Abstract
To study the role of the capsule of Streptococcus suis serotype 2 in virulence, we generated two isogenic mutants disturbed in capsule production. For that purpose, we first cloned and characterized a major part of the capsular polysaccharide biosynthesis (cps) locus of S. suis serotype 2. Based on the established sequence, 14 open reading frames (ORFs), designated Orf2Z, Orf2Y, Orf2X, and Cps2A to Cps2K, were identified. Twelve ORFs belonged to a single transcriptional unit. The gene products of 11 of these ORFs showed similarity to proteins involved in polysaccharide biosynthesis of other gram-positive microorganisms. Nonencapsulated isogenic mutants were generated in the cps2B and cps2EF genes by insertional mutagenesis. In contrast to the wild-type S. suis serotype 2 strain, the nonencapsulated strains were highly sensitive to ingestion by porcine alveolar lung macrophages in vitro. More importantly, the nonencapsulated mutant strains were completely avirulent in young germfree pigs after intranasal inoculation. These observations indicate that the capsule of S. suis serotype 2 plays an essential role in the pathogenesis of S. suis serotype 2 infections.
Streptococcus suis is an important cause of meningitis, septicemia, arthritis, and sudden death in young pigs (4, 38). It can, however, also cause human meningitis (1). S. suis strains are identified by their morphological, biochemical, and serological characteristics. Serological classification is based on the presence of specific epitopes on its polysaccharidic capsule. So far, 35 different serotypes have been described (8, 13). Strains of S. suis can differ in virulence. Some serotypes are more frequently isolated from diseased pigs than others, suggesting that differences in virulence are associated with differences in capsular polysaccharides. In Europe, S. suis serotype 2 is the type most frequently isolated from diseased pigs, followed by serotypes 9 and 1. The idea that the capsule of S. suis serotype 2 plays a role in the pathogenesis was supported by the observation of reduced virulence for transposon mutants of S. suis impaired in capsule production (3). Moreover, it is well known that the levels of virulence of S. suis strains within a single serotype can differ greatly (37, 39). A number of strains of S. suis serotypes 1 and 2 have been shown to be highly virulent in pigs, whereas other strains of serotypes 1 and 2 are completely avirulent (33, 37, 39). Both the virulent and avirulent strains of either serotype seem to be fully encapsulated. This suggests that there is only a minor contribution of the capsule to the virulence of S. suis. Indeed, various bacterial components, such as extracellular and cell membrane-associated proteins, fimbriae, hemagglutinins, and hemolysin, have been suggested as virulence factors (7, 9, 10, 14, 15, 39, 41). However, the precise role of these protein components in the pathogenesis of the disease has not been established (29).
To provide conclusive evidence with regard to the role and contribution of the capsule of S. suis in determining virulence, we identified and characterized a major part of the DNA region encoding the proteins necessary for capsule synthesis. In addition, we generated isogenic mutants in two different capsular genes. Both isogenic mutants were found to be resistant to phagocytosis by alveolar lung macrophages in vitro. In addition, the nonencapsulated mutants were completely avirulent in young germfree pigs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. suis strains were grown in Todd-Hewitt broth (code CM189; Oxoid) and plated on Columbia agar blood base (code CM331; Oxoid) containing 6% (vol/vol) horse blood. Escherichia coli strains were grown in Luria broth (23) and plated on Luria broth containing 1.5% (wt/vol) agar. If required, antibiotics were added to the plates at the following concentrations: spectinomycin, 100 μg/ml for S. suis and 50 μg/ml for E. coli; ampicillin, 50 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CC118 | PhoA− | 21 |
| XL2 blue | Stratagene | |
| S. suis | ||
| 10 | Virulent serotype 2 strain | 40 |
| 10cpsB | Isogenic cpsB mutant of strain 10 | This work |
| 10cpsEF | Isogenic cpsEF mutant of strain 10 | This work |
| Plasmids | ||
| pKUN19 | Replication functions pUC, Ampr | 19 |
| pGEM7Zf(+) | Replication functions pUC, Ampr | Promega Corp. |
| pIC19R | Replication functions pUC, Ampr | 22 |
| pIC20R | Replication functions pUC, Ampr | 22 |
| pIC-spc | pIC19R containing Spcr gene of pDL282 | Lab collection |
| pDL282 | Replication functions of pBR322 and pVT736-1, Ampr, Spcr | 31 |
| pPHOS2 | pIC-spc containing the truncated phoA gene of pPHO7 as a PstI-BamHI fragment | This work |
| pPHO7 | Contains truncated phoA gene | 12 |
| pPHOS7 | pPHOS2 containing chromosomal S. suis DNA | This work |
| pCPS6 | pKUN19 containing 6-kb HindIII fragment of cps operon | This work (Fig. 1) |
| pCPS7 | pKUN19 containing 3.5-kb EcoRI-HindIII fragment of cps operon | This work (Fig. 1) |
| pCPS11 | pCPS7 in which 0.4-kb PstI-BamHI fragment of cpsB gene is replaced by Spcr gene of pIC-spc | This work (Fig. 1) |
| pCPS17 | pKUN19 containing 3.1-kb KpnI fragment of cps operon | This work (Fig. 1) |
| pCPS18 | pKUN19 containing 1.8-kb SnaBI fragment of cps operon | This work (Fig. 1) |
| pCPS20 | pKUN19 containing 3.3-kb XbaI-HindIII fragment of cps operon | This work (Fig. 1) |
| pCPS23 | pGEM7Zf(+) containing 1.5-kb MluI fragment of cps operon | This work (Fig. 1) |
| pCPS25 | pIC20R containing 2.5-kb KpnI-SalI fragment of pCPS17 | This work (Fig. 1) |
| pCPS26 | pKUN19 containing 3.0-kb HindIII fragment of cps operon | This work (Fig. 1) |
| pCPS27 | pCPS25 containing 2.3-kb XbaI (blunt)-ClaI fragment of pCPS20 | This work (Fig. 1) |
| pCPS28 | pCPS27 containing the 1.2-kb PstI-XhoI Spcr gene of pIC-spc | This work (Fig. 1) |
Ampr, ampicillin resistant; Spcr, spectinomycin resistant; and cps, capsular polysaccharide.
Serotyping.
The S. suis strains were serotyped by the slide agglutination test with serotype-specific antibodies (36).
Selection of genes encoding exported proteins.
Chromosomal DNA of S. suis serotype 2 was digested with AluI. The 300- to 500-bp fragments were ligated to SmaI-digested pPHOS2. Ligation mixtures were transformed to PhoA− E. coli CC118. Transformants were plated on Luria broth agar plates supplemented with 5-bromo-4-chloro-3-indolylphosphate (BCIP) (50 μg/ml) (Boehringer, Mannheim, Germany). Blue colonies were purified on fresh Luria broth/BCIP plates to verify the blue phenotype.
DNA techniques and sequence analysis.
Routine DNA manipulations were performed as described by Sambrook et al. (28). DNA sequences were determined on a 373A DNA Sequencing System (Applied Biosystems, Warrington, Great Britain). Samples were prepared by use of an ABI/PRISM dye terminator cycle sequencing ready reaction kit (Applied Biosystems). Sequencing data were assembled and analyzed by using the MacMollyTetra software package. Custom-made sequencing primers were purchased from Life Technologies. The BLAST software package was used to search for protein sequences homologous to the deduced amino acid sequences in the GenBank/EMBL databases.
Construction of gene-specific knock out mutants.
To construct the mutant strains 10cpsΔB and 10cpsΔEF, we electrotransformed pathogenic strain 10 (37, 41) of S. suis serotype 2 with pCPS11 and pCPS28, respectively. In these plasmids the cpsB and cpsEF genes are inactivated by the insertion of a spectinomycin resistance gene. To create pCPS11, the internal 400-bp PstI-BamHI fragment of the cpsB gene in pCPS7 was replaced by the 1,200-bp PstI-BamHI fragment from pIC-spc, containing the spectinomycin resistance gene. To construct pCPS28 we have used pIC20R. Into this plasmid we inserted the KpnI-SalI fragment from pCPS17 (resulting in pCPS25) and the XbaI-ClaI fragment from pCPS20 (resulting in pCPS27). pCPS27 was digested with PstI and XhoI and ligated to the 1,200-bp PstI-XhoI fragment, containing the spectinomycin resistance gene of pIC-spc. The electrotransformation to S. suis was carried out as described before (30).
Southern blotting and hybridization.
Chromosomal DNA was isolated as described by Sambrook et al. (28). DNA fragments were separated on 0.8% agarose gels and transferred to Zeta-Probe GT membranes (Bio-Rad) as described by Sambrook et al. (28). DNA probes were labelled with [α-32P]dCTP (3,000 Ci mmol−1; Amersham) by use of a random primed labelling kit (Boehringer). The DNA on the blots was hybridized at 65°C with the appropriate DNA probes as recommended by the supplier of the Zeta-Probe membranes. After hybridization, the membranes were washed twice with a solution of 40 mM sodium phosphate (pH 7.2), 1 mM EDTA, and 5% sodium dodecyl sulfate for 30 min at 65°C and twice with a solution of 40 mM sodium phosphate (pH 7.2), 1 mM EDTA, and 1% sodium dodecyl sulfate for 30 min at 65°C.
Electron microscopy.
Bacteria were prepared for electron microscopy as described by Wagenaar et al. (42). Shortly, bacteria were mixed with agarose MP (Boehringer) of 37°C to a concentration of 0.7%. The mixture was immediately cooled on ice. Upon gelling, samples were cut into 1- to 1.5-mm-thick slices and incubated in a fixative containing 0.8% glutaraldehyde and 0.8% osmium tetroxide. Subsequently, the samples were fixed and stained with uranyl acetate by microwave stimulation, dehydrated, and embedded in eponaraldite resin. Ultrathin sections were counterstained with lead citrate and examined with a Philips CM 10 electron microscope at 80 kV. In addition, we used the polycationic ferritin method as described by Quessy et al. (26).
Phagocytosis assay.
Porcine alveolar macrophages (AM) were obtained from the lungs of specific-pathogen-free (SPF) pigs. Lung lavage samples were collected as described by van Leengoed et al. (35). Cells were suspended in Eagle’s minimal essential medium (EMEM) containing 6% (vol/vol) SPF pig serum and adjusted to 107 cells per ml. Phagocytosis assays were performed as described by Leij et al. (20). Briefly, 107 S. suis cells were incubated with 6% SPF pig serum for 30 min at 37°C in a head-over-head rotor at 6 rpm, to opsonize the cells. We combined 107 AM and 107 opsonized S. suis cells and incubated them at 37°C under continuous rotation at 6 rpm. At 0, 30, 60, and 90 min, 1-ml samples were collected and mixed with 4 ml of ice-cold EMEM to stop phagocytosis. Phagocytes were removed by centrifugation for 4 min at 110 × g and 4°C. The number of CFU in the supernatants was determined by plating. Control experiments were carried out simultaneously by combining 107 opsonized S. suis cells with EMEM without AM.
Killing assays.
Killing assays were performed as described by Leij et al. (20). AM (107/ml) and opsonized S. suis cells (107/ml) were mixed 1:1 and incubated for 10 min at 37°C under continuous rotation at 6 rpm. Ice-cold EMEM was added to stop further phagocytosis and killing. To remove extracellular S. suis cells, phagocytes were washed twice (4 min, 110 × g, 4°C) and resuspended in 5 ml of EMEM containing 6% SPF pig serum. The resuspended AM were incubated at 37°C under rotation at 6 rpm. After 0, 15, 30, 60, and 90 min, samples were collected and mixed with ice-cold EMEM to stop further killing. The samples were centrifuged for 4 min at 110 × g at 4°C, and the phagocytic cells were lysed in EMEM containing 1% saponin for 20 min at room temperature. The number of CFU in the suspensions was determined by plating.
Experimental infections.
Germfree pigs, crossbreeds of Great Yorkshire and Dutch Landrace, were obtained from sows by cesarean sections. The surgery was performed in sterile flexible film isolators. Pigs were allotted to groups, each consisting of 4 pigs, and were housed in sterile stainless steel incubators. Housing conditions and feeding regimens were as described before (37, 41). Pigs were inoculated intranasally with S. suis serotype 2 as described before (37, 41). To predispose the pigs to infection with S. suis, 5-day-old pigs were inoculated intranasally with about 107 CFU of Bordetella bronchiseptica 92932. Two days later the pigs were inoculated intranasally with S. suis serotype 2 (106 CFU). Pigs were monitored twice daily for clinical signs of disease, such as fever, nervous signs, and lameness. Blood samples were collected three times a week from each pig. Leucocytes were counted with a cell counter. To monitor infection with S. suis and B. bronchiseptica and to check for absence of contaminants, we collected swabs of the nasopharynx and the feces daily. The swabs were plated directly onto Columbia agar containing 6% horse blood. After the pigs were killed, they were examined for pathological changes. Tissue specimens from the central nervous system (CNS), serosae, and joints were examined bacteriologically and histologically as described before (37, 41). Colonization of the serosae was scored positively when S. suis was isolated from the pericardium, thoracic pleura, or peritoneum. Colonization of the joints was scored positively when S. suis was isolated from one or more joints (12 joints per animal were scored).
Nucleotide sequence accession number.
The nucleotide sequence data have been submitted to GenBank under accession no. AF118389.
RESULTS
Identification and isolation of capsule-encoding DNA.
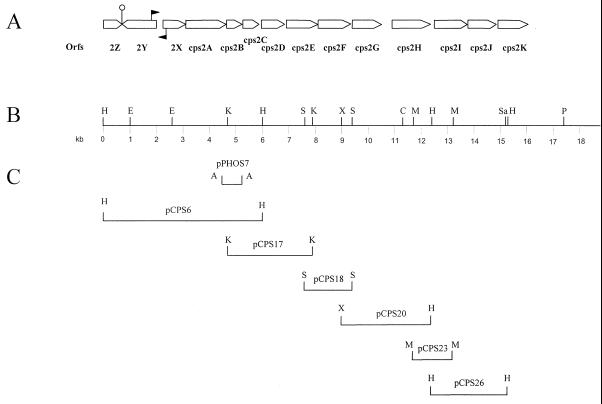
Initially, a part of the capsular locus of S. suis serotype 2 was isolated in an attempt to identify secreted proteins by genetic means (12, 25). For this purpose chromosomal DNA of S. suis serotype 2 was cloned in E. coli in front of a 5′-truncated alkaline phosphatase gene. To do this, we made use of the vector pPHOS2 (Table 1), which contained the truncated alkaline phosphatase gene of pPHO7 (12) as well as a spectinomycin resistance gene (31). A number of E. coli clones displayed a dark blue phenotype when plated on media containing BCIP, indicating that the cloned fragment contained a promoter, a translational start site, and a signal sequence. The deduced amino acid sequence of one of the cloned fragments (on plasmid pPHOS7) showed a high similarity (37% identity) to a protein (Cps14C) involved in capsular synthesis of Streptococcus pneumoniae (18). This strongly suggested that pPHOS7 contained a part of the corresponding cps gene of S. suis serotype 2. Subsequently, the insert of pPHOS7 (Fig. 1C) was used as a probe to identify chromosomal DNA fragments containing flanking cps genes. A 6-kb HindIII fragment was identified and cloned in pKUN19. This yielded clone pCPS6 (Fig. 1C). Sequence analysis of the insert revealed that pCPS6 contained the 5′ end of the cps locus. Sequences of the 3′ end of pCPS6 were, in turn, used to identify a chromosomal fragment containing cps sequences located further downstream. This fragment was also cloned in pKUN19, resulting in pCPS17. Using a similar approach, we subsequently isolated the plasmids pCPS18, pCPS20, pCPS23, and pCPS26 containing downstream cps sequences (Fig. 1C).
FIG. 1.
Organization of the cps2 gene cluster of S. suis type 2. (A) Genetic map of the cps2 gene cluster. The open arrows represent potential ORFs. Gene designations are indicated below the ORFs. The closed arrows indicate the position of the potential promoter sequences. | indicates the position of the potential transcription regulator sequence. (B) Physical map of the cps2 locus. (C) The DNA fragments cloned in the various plasmids. Restriction sites are as follows: A, AluI; C, ClaI; E, EcoRI; H, HindIII; K, KpnI; M, MluI; P, PstI; S, SnaBI; Sa, SacI; X, XbaI.
Analysis of the cps operon.
The complete nucleotide sequences of the cloned fragments were determined. Examination of the compiled sequence revealed the presence of 14 potential open reading frames (ORFs), which were designated Orf2Z, Orf2Y, Orf2X, and Cps2A through Cps2K (Fig. 1A). Orf2Z, located at the 5′ end of the sequence, was incomplete. Compared to the other ORFs, Orf2Y is expressed in the opposite orientation. Two potential promoter sequences were identified. One was located 313 bp (positions 1885 to 1865 and 1884 and 1889) upstream of Orf2X. The other was located 68 bp upstream of Orf2Y (positions 2241 to 2236 and 2216 to 2211). Between Orf2Y and Orf2Z the sequence contained a potential stem-loop structure, which could act as a transcription terminator. Each ORF is preceded by a ribosome-binding site, and the majority of the ORFs are very closely linked. The only significant intergenic gap was that found between Cps2G and Cps2H (389 nucleotides). No obvious promoter sequences or potential stem-loop structures were found in this region. This suggests that Orf2X and Cps2A through Cps2K are part of a single transcriptional unit.
A list of all ORFs with their properties is shown in Table 2. Orf2Z showed similarity to the YitS protein of Bacillus subtilis, a protein with an unknown function. Orf2Y showed homology to the YcxD protein of B. subtilis (43), which is supposed to be a regulatory protein. Orf2X showed homology with the hypothetical YAAA proteins with unknown function of Haemophilus influenzae and E. coli. The products of the cps2A, cps2B, cps2C, and cps2D genes showed significant homologies with the CpsA, CpsC, CpsD, and CpsB proteins of several streptococci (Table 2), suggesting similar functions for these proteins. Hence, Cps2A may have a role in the regulation of the capsular polysaccharide synthesis, Cps2B and Cps2C could be involved in the chain length determination of the type 2 capsule, and Cps2C could play an additional role in the export of the polysaccharide. Cps2D is homologous to Cps proteins of streptococci involved in the polysaccharide or exopolysaccharide synthesis, but it is without a known specific function (18). The proteins encoded by the cps2E, cps2F, cps2G, cps2H, cps2J, and cps2K genes showed homology to proteins with glycosyltransferase activities of several streptococci (5, 16, 17, 18, 32), suggesting that these proteins are involved in the biosynthesis of the type 2 oligosaccharide subunit. The protein encoded by the cps2I gene showed homology to a protein of S. pneumoniae with potential polysaccharide polymerase activity (5).
TABLE 2.
Properties of ORFs in the cps locus of S. suis serotype 2 and similarities to gene products of other bacteria
| ORF | Nucleotide positions in sequence | No. of amino acids | Proposed function of gene producta | Bacterial strain(s) with similar gene product (% identity) | Reference or accession no. |
|---|---|---|---|---|---|
| Orf2Z | 1–719 | 240 | Unknown | B. subtilis YitS (26) | Y09478 |
| Orf2Y | 2079–822 | 419 | Transcription regulation | B. subtilis YcxD (39) | 43 |
| Orf2X | 2202–2934 | 244 | Unknown | H. influenzae YAAA (24) | P43908 |
| Cps2A | 3041–4484 | 481 | Regulation | S. pneumoniae Cps19fA (58) | 11, 24 |
| Cps2B | 4504–5191 | 229 | Chain length determination | S. pneumoniae type 3 Orf1 (58) | 2 |
| Cps2C | 5203–5878 | 225 | Chain length determination/export | S. pneumoniae Cps23fD (63) | 5 |
| Cps2D | 5919–6648 | 243 | Unknown | S. pneumoniae CpsB (62) | 5 |
| Cps2E | 6675–8052 | 459 | Glycosyltransferase | S. pneumoniae Cps14E (56) | 16, 18 |
| Cps2F | 8089–9256 | 389 | Glycosyltransferase | S. pneumoniae Cps23fT (72) | 5 |
| Cps2G | 9262–10417 | 385 | Glycosyltransferase | S. thermophilus EpsF (25) | 32 |
| Cps2H | 10808–12176 | 457 | Glycosyltransferase | S. mutans RGPECb (29) | D1033055 |
| Cps2I | 12213–13443 | 410 | Capsular polysaccharide polymerase | S. pneumoniae Cps23fI (48) | 5 |
| Cps2J | 13583–14579 | 332 | Glycosyltransferase | S. pneumoniae Cps14J (31) | 17 |
| S. pneumoniae Cps 14I (27) | 17 | ||||
| Cps2K | 14574–15401 | 276 | Glycosyltransferase | S. pneumoniae Cps14J (40) | 17 |
Predicted by sequence similarity.
Similarity refers to the amino-terminal part of the gene product.
Construction of mutants impaired in capsule synthesis.
To evaluate the role of the capsule of S. suis serotype 2 in virulence, we constructed two isogenic mutants in which capsule production was disturbed. To construct mutants 10cpsΔB and 10cpsΔEF, the plasmids pCPS11 and pCPS28 were used. pCPS11 and pCPS28 were electrotransformed into strain 10 of S. suis serotype 2, and spectinomycin-resistant colonies were selected. Southern blotting and hybridization experiments were used to select double crossover integration events (data not shown).
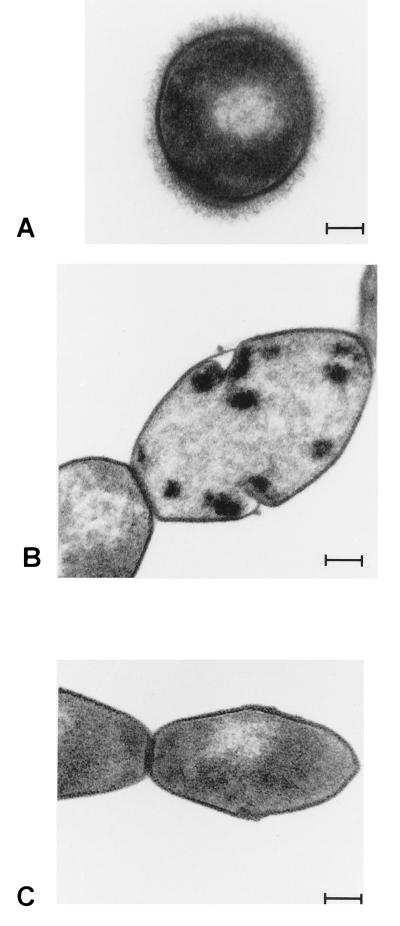
To test whether the capsular structure of the mutant strains 10cpsΔB and 10cpsΔEF was disturbed, we used a slide agglutination test (36). The parent strain, strain 10, of S. suis serotype 2 agglutinated only in type 2-specific serum. The mutant strains, however, agglutinated in sera specific for all S. suis strains, but they also agglutinated in the absence of serotype-specific serum. This indicated that in the mutant strains the capsular structure was disturbed. To confirm this, thin sections of wild-type and mutant strains were compared by electron microscopy. Compared to the wild-type strain (Fig. 2A), the amount of capsule produced by the mutant strains was greatly reduced (Fig. 2B and C). No capsular material could be detected on the surfaces of the mutant strains. Similar results were obtained after the polycationic ferritin method was used (results not shown).
FIG. 2.
Transmission electron micrographs of thin sections of various S. suis strains. Panels: A, wild-type strain, strain 10; B, mutant strain 10cpsΔB; and C, mutant strain 10cpsΔEF. Bar = 100 nm.
Capsular mutants are sensitive to phagocytosis and killing by AM.
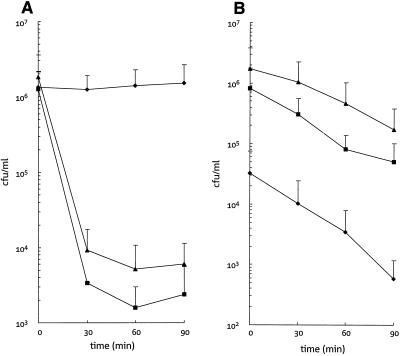
The capsular mutants and the parent strain were tested for the ability to resist phagocytosis by AM in the presence of porcine SPF serum. As shown in Fig. 3A, the wild-type strain (strain 10) is resistant to phagocytosis under the in vitro conditions used (Fig. 3A). In contrast, both mutant strains were efficiently ingested by the macrophages (Fig. 3A). After 90 min, more than 99.7% (strain 10cpsΔB) and 99.8% (strain 10cpsΔEF) of the mutants were ingested by the macrophages. Moreover, as shown in Fig. 3B, the ingested strains were efficiently killed by the macrophages. From 90 to 98% of all ingested cells were killed within 90 min. No differences in killing efficiency could be observed between wild-type and mutant strains. Similar results were obtained after polymorphonuclear leukocytes were used (results not shown). These data indicate that the capsule of S. suis serotype 2 efficiently protects the bacterium from uptake by macrophages in vitro.
FIG. 3.
(A) Kinetics of phagocytosis of wild-type and mutant S. suis strains by porcine AM. Average data of several experiments are presented. Bars are standard deviations. (B) Kinetics of intracellular killing of wild-type and mutant S. suis strains by porcine AM. Average data of several experiments are represented. Bars are standard deviations. ⧫, wild-type strain, strain 10; ▴, mutant strain 10cpsΔB; ■, mutant strain 10cpsΔEF.
Capsular mutants are avirulent in germfree piglets.
The virulence properties of the wild-type and mutant strains were tested by experimental infection of newborn germfree pigs (37, 41). Table 3 shows that specific and nonspecific signs of disease could be observed in all pigs inoculated with the wild-type strain. All pigs inoculated with the wild-type strain died during the course of the experiment or were killed because of serious illness or nervous disorders (Table 3). In contrast, the pigs inoculated with strains 10cpsΔB or 10cpsΔEF showed no specific signs of disease and all of these pigs survived until the end of the experiment. Moreover, we observed significant differences in the fever index and in the leukocyte index between pigs inoculated with wild-type and mutant strains (Table 3). S. suis strains and B. bronchiseptica could be isolated from the nasopharyngeal and fecal swab samples of all pigs from 1 day postinfection until the end of the experiment. Postmortem, the wild-type strain could frequently be isolated from the CNS, kidney, heart, liver, spleen, serosae, joints, and tonsils. Mutant strains could be recovered from the tonsils but were never recovered from the kidney, liver, or spleen. Interestingly, small numbers of the mutant strains could be isolated from the CNS, the serosae, the joints, the lungs, and the heart. Agglutination tests and Southern blot analyses showed that these mutant strains had the unencapsulated phenotype and genotype (results not shown). Taken together, these data demonstrate that mutant S. suis strains impaired in capsule production are avirulent in young germfree pigs.
TABLE 3.
Virulence of wild-type and capsular mutant S. suis strains in germfree pigs
| S. suis straina | No. of pigs | Mortality (%)b | Morbidity (%)c | Clinical indexd of the group (%)
|
Fever index (%)g | Leukocyte index (%)h | No. of pigs in which S. suis was isolated from:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Specific symptomse | Nonspecific symptomsf | CNS | Serosae | Joints | ||||||
| 10 | 4 | 100 | 100 | 11 | 88 | 43 | 44 | 2 | 3 | 4 |
| 10cpsΔB | 4 | 0 | 0 | 0 | 10 | 1 | 3 | 1 | 3 | 2 |
| 10cpsΔEF | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 2 |
Strain 10 is the wild-type strain, and strains 10cpsΔB and 10cpsΔEF are isogenic capsular mutant strains.
Percentage of piglets that died due to infection or had to be killed for animal welfare reasons.
Percentage of pigs with specific symptoms.
Percentage of observations which matched the described criteria.
Ataxia, lameness of at least one joint, and/or stiffness.
Inappetence and/or depression.
Percentage of observations for the experimental group of a body temperature of >40°C.
Percentage of blood samples for the group in which the concentration of granulocytes was >1010/liter.
DISCUSSION
In the present paper we describe the identification and the molecular characterization of a 16-kb DNA fragment containing a major part of the genetic determinant involved in the capsular polysaccharide biosynthesis of S. suis serotype 2. To study the role of the capsule in resistance to phagocytosis and in virulence, we constructed two isogenic mutants in which capsule synthesis was disturbed. In 10cpsΔB, the cps2B gene was disturbed by the insertion of an antibiotic resistance gene, whereas in 10cpsΔEF parts of the cps2E and cps2F genes were replaced by an antibiotic resistance gene. By electron microscopical analysis both mutant strains were found to be completely unencapsulated. Although this finding confirms that the cpsB and cpsEF genes are involved in capsular synthesis, this finding does not give any clues to the function of these proteins, since they form part of an operon structure and polar effects on the expression of downstream genes cannot be excluded. The behavior of the mutants in the in vitro phagocytosis and killing assays clearly showed that the capsular polysaccharide of S. suis serotype 2 is a surface component with antiphagocytic activity. Wild-type encapsulated bacteria were ingested by phagocytes at a very low frequency, whereas the mutant unencapsulated bacteria were efficiently ingested by porcine macrophages. Once ingested, wild-type and mutant strains seemed to be killed with the same relative efficiency. This suggests that the loss of capsular material is associated with loss of capacity to resist uptake by macrophages. This loss of resistance to in vitro phagocytosis was associated with an almost complete attenuation of the virulence of the mutant strains in germfree pigs. All pigs inoculated with the mutant strains survived the experiment and did not show any specific clinical signs of disease. Only some nonspecific clinical signs of disease could be observed. Moreover, small numbers of mutant bacteria could be reisolated from the pigs. This supports the idea that, as in other pathogenic streptococci, the capsule of S. suis acts as an important virulence factor. Our data obtained with the isogenic mutants are in agreement with the data recently reported by Charland et al. (3). They reported that transposon mutants, which are impaired in capsule production, showed reduced virulence in pigs and mice (3). However, to construct these transposon mutants, the authors used the serotype 2 reference strain S735. We previously showed that strain S735 is only weakly virulent for young pigs (40). Moreover, since the insertion sites of the transposon in the mutants were not determined, it could not be concluded that the observed reduction in virulence was a direct consequence of impaired capsule synthesis.
Initially a part of the cps2B gene was cloned by screening for signal sequences. The hydrophobicity profile of the clone showed that the N-terminal part of the sequence resembled the characteristics of a typical signal peptide: a short N-terminal region is followed by a hydrophobic region of 38 amino acids. Apparently, this region was able to translocate alkaline phosphatase across the cellular membrane in E. coli. The hydrophobicity plot of the corresponding Cps14C protein of S. pneumoniae showed two hydrophobic segments, one each at its N and C termini, and a hydrophilic domain in the central part (18). The cellular location of this protein is unknown. The region homologous to the second hydrophobic domain was not cloned in pPHOS7.
The cloned and sequenced region described here contained 14 ORFs. At least 12 of these ORFs belong to a single transcriptional unit, suggesting a coordinated control of the expression of these genes. Based on sequence similarities we could assign putative functions to most of the gene products. We thereby identified gene products involved in regulation (Cps2A), chain length determination (Cps2B, C), export (Cps2C), biosynthesis (Cps2E through Cps2H, Cps2J, and Cps2K), and polymerization (Cps2I). The overall organization is similar to that of the cps and eps gene clusters of a number of gram-positive bacteria (17, 27, 32, 34). A region involved in biosynthesis is preceded by a region containing genes with more common functions. Although, based on sequence similarities, a role of most of the gene products in the polysaccharide biosynthesis could be envisaged, the role of the orf2Z, orf2Y, and orf2X genes remains unclear so far. The incomplete orf2Z gene was located at the 5′ end of the cloned fragment. Orf2Z showed some similarity to the YitS protein of B. subtilis. However, because the function of the YitS protein is unknown, this did not give us any information about the possible function of Orf2Z. Because the orf2Z gene is not a part of the cps operon, a role of this gene in polysaccharide biosynthesis is not expected. The Orf2Y protein showed similarity to the YcxD protein of B. subtilis (43). The YcxD protein was suggested to be a regulatory protein. Similarly, Orf2Y may be involved in the regulation of polysaccharide biosynthesis. The Orf2X protein showed similarity to the YAAA proteins of H. influenzae and E. coli. The function of these proteins is unknown. In S. suis serotype 2 the orf2X gene seems to be the first gene in the cps2 operon. This suggests a role of Orf2X in polysaccharide biosynthesis. In H. influenzae and E. coli, however, these proteins are not associated with capsular gene clusters.
The products encoded by the cps2E, cps2F, cps2G, cps2H, cps2J, and cps2K genes showed similarities to glycosyltransferases of several streptococci (5, 16–18, 32). The cps2E gene product showed strong homology to the Cps14E protein of S. pneumoniae (16, 18). Cps14E is a glucosyl-1-phosphate transferase that links glucose to a lipid carrier (18). In S. pneumoniae this is the first step in the biosynthesis of the oligosaccharide repeating unit. The structure of the S. suis serotype 2 capsule is unknown, but it is composed of glucose, galactose, N-acetylglucosamine, rhamnose, and sialic acid in a ratio of 1:3:1:1:1 (6). Therefore, because the capsule of S. suis serotype 2 does contain glucose (6), we speculate that Cps2E of S. suis could also have glucosyltransferase activity and is probably involved in the linkage of the first sugar to the lipid carrier. The cps2F gene product showed homology to the Cps23fT protein, which has rhamnosyltransferase activity, of S. pneumoniae (5). Because rhamnose is a component of the S. suis serotype 2 polysaccharide (6) Cps2F could have rhamnosyltransferase activity. The cps2G gene encoded a protein that showed moderate similarity to the epsF gene product of Streptococcus thermophilus (32). On the basis of homology epsF is suggested to encode galactosyltransferase activity. Hence, a similar galactosyltransferase activity is proposed for Cps2G. The cps2H gene encodes a protein with an N-terminal region that is similar to the N-terminal region of the RGPEC protein of Streptococcus mutans (D1033055). For this protein a glycosyltransferase activity was suggested. Moreover, the hydrophobicity plots of Cps2H and RGPEC looked very similar in these regions (data not shown). Therefore, Cps2H could have glycosyltransferase activity as well. Cps2J and Cps2K showed homology to Cps14J of S. pneumoniae (17). Cps2J also showed homology to Cps14I of S. pneumoniae. Cps14I has N-acetylglucosaminyltransferase activity, whereas Cps14J possesses a β-1,4-galactosyltransferase activity (17). In S. pneumoniae Cps14I is responsible for the addition of the third sugar and Cps14J is responsible for the addition of the last sugar in the synthesis of the type 14 repeating unit (17). Because the capsule of S. suis serotype 2 contains galactose as well as N-acetylglucosamine components, galactosyltransferase N-acetylglucoaminyltransferase activities could be envisaged for the cps2J and cps2K gene products, respectively. The two conserved regions, DXS and DXDD, which are conserved in several glycosyltransferases (17) and which are proposed to be important for catalytic activity, were also found in Cps2J and Cps2K. The Cps2I protein showed similarity to the Cps23fI protein of S. pneumoniae, which has a capsular polysaccharide polymerase activity (5), suggesting that Cps2I could be involved in the polymerization of the type 2 specific oligosaccharides.
The capsule of S. suis serotype 2 is composed of glucose, galactose, N-acetylglucosamine, rhamnose, and sialic acid (6). Based on sequence homology genes encoding potential glucosyl-, galactosyl-, N-acetylglucosaminyl-, and rhamnosyltransferase activities could be identified. However, we have not found genes homologous to genes involved in the synthesis, activation, and transfer of sialic acid. Moreover, since we do not know whether the cps2K gene is the last gene in the cps2 locus, these genes can be located downstream of cps2K. Therefore, in future experiments we will concentrate on the cloning and characterization of these genes. Moreover, the analysis of isogenic mutants in which the individual genes are interrupted, without disturbing expression of the downstream genes, will give more information about the role of the individual cps2 genes in the polysaccharide biosynthesis of the S. suis serotype 2 capsule.
REFERENCES
- 1.Arends J P, Zanen H C. Meningitis caused by Streptococcus suis in humans. Rev Infect Dis. 1988;10:131–137. doi: 10.1093/clinids/10.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Arrecubieta C, Garcia E, Lopez R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 3.Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology. 1998;144:325–332. doi: 10.1099/00221287-144-2-325. [DOI] [PubMed] [Google Scholar]
- 4.Clifton-Hadley F A. Streptococcus suis type 2 infections. Br Vet J. 1983;139:1–5. doi: 10.1016/s0007-1935(17)30581-x. [DOI] [PubMed] [Google Scholar]
- 5.Coffey T J, Enright M C, Daniels M, Morona J K, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthesis locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 6.Elliott S D, Tai J Y. The type specific polysaccharide of Streptococcus suis. J Exp Med. 1978;148:1699–1704. doi: 10.1084/jem.148.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feder I, Chengappa M M, Fenwick B, Rider M, Staats J. Partial characterization of Streptococcus suis type 2 hemolysin. J Clin Microbiol. 1994;32:1256–1260. doi: 10.1128/jcm.32.5.1256-1260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottschalk M, Lacouture S, Dubreuil J D. Characterization of Streptococcus suis type 2 hemolysin. Microbiology. 1995;141:189–195. doi: 10.1099/00221287-141-1-189. [DOI] [PubMed] [Google Scholar]
- 10.Gottschalk M, Lebrun A, Jacques M, Higgins R. Hemagglutination properties of Streptococcus suis. J Clin Microbiol. 1990;28:2156–2158. doi: 10.1128/jcm.28.9.2156-2158.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidolin A, Morona J M, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guitierrez C, Devedjian J C. Plasmid facilitating in vitro construction of PhoA fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. Description of six new capsular types (28 through 34) of Streptococcus suis. J Vet Diagn Investig. 1995;7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs A A C, Loeffen P L W, van den Berg A J G, Storm P K. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect Immun. 1994;62:1742–1748. doi: 10.1093/benz/9780199773787.article.b00034458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacques M, Gottschalk M, Foiry B, Higgins R. Ultrastructural study of surface components of Streptococcus suis. J Bacteriol. 1990;172:2833–2838. doi: 10.1128/jb.172.6.2833-2838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolkman M A B, Morrison D A, van der Zeijst B A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 18.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 19.Konings R N H, Verhoeven E J M, Peeters B P H. pKUN vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- 20.Leij P C J, van Furth R, van Zwet T L. In vitro determination of phagocytosis and intracellular killing of polymorphonuclear and mononuclear phagocytes. In: Weir D M, Herzenberg L A, Blackwell C, Herzenberg L A, editors. Handbook of experimental immunology. 2. Cellular immunology. Oxford, United Kingdom: Blackwell Scientific Publications; 1986. pp. 46.1–46.21. [Google Scholar]
- 21.Manoil C, Beckwith J. A transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 23.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthesis pathway. Mol Microbiol. 1997;23:761–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 25.Pearce B J, Yin Y B, Masure H R. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol Microbiol. 1993;9:1037–1050. doi: 10.1111/j.1365-2958.1993.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 26.Quessy S, Dubreuil J D, Jacques M, Malouin F, Higgins R. Increase of capsular material thickness following in vivo growth of virulent Streptococcus suis serotype 2 strains. FEMS Microbiol Lett. 1994;115:19–26. doi: 10.1111/j.1574-6968.1994.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 27.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Smith H E, Vecht U, Wisselink H J, Stockhofe-Zurwieden N, Biermann Y, Smits M A. Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect Immun. 1996;64:4409–4412. doi: 10.1128/iai.64.10.4409-4412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith H E, Wisselink H J, Vecht U, Gielkens A L J, Smits M A. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology. 1995;141:181–188. doi: 10.1099/00221287-141-1-181. [DOI] [PubMed] [Google Scholar]
- 31.Sreenivasan P K, LeBlanc D L, Lee L N, Fives-Taylor P. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect Immun. 1991;59:4621–4627. doi: 10.1128/iai.59.12.4621-4627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockhofe-Zurwieden N, Vecht U, Wisselink H J, van Lieshout H, Smith H E. Comparative studies on the pathogenicity of different Streptococcus suis serotype 1 strains. In: Monetti P G, Vignola G, editors. Proceedings of the 14th International Pig Veterinary Society Congress. 1996. p. 299. Bologna, Italy. [Google Scholar]
- 34.van Kranenburg R, Marugg J D, van Swam I I, Willem N J, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 35.van Leengoed L A, Kamp E M, Pol J M A. Toxicity of Haemophilus pleuropneumoniae to porcine lung macrophages. Vet Microbiol. 1989;19:337–349. doi: 10.1016/0378-1135(89)90099-0. [DOI] [PubMed] [Google Scholar]
- 36.van Leengoed L A M G, Vecht U, Verheyen E R M. Streptococcus suis type 2 infections in pigs in The Netherlands (part two) Vet Q. 1987;9:111–117. doi: 10.1080/01652176.1987.9694087. [DOI] [PubMed] [Google Scholar]
- 37.Vecht U, Arends J P, van der Molen E J, van Leengoed L A M G. Differences in virulence between two strains of Streptococcus suis type 2 after experimentally induced infection of newborn germfree pigs. Am J Vet Res. 1989;50:1037–1043. [PubMed] [Google Scholar]
- 38.Vecht U, van Leengoed L A M G, Verheyen E R M. Streptococcus suis infections in pigs in The Netherlands (part one) Vet Q. 1985;7:315–321. doi: 10.1080/01652176.1985.9694005. [DOI] [PubMed] [Google Scholar]
- 39.Vecht U, Wisselink H J, Jellema M L, Smith H E. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect Immun. 1991;59:3156–3162. doi: 10.1128/iai.59.9.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vecht U, Wisselink H J, Stockhofe-Zurwieden N, Smith H E. Characterization of virulence of the Streptococcus suis serotype 2 reference strain Henrichsen S 735 in newborn gnotobiotic pigs. Vet Microbiol. 1996;51:125–136. doi: 10.1016/0378-1135(96)00028-4. [DOI] [PubMed] [Google Scholar]
- 41.Vecht U, Wisselink H J, van Dijk J E, Smith H E. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun. 1992;60:550–556. doi: 10.1128/iai.60.2.550-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagenaar F, Kok G L, Broekhuijsen-Davies J M, Pol J M A. Rapid cold fixation of tissue samples by microwave irradiation for use in electron microscopy. Histochem J. 1993;25:719–725. doi: 10.1007/BF00211767. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, Kumamano M, Kurita K. The 25°-36° region of the Bacillus subtilis chromosome: determination of the sequence of a 146 kb segment and identification of 113 genes. Microbiology. 1996;142:3047–3056. doi: 10.1099/13500872-142-11-3047. [DOI] [PubMed] [Google Scholar]