Abstract
Herein, we reported the synthesis of nineteen novel 1,2,4-triazole derivatives including 1,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-ones (7a-e), 1-(1,3-diphenylpropan-2-yl)-1H-1,2,4-triazole (8a-c) and 1,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-diones (10a-k). The structures of these derivatives were confirmed by spectroscopic techniques like IR, 1H-NMR, Mass spectroscopy and Elemental analysis. The cytotoxic activities of the synthesized compounds were evaluated against three human cancer cell lines including MCF-7, Hela and A549 using MTT assay. Compounds 7d, 7e, 10a and 10d showed a promising cytotoxic activity lower than 12 μM against Hela cell line. The safety of these compounds was also, evaluated on MRC-5 as a normal cell line and relieved that most of the synthesized compounds have proper selectivity against normal and cytotoxic cancerous cell lines. Finally, molecular docking studies were also, done to understand the mechanism and binding modes of these derivatives in the binding pocket of aromatase enzyme as a possible target.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13065-022-00887-x.
Keywords: 1,2,4-Triazole; Anticancer; MTT assay; Molecular docking; ADME
Introduction
Cancer is characterized by the uncontrolled growth and proliferation of abnormal cells and is the second leading cause of morbidity and mortality in the world [1, 2]. According to the global cancer statistics, 9.6 million deaths and also, more than 18 million new cancer occurred in 2018 [3]. It is expected that the cancer mortality rate will rise dramatically in the future [3].
Various internal and external factors cause abnormal cell proliferation, leading to development of various cancers, including genetics, viruses, drugs, diet and smoking [4, 5]. Currently, three strategies including chemotherapy, radiotherapy, and surgery are used for the treatment of cancer. Chemotherapy is the most common treatment for cancer disease, in which various chemotherapeutic agents are utilized to kill the cancer cells with minimum harmful effect on normal cells [4, 6]. However, drug resistance, non-selectivity and toxicity of many anticancer drugs have limited their clinical uses [1, 7]. Hence, the discovery and development of more effective and potent anticancer agents is one of the most clinical challenges in modern medicinal chemistry [8].
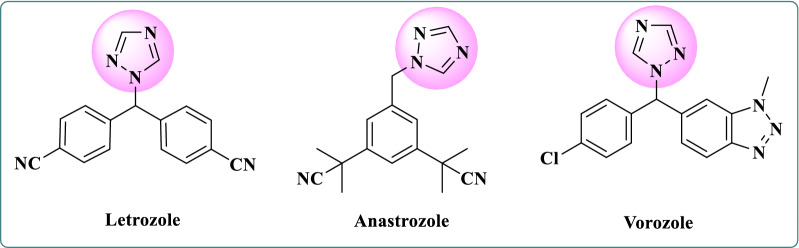
Heterocyclic compounds containing nitrogen atoms, especially heterocyclic rings with three nitrogen atoms, like 1,2,4-triazole ring, are one of the most important active pharmaceutical scaffolds. These scaffolds are able to form hydrogen bonds with different targets, which leads to the improvement of pharmacokinetics, pharmacological, and toxicological properties of compounds [2–4, 9]. Among heterocyclic compounds, 1,2,4-triazole derivatives have attracted much attention because of their various biological activities such as antiviral [10], antibacterial [11], antifungal [12, 13], anti-tubercular [14–16], immunosuppressant [17], antihypertensive [18], anti-inflammatory [19, 20], anticonvulsant [21, 22], analgesic [23], hypoglycemic [24], antidepressant [25, 26] and anticancer [9, 27, 28] activities. Currently, Letrozole, Anastrozole, and Vorozole which are 1,2,4-triazole-based drugs, are widely used in the treatment of estrogen-dependent breast cancer [29, 30] (Fig. 1).
Fig. 1.
Chemical structures of 1,2,4-triazole-based drugs
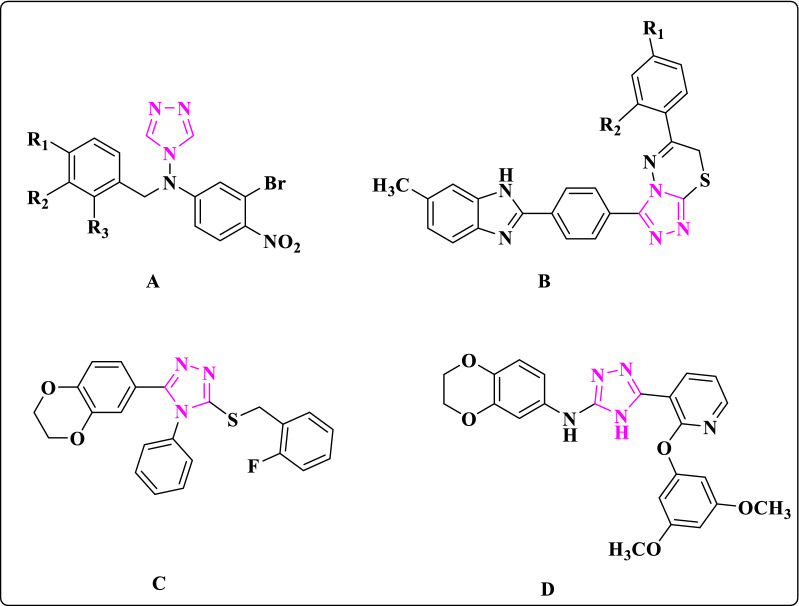
Some Clotrimazole derivatives have been reported as antifungal agents [2, 9, 31–33]. Song et al. synthesized a series of 4-N-nitrophenyl substituted amino-4H-1,2,4-triazole derivatives as promising aromatase inhibitors (Fig. 2, A) [34]. Moreover, Cevik et al. explored a new set of benzimidazole-triazolothiadiazine hybrids with potent aromatase inhibitory activities (Fig. 2, B) [35]. Hou et al. reported a series of 1,2,4-triazole derivatives with potent inhibitory activity against HepG2 cancer cell line (Fig. 2, C) [36]. In addition, X. Ouyang et al. showed that a set of 1,2,4-triazole derivatives, completely inhibited the tubulin polymerization by inducing cell cycle arrest at the G2/M phase of A431 cell line (Fig. 2, D) [37].
Fig. 2.
Structure of 1,2,4-triazole derivatives with anticancer activity
In the present study, three series of 1,2,4-triazole derivatives including 1,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-ones (7a-e), 1-(1,3-diphenylpropan-2-yl)-1H-1,2,4-triazole (8a-c) and 1,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-diones (10a-k) derivatives were designed, synthesized and evaluated for their anticancer activity against three human cancer cell lines (MCF-7, Hela and A549). The cytotoxic activity of all the synthesized compounds were assessed using the standard 3-(4,5-dimethylthiazol-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. Furthermore, molecular docking study was carried out to find the possible interaction mode of these derivatives in the active site of aromatase enzyme as possible target.
Results and discussion
Design
The target 1,2,4-triazole derivatives were designed based on the chemical structures of Letrozole (a), Anastrozole (b) and 4-triazolylflavans (c) which act as aromatase inhibitors. Aromatase is a member of the cytochrome P450 superfamily that catalyzes the estrogen biosynthesis and can be considered as a therapeutic target due to its overexpression in breast cancer. Anastrozole and Letrozole are potent aromatase inhibitors that use in the treatment of ER‐positive breast cancer. In addition, it has been previously reported that 4-triazolylflavans derivatives exhibited aromatase inhibitory effect [38, 39]. In these aromatase inhibitors, nitrogen atoms of 1,2,4-triazole ring bind to the iron in the heme moiety of CYP-450 and phenyl moieties have a key interaction in the active site of enzyme. Furthermore, carbonyl group is incorporated in the designed structures due to its ability to form hydrogen bonds. Therefore, 1,2,4-triazole moiety, phenyl rings and carbonyl groups were incorporated in the designed scaffolds (Fig. 3).
Fig. 3.
Design of new 1,2,4-triazole derivatives
Chemistry
The synthesis of the desired compounds (7a-e, 8a-c, 10a-k) was carried out according to the synthetic pathway illustrated in Fig. 4. The phenacyl chloride derivatives 3a-f were prepared in high yield through Fridel Crafts acylation of mono or di-substituted benzene (1a-f) with chloroacetyl chloride (2) using aluminum trichloride (AlCl3) as a strong Lewis acid catalyst [40, 41]. In the second step, intermediates 5a-f were synthesized from the reaction of intermediates 3a-f with 1,2,4-triazole (4) in the presence of sodium bicarbonate (NaHCO3). Compounds 7a-e were synthesized by treating benzyl bromide (6a) or benzhydryl bromide (6b) with intermediates 5a-f in the presence of NaH as a strong base catalyst in acetonitrile. Subsequently, Huang Minlon reduction of compounds 7a and 7c-d in the presence of N2H4.H2O and KOH yielded compounds 8a-c [42]. Compounds 10a-k were prepared from the reaction of 2-chloro-1,2-diphenylethanone (desyl chloride) or 2-chloro-1-phenylethanone derivatives with key intermediates 5a-f utilizing NaH as base and in acetonitrile as solvent. The structures of the synthesized compounds were confirmed through 1H-NMR, Mass and IR techniques. In the IR spectra of compounds 7a-e, a signal for C = O group was observed at 1690–1723 cm−1, while this signal was removed for compounds 8a-c. The IR absorption spectra of 10a-k were characterized the presence of two signals for C = O groups at 1650–1712 cm−1. 1H-NMR spectrum of compounds 7a-e showed two singlet peaks at 8.12–8.33 and 7.77–7.91 ppm assigned to the 1,2,4-triazole ring. In addition, in compounds 7a and 7b, the CH proton was observed at 6.17–6.24 ppm as a doublet of doublet peak and in compounds 7c-e the CH proton was observed at 6.73–6.79 ppm as a doublet peak. 1H-NMR spectrum of compounds 8a-c showed two singlet peaks at 8.17–8.29 ppm and 7.78–7.85 ppm assigned to the 1,2,4-triazole ring and a multiplet peak at 4.59–4.67 ppm assigned to the CH proton. 1H-NMR spectrum of compounds 10a-k showed two singlet peaks at 7.97–8.35 ppm and 7.79–8.03 ppm assigned to the 1,2,4-triazole ring. Furthermore, in compounds 10a-e with R = H, the CH proton appeared at 6.52–6.69 ppm as a triplet peak whereas in compounds 10f-k with R = Ph, the CH proton appeared at 6.45–6.81 ppm as a doublet peak. All the analytical data were documented in the Additional file 1: Data.
Fig. 4.
Synthesis of target compounds 7a-e, 8a-c and 10a-k. Reagents and conditions: a) AlCl3, dichloromethane, r.t., 24 h, b) NaHCO3, toluene, reflux, 20 h, c) NaH, CH3CN, reflux, 24 h, d) N2H4.H2O, KOH, Ethylene glycol,170 ˚C, 4 h, e) NaH, CH3CN, reflux, 24 h
Evaluation of anticancer activity
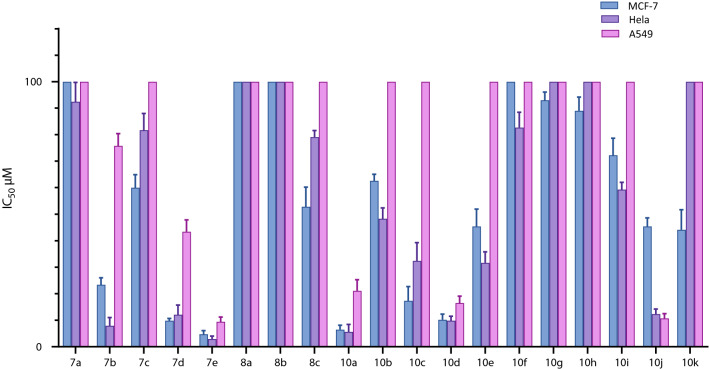
All of the synthetic compounds (7a-e, 8a-c and 10a-k) were screened for their in vitro cytotoxic effects against three human cancerous cell lines (MCF-7, Hela and A549) using MTT assay [43–45]. The biological results were expressed in terms of IC50 (Table 1). Generally, the best anticancer effect was seen on breast (MCF-7) and cervical (Hela) cell lines and less on lung cancer (A549) cell line. In this assay, cis platin was used as positive control [46–48]. As shown in Table 1, some of the compounds such as 7b, 7d-7e, 10a and 10c-d showed better antiproliferative activity compared to cis platin in all studied cancer cell lines. Assessments on propane-1-one derivatives (7a-e), compound 7e bearing X1:X2 = Cl, R = Ph showed significant activity with IC50 = 4.7, 2.9 and 9.4 μM against MCF-7, Hela and A549 cell lines, respectively, followed by compound 7d possessing X1 = Br, X2 = H, R = Ph with IC50 = 9.8, 12.1 and 43.4 μM. The obtained results indicated that the absence of electronegative groups at X position and also, the absence of phenyl ring at R position in analogue 7a (unsubstituted one), dramatically decreased the cytotoxic effects against all studied cancer cell lines. On the other hand, propane-1-yl-derivatives (8a-c), showed less cytotoxicity effect compared to other studied compounds (7a-e and 10a-k), which can be attributed to the reduction of carbonyl group at position 1 of propane chain. Further, a comparison of the cytotoxicity of compounds 7a-e on Hela cell line showed that the anticancer activities of different substitutions on phenyl ring followed the order 2,4-di-Cl > 4-Br > 4-Cl > H, and interestingly, the relative order of the substitution effect on cytotoxicity for compounds 8a-c was 4-Br > 4-Cl > H which was in line with above-mentioned results. In the case of butane-1,4-dione derivatives (10a-k), compound 10a was found to have promising anticancer activity with IC50 = 6.43, 5.6 and 21.1 μM against MCF-7, Hela and A549 cell lines, respectively. The substitution of Br and Cl at X1 and X3 position produced appropriate derivative (10d) with IC50 values of 10.2, 9.8 and 16.5 μM. Besides, the cytotoxic activity of compounds 10a-e on Hela cell line showed that the anticancer activities of different substitutions on the phenyl rings followed the order of H > 4Br > 2,4-di-Cl > 4-Cl. The incorporation of phenyl group at R position decreased cytotoxic activity. Among these compounds, 10j bearing 2,4-diflouro group at the phenyl moiety demonstrated adequate cytotoxic effect. The safety of these compounds was also, evaluated on MRC-5 as normal cell line. The resulted indicated that most of the synthesized compounds have proper selectivity against cancer cell lines (Fig. 5).
Table 1.
Cytotoxicity of the synthesized compounds against MCF-7, Hela and A549 cell lines [IC50 (μM)]
| Compound | X1 | X2 | X3 | Y | R | IC50 (μM ± SEM) | |||
|---|---|---|---|---|---|---|---|---|---|
| MCF-7 | Hela | A549 | MRC-5 | ||||||
| 7a | H | H | – | C = O | H | 154.2 ± 5.8 | 92.4 ± 7.3 | 149.6 ± 5.3 | > 300 |
| 7b | Cl | Cl | – | C = O | H | 23.4 ± 2.6 | 7.9 ± 3.1 | 75.8 ± 4.6 | 72.6 ± 3.4 |
| 7c | Cl | H | – | C = O | C6H5 | 60.0 ± 4.9 | 81.7 ± 6.3 | 177.3 ± 5.9 | 201.2 ± 9.5 |
| 7d | Br | H | – | C = O | C6H5 | 9.8 ± 0.9 | 12.1 ± 3.6 | 43.4 ± 4.5 | 35.6 ± 2.7 |
| 7e | Cl | Cl | – | C = O | C6H5 | 4.7 ± 1.4 | 2.9 ± 1.1 | 9.4 ± 1.8 | 27.8 ± 3.7 |
| 8a | H | H | – | CH2 | H | 158.3 ± 3.5 | 141.5 ± 2.1 | 127.3 ± 4.9 | 279.1 ± 2.5 |
| 8b | Cl | H | – | CH2 | C6H5 | 178.5 ± 2.5 | 198.3 ± 5.5 | 124.1 ± 3.4 | 279.1 ± 2.8 |
| 8c | Br | H | – | CH2 | C6H5 | 52.8 ± 7.4 | 79.1 ± 2.5 | 104.5 ± 7.3 | 104.5 ± 7.3 |
| 10a | H | H | H | – | H | 6.4 ± 1.7 | 5.6 ± 2.8 | 21.1 ± 4.2 | 21.7 ± 1.5 |
| 10b | Cl | H | H | – | H | 62.6 ± 2.5 | 48.3 ± 4.1 | 110.5 ± 1.3 | 259.3 ± 8.3 |
| 10c | Br | H | H | – | H | 17.3 ± 5.4 | 32.4 ± 6.9 | 103.7 ± 1.3 | 57.8 ± 1.7 |
| 10d | Br | H | Cl | – | H | 10.2 ± 2.1 | 9.8 ± 1.7 | 16.5 ± 2.6 | 42.8 ± 2.1 |
| 10e | Cl | Cl | H | – | H | 45.4 ± 6.5 | 31.6 ± 4.2 | 103.7 ± 5.9 | 104.5 ± 7.3 |
| 10f | H | H | H | – | C6H5 | 134.5 ± 4.5 | 82.7 ± 5.8 | 114.5 ± 7.6 | 289.1 ± 10.2 |
| 10 g | F | H | H | – | C6H5 | 93.0 ± 3.1 | 127.9 ± 6.1 | 149.8 ± 4.9 | 257.2 ± 4.3 |
| 10 h | Cl | H | H | – | C6H5 | 89.0 ± 5.2 | 172.3 ± 7.1 | 112.5 ± 10.8 | 249.3 ± 5.8 |
| 10i | Br | H | H | – | C6H5 | 72.2 ± 6.5 | 59.3 ± 2.7 | 134.8 ± 5.3 | 218 ± 3.2 |
| 10 J | F | F | H | – | C6H5 | 45.4 ± 3.2 | 12.3 ± 1.9 | 100.7 ± 1.8 | 101.2 ± 1.3 |
| 10 k | Cl | Cl | H | – | C6H5 | 44.1 ± 7.6 | 121.2 ± 6.8 | 102.5 ± 5.2 | 147.9 ± 6.1 |
| Cis platin | – | – | – | – | – | 36.5 ± 1.9 | 12.3 ± 3.3 | 14.8 ± 0.27 | 45.2 ± 2.5 |
Fig. 5.
Cytotoxic effect of compounds 7a-e, 8a-c and 10a-k on MCF-7, Hela and A549 cell lines Taken to gather, regarding the cytotoxic evaluations on 7a-e and 8a-c derivatives, it can be realizing that 7e was the most potent derivative against all three tested cell lines. The structure activity relationship disclosed that electronegative substitution such as Cl and Br at para position of phenyl ring (X1) and also, the presence of phenyl ring at R position could increase the inhibitory activity significantly in a 7a-e series. Also, the presence of one-carbonyl group showed necessary for pharmacological effect. In addition, propane-1-yl-derivatives (8a-c) had least effect on cytotoxic activity. In the case of 10a-k, replacement of H with Ph moiety at R position led to decreased cytotoxic activity (10f-k) and also, no substituted analogue (10a) had favorable pharmacological effect on MCF-7 and Hela cell lines. The cytotoxicity of all synthesized compounds were shown in Table 1.
Molecular docking
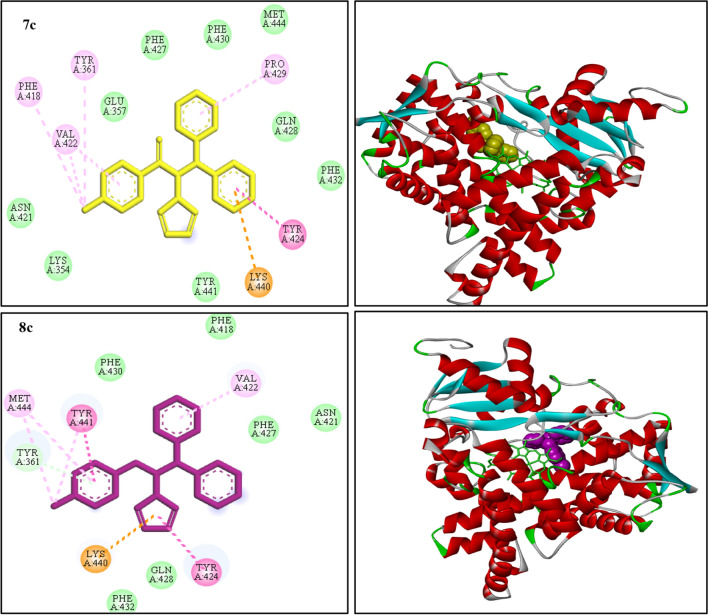
The docking information of four 1,2,4-triazole derivatives possessing the highest (10a and 10d) and lowest (7c and 8c) cytotoxic activity were shown in Fig. 6 and Fig. 7. Redocking of 4-androstene-3–17-dione as co-crystal ligand, was done to assessment the docking results. The RMSD was obtained 0.40 Å in comparison to its coordination in the crystal structure. The all interaction and score binding of all studied compounds was shown in Table 2.
Fig. 6.
2D interaction diagrams representing the docked conformation of compounds 10a and 10d in the human placental aromatase (3EQM). (Van der waals: green, dark pink: π-π, light pink: π-alkyl, purple: π-sigma, orange: π-cation, blue: halogen bond)
Fig. 7.
2D interaction diagrams representing the docked conformation of compounds 7c and 8c in the human placental aromatase (3EQM). (Van der waals: green, dark pink: π-π, light pink: π-alkyl, purple: π-sigma, orange: π-cation, blue: halogen bond)
Table 2.
The bonding energies (kcal/mol) and the detailed interactions of all synthesized compounds on 3EQM target using AutoDock Vina
| Entry | Amino Acid | Ligand involved moiety | Type of interaction | B.E (kcal/mol) | Entry | Amino Acid | Ligand involved moiety | Type of interaction | B.E (kcal/mol) |
|---|---|---|---|---|---|---|---|---|---|
| 7a | Met 374 | benzyl | pi-sulfur | − 9.9 | 10c | HEM 600 | benzoyl | pi-cation | − 9.2 |
| Leu 477, Ile 133, Ala 306, Val 370 | 1,2,4-triazole & benzoyl | pi-alkyl | Phe 134, Phe 221 | 1,2,4-triazole & benzoyl | pi-pi | ||||
| HEM 600 | benzoyl | pi-cation | Leu 372, Ser 478 | 1,2,4-triazole & C = O | Carbon hydrogen bond | ||||
| Thr 310, Arg 115, Val 373, Phe 134, Leu 372, Phe 221, Ser 478, Trp 224 | – | Vander waals | Ala 306, Ile 133, Val 370, Met 374 | benzoyl & 1,2,4-triazole | pi-alkyl & alkyl | ||||
| 7b | Val 370, Leu 477, Val 373, Ala 306, Ile 133, Ile 305, Phe 221, Trp 224 | benzyl & benzoyl & 1,2,4-triazole | pi-alkyl & alkyl | − 10.5 | Arg 115, Trp 224, Val 373, Thr 310, Leu 477, Asp 309 | – | Vander waals | ||
| HEM 600 | benzyl & benzoyl | pi-pi | 10d | Ala 306 | benzoyl | halogen | − 7.9 | ||
| Arg 115 | benzyl | pi-cation | HEM 600 | benzoyl | pi-cation | ||||
| Glu 302, Ser 478, Phe 134, Met 374, Leu 372, Thr 310, Asp 309 | – | Vander waals | Leu 477 | benzoyl | pi-sigma | ||||
| 7c | Tyr 424 | di-phenyl | pi-pi | − 8.2 | Phe 221 | benzoyl | pi-pi | ||
| Lys 440 | di-phenyl | pi-cation | Val 370, Val 369, Leu 372, Met 374, Ile 133, HEM 600, Phe 221 | benzoyl & 1,2,4-triazole | alkyl & pi-alkyl | ||||
| Phe 418, Val 422, Tyr 361, Pro 429 | benzoyl & di-phenyl | pi-alkyl &| alkyl | Asp 309, Ser 478, Phe 134, Trp 224, Thr 310, Val 373, Arg 115 | – | Vander waals | ||||
| Asn 421, Lys 354, Glu 357, Phe 427, Phe 430, Met 444, Gln 428, Phe 432, Tyr 441 | – | Vander waals | 10e | Arg 115 | 1,2,4- triazole | Hydrogen bond | − 9.7 | ||
| 7d | Lys 440 | di-phenyl | pi-alkyl | − 7.9 | Ser 478 | C = O | Carbon hydrogen bond | ||
| Tyr 424, Tyr 441 | benzoyl & di-phenyl | pi-pi | HEM 600 | benzoyl | pi-cation | ||||
| Gln 428 | C = O | Hydrogen bond | HEM 600 | benzoyl | pi-sigma | ||||
| Phe 432, Phe 430, Met 444, Tyr 361, Pro 429, Phe 427, Val 422, Phe 418 | – | Vander waals | Ile 133, Phe 134, Ala 306, Val 370, Met 374, Val 373 | benzoyl & 1,2,4-triazole | alkyl & pi-alkyl | ||||
| 7e | Tyr 441, Phe 430, Met 444, Lys 440, Pro 429 | 1,2,4-triazole & benzoyl | pi-alkyl & alkyl | − 7.3 | Leu 372, Leu 477, Phe 221, Asp 309, Trp 224, Thr 310 | – | Vander waals | ||
| Tyr 424 | di-phenyl | pi-pi | 10f | Lys 440 | C = O | Carbon hydrogen bond | − 8.9 | ||
| Lys 440 | 1,2,4-triazole | pi-cation | Tyr 361 | benzoyl | pi-pi | ||||
| Pro 429 | 1,2,4-triazole | Carbon hydrogen bond | Tyr 424 | phenyl | pi-pi | ||||
| Lys 440 | 1,2,4-triazole | Hydrogen bond | Pro 429 | benzoyl | pi-alkyl | ||||
| Phe 432, Gln 428, Phe 427 | – | Vander waals | Val 422, Phe 427, Phe 432, Gln 428, Met 444, Phe 430, Tyr 441 | – | Vander waals | ||||
| 8a | Arg 115 | benzyl | pi-cation | 9.4 | 10 g | Tyr 424 | phenyl | pi-pi | − 7.5 |
| HEM 600 | benzyl | pi-pi | Pro 429 | benzoyl | pi-alkyl | ||||
| Met 374, Ala 306, Thr 310, Ile 133, Trp 224, Phe 221, Ser 478, Phe 134, Leu 372 | – | Vander waals | Phe 418, Tyr 361, Tyr 441, Lys 440, Phe 430, Phe 432, Gln 428, Val 422, Phe 427 | – | Vander waals | ||||
| Val 373, Val 370, Leu 477 | benzyl | pi-alkyl | 10 h | Glu 357 | benzoyl | pi-anion | -7.2 | ||
| 8b | Phe 432, Tyr 424, Pro 429, Met 444 | benzyl & di-phenyl | pi-alky | − 7.8 | Tyr 424, Tyr 361 | benzoyl & phenyl | pi-pi | ||
| Tyr 424, Tyr 441 | benzyl & di-phenyl | pi-pi | Val 422 | benzoyl | pi-alkyl | ||||
| Tyr 361 | di-phenyl | pi-donor hydrogen bond | Phe 432, Tyr 441, Lys 440, Gln 428, Phe 430, Pro 429, Phe 427, Phe 418, Lys 354, Asn 421 | – | Vander waals | ||||
| Phe 427, Gln 428, Gly 433, Lys 440, Phe 430 | – | Vander waals | 10i | Tyr 244 | 1,2,4-triazole | Hydrogen bond | − 6.7 | ||
| 8c | Met 444, Val 422, Tyr 441 | benzyl & di-phenyl | pi-alkyl & alkyl | − 7.8 | Asp 476 | phenyl | pi-anion | ||
| Tyr 441, Tyr 424 | benzyl & 1,2,4-triazole | pi-pi | Ile 474 | benzoyl | pi-sigma | ||||
| Tyr 361 | benzyl | pi-donor hydrogen bond | Ala 226, Ile 474 | 1,2,4-triazole & benzoyl | alkyl & pi-alkyl | ||||
| Lys 440 | 1,2,4-triazole | pi-cation | His 475, Gln 225, Lys 230, Gly 69, Ile 229, Phe 65, Leu 66, Trp 67 | – | Vander waals | ||||
| Phe 430, Phe 418, Phe 427, Asn 421, Gln 428, Phe 432 | – | Vander waals | 10j | Lys 440, Gln 428 | 1,2,4-triazole & C = O | Hydrogen bond | − 7.8 | ||
| 10a | Ser 478 | C = O | Carbon hydrogen bond | − 9.4 | Lys 440 | 1,2,4-triazole | pi-cation | ||
| HEM 600 | 1,2,4-triazole | pi-cation | Tyr 361 | phenyl | pi-donor hydrogen bond | ||||
| Leu 477 | benzoyl | pi-sigma | Tyr 424, Tyr 441 | benzoyl & 1,2,4-triazole & phenyl | pi-pi | ||||
| HEM 600 | benzoyl | pi-pi | Pro 429, Met 444 | Benzoyl & phenyl | pi-alkyl | ||||
| Val 373, Ile 133, Val 370, Ala 306 | 1,2,4-triazole & benzoyl | pi-alkyl | Phe 432, Val 422, Phe 418, Phe 427, Phe 430, | ––- | Vander waals | ||||
| Arg 115, Met 374, Phe 221, Leu 372, Phe 134, Thr 310, Trp 224 | – | Vander waals | 10 k | Asn 421 | 1,2,4-triazole | Hydrogen bond | − 7.3 | ||
| 10b | Met 374 | benzoyl | pi-sulfur | − 9.3 | Glu 357 | phenyl | pi-anion | ||
| HEM 600 | 1,2,4-triazole | pi-cation | Tyr 361 | phenyl | pi-pi | ||||
| Phe 221, Val 370, Val 369, Ala 306, Ile 133 | 1,2,4-triazole & benzoyl | alkyl & pi-alkyl | Tyr 424, Lys 440, Tyr 441, Phe 430, Pro 429 | benzoyl | alkyl & pi-alkyl | ||||
| Thr 310, Asp 309, Ser 478, Trp 224, Leu 477, Phe 134, Arg 115, Val 373, Leu 372 | – | Vander waals | Met 444, Lys 354, Phe 418, Gln 428, Phe 427, Val 422, | – | Vander waals |
Compound 10a through its carbonyl group interacted with Ser 478 via hydrogen bond and also, formed π-cation and π-π interactions with HEM. The phenyl ring of 10a formed π-sigma bond with Leu 477 and also, some π-alkyl interactions with Val 373, Ile 133, Val 370 and Ala 306 residues were observed. In addition, hydrophobic interactions with Thr 310, Trp 224, Arg 115, Met 374, Phe 221, Leu 372, and Phe 134 residues were existence which led to desire affinity to aromatase enzyme. On the other hand, compound 10d had same similarity interactions with 10a as one of the best compound such as, π-π, π-sigma and π-alkyl with Leu 477, Val 370, Val 369, Phe 221, Leu 372, Met 374 and Ile 133 residues. Compound 10d also, formed halogen bond and π-cation interaction with Ala 306 amino acid residue and HEM group, respectively. These interactions confirmed that compounds 10a and 10d were successfully bound to the aromatase enzyme via most of the previously binding interactions of co-crystal ligand of aromatase enzyme (3EQM) (natural substrate androstenedione (AD)), such as Phe134, Trp224, Val370, Val373, Met374 and HEM [49, 50].
Moreover, compound 7c involved π-π and π-cation interactions with Tyr 424 and Lys 440 residues through its phenyl moiety. In addition, some π-alkyl interactions with Pro 429, Tyr 361, Val 422 and Phe 418 residues were observed. The groups of Glu 357, Phe 427, Phe 430, Met 444, Gln 428, Phe 432, Asn 421, Lys 354 and Tyr 441 residues involved to create a pocket around 7c by van der Waals forces. As it was shown in Fig. 7, the most important residues in binding of 8c was π-π and π-cation interactions between 1,2,4-triazole moiety and Tyr 424 and Lys 440 residues. Also, some π-π and π-alkyl interactions with Tyr 441, Val 422 and Met 444 residues were seen. A pocket of Phe 430, Phe 418, Phe 427, Asn 421, Gln 428 and Phe 432 residue were observed a round of 8c with van der Waals forces.
In silico ADME modeling study
ADME properties of the synthesized compounds were determined using SwissADME online software [51]. As depicted in Table 3, all of the compounds represented admissible molecular weight (MW < 500). They had desire lipophilicity (logP) values. In addition, the hydrogen bond properties including hydrogen bond donor (HBD), hydrogen bond acceptor (HBA) and rotatable bond (RB) are reasonable. According to rule of five, total polar surface area of all compounds are in accepted range ≤ 140 Å. Based on our results, all of the compounds indicated desire potential for oral bioavailability [52].
Table 3.
Physiochemical properties of the synthesized compounds 7a-e, 8a-c and 10a-k
| Entry | MWa | LogPb | HBDc | HBAd | TPSA (Å)e | RBf | Lipinski/Veber violation |
|---|---|---|---|---|---|---|---|
| 7a | 277.32 | 2.39 | 0 | 3 | 47.78 | 5 | 0 |
| 7b | 346.21 | 3.40 | 0 | 3 | 47.78 | 5 | 0 |
| 7c | 387.86 | 4.01 | 0 | 3 | 47.78 | 6 | 0 |
| 7d | 432.31 | 4.11 | 0 | 3 | 47.78 | 6 | 0 |
| 7e | 422.32 | 4.49 | 0 | 3 | 47.78 | 6 | 1 |
| 8a | 263.34 | 3.32 | 0 | 2 | 30.71 | 5 | 0 |
| 8b | 373.88 | 4.93 | 0 | 2 | 30.71 | 6 | 1 |
| 8c | 418.33 | 5.04 | 0 | 2 | 30.71 | 6 | 1 |
| 10a | 305.53 | 1.73 | 0 | 4 | 64.85 | 6 | 0 |
| 10b | 339.78 | 2.23 | 0 | 4 | 64.85 | 6 | 0 |
| 10c | 384.23 | 2.34 | 0 | 4 | 64.85 | 6 | 0 |
| 10d | 418.67 | 2.84 | 0 | 4 | 64.85 | 6 | 0 |
| 10e | 374.22 | 2.72 | 0 | 4 | 64.85 | 6 | 0 |
| 10f | 381.43 | 2.84 | 0 | 4 | 64.85 | 7 | 0 |
| 10 g | 399.42 | 3.22 | 0 | 5 | 64.85 | 7 | 0 |
| 10 h | 415.87 | 3.32 | 0 | 4 | 64.85 | 7 | 0 |
| 10i | 460.32 | 3.42 | 0 | 4 | 64.85 | 7 | 0 |
| 10j | 417.41 | 3.59 | 0 | 6 | 64.85 | 7 | 0 |
| 10 k | 450.32 | 3.79 | 0 | 4 | 64.85 | 7 | 0 |
| Lipinski/Veber’s Rules | ≤ 500 | ≤ 5 | ≤ 5 | ≤ 10 | ≤ 140 | ≤ 10 | ≤ 1 |
aMolecular weight (MW). b Logarithm of partition coefficient between n-octanol and water (LogP). c Number of hydrogen bond donors (HBD). d Number of hydrogen bond acceptors (HBA). e Topological polar surface area (TPSA). f Number of rotatable bonds (RB)
Experimental section
Chemistry
All reagents and solvents with analytical grade were purchased from commercial sources (Merck & Sigma Aldrich) and used without further purification. IR spectra (KBr, cm−1) were recorded using a Perkin Elmer IR instrument. 1H-NMR spectra were recorded in CDCl3 on Bruker 500 MHz spectrophotometer using tetramethylsilane (TMS) as an internal standard. Chemical shift values were expressed in ppm scale (δ) and coupling constants were reported in hertz (Hz). An Agilent spectrometer was used for Mass spectra recordation. All melting points were measured with an Electro-thermal IA 9100 apparatus and were uncorrected. The progress of reactions was monitored using thin layer chromatography (TLC) sheets pre-coated with UV fluorescent silica gel Merck 60F254 and the spots were visualized using UV lamp. The purification of the synthesized compounds was performed by column chromatography.
General procedure for the synthesis of intermediates 3a-f
The appropriate AlCl3 (60 mmol) was added to a stirred solution of phenyl halides (50 mmol) in dichloromethane (30 mL). After stirring for 30 min at room temperature, the mixture was cooled to 0 οC and a solution of chloroacetyl chloride (54 mmol) in dichloromethane (20 mL) was added dropwise to it. The resulting mixture was stirred at room temperature for 24 h. After this time, 50 mL of HCl solution (5%) was slowly added and the reaction mixture was extracted with dichloromethane (3 × 30 mL) and then washed with NaHCO3 (20 mL), water (2 × 20 mL) and brine (20 mL), respectively. The organic layer was dried over anhydrous Na2SO4 and concentrated under vacuum. Finally, the obtained precipitate was recrystallized from n-hexane to give intermediates 3a-f.
General procedure for the synthesis of intermediates 5a-f
1,2,4-triazole (48 mmol) and NaHCO3 (48 mmol) were added to a stirred solution of intermediates 3a-f (40 mmol) in toluene. The reaction mixture was refluxed for 20 h. After this time, the reaction mixture was quenched with an ice bath and extracted with ethyl acetate (3 × 30 mL). Then, the organic phase was washed with water (2 × 20 mL) and brine (10 mL), dried over anhydrous Na2SO4 and concentrated in vacuum. Finally, the residue was recrystallized from diethyl ether to afford the desired compounds 5a-f.
General procedure for the synthesis of compounds 7a-e
A solution of intermediates 5a-f (6 mmol) in acetonitrile (10 mL) was added to a suspension of NaH (8 mmol) in acetonitrile (30 mL) and the resulting mixture was stirred at room temperature for 1 h. Then, a solution of benzhydryl bromide or benzyle bromide (6 mmol) in 10 mL acetonitrile was added dropwise and the mixture was heated under reflux for 24 h. After cooling to room temperature, the solvent was evaporated in vacuum, 50 mL water was added and the mixture was extracted with dichloromethane (3 × 30 mL). In the following, the organic layers were dried over Na2SO4 and purified by column chromatography on silica gel eluting with ethyl acetate and petroleum ether (3:1) to afford desired products 7a-e.
1,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-one (7a)
Yield: 46%. M. P.: 145–148 °C; IR (KBr, cm−1): 3101 (C-H, aromatic), 2928.8 (C-H, aliphatic), 1690.0 (C = O, ketones), 1594.2 (C = N), 1283.7 (C–N stretch, aromatic).1H-NMR (500 MHz, CDCl3) δ (ppm): 8.28 (s, 1H, triazole), 7.92 (d, J = 7.5 Hz, 2H, Ar–H-CO), 7.91 (s, 1H, triazole), 7.59 (t, J = 7.5 Hz, 1H, Ar–H-CO), 7.46 (t, J = 7.5 Hz, 2H, Ar–H-CO), 7.18–7.24 (m, 3H, Ar–H), 7.01 (d, J = 6.8 Hz, 2H, Ar–H), 6.24 (dd, J = 8.8, 5.8 Hz, 1H, CH), 3.55 (dd, J = 14.3, 5.8 Hz, 1H, CH2), 3.41 (dd, J = 14.3, 9.0 Hz, 1H, CH2). 13C-NMR (75 MHz, CDCl3) δ: 193.8, 151.4, 143.6, 135.6, 134.8, 134.7, 129.5, 129.4, 129.3, 129.1, 127.9, 65.5, 39.0. MS m/z (%): 277.2 (5) [M+], 208.1 (45), 105.2 (100), 91.0 (60), 77.1 (100), 51.0 (15). Elem. anal. calcd. For C17H15N3O (277.2); C, 73.63; H, 5.45; N, 15.15. Found: C, 73.60; H, 5.42; N, 15.12.
1-(2,4-dichlorophenyl)-3-phenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-one (7b)
Yield: 52%. M.P.: 121–123 °C; IR (KBr, cm−1): 3133.7 (C–H stretch, aromatic), 2926.1 (C-H, aliphatic), 1723.8 (C = O, ketone), 1587.1 (C = N), 1294.1, 1248.1 (C–N stretch, aromatic), 1149.0 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.18 (s, 1H, triazole), 7.79 (s, 1H, triazole), 7.73 (d, J = 8.6 Hz, 1H, 2,4-diCl-Ar–H, H-6), 7.58 (s, 1H, 2,4-diCl-Ar–H, H-3), 7.38 (d, J = 8.6 Hz, 1H, 2,4-diCl-Ar–H, H-5), 7.18–7.23 (m, 3H, Ar–H), 6.98 (d, J = 6.7 Hz, 2H, Ar–H), 6.18 (dd, J = 8.5, 6.0 Hz, 1H, CH), 3.73 (dd, J = 14.2, 6.0 Hz, 1H, CH2), 3.60 (dd, J = 14.2, 8.6 Hz, 1H, CH2). MS m/z (%): 345.3 (4) [M+], 279.9 (8), 189.9 (30), 144.9 (23), 173.0 (70), 109.0 (22), 91.1 (100), 74.0 (27), 63.1 (18), 50.1 (8). Elem. anal. calcd. For C17H13Cl2N3O (345.3); C, 58.98; H, 3.78; N, 12.14. Found: C, 58.89; H, 3.72; N, 12.10.
1-(4-chlorophenyl)-3,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-one (7c)
Yield: 63%. M.P.: 140–142 °C; IR (KBr, cm−1): 3101.6 (C-H, aromatic), 2926.1 (C-H, aliphatic), 1691.4 (C = O, ketones), 1588.7 (C = N stretch, aromatic), 1275.4 (C–N stretch, aromatic), 1093.1 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.33 (s, 1H, triazole), 7.81 (d, J = 8.2 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.81 (s, 1H, triazole), 7.34 (d, J = 8.3 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.27 (d, J = 7.5 Hz, 2H, Ar–H), 7.23–7.13 (m, 7H, Ar–H), 7.10 (t, J = 7.3 Hz, 1H, Ar–H), 6.79 (d, J = 11.6 Hz, 1H, CH–N), 5.09 (d, J = 11.6 Hz, 1H, CH-(Ph)2). MS m/z (%): 387.1 (12) [M+], 317.1 (45), 242.1 (100), 178.1 (70), 167.3 (100), 152.0 (70), 141.0 (55), 111.0 (100), 91.0 (38), 75.1 (35), 51.1 (7). Elem. anal. calcd. For C23H18ClN3O (387.87); C, 71.22; H, 4.68; N, 10.83. Found: C, 71.20; H, 4.58; N, 10.80.
1-(4-bromophenyl)-3,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-one (7d)
Yield: 58%. M.P.: 110–113 °C; IR (KBr, cm−1): 3103.3 (C-H, aromatic), 2973.0 (C-H, aliphatic), 1692.8 (C = O, ketones), 1583.1 (C = N stretch, aromatic), 1287.8 (C–N stretch, aromatic), 1012.4 (Ar-Br). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.13 (s, 1H, triazole), 7.77 (s, 1H, triazole), 7.71 (d, J = 8.4 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.51 (d, J = 8.4 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.26–7.11 (m, 10 H, Ar–H), 6.73 (d, J = 11.6 Hz, 1H, CH–N), 5.07 (d, J = 11.6 Hz, 1H, CH-(Ph)2). MS m/z (%): 431.1 (5) [M+], 363.0 (25), 248.1 (25), 207.0 (22), 184.9 (75), 167.3 (100), 152.0 (100), 128.0 (8), 91.0 (28), 76.1 (33), 51.0 (7). Elem. anal. calcd. For C23H18BrN3O (433.1); C, 63.90; H, 4.20; N, 9.72. Found: C, 63.85; H, 4.16; N, 9.68.
1-(2,4-dichlorophenyl)-3,3-diphenyl-2-(1H-1,2,4-triazol-1-yl) propan-1-one (7e)
Yield: 48%. M.P.: 118–121˚C. IR (KBr, cm−1): 3130.3 (C–H stretch, aromatic), 2923.9 (C-H, aliphatic), 1700.1 (C = O, ketone), 1578.6 (C = N), 1276.3 (C–N stretch, aromatic), 1137.6 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.12 (s, 1H, triazole), 7.79 (s, 1H, triazole), 7.31 (d, J = 1.8 Hz, 1H, 2,4-diCl-Ar–H, H-6), 7.30 (s, 1H, 2,4-diCl-Ar–H, H-3), 7.23–7.15 (m, 8H, Ar–H), 7.14 (d, J = 1.8 Hz, 1H, 2,4-diCl-Ar–H, H-5), 7.13–7.10 (m, 2H, Ar–H), 6.74 (d, J = 11.6 Hz, 1H, CH–N), 4.94 (d, J = 11.6 Hz, 1H, CH-(Ph)2). MS m/z (%): 421.2 (3) [M+], 351.0 (15), 276.0 (68), 248.1 (45), 165.1 (100), 152.0 (52), 109.0 (20), 91.0 (33), 77.1 (12), 51.1 (5). Elem. anal. calcd. For C23H17Cl2N3O (421.2); C, 65.41; H, 4.06; N, 9.95. Found: C, 65.38; H, 4.02; N, 9.92.
General procedure for the synthesis of compounds 8a-c
A mixture of compound 7a or 7c-d (1.6 mmol), hydrazine monohydrate (8 mmol) and potassium hydroxide (8 mmol) in ethylene glycol (50 mL) was heated at 170 ˚C for 4 h. Then, the reaction mixture was cooled to room temperature, quenched with water (500 mL) and acidified to pH = 1 with concentrated hydrochloric acid and extracted with chloroform (3 × 30 mL). Afterwards, the organic layer was washed with brine and dried over Na2SO4. The crude product was purified by column chromatography on silica gel eluting with ethyl acetate and petroleum ether (1:1) to give pure compounds 8a-c.
1-(1,3-diphenylpropan-2-yl)-1H-1,2,4-triazole (8a)
Yield: 39%. M.P.: 81–83 °C, IR (KBr, cm−1): 3103.7 (C-H, aromatic), 2931.8 (C-H, aliphatic), 1598.8 (C = N), 1285.8 (C–N stretch, aromatic). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.17 (s, 1H, triazole), 7.85 (s, 1H, triazole), 7.29–7.27 (m, 6H, Ar–H, H-2, H-4 and H-6), 7.13–7.11 (m, 4H, Ar–H, H-3 and H-5), 4.64 (m, 1H, CH), 3.70 (d, J = 8.8 Hz, 4H, CH2). MS m/z (%): 263.1 (10) [M+], 201.1 (17), 172.0 (9) 105.2 (100), 91.0 (73), 77.1 (100), 63.1 (12), 51.1 (15). Elem. anal. calcd. For C17H17N3 (263.1); C, 77.54; H, 6.51; N, 15.96. Found: C, 77.35; H, 6.49; N, 15.85.
1-(3-(4-chlorophenyl)-1,1-diphenylpropan-2-yl)-1H-1,2,4-triazole (8b)
Yield: 41%. M.P.: 93–96 °C; IR (KBr, cm−1): 3103.4 (C-H, aromatic), 2959.5 (C-H, aliphatic), 1591.1 (C = N), 1278.6 (C–N stretch, aromatic). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.29 (s, 1H, triazole), 7.78 (s, 1H, triazole), 7.46 (d, c = 8.4 Hz, 2H, 4-Cl-Ar–H, H-2 and H-6), 7.28–7.26 (d, J = 7.1 Hz, 2H, Ar–H), 7.24–7.08 (m, 8H, Ar–H), 7.05 (d, J = 8.4 Hz, 2H, 4-Cl-Ar–H, H-3 and H-5), 4.67 (m, 1H, 4-Cl-Ar-CH2-CH), 3.89 (d, J = 11.2 Hz, 1H, Ar–CH), 3.42 (dd, J = 14.2, 6.1 Hz, 1H, CH2), 3.27 (dd, J = 14.3, 8.5 Hz, 1H, CH2). MS m/z (%): 373.1 (5) [M+], 306.1 (8), 245.1 (15) 173.1 (35), 155.1 (30), 126.0 (100), 111.0 (45), 103.1 (33), 77.0 (25), 63.0 (4), 51.1 (7). Elem. anal. calcd. For C23H20ClN3 (373.13); C, 73.89; H, 5.39; N, 11.24. Found: C, 73.80; H, 5.28; N, 11.19.
1-(3-(4-bromophenyl)-1,1-diphenylpropan-2-yl)-1H-1,2,4-triazole (8c)
Yield: 45%. M.P.: 88–91 °C; IR (KBr, cm−1): 3105.1 (C-H, aromatic), 2970.5 (C-H, aliphatic), 1585.2 (C = N), 1288.1 (C–N stretch, aromatic). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.18 (s, 1H, triazole), 7.81 (s, 1H, triazole), 7.57 (d, J = 8.4 Hz, 2H, 4-Br-Ar–H, H-2 and H-6), 7.28–7.26 (d, J = 7.0 Hz, 2H, Ar–H), 7.27–7.10 (m, 8H, Ar–H), 7.08 (d, J = 8.5 Hz, 2H, 4-Br-Ar–H, H-3 and H-5), 4.59 (m, 1H, 4-Cl-Ar-CH2-CH), 3.75 (d, J = 11.5 Hz, 1H, Ar–CH), 3.50 (dd, J = 14.2, 6.1 Hz, 1H, CH2), 3.37 (dd, J = 14.2, 8.5 Hz, 1H, CH2). MS m/z (%): 417.0 (8) [M+], 363.0 (10), 286.0 (15) 248.1 (25), 184.9 (55), 169.3 (100), 155.0 (48), 128.1 (15), 104.0 (15), 91.0 (23), 76.0 (43), 51.0 (7). Elem. anal. calcd. For C23H20BrN3 (417.0); C, 66.04; H, 4.82; N, 10.04. Found: C, 66.01; H, 4.79; N, 10.02.
General procedure for the synthesis compounds 10a-k
A solution of intermediates 5a-f (6 mmol) in acetonitrile (10 mL) was added to a suspension of NaH (8 mmol) in acetonitrile (30 mL) and the resulting mixture was stirred at room temperature for 1 h. Then, a solution of 2-chloro-2-phenyl acetophenone or halogenated phenacyl chloride (6 mmol) in 10 mL acetonitrile was added dropwise and the mixture was heated under reflux for 24 h. After cooling to room temperature, the solvent was evaporated in vacuum, 50 mL water was added and the mixture was extracted with dichloromethane (3 × 30 mL). In the following, the organic layers were dried over Na2SO4 and purified by column chromatography on silica gel eluting with ethyl acetate and petroleum ether (3:1) to afford desired products 10a-k.
4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10a)
Yield: 55%. M.P.: 115–118 °C, IR (KBr, cm−1): 3093.5 (C–H stretch, aromatic), 2911.0 (C-H, aliphatic), 1691.0, 1668.8 (C = O, ketone), 1595.7 (C = N), 1271.3 (C–N stretch, aromatic). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.29 (s, 1H, triazole), 7.96 (d, J = 7.2 Hz, 4H, Ar–H-CO and Ar′-H–CO, H-2 and H-6), 7.90 (s, 1H, triazole), 7.58 (t, J = 7.1 Hz, 2H, Ar–H-CO and Ar′-H–CO, H-4), 7.48 (d, 4H, Ar–H-CO and Ar′-H–CO, H-3 and H-5), 6.69 (t, J = 6.0 Hz, 1H, CH), 4.15 (dd, J = 18.0, 6.6 Hz, 1H, CH2), 3.83 (dd, J = 17.8, 5.9 Hz, 1H, CH2). MS m/z (%): 305.3 (3) [M+], 236.1 (10), 276.1 (100), 223.1 (80), 200.1 (35), 178.1 (10), 131.0 (8), 105.2 (98), 77.0 (100), 63.0 (5), 51.1 (20). Elem. anal. calcd. For C18H15N3O2 (305.12); C, 70.81; H, 4.95; N, 13.76. Found: C, 70.75; H, 4.92; N, 13.72.
1-(4-chlorophenyl)-4-phenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10b)
Yield: 67%. M.P.: 131–133 °C; IR (KBr, cm−1): 3092.8 (C–H stretch, aromatic), 2920.3 (C-H, aliphatic), 1692.6, 1655.5 (C = O, ketone), 1586.3 (C = N), 1272.1 (C–N stretch, aromatic), 1090.4 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.25 (s, 1H, triazole), 7.96 (d, J = 7.2 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.90 (s, 1H, triazole), 7.83 (d, J = 8.5 Hz, 2H, 4-Cl-Ar–H-CO, H-2 and H-6), 7.44 (t, J = 7.1 Hz, 1H, Ar–H-CO, H-4), 7.39 (d, J = 8.5 Hz, 2H, 4-Cl-Ar–H-CO, H-3 and H-5), 7.30 (d, J = 7.1 Hz, 2H, Ar–H-CO, H-3 and H-5), 6.65 (t, J = 6.2 Hz, 1H, CH), 4.13 (dd, J = 18.0, 6.6 Hz, 1H, CH2), 3.80 (dd, J = 17.9, 5.9 Hz, 1H, CH2). MS m/z (%): 339.1 (8) [M+], 287.1 (13), 263.0 (25), 178.1 (10), 170.0 (18), 139.1 (100), 105.1 (100), 77.0 (83), 51.1 (13). Elem. anal. calcd. For C10H14ClN3O2 (339.1); C, 63.63; H, 4.15; N, 12.37. Found: C, 63.59; H, 4.10; N, 12.35.
1-(4-bromophenyl)-4-phenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10c)
Yield: 44%. M.P.: 149–151 °C. IR (KBr, cm−1): 3073.7 (C–H stretch, aromatic), 2925.4 (C-H, aliphatic), 1697.1, 1659.5 (C = O, ketone), 1589.5 (C = N), 1279.1 (C–N stretch, aromatic), 1009.6 (Ar-Br).1H-NMR (500 MHz, CDCl3) δ (ppm): 8.34 (s, 1H, triazole), 7.96 (d, J = 7.1 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.92 (s, 1H, triazole), 7.74 (d, J = 8.3 Hz, 2H, 4-Br-Ar–H-CO, H-2 and H-6), 7.56 (d, J = 8.3 Hz, 2H, 4-Br-Ar–H-CO, H-3 and H-5), 7.43 (t, J = 7.1 Hz, 1H, Ar–H-CO, H-4), 7.30 (d, J = 7.1 Hz, 2H, Ar–H-CO, H-3 and H-5), 6.69 (t, J = 6.4 Hz, 1H, CH), 4.15 (dd, J = 18.0, 6.7 Hz, 1H, CH2), 3.82 (dd, J = 18.0, 6.1 Hz, 1H, CH2). MS m/z (%): 383.0 (3) [M+], 303.0 (12), 236.1 (15), 200.1 (17), 183.0 (17), 154.9 (10), 105.1 (98), 77.1 (100), 51.1 (18). Elem. anal. calcd. For C18H14BrN3O2 (383.0); C, 56.27; H, 3.67; N, 10.94. Found: C, 56.20; H, 6.60; N, 10.92.
1-(4-bromophenyl)-4-(4-chlorophenyl)-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10d)
Yield: 46%. M.P.: 162–165 °C, IR (KBr, cm−1): 3069.2 (C–H stretch, aromatic), 2922.3 (C-H, aliphatic), 1689.7 (C = O, ketone), 1590.7 (C = N), 1279.3 (C–N stretch, aromatic), 1093.4 (Ar-Cl), 1012.4 (Ar-Br). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.35 (s, 1H, triazole), 8.04 (d, J = 8.4 Hz, 2H, 4-Cl-Ar–H-CO, H-2 and H-6), 7.94 (s, 1H, triazole), 7.88 (d, J = 8.4 Hz, 2H, 4-Br-Ar–H-CO, H-2 and H-6), 7.40 (d, J = 8.4 Hz, 2H, 4-Br-Ar–H-CO, H-3 and H-5), 7.30 (d, J = 8.4 Hz, 2H, 4-Cl-Ar–H-CO, H-3 and H-5), 6.68 (t, J = 6.0 Hz, 1H, CH), 4.15 (dd, J = 17.9, 6.8 Hz, 1H, CH2), 3.82 (dd, J = 18.0, 6.2 Hz, 1H, CH2). MS m/z (%): 416.9 (3) [M+], 335.0 (100), 291.0 (23), 212.0 (30), 182.9 (50), 156.9 (18), 139.10 (100), 111.0 (100), 75.1 (47), 63.0 (10). Elem. anal. calcd. For C18H13BrClN3O2 (416.9); C, 51.64; H, 3.13; N, 10.04. Found: C, 51.60; H, 3.09; N, 10.01.
1-(2,4-dichlorophenyl)-4-phenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10e)
Yield: 52%. M.P.: 153–156 °C, IR (KBr, cm−1): 3098.2 (C–H stretch, aromatic), 2929.0 (C-H, aliphatic), 1710.5, 1673.5 (C = O, ketone), 1581.3 (C = N), 1272.1 (C–N stretch, aromatic), 1137.6 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.31 (s, 1H, triazole), 7.94 (s, 1H, triazole), 7.92 (d, J = 7.2 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.65 (d, J = 8.3 Hz, 1H, 2,4-diCl-Ar–H-CO, H-2), 7.46 (t, J = 7.2 Hz, 1H, Ar–H-CO, H-4), 7.22 (s, 1H, 2,4-diCl-Ar–H, H-5), 7.14 (t, J = 7.1 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.06 (d, J = 8.4 Hz, 1H, 2,4-diCl-Ar–H-CO, H-3), 6.52 (t, J = 6.0 Hz, 1H, CH), 4.12 (dd, J = 18.2, 5.1 Hz, 1H, CH2), 3.79 (dd, J = 18.2, 7.0 Hz, 1H, CH2. MS m/z (%): 373.2 (4) [M+], 338.1 (25), 268.0 (18), 240.0 (5), 172.9 (75), 145.0 (28), 105.2 (100), 91.0 (20), 77.0 (73), 51.0 (13). Elem. anal. calcd. For C18H13Cl2N3O2 (373.2); C, 57.77; H, 3.50; N, 18.95. Found: C, 57.75; H, 3.48; N, 18.91.
4-triphenyl-3-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10f)
Yield: 64%. M.P.: 185–188 °C; IR (KBr, cm−1): 3068.6 (C–H stretch, aromatic), 2924.0 (C-H, aliphatic), 1698.1, 1663.7 (C = O, ketone), 1596.3 (C = N), 1276.8 (C–N stretch, aromatic). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.15 (s, 1H, triazole), 7.99 (d, J = 7.7 Hz, 2H, Ar–H-CO-CHN, H-2 and H-6), 7.90 (d, J = 7.7 Hz, 2H, Ar–H-CO–CH-Ar, H-2 and H-6), 7.87 (s, 1H, triazole), 7.55–7.46 (m, 2H, 2Ar-H–CO, H-4), 7.40 (t, J = 7.4 Hz, 4H, 2Ar-H–CO, H-3 and H-5), 7.23–7.18 (m, 3H, Ar–H, H-3, H-4 and H-5), 7.13 (d, J = 6.5 Hz, 2H, Ar–H, H-2 and H-6), 6.66 (d, J = 10.5 Hz, 1H, CH–N), 5.69 (d, J = 10.5 Hz, 1H, CH-Ph). MS m/z (%): 381.2 (8) [M+], 312.1 (6), 276.1 (100), 209.1 (35), 170.1 (45), 131.0 (18), 104.8 (100), 90.0 (40), 76.9 (100), 63.1 (7), 51.1 (40). Elem. anal. calcd. For C24H19N3O2 (381.4); C, 75.57; H, 5.02; N, 11.02. Found: C, 75.52; H, 5.01; N, 11.02.
1-(4-fluorophenyl)-3,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10 g)
Yield: 34%. M.P.: 154–157 °C; IR (KBr, cm−1): 3094.9 (C–H stretch, aromatic), 2918.0 (C-H, aliphatic), 1695.9, 1658.5 (C = O, ketone), 1598.2 (C = N), 1240.9 (C–N stretch, aromatic), 1275.5 (Ar-F). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.28 (s, 1H, triazole), 7.92 (dd, J = 8.7, 5.3 Hz, 2H, 4-F-Ar–H-CO, H-3 and H-5), 7.79 (s, 1H, triazole), 7.55 (d, J = 7.5 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.23–7.13 (m, 7H, Ar–H and Ar–H-CO, H-3 and H-5), 7.09 (t, J = 7.5 Hz, 1H, Ar–H-CO, H-4), 7.04 (t, J = 8.6 Hz, 2H, 4-F-Ar–H-CO, H-2 and H-6), 6.81 (d, J = 11.6 Hz, 1H, CH–N), 5.09 (d, J = 11.6 Hz, 1H, CH-Ph). MS m/z (%): 399.2 (5) [M+], 365.4 (4), 332.4 (10), 305.3 (22), 295.1 (10), 226.1 (75), 197.2 (10), 172.0 (18), 149.0 (25), 123.2 (100), 95.1 (100), 75.0 (38), 63.1 (22), 51 (9). Elem. anal. calcd. For C24H18FN3O2 (399.2); C, 72.17; H, 4.54; N, 4.76. Found: C, 72.15; H, 4.51; N, 4.75.
1-(4-chlorophenyl)-3,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10 h)
Yield: 40%. M.P.: 174–176 °C; IR (KBr, cm−1): 3100.4 (C–H stretch, aromatic), 2979.5 (C-H, aliphatic), 1696.4, 1659.6 (C = O, ketone), 1588.8 (C = N), 1275.8 (C–N stretch, aromatic), 1092.2 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.11 (s, 1H, triazole), 7.94 (d, J = 7.4 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.89 (d, J = 8.4 Hz, 2H 4-Cl-Ar–H-CO, H-2 and H-6), 7.84 (s, 1H, triazole), 7.57 (d, J = 8.5 Hz, 2H 4-Cl-Ar–H-CO, H-3 and H-5), 7.53 (t, J = 7.4 Hz, 1H, Ar–H-CO, H-4), 7.541 (t, J = 7.4 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.22–21 (m, 3H, Ar–H), 7.12–10 (m, 2H, Ar–H), 6.60 (d, J = 10.1 Hz, 1H, CH–N), 5.66 (d, J = 10.0 Hz, 1H, CH-Ph). MS m/z (%): 415.1 (4) [M+], 397.1 (5), 346.1 (5), 310.1 (15), 276.1 (30), 242.0 (60), 207.0 (25), 197.2 (10), 170.0 (20), 139.1 (100), 104.9 (100), 90.0 (18), 77.1 (100), 63.0 (3), 51.1 (12). Elem. anal. calcd. For C24H18ClN3O2 (415.1); C, 69.31; H, 4.36; N, 10.10. Found: C, 69.29; H, 4.32; N, 10.09.
1-(4-bromophenyl)-3,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10i)
Yield: 48%. M.P.: 168–171 °C; IR (KBr, cm−1): 3069.2 (C–H stretch, aromatic), 2922.4 (C-H, aliphatic), 1696.7, 1657.8 (C = O, ketone), 1584.4 (C = N), 1277.1 (C–N stretch, aromatic), 1007.3 (Ar-Br). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.16 (s, 1H, triazole), 7.98 (d, J = 7.4 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.79 (s, 1H, triazole), 7.76 (d, J = 8.5 Hz, 2H, 4-Br-Ar–H-CO, H-2 and H-6), 7.55 (d, J = 8.5 Hz, 2H, 4-Br-Ar–H-CO, H-3 and H-5), 7.52 (t, J = 7.4 Hz, 1H, Ar–H-CO, H-4), 7.40 (t, J = 7.4 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.22–21 (m, 3H, Ar–H), 7.11–10 (m, 2H, Ar–H), 6.57 (d, J = 10.4 Hz, 1H, CH–N), 5.64 (d, J = 10.4 Hz, 1H, CH-Ph). MS m/z (%): 459.1 (2) [M+], 390.0 (3), 354.1 (7), 286.0 (35), 276.1 (25), 207.1 (18), 182.9 (60), 154.9 (30), 105.1 (100), 91.1 (18), 77.1 (85), 51.1 (10). Elem. anal. calcd. For C24H18BrN3O2 (459.1); C, 62.62; H, 3.94; N, 9.13. Found: C, 62.60; H, 3.90; N, 9.09.
1-(2,4-difluorophenyl)-3,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10j)
Yield: 45%. M.P.: 114–117 °C; IR (KBr, cm−1): 3062.4 (C–H stretch, aromatic), 2922.7 (C-H, aliphatic), 1676.7 (C = O, ketone), 1611.8 (C = N), 1271.9 (Ar-F). 1H-NMR (500 MHz, CDCl3) δ (ppm): 8.21 (s, 1H, triazole), 8.03 (s, 1H, triazole), 7.98 (d, J = 7.3 Hz, 2H, Ar–H-CO, H-2 and H-6), 7.90 (dt, J = 17.1, 9.5 Hz, 1H, 4-F-Ar–H-CO, H-6), 7.71 (t, J = 7.3 Hz, 1H, Ar–H-CO, H-4), 7.46 (t, J = 7.4 Hz, 2H, Ar–H-CO, H-3 and H-5), 7.18–7.14 (m, 3H, Ar–H), 7.08–7.06 (m, 2H, Ar–H), 6.89 (td, J = 8.4, 2.1 Hz, 1H, 4-F-Ar–H-CO, H-3), 6.73 (ddd, J = 10.1, 10.0, 2.2 Hz, 1H, 4-F-Ar–H-CO, H-5), 6.45 (d, J = 10.4 Hz, 1H, CH–N), 5.61 (d, J = 10.3 Hz, 1H, CH-Ph). MS m/z (%): 417.2 (7) [M+], 342.1 (15), 310.1 (15), 276.1 (50), 244.2 (100), 214.0 (35), 191.1 (10), 176.1 (33), 141.2 (100), 105.2 (100), 90.1 (25), 76.9 (100), 63.0 (43), 51.1 (33). Elem. anal. calcd. For C24H17F2N3O2 (417.2); C, 69.06; H, 4.11; N, 10.07. Found: C, 69.05; H, 4.10; N, 10.02.
1-(2,4-dichlorophenyl)-3,4-diphenyl-2-(1H-1,2,4-triazol-1-yl) butane-1,4-dione (10 k)
Yield: 39%. M.P.: 178–181 °C; IR (KBr, cm−1): 3102.8 (C–H stretch, aromatic), 2931.3 (C-H, aliphatic), 1712.0, 1676.4 (C = O, ketone), 1584.3 (C = N), 1276.0 (C–N stretch, aromatic), 1135.7 (Ar-Cl). 1H-NMR (500 MHz, CDCl3) δ (ppm): 7.97 (s, 1H, triazole), 7.88 (s, 1H, triazole), 7.65 (d, J = 8.5 Hz, 1H, 2,4-diCl-Ar–H, H-6), 7.52 (t, 1H, Ar–H-CO, H-4), 7.49 (d, 2H, Ar–H-CO, H-2 and H-6), 7.42 (t, 2H, Ar–H-CO, H-3 and H-5), 7.32 (d, J = 1.7 Hz, 1H, 2,4-diCl-Ar–H, H-3), 7.22 (dd, J = 8.4, 1.7 Hz, 1H, 2,4-diCl-Ar–H, H-5), 7.18–7.16 (m, 3H, Ar–H), 7.12–7.09 (m, 2H, Ar–H), 6.70 (d, J = 10.3 Hz, 1H, CH–N), 5.57 (d, J = 10.3 Hz, 1H, CH-Ph). MS m/z (%): 449.1 (7) [M+], 344.0 (10), 276.0 (85), 241.1 (5), 172.9 (80), 145.0 (35), 105.2 (100), 90.0 (20), 77.0 (100), 51.1 (12). Elem. anal. calcd. For C24H17Cl2N3O2 (449.1); C, 64.01; H, 3.81; N, 9.33. Found: C, 63.98; H, 3.78; N, 9.26.
In vitro cytotoxic assay
Chemicals
Fetal bovine serum (FBS) and RPMI-1640 medium were purchased from Gibco Invitrogen Co. (Scotland, UK) and sigma Aldrich, respectively. Dimethyl sulfoxide (DMSO), doxorubicin, penicillin, streptomycin was also purchased from Sigma Aldrich.
Cell cultures
Cell cultures were obtained from the human lung cancer cell line (A549), human cervical cancer cell line (Hela), human breast cancer cell line (MCF-7) and Normal cells isolated from human lung tissue (MRC-5) taken from the National Cell Bank of Iran (NCBI, Pasteur Institute, Tehran, Iran). All cells were cultured in RPMI-1640 medium supplemented with 10% FBS, antibiotics (penicillin 100 U/mL, streptomycin 100 lg/mL), at 37 ˚C in a humidified atmosphere containing 5% CO2.
MTT assay
Cytotoxic activities of all the synthesized compounds were assessed by standard 3-(4,5-dimethylthiazol-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay [53]. The assay was performed according to a known protocol [54–56]. The cells were seeded and plated in 96-well microplates at a density of 1 × 104 cells per well in 180 μL complete culture media. After 24 h incubation, the cells were treated with 100 mL of different concentrations of synthesized compounds ranging from 1 × 10–4 to 1 × 10–7 M at identical conditions in triplicates. After 72 h, media were replaced with 150 μL media containing 0.5 mg/mL of MTT solution [57]. The plates were incubated at 37 °C for additional four hours. Then, media containing MTT were discarded and 150 μL dimethylsulfoxide (DMSO, > 99%) was added to each well to dissolve the formazan crystals. The absorbance in each well was determined at 570 nm by a BioRad microplate reader (Model 680) and then the IC50 values were calculated and demonstrated as mean ± SEM [58].
Molecular docking study
Molecular docking studies were performed by AutoDock 4.2 program and AutoDock Tools 1.5.4 to investigate the binding mode of derivative in the active site of receptor [59]. The X-ray crystallographic structures of aromatase (PDB: 3EQM) (39) were obtained from the protein data bank (http://www.rcsb.org). Then, all water molecules and co-crystallized ligand were removed and hydrogen atoms were added to the protein and finally saved as pdbqt format. In the following, 3D structures of ligands were drawn and minimized under Molecular Mechanics MM+ and semi-empirical AM1 methods, using HyperChem software and saved as pdbqt format. The box dimensions were set at 65 × 65 × 65 with 0.375 Å grid spacing. Docking validation was done by re-docking the original ligand into the active site of receptor. Finally, conformations with the lowest free energies of binding were selected for analysis.
Conclusion
In summary, we have designed and synthesized nineteen new 1,2,4-triazole-based derivatives starting from different phenyl halide analogues through three or four different steps. Their chemical structures were fully confirmed by IR, 1H-NMR, Mass spectra and elemental analysis. In vitro cytotoxic activity of the synthesized compounds were evaluated against three human cancer cell lines including MCF-7, Hela and A549, using MTT assay. The obtained results indicated that the synthesized compounds possessed relatively high to moderate antiproliferative activities against MCF-7 and Hela cancer cell lines. Compounds 7d, 7e, 10a and 10d were the most potent ones against three tested cell lines. Based on structure activity relationship (SAR) studies, it was found that the presence of electronegative substituents on the phenyl ring, as well as the presence of one-carbonyl group, resulted in a relative increase in the cytotoxic activity of the synthesized compounds. The outcome results relived that these active derivatives can be considered as a lead compound for anticancer treatments.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- ADME
Adsorption Distribution, Metabolism and Discretion
- RMSD
Root Mean Square Deviation
- TPSA
Total polar surface area
- MTT
(3-(4, 5 dimethylthiazol-yl)-2,5-diphenyl-tetrazolium bromide)
Author contributions
LE write the biological section and performed docking study. SS prepared the manuscript, and write the synthesize section. AM contributed to the synthesis of compounds. SKh contributed to the preparation of the manuscript. AS supervise the computational study. HS edit the manuscript. ZF performed the biological study. MF supervised the biological study ZR contributed to the preparation of the manuscript and supervised the study. All authors read and approved the final manuscript.
Funding
Financial assistance from the Shiraz University of Medical Sciences by way of grant number 16388 is gratefully acknowledged.
Availability of data and materials
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request. We have presented all data in the form of Tables and Figure. The PDB code (3EQM) was retrieved from protein data bank (www.rcsb.org). https://www.rcsb.org/structure/3EQM. A549 cells (ATCC No. CCL-185 human lung cancer cell line), Hela cells (ATCC No. CCL-2 human cervical cancer cell line), MCF-7 cells (ATCC No. HTB-22 human breast cancer cell line), MRC-5 cells (ATCC No. CCL-171 human lung tissue),. All the cell lines were purchased from the National Cell Bank of Iran (NCBI, Pasteur Institute, Tehran, Iran).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Leila Emami, Sara Sadeghian and Ayyub Mojaddami contributed equally to this work
References
- 1.Labib MB, Philoppes JN, Lamie PF, Ahmed ER. Azole-hydrazone derivatives: design, synthesis, in vitro biological evaluation, dual EGFR/HER2 inhibitory activity, cell cycle analysis and molecular docking study as anticancer agents. Bioorg Chem. 2018;76:67–80. doi: 10.1016/j.bioorg.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Narsimha S, Nukala SK, Savitha Jyostna T, Ravinder M, Srinivasa Rao M, Vasudeva RN. One-pot synthesis and biological evaluation of novel 4-[3-fluoro-4-(morpholin-4-yl)] phenyl-1 H-1, 2, 3-triazole derivatives as potent antibacterial and anticancer agents. J Heterocycl Chem. 2020;57(4):1655–1665. [Google Scholar]
- 3.Grytsai O, Valiashko O, Penco-Campillo M, Dufies M, Hagege A, Demange L, et al. Synthesis and biological evaluation of 3-amino-1, 2, 4-triazole derivatives as potential anticancer compounds. Bioorg Chem. 2020;104:104271. doi: 10.1016/j.bioorg.2020.104271. [DOI] [PubMed] [Google Scholar]
- 4.Pragathi YJ, Sreenivasulu R, Veronica D, Raju RR. Design, synthesis, and biological evaluation of 1, 2, 4-thiadiazole-1, 2, 4-triazole derivatives bearing amide functionality as anticancer agents. Arab J Sci Eng. 2021;46(1):225–232. doi: 10.1007/s13369-020-04626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaber AA, Bayoumi AH, El-Morsy AM, Sherbiny FF, Mehany AB, Eissa IH. Design, synthesis and anticancer evaluation of 1H-pyrazolo [3, 4-d] pyrimidine derivatives as potent EGFRWT and EGFRT790M inhibitors and apoptosis inducers. Bioorg Chem. 2018;80:375–395. doi: 10.1016/j.bioorg.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Gomaa HA, El-Sherief HA, Hussein S, Gouda AM, Salem OI, Alharbi KS, et al. Novel 1, 2, 4-triazole derivatives as apoptotic inducers targeting p53: synthesis and antiproliferative activity. Bioorg Chem. 2020;105:104369. doi: 10.1016/j.bioorg.2020.104369. [DOI] [PubMed] [Google Scholar]
- 7.Ghoneim AA, El-Farargy AF, Bakr RB. Design, synthesis, molecular docking of Novel substituted pyrimidinone derivatives as anticancer agents. Polycyclic Aromat Compd. 2022;42(5):2538–2554. [Google Scholar]
- 8.Kamel MM, Abdo NYM. Synthesis of novel 1, 2, 4-triazoles, triazolothiadiazines and triazolothiadiazoles as potential anticancer agents. Eur J Med Chem. 2014;86:75–80. doi: 10.1016/j.ejmech.2014.08.047. [DOI] [PubMed] [Google Scholar]
- 9.El-Sherief HA, Youssif BG, Bukhari SNA, Abdelazeem AH, Abdel-Aziz M, Abdel-Rahman HM. Synthesis, anticancer activity and molecular modeling studies of 1, 2, 4-triazole derivatives as EGFR inhibitors. Eur J Med Chem. 2018;156:774–789. doi: 10.1016/j.ejmech.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Cao X, Wang W, Wang S, Bao L. Asymmetric synthesis of novel triazole derivatives and their in vitro antiviral activity and mechanism of action. Eur J Med Chem. 2017;139:718–725. doi: 10.1016/j.ejmech.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 11.Gao F, Wang T, Xiao J, Huang G. Antibacterial activity study of 1, 2, 4-triazole derivatives. Eur J Med Chem. 2019;173:274–281. doi: 10.1016/j.ejmech.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Sadeghpour H, Khabnadideh S, Zomorodian K, Pakshir K, Hoseinpour K, Javid N, et al. Design, synthesis, and biological activity of new triazole and nitro-triazole derivatives as antifungal agents. Molecules. 2017;22(7):1150. doi: 10.3390/molecules22071150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Y-N, Jiang Z-H, Sun L-S, Su Z-Y, Zhang M-M, Li H-L. Synthesis of 1, 2, 4-triazole benzoyl arylamine derivatives and their high antifungal activities. Eur J Med Chem. 2020;200:112463. doi: 10.1016/j.ejmech.2020.112463. [DOI] [PubMed] [Google Scholar]
- 14.Ganesh N, Singh M, Chandrashekar VM, Pujar GV. Antitubercular potential of novel isoxazole encompassed 1, 2, 4-triazoles: design, synthesis, molecular docking study and evaluation of antitubercular activity. Anti-Infect Agents. 2021;19(2):147–161. [Google Scholar]
- 15.Krishna KM, Inturi B, Pujar GV, Purohit MN, Vijaykumar G. Design, synthesis and 3D-QSAR studies of new diphenylamine containing 1, 2, 4-triazoles as potential antitubercular agents. Eur J Med Chem. 2014;84:516–529. doi: 10.1016/j.ejmech.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 16.Keri RS, Patil SA, Budagumpi S, Nagaraja BM. Triazole: a promising antitubercular agent. Chem Biol Drug Des. 2015;86(4):410–423. doi: 10.1111/cbdd.12527. [DOI] [PubMed] [Google Scholar]
- 17.Chinthakindi PK, Sangwan PL, Farooq S, Aleti RR, Kaul A, Saxena AK, et al. Diminutive effect on T and B-cell proliferation of non-cytotoxic α-santonin derived 1, 2, 3-triazoles: a report. Eur J Med Chem. 2013;60:365–375. doi: 10.1016/j.ejmech.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Liu Q, Yang X, Xu S, Zhang H, Bai R, et al. Design, synthesis, and biological evaluation of 1, 2, 4-triazole bearing 5-substituted biphenyl-2-sulfonamide derivatives as potential antihypertensive candidates. Bioorg Med Chem. 2013;21(24):7742–7751. doi: 10.1016/j.bmc.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Abuo-Rahma GE-DA, Abdel-Aziz M, Farag NA, Kaoud TS. Novel 1-[4-(Aminosulfonyl) phenyl]-1H-1, 2, 4-triazole derivatives with remarkable selective COX-2 inhibition: design, synthesis molecular docking, anti-inflammatory and ulcerogenicity studies. Eur J Med Chemis. 2014;83:398–408. doi: 10.1016/j.ejmech.2014.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Abdel-Aziz M, Beshr EA, Abdel-Rahman IM, Ozadali K, Tan OU, Aly OM. 1-(4-Methoxyphenyl)-5-(3, 4, 5-trimethoxyphenyl)-1H-1, 2, 4-triazole-3-carboxamides: synthesis, molecular modeling, evaluation of their anti-inflammatory activity and ulcerogenicity. Eur J Med Chem. 2014;77:155–165. doi: 10.1016/j.ejmech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Ding J, Li M, Hou Z, Geng Y, Li X, et al. Discovery of [1, 2, 4]-triazolo [1, 5-a] pyrimidine-7 (4H)-one derivatives as positive modulators of GABAA1 receptor with potent anticonvulsant activity and low toxicity. Eur J Med Chem. 2020;185:111824. doi: 10.1016/j.ejmech.2019.111824. [DOI] [PubMed] [Google Scholar]
- 22.Plech T, Luszczki JJ, Wujec M, Flieger J, Pizoń M. Synthesis, characterization and preliminary anticonvulsant evaluation of some 4-alkyl-1, 2, 4-triazoles. Eur J Med Chem. 2013;60:208–215. doi: 10.1016/j.ejmech.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Vijesh A, Isloor AM, Shetty P, Sundershan S, Fun HK. New pyrazole derivatives containing 1, 2, 4-triazoles and benzoxazoles as potent antimicrobial and analgesic agents. Eur J Med Chem. 2013;62:410–415. doi: 10.1016/j.ejmech.2012.12.057. [DOI] [PubMed] [Google Scholar]
- 24.Kholodnyak SV, Schabelnyk KP, Shernova GA, Sergeieva TY, Ivchuk VV, Voskoboynik OY, et al. Hydrolytic cleavage of the pyrimidine ring in 2-aryl-[1, 2, 4] triazole [1, 5-c] quinazolines: physico-chemical properties and the hypoglycemic activity of the compounds synthesized. News Pharm. 2015;3(83):9–17. [Google Scholar]
- 25.Chelamalla R, Akena V, Manda S. Synthesis of N′-arylidene-2-(5-aryl-1H-1, 2, 4-triazol-3-ylthio) acetohydrazides as antidepressants. Med Chem Res. 2017;26(7):1359–1366. [Google Scholar]
- 26.Radhika C, Venkatesham A, Sarangapani M. Synthesis and antidepressant activity of di substituted-5-aryl-1, 2, 4-triazoles. Med Chem Res. 2012;21(11):3509–3513. [Google Scholar]
- 27.El-Sherief HA, Youssif BG, Bukhari SNA, Abdel-Aziz M, Abdel-Rahman HM. Novel 1, 2, 4-triazole derivatives as potential anticancer agents: design, synthesis, molecular docking and mechanistic studies. Bioorg Chem. 2018;76:314–325. doi: 10.1016/j.bioorg.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Turky A, Bayoumi AH, Sherbiny FF, El-Adl K, Abulkhair HS. Unravelling the anticancer potency of 1, 2, 4-triazole-N-arylamide hybrids through inhibition of STAT3: synthesis and in silico mechanistic studies. Mol Diversity. 2021;25(1):403–420. doi: 10.1007/s11030-020-10131-0. [DOI] [PubMed] [Google Scholar]
- 29.Panda P, Chakroborty S. Navigating the synthesis of quinoline hybrid molecules as promising anticancer agents. ChemistrySelect. 2020;5(33):10187–10199. [Google Scholar]
- 30.Abdelsalam MA, AboulWafa OM, Badawey ESA, El-Shoukrofy MS, El-Miligy MM, Gouda N. Design and synthesis of some β-carboline derivatives as multi-target anticancer agents. Future Med Chemis. 2018;10(24):2791–814. doi: 10.4155/fmc-2018-0226. [DOI] [PubMed] [Google Scholar]
- 31.Radwan RR, Zaher NH, El-Gazzar MG. Novel 1, 2, 4-triazole derivatives as antitumor agents against hepatocellular carcinoma. Chem Biol Interact. 2017;274:68–79. doi: 10.1016/j.cbi.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Mahanti S, Sunkara S, Bhavani R. Synthesis, biological evaluation and computational studies of fused acridine containing 1, 2, 4-triazole derivatives as anticancer agents. Synth Commun. 2019;49(13):1729–1740. [Google Scholar]
- 33.Turky A, Sherbiny FF, Bayoumi AH, Ahmed HE, Abulkhair HS. Novel 1, 2, 4-triazole derivatives: design, synthesis, anticancer evaluation, molecular docking, and pharmacokinetic profiling studies. Arch Pharm. 2020;353(12):2000170. doi: 10.1002/ardp.202000170. [DOI] [PubMed] [Google Scholar]
- 34.Song Z, Liu Y, Dai Z, Liu W, Zhao K, Zhang T, et al. Synthesis and aromatase inhibitory evaluation of 4-N-nitrophenyl substituted amino-4H-1, 2, 4-triazole derivatives. Bioorg Med Chem. 2016;24(19):4723–4730. doi: 10.1016/j.bmc.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Acar Çevik U, Sağlık BN, Osmaniye D, Levent S, Kaya Çavuşoğlu B, Karaduman AB, et al. Synthesis and docking study of benzimidazole–triazolothiadiazine hybrids as aromatase inhibitors. Arch Pharm. 2020;353(5):e2000008. doi: 10.1002/ardp.202000008. [DOI] [PubMed] [Google Scholar]
- 36.Hou Y-P, Sun J, Pang Z-H, Lv P-C, Li D-D, Yan L, et al. Synthesis and antitumor activity of 1, 2, 4-triazoles having 1, 4-benzodioxan fragment as a novel class of potent methionine aminopeptidase type II inhibitors. Bioorg Med Chem. 2011;19(20):5948–5954. doi: 10.1016/j.bmc.2011.08.063. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang X, Chen X, Piatnitski EL, Kiselyov AS, He H-Y, Mao Y, et al. Synthesis and structure–activity relationships of 1, 2, 4-triazoles as a novel class of potent tubulin polymerization inhibitors. Bioorg Med Chem Lett. 2005;15(23):5154–5159. doi: 10.1016/j.bmcl.2005.08.056. [DOI] [PubMed] [Google Scholar]
- 38.Yahiaoui S, Pouget C, Fagnere C, Champavier Y, Habrioux G, Chulia AJ. Synthesis and evaluation of 4-triazolylflavans as new aromatase inhibitors. Bioorg Med Chem Lett. 2004;14(20):5215–5218. doi: 10.1016/j.bmcl.2004.07.090. [DOI] [PubMed] [Google Scholar]
- 39.Mojaddami A, Sakhteman A, Fereidoonnezhad M, Faghih Z, Najdian A, Khabnadideh S, et al. Binding mode of triazole derivatives as aromatase inhibitors based on docking, protein ligand interaction fingerprinting, and molecular dynamics simulation studies. Res Pharm Sci. 2017;12(1):21. doi: 10.4103/1735-5362.199043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedel C, Crafts JM. Sur une nouvelle méthode générale de synthèse d’hydrocarbures, d’acétones, etc. Compt rend. 1877;84:1392. [Google Scholar]
- 41.Wang W, Sheng C, Che X, Ji H, Miao Z, Yao J, et al. Design, synthesis, and antifungal activity of novel conformationally restricted triazole derivatives. Arch Pharm. 2009;342(12):732–739. doi: 10.1002/ardp.200900103. [DOI] [PubMed] [Google Scholar]
- 42.Minlon H. A simple modification of the Wolff-Kishner reduction. J Am Chem Soc. 1946;68(12):2487–2488. [Google Scholar]
- 43.Özdemir A, Sever B, Altıntop MD, Temel HE, Atlı Ö, Baysal M, et al. Synthesis and evaluation of new oxadiazole, thiadiazole, and triazole derivatives as potential anticancer agents targeting MMP-9. Molecules. 2017;22(7):1109. doi: 10.3390/molecules22071109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Z, Yu W, Jia T, Guo S, Geng W, Peng B. Experimental study on the variation of surface widths of lignite desiccation cracks during low-temperature drying. ACS Omega. 2021;6(30):19409–19418. doi: 10.1021/acsomega.1c01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sever B, Altıntop MD, Çiftçi GA, Özdemir A. A new series of triazolothiadiazines as potential anticancer agents for targeted therapy of non-small cell lung and colorectal cancers: design, synthesis, in silico and in vitro studies providing mechanistic insight into their anticancer potencies. Med Chem. 2021;17(10):1104–1128. doi: 10.2174/1573406416666201021142832. [DOI] [PubMed] [Google Scholar]
- 46.Narsimha S, Kumar NS, Swamy BK, Reddy NV, Hussain SA, Rao MS. Indole-2-carboxylic acid derived mono and bis 1, 4-disubstituted 1, 2, 3-triazoles: synthesis, characterization and evaluation of anticancer, antibacterial, and DNA-cleavage activities. Bioorg Med Chem Lett. 2016;26(6):1639–1644. doi: 10.1016/j.bmcl.2016.01.055. [DOI] [PubMed] [Google Scholar]
- 47.Narsimha S, Battula KS, Nukala SK, Gondru R, Reddy YN, Nagavelli VR. One-pot synthesis of fused benzoxazino [1, 2, 3] triazolyl [4, 5-c] quinolinone derivatives and their anticancer activity. RSC Adv. 2016;6(78):74332–74339. [Google Scholar]
- 48.Battula KS, Narsimha S, Thatipamula RK, Reddy YN, Nagavelli VR. Synthesis and biological evaluation of (n-(3-methoxyphenyl)-4-((aryl-1h-1, 2, 3-triazol-4-yl) methyl) thiomorpholine-2-carboxamide 1, 1-dioxide hybrids as antiproliferative agents. ChemistrySelect. 2017;2(29):9595–9598. [Google Scholar]
- 49.Ghosh D, Lo J, Morton D, Valette D, Xi J, Griswold J, et al. Novel aromatase inhibitors by structure-guided design. J Med Chem. 2012;55(19):8464–8476. doi: 10.1021/jm300930n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neves MA, Dinis TC, Colombo G, Sá e Melo ML. Combining computational and biochemical studies for a rationale on the anti-aromatase activity of natural polyphenols. ChemMedChem: Chemistry Enabling Drug Discovery. 2007;2(12):1750–62. doi: 10.1002/cmdc.200700149. [DOI] [PubMed] [Google Scholar]
- 51.Khabnadideh S, Heidari R, Amiri Zirtol L, Sakhteman A, Rezaei Z, Babaei E, et al. Efficient synthesis of 1, 3-naphtoxazine derivatives using reusable magnetic catalyst (GO-Fe3O4–Ti (IV)): anticonvulsant evaluation and computational studies. BMC Chemis. 2022;16(1):1–18. doi: 10.1186/s13065-022-00836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faghih Z, Emami L, Zomoridian K, Sabet R, Bargebid R, Mansourian A, et al. Aryloxy Alkyl theophylline derivatives as antifungal agents: design, synthesis, biological evaluation and computational studies. ChemistrySelect. 2022;7(25):e202201618. [Google Scholar]
- 53.Emami L, Khabnadideh S, Faghih Z, Solhjoo A, Malek S, Mohammadian A, et al. Novel N-substituted isatin-ampyrone Schiff bases as a new class of antiproliferative agents: design, synthesis, molecular modeling and in vitro cytotoxic activity. J Heterocycl Chem. 2022;59(7):1144–1159. [Google Scholar]
- 54.Fereidoonnezhad M, Kaboudin B, Mirzaee T, Babadi Aghakhanpour R, Golbon Haghighi M, Faghih Z, et al. Cyclometalated platinum (II) complexes bearing bidentate O, O′-Di (alkyl) dithiophosphate ligands: photoluminescence and cytotoxic properties. Organometallics. 2017;36(9):1707–1717. [Google Scholar]
- 55.Fereidoonnezhad M, Niazi M, Ahmadipour Z, Mirzaee T, Faghih Z, Faghih Z, et al. Cyclometalated Platinum (II) Complexes Comprising 2-(Diphenylphosphino) pyridine and various thiolate ligands: synthesis, spectroscopic characterization, and biological activity. Eur J Inorg Chem. 2017;2017(15):2247–2254. [Google Scholar]
- 56.Fereidoonnezhad M, Niazi M, Shahmohammadi Beni M, Mohammadi S, Faghih Z, Faghih Z, et al. Synthesis, biological evaluation, and molecular docking studies on the DNA binding interactions of platinum (II) rollover complexes containing phosphorus donor ligands. ChemMedChem. 2017;12(6):456–465. doi: 10.1002/cmdc.201700007. [DOI] [PubMed] [Google Scholar]
- 57.Hashemi S, Jassbi AR, Erfani N, Kiani R, Seradj H. Two new cytotoxic ursane triterpenoids from the aerial parts of Salvia urmiensis Bunge. Fitoterapia. 2021;154:105030. doi: 10.1016/j.fitote.2021.105030. [DOI] [PubMed] [Google Scholar]
- 58.Hashemi S, Seradj H, Kiani R, Jassbi AR, Erfani N. Salvurmin A and Salvurmin B, Two Ursane Triterpenoids of Salvia Urmiensis Induce Apoptosis and Cell Cycle Arrest in Human Lung Carcinoma Cells. Pharmaceutical Sciences. 2022 doi: 10.34172/PS.2022.32. [DOI] [Google Scholar]
- 59.Sadeghian B, Sakhteman A, Faghih Z, Nadri H, Edraki N, Iraji A, et al. Design, synthesis and biological activity evaluation of novel carbazole-benzylpiperidine hybrids as potential anti Alzheimer agents. J Mol Struct. 2020;1221:128793. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request. We have presented all data in the form of Tables and Figure. The PDB code (3EQM) was retrieved from protein data bank (www.rcsb.org). https://www.rcsb.org/structure/3EQM. A549 cells (ATCC No. CCL-185 human lung cancer cell line), Hela cells (ATCC No. CCL-2 human cervical cancer cell line), MCF-7 cells (ATCC No. HTB-22 human breast cancer cell line), MRC-5 cells (ATCC No. CCL-171 human lung tissue),. All the cell lines were purchased from the National Cell Bank of Iran (NCBI, Pasteur Institute, Tehran, Iran).