Abstract
Maintaining nutrient and energy homeostasis is crucial for the survival and function of cells and organisms in response to environmental stress. Cells have evolved a stress-induced catabolic pathway, named autophagy, to adapt to stress conditions such as starvation. During autophagy, damaged or non-essential cellular structures are broken down in lysosomes, and the resulting metabolites are reused for core biosynthetic processes or energy production. Recent studies have revealed that autophagy can target and degrade different types of nutrient storages and produce a variety of metabolites and fuels, including amino acids, nucleotides, lipids and carbohydrates. Here, we will focus on how autophagy functions to balance cellular nutrient and energy demand and supply, specifically, how energy deprivation switches on autophagic catabolism, how autophagy halts anabolism by degrading the protein synthesis machinery, and how bulk and selective autophagy-derived metabolites recycle and feed into a variety of bioenergetic and anabolic pathways during stress conditions. The recent new knowledge and progress in these areas provide a better understanding on resource mobilization and reallocation to sustain essential metabolic and anabolic activities under unfavorable conditions.
Introduction
In response to nutritional and energy crisis, cells need to make timely decisions to balance demand and supply, and to allocate limited resource to maintain viability and essential functions, at the expense of non-essential cellular structures and activities. Autophagy, derived from ancient Greek for “self-eating”, is a lysosomal catabolic pathway evolved for such function in eukaryotes from yeast cells to humans 1. Compared to another cellular degradation pathway, the ubiquitin-proteasomal pathway, which degrades short-lived proteins and requires substrate proteins to be unfolded to feed into proteasomes, the substrates for autophagy are more versatile, including long-lived proteins, non-protein structures, or protein aggregates and organelles that are too large or unable to unfold for proteasomal degradation. Autophagy occurs at a low basal level under normal conditions, and its activity can be induced by a variety of stressors, such as starvation, ER stress, hypoxia, pathogen infection and exercise 1–3. In response to nutrient deprivation, autophagy, but not the ubiquitin-proteasomal system, is the key mechanism to maintain energy production and cell survival 4.
The final destination of autophagy cargos is the lysosomal lumen wherein acid hydrolases degrade the delivered lysosomal contents to generate amino acids, fatty acids, nucleotides and sugars, which are recycled back to the cytoplasm and reused. Based on the mechanistic differences in cargo transportation and delivery to lysosomes, autophagy is categorized into three types: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). Macroautophagy (hereafter referred to as autophagy) involves the formation of classical double-membrane autophagosomes, which enwrap and transport cytosolic cargos to lysosomes for degradation. The nascent autophagosomal membrane is termed a phagophore. Autophagosomes are not present in microautophagy and CMA. Instead, microautophagy refers to the direct invagination of lysosomal membrane and subsequent degradation of incorporated cargos, which may resemble the formation of endosomal MVBs (multivesicular bodies) and require the ESCRT machinery (endosomal sorting complexes required for transport) 5. CMA targets a specific subset of substrate proteins containing a KFERQ pentapeptide recognition motif, which are transported into lysosomes through the lysosome membrane receptor LAMP2A with the facilitation of a chaperone HSC70 6. All three autophagy pathways play important roles in regulating nutrient mobilization and energy homeostasis during stress times. A variety of key nutrients, including proteins, nucleic acids, carbohydrates and lipids, can be degraded by the three types of autophagy into the constituent amino acids, nucleotides, glucose and fatty acids for recycling and reuse. These catabolic pathways crosstalk with one another to provide building blocks for biosynthetic activities and feed into glycolysis and mitochondria for energy production (Figure 1). In this review, we will focus on the role and mechanism of autophagy-mediated mobilization of different types of nutrients under stress conditions (Table 1).
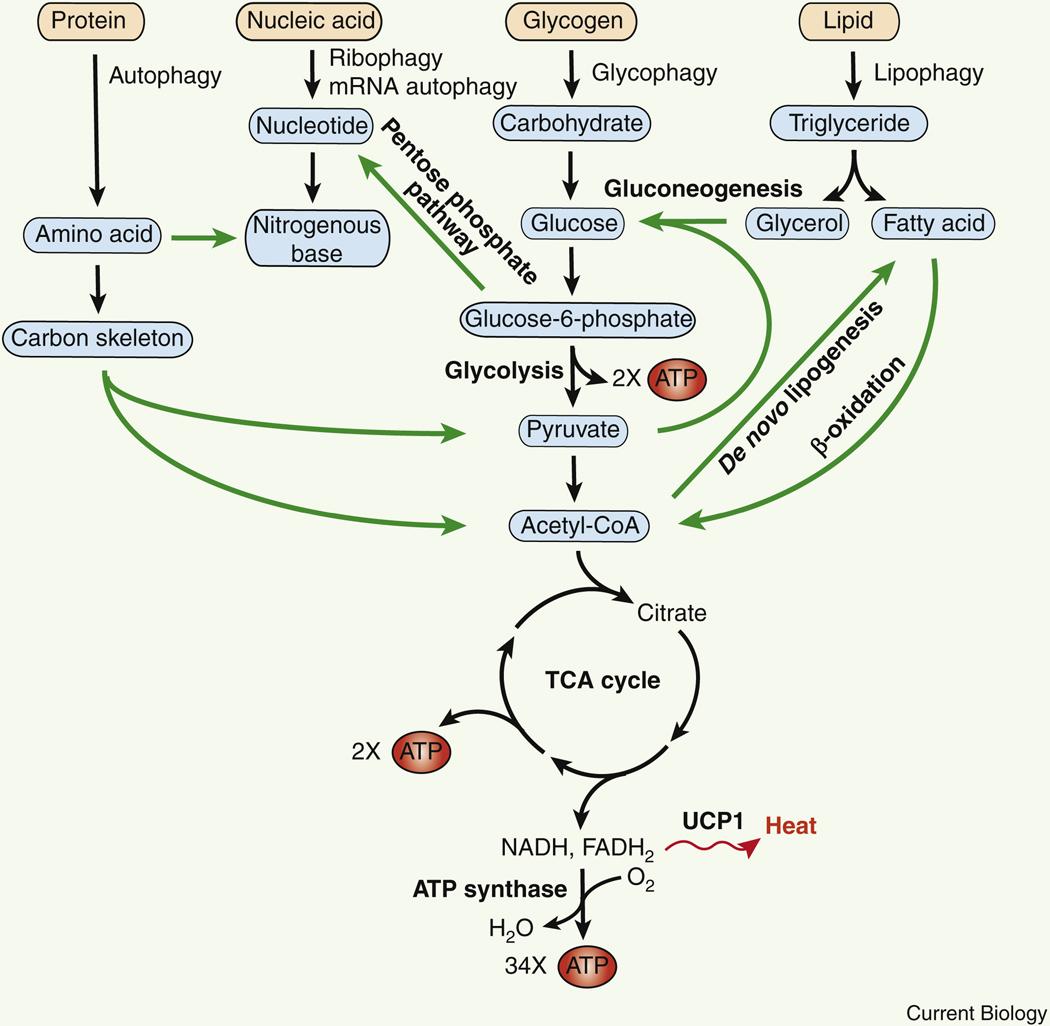
Figure 1. Schematic illustration of autophagic metabolism of proteins, nucleic acids, carbohydrates and lipids, and their crosstalk in energy production.
The amount of energy produced per glucose molecule (or equivalent precusors) is: 2 ATPs from glycolysis, 2 ATPs from the tricarboxylic acid (TCA) cycle, and a maximum of 34 ATPs by the ATP synthase of the mitochondrial electron transport chain. Autophagy-generated energy is also dissipated as heat via mitochondrial UCP1 (Uncoupling protein 1). Communication among different nutrient metabolic pathways is highlighted by green arrows.
Table 1.
Regulatory machinery and physiological outcomes of abnormal autophagy-regulated nutrient mobilization.
| Type of recycled nutrient | Regulatory machinery | Organism/tissue | Disease-relevant phenotypes if defective | References |
|---|---|---|---|---|
| Energy production and consumption | Atg5 | Mouse/brown adipocyte, liver | Reduced β-oxidation and thermogenesis | 36,35 |
| Atg7 | Mouse/POMC neuron | Reduced energy expenditure, hyperglycemia | 37,38 | |
| Atg5, Atg7, Atg12 | Mouse/white and beige adipocytes | Adipocyte “browning”, lean body mass, improved glucose tolerance and insulin sensitivity | 39–41,42 | |
| LAMP2A-mediated CMA | Mouse/liver | HFD-induced hepatosteatosis, lean body mass, enhanced energy expenditure | 43 | |
| Protein → amino acids | Atg1, Atg7 | Yeast | Unable to restore intracellular amino acid levels and survive during starvation | 58,59 |
| Atg5, Atg7 | Mouse/whole body | Neonatal lethality, energy deprivation | 61,62 | |
| Atg7 | Mouse/liver | Low circulating amino acids and glucose | 64,65 | |
| Atg5, Atg7 | Pancreatic stellate cell | Reduced growth of pancreatic ductal adenocarcinoma | 66 | |
| Ribophagy → amino acid + nucleotide | The ubiquitin protease complex Ubp3-Bre5-Cdc48-Ufd3 | Yeast | Uncontrolled ribosome abundance | 68,69 |
| NUFIP1, VPS34, BECN1, ATG7 | Mammalian cells | Reduced cellular nucleotide pools and cell survival during starvation | 72,74,75 | |
| mRNA autophagy → nucleotide nucleotide nucleotide | Atg2, Sorting nexin Atg24-Atg20 and Atg24-Snx41 | Yeast | Elevation of mRNAs encoding amino acid biosynthesis and ribosomal proteins | 70,76 |
| T2 ribonuclease RNST-2 | C. elegans | Defective embryonic and larval development, reduced lifespan | 73 | |
| Lipid → fatty acid | Atg5, Atg7 | Mammal cells | Increased TG accumulation | 80,81 |
| Atg7 | Mouse/AgRP neuron | Reduced food intake and lean phenotype | 81 | |
| LAMP2A-mediated CMA | Mammalian cells | LD accumulation | 91 | |
| Atg39 and ESCRT- mediated microlipophagy | Yeast, hepatocyte | LD accumulation upon ER stress, defects in mitochondria and ER morphology and cell growth upon lipid stress | 92,93,95 | |
| Glycogen→glucose | GAA | Mouse/muscle and brown adipose tissue, drosophila | Pompe disease | 100–102 |
| STBD1 | Mouse/liver | Ameliorated lysosomal glycogen storage | 105,106 |
Autophagy activation: nutrient and energy sensing machinery
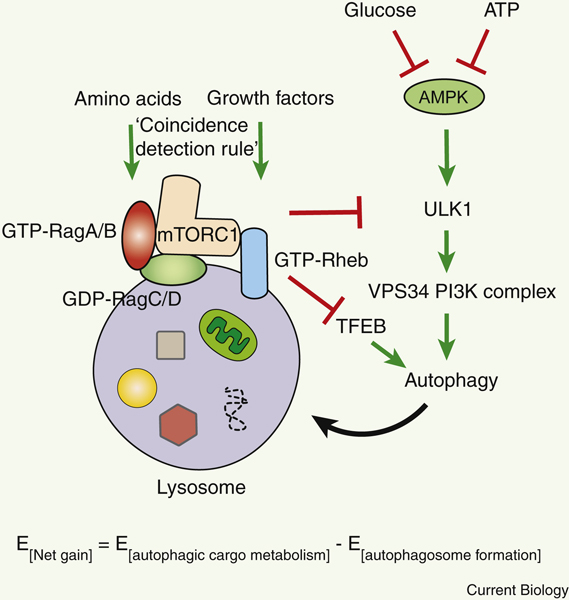
Execution of autophagy costs energy. Thus, the net energy gain from autophagy would be energy required for autophagosome formation subtracted from energy provided by autophagic cargo metabolism (Figure 2), although it is yet unclear how to quantitatively measure the energy expenditure and production during autophagy per time period. The energy needed to maintain the proton gradient and acidification in the lysosomal lumen is not considered in the formula due to the steady state of lysosomal existence. Induction of autophagy is orchestrated by more than 30 autophagy-related (ATG) genes, and originates from a variety of organelles that are in contact with the phagophore 7, including the ER (endoplasmic reticulum), mitochondria, recycling and late endosomes, the plasma membrane and lipid droplets (LDs), with ER subdomains and contact sites with other organelles proposed as the primary origin of autophagosomes thus far 8–11. Autophagosome biogenesis is initiated by the serine-threonine protein kinase Atg1 (yeast)/ULK1 (mammalian, Unc-51 like autophagy activating kinase 1), which phosphorylates the regulatory subunits of the downstream pro-autophagic VPS34 PI3K (class III phosphatidylinositol 3-kinase) complex, including BECN1, ATG14 and AMBRA1 12–16 (Figure 2). Phosphorylation of these subunits by ULK1 leads to activation of the lipid kinase VPS34 and production of phosphatidylinositol-3-phosphate (PI3P) at the phagophore. PI3P is a specific lipid molecule of autophagosome vesicles, and recruits additional PI3P effectors (PI3P-binding autophagy proteins), including Atg18/WIPIs and their binding partner ATG2, to facilitate autophagosome expansion. ATG2 functions in membrane tethering, and also directly transfers phospholipids between membranes via an N-terminal hydrophobic pocket that can bind multiple glycerophospholipids 17–19. Autophagosome membrane expansion also requires two ubiquitin-like conjugation systems, Atg8/LC3-PE (phosphatidylethanolamine) and the ATG12-ATG5-ATG16 complex. LC3 and ATG12 are the ubiquitin-like proteins in each system, and their conjugation requires a common E1-like enzyme, ATG7. Formation of the thioester linkage between the E1 ATG7 and its substrates (the ubiquitin-like proteins LC3 and ATG12) consumes energy (in the form of ATP hydrolysis). The subsequent E2-like enzyme for LC3 lipidation is ATG3, and for ATG12 conjugation is ATG10. The ATG12-ATG5-ATG16 complex promotes the lipidation of LC3 to PE and serves the role of an E3-like enzyme.
Figure 2. mTORC1-mediated inhibition and AMPK-mediated activation of autophagy via nutrient-, growth factor-, and energy-sensing.
Amino acids activate Rag GTPases, and growth factors activate the Rheb GTPase. Active Rag GTPases (GTP-bound RagA/B and GDP-bound RagC/D) and the active Rheb GTPase (GTP-bound Rheb) synergistically recruit and activate mTORC1 on the lysosomal membrane. Active mTORC1 phosphorylates and inhibits the autophagy-initiating kinase ULK1 and the master transcriptional regulator of autophagy TFEB. By contrast, AMPK senses low glucose and ATP levels, and activates ULK1 and the downstream VPS34 PI3K complex to activate autophagy. Autophagy delivers a variety of substrates, including proteins, organelles (such as ribosomes and associated RNAs), lipid droplets (LDs), glycogen and iron storages, to the lysosome for degradation, which promotes nutrient mobilization for biosynthesis and energy production in response to nutrient and energy depletion. E, energy.
Subsequent incorporation of LC3-PE onto the forming autophagosome drives membrane expansion and cargo recruitment. During the last step of autophagy, autophagosomes fuse with lysosomes (or vacuoles in yeast) and form autolysosomes for substrate degradation. As other membrane fusion events in the cell, autophagosome-lysosome fusion requires the function of the SNARE (Soluble N-ethylmaleimide-sensitive factor Attachment protein REceptor) protein complex as a zipper to enclose two target membranes, which overcomes the high energy barrier of membrane fusion. STX17 (Syntaxin 17) is the autophagosomal membrane Q-SNARE, which provides a glutamine (Q) to the SNARE complex, and VAMP7 and VAMP8 are lysosomal membrane R-SNAREs that provide an arginine (R) to the SNARE complex. After digestion of autophagy substrates by lysosomal resident proteases, lipases and nucleotidases, a variety of lysosomal membrane nutrient transporters mediate the efflux of metabolites from the lysosomal lumen back to the cytosol, including amino acid transporters such as SLC38A9, LAAT-1 (also known as PQLC2), SNAT7 (also known as SLC38A7) and yeast Atg22, the sugar transporter SPIN, and ion channels such as MCOLN1 (also known as TRPML1) 20,21, some of which need the proton gradient for the efflux function 22. These transporters play an important role in the maintenance of metabolite storage and nutrient balance in lysosomes.
The metabolic kinases mTORC1 (mammalian target of rapamycin complex 1) and AMPK (AMP-activated protein kinase) are two main upstream regulators of autophagy (Figure 2). mTORC1 is the central negative regulator of autophagy that phosphorylates ULK1 at S757 and suppresses ULK1 activity. mTORC1 also negatively regulates autophagy at the transcriptional level, by phosphorylating the master transcription factor of autophagy and lysosomal genes, TFEB (transcription factor EB), at multiple sites. Phosphorylation of TFEB promotes its nuclear export and shuts off autophagy gene transcription 23,24. Activation of mTORC1 is mediated by small GTPases RagA/B, RagC/D, and Rheb, via a “coincidence detection” mechanism: mTORC1 is activated only when both growth factors (signals) and nutrients (building blocks) are present 21. This is achieved by recruitment of mTORC1 to the lysosomal membrane by GTP-bound RagA/B and GDP-bound RagC/D (activated by cytosolic amino acid sensors and lysosomal cholesterol sensors), and activation of mTORC1 on the lysosomal membrane by GTP-bound Rheb (activated by growth factors). Therefore, deprivation of either growth factors or nutrients fails to activate mTORC1 and is sufficient to switch on autophagy. In mice, although different organs show variable levels of autophagy capacity, autophagy is profoundly induced in most organs by 12–48 hours of nutrient deprivation, and only 2–4 hours of starvation in cell culture and single cell (such as yeast) systems 25–28. AMPK plays a central role in metabolic regulation and is the major positive regulator of autophagy. AMPK is activated by low cellular energy (an increased AMP/ATP ratio) or low glucose availability (such as a low fructose-1,6-bisphosphate level) 29,30 (Figure 2). AMPK further phosphorylates autophagy proteins including ULK1 and BECN1 31–33. Different from mTORC1-mediated phosphorylation, phosphorylation of ULK1 by AMPK occurs at S555, which activates ULK1 and autophagy 33,34. Together, deprivation of nutrients, growth factors, or energy robustly induces autophagy via inhibition of mTORC1 and/or activation of AMPK.
Regulation of bioenergetic balance by autophagy
Autophagy modulates both energy production and expenditure. Depletion of the autophagy protein ATG5 decreases hepatic β-oxidation induced by thyroid hormones 35, suggesting that autophagy positively regulates fatty acid β-oxidation in the liver. In brown adipose tissue, long-term cold stress also induces both autophagy and fatty acid β-oxidation 36, which is likely regulated centrally by anorexigenic POMC (proopiomelanocortin) neurons 37,38. In theory, the best energy return of autophagy is for all autophagy-derived metabolites to be converted to acetyl-CoA and used as substrates in aerobic respiration by mitochondrial ATP synthase (Figure 1). Yet it is not the case in reality. Aside from contributing substrate metabolites to ATP production and biosynthetic pathways, cold stress-induced autophagy also enhances thermogenesis in brown adipose tissues 36. Thermogenesis is a bioenergetic mechanism that homeotherms (warm-blooded animals), including birds and mammals, have evolved to maintain their body temperature and adapt to environmental changes, dissipating energy in the form of heat instead of ATP production. Thermogenesis is carried out by brown adipose tissues and beige adipose tissues that are rich in mitochondria, where protons flow into the mitochondrial matrix via Uncoupling protein 1 (UCP1), bypassing the ATP synthase. Accordingly, the three outcomes for autophagy-derived metabolites are: building blocks for biosynthetic pathways, ATP production, and thermogenesis (Figure 1).
The role of autophagy in controlling energy consumption is complex. Basal autophagy is required for white adipose tissue development 39–41 and autophagy activation promotes beige adipose tissue “whitening” by degrading mitochondria 42. Inhibition of autophagy specifically in beige adipocytes by Atg5 or Atg12 KO is beneficial for beige fat maintenance and protects mice from high-fat diet (HFD)-induced obesity and insulin resistance 42. In addition, although liver-specific inhibition of CMA causes hepatosteatosis after HFD feeding, such inhibition enhances, rather than decreases, hepatic energy expenditure and weight loss 43, which may be attributed to inhibited CMA degradation of metabolic enzymes, or compensatory upregulation of (macro)autophagy activity. Thus, on one hand, autophagy activation can remodel adipose tissue and liver, and render them less thermogenic with reduced energy expenditure; on the other hand, in response to cold stress autophagy enhances thermogenesis in mature brown adipose tissues 36, likely through improving mitochondrial quality control and providing metabolites and substrates. Future quantitative analyses are needed, preferentially in single cells or single populations of homogenous cells, to model in what scenarios autophagy may contribute the highest ATP yield, in what scenarios autophagy may be energetically futile, and in what other scenarios autophagy balances energy demand and supply. In addition, the roles of autophagy in the regulation of resting energy expenditure (or basal metabolic rate) versus activity-induced energy expenditure are unclear and also require further investigation.
Other physiological conditions that dramatically increase energy demand, such as exercise, also activate autophagy. During intense exercise, the metabolic rate in contracting muscle can surge to nearly 100-fold 44, and rapid ATP production is essential to sustain exercise intensity and duration. Recent studies in both rodents and humans have demonstrated that aerobic exercise, as short as 30 min, potently activates autophagy in skeletal muscle 3,45–47. A number of mechanisms have been proposed on how autophagy is activated in muscle during contraction, including energetic stress 48,49, oxidative stress 50, and transcriptional upregulation of autophagy genes by increased intracellular Ca2+ 51. Prolonged endurance exercise results in net breakdown of muscle proteins (protein degradation rate > protein synthesis rate) and increased oxidation of amino acids, especially branched-chain amino acids (BCAAs), as energy source 52. Such protein degradation even occurs during low-intensity exercise in McArdle’s disease patients, who have defective glycogen phosphorylase and limited glycogen utilization as fuels in the muscle, evidenced by a high accumulation of non-metabolized amino acids, such as lysine, threonine and tyrosine that cannot be trans-aminated 53. Thus, these findings suggest that exercise-induced autophagic breakdown is important to combat the negative energy balance during muscle contractions.
Protein catabolism to replenish amino acids, glucose and energy
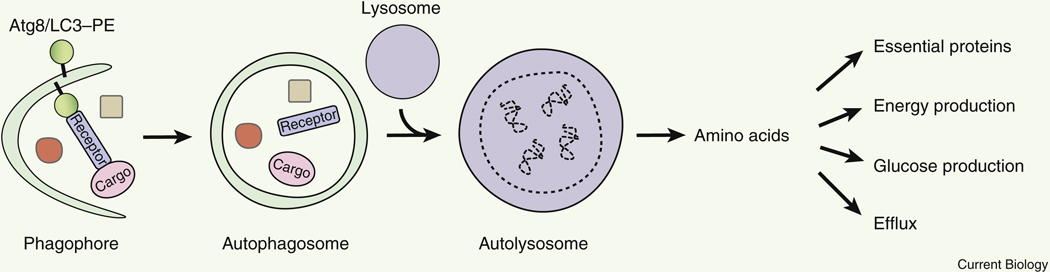
Autophagy mediates both non-selective degradation of bulk proteins, and selective degradation of cargos via a receptor that binds both the autophagosome protein Atg8/LC3 and the cargo proteins (Figure 3, Table 1). Although an autophagosome in mammalian cells is small (0.5–2 μm in diameter) 54 and only occupies up to 0.1% of the cell volume, because the half-life of autophagosomes is short (<10 min), the degradative capacity of autophagy is high 55. In the rat liver, the protein degradation rate is approximately 1.5% of total proteins per hour under normal conditions, compared to 4–5% of total proteins per hour under nutrient starvation conditions; the 3-fold increase in protein degradation is largely attributed to autophagy activation by starvation. In addition, organelle turnover is also mediated by autophagy. For example, mitochondria in hepatocytes have an average half-life of only 2–4 days 56,57, and are turned over by mitophagy (selective autophagic degradation of mitochondria).
Figure 3.
Amino acids produced by non-selective bulk autophagy, or selective autophagy via a receptor that binds both Atg8/LC3 and the cargo, fulfill diverse functions of the cell.
Supplying amino acids and ATPs is an important function of autophagy for cells to survive starvation conditions. As discussed above, autophagy is induced when the cellular amino acid level drops to the threshold that is unable to activate mTORC1, or yeast mTOR homologs Tor1 and Tor2. Indeed, the initial screen for ATG genes was carried out in yeast for loss of viability in response to nitrogen starvation (cultured in a minimal synthetic medium without amino acids and nitrogen salt) 58. Approximately half of autophagy-deficient Atg1-null yeast cells lose viability after cultured for 3 days in the nitrogen starvation medium, whereas wild-type (WT) cells maintain nearly 100% viability after 5 days of cell culture. Furthermore, unlike WT cells, autophagy-deficient Atg7-null yeast cells are unable to restore intracellular amino acid levels after 3 hours of nitrogen starvation 59. Autophagy-derived amino acids are used for protein biosynthesis, traced by 14C-isotope labeled valine, especially for proteins crucial for yeast survival through amino acid starvation, such as Arg1 (argininosuccinate synthetase for arginine synthesis) and Hsp26 (heat shock protein of 26 kD with chaperone activity facilitating protein folding) 59. In particular, autophagy-derived serine can be used to produce one-carbon (C1) unit molecules as starting materials for biosynthesizing various key anabolic precursors, which is essential for mitochondrial protein synthesis and metabolic remodeling for yeast to adapt from glycolytic to respiratory growth60.
In mammals, autophagic degradation is important to maintain amino acid levels in newborns to survive the neonatal starvation period. Autophagy-deficient Atg5-null and Atg7-null mice are born at a normal Mendelian ratio, but die perinatally before milk suckling within one day after birth 61,62, which is partially rescued by force feeding. While not showing differences immediately after birth, the amino acid level in both the plasma and organs (such as heart, liver and brain), especially that of the essential amino acids and BCAAs, significantly drops in 10 hours. Consistently, Atg5-null embryos show early embryonic lethality at the 4–8-cell stage and decreased protein synthesis traced by 35S-labelled methionine 63, suggesting that autophagy is essential for supplying amino acids as building blocks early in life (post-fertilization) during embryogenesis. In addition, AMPK is activated in Atg5-null, but not WT, newborns, suggesting that autophagy deficiency also causes energy deprivation. Notably, other nutrients, including blood glucose and fatty acid levels, are comparable between Atg5-null and Atg5-WT mice. These findings suggest that autophagy-provided amino acids, especially BCAAs, are used as an important energy source in newborns during the neonatal period. Oxidative degradation of BCAAs (such as leucine and isoleucine) generates acetyl-CoA, which can enter the TCA cycle and be oxidized for energy production. In addition to protein synthesis and ATP production, autophagy-produced amino acids are also used in glucose production in the liver via gluconeogenesis. Whole-body inducible or liver-specific Atg7 knockout (KO) adult mice are unable to maintain their blood levels of not only amino acids but also glucose during food deprivation 64,65. Different from normal blood glucose levels in Atg-null newborns, these findings emphasize the importance of autophagy during adulthood in maintaining systemic glucose metabolism. Taken together, autophagy plays a key role in the maintenance of cellular and blood amino acid levels, housekeeping protein synthesis, energy production and survival during starvation.
Aside from synthesizing intracellular proteins, autophagy-derived amino acids can also be secreted, or contribute to the synthesis of secretory proteins. For example, alanine secreted through autophagic degradation in pancreatic stellate cells (myofibroblast cells that regulate extracellular matrix formation in the pancreas) can be utilized by neighboring pancreatic ductal adenocarcinoma (PDAC), which promotes tumor growth under nutrient-deprived conditions 66. In addition, synthesis of secretory proteins, such as Interleukin-6/8, is dependent on the formation of a specialized location termed TASCC (TOR-autophagy spatial coupling compartment) under Ras-induced senescence conditions 67. TASCC is a compartment where autolysosomes, mTORC1 and the secretory apparatus (including the rough ER and trans-Golgi network) are adjacent to one another, and thus is proposed to spatially couple the catabolic and anabolic machineries for rapid protein turnover and resynthesis. Because TASCC forms under nutrient-rich conditions when both amino acids and growth factors are available, it is likely that mTORC1 is recruited to lysosomes by Rag and Rheb smOxidation and autophagy are critical energy providers duringall GTPases (Figure 2), but it is unknown what regulates the connection between the mTORC1-containing lysosomes and the secretory pathway.
Turnover of rRNAs and mRNAs to maintain nucleotide pool
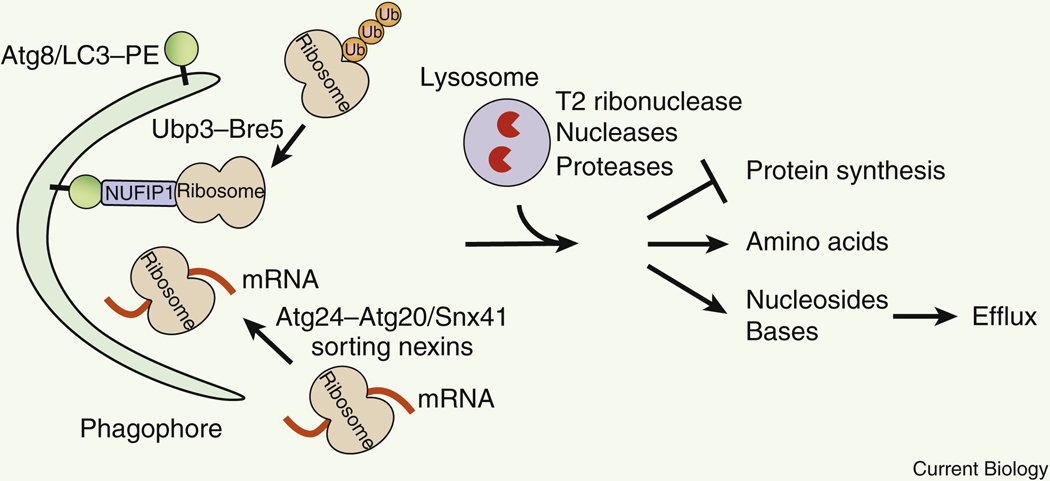
In response to starvation, protein synthesis is halted. Under such conditions, autophagy mediates the degradation of the unnecessary protein translation machinery, including ribosomes and mRNAs 68–70. Ribosomes constitute 30–40% of the cytosolic volume, and rRNA (ribosomal RNA) accounts for 80% of total cellular RNA 71, which can also serve as an abundant reservoir of amino acids and nucleotides. Autophagy-mediated selective degradation of ribosomes is termed as ribophagy, first identified in yeast and then in mammalian cells (Figure 4, Table 1). Ribophagy in both yeast and mammals is induced by starvation or Tor/mTORC1 inhibition, but the mechanisms in yeast versus mammalian cells are unclear and under debate. In yeast, ribophagy requires the core autophagy machinery and involves deubiquitination of ribosomal proteins by the ubiquitin protease Ubp3 and its cofactors Bre5, Cdc48 and Ufd3 68,69 (Figure 4). Yet similar deubiquitination mechanisms have not been reported in mammalian cells. Instead, mammalian ribophagy is dependent on the PI3K components VPS34 and BECN1, but not ATG5 72, resembling non-canonical autophagy that is ATG5-independent 73. Notably, a receptor for ribophagy has recently been identified as NUFIP1 in mammalian cells 74 (Figure 4). NUFIP1 interacts with both ribosomes and the autophagosome protein LC3 to recruit ribosomes to forming autophagosomes, and is essential for ribosomal protein degradation and cell survival during starvation.
Figure 4. Degradation of ribosomal proteins, rRNA and ribosome-bound translating mRNA by ribophagy or autophagy-mediated RNA degradation.
Ribophagy and mRNA autophagy switches off protein synthesis and provides amino acids, nucleosides and bases, which can be further secreted outside of the cells. In yeast, deubiquitination of ribosomal proteins by the Ubp3-Bre5 ubiquitin protease is required for ribophagy. In mammalian cells, NUFIP1 serves as a ribophagy receptor that binds both LC3 and ribosomes. Autophagic sequestration of mRNAs requires Atg24-Atg20 or Atg24-Snx41 sorting nexin complexes in yeast. Ub, ubiquitin.
Autophagy mediates the degradation of RNAs, including rRNA and mRNA, to maintain cellular pools of nucleotides in response to starvation in yeast and higher eukaryotes. During starvation, Atg7 is essential for stabilizing the intracellular level of all four nucleosides (adenosine, guanosine, cytidine and uridine) in lung cancer cells 75. Notably, nitrogen starvation induces a transient increase in nucleoside concentrations in yeast. Such increases peak at 1–2 hour after starvation and dependent on the core autophagy machinery, including Atg2 76. Although autophagy preferentially targets ribosome-bound translating mRNAs for vacuolar delivery 70, the ribophagy ubiquitin protease Ubp3-Bre5 is not required in this process 76, suggesting that autophagic degradation of RNAs is distinct from ribophagy and does not involve deubiquitination of ribosomes. Instead, in yeast, autophagic mRNA degradation requires the Atg24-Atg20 or the Atg24-Snx41 sorting nexin complexes. Via the autophagic regulation, mRNAs encoding proteins essential for anabolism, including ribosomal proteins and amino acid biosynthetic proteins, are degraded 70, which facilitates the shutdown of anabolic activities under stress conditions. Vacuolar/lysosomal RNA degradation is carried out by an evolutionarily conserved vacuolar T2 family acid ribonuclease, Rny1 in yeast and RNST-2 in C. elegans 70,76,77. Loss of RNST-2 in C. elegans leads to lysosomal accumulation of rRNA, defects of embryonic and larval development, and a reduction in lifespan. Altogether, these findings highlight the dual functions of autophagy in the switch-off of protein synthesis in response to starvation, and in the maintenance of nucleotide homeostasis during starvation and development.
Lipid mobilization by macro-, micro-, and chaperone-mediated autophagy
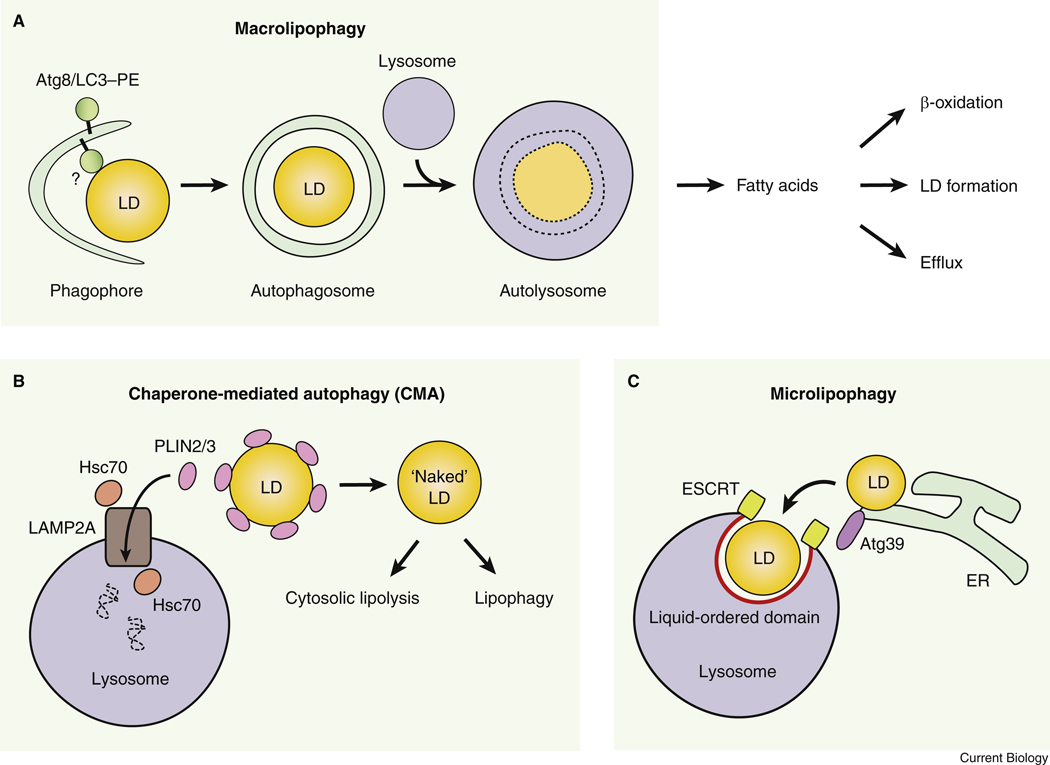
Starvation promotes a shift from carbohydrate metabolism to lipid metabolism for energy production, given glucose is scarce. Starvation induces hydrolysis of LDs to provide free fatty acids (FFAs) for oxidation and ATP generation. LDs are the intracellular lipid storage composed of neutral lipids, triglycerides (TGs) and cholesterol, surrounded by a phospholipid monolayer and coated by Perilipin coating proteins (PLINs). It is accepted that neutral lipids in the core of LDs are hydrolyzed by cytosolic lipases such as ATGL (adipose triglyceride lipase) (lipolysis) 78,79, whereas lysosomal lipases were thought to only degrade endocytosed lipoprotein-associated lipids. The discovery of macrolipophagy (macroautophagic degradation of lipids, hereinafter referred to as lipophagy) and visualization of LC3-labelled, LD-containing, double-membrane autophagosomes, initially in hepatocytes, largely expands our understanding on autophagy in nutrient mobilization 80 (Figure 5A, Table 1). Pharmacological or genetic inhibition of autophagy in vitro or in vivo leads to increased TG accumulation in diverse cell types, including hepatocytes, fibroblasts, vascular endothelial cells and neurons 80,81.
Figure 5. Lipid mobilization by three types of autophagy.
(A) Fatty acids are produced by macrolipophagy (lipophagy) of LDs. It is unclear whether a receptor protein is required in macrolipophagy. (B) Chaperone-mediated autophagy degrades LD coating proteins PLIN2 and PLIN3, and increases the accessibility of the “naked” LDs to both cytosolic lipolysis and lipophagy. (C) Micro-lipophagy mediates the direct engulfment of LDs by the vacuole/lysosome at the liquid-ordered domain. ESCRT proteins, Atg39 and a number of autophagy proteins are involved in micro-lipophagy.
The function and destination of lipophagy-derived FFAs are still not fully understood. Cellular siRNA knockdown of Atg5, or hepatic-specific loss of Atg5 or Atg7, decreases lipid β-oxidation and ketone body production upon starvation 80,82, suggesting that lipophagy is essential for mitochondrial β-oxidation. However, additional evidences suggest that instead of being directly transported and utilized in mitochondria, lipophagy-derived FFAs are used to drive LD formation, or expression of β-oxidation enzymes via PPARα suppression 82,83. Studies in mouse embryonic fibroblasts (MEFs) suggest that under starvation (amino acid- and growth factor-deprived) conditions, cytosolic lipases, rather than lipophagy, are required for the transport of FFAs from LDs to mitochondria 83. By contrast, autophagy is essential for the growth of LDs in MEFs during starvation. Thus, it is plausible that in MEFs, instead of directly being transported to mitochondria for oxidation, lipophagy-derived FFAs are used for the formation of LDs; FFAs are then mobilized from LDs by cytosolic lipases and used in mitochondrial oxidation. Whether this FFA trafficking pathway also occurs in lipid-rich cells with higher lipid metabolism, such as hepatocytes or adipocytes, still awaits to be further investigated. In addition, fatty acids released by lipophagy in hypothalamic AgRP (agouti-related peptide) neurons are important for the expression of the orexigenic neuropeptide AgRP. Accordingly, specific KO of Atg7 in AgRP neurons leads to reduced body weight, fat mass, and food intake upon refeeding after starvation, although whether it impacts systemic glucose homeostasis or insulin sensitivity is yet unclear 81. Another potential function of lipophagy-derived FFAs, other than mitochondrial oxidation, LD formation or gene expression, is to maintain circulating FFA levels and mediate cell-cell signaling, as lipophagy-derived FFAs have been reported to undergo efflux in hepatocytes via lysosomal fusion with the plasma membrane 84 (Figure 5A).
The molecular mechanism that governs LD recognition and degradation by lipophagy is poorly understood. LD size is suggested to be important for lipophagy targeting. In hepatocytes, only small LDs (with diameters less than 1 μm) are associated with the autophagosome protein LC3 and targeted for lipophagy, whereas large LDs are enriched with ATGL and broken down by cytosolic lipolysis 85, or undergo partial sequestration and then piecemeal degradation by lipophagy 80. Given that ATGL is identified as a positive regulator of lipophagy 86, cytosolic lipolysis and lipophagy may synergistically and sequentially function to degrade LDs: ATGL-mediated cytosolic lipolysis occurs before lipophagy to produce smaller LDs to fit in the size of autophagosomes. In the brown adipose tissue and liver, cold stress activates autophagy and promotes the association of LDs with several autophagy proteins, including ATG12-ATG5, BECN1 and LC3 38, suggesting that these proteins may function in autophagosomal incorporation of LDs. Although another autophagy protein ATG2 is also found to localize on the LD surface and Atg2 knockdown causes LD enlargement in mammalian cells, such LD accumulation is not dependent on other components of the autophagy machinery, such as ATG5 87. Given the lipid transferring ability of ATG2 17–19, it is reasonable to speculate that ATG2 may mobilize the phospholipids at the surface of LDs and increase the accessibility for breakdown. After LDs are delivered in autolysosomes, Dyn2 (Dynamin 2), a GTPase functioning in membrane scission and separation, is found to associate with the autolysosomal tubules and promote lysosomal fission and reformation 88. Inhibition of Dyn2 causes elongated autolysosomal structures, ultimate lysosome depletion and impaired LD breakdown under nutrient-deprived conditions in hepatocytes, and is associated with ethanol-induced liver steatosis 88,89. It remains to be determined whether Dyn2-mediated lysosome reformation is also universally important for (auto)lysosomal degradation of diverse substrates.
Two other types of autophagy, CMA and microautophagy, also contribute to lysosome-mediated LD degradation. The LD coating protein PLIN2 contains a KFERQ-like CMA-targeting motif, and is phosphorylated in an AMPK-dependent manner 90 and degraded by CMA during starvation 91 (Figure 5B, Table 1). Mutating the CMA-targeting motif in PLIN2 blocks its degradation by CMA, and leads to accumulation of LDs and inhibition of both cytosolic lipolysis and lipophagy. Thus, CMA plays a key role in providing access of the lipid core to both cytosolic lipases and the autophagy machinery. In addition, the use of CMA-deficient cells may also shed light on the mechanism of lipophagy, as these cells show reduced recruitment of a number of autophagy proteins to LDs, including BECN1, ATG5, LC3 and NBR1 (another autophagy cargo receptor), suggesting that these autophagy proteins may participate in the initiation of lipophagy.
Furthermore, in both yeast and mammalian cells, micro-lipophagy (μ-lipophagy), the selective degradation of LDs by microautophagy, also contributes to lipid utilization and survival in response to lipid stress and nutrient deprivation (Figure 5C, Table 1). In yeast, during adaptation to ER stress or lipid imbalance, for example, defective PC (phosphatidylcholine) biosynthesis, caused by KO of the key enzyme Cho2 (cho2Δ cells), promotes LD formation at the ER to convert phospholipids to TGs as a storage. These LDs are degraded via direct engulfment by the vacuole, representing micro-lipophagy 92,93. Similar to microautophagy, micro-lipophagy does not require the core ATG proteins such as Atg7, but requires the ESCRT complex and an ER membrane protein Atg39 (Autophagy-related protein 39, locus: Ylr312c, also identified as a receptor for reticulophagy and nucleophagy, the selective autophagic degradation of the ER and nucleus). It is likely that Atg39 is a multi-functional receptor in different autophagic pathways targeting various ER-associated structures or domains, including the LDs and nucleus. Yeast micro-lipophagy is also induced by acute glucose restriction via AMPK activation 94. At the vacuolar membrane surface, the PI3K components Atg14 and Atg6 (BECN1 homolog) are enriched at liquid-ordered domains (specific membrane regions with fast diffusion and high-acyl-chain order) to facilitate LD recruitment and membrane invagination. In mammalian hepatocytes, direct contact and piecemeal lipid transfer between LDs (and the LD protein PLIN2) and lysosomes are observed, which morphologically resemble micro-lipophagy 95. Neither the core autophagy protein ATG5, nor the CMA receptor LAMP2A, is required for lipid delivery to lysosomes under such conditions, genetically supporting that it is micro-lipophagy in mammalian systems. The genetic regulators of mammalian micro-lipophagy remain to be discovered, but the ESCRT complex and homologs of the aforementioned yeast autophagy proteins, including BECN1 and ATG14, may serve as candidates for investigation. Altogether, all three autophagy pathways are utilized as an important survival strategy that drives LD turnover and lipid mobilization upon stress.
Regulation of glucose homeostasis by glycophagy
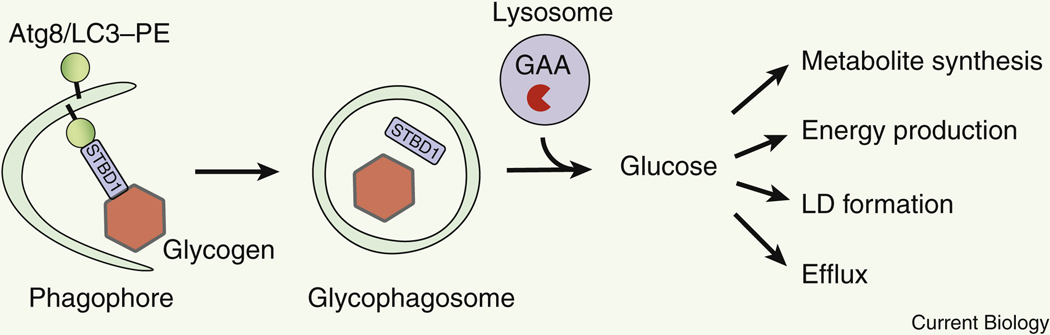
Autophagy plays an important role in glucose metabolism via autophagic degradation of glycogen (glycophagy) 96,97 (Figure 6, Table 1). Glycogen is a polysaccharide of glucose and a main source of intracellular glucose storage in the liver. Similar to LDs, glycogen can be degraded in both the cytosol and the lysosome. In the cytosol, glycogen is broken down via glycogenolysis, the sequential release of glucose-1-phosphate by the cytosolic enzyme, glycogen phosphorylase 98. Cytosolic glucose-1-phosphate is then be isomerized by phosphoglucomutases into glucose 6-phosphate, which can be used intracellularly, or further transported into the ER via the glucose 6-phosphate transporter (G6PT). Inside the ER, glucose 6-phosphate is dephosphorylated by glucose 6-phosphatase to produce free glucose that can be released extracellularly via glucose transporters as fuel for other organs. In comparison, lysosomal glycogen degradation generates non-phosphorylated free glucose that can be readily released. In glycophagy, glycogen is delivered to lysosomes by autophagosomes, and then hydrolyzed into single α-glucose molecules by lysosomal acid α-glucosidase (GAA). Besides liver, muscle also stores glycogen as its energy source. Genetic deficiency in GAA leads to myopathies such as glycogen storage disease type II (also known as Pompe disease), manifested by glycogen accumulation in lysosomes 99.
Figure 6. Selective autophagic degradation of glycogen.
Glycogen is degraded in lysosomes by glycophagy via a receptor STBD1. Glucose is released for many anabolic functions. GAA, lysosomal acid α-glucosidase. LD, lipid droplet.
Glycophagy and cytosolic glycogenolysis can complement each other in glucose production. Glucose released from glycophagy can be further used in the synthesis of fatty acids, nucleotides and nucleic acids, and aromatic amino acids. It should be noted that under normal conditions, lysosomes are responsible for degrading only 1–3% of cellular glycogen, and more glycogen may be degraded by cytosolic glycogenolysis. Because of this reason, energy deprivation or low blood glucose levels are not observed in Pompe patients. However, under stress conditions when the glucose demand increases, glycophagy may play a more important role in glycogen degradation. Indeed, upon 24-hour starvation, glycophagy can almost completely compensate for glycogenolysis in the muscle of drosophila larva 100. In comparison with glucose release from glycogenolysis, which requires coordinated activities of many enzymes and transporters for (de)phosphorylation and trafficking, glycophagy directly produces free glucose and may be a more efficient mechanism to supply glucose to the circulation under stress conditions.
Glycophagosomes (glycogen-containing autophagosomes) are observed in drosophila and mice during development 100,101. In mice, glycophagosomes are found within glycogen granules during mouse embryogenesis in brown adipose tissue, where glycogen breakdown is essential for LD formation. However, because genetic inhibition studies or screens on the autophagy machinery (ATG genes) in glycogen degradation are lacking, the molecular mechanism of glycophagy is still poorly understood. Of note, skeletal muscle-specific KO of Atg5 in GAA KO mice, a Pompe disease mouse model, does not ameliorate lysosomal glycogen accumulation in the muscle 102, suggesting that similar to ribophagy (discussed above), glycophagosome formation and glycophagy may be carried out by the ATG5-independent non-canonical autophagy mechanism 73. Nevertheless, starch binding domain-containing protein 1 (STBD1) has been identified as a receptor for glycophagy in mammalian cells (Figure 6). STBD1 is associated with glycogen and interacts with the Atg8/LC3 family members GABARAP and GABARAPL1, presumably via an LC3-interacting region (LIR) motif 103,104. Yet intriguingly, STBD1 appears to be a liver-specific receptor, because depleting Stbp1 in GAA KO mice reduces lysosomal accumulation of glycogen only in the liver, but not skeletal muscle or heart 105,106. Therefore, it is possible that either glycophagy in muscle tissues is non-selective and does not require a receptor, or there are muscle-specific receptors that mediate glycogen transport and glycophagy in muscle, which are worthy of further investigation as potential therapeutic targets against Pompe disease.
Concluding remarks and unanswered questions
Although autophagy has been shown to play an important role in intracellular nutrient mobilization and re-utilization, there are many unanswered questions. The molecular mechanisms, especially at the cargo recognition and delivery step, of selective autophagic degradation of different nutrient storages, including lipophagy and glycophagy, are not fully understood. There is also a lack of quantitative knowledge on the capacity of autophagy-mediated nutrient storage degradation under stress conditions (compared to cytosolic nutrient breakdown pathways, for example, lipolysis versus lipophagy, and glycogenolysis versus glycophagy), and on the effect of pharmacological autophagy inducers and inhibitors on nutrient mobilization and flow. Besides starvation, how autophagy mediates nutrient mobilization in response to additional stressors, such as exercise and cold stress, is largely unknown and needs further characterization. Furthermore, in addition to direct intracellular nutrient catabolism, the autophagy machinery may carry out a secretory function in higher eukaryotes. Many autophagy proteins have recently been shown to regulate the secretion of a variety of hormones, cytokines and neurotransmitters 107–111. Further understanding of the non-autophagic roles of ATG proteins may shed light on the noncell autonomous intercellular communication in systemic energetics of multicellular organisms. Lastly, the roles of autophagy in different tissues, genders and/or ages in mammalian nutrient mobilization are understudied. For example, in Pompe disease, why liver is not as sensitive as muscle to glycophagy defects is unclear. It is worthy of further study on how diverse tissues adapt to their different metabolic activity and energy demand under healthy and pathological conditions.
Acknowledgement
C.H. is supported by NIH/NIDDK R01 DK113170, R01 DK123447, and BrightFocus Foundation Alzheimer’s Disease Research Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He C, and Klionsky DJ (2009). Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuma A, and Mizushima N (2010). Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin Cell Dev Biol 21, 683–690. [DOI] [PubMed] [Google Scholar]
- 3.Rocchi A, and He C (2017). Regulation of Exercise-Induced Autophagy in Skeletal Muscle. Curr Pathobiol Rep 5, 177–186. 10.1007/s40139-017-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber CA., Sekar K., Tang JH., Warmer P., Sauer U., and Weis K. (2020). beta-Oxidation and autophagy are critical energy providers during acute glucose depletion in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 117, 12239–12248. 10.1073/pnas.1913370117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuck S (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133. 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- 6.Kaushik S, and Cuervo AM (2018). The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19, 365–381. 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biazik J, Yla-Anttila P, Vihinen H, Jokitalo E, and Eskelinen EL (2015). Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 11, 439–451. 10.1080/15548627.2015.1017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364. 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 9.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 10.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, and Lippincott-Schwartz J (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667. 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge L, Melville D, Zhang M, and Schekman R (2013). The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2, e00947. 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191, 155–168. jcb.201002100 [pii] 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, et al. (2018). ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597. 10.1080/15548627.2017.1422851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wold MS, Lim J, Lachance V, Deng Z, and Yue Z (2016). ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington’s disease models. Mol Neurodegener 11, 76. 10.1186/s13024-016-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, et al. (2016). The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 12, 547–564. 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, and Guan KL (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750. 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, and Melia TJ (2019). ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, and Noda NN (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26, 281–288. 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- 19.Maeda S, Otomo C, and Otomo T (2019). The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8. 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Huang J, Geng J, Nair U, and Klionsky DJ (2006). Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 17, 5094–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence RE, and Zoncu R (2019). The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol 21, 133–142. 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Remaileh M., Wyant GA., Kim C., Laqtom NN., Abbasi M., Chan SH., Freinkman E., and Sabatini DM. (2017). Lysosomal metabolomics reveals V-ATPase-and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, Di Malta C, Monfregola J, Medina DL, Lippincott-Schwartz J, and Ballabio A (2018). mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun 9, 3312. 10.1038/s41467-018-05862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Friedrichsen HJ, Andrews S, Picaud S, Volpon L, Ngeow K, Berridge G, Fischer R, Borden KLB, Filippakopoulos P, and Goding CR (2018). A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat Commun 9, 2685. 10.1038/s41467-018-04849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, and Ohsumi Y (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto S, Kuramoto K, Wang N, Situ X, Priyadarshini M, Zhang W, Cordoba-Chacon J, Layden BT, and He C (2018). Autophagy Differentially Regulates Insulin Production and Insulin Sensitivity. Cell Rep 23, 3286–3299. 10.1016/j.celrep.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He C., Song H., Yorimitsu T., Monastyrska I., Yen W-L., Legakis JE., and Klionsky DJ. (2006). Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol 175, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL, and Kiosses WB (2010). Short-term fasting induces profound neuronal autophagy. Autophagy 6, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia D, and Shaw RJ (2017). AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol Cell 66, 789–800. 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin SC, and Hardie DG (2018). AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab 27, 299–313. 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, and Guan KL (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303. 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang L, Chen S, Du F, Li S, Zhao L, and Wang X (2011). Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A 108, 4788–4793. 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Kundu M, Viollet B, and Guan KL (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141. 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, et al. (2012). Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest 122, 2428–2438. 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yau WW, Wong KA, Zhou J, Thimmukonda NK, Wu Y, Bay BH, Singh BK, and Yen PM (2021). Chronic cold exposure induces autophagy to promote fatty acid oxidation, mitochondrial turnover, and thermogenesis in brown adipose tissue. iScience 24, 102434. 10.1016/j.isci.2021.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quan W., Kim HK., Moon EY., Kim SS., Choi CS., Komatsu M., Jeong YT., Lee MK., Kim KW., Kim MS., and Lee MS. (2012). Role of hypothalamic proopiomelanocortin neuron autophagy in the control of appetite and leptin response. Endocrinology 153, 1817–1826. 10.1210/en.2011-1882. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, and Singh R (2016). Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab 23, 113–127. 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, and Jin S (2009). Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A 106, 19860–19865. 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, and Czaja MJ (2009). Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 119, 3329–3339. 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baerga R, Zhang Y, Chen PH, Goldman S, and Jin S (2009). Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5, 1118–1130. 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, and Kajimura S (2016). Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metab 24, 402–419. 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider JL, Suh Y, and Cuervo AM (2014). Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 20, 417–432. 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawley JA, Maughan RJ, and Hargreaves M (2015). Exercise Metabolism: Historical Perspective. Cell Metab 22, 12–17. 10.1016/j.cmet.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 45.Vainshtein A, and Hood DA (2016). The regulation of autophagy during exercise in skeletal muscle. J Appl Physiol (1985) 120, 664–673. 10.1152/japplphysiol.00550.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez AM., Bernardi H., Py G., and Candau RB. (2014). Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol 307, R956–969. 10.1152/ajpregu.00187.2014. [DOI] [PubMed] [Google Scholar]
- 47.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al. (2012). Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Niu Y, Yuan H, Huang J, and Fu L (2015). AMPK binds to Sestrins and mediates the effect of exercise to increase insulin-sensitivity through autophagy. Metabolism 64, 658–665. 10.1016/j.metabol.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Drake JC, Wilson RJ, Laker RC, Guan Y, Spaulding HR, Nichenko AS, Shen W, Shang H, Dorn MV, Huang K, et al. (2021). Mitochondria-localized AMPK responds to local energetics and contributes to exercise and energetic stress-induced mitophagy. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2025932118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo Verso F, Carnio S, Vainshtein A, and Sandri M (2014). Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy 10, 1883–1894. 10.4161/auto.32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17, 288–299. 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rennie MJ, and Tipton KD (2000). Protein and amino acid metabolism during and after exercise and the effects of nutrition. Annu Rev Nutr 20, 457–483. 10.1146/annurev.nutr.20.1.457. [DOI] [PubMed] [Google Scholar]
- 53.Wagenmakers AJ., Coakley JH., and Edwards RH. (1990). Metabolism of branched-chain amino acids and ammonia during exercise: clues from McArdle’s disease. Int J Sports Med 11 Suppl 2, S101–113. 10.1055/s-2007-1024861. [DOI] [PubMed] [Google Scholar]
- 54.Klionsky DJ, and Eskelinen EL (2014). The vacuole versus the lysosome: when size matters. Autophagy 10, 185–187. 10.4161/auto.27367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizushima N, and Klionsky DJ (2007). Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 27, 19–40. 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 56.Miwa S, Lawless C, and von Zglinicki T (2008). Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model. Aging Cell 7, 920–923. 10.1111/j.1474-9726.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, Zhang J, Zong NC, Lam MP, and Ping P (2012). Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics 11, 1586–1594. 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukada M, and Ohsumi Y (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333, 169–174. 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 59.Onodera J, and Ohsumi Y (2005). Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280, 31582–31586. 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 60.May AI, Prescott M, and Ohsumi Y (2020). Autophagy facilitates adaptation of budding yeast to respiratory growth by recycling serine for one-carbon metabolism. Nat Commun 11, 5052. 10.1038/s41467-020-18805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., and Mizushima N. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. [DOI] [PubMed] [Google Scholar]
- 62.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, and Mizushima N (2008). Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- 64.Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, et al. (2011). Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7, 727–736. 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, and White E (2014). Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927. 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. (2011). Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970. 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, and Dargemont C (2010). Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11, 548–554. 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kraft C., Deplazes A., Sohrmann M., and Peter M. (2008). Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10, 602–610. [DOI] [PubMed] [Google Scholar]
- 70.Makino S, Kawamata T, Iwasaki S, and Ohsumi Y (2021). Selectivity of mRNA degradation by autophagy in yeast. Nat Commun 12, 2316. 10.1038/s41467-021-22574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warner JR (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24, 437–440. 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 72.An H, and Harper JW (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. 10.1038/s41556-017-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, and Shimizu S (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658. 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 74.Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, and White E (2016). Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev 30, 1704–1717. 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, and Fukusaki E (2015). Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 34, 154–168. 10.15252/embj.201489083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Zou W., Yang P., Wang L., Ma Y., Zhang H., and Wang X. (2018). Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. Elife 7. 10.7554/eLife.36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zechner R, Strauss JG, Haemmerle G, Lass A, and Zimmermann R (2005). Lipolysis: pathway under construction. Curr Opin Lipidol 16, 333–340. 10.1097/01.mol.0000169354.20395.1c. [DOI] [PubMed] [Google Scholar]
- 79.Grabner GF, Xie H, Schweiger M, and Zechner R (2021). Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nat Metab 3, 1445–1465. 10.1038/s42255-021-00493-6. [DOI] [PubMed] [Google Scholar]
- 80.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, and Singh R (2011). Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 14, 173–183. 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saito T, Kuma A, Sugiura Y, Ichimura Y, Obata M, Kitamura H, Okuda S, Lee HC, Ikeda K, Kanegae Y, et al. (2019). Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat Commun 10, 1567. 10.1038/s41467-019-08829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rambold AS, Cohen S, and Lippincott-Schwartz J (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32, 678–692. 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui W, Sathyanarayan A, Lopresti M, Aghajan M, Chen C, and Mashek DG (2021). Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis. Autophagy 17, 690–705. 10.1080/15548627.2020.1728097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schott MB., Weller SG., Schulze RJ., Krueger EW., Drizyte-Miller K., Casey CA., and McNiven MA. (2019). Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol 218, 3320–3335. 10.1083/jcb.201803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sathyanarayan A, Mashek MT, and Mashek DG (2017). ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep 19, 1–9. 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Velikkakath AK, Nishimura T, Oita E, Ishihara N, and Mizushima N (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23, 896–909. 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, and McNiven MA (2013). Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 203, 315–326. 10.1083/jcb.201306140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rasineni K, Donohue TM Jr., Thomes PG, Yang L, Tuma DJ, McNiven MA, and Casey CA (2017). Ethanol-induced steatosis involves impairment of lipophagy, associated with reduced Dynamin2 activity. Hepatol Commun 1, 501–512. 10.1002/hep4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaushik S, and Cuervo AM (2016). AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 12, 432–438. 10.1080/15548627.2015.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaushik S, and Cuervo AM (2015). Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol 17, 759–770. 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia EJ, Liao PC, Tan G, Vevea JD, Sing CN, Tsang CA, McCaffery JM, Boldogh IR, and Pon LA (2021). Membrane dynamics and protein targets of lipid droplet microautophagy during ER stress-induced proteostasis in the budding yeast, Saccharomyces cerevisiae. Autophagy 17, 2363–2383. 10.1080/15548627.2020.1826691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, and Pon LA (2015). Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell 35, 584–599. 10.1016/j.devcel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seo AY., Lau PW., Feliciano D., Sengupta P., Gros MAL., Cinquin B., Larabell CA., and Lippincott-Schwartz J. (2017). AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6. 10.7554/eLife.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schulze RJ, Krueger EW, Weller SG, Johnson KM, Casey CA, Schott MB, and McNiven MA (2020). Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc Natl Acad Sci U S A 117, 32443–32452. 10.1073/pnas.2011442117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kondomerkos DJ, Kalamidas SA, Kotoulas OB, and Hann AC (2005). Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol 20, 689–696. 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 97.Kalamidas SA, and Kotoulas OB (2000). Glycogen autophagy in newborn rat hepatocytes. Histol Histopathol 15, 1011–1018. 10.14670/HH-15.1011. [DOI] [PubMed] [Google Scholar]
- 98.Adeva-Andany MM, Gonzalez-Lucan M, Donapetry-Garcia C, Fernandez-Fernandez C, and Ameneiros-Rodriguez E (2016). Glycogen metabolism in humans. BBA Clin 5, 85–100. 10.1016/j.bbacli.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morales JA, and Anilkumar AC (2021). Glycogen Storage Disease Type II. In StatPearls. [PubMed] [Google Scholar]
- 100.Zirin J, Nieuwenhuis J, and Perrimon N (2013). Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 11, e1001708. 10.1371/journal.pbio.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mayeuf-Louchart A, Lancel S, Sebti Y, Pourcet B, Loyens A, Delhaye S, Duhem C, Beauchamp J, Ferri L, Thorel Q, et al. (2019). Glycogen Dynamics Drives Lipid Droplet Biogenesis during Brown Adipocyte Differentiation. Cell Rep 29, 1410–1418 e1416. 10.1016/j.celrep.2019.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., and Plotz P. (2008). Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet 17, 3897–3908. 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang S, Wells CD, and Roach PJ (2011). Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun 413, 420–425. 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, and Roach PJ (2010). Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem 285, 34960–34971. 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yi H, Fredrickson KB, Das S, Kishnani PS, and Sun B (2013). Stbd1 is highly elevated in skeletal muscle of Pompe disease mice but suppression of its expression does not affect lysosomal glycogen accumulation. Mol Genet Metab 109, 312–314. 10.1016/j.ymgme.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 106.Sun T, Yi H, Yang C, Kishnani PS, and Sun B (2016). Starch Binding Domain-containing Protein 1 Plays a Dominant Role in Glycogen Transport to Lysosomes in Liver. J Biol Chem 291, 16479–16484. 10.1074/jbc.C116.741397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang M, Kenny SJ, Ge L, Xu K, and Schekman R (2015). Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife 4. 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bel S., Pendse M., Wang Y., Li Y., Ruhn KA., Hassell B., Leal T., Winter SE., Xavier RJ., and Hooper LV. (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuramoto K, Kim YJ, Hong JH, and He C (2021). The autophagy protein Becn1 improves insulin sensitivity by promoting adiponectin secretion via exocyst binding. Cell Rep 35, 109184. 10.1016/j.celrep.2021.109184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim YJ, Kong Q, Yamamoto S, Kuramoto K, Huang M, Wang N, Hong JH, Xiao T, Levine B, Qiu X, et al. (2021). An autophagy-related protein Becn2 regulates cocaine reward behaviors in the dopaminergic system. Sci Adv 7. 10.1126/sciadv.abc8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cadwell K, and Debnath J (2018). Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217, 813–822. 10.1083/jcb.201706157. [DOI] [PMC free article] [PubMed] [Google Scholar]