Abstract
Rationale: Genetic variation has a substantial contribution to chronic obstructive pulmonary disease (COPD) and lung function measurements. Heritability estimates using genome-wide genotyping data can be biased if analyses do not appropriately account for the nonuniform distribution of genetic effects across the allele frequency and linkage disequilibrium (LD) spectrum. In addition, the contribution of rare variants has been unclear. Objectives: We sought to assess the heritability of COPD and lung function using whole-genome sequence data from the Trans-Omics for Precision Medicine program. Methods: Using the genome-based restricted maximum likelihood method, we partitioned the genome into bins based on minor allele frequency and LD scores and estimated heritability of COPD, FEV1% predicted and FEV1/FVC ratio in 11 051 European ancestry and 5853 African-American participants. Measurements and Main Results: In European ancestry participants, the estimated heritability of COPD, FEV1% predicted and FEV1/FVC ratio were 35.5%, 55.6% and 32.5%, of which 18.8%, 19.7%, 17.8% were from common variants, and 16.6%, 35.8%, and 14.6% were from rare variants. These estimates had wide confidence intervals, with common variants and some sets of rare variants showing a statistically significant contribution (P-value < 0.05). In African-Americans, common variant heritability was similar to European ancestry participants, but lower sample size precluded calculation of rare variant heritability. Conclusions: Our study provides updated and unbiased estimates of heritability for COPD and lung function, and suggests an important contribution of rare variants. Larger studies of more diverse ancestry will improve accuracy of these estimates.
Introduction
Chronic obstructive pulmonary disease (COPD), one of the leading causes of death worldwide, is diagnosed by a decrease in two key measurements of spirometry, forced expiratory volume in one second (FEV1) and its ratio to forced vital capacity (FEV1/FVC). Genetic factors are important risk factors for the development of COPD (1). Genome-wide association genomic studies (GWAS) for COPD and lung function measurements have identified hundreds of associated regions with common variants, with relevant effects in functional assays identified in genes such as IREB2, CHRNA3/5, HHIP, FAM13A, DSP, HTR4, LRP1 and CYP2A6 (2–10). Rare variants also affect COPD and related phenotypes as demonstrated in alpha-1 antitrypsin deficiency (1). Recent large-scale whole-genome sequencing (WGS) data from the National Heart, Lung and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program have enabled us to identify a subset of rare variants with putative associations to COPD and related phenotypes, including ARHGEF17 and CRISP1 (11).
Heritability is a measure that can provide relative estimates of the contribution of genetic versus environmental factors. Accurate determinations of heritability are important to determine the relative contribution of genetic variants, the potential performance of genetic risk scores and the contribution of rare variants. In prior twin studies and family studies, estimates of family-based heritability of COPD-related phenotypes ranged from 38% to 66% (12–14). Estimates in unrelated subjects can be obtained from genome-wide array data, and a recent study estimated heritability for COPD-related phenotypes using array data of ~35% (15). In another study using array data in the Framingham Heart Study (FHS), single nucleotide variant (SNV)-based heritability was estimated to be between 50% and 65% using SNVs with minor allele frequency (MAF) > 0.5% (14), with some of family-based heritability recovered from low-frequency SNVs (0.5% < MAF 1%).
1%).
However, these previous studies have some limitations. First, heritability captured by rare variants has not been systematically assessed. Most low-frequency and rare variants are not captured by most genotyping arrays (16–18). In general, SNP arrays do not perform well for detecting, or imputing, rare variants (19,20). Determining the contribution of rare variants requires large subject sizes and WGS data. Second, prior estimates of heritability of lung function and COPD have used a single genetic relationship matrix (GRM). A GRM is calculated based on genetic variants to quantify genetic similarities between distantly related individuals. It has previously been shown that a single GRM approach can lead to biased estimates of heritability when causal variants have a different MAF or linkage disequilibrium (LD) properties than variants used in the analysis. Specifically, if causal variants are rarer (or more common) than variants used in the analysis, the estimate of heritability is biased downward (or upward) (21,22). This can be solved by using MAF-stratified GRMs in a model. In addition, if causal variants are enriched in genomic regions with lower (or higher) LD than average, the heritability estimate is downward (or upward) biased. Similar to the uneven MAF spectrum of causal variants, bias of heritability estimates can be solved by stratifying variants by their LD scores. To address these issues, Yang et al. (21) proposed a statistical method, termed the LD and MAF stratified genome-based restricted maximum likelihood (GREML-LDMS) approach, that creates bins of variants in different allele frequencies and LD thus reducing the bias of heritability estimates.
The recent advent of large-scale WGS data from the NHLBI TOPMed program enables an updated and more comprehensive assessment of the contribution of rare variants (11). Here we report SNV-based heritability of COPD and related phenotypes (FEV1% predicted and FEV1/FVC) using GREML-LDMS in nearly 12 000 unrelated individuals from high-coverage WGS data. Study participants were from four studies including three population-based studies and one COPD-enriched study as part of the TOPMed program. We also leveraged high-coverage WGS data to assess the proportion of phenotypic variance of COPD and related phenotypes explained by rare variants.
Results
Heritability estimates using a single GRM
We first calculated heritability using the standard method of a single component genetic relationship matrix (GREML-SC) (23). We estimated GRMs for rare variants (MAF < 0.01) and common variants (MAF 0.01), then conducted GREML-SC analysis using each GRM. In European ancestry participants (N = 11 501), for variants with MAF < 0.01, none of heritability estimates were statistically significantly >0 (Table 1). For variants with MAF
0.01), then conducted GREML-SC analysis using each GRM. In European ancestry participants (N = 11 501), for variants with MAF < 0.01, none of heritability estimates were statistically significantly >0 (Table 1). For variants with MAF 0.01, we estimated heritability to be 23.3% [standard error (SE) = 3.6%, P-value = 4.5
0.01, we estimated heritability to be 23.3% [standard error (SE) = 3.6%, P-value = 4.5 10−11] for FEV1% predicted, 17.9% (SE = 3.2%, P-value = 8.7
10−11] for FEV1% predicted, 17.9% (SE = 3.2%, P-value = 8.7 10−9) for FEV1/FVC and 18% (SE = 5%, P-value = 1.6
10−9) for FEV1/FVC and 18% (SE = 5%, P-value = 1.6 10−4) for COPD. The heritability estimates of height and body mass index (BMI) were 54.8% (SE = 3.6%, P-value = 8.2
10−4) for COPD. The heritability estimates of height and body mass index (BMI) were 54.8% (SE = 3.6%, P-value = 8.2 10−52) and 21.1% (SE = 3.6%, P-value = 1.5
10−52) and 21.1% (SE = 3.6%, P-value = 1.5 10−9), respectively.
10−9), respectively.
Table 1.
Heritability estimates and SE using GREML-SC in European ancestry
| Phenotype | Estimates | Rare variants | Common variants | |
|---|---|---|---|---|
| FEV1 % predicted |
 (SE)
(SE) |
0.031 (0.041) | 0.233 (0.036) | |
| 95% CI | One-sided | (−0.037, ) ) |
(0.173,  ) )
|
|
| Two-sided | (−0.05, 0.112) | (0.162, 0.303) | ||
| P-value | 0.225 |
4.47  10 –11 10 –11
|
||
| FEV1/FVC |
 (SE)
(SE) |
−0.003 (0.032) | 0.179 (0.032) | |
| 95% CI | One-sided | (−0.057,  ) ) |
(0.126,  ) )
|
|
| Two-sided | (−0.067, 0.06) | (0.117, 0.241) | ||
| P-value | 0.54 |
8.74  10 −9 10 −9
|
||
| COPD |
 (SE)
(SE) |
−0.141 (0.029) | 0.18 (0.05) | |
| 95% CI | One-sided | (−0.189,  ) ) |
(0.098,  ) )
|
|
| Two-sided | (−0.198, −0.083) | (0.082, 0.278) | ||
| P-value | 1 |
1.55  10 −4 10 −4
|
||
| Height |
 (SE)
(SE) |
0.071 (0.044) | 0.548 (0.036) | |
| 95% CI | One-sided | (−0.001,  ) ) |
(0.488,  ) )
|
|
| Two-sided | (−0.015, 0.157) | (0.477, 0.619) | ||
| P-value | 0.052 |
8.20  10 −52 10 −52
|
||
| BMI |
 (SE)
(SE) |
−0.026 (0.036) | 0.211 (0.036) | |
| 95% CI | One-sided | (−0.086,  ) ) |
(0.152,  ) )
|
|
| Two-sided | (−0.097, 0.046) | (0.141, 0.281) | ||
| P-value | 0.758 |
1.52  10 −9 10 −9
|
||
Estimates for FEV1% predicted and FEV1/FVC were adjusted for ascertainment bias. Rare and common variants stand for variants with MAF < 1% and MAF 1%, respectively. Bold indicates statistically significant under the significance level of 0.05. One-sided 95% CI means 95% confidence interval for the alternative hypothesis,
1%, respectively. Bold indicates statistically significant under the significance level of 0.05. One-sided 95% CI means 95% confidence interval for the alternative hypothesis,  . Two-sided 95% CI means 95% confidence interval for the alternative hypothesis,
. Two-sided 95% CI means 95% confidence interval for the alternative hypothesis,  .
.
In African-American participants (N = 5853), for common variants, the estimated heritability was 21.6% (SE = 7.8%, P-value = 0.003) for FEV1% predicted, 25.3% (SE = 7.1%, P-value = 1.8 10−4) for FEV1/FVC, 69% (SE = 7.4%, P-value = 7.9
10−4) for FEV1/FVC, 69% (SE = 7.4%, P-value = 7.9 10−21) for height and 27.7% (SE = 7.8%, P-value = 2.1
10−21) for height and 27.7% (SE = 7.8%, P-value = 2.1 10−4) for BMI (Supplementary Material, Table S1). Heritability estimation for COPD, and for rare variant analysis of height and FEV1/FVC, did not converge because of smaller sample size among African ancestry participants; thus, subsequent analyses using the GREML-LDMS methods (21) were restricted to European Ancestry.
10−4) for BMI (Supplementary Material, Table S1). Heritability estimation for COPD, and for rare variant analysis of height and FEV1/FVC, did not converge because of smaller sample size among African ancestry participants; thus, subsequent analyses using the GREML-LDMS methods (21) were restricted to European Ancestry.
Aggregated heritability estimates using WGS in European ancestry participants
Multiple studies have demonstrated a reduction in bias when variants are partitioned by MAF and degree of LD (21,22,24). Thus, we stratified all variants by eight bins based on their MAF in the dataset (0 MAF < 0.0001, 0.0001
MAF < 0.0001, 0.0001 MAF < 0.001, 0.001
MAF < 0.001, 0.001 MAF < 0.01, 0.01
MAF < 0.01, 0.01 MAF < 0.1, 0.1
MAF < 0.1, 0.1 MAF < 0.2, 0.2
MAF < 0.2, 0.2 MAF < 0.3, 0.3
MAF < 0.3, 0.3 MAF < 0.4, 0.4
MAF < 0.4, 0.4 MAF
MAF  0.5). Each of the eight MAF bins were further stratified into a low-LD bin and a high-LD bin based on the median value of LD scores of each MAF bin. Finally, we generated GRMs for 16 variant bins and performed GREML-LDMS using GCTA software. To get heritability estimates of rare and common variants, we aggregated heritability estimates for GRMs with MAF < 0.01 and MAF
0.5). Each of the eight MAF bins were further stratified into a low-LD bin and a high-LD bin based on the median value of LD scores of each MAF bin. Finally, we generated GRMs for 16 variant bins and performed GREML-LDMS using GCTA software. To get heritability estimates of rare and common variants, we aggregated heritability estimates for GRMs with MAF < 0.01 and MAF 0.01, respectively.
0.01, respectively.
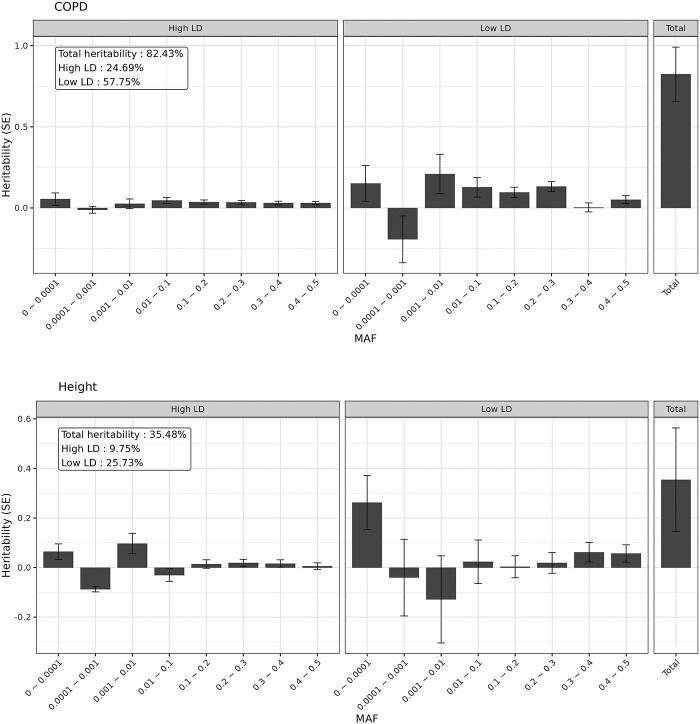
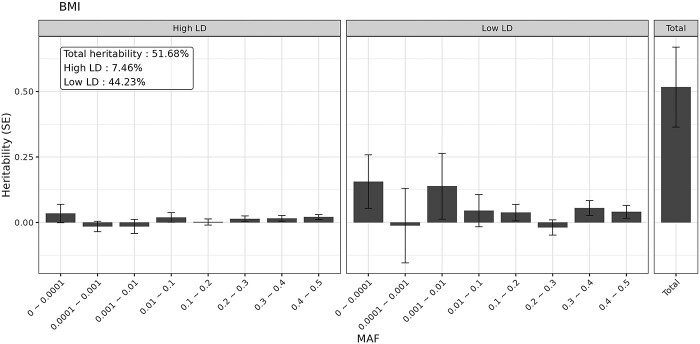
The aggregated estimates of SNV-based heritability using GREML-LDMS are displayed in Table 2 and Supplementary Material, Table S2. To first confirm that our approach in this dataset yielded similar estimates to prior reports, we applied GREML-LDMS to height and BMI, and estimated a heritability of 82.4% (SE = 16.7%, P-value = 3.9 10−7) for height and 51.7% (SE = 15.3%, P-value = 3.5
10−7) for height and 51.7% (SE = 15.3%, P-value = 3.5 10−4) for BMI. These estimates are slightly higher than the previously reported estimates (56–79% for height and 22–40% for BMI, Supplementary Material, Table S3) (21,22,24) that did not include the rarest variant bin.
10−4) for BMI. These estimates are slightly higher than the previously reported estimates (56–79% for height and 22–40% for BMI, Supplementary Material, Table S3) (21,22,24) that did not include the rarest variant bin.
Table 2.
Aggregated heritability estimates and SE for FEV1% predicted, FEV1/FVC and COPD using GREML-LDMS in European ancestry
| Phenotype | LD | Estimates | Rare variants | Common variants | Total | |
|---|---|---|---|---|---|---|
| FEV1 % predicted | High-LD |
 (SE)
(SE) |
0.05 (0.041) | 0.075 (0.023) | 0.125 (0.046) | |
| 95% CId | One-sided | (−0.018,  ) ) |
(0.037,  ) )
|
(0.049,  ) )
|
||
| Two-sided | (−0.03, 0.13) | (0.029, 0.121) | (0.035, 0.215) | |||
| P-value | 0.111 |
6.62  10 −4 10 −4
|
0.003 | |||
| Low-LD |
 (SE)
(SE) |
0.308 (0.152) | 0.122 (0.061) | 0.43 (0.152) | ||
| 95% CI | One-sided |
(0.058,  ) )
|
(0.021,  ) )
|
(0.18,  ) )
|
||
| Two-sided | (0.011, 0.605) | (0.002, 0.242) | (0.133, 0.727) | |||
| P-value | 0.021 | 0.023 | 0.002 | |||
| Total |
 (SE)
(SE) |
0.358 (0.174) | 0.197 (0.06) | 0.556 (0.172) | ||
| 95% CI | One-sided |
(0.071,  ) )
|
(0.098,  ) )
|
(0.272,  ) )
|
||
| Two-sided | (0.017, 0.699) | (0.079, 0.316) | (0.219, 0.892) | |||
| P-value | 0.02 |
5.28  10 −4 10 −4
|
6.06  10 −4 10 −4
|
|||
| FEV1/FVC | High-LD |
 (SE)
(SE) |
0.046 (0.035) | 0.06 (0.022) | 0.105 (0.04) | |
| 95% CI | One-sided | (−0.012,  ) ) |
(0.023,  ) )
|
(0.039,  ) )
|
||
| Two-sided | (−0.023, 0.115) | (0.016, 0.103) | (0.026, 0.185) | |||
| P-value | 0.097 | 0.003 | 0.005 | |||
| Low-LD |
 (SE)
(SE) |
0.101 (0.142) | 0.119 (0.058) | 0.219 (0.142) | ||
| 95% CI | One-sided | (−0.133,  ) ) |
(0.023,  ) )
|
(−0.015,  ) ) |
||
| Two-sided | (−0.177, 0.379) | (0.005, 0.232) | (−0.059, 0.498) | |||
| P-value | 0.239 | 0.02 | 0.061 | |||
| Total |
 (SE)
(SE) |
0.146 (0.162) | 0.178 (0.057) | 0.325 (0.16) | ||
| 95% CI | One-sided | (−0.121,  ) ) |
(0.084,  ) )
|
(0.061,  ) )
|
||
| Two-sided | (−0.171, 0.464) | (0.067, 0.29) | (0.012, 0.638) | |||
| P-value | 0.183 |
8.54  10 −4 10 −4
|
0.021 | |||
| COPD | High-LD |
 (SE)
(SE) |
0.073 (0.042) | 0.024 (0.033) | 0.098 (0.052) | |
| 95% CI | One-sided |
(0.003,  ) )
|
(−0.03,  ) ) |
(0.012,  ) )
|
||
| Two-sided | (−0.01, 0.156) | (−0.04, 0.088) | (−0.004, 0.199) | |||
| P-value | 0.042 | 0.229 | 0.03 | |||
| Low-LD |
 (SE)
(SE) |
0.093 (0.187) | 0.164 (0.087) | 0.257 (0.188) | ||
| 95% CI | One-sided | (−0.215,  ) ) |
(0.02,  ) )
|
(−0.053,  ) ) |
||
| Two-sided | (−0.273, 0.459) | (−0.007, 0.336) | (−0.112, 0.626) | |||
| P-value | 0.309 | 0.03 | 0.086 | |||
| Total |
 (SE)
(SE) |
0.166 (0.211) | 0.188 (0.086) | 0.355 (0.208) | ||
| 95% CI | One-sided | (−0.182,  ) ) |
(0.047,  ) )
|
(0.011,  ) )
|
||
| Two-sided | (−0.248, 0.581) | (0.02, 0.357) | (−0.054, 0.763) | |||
| P-value | 0.215 | 0.014 | 0.044 | |||
Estimates for FEV1% predicted, FEV1/FVC were adjusted for ascertainment bias. Heritability estimates were aggregated by MAF (rare variants and common variants) and LD (high-LD versus low-LD) groups. Rare and common variants stand for variants with MAF < 1% and MAF 1%, respectively. Bold indicates statistically significant under the significance level of 0.05. One-sided 95% CI means 95% confidence interval for the alternative hypothesis,
1%, respectively. Bold indicates statistically significant under the significance level of 0.05. One-sided 95% CI means 95% confidence interval for the alternative hypothesis,  . Two-sided 95% CI means 95% confidence interval for the alternative hypothesis,
. Two-sided 95% CI means 95% confidence interval for the alternative hypothesis,  .
.
For lung function measurements, after adjusting for ascertainment bias, we estimated the heritability of FEV1% predicted to be 55.6% (SE = 17.2%, P-value = 6.1 10−4), and rare variants and common variants accounted for 35.8% (SE = 17.4%, P-value = 0.02) and 19.8% (SE = 6%, P-value = 5.3
10−4), and rare variants and common variants accounted for 35.8% (SE = 17.4%, P-value = 0.02) and 19.8% (SE = 6%, P-value = 5.3 10−4), respectively. The largest contributor to heritability was from rare variants in the low-LD group, though with large SEs (h2 = 30.8%, SE = 15.2%, P-value = 0.021). When we aggregated the heritability by LD groups, 43% (SE = 15.2%, P-value = 0.002) and 12.5% (SE = 4.6%, P-value = 0.003) of heritability were accounted for by variants in the low-LD group and high-LD group, respectively.
10−4), respectively. The largest contributor to heritability was from rare variants in the low-LD group, though with large SEs (h2 = 30.8%, SE = 15.2%, P-value = 0.021). When we aggregated the heritability by LD groups, 43% (SE = 15.2%, P-value = 0.002) and 12.5% (SE = 4.6%, P-value = 0.003) of heritability were accounted for by variants in the low-LD group and high-LD group, respectively.
The overall heritability of FEV1/FVC was estimated to be 32.5% (SE = 16%, P-value = 0.021). A total of 14.6% (SE = 16.2%) and 17.8% (SE = 5.7%, P-value = 8.5 10−4) of the heritability were contributed by rare variants and common variants, respectively. In terms of LD structure, variants in low-LD group and high-LD group explained 21.9% (SE = 14.2%) and 10.5% (SE = 4%, P-value = 0.005) of the heritability, respectively.
10−4) of the heritability were contributed by rare variants and common variants, respectively. In terms of LD structure, variants in low-LD group and high-LD group explained 21.9% (SE = 14.2%) and 10.5% (SE = 4%, P-value = 0.005) of the heritability, respectively.
For COPD, the heritability estimate was 35.5% (SE = 20.8%, P-value = 0.044) and of those, 16.6% (SE = 21.1%) and 18.8% (SE = 8.6%, P-value = 0.014) were explained by rare variants and common variants, respectively. Similar to the lung function measurements, heritability was mostly contributed by variants in the low-LD group (low-LD: h2 = 25.7%, SE = 18.8%, P-value = 0.086; high-LD: h2 = 9.8%, SE = 5.2%, P-value = 0.03). These estimates showed higher SEs compared with lung function measurements due in part to a reduction in sample size from excluding participants who were neither cases nor controls.
Significant heritability estimates of individual MAF and LD bins in European ancestry participants
The heritability estimates for individual MAF and LD bins are shown in Supplementary Material, Table S4 and Figure 1. For FEV1% predicted, variants with 0.001 MAF < 0.1 showed the largest contribution to the phenotype among MAF bins (h2 = 23.8%, SE = 13%, P-value = 0.033). In the high-LD group, heritability estimates of the variants with 0.2
MAF < 0.1 showed the largest contribution to the phenotype among MAF bins (h2 = 23.8%, SE = 13%, P-value = 0.033). In the high-LD group, heritability estimates of the variants with 0.2 MAF < 0.3 and 0.4
MAF < 0.3 and 0.4 MAF
MAF 0.5 were significantly larger than zero (h2 = 2.4%, SE = 1.1%, P-value = 0.017 and h2 = 1.9%, SE = 1%, P-value = 0.023, respectively). In the low-LD group, heritability estimates of the variants with 0.3
0.5 were significantly larger than zero (h2 = 2.4%, SE = 1.1%, P-value = 0.017 and h2 = 1.9%, SE = 1%, P-value = 0.023, respectively). In the low-LD group, heritability estimates of the variants with 0.3 MAF < 0.4 was statistically significant (h2 = 6.8%, SE = 2.8%, P-value = 0.008). For FEV1/FVC, we found two significant heritability estimates of the variants with 0.2
MAF < 0.4 was statistically significant (h2 = 6.8%, SE = 2.8%, P-value = 0.008). For FEV1/FVC, we found two significant heritability estimates of the variants with 0.2 MAF < 0.3 in the high-LD group (h2 = 2.5%, SE = 1.1%, P-value = 0.013) and 0.3
MAF < 0.3 in the high-LD group (h2 = 2.5%, SE = 1.1%, P-value = 0.013) and 0.3 MAF < 0.4 in the low-LD group (h2 = 5.7%, SE = 2.7%, P-value = 0.016). For COPD disease status, a substantial fraction of heritability was captured by the rarest MAF bin (0
MAF < 0.4 in the low-LD group (h2 = 5.7%, SE = 2.7%, P-value = 0.016). For COPD disease status, a substantial fraction of heritability was captured by the rarest MAF bin (0 MAF < 0.0001) with estimates of 32.6% (SE = 13.6%, P-value = 0.008), and heritability estimates for its LD-stratified bins were also statistically significant (high-LD: h2 = 6.4%, SE = 3.2%, P-value = 0.021; low-LD: h2 = 26.2%, SE = 11%, P-value = 0.008). We observed significant heritability estimates of 9.7% among rare variants (0.001
MAF < 0.0001) with estimates of 32.6% (SE = 13.6%, P-value = 0.008), and heritability estimates for its LD-stratified bins were also statistically significant (high-LD: h2 = 6.4%, SE = 3.2%, P-value = 0.021; low-LD: h2 = 26.2%, SE = 11%, P-value = 0.008). We observed significant heritability estimates of 9.7% among rare variants (0.001 MAF < 0.01) in the high-LD group (SE = 4.1%, P-value = 0.009). Compared with the heritability estimates using GREML-SC, the SC method did not capture heritability of rare variants.
MAF < 0.01) in the high-LD group (SE = 4.1%, P-value = 0.009). Compared with the heritability estimates using GREML-SC, the SC method did not capture heritability of rare variants.
Figure 1.
Heritability estimates stratified in 16 bins (eight MAF bins and two LD bins) in European ancestry. The bars display SEs.
Discussion
A genetic contribution to COPD and lung function has been recognized for decades, based on family-based and twin studies and more recently, from genome-wide genotyping data, widely available in population-based samples. However, these methods relied on the assumptions of a normal distribution of genetic variant effect sizes independent of LD and inversely proportional to the MAF. In addition, despite examples of rare variant contributions to complex disease, many studies have not been able to identify and replicate rare variants of statistically significant effect (25–27). Prior studies in COPD and lung function have either not identified statistically significant variants, or identified variants that have not been consistently replicated (28–33).
Our study sought to use comprehensive WGS data available in four cohorts to provide unbiased estimates of heritability and to examine the contribution of rare variants on COPD and lung function. Compared with prior estimates in a COPD-enriched study (15), comprised of smokers with and without COPD, our study of population-based and COPD-enriched participants resulted in a smaller heritability estimate from common variants than the previous study. Compared with a prior study of low-frequency variants (0.5% MAF < 5%) in the UK Biobank, our heritability estimates of rare variant (MAF < 1%) for FEV1/FVC were larger (34) suggesting nonzero contribution of rare variants to these phenotypes. Our heritability estimates of common variants based on GREML-LDMS and GREML-SC indicate that bias of common variants because of heritability partitioning was negligible. However, we identified substantial differences in the estimates of rare variants depending on stratification of GRMs. Although the estimates based on the GREML-SC approach were nearly zero, 35.8%, 14.6% and 16.6% of heritability of rare variants were recovered for FEV1% predicted, FEV1/FVC and COPD by using GREML-LDMS, respectively. Therefore, rare causal variants may lie within specific MAF or LD bins, whereas common causal variants may spread among multiple MAF and LD bins. Another possible explanation is that the genetic similarity between individuals modeled by different common variant bins are more similar to each other than when using rare variant bins. This finding may be because of chance or because of a selection process that we do not model.
MAF < 5%) in the UK Biobank, our heritability estimates of rare variant (MAF < 1%) for FEV1/FVC were larger (34) suggesting nonzero contribution of rare variants to these phenotypes. Our heritability estimates of common variants based on GREML-LDMS and GREML-SC indicate that bias of common variants because of heritability partitioning was negligible. However, we identified substantial differences in the estimates of rare variants depending on stratification of GRMs. Although the estimates based on the GREML-SC approach were nearly zero, 35.8%, 14.6% and 16.6% of heritability of rare variants were recovered for FEV1% predicted, FEV1/FVC and COPD by using GREML-LDMS, respectively. Therefore, rare causal variants may lie within specific MAF or LD bins, whereas common causal variants may spread among multiple MAF and LD bins. Another possible explanation is that the genetic similarity between individuals modeled by different common variant bins are more similar to each other than when using rare variant bins. This finding may be because of chance or because of a selection process that we do not model.
For all phenotypes, heritability estimates were predominantly explained by rare variants in low-LD bins with some, but not all, statistically significant estimates of heritability. These findings are consistent with other studies demonstrating that, in general, rare variants with lower levels of LD have higher levels of heritability (35). These variants also are more enriched in regions of functional annotation. We note that the majority of identified variants in this study, as in other sequencing studies are rare. These findings are consistent with the effects of selection in reducing deleterious alleles over time (35,36).
Heritability is defined between 0 and 1 but its estimate can exceed these values, following the normal distribution. Thus, to get an unbiased estimate, we allowed the heritability estimate to be negative. Indeed, we observed negative estimates with wide confidence interval in several individual MAF and LD bins. We kept negative estimates to obtain unbiased estimates of total heritability, although negative heritability has no meaning in itself and should be interpreted as zero heritability. In practice, we are likely to observe a negative estimate in two situations. First, when the sample size is small, the estimate has a chance to be out of the parameter space because of a large SE. Second, when the true heritability is very small, then we also have a certain probability to see negative estimate even though the sample size is large. Although our sample sizes were large for WGS data for COPD and lung function, the SEs of our estimates are large, resulting in only nominally (P-value < 0.05) significant P-values unadjusted for multiple testing, indicating that even larger sample sizes are needed to accurately identify the contribution of rare variants generally and also to determine the relative contribution of specific rare functional regions. Nevertheless, the estimates of the overall heritability were reliable. In our study, heritability estimation using GREML-LDMS was limited to European ancestry because heritability estimates in African ancestry were imprecise or did not converge because of sample size. Estimating and interpreting SNV-heritability is challenging in diverse populations (37) and GCTA-GREML that is the method estimating heritability from individual-level genetic data can generate biased estimates in the presence of population stratification (38). Our study thus further emphasizes the need to increase the number of participants of non-European ancestry. Finally, heritability is a measure that is specific to a population and set of environmental conditions. We studied lung function and disease status phenotypes available in our sample. In addition, the relative contribution of rare variants to other COPD-related phenotypes, such as those obtained from imaging studies, is unknown.
Overall, our results provide an updated estimate of heritability, in the largest cohort for lung function and COPD to undergo WGS to date. We find evidence for a contribution of rare variants to lung function and COPD and an enrichment in low-LD regions. These findings support the ongoing investigation of rare variants in COPD.
Materials and Methods
Study participants
We selected 11 849 European ancestry participants and 7167 African-American participants from four population-based cohorts [the Atherosclerosis Risk in Communities (ARIC) Study, the FHS, the Multi-Ethnic Study of Atherosclerosis (MESA) and Jackson Heart Study (JHS)] and one COPD-enriched study [the Genetic Epidemiology of COPD (COPDGene) Study] as a part of NHLBI TOPMed program. The ancestry was defined by genetic data. The study descriptions for each cohort are provided in the Supplementary Material, Text S2.
Genetic variants were extracted from the Freeze 5b version of WGS data in the TOPMed program (39). Details on sequencing method of TOPMed are found at https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-project-freeze-5b-phases-1-and-2. Briefly, joint-subject variant calling identified ~ 470M genetic variants for 54 499 participants which is a subset of TOPMed participants on human genome assembly GRCh38 with deep coverage (~30×). Among these variants, we only considered genetic variants carried by at least one participant in the cohorts included in this analysis.
Quality control
The TOPMed callset includes extensive quality control, detailed at https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-project-freeze-5b-phases-1-and-2. We additionally excluded SNVs with missingness rate > 5% in our subsample, or a P-value of the Hardy–Weinberg equilibrium test < 1 10−5. We considered only biallelic SNVs on the autosomes. We also excluded any participants whose genotype missingness rate was >5%. To include only unrelated participants, we estimated kinship coefficients of every pair of participants using PC-Relate (40) and we randomly excluded one of each pair of participants with estimates of kinship coefficient > 0.05. After filtering, we retained 11 501 European ancestry participants and 5853 African ancestry participants with ~123M and ~112M SNVs, respectively (Table 3 and Figure 2). We confirmed that there were no related participants on the reported pedigree after quality control of the samples. All participants that were not European-ancestry or African-American were excluded from the analysis because of sample size. As expected, the COPD-enriched study (COPDGene) showed lower mean values of lung function and higher pack-years than the population-based studies in both ancestries.
10−5. We considered only biallelic SNVs on the autosomes. We also excluded any participants whose genotype missingness rate was >5%. To include only unrelated participants, we estimated kinship coefficients of every pair of participants using PC-Relate (40) and we randomly excluded one of each pair of participants with estimates of kinship coefficient > 0.05. After filtering, we retained 11 501 European ancestry participants and 5853 African ancestry participants with ~123M and ~112M SNVs, respectively (Table 3 and Figure 2). We confirmed that there were no related participants on the reported pedigree after quality control of the samples. All participants that were not European-ancestry or African-American were excluded from the analysis because of sample size. As expected, the COPD-enriched study (COPDGene) showed lower mean values of lung function and higher pack-years than the population-based studies in both ancestries.
Table 3.
Descriptive statistics of study participants after quality control
| Phenotype | Ancestry | Study | ||
|---|---|---|---|---|
| European | African | COPD-enriched | Population-based | |
| Number of subjects | 11 051 | 5853 | 8397 | 8507 |
| COPD subjects | 3234 | 930 | 3120 | 1044 |
| Gender (F/M) | 5460/5591 | 3025/2828 | 3907/4490 | 4578/3929 |
| Age | 61.64 (9.65) | 57.61 (10.43) | 59.5 (9.08) | 60.98 (10.99) |
| Pack-years | 32.04 (29.58) | 22.28 (24.47) | 44.36 (25) | 13.17 (22.16) |
| Height | 169.03 (9.54) | 169.14 (9.8) | 170.2 (9.47) | 167.95 (9.65) |
| BMI | 28.18 (5.63) | 30.07 (6.69) | 28.88 (6.26) | 28.79 (5.91) |
| FEV1 % predicted | 80.24 (24.55) | 86.13 (22.27) | 72.86 (25.58) | 91.57 (17.88) |
| FEV1/FVC | 0.68 (0.14) | 0.75 (0.12) | 0.65 (0.16) | 0.75 (0.09) |
We counted the number of subjects, COPD subjects and each gender. For continuous variables, mean and standard deviation were shown.
Figure 2.

Quality control for the SNVs and participants. Multiple standard quality controls were performed for the raw data to exclude outlier SNVs and participants.
Phenotypes
We estimated the heritability of spirometry measures, COPD affection status, as well as anthropometric values (the latter as ‘positive controls’). For lung function, we used percentage of predicted value of FEV1 (FEV1% predicted) and FEV1/FVC ratio as continuous phenotypes. To ensure harmonized phenotypes across multiple TOPMed population studies, we used phenotypes derived from the protocol of the NHLBI Pooled Cohorts Study (Supplementary Material, Text S3) (41). We calculated percent predicted values of FEV1 using Hankinson’s reference equations (42). We also examined the heritability of height and BMI within our participants, as these measures have been estimated previously in larger samples. Similar to a previous approach (24), to adjust for age, we regressed all continuous phenotypes on age within each sex and study, and residuals were standardized by sex-specific standard deviations. We then pooled the standardized residuals together and applied a rank-based inverse normal transformation to the standardized residuals. For the dichotomous phenotype, we defined COPD status of each participant using GOLD criteria for moderate-to-severe obstruction (43) as follows: (i) cases: FEV1% predicted <80% and FEV1/FVC < 0.7, (ii) controls: FEV1% predicted  80% and FEV1/FVC
80% and FEV1/FVC 0.7. In addition to age and sex, we included pack-years of smoking as a covariate for all phenotypes except for height, as well as sequencing center, and first 10 principal components of genetic ancestry scores as fixed effects.
0.7. In addition to age and sex, we included pack-years of smoking as a covariate for all phenotypes except for height, as well as sequencing center, and first 10 principal components of genetic ancestry scores as fixed effects.
Statistical methods for estimating heritability
The overview of the workflow for the statistical analysis is illustrated in Figure 3. GREML, which is implemented in the GCTA software, is a statistical method to estimate genetic and environmental variance components using a linear mixed model (21,23). For a quantitative trait  , the model of GREML is given by
, the model of GREML is given by
 |
where  is a vector of coefficients of the covariates
is a vector of coefficients of the covariates  such as age, sex or principal component scores,
such as age, sex or principal component scores,  is a random effect of
is a random effect of  th SNV-set with a corresponding GRM
th SNV-set with a corresponding GRM  , which follows
, which follows  and
and  is a vector of residual effects following
is a vector of residual effects following  . For the
. For the  th GRM,
th GRM,  , the genetic relationship between individuals
, the genetic relationship between individuals  and
and  was calculated as follows,
was calculated as follows,
 |
where  is the number of SNVs,
is the number of SNVs,  is a minor allele count of
is a minor allele count of  th SNV for individual
th SNV for individual  and
and  is an MAF of
is an MAF of  th SNV in the dataset. The variance–covariance matrix of a quantitative trait
th SNV in the dataset. The variance–covariance matrix of a quantitative trait  is
is  and we estimate variance components,
and we estimate variance components,  and
and  , using the restricted maximum likelihood approach (44). The proportion of phenotypic variance explained by the
, using the restricted maximum likelihood approach (44). The proportion of phenotypic variance explained by the  th set of SNV and by all the SNVs are defined as
th set of SNV and by all the SNVs are defined as  and
and  , respectively.
, respectively.
Figure 3.

Workflow of statistical analysis for the heritability estimation of COPD-related phenotypes. Overview of the statistical analysis to estimate heritability of the phenotypes is illustrated.
Because our study includes a COPD-enriched population, study participants had lower values of FEV1% predicted or FEV1/FVC than the general population on average. To estimate heritability of FEV1% predicted and FEV1/FVC in the general population, we adjusted the heritability estimates in the ascertained participants by using their relationship expressed as a function of proportions of participants at the specific threshold value of the phenotype in both the general population and the ascertained participants, and the population mean and variance of the phenotype. To adjustment ascertainment bias, we expressed heritability in the participants as a function of the heritability in the population given that the phenotype and genetic factor are jointly normally distributed; the heritability in the population can be obtained by inverting the equation. The detailed derivation is provided in the Supplementary Material, Text S4.
Similarly, heritability for a dichotomous phenotype, COPD status in our case, needs to be adjusted when cases and controls are not a random sample from the general population (45). Here, we assume that cases and controls are determined by the unobserved continuous liability score  and the threshold
and the threshold  . Under the assumption, participants are considered as a case if their liability scores are larger than the threshold
. Under the assumption, participants are considered as a case if their liability scores are larger than the threshold  ; otherwise, they are considered as a control. The threshold
; otherwise, they are considered as a control. The threshold  is chosen to maintain the assumed prevalence q. In the absence of case–control ascertainment, the heritability of a dichotomous trait on the liability scale,
is chosen to maintain the assumed prevalence q. In the absence of case–control ascertainment, the heritability of a dichotomous trait on the liability scale,  , can be expressed with the heritability on the observed 0–1 scale,
, can be expressed with the heritability on the observed 0–1 scale,  , as follows:
, as follows:
 |
where  the population prevalence and
the population prevalence and  the value of the probability density function for the standard normal distribution at the threshold
the value of the probability density function for the standard normal distribution at the threshold 
Under the liability threshold model, the heritability of a dichotomous phenotype on the liability scale,  , can be expressed with the heritability on the observed 0–1 scale,
, can be expressed with the heritability on the observed 0–1 scale,  , as follows:
, as follows:
 |
where  the population prevalence,
the population prevalence,  the proportion of the cases in the sample and
the proportion of the cases in the sample and  the value of the probability density function for the standard normal distribution at the threshold
the value of the probability density function for the standard normal distribution at the threshold  . We assumed a 10% prevalence of COPD in the population and the same prevalence was assumed for all studies (46).
. We assumed a 10% prevalence of COPD in the population and the same prevalence was assumed for all studies (46).
Partitioning heritability
To assess the heritability of COPD and lung function using methods comparable to prior studies and to serve as a baseline to examine the effects of partitioning on heritability, we estimated heritability using single-component GREML analysis (GREML-SC) in GCTA software for common variants (MAF 0.01) and rare variants (MAF < 0.01) with the same fixed effects in the GREML-LDMS analysis.
0.01) and rare variants (MAF < 0.01) with the same fixed effects in the GREML-LDMS analysis.
According to previous studies demonstrating a reduction in bias when variants are partitioned by allele frequency and degree of LD (21,22,24), we stratified all variants by eight bins based on their MAF in the dataset (0 MAF < 0.0001, 0.0001
MAF < 0.0001, 0.0001 MAF < 0.001, 0.001
MAF < 0.001, 0.001 MAF < 0.01, 0.01
MAF < 0.01, 0.01 MAF < 0.1, 0.1
MAF < 0.1, 0.1 MAF < 0.2, 0.2
MAF < 0.2, 0.2 MAF < 0.3, 0.3
MAF < 0.3, 0.3 MAF < 0.4, 0.4
MAF < 0.4, 0.4 MAF
MAF 0.5). We then calculated LD score of the variants on a sliding window of 10 Mb within each MAF bin as follows:
0.5). We then calculated LD score of the variants on a sliding window of 10 Mb within each MAF bin as follows:
 |
where  is a mean
is a mean  between the target variant and all other variants in the window and
between the target variant and all other variants in the window and  is the number of variants in the window. Here, LD score is defined as the sum of
is the number of variants in the window. Here, LD score is defined as the sum of  between a variant and all the variants in a window. We used the GCTA software for the LD score calculation (23). On each chromosome, we further stratified each of the eight MAF bins into a low-LD bin and a high-LD bin based on the median value of LD scores of each MAF bin, and combined all variants in the same MAF and LD bin across chromosomes. The actual median LD scores in MAF bins were provided in Supplementary Material, Table S5. In total, we generated 16 GRMs and performed GREML-LDMS using GCTA software. To estimate aggregated heritabilities of rare and common variants, we summed up all heritability estimates for GRMs with MAF < 0.01 and MAF
between a variant and all the variants in a window. We used the GCTA software for the LD score calculation (23). On each chromosome, we further stratified each of the eight MAF bins into a low-LD bin and a high-LD bin based on the median value of LD scores of each MAF bin, and combined all variants in the same MAF and LD bin across chromosomes. The actual median LD scores in MAF bins were provided in Supplementary Material, Table S5. In total, we generated 16 GRMs and performed GREML-LDMS using GCTA software. To estimate aggregated heritabilities of rare and common variants, we summed up all heritability estimates for GRMs with MAF < 0.01 and MAF 0.01, respectively, and its SE was approximately derived based on the Delta method (Supplementary Material, Text S5).
0.01, respectively, and its SE was approximately derived based on the Delta method (Supplementary Material, Text S5).
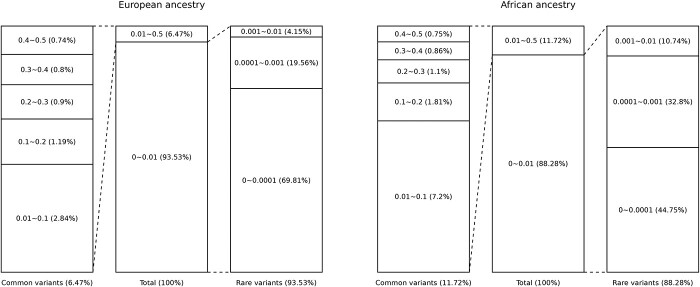
Among ~ 123M SNVs in 11 502 unrelated European ancestry participants, 93.53% were rare variants (MAF < 0.01) and 6.47% were common variants (MAF 0.01). SNVs with MAF < 0.001 consisted of 89.38% of all variants. The remaining MAF bins (0.001
0.01). SNVs with MAF < 0.001 consisted of 89.38% of all variants. The remaining MAF bins (0.001 MAF < 0.01, 0.01
MAF < 0.01, 0.01 MAF < 0.1, 0.1
MAF < 0.1, 0.1 MAF < 0.2, 0.2
MAF < 0.2, 0.2 MAF < 0.3, 0.3
MAF < 0.3, 0.3 MAF < 0.4 and 0.4
MAF < 0.4 and 0.4 MAF
MAF 0.5) each accounted for <5% of total SNVs, with a decreasing proportion of total SNVs with increasing MAF (Fig. 4).
0.5) each accounted for <5% of total SNVs, with a decreasing proportion of total SNVs with increasing MAF (Fig. 4).
Figure 4.
MAF distribution of variants in WGS data. Values indicate MAF and the proportion of variants in the MAF groups (%).
Code Availability
The analysis is readily applicable in other diseases. The code used in the study is provided at https://github.com/wonjikim11/RV_h2_COPD. Detailed information and additional options for GCTA-GREML analysis can be found in the GCTA online manual. (https://yanglab.westlake.edu.cn/software/gcta/#Overview).
Supplementary Material
Acknowledgements
Study-specific acknowledgements are given in the Supplementary Material, Text S1. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the US Department of Health and Human Services. A full list of authors for the NHLBI TOPMed Consortium is provided at https://www.nhlbiwgs.org/topmed-banner-authorship.
Conflict of Interest statement. M.H.C. has received grant support from GSK and Bayer, consulting or speaking fees from Genentech, AstraZeneca and Illumina. E.K.S. has received grant support from GSK and Bayer. B.M.P serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson.
Contributor Information
Wonji Kim, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Julian Hecker, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
R Graham Barr, Departments of Medicine and Epidemiology, Columbia University Medical Center, New York, NY 10032, USA.
Eric Boerwinkle, Department of Epidemiology, Human Genetics and Environmental Sciences, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX 77030, USA; Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA.
Brian Cade, Division of Sleep and Circadian Disorders, Brigham and Women's Hospital, Boston, MA 02115, USA.
Adolfo Correa, Department of Medicine, University of Mississippi Medical Center, Jackson, MS 39216, USA.
Josée Dupuis, Department of Biostatistics, Boston University of Public Health, Boston, MA 02118, USA.
Sina A Gharib, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98101, USA; Division of Pulmonary, Critical Care and Sleep Medicine, Department of Medicine, University of Washington, Seattle, WA 98109, USA.
Leslie Lange, Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO 80045, USA.
Stephanie J London, Epidemiology Branch, National Institute of Environmental Health Sciences, Department of Health and Human Services, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Alanna C Morrison, Human Genetics Center, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
George T O'Connor, Pulmonary Center, Boston University School of Medicine, Boston, MA 02118, USA.
Elizabeth C Oelsner, Departments of Medicine and Epidemiology, Columbia University Medical Center, New York, NY 10032, USA.
Bruce M Psaty, Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98101, USA; Departments of Epidemiology and Health Services, University of Washington, Seattle, WA 98101, USA.
Ramachandran S Vasan, Lung and Blood Institute Framingham Heart Study, Boston University and National Heart, Framingham, MA 01702, USA; Department of Preventive Medicine and Epidemiology, School of Medicine and Public Health, Boston University, Boston, MA 02118, USA.
Susan Redline, Department of Medicine, Brigham and Women's Hospital, Boston, MA 02115, USA.
Stephen S Rich, Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA 22908, USA.
Jerome I Rotter, The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA.
Bing Yu, Department of Epidemiology, Human Genetics and Environmental Sciences, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Christoph Lange, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA; Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA.
Ani Manichaikul, Center for Public Health Genomics, University of Virginia, Charlottesville, VA 22908, USA.
Jin J Zhou, Department of Epidemiology and Biostatistics, University of Arizona, Tucson, AZ 85721, USA.
Tamar Sofer, Division of Sleep and Circadian Disorder, Brigham and Women’s Hospital, Boston, MA 02115, USA.
Edwin K Silverman, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Dandi Qiao, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Michael H Cho, Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Funding
Whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for `NHLBI TOPMed: Atherosclerosis Risk in Communities (ARIC)' (phs001211) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN268201500015C and 3U54HG003273-12S2) and the Broad Institute for MIT and Harvard (3R01HL092577-06S1). WGS for `NHLBI TOPMed: Whole Genome Sequencing and Related Phenotypes in the Framingham Heart Study (FHS)' (phs000974) was performed at the Broad Institute of MIT and Harvard (HHSN268201500014C and 3R01HL092577-06S1). WGS for `NHLBI TOPMed: Multi-Ethnic Study of Atherosclerosis (MESA)' (phs001416) was performed at the Broad Institute of MIT and Harvard (3U54HG003067-13S1 and HHSN268201500014C). WGS for `NHLBI TOPMed: Genetic Epidemiology of COPD (COPDGene) in the TOPMed Program' (phs000951) was performed at the University of Washington Northwest Genomics Center (3R01 HL089856-08S1) and the Broad Institute of MIT and Harvard (HHSN268201500014C). WGS for `NHLBI TOPMed: The Jackson Heart Study (JHS)' (phs000964) was performed at the University of Washington Northwest Genomics Center (HHSN268201100037C). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I). Phenotype harmonization for pulmonary traits was contributed by the NHLBI Pooled Cohorts Study with funding from NIH/NHLBI R21 HL121457, R21 HL129924, K23 HL130627, R01 HL077612. This research was also supported by R01 HL149861, R01 HL135142, R01 HL137927 (M.H.C.) and R01 HL089856 and R01 HL147148 (M.H.C. and E.K.S.), and K01 HL129039 (D.Q.). The COPDGene study (NCT00608764) is also supported by NHLBI grants U01 HL089897 and U01 HL089856, and the COPD Foundation through contributions made to an Industry Advisory Board that has included AstraZeneca, Bayer Pharmaceuticals, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Novartis, Pfizer, and Sunovion.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
References
- 1. Silverman, E.K., Chapman, H.A., Drazen, J.M., Weiss, S.T., Rosner, B., Campbell, E.J., O'Donnell, W.J., Reilly, J.J., Ginns, L. and Mentzer, S. (1998) Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med., 157, 1770–1778. [DOI] [PubMed] [Google Scholar]
- 2. Sakornsakolpat, P., Prokopenko, D., Lamontagne, M., Reeve, N.F., Guyatt, A.L., Jackson, V.E., Shrine, N., Qiao, D., Bartz, T.M., Kim, D.K. et al. (2019) Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat. Genet., 51, 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castaldi, P.J., Cho, M.H., Zhou, X., Qiu, W., Mcgeachie, M., Celli, B., Bakke, P., Gulsvik, A., Lomas, D.A., Crapo, J.D. et al. (2015) Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum. Mol. Genet., 24, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamontagne, M., Couture, C., Postma, D.S., Timens, W., Sin, D.D., Pare, P.D., Hogg, J.C., Nickle, D., Laviolette, M. and Bosse, Y. (2013) Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One, 8, e70220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeMeo, D.L., Mariani, T., Bhattacharya, S., Srisuma, S., Lange, C., Litonjua, A., Bueno, R., Pillai, S.G., Lomas, D.A., Sparrow, D. et al. (2009) Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet., 85, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pillai, S.G., Ge, D., Zhu, G., Kong, X., Shianna, K.V., Need, A.C., Feng, S., Hersh, C.P., Bakke, P., Gulsvik, A. et al. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet., 5, e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho, M.H., Castaldi, P.J., Wan, E.S., Siedlinski, M., Hersh, C.P., Demeo, D.L., Himes, B.E., Sylvia, J.S., Klanderman, B.J., Ziniti, J.P. et al. (2012) A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet., 21, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho, M.H., Boutaoui, N., Klanderman, B.J., Sylvia, J.S., Ziniti, J.P., Hersh, C.P., DeMeo, D.L., Hunninghake, G.M., Litonjua, A.A., Sparrow, D. et al. (2010) Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet., 42, 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. House, J.S., Li, H., DeGraff, L.M., Flake, G., Zeldin, D.C. and London, S.J. (2015) Genetic variation in HTR4 and lung function: GWAS follow-up in mouse. FASEB J., 29, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nichols, C.E., House, J.S., Li, H., Ward, J.M., Wyss, A., Williams, J.G., Deterding, L.J., Bradbury, J.A., Miller, L., Zeldin, D.C. and London, S.J. (2021) Lrp1 regulation of pulmonary function. Follow-Up of Human GWAS in Mice. Am. J. Respir. Cell Mol. Biol., 64, 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao, X., Qiao, D., Yang, C., Kasela, S., Kim, W., Ma, Y., Shrine, N., Batini, C., Sofer, T. and Taliun, S.A.G. (2020) Whole genome sequence analysis of pulmonary function and COPD in 19,996 multi-ethnic participants. Nat. Commun., 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeMeo, D., Carey, V., Chapman, H., Reilly, J., Ginns, L., Speizer, F., Weiss, S. and Silverman, E. (2004) Familial aggregation of FEF25–75 and FEF25–75/FVC in families with severe, early onset COPD. Thorax, 59, 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingebrigtsen, T., Thomsen, S.F., Vestbo, J., van der Sluis, S., Kyvik, K.O., Silverman, E.K., Svartengren, M. and Backer, V. (2010) Genetic influences on chronic obstructive pulmonary disease–a twin study. Respir. Med., 104, 1890–1895. [DOI] [PubMed] [Google Scholar]
- 14. Klimentidis, Y.C., Vazquez, A.I., de los Campos, G., Allison, D.B., Dransfield, M.T. and Thannickal, V.J. (2013) Heritability of pulmonary function estimated from pedigree and whole-genome markers. Front. Genet., 4, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou, J.J., Cho, M.H., Castaldi, P.J., Hersh, C.P., Silverman, E.K. and Laird, N.M. (2013) Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am. J. Respir. Crit. Care Med., 188, 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cortes, A. and Brown, M.A. (2011) Promise and pitfalls of the Immunochip. Arthritis Res. Ther., 13, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Voight, B.F., Kang, H.M., Ding, J., Palmer, C.D., Sidore, C., Chines, P.S., Burtt, N.P., Fuchsberger, C., Li, Y., Erdmann, J. et al. (2012) The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet., 8, e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bahcall, O. and Orli, B. (2013) COGS project and design of the iCOGS array. Nat. Genet., 45, 343. [DOI] [PubMed] [Google Scholar]
- 19. Wright, C.F., West, B., Tuke, M., Jones, S.E., Patel, K., Laver, T.W., Beaumont, R.N., Tyrrell, J., Wood, A.R., Frayling, T.M., Hattersley, A.T. and Weedon, M.N. (2019) Assessing the pathogenicity, penetrance, and expressivity of putative disease-causing variants in a population setting. Am. J. Hum. Genet., 104, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitt, M., Kals, M., Pärn, K., Gabriel, S.B., Lander, E.S., Palotie, A., Ripatti, S., Morris, A.P., Metspalu, A., Esko, T., Mägi, R. and Palta, P. (2017) Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet., 25, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang, J., Bakshi, A., Zhu, Z., Hemani, G., Vinkhuyzen, A.A., Lee, S.H., Robinson, M.R., Perry, J.R., Nolte, I.M. and van Vliet-Ostaptchouk, J.V. (2015) Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet., 47, 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans, L.M., Tahmasbi, R., Vrieze, S.I., Abecasis, G.R., Das, S., Gazal, S., Bjelland, D.W., De Candia, T.R., Goddard, M.E. and Neale, B.M. (2018) Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet., 50, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang, J., Lee, S.H., Goddard, M.E. and Visscher, P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wainschtein, P., Jain, D., Zheng, Z., Cupples, L.A., Shadyab, A.H., McKnight, B., Shoemaker, B.M., Mitchell, B.D., Psaty, B.M. and Kooperberg, C. (2022) Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nature Genetics, 54, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flannick, J., Mercader, J.M., Fuchsberger, C., Udler, M.S., Mahajan, A., Wessel, J., Teslovich, T.M., Caulkins, L., Koesterer, R., Barajas-Olmos, F. et al. (2019) Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature, 570, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jun, G., Manning, A., Almeida, M., Zawistowski, M., Wood, A.R., Teslovich, T.M., Fuchsberger, C., Feng, S., Cingolani, P., Gaulton, K.J. et al. (2018) Evaluating the contribution of rare variants to type 2 diabetes and related traits using pedigrees. Proc. Natl. Acad. Sci., 115, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gibson, G. (2012) Rare and common variants: twenty arguments. Nat. Rev. Genet., 13, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiao, D., Ameli, A., Prokopenko, D., Chen, H., Kho, A.T., Parker, M.M., Morrow, J., Hobbs, B.D., Liu, Y., Beaty, T.H. et al. (2018) Whole exome sequencing analysis in severe chronic obstructive pulmonary disease. Hum. Mol. Genet., 27, 3801–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiao, D., Lange, C., Beaty, T.H., Crapo, J.D., Barnes, K.C., Bamshad, M., Hersh, C.P., Morrow, J., Pinto-Plata, V.M., Marchetti, N. et al. (2016) Exome sequencing analysis in severe, early-onset chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med., 193, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prokopenko, D., Sakornsakolpat, P., Fier, H.L., Qiao, D., Parker, M.M., McDonald, M.-L.N., Manichaikul, A., Rich, S.S., Barr, R.G., Williams, C.J. et al. (2018) Whole-genome sequencing in severe chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol., 59, 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Artigas, M.S., Wain, L.V., Shrine, N., McKeever, T.M., BiLEVE, U., Sayers, I., Hall, I.P. and Tobin, M.D. (2017) Targeted sequencing of lung function loci in chronic obstructive pulmonary disease cases and controls. PLoS One, 12, e0170222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wain, L.V., Sayers, I., Artigas, M.S., Portelli, M.A., Zeggini, E., Obeidat, M.e., Sin, D.D., Bossé, Y., Nickle, D., Brandsma, C.-A. et al. (2014) Whole exome re-sequencing implicates CCDC38 and cilia structure and function in resistance to smoking related airflow obstruction. PLoS Genet., 10, e1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radder, J.E., Zhang, Y., Gregory, A.D., Yu, S., Kelly, N.J., Leader, J.K., Kaminski, N., Sciurba, F.C. and Shapiro, S.D. (2017) Extreme trait whole-genome sequencing identifies PTPRO as a novel candidate gene in emphysema with severe airflow obstruction. Am. J. Respir. Crit. Care Med., 196, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gazal, S., Loh, P.-R., Finucane, H.K., Ganna, A., Schoech, A., Sunyaev, S. and Price, A.L. (2018) Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet., 50, 1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gazal, S., Finucane, H.K., Furlotte, N.A., Loh, P.-R., Palamara, P.F., Liu, X., Schoech, A., Bulik-Sullivan, B., Neale, B.M., Gusev, A. and Price, A.L. (2017) Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat. Genet., 49, 1421–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng, J., De Vlaming, R., Wu, Y., Robinson, M.R., Lloyd-Jones, L.R., Yengo, L., Yap, C.X., Xue, A., Sidorenko, J., McRae, A.F. et al. (2018) Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet., 50, 746–753. [DOI] [PubMed] [Google Scholar]
- 37. Peterson, R.E., Kuchenbaecker, K., Walters, R.K., Chen, C.-Y., Popejoy, A.B., Periyasamy, S., Lam, M., Iyegbe, C., Strawbridge, R.J., Brick, L. et al. (2019) Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell, 179, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Browning, S.R. and Browning, B.L. (2011) Population structure can inflate SNP-based heritability estimates. Am. J. Hum. Genet., 89, 191–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taliun, D., Harris, D.N., Kessler, M.D., Carlson, J., Szpiech, Z.A., Torres, R., Taliun, S.A.G., Corvelo, A., Gogarten, S.M. and Kang, H.M. (2021) Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature, 590, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conomos, M.P., Reiner, A.P., Weir, B.S. and Thornton, T.A. (2016) Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet., 98, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oelsner, E.C., Balte, P.P., Cassano, P.A., Couper, D., Enright, P.L., Folsom, A.R., Hankinson, J., Jacobs, D.R., Jr., Kalhan, R., Kaplan, R. et al. (2018) Harmonization of respiratory data from 9 US population-based cohorts: the NHLBI pooled cohorts study. Am. J. Epidemiol., 187, 2265–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hankinson, J.L., Kawut, S.M., Shahar, E., Smith, L.J., Stukovsky, K.H. and Barr, R.G. (2010) Performance of American Thoracic Society-recommended spirometry reference values in a multiethnic sample of adults: the multi-ethnic study of atherosclerosis (MESA) lung study. Chest, 137, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pauwels, R.A., Buist, A.S., Calverley, P.M., Jenkins, C.R. and Hurd, S.S. (2001) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am. J. Respir. Crit. Care Med., 163, 1256–1276. [DOI] [PubMed] [Google Scholar]
- 44. Patterson, H.D. and Thompson, R. (1971) Recovery of inter-block information when block sizes are unequal. Biometrika, 58, 545–554. [Google Scholar]
- 45. Lee, S.H., Wray, N.R., Goddard, M.E. and Visscher, P.M. (2011) Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet., 88, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buist, A.S., McBurnie, M.A., Vodllmer, W.M., Gillespie, S., Burney, P., Mannino, D.M., Menezes, A.M., Sullivan, S.D., Lee, T.A., Weiss, K.B. et al. (2007) International variation in the prevalence of COPD (the BOLD study): a population-based prevalence study. Lancet, 370, 741–750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.