Abstract
An algorithm for retrospective correction of frequency and phase offsets in MRS data is presented. The algorithm, termed robust spectral registration (rSR), contains a set of subroutines designed to robustly align individual transients in a given dataset even in cases of significant frequency and phase offsets or unstable lipid contamination and residual water signals. Data acquired by complex multiplexed editing approaches with distinct subspectral profiles are also accurately aligned. Automated removal of unstable lipid contamination and residual water signals is applied first, when needed. Frequency and phase offsets are corrected in the time domain by aligning each transient to a weighted average reference in a statistically optimal order using nonlinear least-squares optimization. The alignment of subspectra in edited datasets is performed using an approach that specifically targets subtraction artifacts in the frequency domain. Weighted averaging is then used for signal averaging to down-weight poorer-quality transients. Algorithm performance was assessed on one simulated and 67 in vivo pediatric GABA−/GSH-edited HERMES datasets and compared with the performance of a multistep correction method previously developed for aligning HERMES data. The performance of the novel approach was quantitatively assessed by comparing the estimated frequency/phase offsets against the known values for the simulated dataset or by examining the presence of subtraction artifacts in the in vivo data. Spectral quality was improved following robust alignment, especially in cases of significant spectral distortion. rSR reduced more subtraction artifacts than the multistep method in 64% of the GABA difference spectra and 75% of the GSH difference spectra. rSR overcomes the major challenges of frequency and phase correction.
Keywords: edited MRS, frequency correction, HERMES, phase correction, spectral registration
1 ∣. INTRODUCTION
An essential step in the processing of single-voxel MRS data is the correction of shot-to-shot frequency and phase offsets.1,2 These offsets occur as a result of heating/cooling of scanner hardware elements (which leads to frequency drift in the B0 field) and subject bulk and physiological motion (which lead to abrupt, often transient, offsets in frequency and phase). Left unaccounted for, these errors will cause incoherent signal averaging, resulting in line-broadening, lineshape distortion, and signal loss. Given that in vivo MRS experiments require the collection of tens to hundreds of individual transients to improve the inherently low signal-to-noise ratio (SNR) of metabolite signals, retrospective frequency-and-phase correction (FPC) should be performed as a matter of course during data processing. For spectral-edited experiments, in particular, accurate alignment of subspectra is critical to avoid subtraction artifacts, loss in SNR, and under/overestimation of metabolite levels.3-6
Multiple approaches have been developed for retrospective FPC. For instance, the creatine (Cr)3,7 or residual water peaks8 can be used for alignment in the frequency domain, while another method based on metabolite cycling uses the unsuppressed water signal in each transient to correct frequency and phase errors.9,10 It is also possible to incorporate estimated frequency offsets into simulated metabolite basis sets to account for frequency shifts in individual transients.6 The time-domain-based spectral registration method11 is a simple yet efficient algorithm that accurately aligns single transients and has excellent performance.12 The challenge of correcting frequency and phase shifts in datasets acquired by multiplexed editing schemes such as HERMES (Hadamard encoding and reconstruction of MEGA-edited spectroscopy)13,14 or HERCULES (Hadamard editing resolves chemicals using linear-combination estimation of spectra)15 necessitates more sophisticated routines, which have been addressed recently with a multistep approach incorporating spectral registration.16 Additional challenges that make FPC difficult include instabilities in the residual water, lipid, and baseline signals11,17 and low SNR of individual transients.18,19
This paper describes a novel FPC algorithm built upon spectral registration, which is a hybrid method using information from both the time and frequency domains. It has been designed to overcome some of the common challenges for correcting frequency and phase errors in in vivo MRS data. These include (i) substantial subject motion (common in clinical and pediatric subjects), (ii) substantial B0 field drift, (iii) alignment of subspectra in data acquired by multiplexed editing, and (iv) unstable lipid contamination and residual water signals. The set of subroutines that comprise the algorithm, which is termed robust spectral registration (rSR), is entirely automated, requiring no user input. This algorithm is novel in three key aspects: reference updating (combining the SNR and linewidth benefits of prior approaches), alignment reordering (preferentially favoring good-quality transients during alignment), and weighted averaging (down-weighting poor-quality transients when signal averaging). The performance of rSR was assessed by comparing it against a recently developed method for aligning γ-aminobutyric acid- (GABA−)/glutathione-(GSH−) edited HERMES data16 using both simulated and in vivo datasets.
2 ∣. METHODS
A schematic diagram of the rSR pipeline is shown in Figure 1. Each step is described fully below. The code for the algorithm is provided as part of the open-source Gannet software package,20 which can be found on GitHub (https://github.com/richardedden/Gannet3.1).
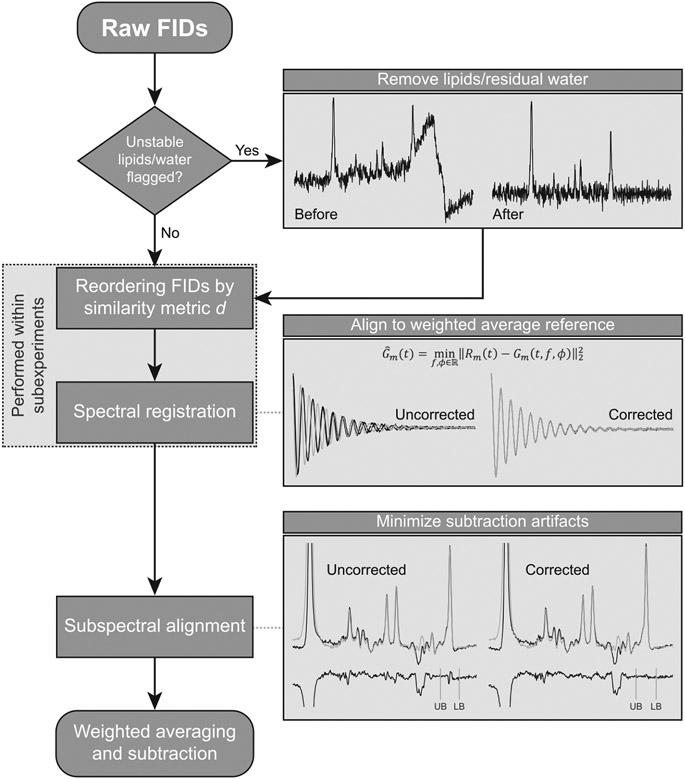
FIGURE 1.
Schematic diagram of the rSR pipeline. Raw FIDs are first assessed for unstable lipid contamination and residual water signals, which are removed if needed. FIDs are then reordered based on their similarity and aligned in the time domain using spectral registration (this is performed within subexperiments). Subspectra are aligned by minimizing subtraction artifacts in the frequency domain. Weighted averaging and subtraction are used to derive the final difference spectra
2.1 ∣. Unstable lipid contamination and residual water signals
Unstable lipid contamination and residual water signals make FPC more challenging because they represent shot-to-shot signal differences that are distinct from the frequency and phase shifts that FPC seeks to rectify.11,17 To minimize their impact, spectral registration can be preceded by automated removal of lipid contamination and residual water signals. To detect substantial lipid contamination automatically, raw free induction decays (FIDs) are Fourier-transformed and averaged to obtain the SUM spectrum. The ratio between the standard deviation of the real-valued lipid signal between 0 and 1.85 ppm and the standard deviation of the real-valued noise signal between 8 and 9 ppm is calculated. Based on prior testing on lipid-contaminated datasets, a ratio above 40 indicates that lipid signals strongly contaminate the data. To automatically detect unstable residual water, the FIDs containing residual water not saturated by an editing pulse are Fourier-transformed (eg, GSH edit-ON transients would not be considered). The integrals of the residual water signals are calculated between 4.43 and 4.93 ppm in the real frequency-domain spectra. The absolute integrals are then normalized to the maximum integral. Based on prior testing on datasets with residual water that was unstable shot to shot, the residual water signal is classified as unstable if more than 10% of the normalized integrals have a value of less than 0.5.
If either of the two above conditions is met, an additional subroutine is called to suppress water and lipid signals, which consists of baseline modeling21,22 and Whittaker smoothing.23 First, as in Reference,22 each FID is Fourier-transformed, and the corresponding real spectra are further transformed by applying a continuous wavelet transform (CWT) using a Haar function as the mother wavelet to obtain the approximate first derivative24,25 (setting the scale parameter to 10 by default). These first-derivative spectra are then converted to power spectra. The regions of the spectra that are classified as baseline are then identified, as in Reference,21 by running a sliding window of length l points (set to 5 by default) across the power spectrum y, where within each window segment signal is considered baseline signal if the range max(yn, …, yn+l − 1) − min(yn, …, yn+l − 1) is less than three times the standard deviation of the noise (estimated between 8 and 9 ppm). If lipids are flagged, the range between −2 and 1.85 ppm is also classified as baseline; if unstable residual water is flagged, signal between 3.6 and 5.5 ppm is also classified as baseline. The baseline of the real frequency-domain spectrum is then modeled (using those points classified as baseline while setting non-baseline points to zero) using a Whittaker smoother with a second-order penalty and a smoothing parameter of 1000. If lipids and/or water are flagged, additional Whittaker smoothing is applied to model the lipid range (smoothing parameter = 10) and/or the water range (smoothing parameter = 0.2). The Hilbert transform is used to calculate the imaginary part of the smoothed baseline from the real-valued smoothed baseline model. The complex smoothed baselines are then subtracted from the corresponding raw spectra, which are then transformed back into the time domain by inverse Fourier transformation. Whittaker smoothing was chosen for removing lipids and residual water in every transient because the implementation uses sparse matrices,23 making it fast and highly computationally efficient even with very large datasets. Removing residual water from one transient digitized with 2048 complex data points takes about 30 ms (2.9 GHz Intel Core i5 CPU, 8 GB RAM). An example illustrating lipid contamination removal is presented in Figure 2.
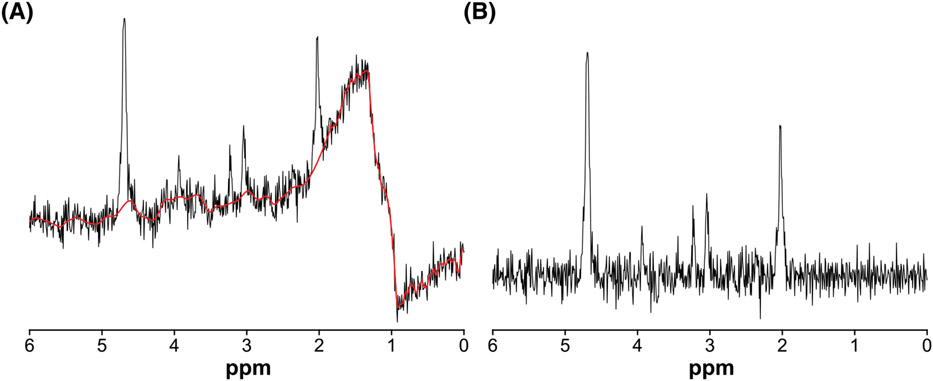
FIGURE 2.
Example of the lipid contamination removal subroutine applied to an individual subspectrum from an in vivo HERMES dataset. A, Raw data with lipid contamination at about 1.0-1.8 ppm. The red line represents the smoothed baseline estimated by baseline modeling and Whittaker smoothing. B, The subspectrum after baseline correction and lipid contamination removal
Spectral registration is then performed on these baseline-corrected, lipid-/residual water-filtered FIDs. Note that the estimated frequency/phase offsets determined by spectral registration are applied to the original FIDs; the baseline-corrected FIDs are not used for further analysis after the estimation of offsets.
2.2 ∣. Spectral registration
Spectral registration corrects frequency and phase offsets in the time domain.11 Each mth FID Sm(t) is aligned to a reference FID R(t) by simultaneous adjustment of its frequency, f (Hz), and phase, ϕ (°), using nonlinear least-squares optimization:
| (1) |
where
| (2) |
Optimization was performed using Isqnonlin in MATLAB (version R2019a; MathWorks, Natick, MA) and the Levenberg-Marquardt algorithm. Although Gm(t, f, ϕ) is complex in Equation (2 (), its real and imaginary parts are concatenated into a one-dimensional real-valued vector in the objective function so that real-valued frequency and phase parameter estimates are derived. The search for a global minimum was aided by setting the starting value of the frequency offset parameter close to the optimum solution, estimated by calculating the frequency difference between Sm(t) and R(t) based on the frequency-domain absolute-maximum of the 3 ppm Cr signal. The starting value for the phase offset parameter was set to zero. For edited datasets, spectral registration is performed separately on FIDs from each subexperiment.16
To decrease optimization time, only the first n points of the FIDs were used during registration, with n conservatively chosen to restrict registration to the high-SNR portions of the FIDs. n was determined by (i) calculating the magnitude FIDs, normalized to twice the standard deviation of noise signal in the last quarter of the FIDs (effectively an FID of point-by-point SNR); (ii) averaging all SNR FIDs; (iii) calculating the point-to-point difference (effectively a high-pass filter); (iv) taking the absolute of this signal; and (v) determining the last point for which this signal was greater than 0.5. To ensure that there were always enough points for the optimization, at least the first 100 ms of each FID was used. Conversely, because it was found that spurious echoes in the latter part of transients can lead to incorrect solutions during registration, no more than the first 200 ms of each FID was used. Lower and upper limits of 100 and 200 ms equate to the first 201 and 401 points, respectively, for an FID digitized with 2048 complex data points and 2000 Hz spectral width.
2.3 ∣. Reference updating and optimal alignment order
The choice of reference transient R(t) is critical for successful spectral registration. A single transient (such as the first acquired in a given dataset) can be used, but the disadvantages include low SNR and the possibility of choosing an FID with signal artifacts. Another option is to use the mean or median of the unaligned M transients,16 which generates a reference with higher SNR. However, averaging before alignment incurs a line-broadening effect because of the misaligned transients, and the spectral profile of such a reference would not match a typical transient.
Here, an alternative approach is proposed, where R(t) is updated during spectral registration using weighted averaging. For the M transients in a dataset, R(t) is updated after each mth transient is aligned such that R(t) is a weighted average between each newly aligned transient and the previous reference. This approach retains some of the SNR benefits of using an averaged reference but without the line-broadening effect. It is helpful to perform this process, not in the order in which data were acquired, but in order of decreasing similarity. FIDs that are more typical of the whole dataset are corrected first, which builds up the SNR of the reference while down-weighting the influence of poorer-quality FIDs.
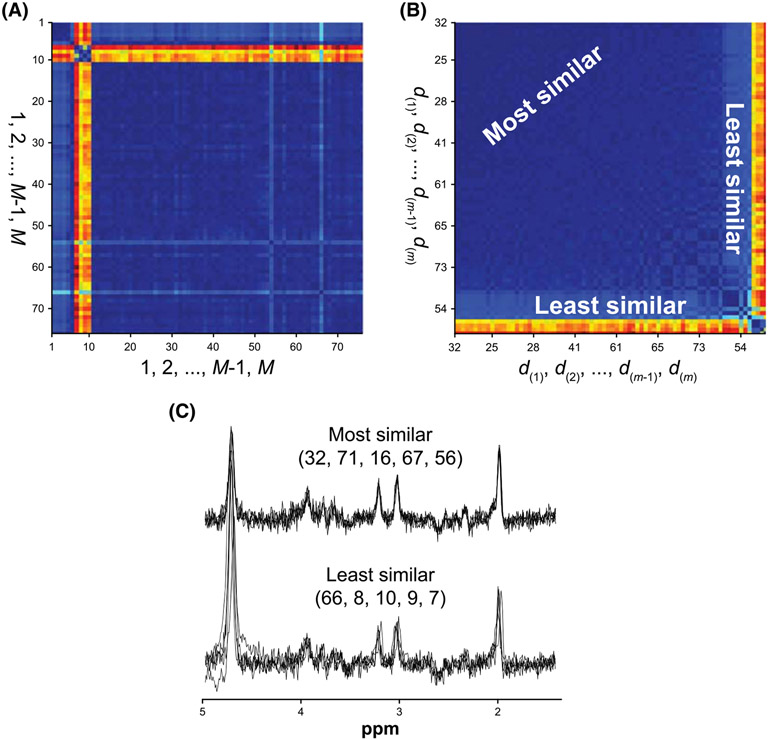
Formally, a similarity matrix is generated by calculating the mean squared error (MSE) between each real-valued unaligned transient and every other real-valued transient. A similarity metric d is calculated as the columnwise median of D. The elements of d are ranked (d(1), d(2), …, d(m−1), d(m)) and the corresponding new rank order is used as the alignment order for spectral registration, such that smaller values in d denote more similar FIDs (which are aligned first) and larger values in d denote less similar FIDs (which are aligned last). An illustration of the optimal alignment order subroutine is presented in Figure 3.
FIGURE 3.
Example of the alignment order subroutine applied to subspectra from an in vivo HERMES dataset. A, Similarity matrix D before rank ordering based on the similarity of FIDs. B, The same similarity matrix after rank ordering. The most similar FIDs are reordered to the first quadrant of the matrix while the least similar are reordered to the fourth quadrant. Spectral registration then proceeds in order of decreasing similarity: d(1) → d(m). C, Individual subspectra corresponding to the most and least similar FIDs identified by the similarity metric. Numbers in parentheses indicate the FID acquisition number
The reference Rm(t) is calculated to correct the mth FID Sm(t), where the index m now follows the rank order determined by d:
| (3) |
where
| (4) |
The weighting factor wm is half the squared Pearson correlation coefficient between each aligned transient and the reference Rm − 1(t) from the previous iteration. This ensures that poorly aligned transients contribute less to the updated reference.
2.4 ∣. Alignment of subspectra
In spectral editing experiments, editing pulses generate subspectra with distinct spectral profiles. Conventional spectral registration is not designed to align spectra with different underlying signal characteristics.16 However, after running spectral registration on the subexperiment FIDs, subspectra can be aligned by minimizing the frequency-domain difference signal within a particular range δLB < δ < δUB known to have identical signal in pairs of subspectra, adjusting the frequency and phase shifts in the time domain using nonlinear least-squares optimization.
For MEGA-PRESS data, the edit-OFF subspectrum is first zero-order phase-corrected by frequency-domain modeling of the Cr and choline (Cho) signals between 2.6 and 3.6 ppm using a two-Lorentzian function with a phase parameter.20 For datasets where editing pulses do not saturate residual water, the edit-ON subspectrum is then aligned to the edit-OFF subspectrum by minimizing the residual water subtraction artifact between 4.46 and 4.9 ppm. (Note that, in cases of strong water suppression, minimizing the residual Cho subtraction artifact between 3.12 and 3.28 ppm can be used instead.) If water is saturated by editing pulses, the N-acetylaspartic acid (NAA) subtraction artifact between 1.83 and 2.19 ppm is minimized.
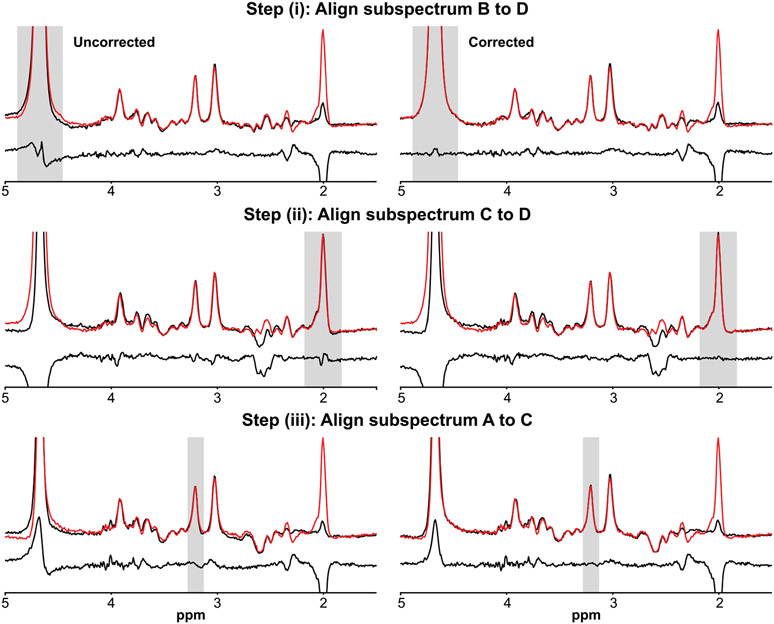
For HERMES data, subspectrum D (GABA-OFF, GSH-OFF) is first zero-order phase-corrected, as above. Alignment is then performed in three successive steps: (i) aligning subspectrum B (GABA-ON, GSH-OFF) to subspectrum D (GABA-OFF, GSH-OFF) by minimizing the residual water subtraction artifact, as above; (ii) aligning the subspectrum C (GABA-OFF, GSH-ON) to subspectrum D (GABA-OFF, GSH-OFF) by minimizing the NAA subtraction artifact, as above; and (iii) aligning subspectrum A (GABA-ON, GSH-ON) to the newly corrected subspectrum C by minimizing the Cho subtraction artifact, as above. The frequency and phase offset estimates determined from subspectrum alignment are then applied back to the individual transients. An illustration of alignment of HERMES subspectra is presented in Figure 4.
FIGURE 4.
Example of the subspectral alignment subroutine applied to subspectra from an in vivo HERMES dataset. Steps (i), (ii), and (iii) are shown. The red subspectra are the references and the grey shading illustrates the frequency bounds where the objective function seeks to minimize subtraction artifacts in the difference spectra. Note that the reference in Step (iii) is the corrected subspectrum C from Step (ii)
Finally, a global frequency correction is applied to all subspectra by determining the difference between the frequency of the Cr signal maximum in the real-valued SUM spectrum and 3.02 ppm and then subsequently conducting a frequency-domain circular shift to all subspectra by the estimated frequency difference.
2.5 ∣. Weighted averaging and subtraction
Once all FIDs have been aligned, 3 Hz exponential apodization is applied. FIDs are then zero-filled to a nominal spectral resolution of 0.061 Hz/point and Fourier-transformed. A weighted averaging approach is used to down-weight subspectra that are corrupted by signal artifacts—this is an important distinction from traditional signal averaging. First, the difference between sequentially acquired pairs of edit-ON and edit-OFF subspectra is calculated. Like the optimal alignment order subroutine, a similarity matrix is obtained by calculating the MSE between each real-valued difference subspectrum p and every other real-valued difference subspectrum (in the range 1.8 to 4.25 ppm). A similarity metric dp is calculated as the columnwise median of D. Normalized weights wp are then derived, , and applied to the difference pairs before summation. It should be noted that previous approaches applying outlier rejection7 can be thought of as weighted averaging with binary weighting.
2.6 ∣. Algorithm performance
The performance of rSR was assessed on both simulated and in vivo GABA−/GSH-edited HERMES data14 and compared against a previously described multistep FPC (msFPC) approach,16 which is considered the current-best method for correcting offsets in multiplexed edited data.
2.6.1 ∣. Simulated data
As previously described,16 a simulated HERMES dataset consisting of 320 FIDs was generated in FID-A26 using ideal excitation/refocusing pulses and shaped editing pulses. Acquisition parameters were TE = 80 ms; 20 ms Gaussian editing pulses; 2048 data points; 2 kHz spectral width. Fourteen metabolites were included in the simulated dataset, plus eight macromolecule resonances and a residual water signal, with signal intensities and lineshapes modeling in vivo values. Complex Gaussian noise was added to each FID (at an SNR level approximating the SNR of a single coil-combined in vivo transient). Known shot-to-shot frequency/phase offsets were added as previously described.11 Frequency offsets consisted of a linear trend with a total drift of 5 Hz over the course of the entire experiment, superposed with normally distributed random shot-to-shot frequency offsets with a standard deviation of 1 Hz. Similarly, phase offsets consisted of a linear trend with a total of −1° of drift, superposed with normally distributed random shot-to-shot phase offsets with a standard deviation of 6°. Finally, to simulate the effect of a “jump” in frequency/phase offsets due to subject head motion, the frequency/phase offset functions were then each superposed with step functions with amplitudes of 5 Hz and 20°, respectively, with the step randomly chosen to occur at the 264th transient. Additional data processing steps included 3 Hz exponential line-broadening, zero-filling to 32k points and removal of the residual water signal using a Hankel singular value decomposition filter.27
For the simulated data, the true frequency/phase offsets were known so it was possible to assess the performance of both FPC approaches by calculating the normalized root sum square error:
| (5) |
where py,m is either the estimated or the true frequency (Δf) or phase offset (Δϕ) of the mth FID. A score of 1 indicates perfect alignment, a score of 0 indicates no correction, and a score less than 0 indicates that alignment is worse than if no alignment were attempted.
2.6.2 ∣. In vivo data
Sixty-seven HERMES datasets from a pediatric cohort consisting of children with Tourette syndrome and age-matched controls (age range: 8-12 years) were collected on a Philips 3 T Ingenia Elition system as part of an ongoing study. Of the 67 datasets, 23 were acquired in the right sensorimotor cortex (SM), 23 were acquired in the supplementary motor area (SMA), and 21 were acquired in the right insula (Ins). Acquisition parameters were the same for all measurements: TE/TR = 80/2000 ms; 304 averages; voxel size = 3 × 3 × 3 cm3; 2 kHz spectral width; 2048 data points. An unsuppressed water reference signal was also acquired from the same voxels for concentration referencing.
GABA+ and GSH were quantified relative to the water reference signal and are presented in institutional units (i.u.). Partial volume tissue correction was not applied. The group-level variance in GABA+ and GSH values for the three voxels was assessed by calculating between-subject coefficients of variation (CVs). The following subtraction-artifact-based metric was used to assess the comparative performance of rSR: , where is the variance of the Cho (3.15-3.285 ppm) or NAA (1.88-2.14 ppm) subtraction artifact in the GABA− or GSH-edited difference spectrum, respectively, following correction by method y (either rSR or msFPC). A score between 0.5 and 1 indicates that rSR resulted in smaller subtraction artifacts, whereas a score between 0 and 0.5 indicates that msFPC resulted in smaller subtraction artifacts. A score of 0.5 indicates that the two methods performed the same. Two-tailed paired-samples t-tests were conducted to test the hypothesis that the performances of msFPC and rSR are significantly different (alpha level = 0.05). Note that to appropriately conduct these tests the groups needed to be balanced. The comparison group was 1 minus the calculated performance score of each GABA− or GSH-edited difference spectrum (ie, the metric was reversed).
3 ∣. RESULTS
Separate timing tests showed that the pipeline takes on average about 5 s to process a single dataset of 320 transients (2.9 GHz Intel Core i5 CPU, 8 GB RAM). If lipid/residual water removal is also run, the runtime increases to about 15 s.
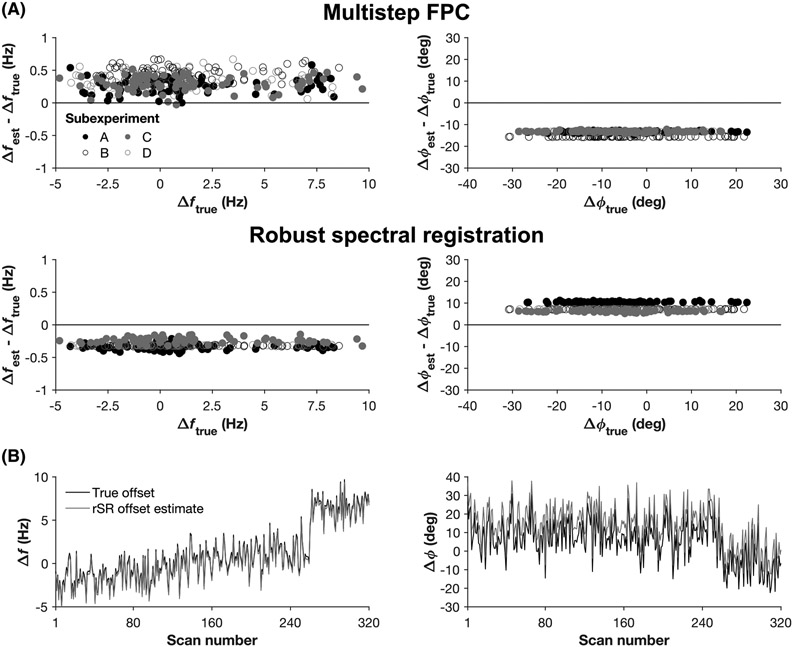
The simulation results are represented in Figure 5 as Bland-Altman plots of the accuracy and biases in the estimation of frequency and phase offsets using msFPC and rSR. The Qp scores for msFPC were 0.89 (frequency) and −0.11 (phase). The Qp scores for rSR were 0.90 (frequency) and 0.36 (phase). The mean differences between the estimated and true frequency and phase offsets were 0.34 Hz and −13.82° for msFPC, respectively, and −0.35 Hz and 7.80° for rSR, respectively.
FIGURE 5.
A, Bland-Altman plots showing the accuracy and biases of msFPC and rSR with respect to estimation of frequency (left column) and phase (right column) offsets in the simulated HERMES dataset. The errors in estimated frequency/phase offsets (Δfest − Δftrue, Δϕest − Δϕtrue) are plotted against the true offsets (Δftrue, Δϕtrue). B, The true frequency (left) and phase (right) offsets (black line) overlaid by the offsets estimated by rSR (grey line) for each simulated FID
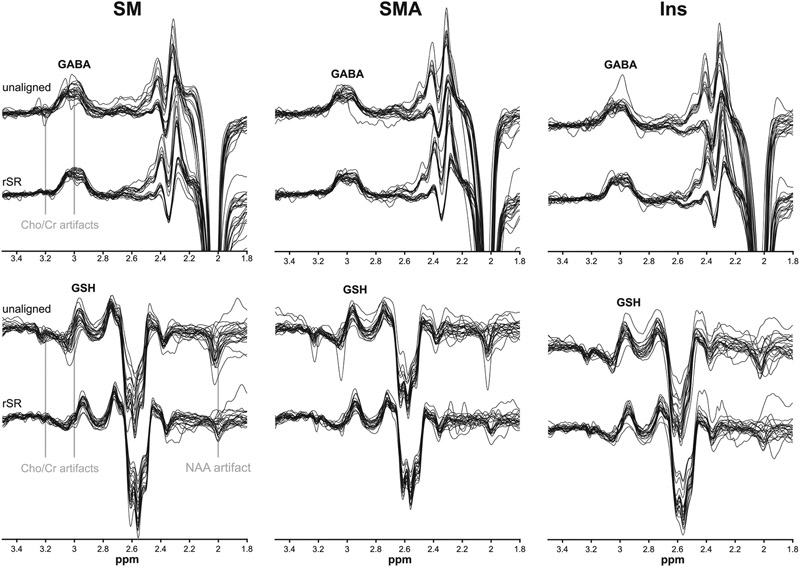
The in vivo SM, SMA, and Ins GABA− and GSH-edited difference spectra acquired in the pediatric subjects are displayed in Figure 6. The unaligned data show clear Cho, Cr, and NAA subtraction artifacts that are reduced after alignment using rSR. Additionally, the frequency and phase corrections led to better-resolved GABA and GSH peaks at about 3 ppm with reduced signal distortion.
FIGURE 6.
In vivo GABA− and GSH-edited difference spectra acquired by HERMES in three brain regions in pediatric subjects with Tourette syndrome and age-matched healthy controls. Each panel shows the spectra before and after alignment by rSR. The vertical gray lines indicate Cho, Cr, and NAA subtraction artifacts
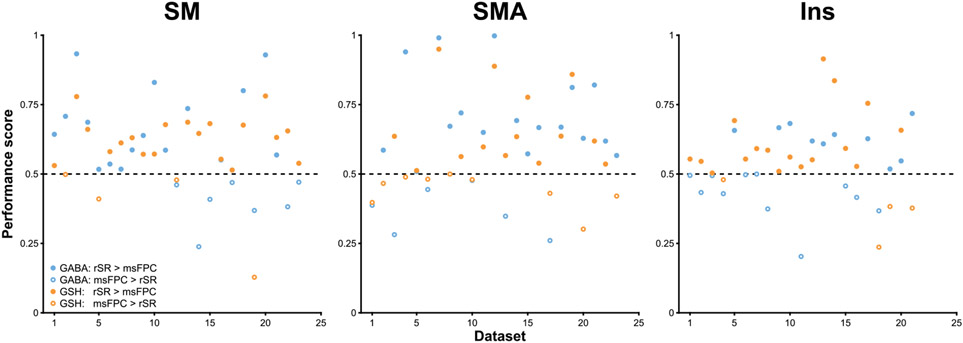
Mean concentration values and CVs are shown in Table 1. The mean performance scores for the three sets of data were SM, 0.59 ± 0.18 (GABA) and 0.59 ± 0.14 (GSH); SMA, 0.62 ± 0.21 (GABA) and 0.58 ± 0.16 (GSH); Ins, 0.52 ± 0.13 (GABA) and 0.57 ± 0.15 (GSH). Across all datasets, the mean performance scores were 0.58 ± 0.18 (GABA) and 0.58 ± 0.15 (GSH). Additionally, rSR produced smaller subtraction artifacts than msFPC in the GABA and GSH difference spectra in 70% and 83% of the SM data, 74% and 61% of the SMA data, and 48% and 81% of the Ins data, respectively (Figure 7). Across all datasets, rSR performed better in 64% of the GABA difference spectra and 75% of the GSH difference spectra. The paired-samples t-tests showed a significant difference in performance scores for both GABA (t(66) = 3.72, p < 0.001) and GSH (t(66) = 4.42, p < 0.001) datasets, indicating that rSR produced significantly better scores.
TABLE 1.
Metabolite quantification results following alignment by msFPC and rSR
| Mean concentration ± 1 s.d. (i.u.) | CV (%) | ||
|---|---|---|---|
| SM GABA+ | msFPC | 1.60 ± 0.29 | 18.3 |
| rSR | 1.52 ± 0.20 | 13.2 | |
| SMA GABA+ | msFPC | 1.39 ± 0.95 | 68.3 |
| rSR | 1.40 ± 0.26 | 18.3 | |
| Ins GABA+ | msFPC | 1.74 ± 0.37 | 21.5 |
| rSR | 1.66 ± 0.33 | 20.1 | |
| SM GSH | msFPC | 0.55 ±0.16 | 29.7 |
| rSR | 0.56 ± 0.14 | 25.0 | |
| SMA GSH | msFPC | 0.50 ± 0.56 | 111.5 |
| rSR | 0.57 ± 0.15 | 26.5 | |
| Ins GSH | msFPC | 0.63 ± 0.17 | 26.7 |
| rSR | 0.69 ± 0.19 | 28.2 |
FIGURE 7.
Dot plots showing the performance scores for every in vivo HERMES dataset. A score above 0.5 signifies that rSR led to reduced subtraction artifacts compared to msFPC, whereas a score below 0.5 signifies that msFPC led to reduced subtraction artifacts compared to rSR
4 ∣. DISCUSSION
rSR has been developed to address the more challenging aspects of correcting frequency and phase offsets in in vivo MRS data. The major strength and novelty of this algorithm is its complement of subroutines that are designed to directly and automatedly deal with large offsets, unstable lipid contamination, unstable residual water, and misaligned subspectra from edited experiments. Tests on a simulated multiplexed-edited HERMES dataset showed that rSR had better performance than msFPC but this was only marginal. However, assessment on empirical HERMES data acquired in (unsedated) children showed that it had significantly better performance over the msFPC algorithm. Additionally, group-level variances in GABA+ and GSH values were lower when compared with the variances after aligning with msFPC. The mean performance score across all datasets for the GABA and GSH difference spectra was about 0.6, meaning that the robust algorithm reduced subtraction artifacts about 1.5 times more on average than msFPC. These results indicate that overall it outperformed this other method.
Significant frequency offsets (>10 Hz) can be problematic for spectral registration11,18 for two main reasons. First, the starting values in the optimization algorithm will have an influence on whether the algorithm finds a local or global minimum. Although the Levenberg-Marquardt algorithm is robust to suboptimal starting values, providing a good initial guess will make it more likely that a global minimum is found. Spectral registration may fail at estimating large frequency offsets because an inappropriate starting frequency offset value leads to convergence on a local minimum. Second, and more importantly, large frequency offsets will significantly impact water suppression and change the lineshape and phase of the residual water signal. Alignment errors are more likely to occur when the residual water signal is unstable.11 rSR overcomes these issues by setting a reasonable starting value for frequency offsets and by removing unstable residual water signals.
Contamination from lipid signals, which can be large, broad, and poorly phased, adversely impacts the baseline and can render data unusable. Wilson17 recently proposed an elegant solution to mitigating the influence of baseline distortion on FPC by incorporating a polynomial baseline basis set into the optimization process using the variable projection method (VARPRO).28 The approach used in rSR is to model and (temporarily) remove the baseline before FPC when lipid contamination is detected. The advantage of this is that it would not be possible to accurately parameterize lipid signals using a polynomial basis set given the variable lineshapes of lipid signals. The disadvantage, however, is that appropriate parameters need to be selected for the CWT and Whittaker smoothing steps. The default parameters used in this study appeared to work well for the majority of datasets tested, but a procedure to determine optimal parameters on a dataset-by-dataset basis could be advantageous. Although lipid contamination and/or the residual water signal are only temporarily removed from transients in rSR, the novel use of weighted averaging of subspectra will counter error propagation. Weighted averaging (of quantification results, not spectra) has already been shown to be beneficial for mitigating the influence of measurement error in MRS measurements and improving statistical power.29 Additionally, while not the intention in this work, using the Whittaker smoother to remove the residual water signal or lipid contamination in fully processed and averaged spectra would be worth exploring. Removing residual water in the time domain in this way has already been implemented in a software package for processing high-resolution NMR data.30 An obvious advantage of the smoother is its simplicity and notably short computation time. Nonetheless, a major challenge would be to ensure that metabolite signal amplitudes are not adversely affected, particularly when strong lipids distort metabolite signal lineshapes.
Although it has been demonstrated that rSR is highly beneficial for improving the spectral quality of edited MRS data, it is important to highlight that retrospective correction of frequency and phase errors does not correct for errors induced by shifted voxel location or recover signal losses from reduced editing efficiency. Quality control of all datasets is essential in order to detect cases where artifacts have made specific acquisitions unusable. It is for this reason that prospective frequency, B0 shim, and motion correction techniques2,5,31-34 remain valuable methods for MRS data acquisition.
The most challenging step in FPC of edited data is the alignment of subspectra with distinct spectral profiles. One proposed approach is to align the averaged edit-OFF spectrum to the averaged edit-ON spectrum by minimizing the L1 norm of the difference so that the smaller-amplitude subtraction artifacts are given more weight in the optimization.19 The solution used in rSR is to focus specifically on the portion of signal where subtraction artifacts arise in the difference spectrum when subspectra are misaligned. A similar approach has been demonstrated previously for correcting offsets in GSH-edited MEGA-PRESS data.17 For data acquired with more complex multiplexed editing schemes, such as the one used in HERCULES,15 the challenge is to choose a portion of signal that has a similar signal profile in pairs of subspectra. Although not shown here, preliminary testing showed that rSR aligns HERCULES data well.
The subroutines and approaches described in this paper are not restricted to implementation in spectral registration. The methods could be easily integrated into other FPC algorithms that may be more appropriate for a given type of data. Moreover, in cases where rSR fails, it would be feasible to align data with multiple alternative approaches to obtain the best result. This outcome-driven approach was first proposed in the msFPC paper.16 Finally, although this paper focuses on edited 1H MRS data acquired in the brain, most of the described subroutines may be applicable to in vivo MRS data acquired in other regions of the body or by non-edited or non-proton MRS techniques. While we have not tested rSR on short-TE spectra, there are no hindrances to applying rSR to such data. Spectral registration is not, however, generally applicable to MRSI data because these data are phase-encoded and as such there is no template that can be used for registration.
A challenge not addressed in this paper is the issue of low-SNR data. The in vivo data that were used for performance testing had relatively high SNR (they were acquired in 27 mL voxels). It is unclear at this stage what the performance of rSR is for data acquired in voxels with volumes less than 27 mL, but it has already been shown that spectral registration tends to estimate frequency offsets poorly at SNR levels below 5 compared with other approaches.11,17,18 However, this could be mitigated by not selecting a single transient as the reference. A mean/median reference or a weighted average reference (as used in rSR) reduces the problem of aligning two noisy measurements to each other. An alternative solution would be to use the Whittaker smoother to filter out noise in individual transients. An approach that has yet to be explored is the use of a simulated or phantom reference transient, which would not only mitigate the issues with noise but also allow signals to be aligned to the intrinsic frequency and phase. The difficulty would be to generate data that match the expected subspectra of the given MRS experiment (i.e., by using identical acquisition parameters and RF pulse timings/waveforms, including all appropriate metabolites and macromolecules in the simulation or phantom solution, and matching in vivo linewidth and lineshape).
5 ∣. CONCLUSION
rSR builds on the strengths of conventional spectral registration and incorporates several novel subroutines to correct “difficult-to-align” datasets that are affected by significant frequency and phase errors and unstable lipid contamination and residual water signals. It can also accurately align the subspectra of datasets acquired by complex multiplexed editing schemes such as HERMES. Implementing rSR in the processing pipeline of MRS data can reduce errors in metabolite quantification and prevent data exclusion (which is highly beneficial when sample sizes are low).
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 EB016089, R01 EB023963, R01 MH106564, R01 NS096207, and P41 EB015909.
Funding information
National Institute of Biomedical Imaging and Bioengineering, Grant/Award Numbers: P41 EB015909, R01 EB016089, R01 EB023963; National Institute of Mental Health, Grant/Award Number: R01 MH106564; National Institute of Neurological Disorders and Stroke, Grant/Award Number: R01 NS096207
Abbreviations:
- Cr
creatine
- CV
coefficient of variation
- CWT
continuous wavelet transform
- FID
free induction decay
- FPC
frequency-and-phase correction
- GABA
γ-aminobutyric acid
- GSH
glutathione
- HERCULES
Hadamard editing resolves chemicals using linear-combination estimation of spectra
- HERMES
Hadamard encoding and reconstruction of MEGA-edited spectroscopy
- Ins
insula
- i.u.
institutional units
- MSE
mean squared error
- msFPC
multistep FPC
- NAA
N-acetylaspartic acid
- rSR
robust spectral registration
- SM
sensorimotor cortex
- SMA
supplementary motor area
REFERENCES
- 1.Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2020;e4257. 10.1002/nbm.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saleh MG, Edden RAE, Chang L, Ernst T. Motion correction in magnetic resonance spectroscopy. Magn Reson Med. 2020:1–15. 10.1002/mrm.28287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans CJ, Puts NAJ, Robson SE, et al. Subtraction artifacts and frequency (mis-)alignment in J-difference GABA editing. J Magn Reson Imaging. 2013;38(4):970–975. 10.1002/jmri.23923 [DOI] [PubMed] [Google Scholar]
- 4.Harris AD, Glaubitz B, Near J, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72(4):941–948. 10.1002/mrm.25009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edden RAE, Oeltzschner G, Harris AD, et al. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J Magn Reson Imaging. 2016;44(6):1474–1482. 10.1002/jmri.25304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Veen JW, Marenco S, Berman KF, Shen J. Retrospective correction of frequency drift in spectral editing: the GABA editing example. NMR Biomed. 2017;30(8):e3725. 10.1002/nbm.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. 2007;25(7):1032–1038. 10.1016/j.mri.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helms G, Piringer A. Restoration of motion-related signal loss and line-shape deterioration of proton MR spectra using the residual water as intrinsic reference. Magn Reson Med. 2001;46(2):395–400. 10.1002/mrm.1203 [DOI] [PubMed] [Google Scholar]
- 9.Dreher W, Leibfritz D. New method for the simultaneous detection of metabolites and water in localized in vivo 1H nuclear magnetic resonance spectroscopy. Magn Reson Med. 2005;54(1):190–195. 10.1002/mrm.20549 [DOI] [PubMed] [Google Scholar]
- 10.Giapitzakis I-A, Shao T, Avdievich N, Mekle R, Kreis R, Henning A. Metabolite-cycled STEAM and semi-LASER localization for MR spectroscopy of the human brain at 9.4T. Magn Reson Med. 2018;79(4):1841–1850. 10.1002/mrm.26873 [DOI] [PubMed] [Google Scholar]
- 11.Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. 2015;73(1):44–50. 10.1002/mrm.25094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowland BC, Liao H, Adan F, Mariano L, Irvine J, Lin AP. Correcting for frequency drift in clinical brain MR spectroscopy. J Neuroimaging. 2017;27(1):23–28. 10.1111/jon.12388 [DOI] [PubMed] [Google Scholar]
- 13.Chan KL, Puts NAJ, Schär M, Barker PB, Edden RAE. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016;76(1):11–19. 10.1002/mrm.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleh MG, Oeltzschner G, Chan KL, et al. Simultaneous edited MRS of GABA and glutathione. NeuroImage. 2016;142:576–582. 10.1016/j.neuroimage.2016.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeltzschner G, Saleh MG, Rimbault D, et al. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: principles of HERCULES. NeuroImage. 2019;185:181–190. 10.1016/j.neuroimage.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen M, Saleh MG, Near J, et al. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magn Reson Med. 2018;80(1):21–28. 10.1002/mrm.27027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson M. Robust retrospective frequency and phase correction for single-voxel MR spectroscopy. Magn Reson Med. 2019;81(5):2878–2886. 10.1002/mrm.27605 [DOI] [PubMed] [Google Scholar]
- 18.Wiegers EC, Philips BWJ, Heerschap A, van der Graaf M. Automatic frequency and phase alignment of in vivo J-difference-edited MR spectra by frequency domain correlation. Magn Reson Mater Phys Biol Med. 2017;30(6):537–544. 10.1007/s10334-017-0627-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleve M, Krämer M, Gussew A, Reichenbach JR. Difference optimization: automatic correction of relative frequency and phase for mean non-edited and edited GABA 1H MEGA-PRESS spectra. J Magn Reson. 2017;279:16–21. 10.1016/j.jmr.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 20.Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ. Gannet: a batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40(6):1445–1452. 10.1002/jmri.24478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golotvin S, Williams A. Improved baseline recognition and modeling of FT NMR spectra. J Magn Reson. 2000;146(1):122–125. 10.1006/jmre.2000.2121 [DOI] [PubMed] [Google Scholar]
- 22.Cobas JC, Bernstein MA, Martin-Pastor M, Tahoces PG. A new general-purpose fully automatic baseline-correction procedure for 1D and 2D NMR data. J Magn Reson. 2006;183(1):145–151. 10.1016/j.jmr.2006.07.013 [DOI] [PubMed] [Google Scholar]
- 23.Eilers PHC. A perfect smoother. Anal Chem. 2003;75(14):3631–3636. 10.1021/ac034173t [DOI] [PubMed] [Google Scholar]
- 24.Leung AK, Chau F, Gao J. Wavelet transform: a method for derivative calculation in analytical chemistry. Anal Chem. 1998;70(24):5222–5229. 10.1021/ac9803737 [DOI] [Google Scholar]
- 25.Shao X, Pang C, Su Q. A novel method to calculate the approximate derivative photoacoustic spectrum using continuous wavelet transform. Fresenius J Anal Chem. 2000;367(6):525–529. 10.1007/s002160000404 [DOI] [PubMed] [Google Scholar]
- 26.Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—an open source, MATLAB-based toolkit. Magn Reson Med. 2017;77(1):23–33. 10.1002/mrm.26091 [DOI] [PubMed] [Google Scholar]
- 27.Barkhuijsen H, de Beer R, van Ormondt D. Improved algorithm for noniterative time-domain model fitting to exponentially damped magnetic resonance signals. J Magn Reson. 1987;73(3):553–557. 10.1016/0022-2364(87)90023-0 [DOI] [Google Scholar]
- 28.van der Veen JWC, de Beer R, Luyten PR, van Ormondt D. Accurate quantification of in vivo 31P NMR signals using the variable projection method and prior knowledge. Magn Reson Med. 1988;6(1):92–98. 10.1002/mrm.1910060111 [DOI] [PubMed] [Google Scholar]
- 29.Miller JJ, Cochlin L, Clarke K, Tyler DJ. Weighted averaging in spectroscopic studies improves statistical power. Magn Reson Med. 2017;78(6):2082–2094. 10.1002/mrm.26615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M, Legat B, Leenders J, et al. PepsNMR for 1H NMR metabolomic data pre-processing. Anal Chim Acta. 2018;1019:1–13. 10.1016/j.aca.2018.02.067 [DOI] [PubMed] [Google Scholar]
- 31.Zaitsev M, Speck O, Hennig J, Büchert M. Single-voxel MRS with prospective motion correction and retrospective frequency correction. NMR Biomed. 2010;23:325–332. 10.1002/nbm.1469 [DOI] [PubMed] [Google Scholar]
- 32.Lange T, Zaitsev M, Buechert M. Correction of frequency drifts induced by gradient heating in 1H spectra using interleaved reference spectroscopy. J Magn Reson Imaging. 2011;33(3):748–754. 10.1002/jmri.22471 [DOI] [PubMed] [Google Scholar]
- 33.Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. NeuroImage. 2014;103:290–302. 10.1016/j.neuroimage.2014.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh MG, Alhamud A, Near J, van der Kouwe AJW, Meintjes EM. Volumetric navigated MEGA-SPECIAL for real-time motion and shim corrected GABA editing. NMR Biomed. 2016;29(3):248–255. 10.1002/nbm.3454 [DOI] [PubMed] [Google Scholar]