriched at P2 included negative regulation of transcription and nucleosome assembly, which contained genes encoding histone acetyltransferases (e.g. Ep300), histone demethylases (e.g. Kdm5c) and subunit of Polycomb Repressive Complex 2 (e.g. Suz12) (Supplementary Material, Fig. S2D). At P4, we identified the enrichment of genes associated with proliferation, survival and rod morphogenesis (e.g. Bbs2, Dctn1, Ttc8) (Supplementary Material, Fig. S2E). Terms associated with functional differentiation of rods, including visual perception (e.g. Rho, Gnat1, Pde6b), showed enrichment at P10 (Fig. 1H), whereas mature rods at P28 uncovered NRL-binding to genes primarily involved in photoreceptor cell maintenance (e.g. Clcn3, Esrrb, Mdm1), regulation of mitochondrial function, as well as endoplasmic reticulum overload response (Supplementary Material, Fig. S2F). Thus, the developmental transition of the global binding profile of NRL correlates with the milestone events during rod photoreceptor differentiation.
We then examined whether the temporal change of NRL occupancy associates with that of chromatin architecture. We focused on three rod-specific genes (Rho, Gnat1, Pde6b) and examined NRL-binding dynamics with the transition of epigenetic landscape observed in the whole retina (56) (Supplementary Material, Fig. S3A-D). The qualitative and quantitative gain of NRL peaks (P2-P10) were associated with switching of chromatin architecture to a more active state, but the reduced NRL binding (P28) was not always associated with an altered chromatin state.
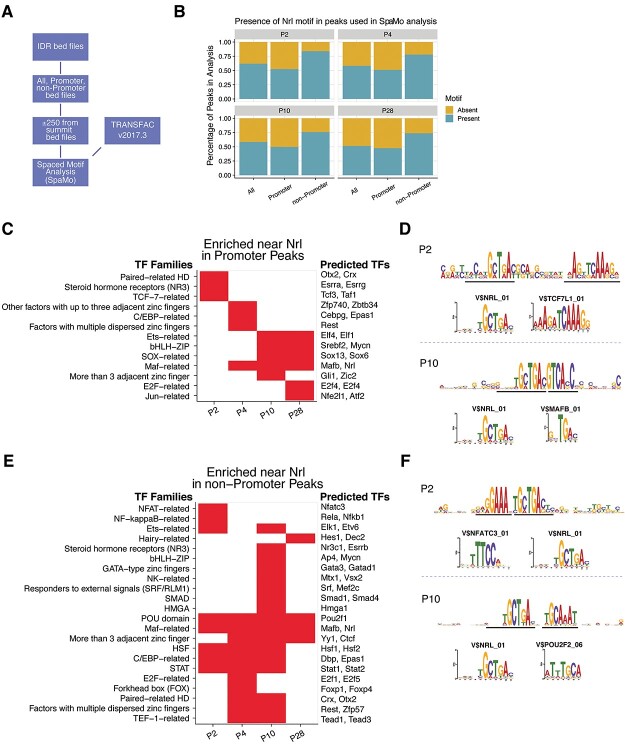
Enrichment of distinct TF binding motifs within NRL CUT&RUN peaks
We predicted that the presence of NRL-binding sites within the peaks and TF motif neighborhoods near NRL-binding sites change during development and might be distinctive between promoter-annotated (−1000 to +500 bases from the TSS) and non-promoter (potential enhancer) peaks. We employed the spaced motif (SpaMo) analysis algorithm in the MEME Suite of tools to predict the enrichment of TF binding motifs neighboring NRL-binding motifs (Fig. 2A). In addition to a generic motif enrichment, the SpaMo algorithm takes the distance between the primary and secondary motifs into account when determining significance. By examining how often SpaMo finds an NRL-binding motif in the peaks, we observed that the percentage of the peaks containing the motif did not change substantially with age, with 50.5% of promoter peaks and 78% of non-promoter peaks containing the NRL motif (Fig. 2B). We would not predict every peak to contain an NRL motif since DNA folding can bring disparate genomic regions together where NRL only needs to be a member of the complex. We then investigated the motif enrichment from the promoter peaks based on the TF family designation to reduce motif redundancy and observed temporal differential enrichment of distinct families (Fig. 2C, Supplementary Material, Table S2). Paired-related homeodomain (HD) factors (OTX2, CRX) and TCF-7-related (TCF7L1, TCF3) motifs were enriched only at P2. The OTX2/CRX-binding motif was detected 61 base pairs upstream (−strand) and 61 base pairs downstream (+strand) from the NRL motif. The proximity of the enrichment of the TCF7L1-binding motif was 10 base pairs from the NRL motif (Fig. 2D). The TCF/LEF (TCF3 and TAF1) family proteins are active in retinal progenitor cells leading to several cell types, including cone photoreceptors, in a β-catenin/Wnt independent manner (57). The MAF-related factors (such as MAFB, NRL) showed enrichment from P4 through P28 with the MAFB motif being directly adjacent, but on the opposite strand, from the NRL motif, indicative of an NRL-homodimer or NRL-MAFB heterodimer. The non-promoter NRL peaks revealed a much different motif enrichment than the promoter peaks. Several TF families were observed only at P2, including NFAT-related (NFATC3), NF-kappa-B-related, and Ets-related factors (Fig. 2E). NFAT-related motifs showed the most significant enrichment at P2. NFATs are known to cooperatively bind with AP-1 (Fos/Jun) heterodimers in immune cells to form composite NFAT/AP-1 sites in regulatory regions of inducible genes important to immune cells (58,59). The NFAT motif is present directly adjacent to NRL motifs (Fig. 2F). Notably, of the five members of the NFAT family, only two are expressed in rod photoreceptors, NFATC3 and NFAT5; however, NFAT5 does not form complexes with AP-1 proteins. Paired-related HD (OTX2/CRX) were enriched during the P4/P10 transition in rod development (Fig 2E). The motifs for the two important factors for rod maturation, ESRRB and MEF2C, were enriched only at the P10 timepoint. Interestingly, ESRRB and MEF2C motifs are also enriched in rod-specific open chromatin sites (60), indicating their potential role as members of rod-specific regulatory hubs. The POU domain (POU2 subfamily) and Maf-related factors were enriched at every timepoint, and POU2 motifs were evident in two base pairs downstream of the NRL motif (Fig. 2F). POU2F1 and POU2F2 have been shown to promote cone cell genesis by repressing Nrl expression (61).
Figure 2.
Temporal transcription factor spaced-motif enrichment. (A) Spaced motif (SpaMo) TF enrichment analysis process overview. NRL motif was used as the primary motif, and all other TRANSFAC vertebrate motifs were considered secondary motifs. (B) Presence of NRL-binding motif in consensus peaks identified in SpaMo analysis. All peaks and peaks annotated as promoter or non-promoter were analyzed at each timepoint. The presence and absence of NRL motif is presented as a percentage of peaks investigated. (C, E) Secondary TF motifs enriched within 150 bases flanking the NRL motif in promoter and non-promoter peaks, respectively. Motif redundancy reduction was achieved by presenting results at the TF family classification level. Individual genes listed on the right of the heatmap are examples of family members expressed in rod photoreceptors. The first gene in the list is the highest-scoring motif in the family, whereas the other gene/s listed are potential candidates based on their ability to bind the same motif and their expression level in the rod photoreceptors. (D, F) Examples of NRL and secondary motif enrichments in promoter and non-promoter peaks, respectively. Enrichment sequence logos of the SpaMo results along with the Transfac motif sequence logos for the highest scoring secondary motifs. Black lines indicate the motif-binding site of enrichment for the primary and secondary motifs. Transfac V$TCF7l1_01 and V$NFATC3_01 motifs are presented from the P2 results (upper panels), whereas V$MAFB_01 and V$POU2F2_06 are shown from the P10 results (lower panels).
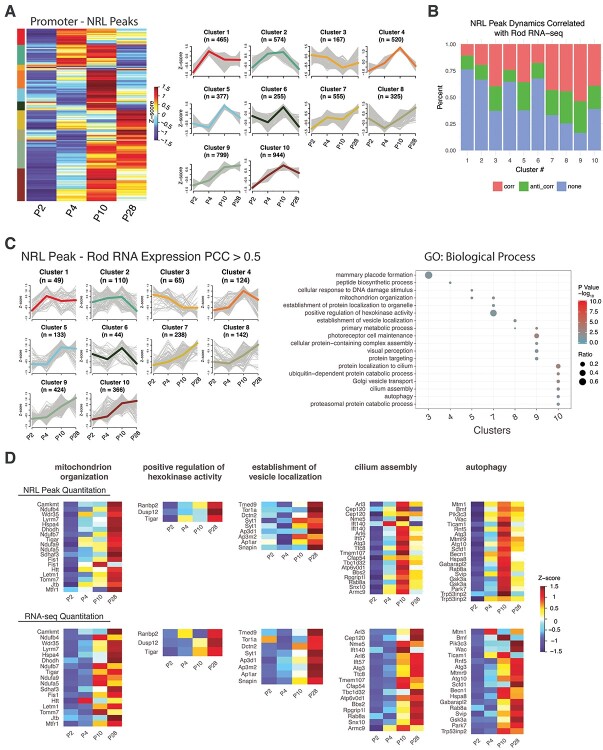
Association of temporal dynamics of NRL occupancy with gene expression
Next, we investigated whether and how the binding of NRL correlates with the transcription levels of the target gene and performed quantification of the peaks within promoters across all time points. Supervised hierarchical clustering identified 10 patterns of temporal dynamics (Fig. 3A, Supplementary Material, Table S3). For each cluster, we correlated the peak intensity changes with that of the expression of corresponding genes in rods (45). These clusters contained a varying percentage of peaks whose temporal dynamics of intensity either correlated (Pearson correlation coefficient [PCC] > 0.5) or anti-correlated (PCC < −0.5) with the gene expression pattern in developing rods (Fig. 3B). GO term enrichment of correlated genes in each cluster revealed distinct biological processes (Fig. 3C, Supplementary Material, Table S4). Specifically, we noted the enrichment of genes involved in mitochondrion organization (e.g. Camkmt, Ndufb4, Wdr35, Lyrm7) and positive regulation of hexokinase activity (e.g. Ranbp2, Dusp12, Tigar) from Cluster 7 (Fig. 3C, D). The continuous upregulation of NRL binding and their expression across developmental time points are consistent with an increased demand of mitochondria-associated metabolic functions during rod maturation. Cluster 8 included genes responsible for establishment of vesical localization (e.g. Tmed9, Tor1a, Dctn2, Syt1) (Fig. 3C, D). The turning point of the dynamics at P4 could be interpreted by the vesicle transport activities during rod mitosis and during ciliogenesis (62,63), which are respectively terminated and initiated after P4. Signature biological processes associated with photoreceptors, such as visual perception, cilium assembly (e.g. Arl3, Cep120, Nme5, Ift140) and autophagy (e.g. Mtm1, Rnf5, Hspa8, Gabarapl2), are enriched in Clusters 9 and 10 (Fig. 3C, D). The patterns of NRL peak and gene expression for these two clusters suggested a reduced requirement of NRL in sustaining their high-level expression in mature rods. We also performed the GO term analysis for anti-correlated genes for each cluster (Supplementary Material, Fig. S4A, Supplementary Material, Table S5). Genes involved in chromatin organization, encoding components of histone-modifying complexes (e.g. Cxxc1, Cbx8, Ing5, Hcfc1, Trrap, Brpf3), are enriched in Cluster 1 (Supplementary Material, Fig. S4B, Supplementary Material, Table S5), whereas mitotic processes are detected within Clusters 5 and 10.
Figure 3.
Dynamic NRL peak quantitation analysis. (A) Promoter peak quantitation at each timepoint was hierarchically clustered and dendrogram tree cut to yield 10 clusters. Per peak Z-scores were used in the clustering and are shown in heatmap (left panel) and parallel plot (right panel) showing all peak Z-scores with the mean Z-score highlighted. The number of peaks per cluster is indicated in the parallel plot (n). (B) PCC was calculated between promoter peak Z-scores and the corresponding gene-level RNA-seq expression Z-scores in rod photoreceptors (Kim et al. [45]). Percentages of correlated (PCC > 0.5, pink), anti-correlated (PCC < −0.5, green) and no correlation (0.5 > PCC > −0.5, blue) scores are shown for each peak cluster from panel A. (C) Left panel: Parallel plot of cluster-wise rod photoreceptor RNA-seq expression Z-scores from genes found correlated (PCC > 0.5) with peak quantitation Z-scores from B. Highlighted Z-score is mean of cluster Z-scores. Right panel: Functional gene enrichment (GO: Biological Process gene set) of genes in clusters from left panel. (D) Heatmaps of NRL peak Z-scores (top row) or rod RNA-seq expression Z-scores (bottom row) for genes in selected pathways found in panel C.
Direct targets of NRL identified by integrating NRL occupancy with gene expression
To identify developmental stage-specific transcriptional targets of NRL, we integrated the CUT&RUN data with the genes that are differentially expressed (DE) between wild-type rods and Nrl−/− S-cone-like photoreceptors at corresponding time points (45) (Fig. 4A, Supplementary Material, Tables S6,S7). As predicted, NRL occupancy contributed to both transcriptional activation and repression (Fig. 4B). GO Biological Process analysis of direct NRL-activated targets at P2 revealed the enrichment of genes involved in the modification of postsynaptic actin cytoskeleton (e.g. Myh10 and Tiam1) (Fig. 4C, Supplementary Material, Fig. S5A, and Supplementary Material, Table S6), and those at P4 belonged to retinal development in camera-type eyes (e.g. Gnb1, Nr2e3) (Fig. 4C, Supplementary Material, Table S6). At P10, we detected activation of NRL-target genes responsible for phototransduction, translation (e.g. Rpl10a, Rplp1, Rps2) as well as ATP synthesis (e.g. Coq7, Cycs, Cox8a, Ndufv1) (Fig. 4C, Supplementary Material, Fig. S5B, Supplementary Material, Table S6). Genes linked to metabolic process (e.g. Calr, Maz, Derl2, Rnf5) and clathrin coat assembly (e.g. Ap1b1, Epn1, Cltc, Hip1) are enriched in mature rods at P28 (Fig. 4C and Supplementary Material, Fig. S5C, Supplementary Material, Table S6). In parallel, GO Cellular Component analysis of direct activated targets of NRL revealed AP-1 transcription factor complex at P2 (e.g. Jun, Fos). Photoreceptor outer segment genes are included among NRL targets at P4 (e.g. Bbst, Rom1, Rp1), whereas P10 targets of NRL showing induction of gene expression corresponded to dendrite (e.g. Abhd12, Actn1), axon, pre-synapse and mitochondria. At P28, we observed an enrichment of NRL-activated target genes that are part of cellular metabolism and transport (Fig. 4D, Supplementary Material, Fig. S5D, Supplementary Material, Table S6). GO enrichment analysis of NRL-repressed genes could be linked to separate biological processes in developing and mature rod photoreceptors (Fig. 4E and Supplementary Material, Table S7). Curiously, genes involved in circadian regulation of translation (e.g. Hnrnpd, Per1, Rbm4) are repressed by NRL at P28.
Figure 4.
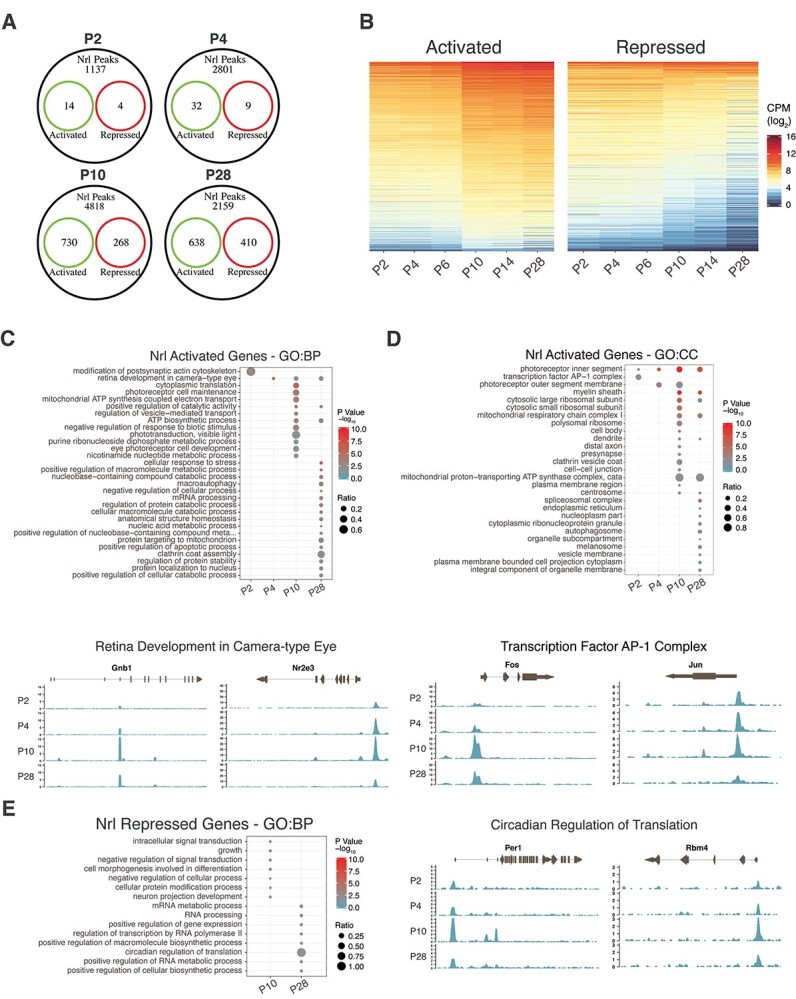
Association of NRL peaks with the expression of genes potentially activated or repressed by NRL. Genes activated or repressed by NRL were defined as genes whose expression was significantly differentially expressed in Nrl−/− rods at each timepoint investigated by CUT&RUN. (A) Overlap of annotated NRL IDR peaks with genes differentially expressed in rods at each timepoint. Activated (green): genes downregulated in Nrl−/−, Repressed (red): genes upregulated in Nrl−/−. (B) Expression dynamics with time of genes from wild-type rods having associated NRL bound loci in potentially activated (left panel) or repressed (right panel) genes found in (A). CPM: counts per million reads. (C) Upper panel: Functional gene enrichment (GO Biological Process gene set) at each timepoint of potentially activated genes. Lower panel: NRL bound reads in the near genomic region (± 2 kb) at each timepoint for selected genes of the Retinal Development in Camera-type Eye pathway. (D) Top panel: Functional gene enrichment (GO Cellular Component gene set) at each timepoint of potentially activated genes. Bottom panel: NRL bound reads in the near genomic region (± 2 kb) at each timepoint for selected genes of the Transcription Factor AP-1 Complex gene set. (E) Left panel: Functional gene enrichment (GO Biological Process gene set) at each timepoint of potentially repressed genes. Right panel: NRL bound reads in the near genomic region (± 2 kb) at each timepoint for selected genes of the Circadian Regulation of Translation pathway.
C-Jun as a direct transcriptional target of NRL in rods
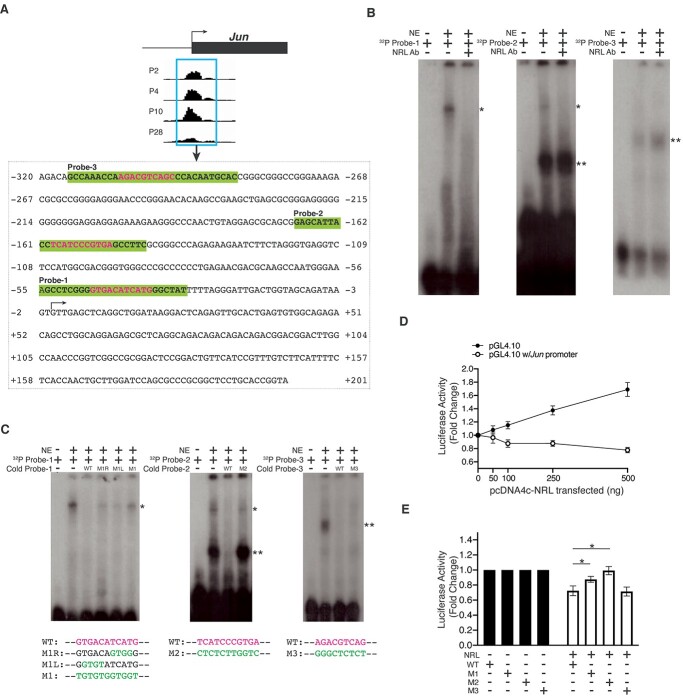
To identify NRL targets that could modulate NRL function, especially in newborn and developing rods, we centered on genes encoding transcription regulatory factors (Supplementary Material, Tables S6 and S7) and discovered the binding of NRL to AP-1 complex genes c-Jun and c-Fos (Fig. 4D). Given that c-Fos does not bind DNA directly or homodimerize (54), we focused on the distinctive characteristics of c-Jun. The expression of c-Jun in post-mitotic rods decreased by almost 10-fold (from P2 to P28) during differentiation, whereas NRL transcripts increased to a similar extent in the corresponding period (45). Nonetheless, NRL occupied the promoter of c-Jun at all time points (Fig. 4D).
In silico examination of the NRL peak region (−320 to +201) within the c-Jun promoter revealed three putative binding motifs (Fig. 5A), and we, therefore, examined the direct binding of NRL to these motifs by electrophoretic mobility shift assay (EMSA). Positive control using an oligonucleotide probe derived from the mouse Rhodopsin promoter that encompasses a validated NRL-binding site demonstrated the binding of nuclear proteins from adult mouse retina (64,65), and this binding could be abolished by pre-incubating the nuclear extracts with another NRL antibody previously used for ChIP-seq (66) but not our custom-generated antibody (Supplementary Material, Fig. S6). The addition of an excess of unlabeled probe with wild-type but not mutant NRL-binding motif eliminated the shifted band. With respect to c-Jun promoter, nuclear proteins bound to the three oligonucleotide probes including the putative NRL-binding site (Fig. 5B). Pre-incubation of nuclear extracts with the NRL antibody abolished the shifted band for Probe-1 and Probe-2, but not Probe-3. We then altered specific nucleotides within the putative NRL-binding sequence of each probe by site-directed mutagenesis (Fig. 5C). The shifted bands associated with all three probes could be abrogated by the addition of an excess of corresponding wild-type unlabeled probe, whereas unlabeled mutant Probe-1 and Probe-2 (M1 and M2, respectively) failed to compete with the respective 32P-labeled probes. The mutant Probe-3 oligonucleotide had no effect on the shifted band in EMSA. These results validate the binding of NRL to indicated sequence elements within Probe-1 and Probe-2.
Figure 5.
Direct regulation of Jun transcription by NRL in vitro. (A) Jun proximal promoter sequence encompassing the NRL peak, with the probe sequences highlighted in bold and shaded in green and the putative NRL motifs in magenta. The TSS is indicated with an arrow. (B and C) NRL directly binds to Jun proximal promoter in vitro. 32P-labeled probes containing putative NRL-binding sites were incubated with nuclear extract (NE) of P28 mouse retina. The shift due to protein-DNA binding (indicated by a single asterisk) was eliminated by the addition of a specific antibody against NRL into the nuclear extract prior to incubation with the probes (B), or by the addition of unlabeled probes with wild-type (magenta) but not mutated sequences (green), in 300x molar excess (C). Double asterisks denote a non-specific shift. (D) NRL represses Jun promoter activity in a dose-dependent manner in vitro. Fold change in relative luciferase activity driven by the Jun promoter in the presence of NRL of increasing concentration as indicated. The relative luciferase activity with the absence of the Jun promoter at each concentration of NRL serves as a reference. Each condition includes three biological replicates and error bars represent the standard error of the mean. (E) NRL-mediated repression of Jun promoter activity requires the functional NRL-binding sites. Fold change in relative luciferase activity driven by the Jun promoter with wild-type or mutated NRL-binding site, in the absence or presence of NRL. Each condition includes five biological replicates and error bars represent the standard error of the mean; *indicate P < 0.05 by Student’s t-test.
We then cloned the 521-bp NRL peak region from c-Jun promoter upstream of a luciferase reporter gene in pGL4.10 vector and co-transfected the c-Jun-Luc with NRL-expression construct in HEK293 cells. We observed dose-dependent activation of control pGL4.10 vector by NRL potentially due to the presence of cryptic regulatory elements as reported (67–71). Nonetheless, Jun promoter activity was repressed by NRL in a dose-dependent manner (Fig. 5D). Mutagenesis of NRL-binding sites in M1 and M2 mutants, but not M3, compromised the NRL-dependent repression (Fig. 5E). Taken together, our EMSA and luciferase activity assays demonstrate that NRL binds to two distinct sequence elements in c-Jun promoter and can repress its transcriptional activity in vitro.
Interaction of NRL with c-Jun in HEK293 cells and in developing retina
Given that purified NRL and c-Jun polypeptides can heterodimerize and bind to AP-1 sites (55), we wanted to further examine their interaction. NRL and c-Jun expression constructs were first co-transfected in HEK293 cells. Immunoprecipitation of transfected cell extracts with anti-c-Jun antibody detected NRL by immunoblot analysis (Fig. 6A, left panel), and we could co-immunoprecipitate c-Jun with the antibody against NRL (Fig. 6A, right panel). Next, we performed co-immunoprecipitation using mouse retinas. Whole retina immunoblot analysis showed NRL and c-Jun expression both during development and in adult tissues (Fig. 6B, upper panels); however, NRL could be co-immunoprecipitated with c-Jun from P4 and to a lesser extent from P10 retinas, but not from mature P28 retina (Fig. 6B, lower left panel). Reciprocal co-immunoprecipitation also demonstrated c-Jun association with NRL only at P4 but not at P10 or P28 (Fig. 6B, lower right panel). These results suggest a developmental stage- and/or context-specific functional interaction of c-Jun and NRL.
Figure 6.
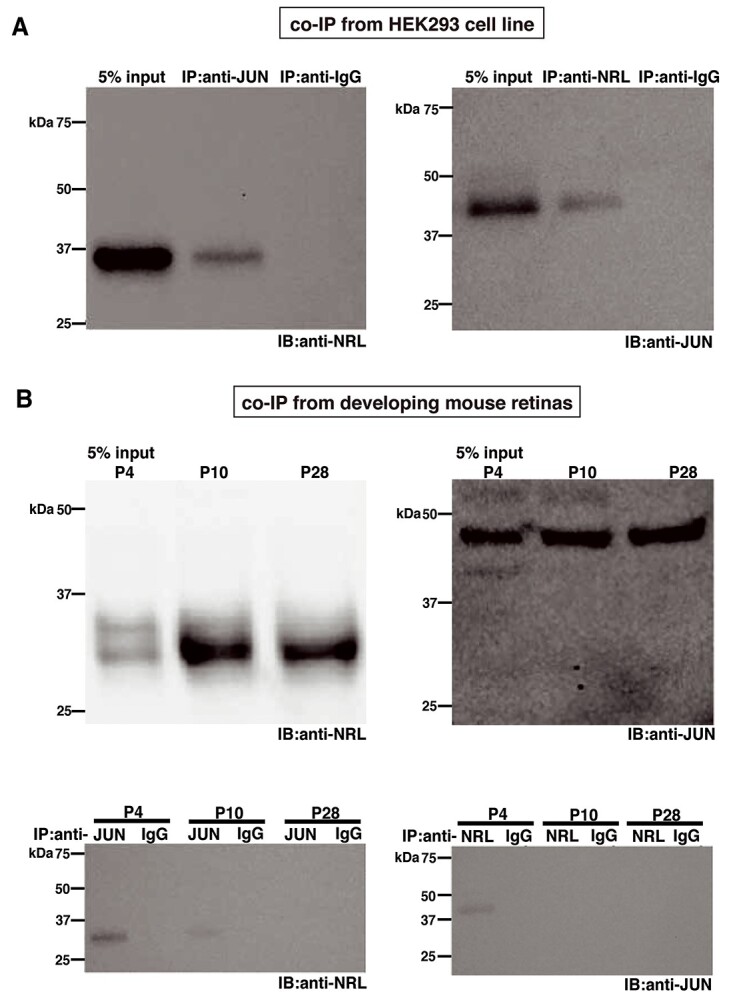
Co-immunoprecipitation of NRL and c-Jun from HEK293 cells and developing mouse retina. (A) Immunoblots (IB) with anti-NRL (left) or anti-JUN (right) antibody of 5% input or eluate of immunoprecipitation (IP) with anti-JUN (left) or anti-NRL (right) antibody or non-specific IgG from HEK293 cells co-transfected with NRL- and JUN-expressing vectors. (B) Upper panels: immunoblots with anti-NRL (left) or anti-JUN (right) antibody from 5% input of mouse retinas of postnatal (P) days as indicated. Lower panels: immunoblots with anti-NRL (left) or anti-JUN (right) antibody of eluate of immunoprecipitation with anti-JUN (left) or anti-NRL (right) antibody or non-specific IgG, from mouse retinas of postnatal days as indicated. Note that our custom-generated NRL antibody was used for co-immunoprecipitation.
Modulation of NRL-directed transcriptional program by c-Jun in developing rods
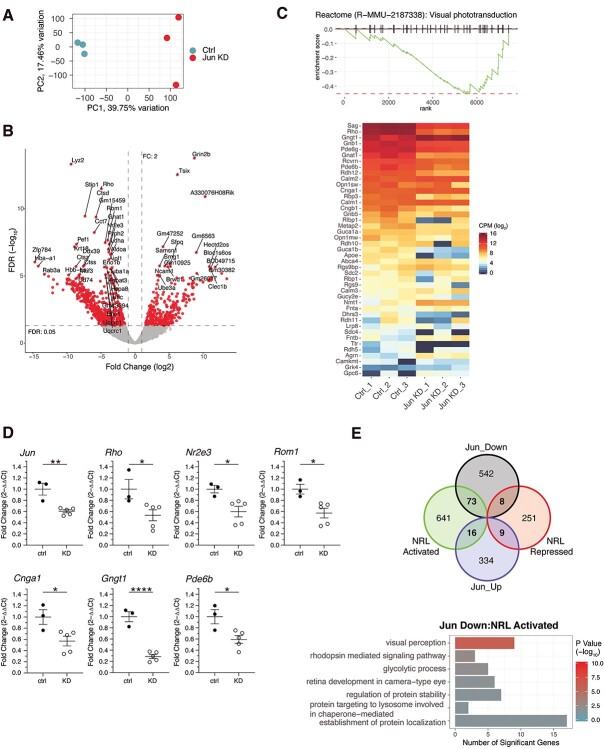
Based on interaction studies, we predicted that the activity of NRL could be modulated by c-Jun during the early postnatal stages of rod development when NRL levels are relatively low. To test this hypothesis, we performed shRNA-mediated knockdown of c-Jun in developing mouse retina. The construct encoding control shRNA or shRNA against mouse c-Jun was co-electroporated with Nrlp-EGFP plasmid (41) into the subretinal space of P0 mice, as described (72). We then sorted the rod photoreceptors from P10 control and c-Jun shRNA retinas based on GFP expression (0.4% and 0.3% GFP+ rods from control and shRNA group, respectively) and performed RNA-seq. Principal component analysis indicated that the largest variance (39.75%) in the data showed the separation of control samples from the knockdown (Fig. 7A). c-Jun knockdown resulted in upregulation of 359 and downregulation of 623 genes (FC cutoff of 2; 5% FDR) (Fig. 7B, Supplementary Material, Table S8). Gene set enrichment analysis revealed downregulation of many genes crucial for rod morphogenesis and phototransduction (Fig. 7C) and RT-qPCR analysis confirmed the changes in the mRNA level of these genes (Fig. 7D). These data provided evidence in support of c-Jun modulating NRL-mediated gene transcription. To further study the extent to which c-Jun contributes to the expression of NRL-regulated genes, we compared the DE genes identified by c-Jun knockdown with the P10 direct NRL targets. Of the 623 downregulated genes, 81 genes were ascertained as direct targets of NRL (73 activated and 8 repressed) (Fig. 7E). In parallel, 25 of the 359 upregulated genes were direct NRL targets (16 activated and 9 repressed). GO term analysis showed that genes co-activated by NRL and c-Jun belonged to visual perception, glycolysis and establishment of protein localization (Fig. 7E, Supplementary Material, Table S9).
Figure 7.
Differential gene expression in Jun knock-down retinas. (A) Principal component analysis of expressed gene used in the analysis. Samples marked Ctrl (blue) are scrambled shRNA probe controls, whereas Jun KD samples (red) are Jun shRNA knock down. (B) Volcano plot of differential expression results. Red dots correspond to genes considered significantly differentially expressed with a fold change greater than two and an FDR value lower than 5%. FDR: false discovery rate (C) Upper panel: Gene set enrichment analysis (GSEA) of the Visual Phototransduction (Reactome pathway) using fold change as the ranking value. Lower panel: Gene expression heatmap of the genes composed of the Visual phototransduction pathway. CPM: counts per million reads (D) RT-qPCR validation of selected genes from panel C. ctrl: scrambled probe shRNA, KD: Jun shRNA. Error bars represent standard error of the mean; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Student’s t-test. (E) Upper panel: Overlap of differentially expressed genes in Jun KD and genes with NRL bound loci and either activated or repressed at P10 in rods (Fig. 3A). Lower panel: Gene set enrichment analysis (GO Biological Process gene set) of genes (73 genes) that were downregulated in Jun KD, having an associated NRL bound loci and potentially activated by NRL based on rod photoreceptor gene expression studies.
Discussion
Transcription factors integrate the cellular response to multiple signals and control spatio-temporally precise expression of genes during the development of divergent cell types in metazoans. Combinatorial synergistic or antagonistic interactions among TFs, together with epigenomic modifications, influence DNA-binding characteristics and the formation of a productive transcription initiation complex at gene promoters. Relocation of TFs to different cis-regulatory elements is a common mechanism that drives cellular differentiation and organogenesis, as exemplified by SOX2 during the transition of embryonic stem cells to neuronal differentiation (73), Runx2 among major stages of osteoblastogenesis (74) and MADS-domain TFs during Arabidopsis flower development (75). Here we demonstrate a dynamic and developmental stage-specific association of Maf-family bZIP protein NRL to distinct cis-regulatory regions by mapping the genome-wide occupancy in the retina using a sensitive and precise CUT&RUN method. Detection of multiple NRL isoforms by our custom antibody consistently across the retinal developmental period demonstrated that the capture of temporal distinct binding regions was the result of a shift in NRL localization rather than a change of antibody reactivity. The question then is what is the driving force for the temporal change of NRL occupancy? Non-specific binding of TF can facilitate the search of specific sites by 3D diffusion or local motions (76,77); therefore, NRL might navigate the low specificity regions before being stabilized at high specificity sites upon rod maturation. However, our analysis reveals a comparable percentage of peaks harboring the NRL motif across the time points and does not support the hypothesis of increased binding specificity. Alternatively, our data show an association between the transition of NRL binding and that of chromatin states (56); however, we cannot differentiate whether the qualitative and quantitative changes in NRL binding are the cause or consequence of the dynamic epigenetic landscape.
A TF can bind to numerous genomic sequences in a non-specific manner; however, selectivity and kinetics of binding (such as residence time) at target cis-regulatory elements vary depending upon TF concentration and interaction with other proteins, among others (78,79). Low- and high-affinity binding sequences can exhibit distinct TF dynamics, and therefore quantitative differences in local TF concentration within the nucleus can impact the stoichiometry of regulatory proteins at gene promoters influencing the transcription rate and expression levels (80). For example, different concentrations of NF-κB or MYC lead to distinct promoter occupancies and transcriptional profiles (81,82). NRL is critical for rod cell fate determination, yet its expression increases dramatically during differentiation and is then maintained at high levels. Our CUT&RUN results demonstrate common as well as distinct targets of NRL at different stages of photoreceptor development, suggesting gradual acquisition of biological components that determine rod morphology and function. As evident from our study, in addition to rod-specific genes, NRL controls the expression of many widely expressed genes including those involved in protein translation, mitochondrial ATP synthesis and vesicle-mediated transport. How and why is the expression of such genes controlled by a cell type specific TF, such as NRL? We suggest that high metabolic activities of photoreceptors require multiple levels of control mechanisms and coordination of gene expression for maintaining energy homeostasis and functional requirements. Alternative promoter usage can provide one possible mechanism for exerting a specific control, as in the case of Frmpd1 (83). We also note that the expression of many genes bound by NRL in CUT&RUN assays is not impacted by the loss of Nrl, reflecting low-affinity NRL binding, unfavorable genome topology in photoreceptors and/or functional redundancy among transcriptional regulators (84). Non-functional binding of TFs may arise from the rewiring of transcriptional networks and enhance adaption to environmental perturbations by bringing about evolutionary diversity (85).
NRL is a photoreceptor-specific bZIP transcription factor that belongs to Maf-subfamily within the AP-1 complex superfamily (86). AP-1 functions as a dimer, comprising different combinations of JUN, FOS, MAF and ATF subfamily of bZIP proteins that can form homo- or heterodimers and control divergent biological processes from cell differentiation to death (54). JUN proteins can bind DNA as homo- and heterodimers, with the latter being strong transcriptional regulators. Curiously, NRL bound to the promoter sequences of c-Jun, which is a positive regulator of cell growth and proliferation. EMSA validated the binding of NRL to two distinct sites in the c-Jun promoter. The NRL motif within Probe-2 in our assay had an identical sequence as the AP-1 site within the human C-JUN promoter (−190 to −183), which bound to heterodimers of c-Jun and ATF-2 in vitro (87–89). Thus, NRL occupies the same AP-1 element in the c-Jun promoter in rod photoreceptors as a homo- or heterodimer. Co-transfection assays in HEK293 cells suggest that NRL as a homodimer likely suppresses c-Jun promoter activity. c-Jun is highly expressed in HEK293 cell line according to BioGPS Cell Line Expression Profiles and can autoactivate its own transcription (90); as such, the downregulated c-Jun promoter activity could be the result of both NRL-mediated repression and the reduced expression of endogenous c-Jun. These observations are consistent with the reciprocal and dramatic changes in expression of c-Jun and Nrl from P2 to P28 as rod maturation proceeds (45). We also note that c-Fos could not be immunoprecipitated from mouse retinal extracts with anti-NRL antibody (data not shown) despite their interaction in vitro (55), suggesting a specific and biologically relevant interaction of c-Jun and NRL interaction in early developing rod photoreceptors.
We were intrigued by the extensive differential occupancy of NRL during development and wondered whether changes in NRL concentration alone define promoter affinities and consequently expression levels. NRL collaborates with CRX, NR2E3 and other proteins to fine-tune rod-specific gene expression patterns (17,18). Our data show that c-Jun and NRL (both AP-1 superfamily bZIP proteins) can likely heterodimerize as shown by co-immunoprecipitation in HEK293 as well as from mouse retina extract. Reduced expression of many direct targets of NRL upon c-Jun knockdown in developing mouse retina in vivo provides strong evidence of cooperation between the two TFs in priming rod-specific gene expression. Hence, the kinetics of NRL binding is likely governed by local TF concentration as well as spatiotemporal availability of regulatory proteins that form a productive transcription complex at selective promoters to generate quantitatively distinct expression of genes. We propose that NRL binding to the c-Jun promoter is likely unrelated to NRL-c-Jun-mediated gene regulation. NRL homodimers could occupy c-Jun promoter at all developmental stages and repress its activity in a concentration-dependent manner. We suggest that c-Jun-NRL heterodimers are stronger activators of rod genes and needed to prime transcription until other transcriptional regulators are present at relevant concentration after P6, eventually leading to the major detectable shift in rod transcriptome (45). However, our study does not rule out the possibility that the reduced expression of NRL targets in c-Jun knockdown might be caused by NRL-independent mechanisms. For example, c-Jun is reported to regulate cellular stress and glucose metabolism (91–96), and both stress response and metabolic products can modulate gene transcription (97–100). One limitation of our current study is the lack of direct evidence showing the binding of c-Jun to NRL transcriptional targets, as our attempts to map the genome-wide occupancy of c-Jun in P4 or P10 flow-sorted rods by CUT&RUN were unsuccessful, with very few IDR peaks identified. Further optimization of CUT&RUN protocol or exploration of other approaches such as CUT&Tag (101) may be needed to elucidate the genomic binding landscape of c-Jun in rod photoreceptors. In addition, a comprehensive understanding of c-Jun’s role in rod differentiation would require the creation of more critical resources including a conditional knockout; nonetheless, our findings serve as a starting point for future research.
In conclusion, our study expands the NRL-directed transcriptional regulatory landscape that controls rod photoreceptor morphogenesis and functional maturation and proceeds over a period of three weeks during mouse retinal development. We hypothesize that combinatorial and synergistic (or at times antagonistic) actions of NRL with c-Jun and other widely expressed transcriptional regulatory proteins provide target-specific spatio-temporal as well as quantitative control over rod gene expression and permit integration of divergent signaling pathways.
Materials and Methods
Mice
All mice involved in the current studies were treated with protocols approved by the Institutional Animal Care and Use Committee of the National Eye Institute (ASP#650). C57BL/6 J mice were used for CUT&RUN, and CD-1 mice (Charles River, Wilmington, MA, USA) were used for in vivo electroporation experiments.
Antibodies
For CUT&RUN and immunoblot, a custom antibody generated as follows was used: a domain that encodes the mouse NRL amino acids 23–130 was expressed in Escherichia coli and used as the antigen to immunize rabbits and antibodies were purified from the rabbit serum using affinity column chromatography. For EMSA, a rabbit polyclonal anti-NRL (Cat#ab-137 193, Abcam, Cambridge, UK) was used. All other antibodies were purchased from commercial sources: mouse monoclonal anti-GAPDH (Cat#G8795, MilliporeSigma, Burlington, MA, USA), mouse monoclonal anti-JUN (Cat#sc-74 543, Santa Cruz, Dallas, TX, USA), anti-mouse and anti-rabbit light chain specific horseradish peroxidase conjugated anti-IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, USA).
Plasmid DNA constructs and mutagenesis
Promoter sequence of mouse Jun (521 base-pair) was amplified with Q5 High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) from C57BL/6 J mouse genomic DNA and was subcloned into pGL4.10[luc2] (Promega, Madison, WI, USA). Mutagenesis was performed with Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA). Mouse Jun cDNA sequence was amplified from mouse retinal cDNA library with the reagents as described above and was subcloned into pcDNA4c His/MaxC (Thermo Fisher Scientific, Rockford, IL, USA). Human NRL-expressing construct was described earlier (39). shRNA-mediated knockdown of mouse Jun was performed with shRNA-expressing vector (TRC Clone ID TRCN0000042695, MilliporeSigma, Burlington, MA, USA) and control pLKO.1-Scrambled vector (Addgene, Watertown, MA, USA). The effectiveness of knockdown of mouse Jun was tested in HEK293 cells. Nrlp-EGFP vector was adopted from (102). Primers are listed in Supplementary Material, Table S10.
In vivo electroporation
Neonatal mice at P0 were used for in vivo electroporation as previously described (72,102) with shRNA vectors (2.5 μg/μL, 0.4 μL) and the Nrlp-EGFP vector (1.5 μg/μL, 0.4 μL).
Cut&run
CUT&RUN protocol was adopted and modified from the previous description (51). In brief, retinas were dissected at P2, P4, P10 and P28 and cells were dissociated using dissociation solution (30 U/mL papain, HBSS pH 7.4, 1 mg/mL glucose, 10 mM Hepes, 100 U/mL DNase I, 5 μg/mL superoxide dismutase, 5 μg/mL catalase, 10 μg/mL D-alpha-tocopherol acetate, 1 mg/mL glucoscysteine-HCl, 5 μg/mL superoxide dismutase, 50 μg/mL gentamycin). Cells were pelleted at 200 g for 5 min, washed and re-suspended in Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl and 0.5 mM Spermidine) with Complete Protease Inhibitor tablet (MilliporeSigma, Burlington, MA, USA). Concanavalin A–coated beads (Polysciences, Warrington, PA, USA) were incubated with the cell suspension containing 250 000 cells for 10 min, washed to remove unbound cells and incubated with NRL antibody (1:200) in Antibody Buffer (20 mM EDTA and 0.02% digitonin in Wash Buffer) for overnight. The next day beads were washed with Wash Buffer containing 0.02% digitonin and incubated with 700 ng/mL pA-MNase for 1 h at 4°C. Beads were washed, re-suspended in 100 μL of Wash Buffer containing 0.02% digitonin and incubated with 2 mM CaCl2 at 0°C for 30 min. Cleavage reaction was quenched by adding 100 μL 2X Stop Buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.02% digitonin, 25 μL RNase A, 0.05 mg glycogen and 10 pg heterologous spike-in DNA). DNA fragments were released from insoluble nuclear chromatin by incubation at 37°C for 10 min and extracted by spin column (Qiagen, Hilden, Germany). Libraries were prepared with ThruPLEX DNA-seq kit (Takara Bio USA, Inc, Mountain View, USA).
EMSA
Probes were produced by annealing complementary oligonucleotides in duplex buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 50 mM NaCl), heated at 95°C for 2 min and cooled to room temperature over 60 min in thermocycler. Probes were labeled with γ32P ATP (30 pmol probe, 1X T4 polynucleotide kinase reaction buffer, 20 units T4 PNK [New England Biolabs, Ipswich, USA], 16.5 pmol γ32P ATP [PerkinElmer, Waltham, USA]) at 37°C for 30 min followed by heat inactivation at 65°C for 20 min. 32P labeled probes were purified with Illustra MicroSpin™ G-25 Columns (GE Healthcare, Chicago, USA) and radioactivity was measured. EMSA was performed in binding buffer (LightShift, Thermo Fisher Scientific, Rockford, IL, USA) with 5 mM MgCl2, 50 ng/μL Poly dI•dC, 100 000 dpm of 32P-labeled probe and 20 μg nuclear protein. Unlabeled wild-type and mutant probes (300 molar excess) were used for the competition assay. NRL-specific binding was tested by adding 1 μg NRL antibody to the nuclear extracts and incubating for 1 h prior to the addition of probes. Reactions were performed for 1 h at room temperature and resolved on a 6% DNA Retardation gel (Invitrogen, Carlsbad, USA) in TBE buffer (Thermo Fisher Scientific, Waltham, MA, USA). Gels were dried and exposed to radiographic film on an amplifying screen cassette overnight at −80°C.
Dual luciferase reporter assays
HEK293 cells were seeded in 48-well plates in Dulbecco’s modified Eagles’s medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) that contains 10% fetal calf serum. Transfection was performed with X-tremeGENE 9 DNA Transfection Reagent (MilliporeSigma, Burlington, MA, USA). Cells at 70–80% confluence were co-transfected with 2.5 ng CMV-Renilla, 100 ng wild-type or mutant mouse Jun promoter-driving firefly luciferase, and a varying amount of NRL-expressing construct as indicated. Empty pcDNA4C vector was used to adjust the total amount of DNA. Cells were harvested after 48 h and reporter assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly and Renilla luciferase activities were measured with a modulus microplate luminometer (Turner Biosystems, Sunnyvale, CA). All experiments were performed in biological triplicates and statistical significance was determined by Students’ t-test.
Co-immunoprecipitation
For co-immunoprecipitation from HEK293 cells, cells were seeded in 6-well plates and co-transfected with 500 ng human NRL and mouse c-Jun expressing vectors as previously described. For co-immunoprecipitation from developing mouse retinas, retinas were collected at the time points indicated. In both cases, protein extracts were prepared in IP buffer (40 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA and 0.2% NP-40) in the presence of Complete Protease Inhibitor tablet. Protein A (or G) Dynabeads (Thermo Fisher Scientific, Waltham, MA, USA) were incubated with 1 μg of the desired antibodies, washed and incubated with 500 μg protein extracts overnight at 4°C. Beads were then washed and bound protein complexes were eluted in Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA).
Immunoblot
Protein samples in Laemmli sample buffer were boiled at 95°C for 10 min and were resolved by SDS-PAGE and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked in 5% skim milk dissolved in 1x Tris-buffered saline (20 mM Tris, 150 mM NaCl) with 0.1% Tween-20 (TBST) for 1 hr, and incubated with the desired primary antibodies (1:1000) overnight at 4°C. The membranes were washed in 1x TBST and incubated with conjugated-HRP secondary antibody (1:5000) for 2 h at room temperature. The membranes were washed again in 1x TBST and subjected to visualization of protein bands by enhanced chemiluminescence plus (Thermo Fisher Scientific, Waltham, MA, USA).
RT-qPCR
EGFP positive electroporated retinal areas were excised under a fluorescence dissecting scope. Total RNA was extracted with TriPure Isolation Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and first-strand cDNA synthesis was performed with SuperScript II Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA). Quantitative PCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA) in multiple biological and technical replicates. Primers are listed in Supplementary Material, Table S10.
CUT&RUN sequencing data analysis
CUT&RUN libraries generated of quadruplicate biological replicates at each time point were paired-end sequenced to a length of 101 bases on a HiSeq 2500 System (Illumina, San Diego, CA, USA). Sequencing reads were trimmed using Trimmomatic v0.38 (103) for library adapters, 6-base tail cropping, end-wise PHRED 20 quality score, and requiring a minimum length of 25 bases for further analysis using the following settings: ILLUMINACLIP:(adapter):2:15:4:4:true TAILCROP:6 LEADING:20 TRAILING:20 MINLEN:25. Trimmed sequencing reads were aligned to a chimeric index using bowtie2 v2.3.5.1 (104) with the following settings: —end-to-end—dovetail -I 10 -X 700—very-sensitive—no-unal—no-mixed—no-discordant -q—phred33. A chimera bowtie2 index was created by concatenating genomic DNA fasta files of Mus musculus GRCm38 (http://sep2019.archive.ensembl.org/Mus_musculus/Info/Index), Saccharomyces cerevisiae S288C (https://www.ncbi.nlm.nih.gov/assembly/GCF_000146045.2) and E. coli ASM584v2 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000005845.2) prior to running the bowtie2 index function. Quality alignments were kept if MAPQ scores were greater than 20 using samtools v1.9 (105). Duplicate aligned reads were removed using picard v2.20.8 (https://broadinstitute.github.io/picard/) MarkDuplicates algorithm. Peaks were identified using MACS2 v2.2.5 (106). Called peaks overlapping ENCODE blacklist regions (107) were removed using Bedtools v2.29.0 (108). The IDR approach was used to determine consensus peaks at each timepoint using IDR v2.0.3 (https://doi.org/10.1214/11-AOAS466) with an IDR threshold of 0.05. IDR was performed using two replicates with consensus peaks passing the IDR threshold. For each timepoint, IDR was performed using all possible pair combinations of the replicates. The consensus peaks determined from the pair of replicates yielding the highest number of consensus peaks was used for further analysis. Annotation was assigned to each peak using the ‘annotatePeak’ function of ChipSeeker v1.22.1 package. Functional gene enrichment analysis throughout the manuscript was performed using gProfiler v0.7.0 (https://biit.cs.ut.ee/gprofiler/gost) using GO Biological Process gene sets.
Spaced motif enrichment analysis
Spaced motif analysis was performed with MEME Suite module SpaMo v5.4.1 (109) using transcription factor motifs from TRANSFAC v2017.3 (110). Motif V$NRL_01 was used as the primary motif whereas the vertebrate motifs were used as secondary motifs. Fasta files for input were generated using Bedtools from bed files containing promoter, non-promoter or all peaks at each timepoint. Bed files were modified so that every genomic region was 500 bases flanking the peak summit. Motif redundancy from the SpaMo results was performed at the transcription factor Family rank denomination using the classifications defined in TFClass (111).
Chromatin state mapping
Fastq files from the retina data, as reported (56), were processed through the ENCODE Data Coordinating Center Uniform Processing Pipelines (ENCODE 4) ChIP-seq pipelines. Output bam files were used to generate chromatin state maps using ChromHMM v1.22 (112) generating 16 chromatin states. Annotation of the chromatin state was based on descriptive titles of chromatin states in Gorkin et al. (2020) (113).
Temporal peak quantitation analysis
Temporal peak quantitation was performed in R v3.6.1 (https://www.r-project.org/) using the consensus peaks for each timepoint. Overlapping peaks in each consensus peak were merged using the ‘reduce’ function in IRanges v2.20.2 (114) package to generate a common peak list for quantitating peaks. Each sample bam file created in the analysis was used for quantifying the peaks using the ‘regionCounts’ function in the csaw v1.20.0 (115) package. Normalization factors for each sample were generated using ‘windowCounts’ function binning the reads into 10 000 base windows with a maximum fragment size of 400 bases. Z-score of the average normalized log counts-per-million value for each timepoint was used to cluster the change in temporal peak score into 10 clusters using Ward’s D2 hierarchical clustering method.
RNA-seq analysis
EGFP positive rods electroporated with control and Jun shRNA constructs were flow-sorted as previously described (45). Full length cDNA was generated using SMART-seq v4 Ultra Low Input RNA Kit (Takara, Kusatsu, Shiga, Japan) from 200 to 2000 cells per sample. Library generation from cDNA was prepared using Nextera XT DNA Library Kit (Illumina, San Diego, CA, USA) and paired-end sequenced to a length of 126 bases on a HiSeq-2500 System (Illumina, San Diego, CA, USA). Fastq files were generated from reads passing chastity filter and used for further analysis. Illumina adapter, polyA and polyT sequence trimming was performed with Trimmomatic v0.39. Transcript level quantitation was performed using Kallisto v0.44.0 (116) employing a merged transcript cDNA and ncRNA FASTA file downloaded from Ensembl v94 (http://oct2018.archive.ensembl.org/index.html). Gene level quantification performed in R (https://www.r-project.org/) was generated by summarization of the transcript level quantitation using tximport v1.14.0 (117) with the option ‘countFromAbundance = lengthScaledTPM’. The gene level count values were TMM normalized and differential expression was performed using the ‘exactTest’ function in edgeR v3.28.1 package (118).
Data Availability
All raw and processed data are available through NCBI’s Gene Expression Omnibus: GSE197421 (http://www/ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197421).
Supplementary Material
Acknowledgements
We are grateful to Jacob Nellissery, Rafael Villasmil and Linn Gieser for advice and assistance. This research utilized the high-performance computational capabilities of the Biowulf Linux cluster at National Institutes of Health (http://biowulf.nih.gov). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest statement. The authors declare no conflicts of interest with the contents of this article.
Contributor Information
Xulong Liang, Neurobiology, Neurodegeneration and Repair Laboratory, National Eye Institute, National Institutes of Health, 6 Center Drive, MSC0610, Bethesda, MD 20892, USA.
Matthew J Brooks, Neurobiology, Neurodegeneration and Repair Laboratory, National Eye Institute, National Institutes of Health, 6 Center Drive, MSC0610, Bethesda, MD 20892, USA.
Anand Swaroop, Neurobiology, Neurodegeneration and Repair Laboratory, National Eye Institute, National Institutes of Health, 6 Center Drive, MSC0610, Bethesda, MD 20892, USA.
Funding
Intramural Research Program of the National Eye Institute (ZIAEY000450).
References
- 1. Pope, S.D. and Medzhitov, R. (2018) Emerging principles of gene expression programs and their regulation. Mol. Cell, 71, 389–397. [DOI] [PubMed] [Google Scholar]
- 2. Goldman, J.A. and Poss, K.D. (2020) Gene regulatory programmes of tissue regeneration. Nat. Rev. Genet., 21, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furlong, E.E.M. and Levine, M. (2018) Developmental enhancers and chromosome topology. Science (New York, N.Y.), 361, 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cramer, P. (2019) Organization and regulation of gene transcription. Nature, 573, 45–54. [DOI] [PubMed] [Google Scholar]
- 5. Haberle, V. and Stark, A. (2018) Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol., 19, 621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soutourina, J. (2018) Transcription regulation by the mediator complex. Nat. Rev. Mol. Cell Biol., 19, 262–274. [DOI] [PubMed] [Google Scholar]
- 7. Gil, N. and Ulitsky, I. (2020) Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet., 21, 102–117. [DOI] [PubMed] [Google Scholar]
- 8. Talbert, P.B., Talbert, P.B. and Meers, M.P. (2019) Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat. Rev. Genet., 20, 283–297. [DOI] [PubMed] [Google Scholar]
- 9. Bonev, B., Mendelson Cohen, N., Szabo, Q., Fritsch, L., Papadopoulos, G.L., Lubling, Y., Xu, X., Lv, X., Hugnot, J.P., Tanay, A. et al. (2017) Multiscale 3D genome rewiring during mouse neural development. Cell, 171, 557–572 e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yadav, T., Quivy, J.P. and Almouzni, G. (2018) Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Science (New York, N.Y.), 361, 1332–1336. [DOI] [PubMed] [Google Scholar]
- 11. Miesfeld, J.B. and Brown, N.L. (2019) Eye organogenesis: a hierarchical view of ocular development. Curr. Top. Dev. Biol., 132, 351–393. [DOI] [PubMed] [Google Scholar]
- 12. Lee, T.I. and Young, R.A. (2013) Transcriptional regulation and its misregulation in disease. Cell, 152, 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karnuta, J.M. and Scacheri, P.C. (2018) Enhancers: bridging the gap between gene control and human disease. Hum. Mol. Genet., 27, R219–R227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corso-Diaz, X., Jaeger, C., Chaitankar, V. and Swaroop, A. (2018) Epigenetic control of gene regulation during development and disease: a view from the retina. Prog. Retin. Eye Res., 65, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anania, C. and Lupianez, D.G. (2020) Order and disorder: abnormal 3D chromatin organization in human disease. Brief. Funct. Genomics, 19, 128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janssen, S.M. and Lorincz, M.C. (2021) Interplay between chromatin marks in development and disease. Nat. Rev. Genet., 23, 137–153. [DOI] [PubMed] [Google Scholar]
- 17. Swaroop, A., Kim, D. and Forrest, D. (2010) Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci., 11, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brzezinski, J.A. and Reh, T.A. (2015) Photoreceptor cell fate specification in vertebrates. Development, 142, 3263–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Norrie, J.L., Lupo, M.S., Xu, B., Al Diri, I., Valentine, M., Putnam, D., Griffiths, L., Zhang, J., Johnson, D., Easton, J. et al. (2019) Nucleome dynamics during retinal development. Neuron, 104, 512–528 e511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamb, T.D. (2013) Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res., 36, 52–119. [DOI] [PubMed] [Google Scholar]
- 21. Nishida, A., Furukawa, A., Koike, C., Tano, Y., Aizawa, S., Matsuo, I. and Furukawa, T. (2003) Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci., 6, 1255–1263. [DOI] [PubMed] [Google Scholar]
- 22. Brzezinski, J.A.T., Lamba, D.A. and Reh, T.A. (2010) Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development, 137, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghinia Tegla, M.G., Buenaventura, D.F., Kim, D.Y., Thakurdin, C., Gonzalez, K.C. and Emerson, M.M. (2020) OTX2 represses sister cell fate choices in the developing retina to promote photoreceptor specification. elife, 9, e54279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roger, J.E., Hiriyanna, A., Gotoh, N., Hao, H., Cheng, D.F., Ratnapriya, R., Kautzmann, M.A., Chang, B. and Swaroop, A. (2014) OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest., 124, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furukawa, T., Morrow, E.M. and Cepko, C.L. (1997) Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell, 91, 531–541. [DOI] [PubMed] [Google Scholar]
- 26. Chen, S., Wang, Q.L., Nie, Z., Sun, H., Lennon, G., Copeland, N.G., Gilbert, D.J., Jenkins, N.A. and Zack, D.J. (1997) Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron, 19, 1017–1030. [DOI] [PubMed] [Google Scholar]
- 27. Corbo, J.C., Lawrence, K.A., Karlstetter, M., Myers, C.A., Abdelaziz, M., Dirkes, W., Weigelt, K., Seifert, M., Benes, V., Fritsche, L.G. et al. (2010) CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res., 20, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mears, A.J., Kondo, M., Swain, P.K., Takada, Y., Bush, R.A., Saunders, T.L., Sieving, P.A. and Swaroop, A. (2001) Nrl is required for rod photoreceptor development. Nat. Genet., 29, 447–452. [DOI] [PubMed] [Google Scholar]
- 29. Oh, E.C., Khan, N., Novelli, E., Khanna, H., Strettoi, E. and Swaroop, A. (2007) Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc. Natl. Acad. Sci. U. S. A., 104, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim, J.W., Yang, H.J., Oel, A.P., Brooks, M.J., Jia, L., Plachetzki, D.C., Li, W., Allison, W.T. and Swaroop, A. (2016) Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev. Cell, 37, 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng, L., Lu, A., Swaroop, A., Sharlin, D.S., Swaroop, A. and Forrest, D. (2011) Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J. Neurosci., 31, 11118–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oh, E.C., Cheng, H., Hao, H., Jia, L., Khan, N.W. and Swaroop, A. (2008) Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res., 1236, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corbo, J.C. and Cepko, C.L. (2005) A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet., 1, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng, H., Aleman, T.S., Cideciyan, A.V., Khanna, R., Jacobson, S.G. and Swaroop, A. (2006) In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet., 15, 2588–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng, H., Khanna, H., Oh, E.C., Hicks, D., Mitton, K.P. and Swaroop, A. (2004) Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet., 13, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 36. Bessant, D.A., Payne, A.M., Mitton, K.P., Wang, Q.L., Swain, P.K., Plant, C., Bird, A.C., Zack, D.J., Swaroop, A. and Bhattacharya, S.S. (1999) A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat. Genet., 21, 355–356. [DOI] [PubMed] [Google Scholar]
- 37. Nishiguchi, K.M., Friedman, J.S., Sandberg, M.A., Swaroop, A., Berson, E.L. and Dryja, T.P. (2004) Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc. Natl. Acad. Sci. U. S. A., 101, 17819–17824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wright, A.F., Reddick, A.C., Schwartz, S.B., Ferguson, J.S., Aleman, T.S., Kellner, U., Jurklies, B., Schuster, A., Zrenner, E., Wissinger, B. et al. (2004) Mutation analysis of NR2E3 and NRL genes in enhanced S cone syndrome. Hum. Mutat., 24, 439. [DOI] [PubMed] [Google Scholar]
- 39. Kanda, A., Friedman, J.S., Nishiguchi, K.M. and Swaroop, A. (2007) Retinopathy mutations in the bZIP protein NRL alter phosphorylation and transcriptional activity. Hum. Mutat., 28, 589–598. [DOI] [PubMed] [Google Scholar]
- 40. Carter-Dawson, L.D. and LaVail, M.M. (1979) Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol., 188, 263–272. [DOI] [PubMed] [Google Scholar]
- 41. Akimoto, M., Cheng, H., Zhu, D., Brzezinski, J.A., Khanna, R., Filippova, E., Oh, E.C., Jing, Y., Linares, J.L., Brooks, M. et al. (2006) Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U. S. A., 103, 3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blanks, J.C., Adinolfi, A.M. and Lolley, R.N. (1974) Synaptogenesis in the photoreceptor terminal of the mouse retina. J. Comp. Neurol., 156, 81–93. [DOI] [PubMed] [Google Scholar]
- 43. Obata, S. and Usukura, J. (1992) Morphogenesis of the photoreceptor outer segment during postnatal development in the mouse (BALB/c) retina. Cell Tissue Res., 269, 39–48. [DOI] [PubMed] [Google Scholar]
- 44. De Robertis, E. (1956) Morphogenesis of the retinal rods; an electron microscope study. J. Biophys. Biochem. Cytol., 2, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim, J.W., Yang, H.J., Brooks, M.J., Zelinger, L., Karakulah, G., Gotoh, N., Boleda, A., Gieser, L., Giuste, F., Whitaker, D.T. et al. (2016) NRL-regulated transcriptome dynamics of developing rod photoreceptors. Cell Rep., 17, 2460–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mitton, K.P., Swain, P.K., Chen, S., Xu, S., Zack, D.J. and Swaroop, A. (2000) The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem., 275, 29794–29799. [DOI] [PubMed] [Google Scholar]
- 47. Hennig, A.K., Peng, G.H. and Chen, S. (2008) Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res., 1192, 114–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. White, M.A., Kwasnieski, J.C., Myers, C.A., Shen, S.Q., Corbo, J.C. and Cohen, B.A. (2016) A simple grammar defines activating and repressing cis-regulatory elements in photoreceptors. Cell Rep., 17, 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Assawachananont, J., Kim, S.Y., Kaya, K.D., Fariss, R., Roger, J.E. and Swaroop, A. (2018) Cone-rod homeobox CRX controls presynaptic active zone formation in photoreceptors of mammalian retina. Hum. Mol. Genet., 27, 3555–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hao, H., Kim, D.S., Klocke, B., Johnson, K.R., Cui, K., Gotoh, N., Zang, C., Gregorski, J., Gieser, L., Peng, W. et al. (2012) Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet., 8, e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skene, P.J., Henikoff, J.G. and Henikoff, S. (2018) Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc., 13, 1006–1019. [DOI] [PubMed] [Google Scholar]
- 52. Meers, M.P., Bryson, T.D., Henikoff, J.G. and Henikoff, S. (2019) Improved CUT&RUN chromatin profiling tools. elife, 8, e46314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Angel, P. and Karin, M. (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta, 1072, 129–157. [DOI] [PubMed] [Google Scholar]
- 54. Shaulian, E. and Karin, M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol., 4, E131–E136. [DOI] [PubMed] [Google Scholar]
- 55. Kerppola, T.K. and Curran, T. (1994) Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene, 9, 675–684. [PubMed] [Google Scholar]
- 56. Aldiri, I., Xu, B., Wang, L., Chen, X., Hiler, D., Griffiths, L., Valentine, M., Shirinifard, A., Thiagarajan, S., Sablauer, A. et al. (2017) The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron, 94, 550–568.e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fuhrmann, S., Riesenberg, A.N., Mathiesen, A.M., Brown, E.C., Vetter, M.L. and Brown, N.L. (2009) Characterization of a transient TCF/LEF-responsive progenitor population in the embryonic mouse retina. Invest. Ophthalmol. Vis. Sci., 50, 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rao, A., Luo, C. and Hogan, P.G. (1997) Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol., 15, 707–747. [DOI] [PubMed] [Google Scholar]
- 59. Chen, L., Glover, J.N., Hogan, P.G., Rao, A. and Harrison, S.C. (1998) Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature, 392, 42–48. [DOI] [PubMed] [Google Scholar]
- 60. Hughes, A.E., Enright, J.M., Myers, C.A., Shen, S.Q. and Corbo, J.C. (2017) Cell type-specific Epigenomic analysis reveals a uniquely closed chromatin architecture in mouse rod photoreceptors. Sci. Rep., 7, 43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Javed, A., Mattar, P., Lu, S., Kruczek, K., Kloc, M., Gonzalez-Cordero, A., Bremner, R., Ali, R.R. and Cayouette, M. (2020) Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina. Development, 147, dev188730. [DOI] [PubMed] [Google Scholar]
- 62. Fremont, S. and Echard, A. (2018) Membrane traffic in the late steps of cytokinesis. Curr. Biol., 28, R458–R470. [DOI] [PubMed] [Google Scholar]
- 63. Long, H. and Huang, K. (2020) Transport of ciliary membrane proteins. Front. Cell Dev. Biol., 7, 381–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim, J.W., Jang, S.M., Kim, C.H., An, J.H. and Choi, K.H. (2012) Transcriptional activity of neural retina leucine zipper (Nrl) is regulated by c-Jun N-terminal kinase and Tip60 during retina development. Mol. Cell. Biol., 32, 1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee, J., Myers, C.A., Williams, N., Abdelaziz, M. and Corbo, J.C. (2010) Quantitative fine-tuning of photoreceptor cis-regulatory elements through affinity modulation of transcription factor binding sites. Gene Ther., 17, 1390–1399. [DOI] [PubMed] [Google Scholar]
- 66. Cherry, T.J., Yang, M.G., Harmin, D.A., Tao, P., Timms, A.E., Bauwens, M., Allikmets, R., Jones, E.M., Chen, R., De Baere, E. et al. (2020) Mapping the cis-regulatory architecture of the human retina reveals noncoding genetic variation in disease. Proc. Natl. Acad. Sci. U. S. A., 117, 9001–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schüle, R., Muller, M., Kaltschmidt, C. and Renkawitz, R. (1988) Many transcription factors interact synergistically with steroid receptors. Science, 242, 1418–1420. [DOI] [PubMed] [Google Scholar]
- 68. Thirunavukkarasu, K., Miles, R.R., Halladay, D.L. and Onyia, J.E. (2000) Cryptic enhancer elements in luciferase reporter vectors respond to the osteoblast-specific transcription factor Osf2/Cbfa1. BioTechniques, 28, 506–510. [DOI] [PubMed] [Google Scholar]
- 69. Hong, S.J., Chae, H. and Kim, K.S. (2002) Promoterless luciferase reporter gene is transactivated by basic helix-loop-helix transcription factors. BioTechniques, 33, 1236–1240 1240. [DOI] [PubMed] [Google Scholar]
- 70. Grimm, S.L. and Nordeen, S.K. (1999) Luciferase reporter gene vectors that lack potential AP-1 sites. BioTechniques, 27, 220–222. [DOI] [PubMed] [Google Scholar]
- 71. Sandkam, B.A., Campello, L., O'Brien, C., Nandamuri, S.P., Gammerdinger, W.J., Conte, M.A., Swaroop, A. and Carleton, K.L. (2020) Tbx2a modulates switching of RH2 and LWS opsin gene expression. Mol. Biol. Evol., 37, 2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Matsuda, T. and Cepko, C.L. (2004) Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A., 101, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bunina, D., Abazova, N., Diaz, N., Noh, K.M., Krijgsveld, J. and Zaugg, J.B. (2020) Genomic rewiring of SOX2 chromatin interaction network during differentiation of ESCs to Postmitotic neurons. Cell Syst, 10, 480–494 e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu, H., Whitfield, T.W., Gordon, J.A., Dobson, J.R., Tai, P.W., van Wijnen, A.J., Stein, J.L., Stein, G.S. and Lian, J.B. (2014) Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol., 15, R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pajoro, A., Madrigal, P., Muiño, J.M., Matus, J.T., Jin, J., Mecchia, M.A., Debernardi, J.M., Palatnik, J.F., Balazadeh, S., Arif, M. et al. (2014) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol., 15, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Berg, O.G., Winter, R.B. and von Hippel, P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry, 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- 77. Suter, D.M. (2020) Transcription factors and DNA play Hide and Seek. Trends Cell Biol., 30, 491–500. [DOI] [PubMed] [Google Scholar]
- 78. Chen, J., Zhang, Z., Li, L., Chen, B.C., Revyakin, A., Hajj, B., Legant, W., Dahan, M., Lionnet, T., Betzig, E. et al. (2014) Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell, 156, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mazzocca, M., Colombo, E., Callegari, A. and Mazza, D. (2021) Transcription factor binding kinetics and transcriptional bursting: what do we really know? Curr. Opin. Struct. Biol., 71, 239–248. [DOI] [PubMed] [Google Scholar]
- 80. Kribelbauer, J.F., Rastogi, C., Bussemaker, H.J. and Mann, R.S. (2019) Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu. Rev. Cell Dev. Biol., 35, 357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee, R.E., Walker, S.R., Savery, K., Frank, D.A. and Gaudet, S. (2014) Fold change of nuclear NF-kappaB determines TNF-induced transcription in single cells. Mol. Cell, 53, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lorenzin, F., Benary, U., Baluapuri, A., Walz, S., Jung, L.A., von Eyss, B., Kisker, C., Wolf, J., Eilers, M. and Wolf, E. (2016) Different promoter affinities account for specificity in MYC-dependent gene regulation. elife, 5, e15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Campla, C.K., Mast, H., Dong, L., Lei, J., Halford, S., Sekaran, S. and Swaroop, A. (2019) Targeted deletion of an NRL- and CRX-regulated alternative promoter specifically silences FERM and PDZ domain containing 1 (Frmpd1) in rod photoreceptors. Hum. Mol. Genet., 28, 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Spivakov, M. (2014) Spurious transcription factor binding: non-functional or genetically redundant? BioEssays, 36, 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsu, P.C., Lu, T.C., Hung, P.H., Jhou, Y.T., Amine, A.A.A., Liao, C.W. and Leu, J.Y. (2021) Plastic rewiring of Sef1 transcriptional networks and the potential of nonfunctional transcription factor binding in facilitating adaptive evolution. Mol. Biol. Evol., 38, 4732–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Eychene, A., Rocques, N. and Pouponnot, C. (2008) A new MAFia in cancer. Nat. Rev. Cancer, 8, 683–693. [DOI] [PubMed] [Google Scholar]
- 87. Rozek, D. and Pfeifer, G.P. (1993) In vivo protein-DNA interactions at the c-Jun promoter: preformed complexes mediate the UV response. Mol. Cell. Biol., 13, 5490–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stein, B., Angel, P., van Dam, H., Ponta, H., Herrlich, P., van der Eb, A. and Rahmsdorf, H.J. (1992) Ultraviolet-radiation induced c-Jun gene transcription: two AP-1 like binding sites mediate the response. Photochem. Photobiol., 55, 409–415. [DOI] [PubMed] [Google Scholar]
- 89. van Dam, H., Duyndam, M., Rottier, R., Bosch, A., de Vries-Smits, L., Herrlich, P., Zantema, A., Angel, P. and van der Eb, A.J. (1993) Heterodimer formation of cJun and ATF-2 is responsible for induction of c-Jun by the 243 amino acid adenovirus E1A protein. EMBO J., 12, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Angel, P., Hattori, K., Smeal, T. and Karin, M. (1988) The Jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell, 55, 875–885. [DOI] [PubMed] [Google Scholar]
- 91. Leppä, S. and Bohmann, D. (1999) Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene, 18, 6158–6162. [DOI] [PubMed] [Google Scholar]
- 92. Wisdom, R., Johnson, R.S. and Moore, C. (1999) C-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J., 18, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qi, X., Borowicz, S., Pramanik, R., Schultz, R.M., Han, J. and Chen, G. (2004) Estrogen receptor inhibits c-Jun-dependent stress-induced cell death by binding and modifying c-Jun activity in human breast cancer cells. J. Biol. Chem., 279, 6769–6777. [DOI] [PubMed] [Google Scholar]
- 94. Lukey, M.J., Greene, K.S., Erickson, J.W., Wilson, K.F. and Cerione, R.A. (2016) The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun., 7, 11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tang, C., Yeung, L.S.N., Koulajian, K., Zhang, L., Tai, K., Volchuk, A. and Giacca, A. (2018) Glucose-induced β-cell dysfunction in vivo: evidence for a causal role of C-Jun N-terminal kinase pathway. Endocrinology, 159, 3643–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xiao, F., Guo, Y., Deng, J., Yuan, F., Xiao, Y., Hui, L., Li, Y., Hu, Z., Zhou, Y., Li, K. et al. (2019) Hepatic c-Jun regulates glucose metabolism via FGF21 and modulates body temperature through the neural signals. Mol Metab, 20, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dai, Z., Ramesh, V. and Locasale, J.W. (2020) The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet., 21, 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li, X., Egervari, G., Wang, Y., Berger, S.L. and Lu, Z. (2018) Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol., 19, 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jolly, C. and Morimoto, R.I. (1999) Stress and the cell nucleus: dynamics of gene expression and structural reorganization. Gene Expr., 7, 261–270. [PMC free article] [PubMed] [Google Scholar]
- 100. Himanen, S.V. and Sistonen, L. (2019) New insights into transcriptional reprogramming during cellular stress. J. Cell Sci., 132, jcs238402. [DOI] [PubMed] [Google Scholar]
- 101. Kaya-Okur, H.S., Wu, S.J., Codomo, C.A., Pledger, E.S., Bryson, T.D., Henikoff, J.G., Ahmad, K. and Henikoff, S. (2019) CUT&tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun., 10, 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kautzmann, M.A., Kim, D.S., Felder-Schmittbuhl, M.P. and Swaroop, A. (2011) Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. J. Biol. Chem., 286, 28247–28255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bolger, A.M., Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Langmead, B. and Salzberg, S.L. (2012) Fast gapped-read alignment with bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R. and Genome Project Data Processing, S (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nusbaum, C., Myers, R.M., Brown, M., Li, W. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Amemiya, H.M., Kundaje, A. and Boyle, A.P. (2019) The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep., 9, 9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Quinlan, A.R. and Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Whitington, T., Frith, M.C., Johnson, J. and Bailey, T.L. (2011) Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res., 39, e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wingender, E. (1988) Compilation of transcription regulating proteins. Nucleic Acids Res., 16, 1879–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wingender, E., Schoeps, T. and Dönitz, J. (2013) TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res., 41, D165–D170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ernst, J. and Kellis, M. (2017) Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc., 12, 2478–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gorkin, D.U., Barozzi, I., Zhao, Y., Zhang, Y., Huang, H., Lee, A.Y., Li, B., Chiou, J., Wildberg, A., Ding, B. et al. (2020) An atlas of dynamic chromatin landscapes in mouse fetal development. Nature, 583, 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lawrence, M., Huber, W., Pages, H., Aboyoun, P., Carlson, M., Gentleman, R., Morgan, M.T. and Carey, V.J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lun, A.T. and Smyth, G.K. (2016) Csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res., 44, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bray, N.L., Pimentel, H., Melsted, P. and Pachter, L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527. [DOI] [PubMed] [Google Scholar]
- 117. Soneson, C., Love, M.I. and Robinson, M.D. (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research, 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Robinson, M.D., McCarthy, D.J. and Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed data are available through NCBI’s Gene Expression Omnibus: GSE197421 (http://www/ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197421).