Wenfan Wei and Biyu Zheng et al. report the crucial role of the Cdc42 GTPase-activating protein in dictating the monopolar growth of spores and demonstrate that the specific function of Rga6 depends on its characteristic localization on the plasma membrane.
Abstract
The molecular mechanisms underlying the establishment of the monopolar growth of fission yeast spores have been less characterized. Here, we report that the Cdc42 GTPase-activating protein (GAP) Rga6 is required for promoting monopolar growth during spore germination. The absence of Rga6 increases the number of spores that grow in a bipolar fashion. Rga6 decorates the non-growing cortical region, binds phosphatidylinositol 4,5-bisphosphate, and colocalizes with the phosphatidylinositol 4,5-bisphosphate-binding protein Opy1. Overexpression of Opy1 diminishes the cortical localization of Rga6. The characteristic localization of Rga6 on the cell cortex depends on the C-terminal PBR region of Rga6. Moreover, engineered chimera composed of the Rga6 C-terminal PBR region fused to the GAP domain of Rga3 or Rga4 are sufficient to rescue the spore growth phenotype caused by the absence of Rga6. Hence, our work establishes a paradigm in which the lipid composition of the plasma membrane directs polarized cell growth by specifying the cortical localization of a GAP protein.
Introduction
Asymmetric cell division dictates cell fate (Horvitz and Herskowitz, 1992; Roegiers and Jan, 2004) and generates cell diversity. For example, asymmetric division of stem cells leads to one mother cell capable of self-renewal and one differentiated daughter cell (Chen et al., 2016). Spores of the fission yeast Schizosaccharomyces pombe also undergo asymmetric cell division during germination (Bonazzi et al., 2014; Hatanaka and Shimoda, 2001; Plante and Labbe, 2019). The germinating fission yeast cell is bottle-shaped once outgrowth takes place and grows in a monopolar fashion (Bonazzi et al., 2014). The first division, taking place near the bottle neck, then generates a mother cell capable of growing in a monopolar fashion for several cell cycles (Bonazzi et al., 2014) and a daughter cell that undergoes vegetative growth. The physiological significance of this stem cell-like behavior of spore germination is not known, and the mechanism underlying the asymmetric division of germinating fission yeast cells is not well understood.
Polarized cell growth is a fundamental process in many eukaryotic cells (Brand and Gow, 2009; Chiou et al., 2017; Ghose et al., 2022; Martin-Belmonte and Mostov, 2008; Merlini et al., 2013) and is mediated by the small Rho GTPase Cdc42 (Adams et al., 1990; Genova et al., 2000; Johnson and Pringle, 1990; Kay and Hunter, 2001; Lin et al., 2000; Martin, 2015; Woods and Lew, 2019). Cdc42 cycles between the active GTP-bound form (Cdc42-GTP) and the inactive GDP-bound form (Cdc42-GDP; Etienne-Manneville, 2004; Johnson and Pringle, 1990). While guanine nucleotide exchange factors (GEFs) promote the switch of Cdc42 from a GDP-bound form to a GTP-bound form, GTPase-activating proteins (GAPs) function oppositely (Etienne-Manneville and Hall, 2002). In general, cell polarization requires local activation of Cdc42, depending on positive feedback pathways (Bendezu et al., 2015; Butty et al., 2002; Goryachev and Pokhilko, 2008; Haupt et al., 2018; Irazoqui et al., 2003; Kokkoris et al., 2014; Kozubowski et al., 2008; Lamas et al., 2020; Martin, 2015; Woods et al., 2015; Zhou et al., 2003). Moreover, negative feedback pathways and mechanical signaling are also required to regulate the stability and distribution of Cdc42-GTP (Davi et al., 2018; Howell et al., 2012; Okada et al., 2013; Tong et al., 2007; Wu and Lew, 2013). Despite intensive studies of the spatiotemporal regulation of Cdc42-GTP, how Cdc42 is orchestrated during fission yeast spore germination to promote characteristic monopolar growth has remained elusive.
In the fission yeast S. pombe, multiple GEFs and GAPs have been shown to regulate Cdc42 activity. For example, Gef1 and Scd1 function as global and local Cdc42 GEFs, respectively (Chang et al., 1994; Coll et al., 2003; Tay et al., 2018), and the absence of both Gef1 and Scd1 is lethal (Coll et al., 2003). In general, these GEFs colocalize with Cdc42 to activate Cdc42 locally (Coll et al., 2003; Das et al., 2012; Kelly and Nurse, 2011). Inactivation of Cdc42 requires GAPs, and Cdc42 inactivation is a mechanism to prevent the spread of Cdc42-GTP to the non-growing regions of cells (Das et al., 2012; Gallo Castro and Martin, 2018; Pino et al., 2021). In fission yeast, Rga3 (Gallo Castro and Martin, 2018), Rga4 (Tatebe et al., 2008), and Rga6 (Revilla-Guarinos et al., 2016) have been identified as Cdc42 GAPs. Simultaneous deletion of the three Cdc42 GAPs abolishes the polarity of vegetative cells, leading to round-shaped cells (Gallo Castro and Martin, 2018). By contrast, deletion of a single Cdc42 GAP only slightly affects the length and width of vegetative cells (Gallo Castro and Martin, 2018; Revilla-Guarinos et al., 2016; Tatebe et al., 2008). During vegetative growth, Rga3 colocalizes with active Cdc42 at the growing cell tip (Gallo Castro and Martin, 2018) while both Rga4 and Rga6 localize to the cell sides and non-growing zones (Revilla-Guarinos et al., 2016; Tatebe et al., 2008). Rga3 has also been shown to promote instability of Cdc42-GTP zones to regulate cell mating (Gallo Castro and Martin, 2018). How the Cdc42 GAPs are involved in regulating the monopolar growth of spore germination has not been tested.
Spores are dormant under unfavorable environmental conditions and germinate once environmental conditions become favorable (Bonazzi et al., 2014; Kono et al., 2005; Pandey et al., 2013; Plante and Labbe, 2019; Tanaka and Hirata, 1982; Wallace et al., 2011). During germination, the fission yeast spore doubles its volume (i.e., isotropic swelling) and hatches out a polar tube from the outer spore wall (i.e., outgrowth; Bonazzi et al., 2014). Therefore, spore germination is an outstanding model system for studying polarized cell growth and asymmetric cell division. It has been shown that the mechanical properties of the outer spore wall play a crucial role in dictating spore outgrowth which also requires a polar cap composed of polarity factors, including Cdc42, Bud6, and Bgs4 (Bonazzi et al., 2014). Therefore, it is conceivable that the proteins involved in spore wall organization play important roles in spore outgrowth (Bonazzi et al., 2014). Interestingly, the proteins in the microtubule-dependent polarity (e.g., Tip1, Tea1, and Tea4) and actin assembly (e.g., For3) pathways do not appear to contribute to the maintenance of the polar cap at the outgrowth zone (Bonazzi et al., 2014). Therefore, in addition to the proteins related to the spore wall, the proteins responsible for promoting monopolar cell growth during spore germination remain to be identified.
Here, we show that the absence of the Cdc42 GAP Rga6 causes abnormal bipolar outgrowth of spores. Moreover, Rga6 binds phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2) and decorates the non-growing zones on the plasma membrane of outgrowing spores. In addition, the role of Rga6 in regulating monopolar spore outgrowth depends on its GAP domain and the polybasic region required for localizing Rga6 to the plasma membrane. Hence, our work supports a model in which the characteristic cortical localization of Rga6 functions to restrict Cdc42-GTP at the growing zone to promote efficient monopolar spore outgrowth.
Results
Characterization of the growth pattern of germinating and vegetative fission yeast cells
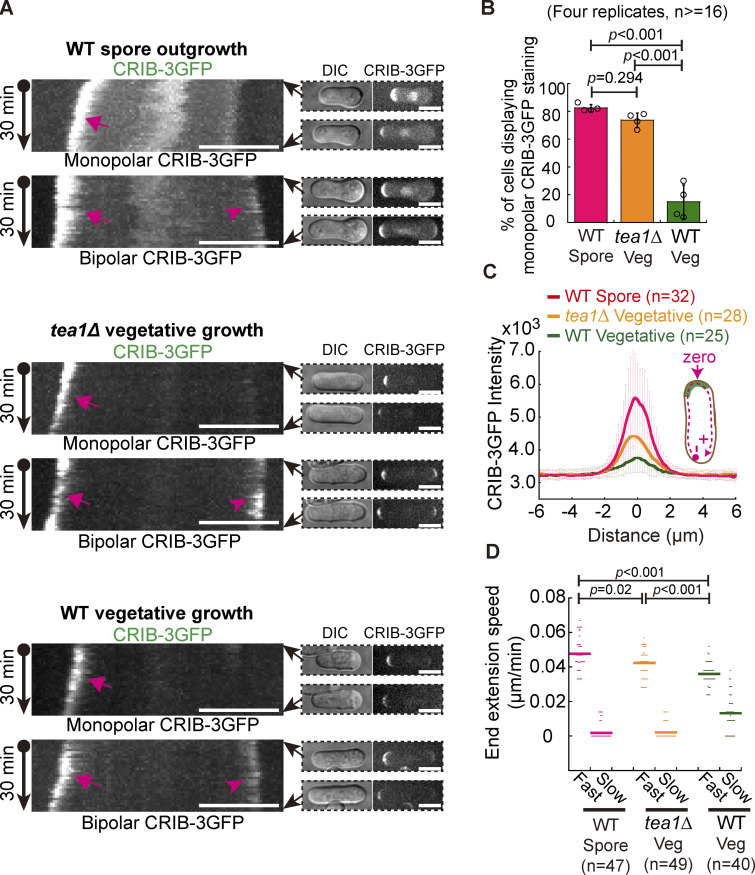
To delineate the molecular mechanisms underlying monopolar spore germination and bipolar vegetative cell growth, we carefully examined the localization of active Cdc42 (referred to as Cdc42-GTP, reported by CRIB-3GFP), the master regulator of cell polarity (Chiou et al., 2017; Das et al., 2012; Martin, 2015), in wild-type germinating and vegetative cells by live-cell microscopy. As shown in Fig. 1, A and B, CRIB-3GFP predominantly resided at the growing ends of the bottle-shaped germinating cells but localized to both ends of most rod-shaped vegetative cells. Quantitative analyses showed that 82.5% of wild-type germinating spores displayed monopolar staining of CRIB-3GFP whereas only 15.0% of wild-type vegetative cells displayed monopolar staining of CRIB-3GFP (Fig. 1 B). In addition, we noticed that the distribution of CRIB-3GFP at the growing end of germinating cells was generally brighter and more stable than those at the cell ends of vegetative cells (Fig. 1, A and C). Interestingly, measurements of the extension speed of cell ends showed that the two ends of both wild-type germinating and vegetative cells grew in an asymmetric manner (Fig. 1 D). One end of wild-type vegetative cells grew 2.7 times faster (referred to as Fast end; average speed: 0.036 µm/min) than the other end (referred to as Slow end; average speed: 0.013 µm/min). By contrast, the fast-growing end of germinating cells grew at an average speed of 0.048 µm/min, ∼1.3 times faster than the extension speed of the fast-growing end of vegetative cells, and the slow-growing end of wild-type germinating cells showed almost no measurable extension. This is consistent with the finding shown in Fig. 1 C that CRIB-3GFP more strongly decorated the growing end of the wild-type germinating spore than the growing ends of wild-type vegetative cells. Together, these results indicate that different mechanisms may be required for directing the characteristic localization of Cdc42-GTP in germinating and vegetative cells to promote monopolar and bipolar cell growth, respectively.
Figure 1.
CRIB-3GFP dynamics in fission yeast WT germinating spores and WT and tea1Δ vegetative cells. (A) Maximum projection images of CRIB-3GFP (marking Cdc42-GTP) dynamics in WT germinating spores and WT and tea1-deletion (tea1Δ) vegetative cells. Movies of 30 min (1-min intervals) were acquired for the indicated cells and were used to construct kymograph graphs (shown on the left panel). Arrows and arrowheads on the kymograph graphs indicate the fast- and slow-growing ends that displayed CRIB-3GFP staining, respectively. Images at 0 and 30 min are shown on the right of the kymograph graphs. Scale bar, 10 μm. (B) Quantification of monopolar CRIB-3GFP staining in WT germinating spores and WT and tea1Δ vegetative cells. Independent experiments were repeated four times, and the number of cells analyzed for each experiment was not less than 16. One-way ANOVA with Tukey HSD post hoc test was used to calculate the P values. (C) Plot of CRIB-3GFP intensity measured along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars are the mean ± SD. The number (n) of cells analyzed is indicated. (D) Extension speed of the fast- and slow-growing ends of the indicated cells. Bars represent the mean. The Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. The number (n) of cells analyzed is indicated.
The polarisome component Tea1 plays a crucial role in regulating cell polarity (Chang and Martin, 2009), and the absence of Tea1 causes monopolar growth of vegetative cells (Glynn et al., 2001; Taheraly et al., 2020). Therefore, we also compared the monopolar growth of wild-type germinating cells and tea1-deletion (tea1∆) vegetative cells. We confirmed that most of the tea1∆ vegetative cells (73.6 vs. 82.5% for wild-type germinating cells) displayed monopolar CRIB-3GFP staining at one cell end (Fig. 1, A and B). Interestingly, the average intensity of CRIB-3GFP at the cell end was higher in tea1∆ vegetative cells than in wild-type vegetative cells, but lower in tea1∆ vegetative cells than in wild-type outgrowing cells (Fig. 1 C). Consistently, the fast-growing end of tea1∆ vegetative cells grew at an average speed of 0.042 µm/min, which was higher than that of wild-type vegetative cells but lower than that of wild-type outgrowing cells (Fig. 1 D). The slow-growing end of tea1∆ vegetative cells showed almost no measurable extension (Fig. 1 D). These results are consistent with the finding that the growth rate of the cell surface plays a crucial role in controlling the stability of Cdc42-GTP domains (Haupt et al., 2018) and support the model proposed previously that growing ends compete for a pool of factors regulating cell growth (Taheraly et al., 2020). Together, these findings highlight the differential regulation of polarized growth between germinating and vegetative cells.
The Cdc42 GTPase-activating protein Rga6 dictates the monopolar growth of germinating cells
Rga3, Rga4, and Rga6 have been identified as Cdc42 GTPase-activating proteins (GAPs) in fission yeast cells (Gallo Castro and Martin, 2018; Revilla-Guarinos et al., 2016), and these Cdc42 GAPs work in concert to restrict Cdc42-GTP to the growing cell ends during vegetative growth. We hypothesized that the characteristic localization of Cdc42-GTP in germinating cells may be regulated by a different combined effect of the three Cdc42 GAPs to promote monopolar cell growth.
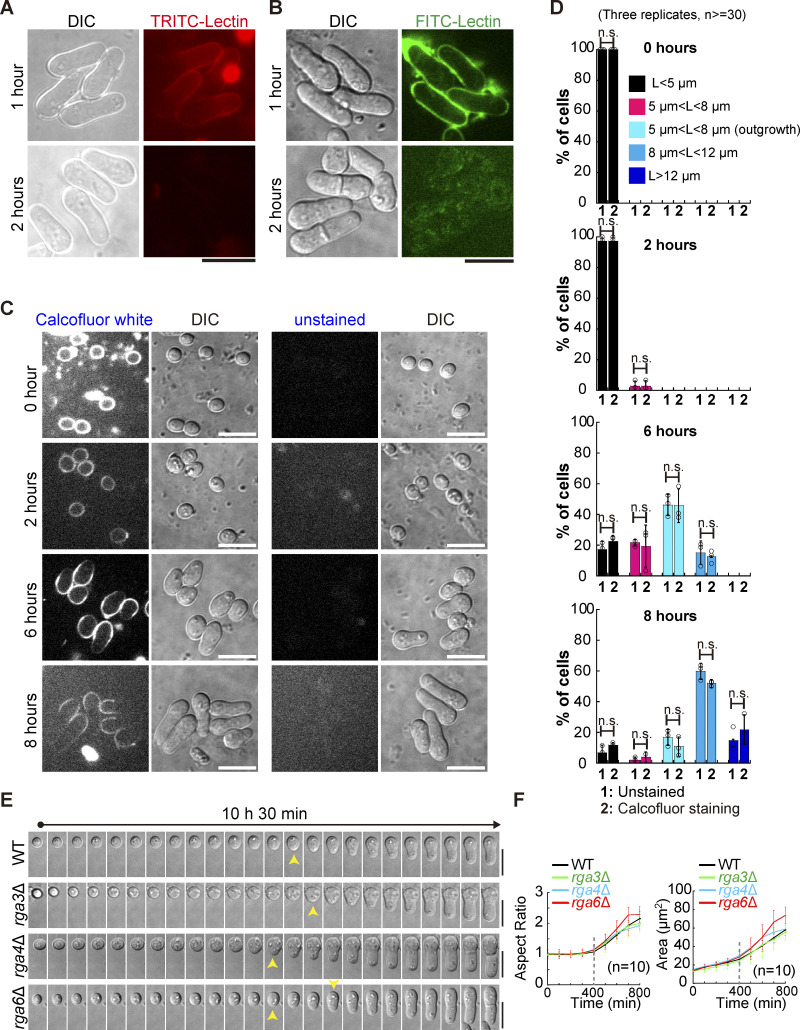
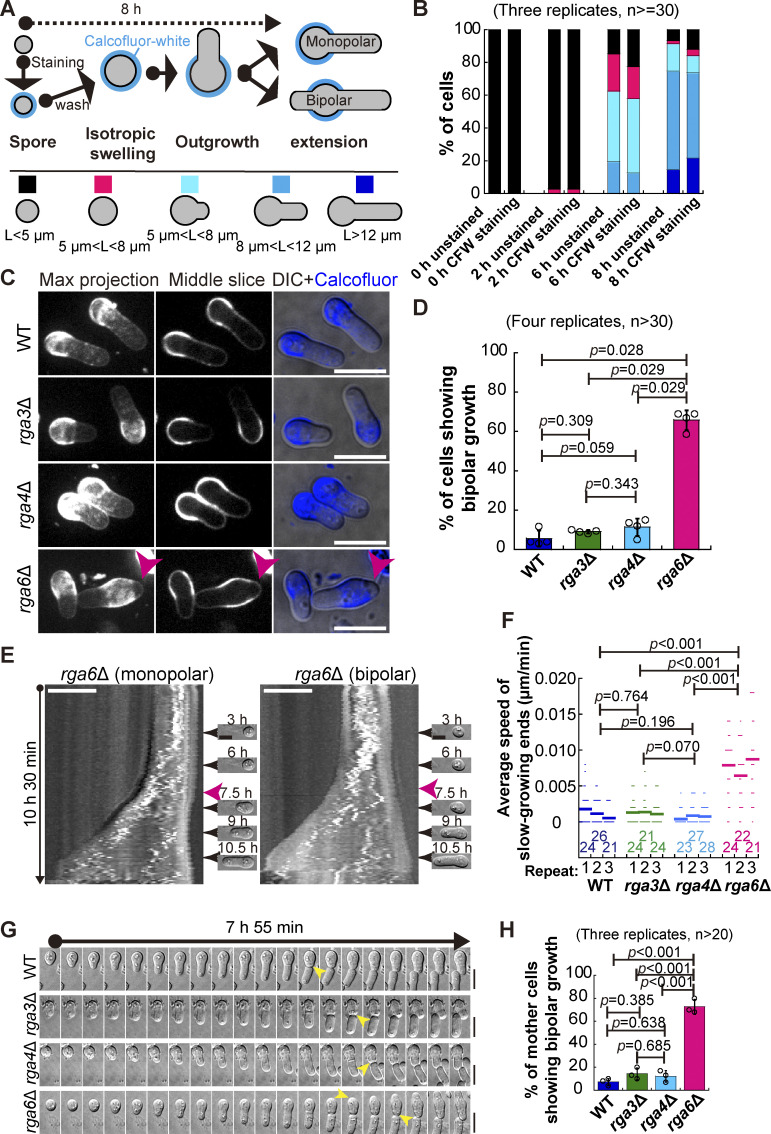
To test this hypothesis, reliable fluorescence-based microscopic approaches are necessary for examining the growth pattern of germinating cells in a quantitative manner. In general, spores undergo outgrowth between 6 and 8 h after exiting the dormancy stage, as reported previously (Bonazzi et al., 2014; Plante et al., 2017) and shown in the present study (see below). Therefore, to study spore outgrowth, fluorescent staining of cells should be stable for at least 8 h. We first tested the stability of staining with TRITC-conjugated or FITC-conjugated Lectin. As shown in Fig. S1 A, after only 2 h of staining, fluorescence was almost completely lost from the cell wall, making it impossible to use Lectin staining to study spore outgrowth. We then tested the stability of Calcofluor-white staining, which has been developed previously for monitoring the stage of spore germination (Plante and Labbe, 2019; Plante et al., 2017). Briefly, dormant spores were first stained with Calcofluor-white, and after washing, the stained spores were cultured in Calcofluor-free rich medium for microscopic analysis (Fig. 2 A). This approach allowed for convenient separation of spores that grow in monopolar and bipolar manners (i.e., displaying Calcofluor-white staining at only one end or no Calcofluor-white staining at both ends), respectively (Fig. 2 A). To assess the potential detrimental effect of Calcofluor-white staining on spore growth, we included a group of spores that were not stained in the experiments. As shown in Fig. 2 B and Fig. S1, C and D, fluorescence of Calcofluor-white staining was still visible 8 h after the stained spores were cultured in Calcofluor-free rich medium. Quantification results further revealed that the Calcofluor staining procedure did not appear to affect spore growth because the percentage of spores stained with or without Calcofluor-white at the indicated growth stages was comparable (Fig. 2 B; and Fig. S1, C and D). It is possible that the special properties of the outer spore wall (Taharaly et al., 2020) protect spores against the potential detrimental effects of Calcofluor-white staining. Therefore, the Calcofluor-white staining method, established previously (Plante and Labbe, 2019; Plante et al., 2017) and used in the present study, can be used to reliably study spore outgrowth.
Figure S1.
Assessments of spore staining with Lectin and Calcofluor-white and the spore growth by long time-lapse light microscopy (Related to Fig. 2). (A and B) Staining of germinating spores with TRITC-conjugated (A) or FITC-conjugated (B) Lectin. The stained spores were cultured in YE5S liquid medium for 1 or 2 h, followed by confocal microscopic observation. Scale bar, 10 μm. (C) Z-slice (middle) and DIC images of spores stained with or without Calcofluor-white (CFW). The time after the stained spores were cultured in liquid YE5S medium is indicated. Scale bar, 10 μm. (D) Quantification of the indicated category (also see Fig. 2 A) of spores at the indicated time points after the stained spores were cultured in liquid YE5S medium (as shown in C; related to Fig. 2 B). Three independent experiments were carried out, and the number of spores analyzed is not less than 30 for each replicate. The top of the column and the error bar represent the mean and SD, respectively. Statistical analysis was performed by Student’s t test. (E) Time-lapse differential interference contrast (DIC) images of germinating WT, rga3Δ, rga4Δ, and rga6Δ spores (related to Fig. 2, E and F). Yellow arrowheads indicate the start of outgrowth. Note that images were acquired once spores were cultured on YE5S medium pads. Scale bar, 10 μm. (F) Aspect ratio (left) and area (right) of the indicated germinating spores in E over time. Note that outgrowth was detected at ∼400 min after spores were grown on YE5S medium pads. The number (n) of cells analyzed is indicated. Lines and error bars represent the mean and SD, respectively.
Figure 2.
Characterization of the Cdc42 GTPase-activating proteins Rga3, Rga4, and Rga6 in regulating monopolar growth of germinating cells. (A) Diagrams illustrating the method of using Calcofluor-white staining to determine the growth pattern of germinating cells (i.e., monopolar or bipolar outgrowth). (B) Shown below are the category of spore stages, and quantification in B was based on these categories. L indicates spore length. Average percentage of the indicated categories (in A) of the unstained spores or the spores stained with Calcofluor-white (CFW) at the indicated time after culture in Calcofluor-white–free rich medium. Three independent experiments were carried out, and the number of cells analyzed for each experiment was not less than 30. Additionally, see Fig. S2, C and D, for details. (C) Z-slice (indicated by Middle) and maximum projection images of WT, rga3Δ, rga4Δ, and rga6Δ germinating cells that were stained with Calcofluor-white and cultured in rich medium for 8 h. Pink arrowheads indicate monopolar growth of outgrowing cells. DIC indicates differential interference contrast images. Scale bar, 10 μm. (D) Plot of the percentage of germinating WT, rga3Δ, rga4Δ, and rga6Δ cells (indicated in C) that underwent bipolar growth. Note that four independent experiments were carried out for quantification, and >30 cells were analyzed for each replicate. The top of the column and the error bar are the mean ± SD. The Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. (E) Kymograph graphs of representative rga6Δ spores growing in monopolar and bipolar manners. Images were acquired by time-lapse light microscopy for 10.5 h, and representative DIC images at the indicated timepoint are shown on the right. Pink arrowheads indicate spore outgrowth. Scale bar, 5 μm. (F) Average extension speed of the slow-growing end of WT, rga3Δ, rga4Δ, and rga6Δ spores. The speed was measured using kymograph graphs, as indicated in E, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed are shown. Bars represent the mean. For comparisons between groups, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. (G) Time-lapse DIC images of WT, rga3Δ, rga4Δ, and rga6Δ spores. Note that the pattern of monopolar growth of WT, rga3Δ, and rga4Δ parental cells, but not rga6Δ parental cells, was maintained after the first cell. Yellow arrowheads indicate outgrowth. Scale bar, 10 μm. (H) Quantification of the percentage of WT, rga3Δ, rga4Δ, and rga6Δ parental cells showing bipolar outgrowth after the first cell division. Three independent experiments were carried out, and the number of cells analyzed for each experiment was not less than 20. The top of the column and the error bar are the mean ± SD. One-way ANOVA with Tukey HSD post hoc test was used to calculate the P values.
Given that spore outgrowth took place in >80% of spores after 8 h of culturing spores in rich medium, i.e., liquid YE medium (Fig. 2 B; and Fig. S1, C and D), we analyzed growth polarity after the spores had been grown in rich medium for 8 h. Microscopic analysis and quantification showed that ∼66% of rga6∆ spores grew in an abnormal bipolar manner whereas only ∼6, ∼9, and ∼11% of WT, rga3∆, and rga4∆ spores grew in an abnormal bipolar manner (Fig. 2, C and D). To complement the Calcofluor-white staining approach, we further employed long time-lapse light microscopy to observe the growth of unstained spores (Fig. 2 E). Since the growth of the fast-growing end was pronounced, measurement of the average speed of the slow-growing end should be accurate for determining the pattern of growth polarity. Measurements showed that the average speed of the slow-growing end of rga6∆ spores was significantly higher than that of WT, rga3∆, and rga4∆ spores, which was unnoticeable or zero (Fig. 2, E and F). This result confirmed the above finding that rga6∆, but not WT, rga3∆, and rga4∆, spores grew in an abnormal bipolar manner. Quantification of the time-lapse light microscopic data also confirmed the previous finding (Bonazzi et al., 2014) that outgrowth generally took place between 6–8 h after germination as the aspect ratio and cell area increased significantly at ∼7 h (Fig. S1, E and F). The time for the abrupt change in aspect ratio and cell area was similar in WT, rga3∆, rga4∆, and rga6∆ cells, suggesting that the contribution of the three Cdc42 GAPs to regulating the timing of spore outgrowth is unlikely. Instead, the results suggest that among the three Cdc42 GAPs, Rga6 plays a crucial role in directing monopolar outgrowth of spores.
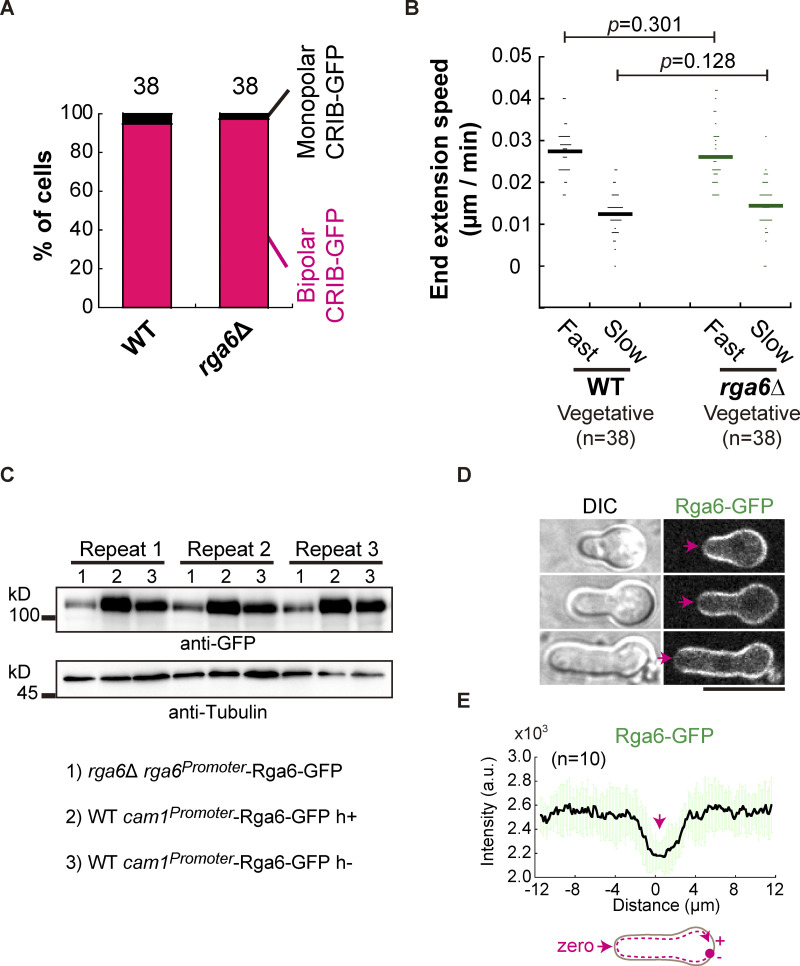
We then asked whether the prominent role of Rga6 in regulating monopolar growth is specific during spore germination. To address this question, we further compared the localization patterns of CRIB-GFP and measured the speed of end extension of wild-type and rga6∆ vegetative cells. No significant difference was found between wild-type and rga6∆ vegetative cells (Fig. S2, A and B), suggesting that Rga6 may play a very minor role in regulating the polarized growth of vegetative cells. Consistently, it has been reported that the absence of Rga6 only mildly affects the growth polarity of vegetative cells (Revilla-Guarinos et al., 2016). By contrast, it has been reported that overexpression of Rga6 causes pronounced monopolar growth of vegetative cells (Revilla-Guarinos et al., 2016). This finding is consistent with the role of Rga6 in promoting monopolar outgrowth during germination. Therefore, it is conceivable that overexpression of Rga6 did not appear to affect the monopolar outgrowth of spores (Fig. S2, C–E). Hence, it is highly likely that Rga6 plays a specific role in directing monopolar growth polarity during spore germination.
Figure S2.
The effect of the absence of Rga6 on vegetative cell growth and the effect of Rga6 overexpression on spore outgrowth. (A) Quantification of staining of the bipolar and monopolar CRIB-GFP at cell ends in WT and rga6Δ vegetative cells. The number of cells analyzed is indicated. (B) Extension speed of the fast- and slow-growing ends of WT and rga6Δ vegetative cells. Bars represent mean, and the number (n) of cells analyzed is indicated. Statistical analysis was performed by Student’s t test. (C) Western blotting analysis of Rga6-GFP expressed from its own promoter in rga6∆ cells or from a cam1 promoter in WT cells (i.e., overexpression). Repeat indicates the three replicates. Antibodies against GFP and Tubulin were used. (D) Z-slice (middle) images of outgrowing spores expressing Rga6-GFP from the cam1 promoter. Arrows indicate the growing tip where relatively less signals of Rga6-GFP were detected. Scale bar, 10 μm. (E) Plots of the signal intensity of Rga6-GFP along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively. Source data are available for this figure: SourceData FS2.
Since the germinating mother cell has been shown to undergo monopolar growth for several cell cycles (Bonazzi et al., 2014), we sought to test how Cdc42 GAPs affect the monopolar growth of the germinating mother cell by time-lapse light microscopy. As shown in Fig. 2 G, the absence of Rga6 resulted in bipolar growth of the germinating mother cell during the second division. Quantification confirmed that only ∼27% of the germinating rga6∆ mother cells still displayed monopolar growth during the second division but ∼93, ∼86, and ∼88% of the germinating WT, rga3∆, and rga4∆ mother cells, respectively, displayed monopolar growth (Fig. 2 H). Thus, we reasoned that Rga6 also plays a crucial role in maintaining the continual monopolar growth of the germinating mother cell.
Rga6 restricts bipolar outgrowth but not the spore wall during germination
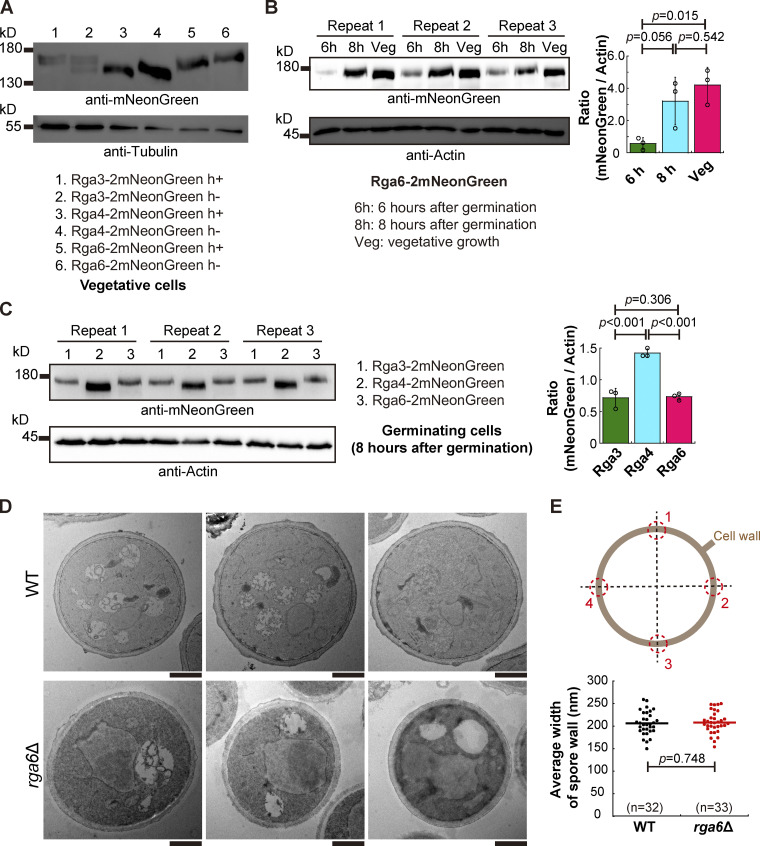
How is Rga6 involved in regulating monopolar growth during spore germination? Is the role of Rga6 in inhibiting bipolar outgrowth during spore germination due to a high level of Rga6 expression? To address these questions, Rga6, along with Rga3 and Rga4, was tagged with 2mNeonGreen and expressed endogenously (Fig. 3 A). We noticed that the size of Rga6-2mNeonGreen, as shown on the blot, was larger than expected, hinting existence of posttranslational modification (Fig. 3 A). Rga6-2mNeonGreen-expressing vegetative cells and Rga6-2mNeonGreen-expressing spores after 6 or 8 h of culture in rich medium were then collected for analysis of Rga6-2mNeonGreen expression by Western blotting assays. As shown in Fig. 3 B, the level of Rga6 expression was generally lower during spore germination than that of vegetative growth. In addition, among the three Cdc42 GAPs, Rga4, but not Rga6, exhibited the highest expression during spore germination (Fig. 3 C). Thus, we concluded that Rga6 inhibits bipolar outgrowth during spore germination independent of its increased expression level.
Figure 3.
Endogenous expression of Rga3-2mNeonGreen, Rga4-2mNeonGreen, and Rga4-2mNeonGreen, and the effect of the absence of Rga6 on the spore wall. (A) Western blotting analysis of Rga3-2mNeoGreen, Rga4-2mNeoGreen, and Rga6-2mNeoGreen expressed in the indicated vegetative cells. Antibodies against mNeonGreen and Tubulin were used. (B) Western blotting analysis of Rga6-2mNeoGreen in germinating spores (6 and 8 h after germination) and in vegetative cells. Three independent experiments were carried out, and antibodies against mNeonGreen and actin were used. The ratio of the band intensity of mNeonGreen to that of Actin is shown on the right. One-way ANOVA with Tukey HSD Post Hoc test was used to calculate the P values. (C) Western blotting analysis of Rga3-2mNeoGreen, Rga4-2mNeoGreen, and Rga6-2mNeoGreen in germinating spores (8 h after germination). Antibodies against mNeonGreen and actin were used. The intensity ratio of mNeonGreen to actin is shown. Three independent experiments were carried out. One-way ANOVA with Tukey HSD Post Hoc test was used to calculate the P values. (D) Transmission electron microscopic (TEM) images of WT and rga6∆ spores growing at the stage of isotropic swelling (3 h after germination). Scale bar, 1 μm. (E) Diagrams showing the four junctions selected for measuring the width of the spore wall (gray circle). The average width of WT and rga6Δ spore cells (i.e., the average width of the spore wall at the four junctions indicated in the diagram) was measured and plotted. Student’s t test was used to calculate the P value. The number (n) of spores analyzed is indicated. Source data are available for this figure: SourceData F3.
The rigid outer spore wall plays a critical role in dictating spore outgrowth (Bonazzi et al., 2014). This finding prompted us to examine whether the outer spore wall was impaired in rga6∆ spores. After culture in rich medium for 3 h, spores that had exited the dormancy stage (presumably undergoing isotropic swelling) were analyzed by transmission electron microscopy. We noticed that thin electron-dense structures, presumably the outer spore wall, were present at the outmost layer of the WT and rga6∆ spores (Fig. 3 D). However, the thin electron-dense structures were amorphous, making it challenging to quantify the thickness of the outer spore wall. Therefore, we sought to examine the spore wall carefully. Examination revealed no discernable difference in the spore wall between WT and rga6∆ spores (Fig. 3 D). Measurements further showed that the average wall width of WT and rga6∆ spores was comparable (Fig. 3 E). Therefore, it is unlikely that the absence of Rga6 causes bipolar outgrowth of spores by impairing the spore wall.
The absence of Rga6 causes Cdc42-GTP to localize to the two ends of outgrowing spores
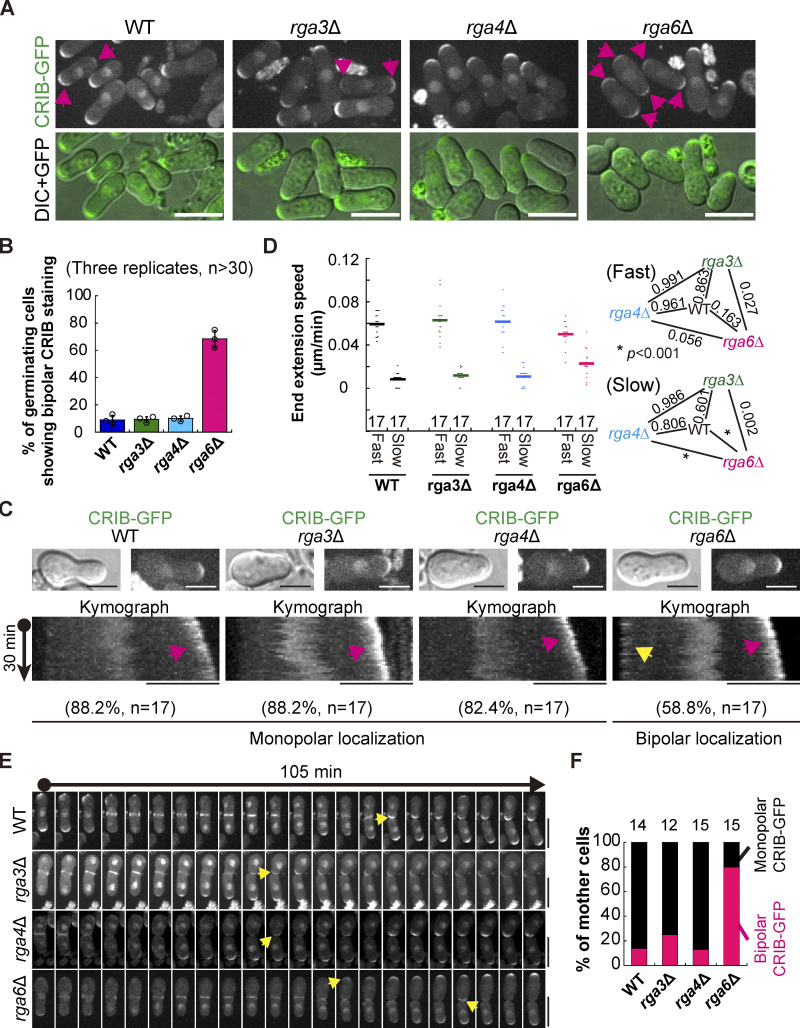
We then examined the localization of Cdc42-GTP, the master regulator of cell polarity, in outgrowing WT, rga3∆, rga4∆, and rga6∆ spores. As shown in Fig. 4, A and B, ∼68% of germinating rga6∆ cells displayed bipolar staining of Cdc42-GTP (marked by CRIB-GFP) whereas only ∼9, ∼9, and ∼10% of germinating WT, rga3∆, and rga4∆ cells, respectively, displayed bipolar staining of Cdc42-GTP. This result suggests that Rga6 regulates monopolar outgrowth of spores likely by confining Cdc42-GTP to one cell end. Time-lapse microscopic analysis further revealed that Cdc42-GTP constantly resided at the fast-growing end of rga6∆ spores but localized to the slow-growing end of rga6∆ spores in a back-and-forth manner (Fig. 4 C). Localization of Cdc42-GTP to the fast-growing end of WT, rga3∆, and rga4∆ spores was pronounced but localization of Cdc42-GTP to the slow-growing end of the cells was rare (Fig. 4 C). Consistently, measurements of the extension speed of the cell end confirmed that the extension speed of the fast-growing end of WT, rga3∆, rga4∆, and rga6∆ spores was comparable but the extension speed of the slow-growing end was significantly higher in rga6∆ spores than in WT, rga3∆, and rga4∆ spores (Fig. 4 D). We then monitored the dynamic localization of Cdc42-GTP in mother cells after the first cell division. As shown in Fig. 4, E and F, 80, 13, 25, and 14% of rga6∆, rga4∆, rga3∆, and WT mother cells, respectively, exhibited dual-end localization of Cdc42-GTP after the first division. Thus, we concluded that Rga6 plays a crucial role in directing the monopolar localization of Cdc42-GTP in germinating cells to promote monopolar growth.
Figure 4.
Rga6, but not Rga3 or Rga4, specifies the localization of CRIB-GFP in outgrowing spores. (A) Maximum projection images of outgrowing WT, rga3Δ, rga4Δ, and rga6Δ cells expressing CRIB-GFP. Images were acquired after spores were cultured in YE5S liquid medium for 8 h. Pink arrows indicate cells showing bipolar CRIB-GFP staining. Scale bar, 10 μm. (B) Quantification of the percentage of outgrowing WT, rga3Δ, rga4Δ, and rga6Δ cells showing bipolar CRIB-GFP localization. Three independent experiments were carried out, and more than 30 cells were analyzed for each replicate. (C) Kymograph graphs of CRIB-GFP of representative outgrowing WT, rga3Δ, rga4Δ, and rga6Δ cells. Maximum projection and DIC images of the cell used for constructing the corresponding kymograph graph are shown at the top. Pink and yellow arrows indicate CRIB-GFP at the fast- and slow-growing ends, respectively. Scale bar, 5 μm. Shown below is the percentage of outgrowing cells displaying monopolar CRIB-GFP localization (WT, rga3Δ, and rga4Δ) and bipolar CRIB-GFP localization (rga6Δ), and the number (n) of cells analyzed is indicated. (D) Dot plots of the end extension speed of the indicated outgrowing cells in C. Bars represent the mean, and one-way ANOVA with Tukey HSD post hoc test was used to calculate the P values. The number of cells analyzed is indicated on the x-axis. (E) Time-lapse maximum projection images of outgrowing spores that were completing the first cell division. Yellow arrows indicate the emergence of CRIB-GFP. Scale bar, 10 μm. (F) Quantification of the growth pattern of the indicated parental cells in E. The number (n) of cells analyzed is indicated on each column.
Rga6 colocalizes with the PI(4,5)P2 sensor Opy1 on the cell cortex during spore germination
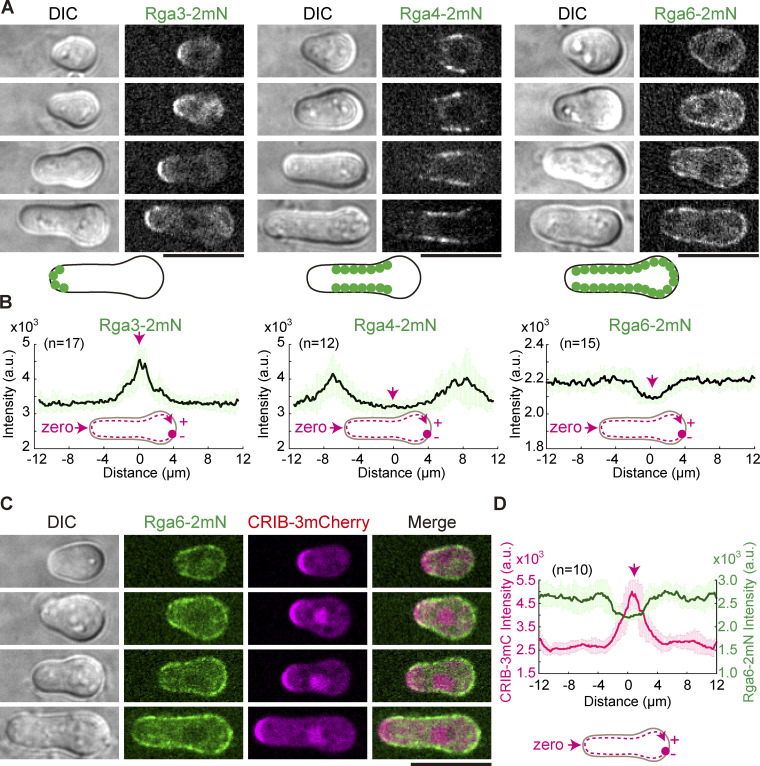
We next employed live-cell microscopy to examine the localization of Rga6-2mNeonGreen, Rga3-2mNeonGreen, and Rga4-2mNeonGreen in outgrowing spores. Consistent with the previously reported data obtained using vegetative cells (Gallo Castro and Martin, 2018; Revilla-Guarinos et al., 2016), Rga3 mainly localized to the growing end of the outgrowing spores while Rga4 mainly localized to the lateral periphery of the outgrowing spores by membrane flows (Gerganova et al., 2021; Fig. 5, A and B). Interestingly, Rga6 strongly decorated the cortical regions, except the growing end, of the outgrowing spores (Fig. 5, A and B). Further colocalization analysis of Cdc42-GTP (marked by CRIB-3mCherry) and Rga6-2mNeonGreen confirmed that the two proteins complement each other in localization on the cell cortex (Fig. 5, C and D). This localization pattern of Rga6 resembled the localization pattern of Opy1, a sensor specifically recognizing phosphatidylinositol-4,5-bisphosphate (PI[4,5]P2; Snider et al., 2020; Fig. 6, A and B). A polybasic region is present at the extreme C-terminus of Rga6 and is required for the cortical localization of Rga6 (Revilla-Guarinos et al., 2016; Zheng et al., 2022). Together, these findings raise a possibility that Rga6 localizes to the non-growing cortical region of the outgrowing spore by interacting with PI(4,5)P2.
Figure 5.
Cortical localization of Rga3, Rga4, and Rga6 in outgrowing cells. (A) Z-slice images of outgrowing cells expressing Rga3-2mNeonGreen, Rga4-2mNeonGreen, or Rga6-2mNeonGreen. Scale bar, 10 μm. Diagrams illustrate the cortical localization of 2mNeonGreen-marked proteins (green filled circles). (B) Plots of the average signal intensity (from the indicated number [n] of cells) of Rga3-2mNeoGreen, Rga4-2mNeoGreen, and Rga6-2mNeoGreen along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively. (C) Z-slice images of outgrowing cells expressing CRIB-3mCherry and Rga6-2mNeonGreen. Scale bar, 10 μm. (D) Plots of the average signal intensity (from 10 cells) of CRIB-3mCherry and Rga6-2mNeoGreen along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively.
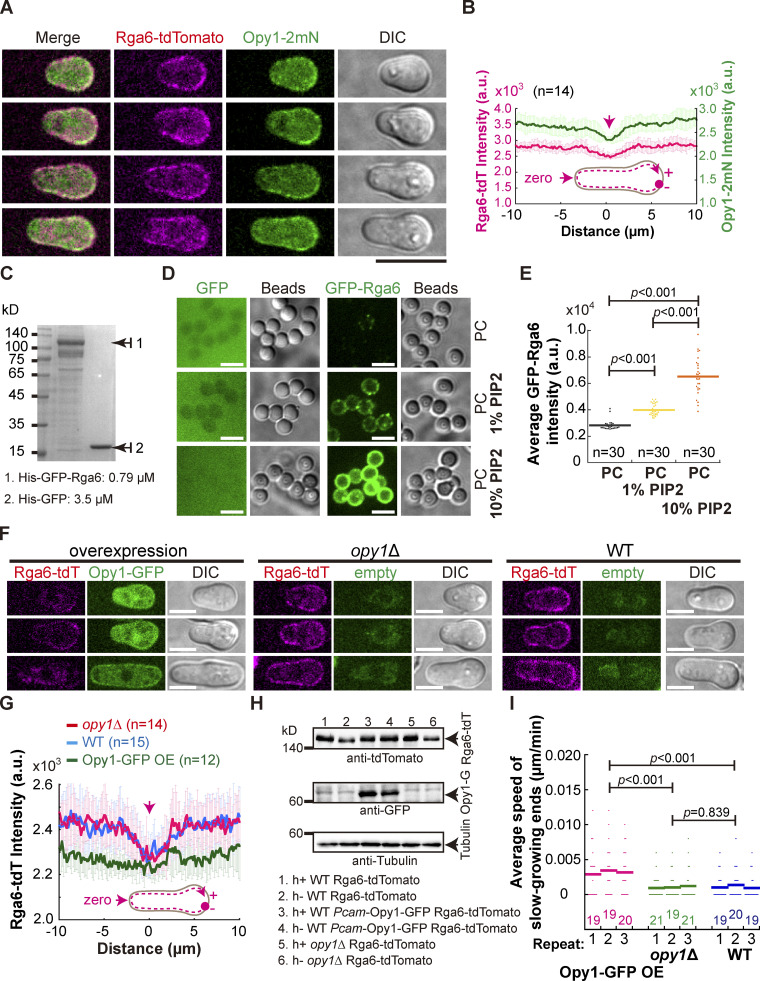
Figure 6.
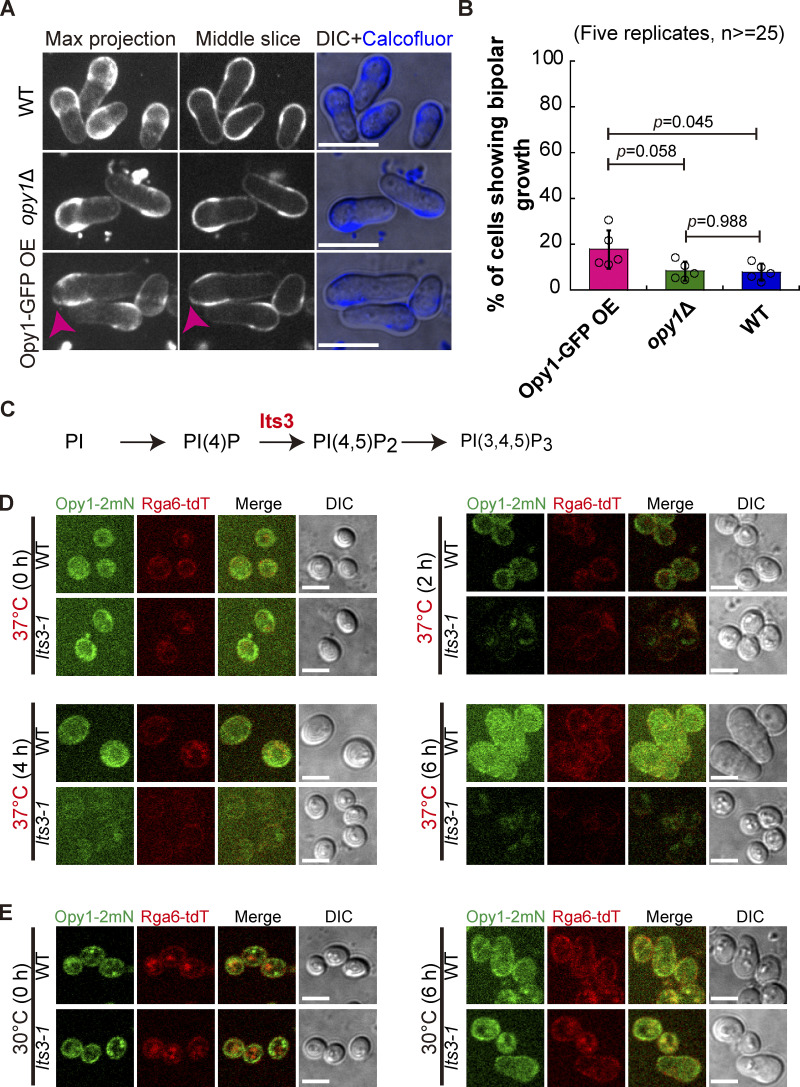
Rga6 colocalizes with Opy1 on the cell cortex in outgrowing cells. (A) Maximum projection images of outgrowing cells expressing Rga6-tdTomato and Opy1-2mNeonGreen (marking PI[4,5]P2). Scale bar, 10 μm. (B) Plots of the average signal intensity (from 14 cells) of Rga6-tdTomato and Opy1-2mNeonGreen along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively. (C) Coomassie-blue staining analysis of recombinant proteins (His-GFP and His-GFP-Rga6) used in the liposome reconstitution assay. Protein concentration is indicated. (D) Representative Z-slice images of lipid-coated silica microsphere beads incubated with the recombinant protein His-GFP or His-GFP-Rga6. Note that liposomes were reconstituted with PC alone, PC+ 1% PI(4,5)P2, or PC+ 10% PI(4,5)P2. Scale bar, 5 μm. (E) Quantification of the intensity of Rga6-GFP signals on the lipid-coated microspheres. Bars represent the mean, and the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. The number (n) of microspheres analyzed is indicated. a.u., arbitrary units. (F) Z-slice images of the indicated cells (i.e., overexpression of Opy1-GFP, opy1Δ, and WT) expressing Rga6-tdTomato. Scale bar, 5 μm. (G) Average intensity of Rga6-tdTomato along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively. The number (n) of cells analyzed is indicated. (H) Western blotting analysis of the indicated cells below the blot graphs. Antibodies against tdTomato, GFP, and tubulin were used. (I) Average extension speed of the slow-growing ends of the three types of cells indicated in F. The speed was measured using kymograph graphs, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed are shown. Bars represent the mean. For comparisons between groups, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. Source data are available for this figure: SourceData F6.
We next sought to examine the affinity of Rga6 for PI(4,5)P2 directly by liposome reconstitution assays, as developed previously (Bridges et al., 2016; Zheng et al., 2022). Briefly, we first coated silica microspheres with a mixture of lipids containing phosphatidylcholines (PC) and PI(4,5)P2 at a different ratio and then incubated the lipid-coated microspheres with recombinant proteins His-GFP and His-GFP-Rga6 (Fig. 6, C and D). Confocal microscopic analysis showed that His-GFP did not appear to bind lipid-coated microspheres whereas His-GFP-Rga6 efficiently bound the lipid-coated microspheres carrying PI(4,5)P2 (Fig. 5 D). Intriguingly, the affinity of His-GFP-Rga6 for lipid-coated microspheres was increased as the percentage of PI(4,5)P2 increased in the lipid mixture (Fig. 6, D and E). These results suggest that Rga6 binds PI(4,5)P2.
Given the similar affinity of Opy1 and Rga6 for PI(4,5)P2, overexpression of Opy1 was expected to affect the cortical localization of Rga6. Consistently, overexpression of Opy1-GFP impaired the cortical localization of Rga6, indicative of a competitive mode of binding Opy1 and Rga6 to PI(4,5)P2 (Fig. 6, F and G), and the expression of Opy1-GFP and Rga6-tdTomato was confirmed by Western blotting analysis (Fig. 6 H). In addition, we examined the localization of Rga6-tdTomato in spores lacking Opy1. Interestingly, the absence of Opy1 did not appear to affect the cortical localization of Rga6-tdTomato (Fig. 6, F and G). The localization patterns of Rga6 in Opy1-GFP-overexpressing and opy1∆ spores were consistent with the phenotypes of spore outgrowth. Specifically, Opy1-GFP-overexpressing spores displayed a low but noticeable speed of slow-growing ends, whereas WT and opy1∆ spores displayed an unnoticeable or zero speed of slow-growing ends (Fig. 6 I). Similar results were obtained by the Calcofluor staining method (Fig. S3, A and B). its3 is an essential gene encoding phosphatidylinositol-4-phosphate 5-kinase responsible for the synthesis of PI(4,5)P2 (Zhang et al., 2000). Therefore, we took opportunity of an existing temperature-sensitive mutant of its3, i.e., its3-1, which was used previously for studying PI(4,5)P2 (Snider et al., 2018; Zhang et al., 2000), and tested how deficient synthesis of PI(4,5)P2 affects the cortical localization of Rga6 and spore outgrowth. As shown in Fig. S3, C–E, the malfunction of Its3 in its3-1 spores significantly impaired the cortical localization of both Opy1-2mNeonGreen and Rga6-tdTomato. Because of the essential role of PI(4,5)P2 in promoting outgrowth (Fig. S3 D), it was impossible to test the effect of the impaired localization of Rga6 by deficient synthesis PI(4,5)P2 on spore outgrowth. Nonetheless, these data are consistent with the finding that both Rga6 and Opy1 have an affinity for PI(4,5)P2.
Figure S3.
The effect of the absence of Opy1 or overexpression of Opy1 on spore outgrowth and the localization of Opy1-2mNeonGreen and Rga6-tdTomato in its3-1 spores (Related to Fig. 6). (A) Maximum projection and Z-slice (Middle) images of WT, opy1Δ, and Opy1-overexpressing spores that were stained with Calcofluor-white. Note that stained spores were cultured in Calcofluor-white-free rich medium for 8 h before microscopic analysis. Pink arrowheads indicate bipolar growth of outgrowing spores. Scale bar, 10 μm. (B) The percentage of germinating WT, opy1Δ, and Opy1-overexpressing spores that underwent bipolar growth, as shown in A. Five independent experiments were carried out, and ≥ 25 spores were analyzed for each replicate. The top of the column and the error bar are the mean ± SD. One-way ANOVA with Tukey HSD post hoc test was used to calculate the P values. (C) Diagram showing the function of Its3 in synthesizing PI(4,5)P2. (D) Maximum projection images of Opy1-2mNeonGreen and Rga6-tdTomato-expressing WT and its3-1 spores cultured at the restricted temperature 37°C for the indicated time. Scale bar, 5 μm. (E) Maximum projection images of Opy1-2mNeonGreen and Rga6-tdTomato-expressing WT and its3-1 spores cultured at the permissive temperature 30°C for the indicative time. Note that both WT and its3-1 spores germinated at the permissive temperature. Scale bar, 5 μm.
In summary, the above findings support a model in which membrane composition, particularly PI(4,5)P2, dictates the localization of Rga6 on the cell cortex to specify the localization of Cdc42-GTP at cell ends.
Both the PBR region and the GAP domain of Rga6 are required for promoting monopolar growth of outgrowing spores
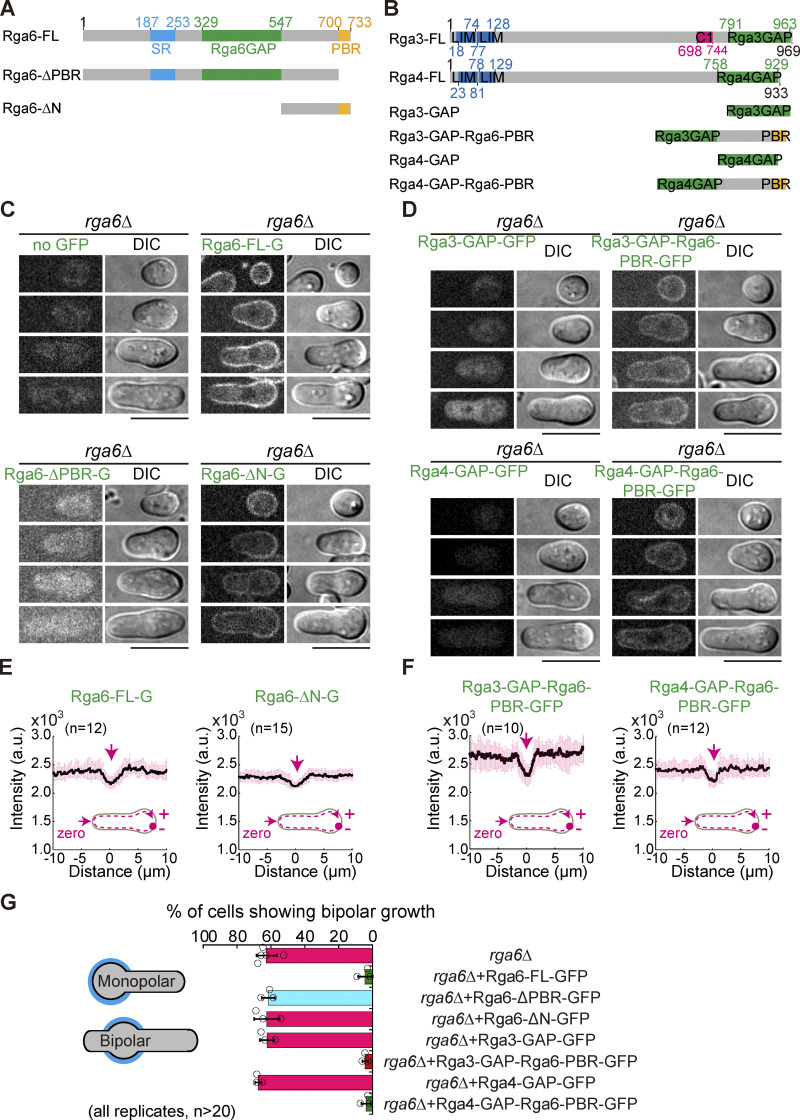
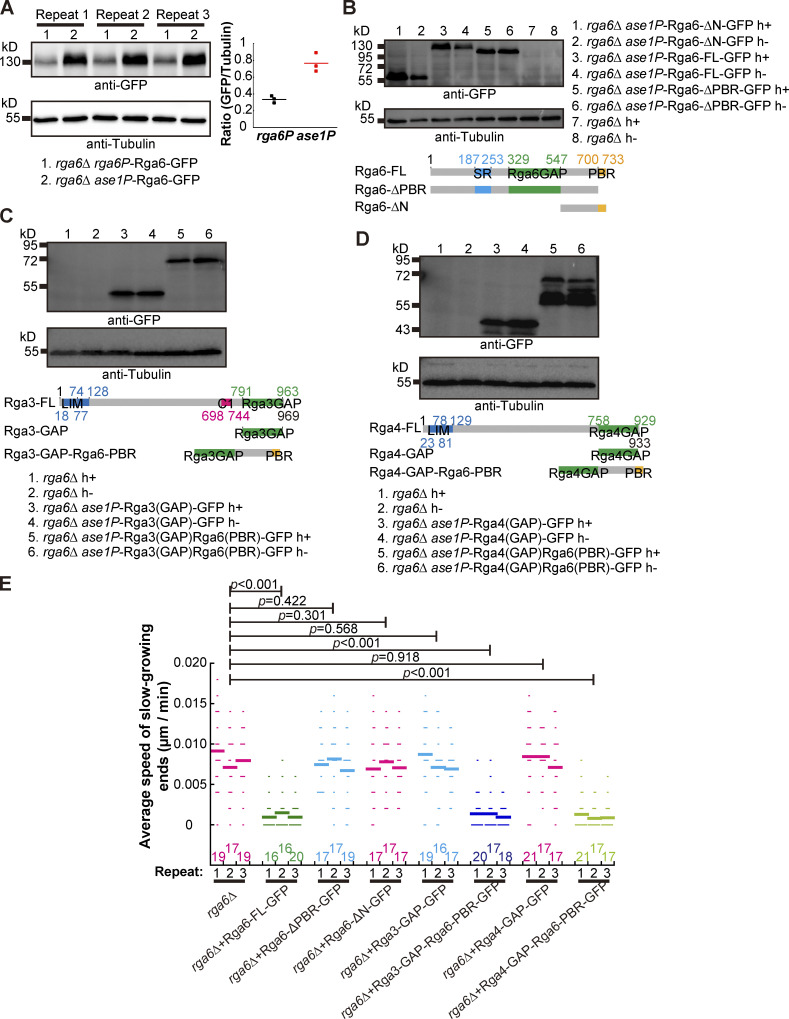
It has been reported that the membrane affinity of Rga6 depends on the C-terminal PBR (polybasic region) region of Rga6 and that the GAP domain is present in the middle of Rga6 (Revilla-Guarinos et al., 2016). To test whether the cortical localization of Rga6 is important for executing Rga6’s function, we expressed GFP-tagged full-length Rga6 (i.e., Rga6-FL), Rga6 lacking the N-terminus (containing a GAP domain; i.e., Rga6-∆N), and Rga6 lacking the PBR region (i.e., Rga6-∆PBR) in spores lacking Rga6 (Fig. 7, A and C). The expression level of Rga6 is quite low from its own promoter (Zheng et al., 2022). For the convenience of microscopic observation, we expressed the GFP-tagged Rga6 variants from the ase1 promoter, the expression levels of which were approximately three times greater than the levels from the rga6 promoter (Fig. S4 A). The expression of all proteins was confirmed by Western blotting analysis (Fig. S4 B). Microscopic observation revealed that Rga6-FL and Rga6-∆N, but not Rga6-∆PBR, decorated the cell cortex (Fig. 7 C), and line-scan analysis showed that both Rga6-FL and Rga6-∆N decorated significantly less at the growing zone of outgrowing spores (Fig. 7 E). Quantification of Calcofluor-white–stained outgrowing spores and live-cell light microscopic data further revealed that expression of only Rga6-FL, but not Rga6-∆N and Rga6-∆PBR, restored the monopolar outgrowth of rga6∆ spores (Fig. 7 G and Fig. S4 E). Taken together, these results suggest that both the GAP activity and the cortical localization of Rga6 are required for promoting monopolar outgrowth of spores.
Figure 7.
Chimera proteins consisting of a GAP domain and the Rga6 PBR domain rescue the growth defect of spores caused by the absence of Rga6. (A and B) Domain structures of Rga6 (A), Rga3 (B), and Rga4 (B). Truncation and chimera mutants are also indicated. (C) Z-slice images of outgrowing rga6Δ spores expressing Rga6-FL-GFP (full-length Rga6), Rga6-ΔPBR-GFP (Rga6 lacking the C-terminal PBR domain), or Rga6-ΔN-GFP (Rga6 lacking the N-terminal region including the SR and GAP domains) from the ase1 promoter. Scale bar, 10 μm. (D) Z-slice images of outgrowing rga6Δ spores expressing Rga3-GAP-GFP (Rga3 GAP domain alone tagged with GFP), Rga3-GAP-Rga6-PBR-GFP (Rga3 GAP domain fused with the PBR domain derived from Rga6), Rga4-GAP-GFP (Rga4 GAP domain alone tagged with GFP), or Rga4-GAP-Rga6-PBR-GFP (Rga4 GAP domain fused with the PBR domain derived from Rga6) from the ase1 promoter. Scale bar, 10 μm. (E and F) Plots of average intensity (from the indicated number [n] of cells) of GFP signals along the cell contour as illustrated by the pink dashed line in the diagram. Zero indicates the geometric center of the growing end while “+” and “−” indicate the positive and negative distances on the x-axis, respectively. Thick lines and error bars represent the mean and SD, respectively. Black lines and error bars represent the mean and SD, respectively. (G) Quantification of the percentage of the indicated cells showing bipolar outgrowth during germination, determined by Calcofluor-white staining.
Figure S4.
Testing the expression of Rga6 truncation mutants and chimera constructs and the effect of the Rga6 truncation mutants and chimeras on spore outgrowth (Related to Fig. 7). (A) Western blotting analysis of expression of Rga6-FL from its own promoter and from an ase1 promoter in rga6∆ cells. Three cultures for rga6∆ rga6p-Rga6-GFP and rga6∆ ase1p-Rga6-GFP cells were collected for analysis. Antibodies against GFP and tubulin were used. Intensity ratio of GFP over Tubulin is shown on the right of the blot. (B) Western blotting analysis of expression of Rga6-FL, Rga6-∆N, and Rga6-∆PBR in rga6∆ cells. Antibodies against GFP and Tubulin were used. (C) Western blotting analysis of expression of Rga3(GAP) and Rga3(GAP)Rga6PBR in rga6∆ cells. Antibodies against GFP and Tubulin were used. (D) Western blotting analysis of expression of Rga4(GAP) and Rga4(GAP)Rga6PBR in rga6∆ cells. Antibodies against GFP and Tubulin were used. (E) Average extension speed of the slow-growing ends of the indicated germinating cells shown in Fig. 7. The speed was measured using kymograph graphs, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed is shown. Bars represent the mean. For comparisons with rga6∆ spores, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. Source data are available for this figure: SourceData FS4.
We further asked whether artificially targeting a Cdc42 GAP domain to the cell cortex is enough to promote monopolar outgrowth of spores. To this end, we engineered chimeric proteins containing a GAP domain and the PBR region of Rga6 and tested the effect of these chimeras on spore outgrowth. Since Rga3, Rga4, and Rga6 are Cdc42 GAPs (Gallo Castro and Martin, 2018), we fused the GAP domains of Rga3 and Rga4, respectively, to the N-terminus of the PBR region of Rga6 to create chimeras Rga3-GAP-Rga6-PBR and Rga4-GAP-Rga6-PBR, respectively (Fig. 7 B). These two chimeric proteins, as well as the control truncation proteins Rga3-GAP (Rga3 GAP domain alone) and Rga4-GAP (Rga4 GAP domain alone), were expressed from the ase1 promoter in rga6∆ cells (Fig. 7 D). Western blotting analysis was carried out to confirm the expression of the GFP-tagged proteins (Fig. S4, C and D). Microscopic observation showed that both Rga3-GAP-Rga6-PBR and Rga4-GAP-Rga6-PBR, but not Rga3-GAP and Rga4-GAP, localized to the cell cortex (Fig. 7 D), and quantification showed that both Rga3-GAP-Rga6-PBR and Rga4-GAP-Rga6-PBR decorated significantly less at the growing zone of cells (Fig. 7 F). Intriguingly, quantification of Calcofluor-white–stained outgrowing spores and live-cell light microscopic data revealed that expression of Rga3-GAP-Rga6-PBR or Rga4-GAP-Rga6-PBR, but not Rga3-GAP and Rga4-GAP, restored monopolar outgrowth of rga6∆ spores (Fig. 7 G and Fig. S4 E). Thus, we concluded that Rga6 exerts its function through the C-terminal PBR for spatial localization and the Cdc42 GAP domain for promoting monopolar growth of outgrowing spores.
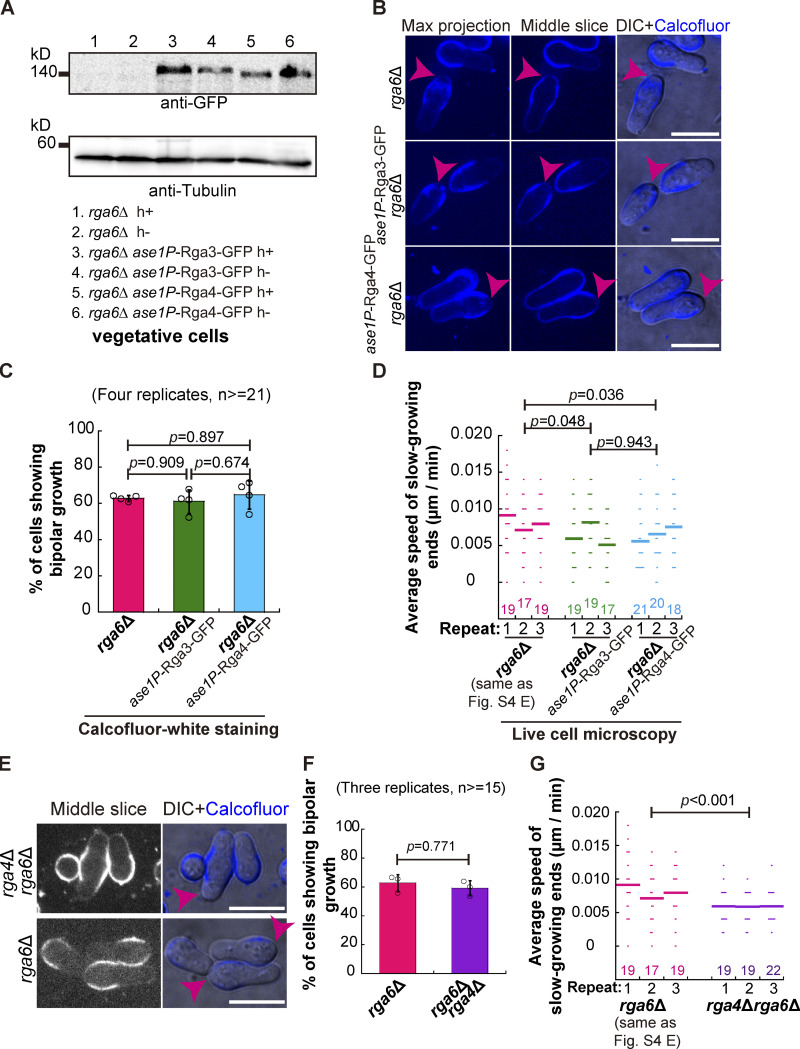
Finally, we asked whether overexpression of either Rga3 or Rga4 could rescue the phenotype of bipolar spore outgrowth caused by the absence of Rga6. To address this question, Rga3-GFP and Rga4-GFP were expressed in rga6∆ cells from the ase1 promoter, and the expression was confirmed by Western blotting analysis (Fig. S5 A). Quantification of Calcofluor-white–stained outgrowing spores revealed that the percentage of all tested spores displaying bipolar outgrowth was comparable (Fig. 5, B and C). Similar results were obtained by long time-lapse light microscopic analysis of spores that were not stained with Calcofluor-white (Fig. S5 D). Moreover, strains lacking both Rga4 and Rga6 were generated. Quantification of Calcofluor-white–stained outgrowing spores and long time-lapse light microscopic analysis consistently revealed that rga6∆ and rga4∆rga6∆ spores displaying bipolar outgrowth were comparable (Fig. S5, E–G). Together, these results further underscore the specific role of Rga6 in regulating monopolar outgrowth of spores.
Figure S5.
The effects of overexpression of Rga3 or Rga4 and the absence of both Rga4 and Rga6 on spore outgrowth. (A) Western blotting analysis of expression of Rga3-GFP and Rga4-GFP from the ase1 promoter in rga6∆ cells. Antibodies against GFP and Tubulin were used. (B) Maximum projection and Z-slice (Middle) images of rga6Δ and Rga3-GFP-overexpressing or Rga4-GFP-overexpressing rga6∆ spores that were stained with Calcofluor-white. Images were acquired after the stained spores were cultured in liquid YE5S medium for 8 h. Pink arrowheads indicate bipolar growth of outgrowing spores. Scale bar, 10 μm. (C) The percentage of the indicated spores that underwent bipolar growth, as shown in B. Note that four independent experiments were carried out, and ≥21 cells were analyzed for each replicate. One-way ANOVA with Tukey HSD Post Hoc test was used to calculate the P values. (D) Average extension speed of the slow-growing ends of the indicated cells. The speed was measured using kymograph graphs, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed is shown. Bars represent the mean. For comparisons between groups, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values, and for convenient comparison, and the data for rga6∆ in Fig. S4 E was used here. (E) Z-slice (Middle) images of rga6Δ and rga4Δrga6Δ spores that were stained with Calcofluor-white. Images were acquired after the stained spores were cultured in liquid YE5S medium for 8 h. Pink arrowheads indicate bipolar growth of outgrowing spores. Scale bar, 10 μm. (F) The percentage of the indicated spores that underwent bipolar growth, as shown in E. Note that three independent experiments were carried out, and ≥ 15 cells were analyzed for each replicate. Student’s t test was used to calculate the P value. (G) Average extension speed of the slow-growing ends of the indicated cells. The speed was measured using kymograph graphs, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed is shown. Bars represent the mean. For comparisons between groups, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values, and for convenient comparison, the data for rga6∆ in Fig. S4 E was used here. Source data are available for this figure: SourceData FS5.
Discussion
Fission yeast spores undergo monopolar growth upon outgrowth during germination (Bonazzi et al., 2014; Plante and Labbe, 2019). The first division of the germinating spore leads to a mother cell capable of monopolar growth for several cell cycles and a daughter cell undergoing vegetative growth (Bonazzi et al., 2014). This characteristic asymmetric cell division and polarized growth necessitate maintenance of Cdc42-GTP at the outgrowth zone on the spore plasma membrane. The underlying mechanism of Cdc42-GTP maintenance is not known. In the present study, we demonstrate that the Cdc42 GAP Rga6, but not Rga3 and Rga4, plays a crucial role in maintaining Cdc42-GTP at the outgrowth zone to promote monopolar growth during germination.
The localization of Cdc42-GTP in cells undergoing vegetative growth and germination shows different characteristics. During vegetative growth, Cdc42-GTP localizes to both ends of the cell and undergoes anti-correlated and out-of-phase oscillations before and after NETO (new end take off) between the two cell ends, respectively (Das et al., 2012; Das and Verde, 2013). By contrast, Cdc42-GTP localized to only one end of the germinating cells (>80%; Fig. 1 B), and the intensity of Cdc42-GTP was much brighter at the cell end in germinating cells than in cells undergoing vegetative growth (Fig. 1 C). These different characteristics of Cdc42-GTP localization and dynamics in vegetative and germinating cells imply a diversity of Cdc42-GTP regulation in a cell-type dependent manner. In vegetative cells, the combined effects of Rga3, Rga4, and Rga6 may be important for orchestrating the localization and dynamics of Cdc42-GTP to control proper cell length and width since deletion of all three Cdc42 GAPs causes Cdc42-GTP to spread all over the cell cortex and results in round-shaped cells and deletion of one or either two of the Cdc42 GAPs slightly alters cell shape (Gallo Castro and Martin, 2018). In germinating cells, among the three Cdc42 GAPs, Rga6 plays a dominant role in dictating the monopolar localization of Cdc42-GTP since deletion of rga6 led to bipolar localization of Cdc42-GTP in ∼68% of the outgrowing spores (Fig. 4, A and B) and bipolar growth of ∼66% of the germinating cells (Fig. 2, C and D). Similar to the specific role of Rga6 in promoting monopolar outgrowth during germination, Rga3 has a specific role in promoting the exploratory dynamics of Cdc42-GTP to confer an advantage in mating during sexual reproduction (Gallo Castro and Martin, 2018). Hence, although the three Cdc42 GAPs work in concert to direct vegetative cell growth, specific functional involvement of each Cdc42 GAP is likely tailored to meet the temporal requirements of the processes.
Intriguingly, continual monopolar growth of the mother germinating cell after the first division also requires Rga6 (Fig. 2, G and H). It is likely that Rga6 provides spatial cues to specify the outgrowth zones of the mother germinating cell to promote monopolar growth. The spatial cues may stem from the characteristic localization of Rga6 on the cell cortex.
As evident by microscopic data, Rga6 decorates the plasma membrane except the outgrowth zones of germinating cells, while Rga4 and Rga3 localize to the sides and the outgrowth zones of germinating cells, respectively (Fig. 5, A and B). Similar localization patterns were reported for Rga3, Rga4, and Rga6 in vegetative cells (Gallo Castro and Martin, 2018; Revilla-Guarinos et al., 2016). Rga6 colocalizes with Opy1, which has been shown to be an endogenous sensor of PI(4,5)P2 (Snider et al., 2020), on the spore plasma membrane except the outgrowth zone (Fig. 6, A and B). We noticed that Opy1 and Rga6 were enriched at the non-growing zones but were reduced at the opposite outgrowth zone (Fig. 6, A and B). Moreover, overexpression of Opy1-GFP significantly diminished the cortical localization of Rga6-tdTomato (Fig. 6, F and G) and caused bipolar outgrowth of spores (Fig. 6 I; and Fig. S3, A and B). Importantly, Rga6 is able to bind PI(4,5)P2 in vitro (Fig. 6, C–E). These findings, together with the phenotype that the absence of Rga6 leads to bipolar outgrowth during germination (Figs. 2 and 4), prompted us to hypothesize that lipid composition, particularly PI(4,5)P2, may dictate the cortical localization of Rga6 and consequently lead to the confinement of Cdc42-GTP to the outgrowth zone to promote monopolar outgrowth of spores. Intriguingly, it has been shown recently that the side localization of Rga4 is mediated by membrane flows induced by exocytosis (Gerganova et al., 2021). Although Rga4 and Rga6 similarly localize to the side of germinating spores, Rga6, but not Rga4, decorates the entire non-growing zones on the cell cortex (Fig. 5, A and B). This finding indicates that different mechanisms may underlie the localization of Rga4 and Rga6 to the cell cortex but does not exclude the possibility that exocytosis-induced membrane flows also contribute to the establishment of the characteristic localization of Rga6 in outgrowing spores.
The specific function of Rga6 in promoting monopolar outgrowth of spores depends on its RhoGAP domain and the PBR region, and the absence of either one phenocopies the phenotype of bipolar growth caused by the absence of Rga6 (Fig. 7 G and Fig. S4 E). This finding defines a minimal system possessing two properties (i.e., Cdc42 GAP enzymatic activity and membrane binding affinity) required for controlling monopolar outgrowth of spores. Intriguingly, we found that Cdc42 GAP activity in the minimal system could be contributed by one of the three Cdc42 GAPs (Fig. 7, B, D, F, and G). Nonetheless, the Cdc42 GAP domain alone, derived either from Rga3 or from Rga4, does not restore the monopolar outgrowth of germinating rga6∆ spores. Despite the specific function of Rga6 in promoting monopolar spore outgrowth, how the bipolar outgrowth arises in rga6∆ spores is unclear. Whether the absence of Rga6 affects the mechanical feedback between the spore wall and cell polarity remains to be further tested.
In sum, we propose that the lipid composition of the spore plasma membrane may create spatial cues to specify the characteristic cortical localization of Rga6. As a consequence, Cdc42-GTP is confined to the outgrowth zone to sustain monopolar outgrowth during germination.
Materials and methods
Yeast genetics and spore preparation
Table S1 lists the strains used in this study. The strains were created either by the random spore digestion method or by tetra-dissection analysis. Gene deletion and tagging were performed by the PCR-based method using the pFA6a plasmids, and yeast transformation was carried out by the lithium acetate method. To obtain spores, strains carrying two opposite mating types were crossed and cultured on malt extract (ME) medium plates for 3 d, and the crossed cells were then incubated in 1% snailase (www.sangon.com; A600870) solution at room temperature for 12 h to remove vegetative cells. Spores were washed with ddwater 3 times. For microscopic and biochemical analyses, the cleaned spores were cultured in yeast extract media containing five supplements (YE5S): adenine, leucine, uracil, histidine, and lysine (0.225 g/liter each; www.formedium.com).
Molecular cloning
The plasmids used in this study are listed in Table S2. The restriction enzymes used for molecular cloning were purchased from NEB (www.neb.com). For generating full-length rga6 and other rga6 variants, corresponding rga6 inserts obtained by PCR were digested with BglII and NotI and ligated into a pJK148 vector with a T4 DNA ligase (www.neb.com). The ClonExpress II One Step Cloning Kit (www.vazymebiotech.com) was used to construct the chimera plasmids: pJK148-ase1p-rga3gap-gfp, pJK148-ase1p-rga4gap-gfp, pJK148-ase1p-rga3gap-rga6∆N-gfp, and pJK148-ase1p-rga4gap-rga6∆N -gfp.
Calcofluor-white staining of spores
Spores were collected and incubated in 100 μg/ml Calcofluor-white solution (www.sigmaaldrich.cn; 18909) at room temperature for 30 min. After washing (3 times, centrifugation at 1,000 rpm for 2 min), stained spores were cultured in Calcofluor-white-free YE5S medium at 30°C for the indicated times. Since Calcofluor-white constitutively binds the spore wall, newly generated regions on the cell wall are not marked by Calcofluor-white.
Lectin staining
Cells were collected and incubated in 5 μg/ml TRITC/FITC-Lectin (www.sigmaaldrich.cn; L5264, L9381) at room temperature for 30 min. After washing (3 times, centrifugation at 1,000 rpm for 2 min), the cells were cultured in YE5S medium at 30°C for the indicated times.
Microscopy and data analysis
For microscopic observation, spores or exponentially growing vegetative cells were sandwiched between a 3% agarose YE5S pad and a coverslip. A PerkinElmer Ultraview Spinning Disk confocal microscope equipped with a Nikon Apochomat TIRF 100× 1.49NA objective and a Hamamatasu C9100-23B EMCCD camera was used to acquire images. The images were acquired at room temperature by Volocity (PerkinElmer). For time-lapse microscopy, images were acquired every 1 or 5 min. For stack images, 11 planes with 0.5 μm spacing were acquired. To analyze CRIB-GFP/3GFP dynamics, stack images containing 5 planes with 0.5 μm spacing were acquired every 1 min. ImageJ 1.5 (imagej.nih.gov) and MetaMorph 7.7 (www.moleculardevices.com) were used to analyze the microscopic data. Plot graphs were created with KaleidaGraph 4.5 (www.synergy.com).
Statistical analysis
Statistical analysis was performed with KaleidaGraph 4.5, and normality test was performed with Origin (www.originlab.com; OriginPro). For the data that are normally distributed, One-way ANOVA with Tukey HSD post hoc test was used; for the data that are not normally distributed, the Wilcoxon-Mann-Whitney Rank Sum test was used.
Analysis of protein expression
To assess protein expression, exponential vegetative cells or germinating spores (cultured for 6 or 8 h in YE5S medium) were collected, and cell lysates were prepared by the TCA (trichloroacetic acid) method. Specifically, the cells were collected and washed one time with 5 ml of sterilized water. The collected cells/spores were resuspended in 50 μl 20% TCA (www.sinopharm.com), followed by the addition of 200-μl glass beads (www.biospec.com; 11079105). Cells were then disrupted using the mixer mill Retsch MM400 (www.retsch.com). After separating samples from glass beads by pipetting, 50 μl 20% TCA was added to cell lysates, and samples were vortexed before an additional 400 μl 5% TCA was added.
The samples were centrifuged at 13,000 rpm for 10 min at 4°C to remove the supernatant. Finally, the pellet was resuspended in 175 μl 1X SDS sample buffer and 25 μl 1.5 M Tris (pH = 8.0), followed by boiling at 100°C for 5 min. Protein samples were analyzed by SDS‒PAGE and Western blotting with PVDF membranes (Millipore; www.merckmillopore.com; HATF00010) and antibodies. The antibodies used in this study are as follows: anti-GFP (www.rockland-inc.com; 600-101-215; dilution factor, 1:3,000), anti-tdTomato (www.genscript.com.cn; Home-made; dilution factor, 1:3,000); anti-mNeonGreen (www.genscript.com.cn; Home-made; dilution factor, 1:3,000), anti-tubulin (www.bioacademia.co.jp; 63-160; dilution factor, 1:10,000), and anti-actin (www.abcam.cn; ab8224; dilution factor, 1:2,000). The secondary antibodies used in this study were as follows: goat anti-mouse-HP Conjugate (www.abclonal.com; AS003; dilution factor, 1:10,000) and rabbit anti-goat-HP-conjugated (www.abclonal.com; AS029; dilution factor, 1:10,000). ECL substrate (www.biotanon.com; 180-501) was used in the Western blotting assays, and detection was performed with a chemiluminescence imaging system (www.clinx.cn; Chemiscope 6000). To quantify the Western-blot results, we used tubulin or actin as reference and calculated the ratio of the band intensity.
Liposome reconstitution assays
To prepare small unilamellar vesicles (SUVs), we used a lipid composition of phosphatidylcholine (PC) alone, or PC plus 1 or 10% phosphatidylinositol-bis-4,5-phosphate (PI[4,5]P2; www.avantilipids.com). Briefly, lipids were mixed in chloroform solvent (100 µl 10 mg/ml PC for PC alone; 90 µl 10 mg/ml PC plus 10 µl 1 mg/ml PI[4,5]P2 for 1% PI[4,5]P2 , 47 µl 10 mg/ml PC plus 53 µl 1 mg/ml PI[4,5]P2 for 10% PI[4,5]P2), dried under a stream of nitrogen for 5 min, and dissolved with 1 ml liposome buffer (20 mM Tris-HCl pH 8.0, 300 mM KCl, and 1 mM MgCl2) to hydrate for 30 min at 37°C. The lipid mixtures were then sonicated for 5 min to allow the formation of SUVs. Afterwards, 75 µl SUVs were mixed with 0.002 g silica microsphere beads (3.17 μm in a mean diameter; www.bangslabs.com; SSD5001), and then the mixture was rotated gently for 1 h at room temperature. After rotation, excess SUVs were removed by centrifugation for 1 min at 900 g, followed by three washes with wash buffer (50 mM Tris-HCl pH 8.0, and 100 mM KCl). To test the binding of proteins to the liposome-coated microspheres, 25 µl protein samples at the indicated concentration were added to 75 µl microsphere solution (100 mM KCl, 50 mM Tris-HCl pH 8.0, 0.1% BSA and 1 mM DTT), and the mixture was incubated for 60–90 min at room temperature. After the incubation, the protein-containing mixture was then mixed by pipetting, and 10 µl of the mixture was added to a glass profusion chamber for imaging with a spinning-disk confocal microscope.
Generation of the temperature-sensitive mutant strain its3-1
The temperature-sensitive mutant strain its3-1 (G281D), reported previously (Zhang et al., 2000), was generated by the CRISPR/Cas9 method (Zhang et al., 2018). The sgRNA target sequence is 5′-ATGCTTACTGGAATTCGCGT-3′. The donor DNA (254 bp) carrying the intended mutation site (G281D) was synthesized by the company General Biol (www.generalbiol.com).
Transmission electron microscopy
WT and rga6Δ spores that had been cultured in liquid YE5S medium for 3 h were harvested and washed with distilled water. The collected spores were fixed with 1% glutaraldehyde (www.emsdiasum.com) and 4% KMnO4 (www.sinopharm.com). After fixation, the samples were dehydrated with a series of graded concentrations of ethanol (50, 70, 90, and 100%). The dehydrated samples were then embedded in Spurr’s resins (www.emsdiasum.com). Thin sections of 70 nm were prepared using an Ultramicrotome Leica EM UC7 (www.leica-microsystems.com) and were examined using a Hitachi HT7700 transmission electron microscope equipped with an AMT Model XR81-B-M1-FX camera.
Online supplemental material
Fig. S1 shows the reliability of employing the Calcofluor-white staining to assess spore growth during germination; the result of employing lectin-staining to assess spore growth is also shown; representative long time-lapse light microscopic data illustrating the monopolar and bipolar growth patterns of WT, rga3∆, rga4∆, and rga6∆ spores and the plots of spore aspect ratio and area against time are shown at the bottom of the figure. Fig. S2 shows the results of assessing effects of the absence of Rga6 on the growth polarity of vegetative cells and on the end extension speed of vegetative cells; the figure also shows the cortical localization of Rga6-GFP once Rga6-GFP is overexpressed from the cam1 promoter. Fig. S3 shows the results of assessing the polarity patterns of WT, opy1∆, and Opy1-GFP-overexpressing spores; the effects of the impairment of Its3, a protein responsible for generating phosphatidylinositol 4,5-bisphosphate, on the cortical localization of Opy1-2mNeonGreen and Rga6-tdTomato and on spore outgrowth are also shown. Fig. S4 shows the following results: (1) the comparison of the expression level of Rga6-GFP from its own promoter and the ase1 promoter, (2) the confirmation of the expression of the Rga6 truncated proteins and the chimeric proteins indicated in Fig. 7, and (3) the growth polarity of the spores indicated in Fig. 7. Fig. S5 shows the following results: (1) the effect of Rga3-overexpression or Rga4-overexpression on the outgrowth polarity of rga6∆ spores, and (2) the effect of the absence of Rga6 or both Rga6 and Rga4 on the outgrowth polarity of spores. Table S1 lists the strains used in the present work. Table S2 lists the plasmids used in the present work.
Supplementary Material
shows yeast strains
shows plasmids
is the source file for Fig. 3.
is the source file for Fig. 6.
is the source file for Fig. S2.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We would like to thank Drs. Phong Tran (UPENN) and Sophie Martin (University of Lausanne) and NBRP (Japan) for providing strains. We would like to also thank members in the Fu laboratory for insightful discussions.
This work is supported by grants from the National Key Research and Development Program of China (2017YFA0503600), the National Natural Science Foundation of China (32070707, 31871350, and 31621002), and the Fundamental Research Funds for the Central Universities (WK9110000151).
The authors declare no competing financial interests.
Author contributions: C. Fu conceived the project. C. Fu, X. Ma, and X. Yao supervised the project. W. Wei, B. Zheng, S. Zhang, D. Wu, S. Zheng, D. Wang, and Y. Chu performed the experiments, W. Wei and B. Zheng analyzed the data. W. Wei, B. Zheng, X. Liu, X. Yao, and C. Fu wrote the paper. All authors made comments.
References
- Adams, A.E., Johnson D.I., Longnecker R.M., Sloat B.F., and Pringle J.R.. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131–142. 10.1083/jcb.111.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu, F.O., Vincenzetti V., Vavylonis D., Wyss R., Vogel H., and Martin S.G.. 2015. Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 13:e1002097. 10.1371/journal.pbio.1002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi, D., Julien J.D., Romao M., Seddiki R., Piel M., Boudaoud A., and Minc N.. 2014. Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Dev. Cell. 28:534–546. 10.1016/j.devcel.2014.01.023 [DOI] [PubMed] [Google Scholar]
- Brand, A., and Gow N.A.. 2009. Mechanisms of hypha orientation of fungi. Curr. Opin. Microbiol. 12:350–357. 10.1016/j.mib.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, A.A., Jentzsch M.S., Oakes P.W., Occhipinti P., and Gladfelter A.S.. 2016. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 213:23–32. 10.1083/jcb.201512029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty, A.C., Perrinjaquet N., Petit A., Jaquenoud M., Segall J.E., Hofmann K., Zwahlen C., and Peter M.. 2002. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 21:1565–1576. 10.1093/emboj/21.7.1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, E.C., Barr M., Wang Y., Jung V., Xu H.P., and Wigler M.H.. 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 79:131–141. 10.1016/0092-8674(94)90406-5 [DOI] [PubMed] [Google Scholar]
- Chang, F., and Martin S.G.. 2009. Shaping fission yeast with microtubules. Cold Spring Harb. Perspect. Biol. 1:a001347. 10.1101/cshperspect.a001347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Fingerhut J.M., and Yamashita Y.M.. 2016. The ins(ide) and outs(ide) of asymmetric stem cell division. Curr. Opin. Cell Biol. 43:1–6. 10.1016/j.ceb.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, J.G., Balasubramanian M.K., and Lew D.J.. 2017. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 33:77–101. 10.1146/annurev-cellbio-100616-060856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, P.M., Trillo Y., Ametzazurra A., and Perez P.. 2003. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol. Biol. Cell. 14:313–323. 10.1091/mbc.e02-07-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Drake T., Wiley D.J., Buchwald P., Vavylonis D., and Verde F.. 2012. Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science. 337:239–243. 10.1126/science.1218377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., and Verde F.. 2013. Role of Cdc42 dynamics in the control of fission yeast cell polarization. Biochem. Soc. Trans. 41:1745–1749. 10.1042/BST20130241 [DOI] [PubMed] [Google Scholar]
- Davi, V., Tanimoto H., Ershov D., Haupt A., De Belly H., Le Borgne R., Couturier E., Boudaoud A., and Minc N.. 2018. Mechanosensation dynamically coordinates polar growth and cell wall assembly to promote cell survival. Dev. Cel. 45:170–182.e7. 10.1016/j.devcel.2018.03.022 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S. 2004. Cdc42–the centre of polarity. J. Cell Sci. 117:1291–1300. 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall A.. 2002. Rho GTPases in cell biology. Nature. 420:629–635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Gallo Castro, D., and Martin S.G.. 2018. Differential GAP requirement for Cdc42-GTP polarization during proliferation and sexual reproduction. J. Cell Biol. 217:4215–4229. 10.1083/jcb.201806016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova, J.L., Jong S., Camp J.T., and Fehon R.G.. 2000. Functional analysis of Cdc42 in actin filament assembly, epithelial morphogenesis, and cell signaling during Drosophila development. Dev. Biol. 221:181–194. 10.1006/dbio.2000.9671 [DOI] [PubMed] [Google Scholar]
- Gerganova, V., Lamas I., Rutkowski D.M., Vjestica A., Castro D.G., Vincenzetti V., Vavylonis D., and Martin S.G.. 2021. Cell patterning by secretion-induced plasma membrane flows. Sci. Adv. 7:eabg6718. 10.1126/sciadv.abg6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose, D., Elston T., and Lew D.. 2022. Orientation of cell polarity by chemical gradients. Annu. Rev. Biophys. 51:431–451. 10.1146/annurev-biophys-110821-071250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J.M., Lustig R.J., Berlin A., and Chang F.. 2001. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11:836–845. 10.1016/s0960-9822(01)00235-4 [DOI] [PubMed] [Google Scholar]
- Goryachev, A.B., and Pokhilko A.V.. 2008. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582:1437–1443. 10.1016/j.febslet.2008.03.029 [DOI] [PubMed] [Google Scholar]
- Hatanaka, M., and Shimoda C.. 2001. The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast. 18:207–217. [DOI] [PubMed] [Google Scholar]
- Haupt, A., Ershov D., and Minc N.. 2018. A positive feedback between growth and polarity provides directional persistency and flexibility to the process of tip growth. Curr. Biol. 28:3342–3351.e3. 10.1016/j.cub.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Horvitz, H.R., and Herskowitz I.. 1992. Mechanisms of asymmetric cell division: Two bs or not two Bs, that is the question. Cell. 68:237–255. 10.1016/0092-8674(92)90468-r [DOI] [PubMed] [Google Scholar]
- Howell, A.S., Jin M., Wu C.F., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 149:322–333. 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui, J.E., Gladfelter A.S., and Lew D.J.. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062–1070. 10.1038/ncb1068 [DOI] [PubMed] [Google Scholar]
- Johnson, D.I., and Pringle J.R.. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111:143–152. 10.1083/jcb.111.1.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, A.J., and Hunter C.P.. 2001. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11:474–481. 10.1016/s0960-9822(01)00141-5 [DOI] [PubMed] [Google Scholar]
- Kelly, F.D., and Nurse P.. 2011. Spatial control of Cdc42 activation determines cell width in fission yeast. Mol. Biol. Cell. 22:3801–3811. 10.1091/mbc.E11-01-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkoris, K., Gallo Castro D., and Martin S.G.. 2014. The Tea4-PP1 landmark promotes local growth by dual Cdc42 GEF recruitment and GAP exclusion. J. Cell Sci. 127:2005–2016. 10.1242/jcs.142174 [DOI] [PubMed] [Google Scholar]
- Kono, K., Matsunaga R., Hirata A., Suzuki G., Abe M., and Ohya Y.. 2005. Involvement of actin and polarisome in morphological change during spore germination of Saccharomyces cerevisiae. Yeast. 22:129–139. 10.1002/yea.1205 [DOI] [PubMed] [Google Scholar]
- Kozubowski, L., Saito K., Johnson J.M., Howell A.S., Zyla T.R., and Lew D.J.. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18:1719–1726. 10.1016/j.cub.2008.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas, I., Merlini L., Vjestica A., Vincenzetti V., and Martin S.G.. 2020. Optogenetics reveals Cdc42 local activation by scaffold-mediated positive feedback and Ras GTPase. PLoS Biol. 18:e3000600. 10.1371/journal.pbio.3000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., and Pawson T.. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. 10.1038/35019582 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte, F., and Mostov K.. 2008. Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20:227–234. 10.1016/j.ceb.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Martin, S.G. 2015. Spontaneous cell polarization: Feedback control of Cdc42 GTPase breaks cellular symmetry. Bioessays. 37:1193–1201. 10.1002/bies.201500077 [DOI] [PubMed] [Google Scholar]
- Merlini, L., Dudin O., and Martin S.G.. 2013. Mate and fuse: How yeast cells do it. Open Biol. 3:130008. 10.1098/rsob.130008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, S., Leda M., Hanna J., Savage N.S., Bi E., and Goryachev A.B.. 2013. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell. 26:148–161. 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R., Ter Beek A., Vischer N.O., Smelt J.P., Brul S., and Manders E.M.. 2013. Live cell imaging of germination and outgrowth of individual bacillus subtilis spores; the effect of heat stress quantitatively analyzed with SporeTracker. PLoS One. 8:e58972. 10.1371/journal.pone.0058972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino, M.R., Nunez I., Chen C., Das M.E., Wiley D.J., D'Urso G., Buchwald P., Vavylonis D., and Verde F.. 2021. Cdc42 GTPase activating proteins (GAPs) regulate generational inheritance of cell polarity and cell shape in fission yeast. Mol. Biol. Cell. 32:ar14. 10.1091/mbc.E20-10-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante, S., and Labbe S.. 2019. Spore germination requires ferrichome biosynthesis and the siderophore transporter Str1 in Schizosaccharomyces pombe. Genetics. 211:893–911. 10.1534/genetics.118.301843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante, S., Normant V., Ramos-Torres K.M., and Labbe S.. 2017. Cell-surface copper transporters and superoxide dismutase 1 are essential for outgrowth during fungal spore germination. J. Biol. Chem. 292:11896–11914. 10.1074/jbc.M117.794677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Guarinos, M.T., Martin-Garcia R., Villar-Tajadura M.A., Estravis M., Coll P.M., and Perez P.. 2016. Rga6 is a fission yeast Rho GAP involved in Cdc42 regulation of polarized growth. Mol. Biol. Cell. 27:1524–1535. 10.1091/mbc.E15-12-0818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers, F., and Jan Y.N.. 2004. Asymmetric cell division. Curr. Opin. Cell Biol. 16:195–205. 10.1016/j.ceb.2004.02.010 [DOI] [PubMed] [Google Scholar]
- Snider, C.E., Willet A.H., Brown H.T., Chen J.S., Evers J.M., and Gould K.L.. 2020. Fission yeast Opy1 is an endogenous PI(4, 5)P2 sensor that binds to the phosphatidylinositol 4-phosphate 5-kinase Its3. J. Cell Sci. 133:jcs247973. 10.1242/jcs.247973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, C.E., Willet A.H., Brown H.T., and Gould K.L.. 2018. Analysis of the contribution of phosphoinositides to medial septation in fission yeast highlights the importance of PI(4, 5)P2 for medial contractile ring anchoring. Mol. Biol. Cell. 29:2148–2155. 10.1091/mbc.E18-03-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheraly, S., Ershov D., Dmitrieff S., and Minc N.. 2020. An image analysis method to survey the dynamics of polar protein abundance in the regulation of tip growth. J. Cell Sci. 133:jcs252064. 10.1242/jcs.252064 [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and Hirata A.. 1982. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J. Cell Sci. 56:263–279. 10.1242/jcs.56.1.263 [DOI] [PubMed] [Google Scholar]
- Tatebe, H., Nakano K., Maximo R., and Shiozaki K.. 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18:322–330. 10.1016/j.cub.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, Y.D., Leda M., Goryachev A.B., and Sawin K.E.. 2018. Local and global Cdc42 guanine nucleotide exchange factors for fission yeast cell polarity are coordinated by microtubules and the Tea1-Tea4-Pom1 axis. J. Cell Sci. 131:jcs216580. 10.1242/jcs.216580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z., Gao X.D., Howell A.S., Bose I., Lew D.J., and Bi E.. 2007. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 179:1375–1384. 10.1083/jcb.200705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, S., Fleming A., Wellman C.H., and Beerling D.J.. 2011. Evolutionary development of the plant and spore wall. AoB Plants. 2011:plr027. 10.1093/aobpla/plr027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, B., Kuo C.C., Wu C.F., Zyla T.R., and Lew D.J.. 2015. Polarity establishment requires localized activation of Cdc42. J. Cell Biol. 211:19–26. 10.1083/jcb.201506108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, B., and Lew D.J.. 2019. Polarity establishment by Cdc42: Key roles for positive feedback and differential mobility. Small GTPases. 10:130–137. 10.1080/21541248.2016.1275370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.F., and Lew D.J.. 2013. Beyond symmetry-breaking: Competition and negative feedback in GTPase regulation. Trends Cell Biol. 23:476–483. 10.1016/j.tcb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.R., He J.B., Wang Y.Z., and Du L.L.. 2018. A cloning-free method for CRISPR/Cas9-Mediated genome editing in fission yeast. G3. 8:2067–2077. 10.1534/g3.118.200164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Sugiura R., Lu Y., Asami M., Maeda T., Itoh T., Takenawa T., Shuntoh H., and Kuno T.. 2000. Phosphatidylinositol 4-phosphate 5-kinase Its3 and calcineurin Ppb1 coordinately regulate cytokinesis in fission yeast. J. Biol. Chem. 275:35600–35606. 10.1074/jbc.M005575200 [DOI] [PubMed] [Google Scholar]
- Zheng, S., Zheng B., Liu Z., Ma X., Liu X., Yao X., Wei W., and Fu C.. 2022. The Cdc42 GTPase-activating protein Rga6 promotes the cortical localization of septin. J. Cell Sci. 135:jcs259228. 10.1242/jcs.259228 [DOI] [PubMed] [Google Scholar]
- Zhou, R., Guo Z., Watson C., Chen E., Kong R., Wang W., and Yao X.. 2003. Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell. 14:1097–1108. 10.1091/mbc.e02-07-0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows yeast strains
shows plasmids
is the source file for Fig. 3.
is the source file for Fig. 6.
is the source file for Fig. S2.
is the source file for Fig. S4.
is the source file for Fig. S5.