Abstract
In vitro fertilization (IVF) is associated with DNA methylation abnormalities and a higher incidence of adverse pregnancy outcomes. However, which exposure(s), among the many IVF interventions, contributes to these outcomes remains unknown. Frozen embryo transfer (ET) is increasingly utilized as an alternative to fresh ET, but reports suggest a higher incidence of pre-eclampsia and large for gestational age infants. This study examines DNA methylation in human placentas using the 850K Infinium MethylationEPIC BeadChip array obtained after 65 programmed frozen ET cycles, 82 fresh ET cycles and 45 unassisted conceptions. Nine patients provided placentas following frozen and fresh ET from consecutive pregnancies for a paired subgroup analysis. In parallel, eight mouse placentas from fresh and frozen ET were analyzed using the Infinium Mouse Methylation BeadChip array. Human and mouse placentas were significantly hypermethylated after frozen ET compared with fresh. Paired analysis showed similar trends. Sex-specific analysis revealed that these changes were driven by male placentas in humans and mice. Frozen and fresh ET placentas were significantly different from controls, with frozen samples hypermethylated compared with controls driven by males and fresh samples being hypomethylated compared with controls, driven by females. Sexually dimorphic epigenetic changes could indicate differential susceptibility to IVF-associated perturbations, which highlights the importance of sex-specific evaluation of adverse outcomes. Similarities between changes in mice and humans underscore the suitability of the mouse model in evaluating how IVF impacts the epigenetic landscape, which is valuable given limited access to human tissue and the ability to isolate specific interventions in mice.
Introduction
Infants conceived with the aid of assisted reproductive technologies (ART) account for 2% of all births (1,2). Of all ART procedures, in vitro fertilization (IVF), followed by embryo transfer (ET) is the most effective at achieving pregnancy, and improved clinical laboratory methodologies have contributed to significant improvement in delivery rates. However, epidemiological data suggest that ART procedures, even those resulting in singleton births, are associated with an increased risk for adverse pregnancy outcomes, including low birth weight (LBW), hypertensive disorders of pregnancy (such as pre-eclampsia) and preterm birth (PTB) (3–6).
Though the molecular and cellular mechanisms responsible for these adverse outcomes are still being elucidated, data from our group and others have demonstrated that IVF leads to epigenetic changes in the placenta, which may play a role in the development of the adverse outcomes associated with human IVF (7–10). It was initially hypothesized that at least some of these epigenetic perturbations could be a consequence of subfertility. However, a study examining placental DNA methylation obtained from pregnancies using donor oocytes in the absence of other infertility diagnoses confirmed that some components of the IVF process were at least in part responsible for the observed changes (9). In addition, multiple animal studies have confirmed that various procedures used in IVF result in phenotypic and epigenetic perturbations in both fetal and placental tissue (even in the absence of underlying infertility) (11–17).
IVF involves multiple laboratory and clinical manipulations, including hormonal stimulation, gamete manipulation, embryo culture, embryo cryopreservation and ET, as well as exposures, including changes in the hormonal environment, temperature, pH and oxygen tension. Understanding which of these interventions influence perinatal outcomes, and elucidating the mechanisms underlying the development of such adverse outcomes are necessary to implement modifications to current protocols and thus minimize adverse outcomes. One ART procedure used with increasing frequency (owing to an improvement in embryo culture and freezing techniques) is embryo vitrification, a cryopreservation technique that allows for embryo freezing without the formation of intracellular ice crystals. The resulting comparable pregnancy rates have led to increased utilization of frozen ET. However, epidemiologic data demonstrate some elevated perinatal risks, including a significant increase in the incidence of large for gestational age (LGA) infants and pre-eclampsia (18–22). Interestingly, several recent studies suggest that male offspring have a higher birth weight after frozen ET compared with female infants and are more likely to be born LGA following frozen ET than female offspring (23). These increased adverse outcomes are seen following frozen ET in contrast with other adverse outcomes, such as small for gestational age (SGA), PTB, LBW and perinatal mortality, all of which show higher incidence after fresh ET (18,22,24–27). Critically, the specific intervention(s) utilized during IVF responsible for these effects, and the mechanism(s) underlying the development of these adverse outcomes are still unclear.
To isolate the effects of individual components of the ART procedures, without the confounding effect of underlying infertility, animal models of ART have been used. The epigenetic impact of ART has particularly been explored in mouse models, which show similar perturbations in DNA methylation after ART procedures to those found in humans (11–13,15–17,28–31). Animal models provide a specific advantage in studying IVF because specific clinical or laboratory interventions contributing to epigenetic changes can be isolated for study (such as embryo culture or ET). Obviously, such independent component evaluation cannot be isolated and undertaken in humans. As a result, parallel studies in humans and murine models are necessary to allow us to reach more meaningful and translatable conclusions (32,33).
In this study, we examine the effect of embryo cryopreservation on genome-wide epigenetic profiles in human and mouse placental samples, specifically focusing on DNA methylation owing to its discrete nature and availability of tools to interrogate localized and global changes. As many adverse outcomes following ART are thought to occur owing to abnormalities in placentation, and placental tissue appears to be particularly susceptible to epigenetic perturbations after ART, we focused our study on the interrogation of DNA methylation changes in placentas (34). For our human studies, placentas were obtained at the time of delivery from pregnancies conceived by IVF following either fresh ET or frozen–thawed ET into uteri primed with estrogen and progesterone. Placental tissue was collected and profiled for changes in DNA methylation (Fig. 1), and changes in placental DNA methylation following frozen versus fresh ET were compared. In addition, to determine if either frozen or fresh ET more closely resembled unassisted conception, DNA methylation in each group was individually compared with placentas from unassisted, natural conceptions.
Figure 1.
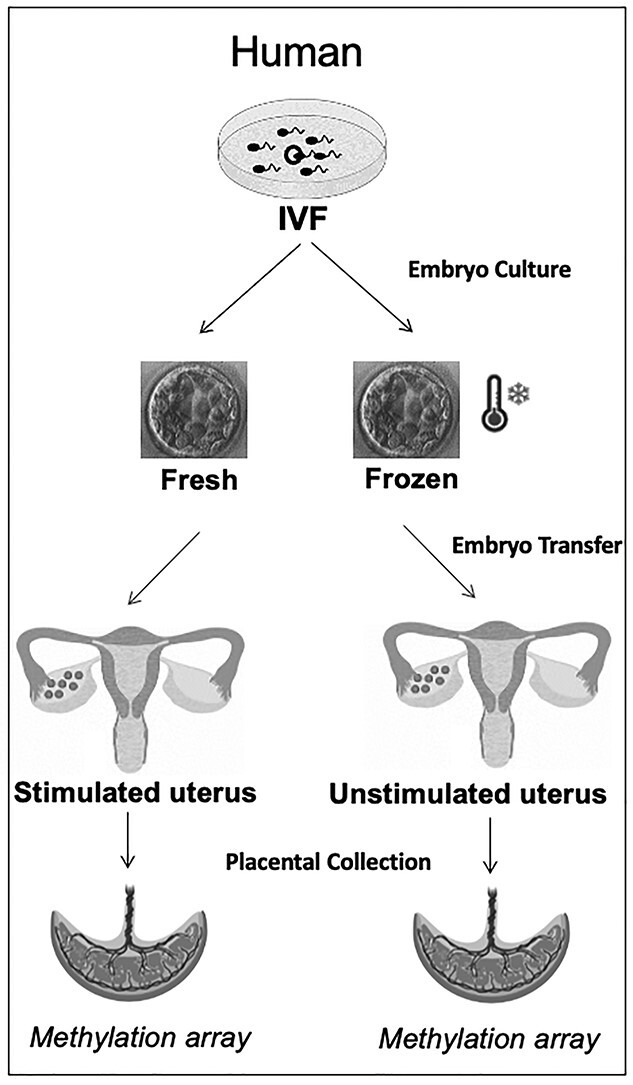
IVF was performed per clinical protocols and embryos were either transferred fresh following a gonadotropin-stimulated cycles or cryopreserved and transferred following endometrial preparation. Placentas were collected at the time of delivery. DNA was isolated from the placentas, bisulfite treated and sent for DNA methylation analysis.
In parallel, because the effects of a stimulated hormonal environment cannot be separated from embryo vitrification in humans, we addressed this question in a mouse model. The recent introduction of the Illumina Infinium Mouse Methylation Array (35) enables the analysis of multiple samples at CpGs across the mouse genome, as we have done with the corresponding human array. We employed this newly developed array that profiles 285 000 CpGs throughout the genome, to examine changes in methylation owing to vitrification in the mouse model. In our unique model, we transfer green fluorescent protein (GFP) tagged or untagged, fresh or frozen blastocysts, into the same unstimulated pseudopregnant host, which allows us to isolate vitrification as the sole exposure (Fig. 2). Using this model, we analyzed placental DNA methylation and histology as well as fetal and placental weight. Through these parallel investigations, we demonstrate that DNA methylation is perturbed in both human and mouse placentas derived from embryos after frozen compared with fresh ET. Importantly, male embryos appear more susceptible to the effects of embryo vitrification than female embryos, potentially providing a mechanism for the increased risk of LGA in males conceived following FET.
Figure 2.
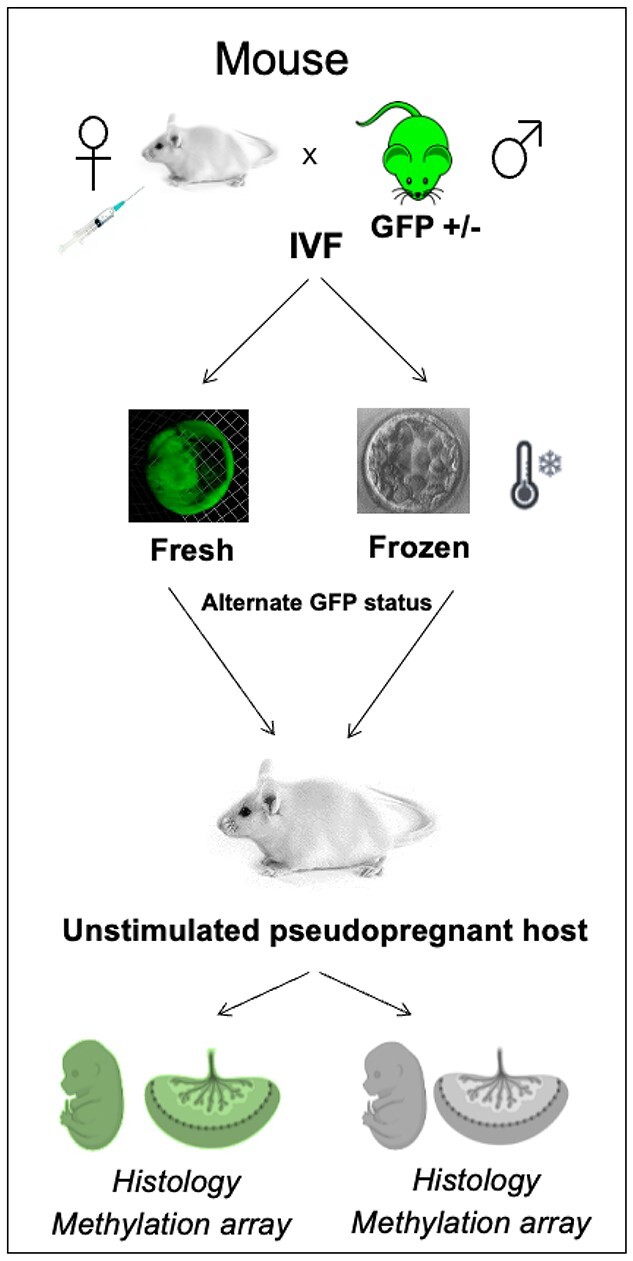
IVF was performed using mouse eggs obtained following gonadotropin stimulation and sperm collected from the epididymis of mice heterozygous for the GFP transgene. Embryos were cultured to the blastocyst stage and embryos were either transferred or vitrified. Ten blastocysts (five fresh, five vitrified, differing in their GFP status) were non-surgically transferred in a single horn of a pseudopregnant female. GFP status was used to distinguish fresh from frozen embryos. Pregnant dams were sacrificed on E12.5 and E18.5. DNA was isolated from E18.5 placentas, bisulfite treated and sent for DNA methylation analysis.
Results
Human sample characteristics
Placental samples were collected from a total of 192 singleton pregnancies [unassisted conception (controls; n = 45), IVF followed by fresh ET (n = 82) and IVF followed by programmed frozen ET (n = 65)]. The demographic profiles and relevant clinical characteristics are shown in Table 1. The frozen and fresh ET groups differed significantly only in the ET stage, with slightly more cleavage stage embryos transferred in the fresh ET group (18%) compared with the frozen ET group (5%). As expected, birth weight in the frozen ET group was significantly higher compared with both fresh ET and controls, and these birth weight differences were driven by male infants, as there were no significant differences in birth weight in female infants. Nine women had two pregnancies conceived by ART with placentas banked, one from a fresh ET and one from a frozen ET. These 18 samples were analyzed independently and the demographics for these women are shown in Supplementary Material, Tables S1 and S2.
Table 1.
Demographic profile and clinical characteristics of the study subjects
| Fresh ET (n = 82) | Frozen ET (n = 65) | Unassisted conception (n = 45) | P-values | ||
|---|---|---|---|---|---|
| Maternal age [median (IQR)] | 35 (32–38) | 34 (33–36) | 34 (30–35) | 0.016a,e,f | |
| Race | White Black Other |
64 (78%) 9 (11%) 9 (11%) |
46 (71%) 12 (18%) 7 (11%) |
26 (58%) 17 (38%) 2 (4%) |
0.01b |
| Ethnicity | Hispanic Non-Hispanic |
2 (2%) 80 (98%) |
2 (3%) 63 (97%) |
1 (2%) 44 (98%) |
1.00b |
| Insemination method | Conventional Some/all ICSI |
26 (32%) 56 (68%) |
19 (29%) 46 (71%) |
N/A | 0.75c |
| Oxygen Tension |
5% 20% |
76 (93%) 6 (7%) |
61 (94%) 4 (6%) |
N/A | 1.00b |
| ET stage | Cleavage Stage Blastocyst stage |
15 (18%) 67 (82%) |
3 (5%) 62 (95%) |
N/A | 0.01b |
| Gestational age [weeks, median (IQR)] | 39.3 (37.6–40) | 39.3 (37.7–40.6) | 38.1 (34.3–39) | 0.001a,e,f | |
| Birth weight [g, median (IQR)] | 3221 (2721–3630) | 3510 (3120–3800) | 3120 (2175–3430) | 0.009a.d,f | |
| Fetal sex | Male Female |
42 (51%) 40 (49%) |
31 (48%) 34 (52%) |
22 (49%) 23 (51%) |
0.91c |
| Fetal weight [g, median (IQR)] | Male | 3260 (2780–3655) | 3540 (3030–3850) | 2955 (2480–3345) | 0.013a,f |
| Female | 3160 (2615–3540) | 3478 (3150–3755) | 3120 (1760–3640) | 0.05 | |
aKruskal–Wallis test.
bFisher exact test.
cChi-square test.
dFresh ET versus frozen ET Rank Sum test, P < 0.01.
eFresh ET versus unassisted conception Rank Sum test, P < 0.01.
fFrozen ET versus unassisted conception Rank Sum test, P < 0.01.
Frozen ET leads to hypermethylation of human placenta
Because the primary objective of this study was to examine how embryo cryopreservation could impact DNA methylation, we first compared placental methylation status in placentas after frozen ET to that after fresh ET. Using a mean difference in methylation of greater than 5% and a P-value less than 0.05 in two-tailed unpaired t-tests, we identified 4402 differentially methylated CpG sites, of which 4124 were hypermethylated in placentas after frozen ET compared with fresh ET, while 278 were hypomethylated. These differentially methylated CpGs fell within 1600 genes, of which 385 showed more than 1 significantly different CpG, 156 genes showed more than 2 affected CpGs and 87 genes contained more than 3 significantly different CpGs (Fig. 3A, Supplementary Material, Table S3). Figure 3 highlights two candidate genes that have potential biologically relevant roles in placentation, gamma-aminobutyric acid A receptor, γ3 (GABRG3), which contained seven differentially methylated CpGS; and adenosine deaminase RNA-specific B2 (ADARB2), which contained nine differentially methylated CpGs. For both genes, placentas after frozen ET were significantly hypermethylated compared with placentas after fresh ET (Fig. 3B and C, one representative CpG per gene shown).
Figure 3.
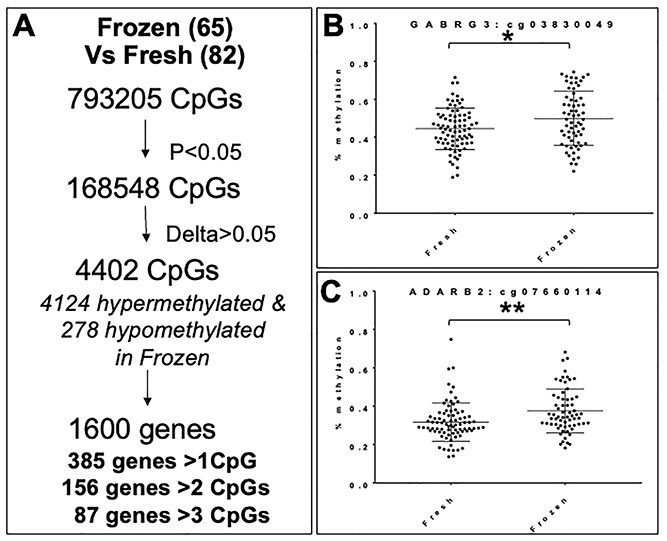
(A) Methylation differences in human placentas from frozen ET compared with placentas from fresh ET. (B and C) DNA methylation in a representative CpGs from two candidate genes: GABRG3 (B) and ADARB2 (C). *P = 0.02, **P = 0.0031.
To begin examining the biological significance of CpGs changed owing to frozen ET, we performed gene ontology (GO) enrichment analysis on differentially methylated CpGs when samples after frozen and fresh ET were compared. Using methylGSA (36), a tool that adjusts for CpG distribution bias owing to gene length, we identified several pathways that were over-represented in our dataset. These included pathways involved in cell cycle regulation; organ morphogenesis; cell-matrix adhesion; metabolism and inflammation (Supplementary Material, Table S4). As an alternative approach to understanding the biological significance of changes owing to frozen ET, we used eFORGE, a tool that examines overlaps in DNAse 1 hypersensitivity sites of specific cell types with differentially methylated probes to identify tissue-specific enrichment (37). However, q-values obtained after inputting the top 1000 changed CpGs did not show an eFORGE signal, suggesting no discernable enrichment pattern. In addition, as ART has been associated with an increased prevalence of imprinting disorders and the manipulations utilized during ART coincide with critical periods of imprint establishment, we examined the effect of cryopreservation on imprinted genes. In our dataset, we found 11 imprinted genes with at least 1 differentially methylated CpGs in frozen ET placenta compared with fresh ET. Among these 11 genes, 10 genes had all of their differentially methylated CpGs hypermethylated, while one gene, KCNQ1DN, had one differentially methylated CpG, which was hypomethylated (Supplementary Material, Table S3).
Both frozen ET and fresh ET have been associated with specific adverse pregnancy outcomes. Therefore, we compared frozen ET and fresh ET placental samples independently to unassisted conceptions (controls), to see if one of these methods had fewer perturbations in DNA methylation. We identified 5600 differentially methylated CpGs when placentas from frozen ET were compared with controls, corresponding to 3979 hypermethylated and 1621 hypomethylated CpGs (Fig. 4A). These CpGs fell within 505 genes with 71 genes having more than 3 differentially methylated CpGs (Supplementary Material, Table S5). When placentas from fresh ET were compared with controls, we identified 4096 differentially methylated CpGs, with 717 hypermethylated and 3379 hypomethylated CpGs (Fig. 4B). These CpGs fell within 1914 genes, of which 51 genes contained more than 3 differentially methylated CpGs (Supplementary Material, Table S6). We also examined the overlap between both techniques when compared with unassisted control placentas and found 1004 CpGs differentially methylated in both datasets. Of those 1004 CpGs, 1003 were differentially methylated in the same direction: 364 were hypermethylated in both fresh and frozen ET and 639 were hypomethylated in both fresh and frozen ET compared with controls. Only one CpG site was hypomethylated in fresh ET and hypermethylated in frozen ET (Fig. 4C).
Figure 4.
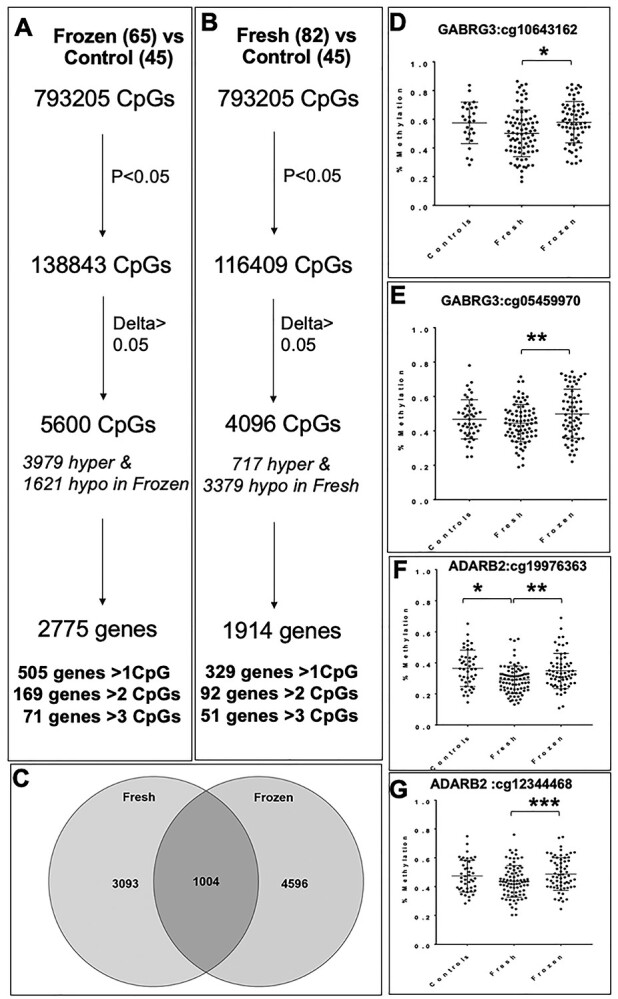
(A) Methylation differences in human placentas from frozen ET compared with placentas from unassisted conceptions (controls) (B). Methylation differences in human placentas from fresh ET compared with placentas from unassisted conceptions. (C) Number of CpGs differentially methylated in fresh ET versus control, frozen ET versus control and overlapping between the two groups. (D and E) DNA methylation in two representative CpGs from GABRG3: *P = 0.0076, **P = 0.0001 and (F and G) ADARB2: *P = 0.006, **P = 0.037; ***P = 0.01.
GO analysis using methylGSA identified several pathways impacted in both frozen versus control (Supplementary Material, Table S7) and fresh versus control (Supplementary Material, Table S8) datasets, which included cell cycle regulation, DNA damage response, cellular response to external stimuli, and RNA regulation. Processes more highly impacted in the frozen versus control dataset compared with the fresh versus control dataset included cardiac development, signaling in innate immunity and inflammation, steroid hormone response and epigenetic regulation.
We also compared DNA methylation at specific CpG sites within our candidate genes to determine if either frozen or fresh ET samples were more likely to resemble control unassisted conceptions. However, results were inconsistent with certain CpGs within our two candidate genes more similar between frozen ET and controls (Fig. 4D and F), and others more similar between fresh ET and controls (Fig. 4E and G).
Collectively, these data indicate that placentas after frozen ET are hypermethylated compared with fresh ET-derived placenta and that frozen and fresh ET samples differ in terms of methylation when compared with placentas following unassisted conceptions, with no one technique leading to methylation levels more similar to unassisted conceptions.
Controlling for the host, frozen ET results in hypermethylation in human placenta
Genetic variability in humans has led to the question of whether observed epigenetic changes are truly the result of ART interventions, or whether the observed changes are owing to genetic variation or susceptibility related to the host/uterine environment. We, therefore, analyzed nine pairs of siblings in pregnancies achieved in the same patients, with one offspring born following fresh ET and another born after frozen ET. An independent paired analysis of these 18 samples was carried out. Among pairs, there were no significant differences in maternal age, a number of cleavage stages versus blastocyst transfers, oxygen tension of embryo culture, insemination [intracytoplasmic sperm injection (ICSI) vs. conventional] or birth outcomes between the pregnancies conceived following fresh or frozen ET (Supplementary Material, Tables S1 and S2). In this subanalysis, we interrogated 732 466 CpGs (note that some CpGs were excluded owing to the small sample size) and found 4855 differentially methylated CpGs (P < 0.05 and delta ß > 0.05), with 2837 hypermethylated CpGs and 2018 hypomethylated CpGs in frozen compared with fresh ET placentas. These differentially methylated CpGs fell within 2152 genes, of which 407 genes showed more than 1 significantly different CpG, 154 genes showed more than 2 affected CpGs and 67 genes contained more than 3 significantly differentially methylated CpGs (Fig. 5A). Analysis of individual CpG methylation within the two candidate genes examined in the initial large cohort, confirmed significant hypermethylation in frozen compared with fresh ET samples (Fig. 5B and C), validating the findings from the larger cohort (Supplementary Material, Table S3). Even in this relatively small cohort, we found 433 genes that overlapped with those identified in the larger cohort. Of these genes, 398 genes had CpGs all hypermethylated in the larger cohort and 228 of these genes had all CpGs hypermethylated in the pair analysis (the remainder had both hyper- and hypomethylation in the paired analysis). Of the 24 genes that had all the CpGs hypomethylated in the larger cohort, 16 were similarly hypomethylated in the paired analysis. These findings indicate that changes in methylation occur following frozen ET despite controlling for maternal genetics and uterine environment.
Figure 5.
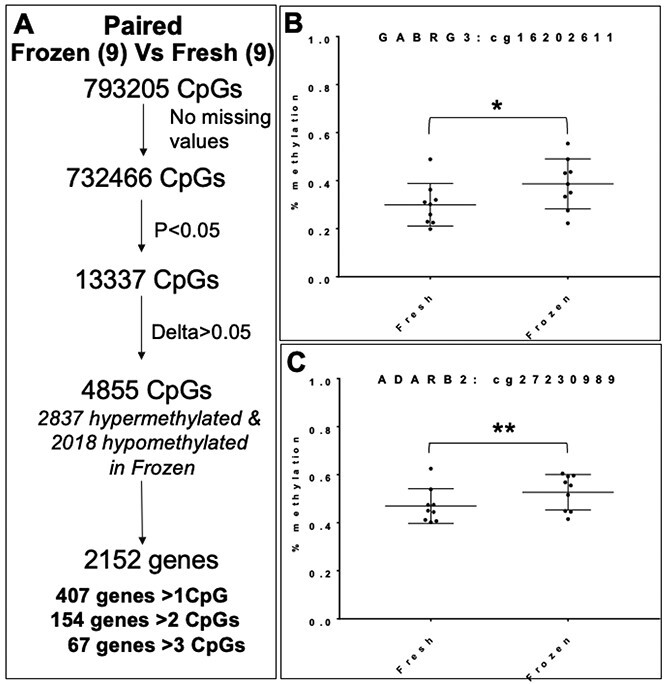
Analysis of fresh versus frozen ET in the same patient/uterus: (A) Paired analysis of siblings showing methylation differences in human placentas from frozen ET compared with placentas from fresh ET. (B and C) Paired analysis of DNA methylation in representative CpGs from two candidate genes: GABRG3 (B) and ADARB2 (C). *P = 0.04, **P = 0.02.
Hypermethylation in placenta after frozen ET is driven by changes in male placentas
We next analyzed methylation array data in a sex-specific manner to identify differences in male compared with female placentas. From our existing cohort of samples and methylation data, we performed a second subanalysis on female (39) and male (41) placentas after fresh ET; and female (33) and male (30) placentas after frozen ET. After filtering for significantly different CpGs, female placentas showed 7441 differentially methylated CpGs, of which 6118 CpGs were hypermethylated and 1323 were hypomethylated (Fig. 6A) when frozen ET placentas were compared with fresh ET. Male placentas showed a considerably larger effect with more than twice as many differentially methylated CpGs: 15572 CpGs were differentially methylated, of which 15 273 CpGs were hypermethylated and 299 CpGs were hypomethylated in male placentas following frozen ET compared with fresh ET (Fig. 6B). Accordingly, a larger number of affected genes were found in the male placenta: 4399 genes overall, with 1587 genes showing at least 1 affected CpG, 791 genes showing at least 2 affected CpGs and 488 genes showing methylation differences in at least 3 CpGs. Female placentas had 3070 genes impacted, with 774 genes containing at least 1 affected CpG, 326 genes with at least 2 CpGs affected and 155 genes with at least 3 CpGs affected. We examined these trends in our two chosen candidate genes described earlier and found that the significant differences in methylation were driven by the male placentas (Fig. 6C and D). In ADARB2, when frozen and fresh ET were compared, nine CpGs were significantly different when placentas from male and female offspring were analyzed together. However, in our sex-specific analysis, 49 CpGs were significantly differentially methylated in male samples while only 4 were significantly different in female placentas. Similarly, in GABRG3, 7 CpGs were differentially methylated in the combined cohort; however, when analyzed by sex, 19 CpGs were differentially methylated in male placentas, while only 2 were differentially methylated in female placentas (Supplementary Material, Table S3). This was true for additional genes as well: for example, in LARGE, 18 CpGs were differentially methylated between placentas following fresh and frozen ETs when all samples were examined, but sex stratification yielded 25 significant CpGs in male placentas and 5 differentially methylated CpGs in female placentas (Supplementary Material, Table S3). Overall, of the 156 genes with at least 2 significantly changed CpGs (P < 0.05, delta or mean methylation difference > 0.05) when male and female placentas were analyzed together (Fig. 3A), 118 genes contained more significantly differentially methylated CpGs in male placentas, 29 genes were more significantly changed in female placentas and 9 contained equal numbers of affected CpGs in male and female placentas.
Figure 6.
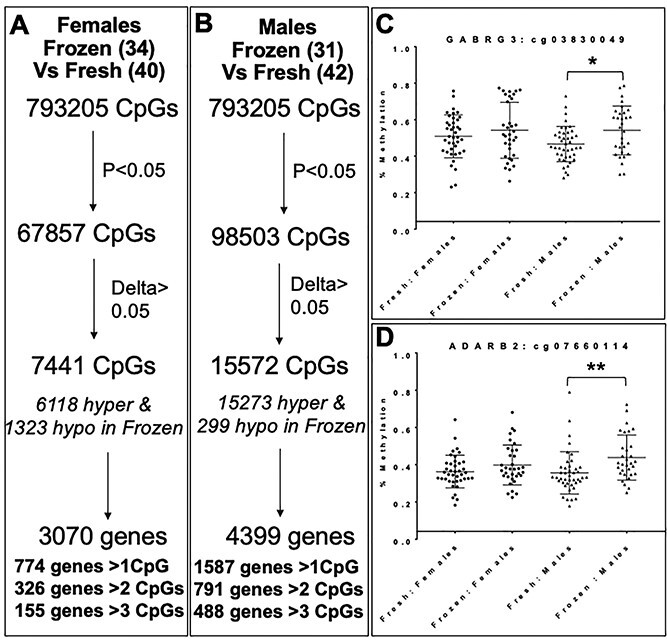
Sex-specific analysis: (A and B) Methylation differences in human placentas from frozen ET compared with placentas from fresh ET in female (A) and male (B) offspring. (C and D) DNA methylation in females and males at a representative CpGs from two candidate genes: GABRG3 (C) and ADARB2 (D). *P = 0.02, **P = 0.006.
According to GO analysis, CpGs differentially methylated in male placentas after frozen ET (Supplementary Material, Table S9) were highly enriched in processes associated with endothelial cell migration, synaptic signaling and glucose metabolism, while CpGs from female placentas after frozen ET were involved in cell cycle and cell death processes (Supplementary Material, Table S10).
We also carried out a sex-specific analysis on placentas after frozen ET and fresh ET when compared with unassisted control placentas (Fig. 7). In female placentas after frozen ET, we found 9082 hypermethylated and 9541 hypomethylated CpGs, which fell within 6054 genes, of which 723 genes contained more than 3 CpGs that were differentially methylated (Fig. 7A). In male placentas, differentially methylated CpGs were overwhelmingly hypermethylated (15064), while only 2122 CpGs were hypomethylated. These CpGs occurred within 4970 genes, of which 718 genes contained more than 3 differentially methylated CpGs (Fig. 7B). Female placentas after fresh ET, on the other hand, contained 10 452 hypomethylated CpGs, while only 3509 CpGs were hypermethylated when compared with control placenta. These CpGs fell within 3531 genes, of which 636 genes contained more than 3 differentially methylated CpGs (Fig. 7C). Male placenta showed a more even distribution of hypomethylated (6748) and hypermethylated (8116) CpGs, which occurred within 4225 genes of which 644 genes contained more than 3 differentially methylated CpGs (Fig. 7D).
Figure 7.
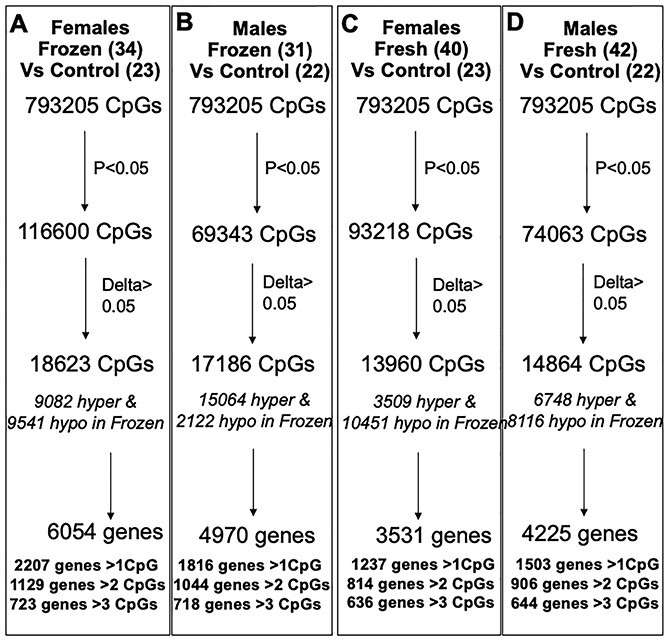
Sex-specific analysis: (A and B) Methylation differences in human placentas from frozen ET compared with control placentas in female (A) and male (B) offspring. (C and D) Methylation differences in human placentas from fresh ET compared with control placentas in female (C) and male (D) offspring.
Differentially methylated CpGs in male placentas after frozen ET were enriched in gene sets associated with methyltransferase activity and response to external stimuli such as toxic substances, reactive oxygen species and nutrient levels (Supplementary Material, Table S11). Frozen ET in female placenta compared with control placenta impacts genes involved with RNA regulation and checkpoint signaling (Supplementary Material, Table S12). After fresh ET, differentially methylated CpGs in female placentas were associated with lipid biosynthesis and regulation of innate immune response among others (Supplementary Material, Table S13) while CpGs in male placentas were associated with cytokine signaling and mitochondrial biogenesis (Supplementary Material, Table S14).
Validation of hypermethylation in placenta after frozen ET
We validated our array findings in an independent cohort of placentas following 77 frozen ET and 79 fresh ET. We selected one gene of interest, GABRG3, and ran a pyrosequencing assay for CpGs in close proximity (within 50 bp) of a significant CpG (cg19843449) that was hypermethylated in male frozen ET placental samples. Our pyrosequencing assay covered two CpGs and one of these CpGs showed a significant difference (P = 0.0029) between frozen (mean methylation = 72.87) versus fresh samples (mean methylation = 64.71). Interestingly, this CpG also showed significant (P = 0.0052) hypermethylation (mean methylation difference = 10.67) in male frozen ET placental samples (n = 33) compared with male fresh ET placental samples (n = 37). However, no significant difference (P = 0.1251) was observed for female frozen ET (n = 44) versus female fresh ET (n = 42) placental samples.
Mouse model studies - Embryo vitrification leads to mouse placental hypermethylation, particularly in male placenta
In parallel, we conducted analogous experiments in the mouse, which eliminated confounders of infertility, maternal age, genetic background and variabilities in clinical and laboratory methodologies. We first compared DNA methylation in E18.5 placentas following frozen ET (n = 8) and fresh ET (n = 8) and identified 589 significantly differentially methylated regions (DMRs) in placental samples. Similar to findings from our human samples, the majority of these DMRs (509) were hypermethylated while only 80 DMRs were hypomethylated in placentas following frozen ET compared with fresh ET. These DMRs were located within 445 genes (Fig. 8A, Supplementary Material, Table S15). GO analysis using the GOnet tool (38) on genes corresponding to DMRs highlighted potential roles in the regulation of growth and in the response to external stimuli (Supplementary Material, Table S16).
Figure 8.
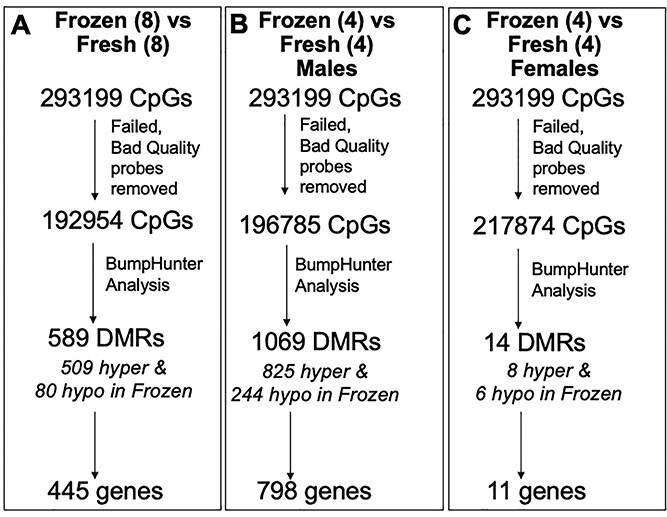
(A) Methylation differences in E18.5 mouse placentas from frozen ET compared with placentas from fresh ET. (B and C) Sex-specific analysis of DNA methylation in mouse placentas. DMR: Differentially methylated regions.
Sex-specific analysis in mice demonstrated even more significant differences between male and female placentas. In the placentas of male offspring, there were 1069 DMRs (825 hypermethylated and 244 hypomethylated following frozen compared with fresh ET) within 798 genes (Fig. 8B, Supplementary Material, Table S15). In female placentas, however, only 14 DMRs were significantly different (8 hypermethylated and 6 hypomethylated following frozen compared with fresh ET) and these fell within 11 genes (Fig. 8C, Supplementary Material, Table S15). GO analysis on genes that contained DMRs in male samples suggested similar functions as those seen in the larger dataset—developmental growth, morphogenesis, cell migration and cell differentiation were among the top GO terms identified (Supplementary Material, Table S17). The number of DMRs changed in female samples was too small to run GO analysis on. We next investigated whether the effects of vitrification on changes in methylation also resulted in changes in gene expression in candidate genes known to be disrupted by ART: in Mest/Peg1 and Tcf4, which were chosen based on highly significant differences in methylation and biological plausibility, we did not see any significant changes in gene expression in male term placenta.
To examine if frozen or fresh ET in mice led to methylation levels that were more similar to control placentas, we conducted independent comparisons of placentas following frozen (n = 8) or fresh (n = 8) ET to placentas following natural conception (n = 8). Frozen ET compared with controls produced a higher number DMRs (1787), of which 674 were hypermethylated and 1139 were hypomethylated. These DMRs were within 1319 genes (Supplementary Material, Fig. S1A, Supplementary Material, Table S18). Comparison of fresh ET placentas relative to control placenta revealed 1119 DMRs, with 114 hypermethylated and 1005 hypomethylated, corresponding to 808 genes (Supplementary Material, Fig. S1B, Supplementary Material, Table S19). There was substantial overlap between the DMRs identified when comparing placentas following fresh and frozen ET to controls, with 294 genes exhibiting disruptions in methylation following both fresh and frozen ET when compared with controls (Supplementary Material, Fig. S1C). GO analysis identified processes associated with cell motility, cell migration and growth regulation when frozen ET placentas were compared with control placentas (Supplementary Material, Table S20). Examining genes disrupted in fresh ET placentas compared with control placentas suggested different potential functions, with nervous system development, transcription and signaling featuring highly among GO terms identified (Supplementary Material, Table S21).
We also carried out a sex-stratified analysis on these datasets and found that male placentas after frozen ET compared with controls contained 882 DMRs in 717 genes, and male placentas after fresh ET compared with controls contained 92 DMRs in 69 genes (Supplementary Material, Fig. S2A and C, Supplementary Material, Tables S18 and S19). In this subanalysis, female placentas showed more perturbation than male placentas by both frozen (1363 DMRs, 1109 genes) and fresh ET (1118 DMRs, 812 genes) (Supplementary Material, Fig. S2B and D, Supplementary Material, Tables S18 and S19). All comparisons showed more hypomethylated versus hypermethylated DMRs. Male placentas after frozen ET showed disruptions in genes involved in the regulation of metabolic processes and gene expression through RNA biosynthesis and transcription (Supplementary Material, Table S22). There were too few genes impacted in male placentas after fresh ET to run GO analysis on. Female placentas after frozen ET contained DMRs within genes associated with nervous system development, cell differentiation and morphogenesis (Supplementary Material, Table S23), and female placentas after fresh ET showed genes associated with remarkably similar functions—nervous system development, cell signaling and differentiation and regulation of RNA biosynthesis and transcription (Supplementary Material, Table S24).
While the effects of IVF on placental methylation in mice have been closely examined by several laboratories in a gene-specific manner, a genome-wide unbiased examination is still lacking. We therefore also examined how IVF as a whole impacts DNA methylation in a mouse model. When comparing placentas derived after IVF (both fresh and frozen ET, n = 16) to control placentas (n = 8), we found 89 DMRs, of which 23 regions were hypermethylated while 66 regions were hypomethylated. These DMRs are localized to 74 genes (Supplementary Material, Fig. S1C, Supplementary Material, Table S25). GO terms identified included cell adhesion, cell signaling, growth and blood vessel development (Supplementary Material, Table S26). Male placentas after IVF showed only 51 DMRs within 41 genes, while female placentas contained a significantly larger number of DMRs (5081, within 3492 genes) (Supplementary Material, Fig. S2, Supplementary Material, Table S25). Male and female placentas showed a larger number of hypomethylated DMRs. While few genes were found impacted in male placenta after IVF, GO terms identified all pointed to a disruption in cell adhesion (Supplementary Material, Table S27). Interestingly, no GO terms were identified in the female placenta, despite a large number of genes identified that contained DMRs.
The role of ART and individual procedures in epigenetic disruption of imprinted genes has been very well characterized in the mouse model (39,40). We therefore also examined how vitrification and IVF as a whole impacted imprinted genes specifically when an epigenetic change was surveyed in a genome-wide manner. When placentas from fresh ET were compared with the placenta from frozen ET, we found significant methylation changes in 12 imprinted genes, representing 3% of all genes identified. Further, 11/12 of these genes were hypermethylated (Supplementary Material, Table S28). The percentage of genes impacted in females was markedly higher—18% though absolute numbers were low (two genes) owing to the minimal change seen in female samples overall. Both genes were hypomethylated (Supplementary Material, Table S28). As many as 21 imprinted genes were impacted in the male placenta, which represented 3% of all genes changed. All but three of these genes were hypermethylated (Supplementary Material, Table S28). When samples after frozen ET were compared with control placenta, 24 imprinted genes (2%) were impacted when all samples were examined; 21 genes (2%) when only female samples were examined and 21 genes (1%) when only male samples were examined. The vast majority of genes impacted by frozen ET in all the three datasets were hypomethylated (Supplementary Material, Table S28). Fresh ET resulted in similar effects on imprinted genes when all samples—19 genes (2%) and female samples—19 genes (2%) were considered, but male samples showed a markedly higher proportion of imprinted genes changed—9 genes, 14% of total genes. The majority of these genes were hypomethylated (Supplementary Material, Table S28).
Individual imprinted genes have been studied in great detail in the context of ART in mouse models. In this first genome-wide screen of genes impacted by IVF, we found seven imprinted genes, representing 9% of all genes containing DMRs, the majority of which were hypermethylated in frozen ET compared with fresh ET. Female samples contained 41 genes (1%), and male samples contained 2 genes (5%); all of which were hypomethylated (Supplementary Material, Table S28).
Weight changes owing to vitrification seen at E12.5 are resolved by E18.5
The use of the mouse also enabled us to investigate additional indicators of development. The phenotypical analysis included fetal and placental weight at mid-gestation (E12.5) immediately after the formation of the mature placenta and invasion of the trophoblast giant cells and at term (E18.5). Three groups were investigated at each time point: control (E12.5, n = 41; E18.5, n = 45); fresh ET (E12.5, n = 30; E18.5, n = 31) and frozen ET (E12.5, n = 28; E18.5, n = 26). Consistently high rates of fertilization, blastocyst development, freeze–thaw survival, pregnancy and implantation indicated IVF and embryo culture success and comparability among experiments (Supplementary Material, Fig. S3A). Because sex-specific analysis of DNA methylation in human placenta showed differences, we also investigated these measures stratified by sex, with comparable numbers of male and female embryos and placentas at each time point (Supplementary Material, Fig. S3B). At E12.5, and consistent with prior studies, the use of IVF and embryo culture overall led to significant decreases in fetal weight (Fig. 9A). While no differences were seen owing to vitrification overall, examination by sex indicated a female-specific decrease in fetal weight owing to vitrification (Fig. 9B). Placental weight at E12.5 was only significantly increased in fresh ET compared with control placenta (Fig. 9C), and separation by sex indicated that this was driven by increases in male placental weight (Fig. 9D). Both fresh and frozen ET led to a significant increase in placental weight compared with unassisted natural conception. As a measure of placental efficiency, we also investigated the fetal to placental weight ratio and found a significant decrease owing to IVF, which was consistent when examined by sex. At E18.5, the sex-specific effects of vitrification on fetal weight had resolved, and no significant changes owing to frozen ET were seen (Fig. 9E and F). However, an overall increase in placental weight was observed in both fresh and frozen ET compared with controls, indicating adaptation of the placenta to the effects of IVF and embryo culture (Fig. 9G and H).
Figure 9.
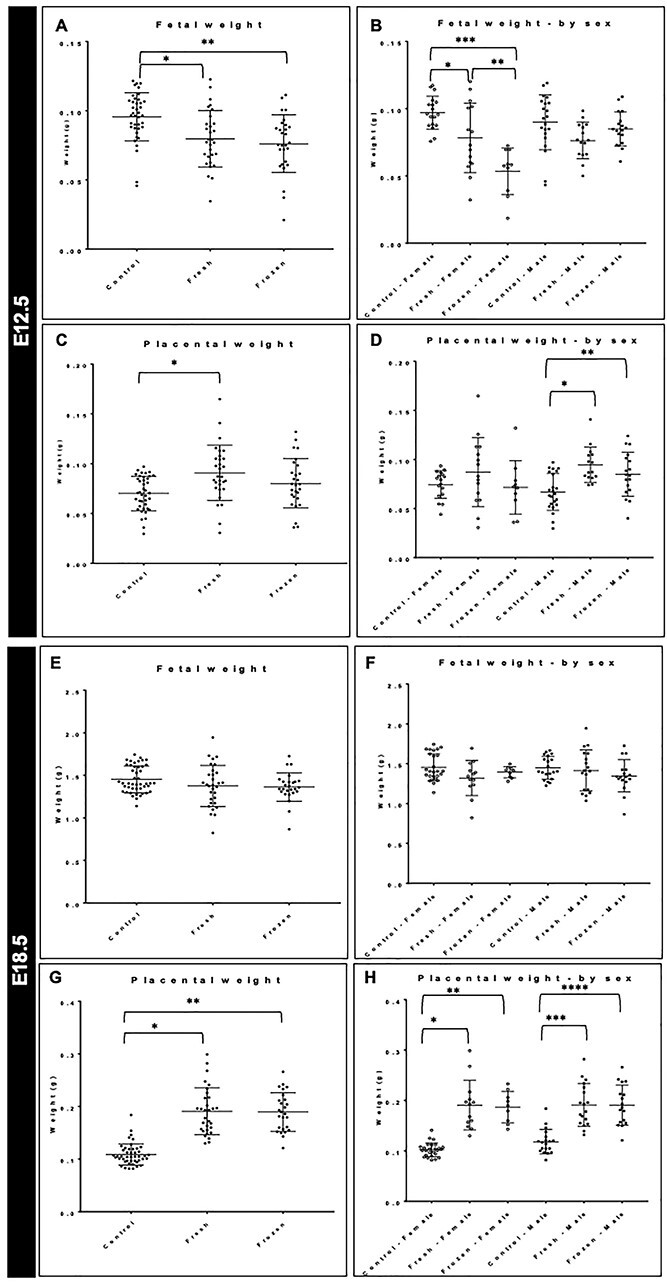
(A and B) Fetal weight in control, fresh and frozen ET at E12.5. *P = 0.003, **P = 0.0003 (A) and stratified by sex. *P = 0.03;***P < 0.0001; **P = 0.01 (B). (C and D) Placental weight in control, fresh and frozen ET at E12.5. *P = 0.0009 (C) and stratified by sex. *P = 0.0003; **P = 0.01 (D). (E and F) Fetal weight in control, fresh and frozen ET at E18.5 (E) and stratified by sex (F). (G and H) Placental weight in control, fresh and frozen ET at E18.5. *P < 0.0001; **P < 0.0001 (G) and stratified by sex. *P < 0.0001; **P < 0.0001; ***P < 0.0001; ****P < 0.0001 (H).
To further examine structural changes within the placenta, we measured cross-sectional areas of the junctional and labyrinth zones and determined the ratio of the junctional to labyrinth zone at E12.5 and E18.5, respectively. No significant changes in these ratios owing to embryo vitrification were seen, although both fresh and frozen ET differed from the control placenta at both time points (Supplementary Material, Fig. S4A, B, E and F). Previous work from our laboratories has shown an effect of hormonal stimulation utilized during IVF on placental microvessel density (15,29). We examined this measure in our current study by staining of placental histological sections with CD31, a marker of blood vessels. While a significant difference in microvessel density was observed particularly in female samples after vitrification, this difference was resolved by E18.5, when no significant changes owing to vitrification were noted (Supplementary Material, Fig. S4C, D, G and H).
Discussion
The use of ART for the treatment of infertility has increased dramatically with adjustments in clinical protocols and laboratory procedures leading to improvements in both pregnancy and live-birth rates. Importantly, the great majority of ART cycles result in uncomplicated pregnancies and apparently healthy offspring. Even though adverse perinatal outcomes following ART are relatively rare, they appear to be more common than those following natural conceptions. Moreover, critical outstanding questions remain concerning the roles: (1) of modifiable clinical and laboratory factors and (2) of the peri-conceptional environment in the development of such adverse outcomes. Understanding the mechanisms involved in the pathogenesis of adverse perinatal outcomes will lead to further optimization of the modifiable laboratory interventions and the peri-conceptional environment, both of which should lead to not only a further improvement of pregnancy rates but also to a reduction, if not elimination, of the adverse perinatal outcomes associated with ART.
In this study, we examined the effects of frozen ET on genome-wide methylation by comparing placentas resulting from the transfer of vitrified-warmed embryos during a programmed ET cycle to placentas resulting from fresh ET into a hormonally hyperstimulated environment. When examining global DNA methylation in placentas, we identified predominant hypermethylation of samples after frozen ET relative to fresh ET. Moreover, DNA methylation of placentas following a frozen ET in the same patient/uterus showed similar trends in methylation as the ones observed in the entire cohort of fresh versus frozen ET. This suggests that our findings are the result of perturbations related to the ART procedures and/or periconceptional hormonal milieu rather than maternal genetics or the uterine structural environment. The comparison of samples after either fresh or frozen ET to placentas obtained following unassisted conception showed epigenetic perturbations in both ART groups. This observation is significant because it suggests that neither ART procedure results in methylation patterns similar to unassisted natural conception and provides some explanation that both procedures result in epigenetic changes that may explain a resultant adverse perinatal outcome.
Although the rates of ongoing pregnancy and live births are approximately equivalent following fresh or frozen ET strategies (41), embryo cryopreservation has gained increasing favor owing to lower risks of ovarian hyperstimulation syndrome and the ability to carry out preimplantation genetic testing of embryos (42–45). However, both fresh and frozen ET results in an increased risk for adverse outcomes when compared with unassisted conceptions. While fresh ET results in higher rates of PTB, LBW and SGA compared with frozen ET, programmed frozen ET has been associated with higher rates of hypertensive disorders of pregnancy such as pre-eclampsia, high birth weight and LGA neonates when compared with fresh ET (42). DNA methylation has been used as a diagnostic and prognostic marker, and as a marker to track treatment and disease progression in multiple disease states including cancer, neurological and immunological conditions (46). Using a similar strategy, genes identified as perturbed following frozen ET compared with fresh ET may offer insights into molecular pathways and/or candidate genes playing a role in the pathogenesis of adverse perinatal outcomes observed following conception with ART. To this end, we found disruptions in methylation in genes with potential roles in early placentation that we chose to examine further: GABRG3 is present in a region associated with Prader–Willi and Angelman syndromes, disorders associated with defective imprinting of genes within the 15q11-q13 region (47). Other members of the GABA A receptor family have been associated with apoptosis of invasive trophoblast cells, and this receptor pathway was recently identified from multiple microarray datasets as being involved in the development of pre-eclampsia, a complication associated with ART and, more recently associated with pregnancies following frozen ET in hormonally programmed ET cycles. ADARB2 is an RNA-editing enzyme that binds to double-stranded RNA and can impact global gene expression (48). RNA editing has been found to be dysregulated in placental disorders such as pre-eclampsia (49). The methylation status of ADARB2 was recently identified as a marker for higher BMI (50), commonly seen in adults who were born LGA (51). Apart from individual genes, GO analysis of pathways impacted by frozen ET when compared with fresh ET or control samples revealed terms related to organ development, cell cycle regulation and cell-matrix adhesion. Soon after embryo implantation, fetal trophoblast invasion and remodeling of maternal spiral arteries occurs, which requires carefully regulated interactions between invading cells and the extracellular matrix (52). Pre-eclampsia, which occurs at a higher rate after frozen ET, is associated with inadequate invasion and remodeling, suggesting that genes involved in the regulation of cell adhesion could play a role in the pathophysiology of this disease (53). The molecular mechanisms underlying the development of macrosomia, or high birth weight, remain unknown, though recent studies have found a role in the regulation of fetal trophoblast proliferation suggesting an important role for cell cycle regulation in normal growth (54,55).
Human frozen ET differs from fresh ET not only in the exposure of the fertilized embryo to the freeze/thaw process but also in the hormonal environment during embryo implantation. Following fresh ET, there is a supraphysiologic hormonal environment that persists during the early implantation period. Therefore, it is difficult to assess if the methylation changes seen in the frozen ET cycles are secondary to the freeze/thaw process or the maternal peri-implantation hormonal environment which may directly or indirectly perturb the implantation and placentation processes. By paralleling our human studies with those using a mouse model, we have been able to isolate the role of the freeze–thaw process, removing factors that may confound the human data. It should be pointed out that other confounders, such as differences in IVF protocols, type of insemination (conventional or ICSI), oxygen tension during embryo culture and day of ET, can also potentially affect the results in humans. Using our unique mouse model, both fresh and vitrified/warmed mouse embryos, exposed to otherwise identical conditions both in vitro and following ET, can be combined and transferred into pseudopregnant (unstimulated) dams. Using new genome-wide methylation array technology recently released by Illumina, we demonstrate that in the mouse, embryo vitrification alone leads to significant changes in DNA methylation in the placenta when compared with embryos not subjected to the vitrification/warming process prior to ET. In the mouse placentas, as in human placentas, the majority of the DMRs (86%) were hypermethylated (509) in the placentas following frozen ET, suggesting that the vitrification/warming process was responsible for the increase in DNA methylation observed following fresh ET.
Though changes in DNA methylation following ART appear to be primarily stochastic, in our mouse model, as in the humans, we found changes in DNA methylation in regions critical to regulating fetal growth and placentation. For example, we identified significant changes in methylation at the Mest/Peg1 locus in placentas from vitrified embryos compared with fresh embryos. Multiple studies have identified this locus as particularly susceptible to changes in DNA methylation following ART (56,57). MEST/PEG1 plays a role in the regulation of fetal growth in mice and humans and was recently found to be downregulated in the placentas of women with early onset pre-eclampsia (58–60). Additionally, via GO analysis, several terms related to growth and developments were identified. These results reinforce the validity of using genome-wide methods for the discovery of genes and pathways playing a role in adverse pregnancy complications following frozen ET.
Most significantly, our mouse studies demonstrate significantly more perturbations in placental DNA methylation in male than in female offspring, paralleling what we uncovered in our human data when placentas after frozen ET were compared with fresh ET. Sex-stratified analysis when placentas after fresh or frozen ET were compared with placentas after unassisted conception did not show particular sensitivity of either males or females to either ET type when human samples were examined. However, the predominance of hypermethylation particularly in male frozen samples reiterates the susceptibility of male placentas to embryo cryopreservation, while the changed hormonal environment that occurs during a fresh ET cycle appears to result in hypomethylation in female placentas. Mouse samples show a bias for methylation perturbation in females when fresh and frozen ET are compared with controls. This could be a real difference between the systems or could result from the variability in human samples resulting from genetic background and procedures which could make it more difficult to observe sex-specific differences. This difference may also be a consequence of the vastly different representation of CpGs on the two arrays. Trends observed in both human and mouse samples were not specific to any particular genomic feature, mimicking what was represented on the arrays, and were consistent when imprinted and non-imprinted genes were examined. Sexual dimorphism in terms of differences in embryo metabolism, gene expression and epigenetics begins during the preimplantation period at the blastocyst stage and occurs in response to environmental stressors and manipulations such as those employed during ART (61). With respect to ART, epidemiologic data have found differences in birth weight between babies born following frozen ET and fresh ET by sex, with male infants more likely to be born LGA compared with female infants following frozen ET (62,63). In support of these initial findings, studies have found that the placenta is a major source of sexual dimorphism both physiologically in terms of transcriptional, growth and nutrient efficiency, and in response to environmental stressors, with the female placenta being more adaptive (64,65). Further work is necessary to understand the mechanism via which sexually dimorphic responses are elicited after the use of ART, by exploring sex-linked genes impacted in our dataset or by considering maternally secreted factors. This finding also highlights the importance of examining and analyzing adverse outcomes and epigenetic perturbations seen after ART in a sex-specific manner.
In addition to examining the difference in DNA methylation following frozen and fresh ET, we analyzed a subset of placentas from unassisted term conceptions to determine whether frozen or fresh ET led to more perturbations in DNA methylation. Evaluation of both human and mouse placentas, both frozen and fresh ET had significant effects on DNA methylation and both differed from DNA methylation in placentas following unassisted conception. This finding is not surprising as both interventions have independently been linked to increased risks of adverse outcomes observed following these ART procedures. These data reinforce the need to continue investigating the effects of ART interventions before altering clinical protocols.
Finally, our work offers a database of epigenetic changes observed in human and mouse placenta following fresh ET and frozen/thawed ET. Our findings help identify potential genes and pathways important to implantation and placentation and offer insight toward understanding the development of complications observed following ART treatments. Ultimately, our improved understanding of molecular mechanisms coupled with improved ART methodologies to mitigate DNA methylation changes introduced by procedures could offer parallel strategies to reduce adverse perinatal outcomes associated with ART.
Materials and Methods
Compliance with ethical regulations
The research performed complies with ethical regulations set forward by the University of Pennsylvania. Human tissue was collected at the Hospital of the University of Pennsylvania under an IRB-approved protocol (#804530). All patients underwent an established protocol for reviewing and obtaining informed consent. Specifically, the informed consent form was reviewed in detail with the patient and signed by both the patient and the study coordinator. In accordance with current regulations, no compensation was provided for obtaining human placental tissue. All animal studies were carried out in accordance with the principles and procedures of the Institutional Animal Care and Use Committee (#803523).
Human patient recruitment and sample collection
Placentas were acquired from term deliveries resulting from IVF pregnancies and unassisted conceptions (controls). For IVF pregnancies, all patients underwent fertility treatment at Penn Fertility Care between 2012 and 2020 using standard clinical and laboratory protocols. Superovulation was performed using recombinant follicle-stimulating hormone and/or urinary human menopausal gonadotropins. Gonadotropin dose and protocol were chosen based on patient characteristics and were adjusted during stimulation as clinically indicated based on patient’s response. Final oocyte maturation was induced with human chorionic gonadotropin and/or leuprolide acetate followed by transvaginal egg retrieval 36 h later. Fertilization was either via conventional insemination or by ICSI (performed for either male factor or, in some cases, unexplained infertility as clinically indicated). Embryo culture was performed in microdroplets under oil. Embryos were either transferred to the uterus on days 3 or 5 or frozen by vitrification on days 5 or 6. Three patients were included that had embryos frozen at the two pronuclear (zygote) stage followed by cleavage stage-ET. Luteal support was provided by intramuscular progesterone (50 mg daily). Frozen ET cycles were performed in a hormonally programmed cycle with increasing doses of oral micronized estradiol (2–6 mg daily) followed by intramuscular progesterone to induce a receptive for implantation of the endometrium. Control placentas from unassisted conceptions were collected from the Labor and Delivery Unit of the Hospital of the University of Pennsylvania, from patients with no comorbidities. Placentas were collected at delivery and placental tissue was excised from the maternal side opposite the cord insertion site, rinsed and stored at −80°C as previously described (9,10,66).
DNA and RNA extraction
Placental genomic DNA was extracted using the Invitrogen PureLink™ Genomic DNA kit (Thermo Fischer Scientific) according to the manufacturer’s instructions. The isolated DNA was dissolved in 50 μl elution buffer and stored at −20°C until further use. RNA was extracted using RNeasy plus mini kit (Qiagen), following the manufacturer’s guidelines. The isolated RNA was resuspended in Milli-Q purified water and stored at −80°C until further use.
Human Illumina 850 K methylation array
Sample preparation
To interrogate genome-wide differences in placental DNA methylation between frozen and fresh ET pregnancies, we utilized the Illumina MethylationEPIC BeadChip which examines over 850 000 CpGs representing gene bodies; regulatory regions including promoters, 5′/3′untranslated regions and enhancers; and intergenic regions. DNA samples were processed at the Penn State University Genome Sciences Facility. The genomic DNA was converted to bisulfite DNA using the Zymo EZ DNA methylation kit using the manufacturer’s protocol for sodium bisulfite conversion of unmethylated cytosines in DNA from low concentration solutions.
Array
The automated protocol for the Infinium HD methylation assay was used for the array run. Briefly, the bisulfite-converted DNA was amplified and then enzymatically fragmented. The fragmented DNA was then precipitated and resuspended followed by hybridization to BeadChips. The unhybridized and non-specifically hybridized DNA sample was washed. The primers hybridized to the DNA were extended using labeled nucleotides and stained. The BeadChip was imaged using the iScan System. The project file was generated from the raw IDAT files using the GenomeStudio version 2011.1. MethylationEPIC_v-1-0_B4.bpm manifest file was used for the downstream data processing.
Human data analysis
The study samples were run in three batches on EPIC arrays. All the samples passed quality control with a mean detection P-value < 0.01. CpG sites failing in more than 20% of the samples were excluded. We provided the sex of the samples to the methylation array facility and the sex was confirmed by the facility. No discordance in the sex was observed for any samples. Probes located on sex chromosomes and probes overlapping/close to SNPs were included in the analyses. A total of 831 858 CpGs were included for further analysis. Mean values were calculated for the frozen and fresh ET groups in each of the three arrays. Bland Altman plots were generated for comparison of mean methylation values between the arrays in fresh and frozen groups. CpG sites not in Bland Altman agreement between the arrays for both fresh and frozen groups were excluded from the analysis. A total of 793 205 CpGs were included for differential methylation analysis between the fresh and frozen groups. Differentially methylated CpGs with a P-value less than 0.05 in the two-tailed t-test and a mean methylation difference of greater than 0.05 were considered to be differentially methylated between the frozen and the fresh groups. GO analysis was done using the methylGSA package in R, and overlap analysis for DNase 1 hypersensitivity sites was carried out using the eFORGE web-based tool (https://eforge.altiusinstitute.org).
Validation by pyrosequencing
We selected one of the genes of interest, GABRG3, for validation by pyrosequencing. We prepared the samples (DNA extraction and bisulfite conversion) as described earlier. Pyrosequencing primers were designed using PyroMark Assay Design Software 2.0. Bisulfite-converted DNA was amplified using designed polymerase chain reaction (PCR) primers followed by pyrosequencing using a designed sequencing primer in PyroMark Q48 as per the manufacturer’s protocol.
Human gene expression
RNA was converted to cDNA using the SuperScript IV first-strand synthesis system (Invitrogen) according to the manufacturer’s protocol. Predesigned TaqMan assays and TaqMan gene expression master mix were used for quantitative real-time RT PCR. Gene expression was quantified on QuantStudio 3 real-time PCR system using comparative cycle threshold (CT) analysis. The geometric mean of three housekeeping genes (TBP, SDHA and YWHAZ) was used to normalize the expression of genes of interest.
Mouse embryo collection and transfer
IVF and embryo culture
Six-week-old mice were obtained and housed in a temperature- and light-controlled environment with a 12 h dark/12 h light cycle and fed ad libitum. Female CF1 mice (Envigo Laboratories, Indianapolis, IN) were superovulated with intraperitoneal injections of 5 IU of pregnant mare serum gonadotropin (PMSG, Lee Biosolutions, Maryland Heights, MO) followed by 5 IU of hCG (Sigma-Aldrich, St. Louis, MO) 48 h later. Approximately 14 h post-hCG injection, cumulus–egg complexes were harvested from the oviducts and placed in TYH (Toyoda, Yokoyama, Hoshi, modified Krebs-Ringer Solution) medium overlayed with mineral oil (Fujifilm/Irvine Scientific, Santa Ana, CA). Mature sperm were collected from the cauda epididymides of male mice (SJLB6) heterozygous for GFP. Sperm were capacitated for 90 min in the TYH medium under mineral oil and then added to the egg droplets. Eggs were incubated with sperm for 3 h at 37°C in a humidified atmosphere of 5% O2, 5% CO2 and 90% N2. Following insemination, eggs were rinsed in drops of HEPES-buffered MHM (Fujifilm/Irvine Scientific, Santa Ana, CA) to remove the sperm and placed in KSOM with amino acids (Sigma-Aldrich, St. Louis, MO) for culture to the blastocyst stage.
Blastocyst vitrification and thawing/warming
On day 3.5 of culture, embryos that had reached the blastocyst stage were sorted by GFP status. Embryos were either vitrified (for up to 9 months) or transferred fresh. Vitrification was performed using Vit Kit-Freeze NX per the manufacturer’s instructions (Fujifilm/Irvine Scientific, Santa Ana, CA). Briefly, blastocysts were held in equilibration solution for 5 min before being rinsed quickly through three drops of vitrification solution and transferred directly to liquid nitrogen using a cryolock vitrification device (Fujifilm/Irvine Scientific, Santa Ana, CA). Embryos were stored in a liquid nitrogen tank (Taylor-Wharton, Minnetonka, MN) until thawing/warming for transfer. Blastocysts were warmed using a Vit Kit-Warm NX (Fujifilm/Irvine Scientific, Santa Ana, CA). Cryolocks with blastocysts were placed in TS (warming solution), the embryos recovered and after 1 min placed in dilution solution to equilibrate for 4 min. The blastocysts were subsequently passed through two wash steps of 4 min each, then placed in equilibrated KSOM with amino acids under oil to recover before transfer.
Embryo transfer
Day 3.5 blastocysts were non-surgically transferred into pseudopregnant CF1 female mice derived through natural mating to vasectomized B6D2F1/J males (Jackson Laboratory, Bar Harbor, ME). The presence of a copulatory plug on post-coitum day 0.5 confirmed mating. Ten blastocysts, 5 fresh and 5 vitrified/warmed, were transferred into a single horn of each pseudopregnant female on post-coital day 3.5 using the Non-Surgical Embryo Transfer Device (Paratechs, Lexington, KY) per the manufacturer’s protocol. GFP status was used to distinguish whether the blastocysts were fresh or vitrified/warmed. Reciprocal transfers were performed to ensure that the observed changes were not owing to the GFP transgene.
Naturally conceived controls
Naturally conceived controls were generated by mating CF1 females with male mice (SJLB6- heterozygous for GFP). The morning that a vaginal plug was observed was designated as day 0.5.
Mouse tissue collection and evaluation
Pregnant mice, following either natural mating or ET, were sacrificed on either embryonic day (E) 12.5 or 18.5. The number of implantation sites and resorption sites were counted for each recipient, the amniotic membranes ruptured and the concepti (fetus and placenta) were carefully dissected from the uterine horn. The fetus and placenta were separated and weighed. Each placenta was bisected through the mid-placental plane, and half the placenta was placed in 10% phosphate-buffered formalin for histologic examination. The other half of the placenta was snap-frozen and stored at −80°C. GFP status of the fetus was determined by PCR to ascertain whether it originated from fresh or vitrified/warmed blastocysts.
DNA and RNA extraction: mouse samples
Placental halves were homogenized in a lysis buffer and the lysate was divided and allocated for either RNA or DNA extraction. Proteinase K (Denville Sci, South Plainfield, NJ) was added to the lysate for DNA extraction at a concentration of 10 μg/ml and the lysate was incubated for 1 h at 55°C. An equal volume of phenol–chloroform/isoamyl alcohol (Sigma Aldrich, St. Louis, MO) was added, mixed well and centrifuged for 5 min at room temperature. The aqueous layer was collected, and isopropanol and 7.5 M ammonium acetate were added to precipitate the DNA. After a 10-min incubation, the sample was centrifuged for 5 min, and the DNA pellet was washed with 80% ethanol. The ethanol was removed, and the DNA was air dried and resuspended in nuclease-free water. RNA was isolated using an RNEasy Plus Mini Kit. The extracted RNA was quantified using a NanoDrop 2000 (ThermoFisher Scientific).
Infinium mouse methylation BeadChip array
DNA (250–750 ng) was treated with sodium bisulfite using the Qiagen EpiTect Bisulfite Conversion kit. DNA methylation was quantified using the Illumina Infinium MouseMethylation-12v1–0 BeadChip (Illumina, CA) run on an Illumina iScan System (Illumina, CA) using the manufacturer’s standard protocol. The samples were processed at the Center for Applied Genomics Genotyping Core at the Children’s Hospital of Philadelphia. The Illumina Infinium Mouse Methylation-12v1–0 BeadChip includes 284 760 CG probes, 1352 rs probes and 938 Ch probes (287 050 probes total) as defined in ‘Infinium Mouse Methylation v1.0 A1 GS Manifest File.bpm’.
Mouse data analysis
Raw IDAT files were processed with the SeSAMe R package (v1.10.4) and the MM285 array manifest file that comes with the package (vM25). Briefly, SeSAMe sets quality masks for probes, computes detection P-value using out-of-band probe’s empirical distribution and produces methylation β-values, which were used for all downstream analyses. To compare different experimental groups, two-tailed t-tests were applied for each probe. We defined significance as probes whose nominal P-value was less than 0.05 and methylation difference was greater than 5%. Nearby genes were assigned to probes based on proximity using the Rpackage annotator (67). Downstream analysis was conducted using the mm10/GRCm10 mouse genome assembly.
Because considerably fewer samples were analyzed in the mouse studies compared with the human, we utilized BumpHunter for the analysis. This methodology identifies contiguous, DMRs of interest, which are all either hypo- or hypermethylated, instead of focusing on individual CpGs (68). We performed 100 permutations using a maximum pairwise distance of 250 bp and used the 95th percentile of calculated effect size estimates as the threshold (coef = 1, cut-off = 0.1, B = 100, maxGap = 250, pickCutoffQ = 0.95). DMRs annotations were obtained using the annotatePeaks function of the Rpackage ChIPSeeker (69) (version 1.22.1; parameter: TxDb = TxDb.Mmusculus.UCSC.mm10.knownGene). Additional annotations and chromosomal distributions were obtained using the Rpackage annotator (67) modified for mouse genome analyses. GOnet (38): a tool for interactive GO analysis was utilized for GO analysis.
Mouse gene expression
We used 73 (24 natural, 27 fresh IVF and 22 ET frozen) samples from our total mouse cohort to analyze gene expression as previously described (17). Briefly, 1500 ng of RNA was treated with 1.5 μl of DNase followed by first-strand synthesis using Superscript III reverse transcriptase (Invitrogen, CA, USA) and random hexamer primers (Roche, Germany). All real-time PCR reactions were performed using 10 μl reactions consisting of 5.0 μl of SYBR green master mix (Applied Biosystems, MA, USA), 0.2 μl of 10 μM forward primer, 0.2 μl of 10 μM reverse primer and 4.6 μl of cDNA (final concentration: 1.09 ng/μl). CTs were detected using QuantStudio 7 Flex Real-Time PCR System (Life Technologies, CA, USA). Reaction efficiency (E) was estimated for each pair of primers using a standard curve and expression levels were quantified by measuring the CTs for each sample using the E(-Cts) method. All samples were run in triplicate. Relative expression was calculated using the quantified expression from the endogenous control B2M that had stable expression levels in mouse placentas across multiple samples and experimental groups.
Mouse histological analysis
Placentas were kept in formalin overnight at 4°C, dehydrated and embedded in a paraffin block with the placenta oriented to obtain appropriate cross-sections. Tissue sections of 4 μm thickness were mounted on glass slides, then deparaffinized and stained with hematoxylin and eosin, or with monoclonal antibody to PLVAP (MECA-32) (Bio-Rad, Raleigh, NC). Aperio ImageScope software (Leica Biosystems, Vista, CA) was used to view images, outline each placental zone and calculate areas and microvessel density.
Statistical analysis
All numerical data are represented as mean ± SD. All statistical analyses were performed using RStudio version 1.3.1093. GraphPad Prism version 9.1.2 was used for all the graphical representations.
Supplementary Material
Acknowledgements
We thank Richard Schultz, Jeremy Wang and Lacey Luense for their input.
Conflict of Interest statement. None declared.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Contributor Information
Sneha Mani, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Jayashri Ghosh, Cancer and Cellular Biology, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA.
Eric A Rhon-Calderon, Department of Cell and Developmental Biology, Epigenetics Institute, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
Yemin Lan, Department of Cell and Developmental Biology, Epigenetics Institute, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
Teri Ord, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Charikleia Kalliora, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Joe Chan, Cancer and Cellular Biology, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA.
Bryant Schultz, Cancer and Cellular Biology, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA.
Elaine Vaughan-Williams, Cancer and Cellular Biology, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA.
Christos Coutifaris, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Carmen Sapienza, Cancer and Cellular Biology, Fels Cancer Institute for Personalized Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA.
Suneeta Senapati, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Marisa S Bartolomei, Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Cell and Developmental Biology, Epigenetics Institute, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
Monica Mainigi, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Research on Reproduction and Women’s Health, University of Pennsylvania, Philadelphia, PA 19104, USA.
Funding
National Institutes of Health (P50 HD068157-06A1).
Author’s roles
S.M., J.G., T.O. and E.R.C. performed the experiments and analyzed the data. C.K. sourced, collected and logged human samples and carried out demographic analysis. J.C., B.S. and E.V-W. performed experiments and contributed data. S.M., C.C., C.S., S.S., M.S.B. and M.M. designed the research, analyzed the data and wrote the manuscript.
Data accessibility
The data underlying this article is available in the Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi and can be accessed using the deposition number GSE201496 for human samples and GSE196547 for mouse samples.
References
- 1. Sunderam, S., Kissin, D.M., Zhang, Y., Jewett, A., Boulet, S.L., Warner, L., Kroelinger, C.D. and Barfield, W.D. (2020) Assisted Reproductive Technology Surveillance-United States, 2017. MMWR Surveill. Summ., 69, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Services, U.D.o.H.a.H . (2018), in press.
- 3. Thomopoulos, C., Tsioufis, C., Michalopoulou, H., Makris, T., Papademetriou, V. and Stefanadis, C. (2013) Assisted reproductive technology and pregnancy-related hypertensive complications: a systematic review. J. Hum. Hypertens., 27, 148–157. [DOI] [PubMed] [Google Scholar]
- 4. Pandey, S., Shetty, A., Hamilton, M., Bhattacharya, S. and Maheshwari, A. (2012) Obstetric and perinatal outcomes in singleton pregnancies resulting from IVF/ICSI: a systematic review and meta-analysis. Hum. Reprod. Update, 18, 485–503. [DOI] [PubMed] [Google Scholar]
- 5. Sullivan-Pyke, C.S., Senapati, S., Mainigi, M.A. and Barnhart, K.T. (2017) In Vitro fertilization and adverse obstetric and perinatal outcomes. Semin. Perinatol., 41, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson, R.A., Gibson, K.A., Wu, Y.W. and Croughan, M.S. (2004) Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstet. Gynecol., 103, 551–563. [DOI] [PubMed] [Google Scholar]
- 7. Ghosh, J., Coutifaris, C., Sapienza, C. and Mainigi, M. (2017) Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin. Epigenetics, 9, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh, J., Mainigi, M., Coutifaris, C. and Sapienza, C. (2016) Outlier DNA methylation levels as an indicator of environmental exposure and risk of undesirable birth outcome. Hum. Mol. Genet., 25, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song, S., Ghosh, J., Mainigi, M., Turan, N., Weinerman, R., Truongcao, M., Coutifaris, C. and Sapienza, C. (2015) DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin. Epigenetics, 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katari, S., Turan, N., Bibikova, M., Erinle, O., Chalian, R., Foster, M., Gaughan, J.P., Coutifaris, C. and Sapienza, C. (2009) DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet., 18, 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doherty, A.S., Mann, M.R., Tremblay, K.D., Bartolomei, M.S. and Schultz, R.M. (2000) Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod., 62, 1526–1535. [DOI] [PubMed] [Google Scholar]
- 12. Mann, M., Lee, S., Doherty, A., Verona, R., Nolen, L., Schultz, R. and Bartolomei, M. (2004) Selective loss of imprinting in the placenta following preimplantation development in culture. Development, 131, 3727–3735. [DOI] [PubMed] [Google Scholar]
- 13. Rivera, R., Stein, P., Weaver, J., Mager, J., Schultz, R. and Bartolomei, M. (2008) Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Genet., 17, 1–14. [DOI] [PubMed] [Google Scholar]
- 14. de Waal, E., Vrooman, L.A., Fischer, E., Ord, T., Mainigi, M.A., Coutifaris, C., Schultz, R.M. and Bartolomei, M.S. (2015) The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum. Mol. Genet., 24, 6975–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weinerman R, Ord T, Bartolomei MS, Coutifaris C, Mainigi M. (2017) The superovulated environment, independent of embryo vitrification, results in low birthweight in a mouse model. Biol Reprod., 97, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doherty, A.S., Mann, M.R., Tremblay, K.D., Bartolomei, M.S. and Schultz, R.M. (2012) Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod., 62, 1526–1535. [DOI] [PubMed] [Google Scholar]
- 17. Vrooman, L.A., Rhon-Calderon, E.A., Chao, O.Y., Nguyen, D.K., Narapareddy, L., Dahiya, A.K., Putt, M.E., Schultz, R.M. and Bartolomei, M.S. (2020) Assisted reproductive technologies induce temporally specific placental defects and the preeclampsia risk marker sFLT1 in mouse. Development, 147, dev186551. 10.1242/dev.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maheshwari, A., Pandey, S., Shetty, A., Hamilton, M. and Bhattacharya, S. (2012) Obstetric and perinatal outcomes in singleton pregnancies resulting from the transfer of frozen thawed versus fresh embryos generated through in vitro fertilization treatment: a systematic review and meta-analysis. Fertil. Steril., 98(368–377), e361–e369. [DOI] [PubMed] [Google Scholar]
- 19. Wei, D., Liu, J.Y., Sun, Y., Shi, Y., Zhang, B., Liu, J.Q., Tan, J., Liang, X., Cao, Y., Wang, Z. et al. (2019) Frozen versus fresh single blastocyst transfer in ovulatory women: a multicentre, randomised controlled trial. Lancet, 393, 1310–1318. [DOI] [PubMed] [Google Scholar]
- 20. Pinborg, A., Henningsen, A.A., Loft, A., Malchau, S.S., Forman, J. and Andersen, A.N. (2014) Large baby syndrome in singletons born after frozen embryo transfer (FET): is it due to maternal factors or the cryotechnique? Hum. Reprod., 29, 618–627. [DOI] [PubMed] [Google Scholar]
- 21. Sazonova, A., Kallen, K., Thurin-Kjellberg, A., Wennerholm, U.B. and Bergh, C. (2012) Obstetric outcome in singletons after in vitro fertilization with cryopreserved/thawed embryos. Hum. Reprod., 27, 1343–1350. [DOI] [PubMed] [Google Scholar]
- 22. Roque, M., Lattes, K., Serra, S., Sola, I., Geber, S., Carreras, R. and Checa, M.A. (2013) Fresh embryo transfer versus frozen embryo transfer in in vitro fertilization cycles: a systematic review and meta-analysis. Fertil. Steril., 99, 156–162. [DOI] [PubMed] [Google Scholar]
- 23. Coetzee, K., Ozgur, K., Bulut, H., Berkkanoglu, M. and Humaidan, P. (2020) Large-for-gestational age is male-gender dependent in artificial frozen embryo transfers cycles: a cohort study of 1295 singleton live births. Reprod. BioMed. Online, 40, 134–141. [DOI] [PubMed] [Google Scholar]
- 24. Kalra, S.K., Ratcliffe, S.J., Coutifaris, C., Molinaro, T. and Barnhart, K.T. (2011) Ovarian Stimulation and Low Birth Weight in Newborns Conceived Through In Vitro Fertilization. Obstet. Gynecol., 118, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kansal Kalra, S., Ratcliffe, S.J., Milman, L., Gracia, C.R., Coutifaris, C. and Barnhart, K.T. (2011) Perinatal morbidity after in vitro fertilization is lower with frozen embryo transfer. Fertil. Steril., 95, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu, S.Y., Teng, B., Fu, J., Li, X., Zheng, Y. and Sun, X.X. (2013) Obstetric and neonatal outcomes after transfer of vitrified early cleavage embryos. Hum. Reprod., 28, 2093–2100. [DOI] [PubMed] [Google Scholar]
- 27. Pelkonen, S., Koivunen, R., Gissler, M., Nuojua-Huttunen, S., Suikkari, A.M., Hyden-Granskog, C., Martikainen, H., Tiitinen, A. and Hartikainen, A.L. (2010) Perinatal outcome of children born after frozen and fresh embryo transfer: the Finnish cohort study 1995-2006. Hum. Reprod., 25, 914–923. [DOI] [PubMed] [Google Scholar]
- 28. de Waal, E., Mak, W., Calhoun, S., Stein, P., Ord, T., Krapp, C., Coutifaris, C., Schultz, R.M. and Bartolomei, M.S. (2014) In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol. Reprod., 90, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weinerman, R., Ord, T., Bartolomei, M.S., Coutifaris, C. and Mainigi, M. (2017) The superovulated environment, independent of embryo vitrification, results in low birthweight in a mouse model. Biol. Reprod., 97, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sullivan-Pyke, C., Mani, S., Rhon-Calderon, E.A., Ord, T., Coutifaris, C., Bartolomei, M.S. and Mainigi, M. (2020) Timing of exposure to gonadotropins has differential effects on the conceptus: evidence from a mouse modeldagger. Biol. Reprod., 103, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Waal, E., Yamazaki, Y., Ingale, P., Bartolomei, M.S., Yanagimachi, R. and McCarrey, J.R. (2012) Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum. Mol. Genet., 21, 4460–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanna, C.W., Demond, H. and Kelsey, G. (2018) Epigenetic regulation in development: is the mouse a good model for the human? Hum. Reprod. Update, 24, 556–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt, A., Morales-Prieto, D.M., Pastuschek, J., Fröhlich, K. and Markert, U.R. (2015) Only humans have human placentas: molecular differences between mice and humans. J. Reprod. Immunol., 108, 65–71. [DOI] [PubMed] [Google Scholar]
- 34. Choux, C., Carmignac, V., Bruno, C., Sagot, P., Vaiman, D. and Fauque, P. (2015) The placenta: phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin. Epigenet., 7, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou, W., Hinoue, T., Barnes, B., Mitchell, O., Iqbal, W., Lee, S.M., Foy, K.K., Lee, K.-H., Moyer, E.J., VanderArk, A. et al. (2022) DNA Methylation Dynamics and Dysregulation Delineated by High-Throughput Profiling in the Mouse. bioRxiv. 10.1101/2022.03.24.485667. [DOI] [PMC free article] [PubMed]
- 36. Ren, X. and Kuan, P.F. (2019) methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics, 35, 1958–1959. [DOI] [PubMed] [Google Scholar]
- 37. Breeze, C.E., Paul, D.S., van Dongen, J., Butcher, L.M., Ambrose, J.C., Barrett, J.E., Lowe, R., Rakyan, V.K., Iotchkova, V., Frontini, M. et al. (2016) eFORGE: A Tool for Identifying Cell Type-Specific Signal in Epigenomic Data. Cell Rep., 17, 2137–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pomaznoy, M., Ha, B. and Peters, B. (2018) GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics, 19, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rhon-Calderon, E.A., Vrooman, L.A., Riesche, L. and Bartolomei, M.S. (2019) The effects of Assisted Reproductive Technologies on genomic imprinting in the placenta. Placenta, 84, 37–43. [DOI] [PubMed] [Google Scholar]
- 40. Mani, S. and Mainigi, M. (2018) Embryo Culture Conditions and the Epigenome. Semin. Reprod. Med., 36, 211–220. [DOI] [PubMed] [Google Scholar]
- 41. Stormlund, S., Sopa, N., Zedeler, A., Bogstad, J., Praetorius, L., Nielsen, H.S., Kitlinski, M.L., Skouby, S.O., Mikkelsen, A.L., Spangmose, A.L. et al. (2020) Freeze-all versus fresh blastocyst transfer strategy during in vitro fertilisation in women with regular menstrual cycles: multicentre randomised controlled trial. BMJ, 370, m2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maheshwari, A., Pandey, S., Amalraj Raja, E., Shetty, A., Hamilton, M. and Bhattacharya, S. (2018) Is frozen embryo transfer better for mothers and babies? Can cumulative meta-analysis provide a definitive answer? Hum. Reprod. Update, 24, 35–58. [DOI] [PubMed] [Google Scholar]
- 43. Coutifaris, C. (2017) Freeze-only in vitro fertilization cycles for all? Fertil. Steril., 108, 233–234. [DOI] [PubMed] [Google Scholar]
- 44. Coutifaris, C. (2016) "Freeze Only"--An Evolving Standard in Clinical In Vitro Fertilization. N. Engl. J. Med., 375, 577–579. [DOI] [PubMed] [Google Scholar]
- 45. Coutifaris, C. (2019) Elective frozen embryo transfer for all? Lancet, 393, 1264–1265. [DOI] [PubMed] [Google Scholar]
- 46. Ehrlich, M. (2019) DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics, 14, 1141–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee, S. and Wevrick, R. (2000) Identification of novel imprinted transcripts in the Prader-Willi syndrome and Angelman syndrome deletion region: further evidence for regional imprinting control. Am. J. Hum. Genet., 66, 848–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishikura, K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem., 79, 321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang, X., Yang, J., Liang, X., Chen, Q., Jiang, S., Liu, H., Gao, Y., Ren, Z., Shi, Y.W., Li, S. et al. (2020) Landscape of Dysregulated Placental RNA Editing Associated With Preeclampsia. Hypertension, 75, 1532–1541. [DOI] [PubMed] [Google Scholar]
- 50. Samblas, M., Milagro, F.I. and Martinez, A. (2019) DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics, 14, 421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Derraik, J.G.B., Maessen, S.E., Gibbins, J.D., Cutfield, W.S., Lundgren, M. and Ahlsson, F. (2020) Large-for-gestational-age phenotypes and obesity risk in adulthood: a study of 195,936 women. Sci. Rep., 10, 2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Damsky, C.H., Fitzgerald, M.L. and Fisher, S.J. (1992) Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Invest., 89, 210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou, Y., Damsky, C.H. and Fisher, S.J. (1997) Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J. Clin. Invest., 99, 2152–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li, J., Fu, Z., Jiang, H., Chen, L., Wu, X., Ding, H., Xia, Y., Wang, X., Tang, Q. and Wu, W. (2018) IGF2-derived miR-483-3p contributes to macrosomia through regulating trophoblast proliferation by targeting RB1CC1. Mol. Hum. Reprod., 24, 444–452. [DOI] [PubMed] [Google Scholar]
- 55. Guo, D., Jiang, H., Chen, Y., Yang, J., Fu, Z., Li, J., Han, X., Wu, X., Xia, Y., Wang, X. et al. (2018) Elevated microRNA-141-3p in placenta of non-diabetic macrosomia regulate trophoblast proliferation. EBioMedicine, 38, 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lazaraviciute, G., Kauser, M., Bhattacharya, S., Haggarty, P. and Bhattacharya, S. (2014) A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update, 20, 840–852. [DOI] [PubMed] [Google Scholar]
- 57. Huntriss, J.D., Hemmings, K.E., Hinkins, M., Rutherford, A.J., Sturmey, R.G., Elder, K. and Picton, H.M. (2013) Variable imprinting of the MEST gene in human preimplantation embryos. Eur. J. Hum. Genet., 21, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Deyssenroth, M.A., Marsit, C.J., Chen, J. and Lambertini, L. (2020) In-depth characterization of the placental imprintome reveals novel differentially methylated regions across birth weight categories. Epigenetics, 15, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lefebvre, L., Viville, S., Barton, S.C., Ishino, F., Keverne, E.B. and Surani, M.A. (1998) Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet., 20, 163–169. [DOI] [PubMed] [Google Scholar]
- 60. Deyssenroth, M.A., Li, Q., Escudero, C., Myatt, L., Chen, J. and Roberts, J.M. (2020) Differences in Placental Imprinted Gene Expression across Preeclamptic and Non-Preeclamptic Pregnancies. Genes, Basel, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hansen, P.J., Dobbs, K.B., Denicol, A.C. and Siqueira, L.G.B. (2016) Sex and the preimplantation embryo: implications of sexual dimorphism in the preimplantation period for maternal programming of embryonic development. Cell Tissue Res., 363, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Litzky, J.F., Boulet, S.L., Esfandiari, N., Zhang, Y., Kissin, D.M., Theiler, R.N. and Marsit, C.J. (2018) Effect of frozen/thawed embryo transfer on birthweight, macrosomia, and low birthweight rates in US singleton infants. Am. J. Obstet. Gynecol., 218, 433 e431–433 e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaartinen, N.M., Kananen, K.M., Rodriguez-Wallberg, K.A., Tomas, C.M., Huhtala, H.S. and Tinkanen, H.I. (2015) Male gender explains increased birthweight in children born after transfer of blastocysts. Hum. Reprod., 30, 2312–2320. [DOI] [PubMed] [Google Scholar]
- 64. Tarrade, A., Panchenko, P., Junien, C. and Gabory, A. (2015) Placental contribution to nutritional programming of health and diseases: epigenetics and sexual dimorphism. J. Exp. Biol., 218, 50–58. [DOI] [PubMed] [Google Scholar]
- 65. Sandman, C.A., Glynn, L.M. and Davis, E.P. (2013) Is there a viability-vulnerability tradeoff? Sex differences in fetal programming. J. Psychosom. Res., 75, 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Turan, N., Turan, N., Katari, S., Gerson, L.F., Chalian, R., Foster, M.W. and Gaughan, J.P. (2010) Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet., 6, e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cavalcante, R.G. and Sartor, M.A. (2017) annotatr: genomic regions in context. Bioinformatics, 33, 2381–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jaffe, A.E., Murakami, P., Lee, H., Leek, J.T., Fallin, M.D., Feinberg, A.P. and Irizarry, R.A. (2012) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol., 41, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu, G., Wang, L.G. and He, Q.Y. (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics, 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article is available in the Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi and can be accessed using the deposition number GSE201496 for human samples and GSE196547 for mouse samples.


