Abstract
The homodimeric diphtheria toxin repressor (DtxR) uses Fe2+ as a corepressor, binds to iron-regulated promoters, and negatively regulates the syntheses of diphtheria toxin, corynebacterial siderophore, and several other Corynebacterium diphtheriae products. The crystal structure of DtxR shows that the second domain of each monomer has two binding sites for Fe2+ or certain other divalent metal ions. In addition, site 1 binds a sulfate or phosphate anion, suggesting that phosphate may function intracellularly as a co-corepressor. The effects of alanine substitutions for selected residues in sites 1 and 2 were determined by measuring the β-galactosidase activities of a tox operator/promoter-lacZ reporter construct in Escherichia coli strains expressing each DtxR variant. Our studies demonstrated that single alanine substitutions for the anion-binding residues in site 1 (R80A, S126A, or N130A) caused severely decreased DtxR activity, similar to the effects of alanine substitutions for metal-binding residues in site 2 (C102A, E105A, or H106A) and greater than the effects of alanine substitutions for metal-binding residues in site 1 (H79A, E83A, or H98A) reported previously by other investigators. Various combinations of alanine substitutions for site 1 and site 2 residues were also analyzed to further elucidate the roles of these cation- and anion-binding ligands in DtxR activity. Furthermore, the interaction between residue E20 in the DNA binding domain and R80 in anion/cation binding site 1 was analyzed, and the E20A variant of DtxR was shown to have a phenotype indistinguishable from that of the R80A variant. Our data demonstrated for the first time that the anion-binding residues R80, S126, and N130 at site 1 are essential for DtxR activity. The data also showed that the interaction of E20 in domain 1 with R80 in domain 2, first revealed by X-ray crystallography in apo-DtxR and holo-DtxR, is a structural feature of DtxR that is important for its repressor activity.
Diphtheria toxin (DT) production by Corynebacterium diphtheriae is mediated by phage conversion (6, 7), and the structural gene for DT, tox, is present in the genomes of corynebacteriophages such as phage β (1, 37). DT production is influenced by the amount of iron in the growth medium (11, 16), is maximal under conditions of iron starvation, and is significantly decreased under high-iron conditions (5, 13). The DT repressor (DtxR) is a negative regulator that controls the expression of DT and the high-affinity iron uptake system of C. diphtheriae, in addition to other C. diphtheriae gene products (10, 15, 24, 25, 28, 31–33, 35).
The formation of DtxR homodimers is mediated by protein-protein interactions between the monomers, and binding of divalent cations is required for the activation of repressor activity (2, 25, 27, 29, 33, 36, 39). Numerous genetic studies have focused on defining the metal binding domain(s) of DtxR and the mechanism(s) by which divalent cations activate the repressor (3, 34, 39). Saturation site-directed mutagenesis demonstrated that C102 is essential for the activation of DtxR (34), and subsequent random and site-directed mutagenesis studies identified other residues that are important for function of the DNA-binding motif in domain 1 and the two metal binding sites in domain 2 (3, 39).
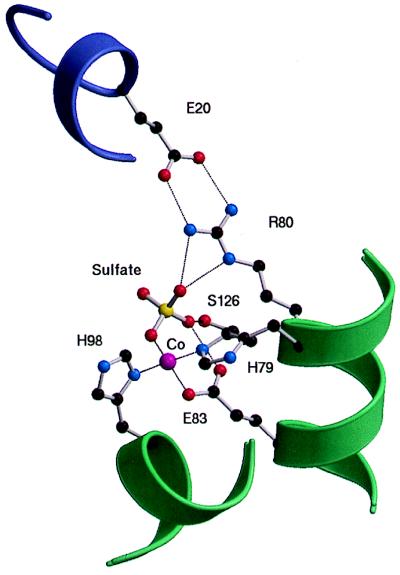
The crystal structure of dimeric DtxR was initially solved at a 2.8-Å resolution in complex with six different divalent transition metals (19). Each subunit was shown to possess an amino-terminal domain that includes a DNA-binding helix-turn-helix motif; an interface domain containing two distinct metal binding sites, which lie 10 Å apart; and a third, flexible carboxyl-terminal domain, which was later shown to be an SH3-like domain that did not appear to be involved in either DNA or metal binding by DtxR (20). High-resolution structures of DtxR complexed with cobalt (2.0 Å) and manganese (2.2 Å) revealed that metal binding site 1 was well occupied in both structures and demonstrated that a sulfate ion served as the fourth ligand for the divalent cation at metal binding site 1 (20). The sulfate anion was also shown to participate in an extensive network of hydrogen bonds with other ligands in DtxR (20). DtxR was crystallized with cobalt at 100 K with a 1.85-Å resolution, providing the highest resolution to date, and at room temperature with zinc at a 2.4-Å resolution, revealing no significant differences between the two structures but providing the most accurate view of the anion/cation binding site (17). In the 1.85-Å cobalt-sulfate-DtxR structure, metal binding site 1 was coordinated tetrahedrally by the Nɛ2 of H79, the Oɛ1 of E83, the Nδ1 of H98, and a sulfate ion. In addition, the Nɛ of R80, the Oγ of S126, and the Oδ1 of N130 were involved in the coordination of the sulfate ion (Fig. 1). The zinc-DtxR crystal was prepared in the absence of sulfate, yet a tetrahedral molecule consistent with a phosphate anion was present in the structure at the anion binding site (17). These results suggested that under physiological conditions phosphate may function as a co-corepressor for DtxR.
FIG. 1.
Close-up view of the anion/cation binding site in the cobalt-DtxR structure determined at a 1.85-Å resolution (17) showing the coordination of the Co2+ and sulfate ions in addition to the interaction between E20 in domain 1 and R80 in the anion/cation binding site of domain 2. Hydrogen bonds are depicted as dashed lines. The helices depicted in green belong to the dimerization domain, and the helix shown in violet represents the DNA binding domain of DtxR. One of the sulfate-coordinating ligands, N130, was omitted (17) to show more clearly the E20-R80 interaction.
Metal binding site 1 was fully occupied in all six of the DtxR-cation complexes, as described above (19, 20). In contrast, metal binding site 2 was highly occupied in wild-type DtxR only when it was in complex with CdCl2, and the metal-binding ligands included the carbonyl group of C102, the Oɛ1 atom of E105, the Nɛ2 atom of H106, and a solvent molecule. The side chain of C102 did not interact with the metal at site 2 but rather reacted with the imidazole ring of H98, one of the direct ligands of metal binding site 1 (19). In these crystals, however, the SH group of C102 appeared to be oxidized (19, 20) or covalently modified (17), which may possibly have altered its capacity to interact with the divalent metal ion at site 2. The structure of a C102D substitution variant of DtxR that retained biological activity was also determined when it was in complex with Ni2+ (3). Both sites 1 and 2 exhibited high occupancy by metal ions in DtxR-C102D, and the metal-binding ligands at site 2 were reported to include the side chains of D102 and M10 and a solvent molecule in addition to the carbonyl oxygen of D102 and the side chains of E105 and H106.
The contributions of the two metal binding sites to the activity of DtxR have been controversial. Variants with alanine substitutions for each of the metal-binding ligands at site 1 and site 2 in DtxR were constructed by site-directed mutagenesis, and only the substitutions at site 2 ligands were found to cause dramatic inactivation of DtxR activity in an Escherichia coli reporter system (3). More recently, the structure of metal-activated DtxR-C102D in complex with an oligonucleotide containing the tox operator sequence was reported at a 3-Å resolution (40). Two dimers of the activated repressor bind to the operator on opposite faces of the DNA, and the main-chain oxygen of residue L4 forms a hydrogen bond with a solvent molecule which is itself a ligand of the metal ion at site 2. Based on these findings, it was proposed that metal binding at site 2 is primarily responsible for activating the repressor activity of DtxR, and the conformational change resulting in activation was proposed to involve a caliper-like rigid body rotation of the DtxR monomers with respect to one another, resulting in decreased distance between their DNA-binding motifs, as well as a helix-to-coil transition at the extreme amino terminus of each DtxR monomer (3, 40).
Both apo-DtxR and holo-DtxR can occur in either of two crystal forms, and structures of form 1 and form 2 crystals of apo-DtxR and holo-DtxR in the presence of zinc were recently determined and compared at resolutions of 2.2 to 2.4 Å (18). The N-terminal DNA binding domain and the last 20 amino acids of the dimerization domain of each subunit were shown to exhibit significant movement with respect to the immobile dimer core as a consequence of binding divalent transition metals. Activation of DtxR, therefore, involves a change in the tertiary structure of DtxR (18) rather than a change in the quaternary structure as proposed by other investigators (3, 23). In addition, both metal binding sites are occupied in activated forms of DtxR; as determined at resolutions or 2.2 Å or less, an anion is present at site 1 in structures of DtxR in complex with metal ions; the R80 residue of binding site 1 interacts with E20 of the DNA binding domain of DtxR (20) (Fig. 1); and the anion-binding ligands Arg80, Ser126, and Asn130 are all conserved in the reported homologs of DtxR (4, 8, 14, 30). Taken together, these findings suggest that anion/cation binding site 1 is an important structural feature of DtxR, and it remains likely that sites 1 and 2 in domain 2 are both important for DtxR function, notwithstanding the conclusions reported previously (3, 40).
In the present study, we used site-directed mutagenesis to construct variants of DtxR with alanine substituted for each of the anion-binding residues at site 1, and we compared the repressor activities of these variants with those of DtxR variants containing alanine substitutions for each of the metal-binding residues at site 1 or site 2 as controls. We also determined the effects of multiple alanine substitutions for these residues, investigated the importance of the interaction between R80 and E20 for DtxR repressor activity, and established the effects of the single or multiple alanine substitutions on the intracellular levels of the variant forms of DtxR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were cultured at 30°C in Luria-Bertani (LB) broth or on LB agar with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), or tetracycline (25 μg/ml) as needed. The ClpP protease-deficient E. coli strain SG22098 was constructed by Susan Gottesman at the National Institutes of Health and was kindly provided by Dorothy E. Pierson at the University of Colorado Health Sciences Center.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | F−supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco BRL |
| CJ236 | dut ung thi relA; pCJ105 (Cmr) | Bio-Rad |
| SG22098 | MC4100 clpP::cat (Cmr) | S. Gottesman |
| Plasmids | ||
| pBluescript SKII(−) | Wild type; Apr | United States Biochemicals |
| pDtxR-7 | dtxR under the transcriptional control of the T7 promoter in pT7-7 | 27 |
| pSKIIdtxR | 1.1 kb BglII-EcoRI fragment from pDtxR-7 in pBluescript SKII(−) | This study |
| pWKS30 | Wild type; Apr | 38 |
| pCMZ100 | tox-lacZ translational fusion in E. coli-C. diphtheriae shuttle vector pCM2.6; Cmr | 26 |
| pTXZ184 | Derivative of pACYC184 carrying tox-lacZ translational fusion; Tcr | 25 |
Construction of recombinant plasmids. (i) Site-specific mutagenesis.
Oligonucleotides containing specific nucleotide substitutions, with respect to the coding sequence for wild-type dtxR, were purchased from Gibco BRL, Gaithersburg, Md., and are shown in Table 2. For single amino acid substitutions, site-specific mutagenesis was performed with the Muta-Gene in vitro mutagenesis kit (Bio-Rad, Hercules, Calif.) and single-stranded DNA generated from pSKIIdtxR in E. coli CJ236. To introduce multiple mutations, single-stranded DNA from clones known to have specific single mutations was subjected to one or more additional rounds of site-directed mutagenesis with different mutagenic oligonucleotides. Putative mutant clones were analyzed by DNA sequencing to confirm the presence of the predicted mutation(s).
TABLE 2.
Mutant oligonucleotides and corresponding DtxR variants
| Oligonucleotidea | DtxR variant |
|---|---|
| Cation-binding ligands of site 1 | |
| 5′-ATGCGTAAAGCTCGCTTAGCT-3′ | H79A |
| 5′-CGCTTAGCTGCGCGCCTTCTT-3′ | E83A |
| 5′-AATAAAGTTGCCGATGAAGCC-3′ | H98A |
| Anion-binding ligands of site 1 | |
| 5′-CGTAAACATGCCTTAGCTGAG-3′ | R80A |
| 5′-GTCAGTCGGGCCCCCTTCGGA-3′ | S126A |
| 5′-CCCTTCGGAGCCCCAATTCCA-3′ | N130A |
| 5′-GAGCTGGAAGCAGAGGGAGTC-3′ | E20A |
| Metal binding site 2 | |
| 5′-CGATGAAGCCGCCCGCTGGGAAC-3′ | C102A |
| 5′-TGCCGCTGGGCACACGTTATG-3′ | E105A |
| 5′-CCGCTGGGAAGCCGTTATGAGTG-3′ | H106A |
| Multiple mutations | |
| 5′-TGCGTAAAGCTGCCTTAGCTGA-3′ | H79A/R80A |
| 5′-GCCTTAGCTGCGCGCCTTCTT-3′ | R80A/E83A |
Boldface print indicates those nucleotides that were changed relative to the wild-type dtxR sequence.
(ii) T7 expression constructs.
For expression in E. coli, wild-type or mutagenized variants of dtxR were transferred into the low-copy-number plasmid vector pWKS30 (38). The resulting constructs possessed various dtxR alleles under the transcriptional control of two T7 promoters. Such constructs express DtxR constitutively at low levels in E. coli in the absence of T7 RNA polymerase (unpublished results).
DNA sequencing and analysis.
Double-stranded plasmid DNA for sequencing was isolated from E. coli DH5α clones, and for each clone the sequence of the segment of the dtxR gene that had been subjected to mutagenesis was determined by the dideoxy chain termination method of Sanger et al. (22). Dideoxy chain termination reactions were performed with T7 polymerase (Sequenase 2.0; United States Biochemicals, Cleveland, Ohio) by using primers MW-1 (5′[326]-CTCATAACGTGTTCCCAGC-3′[308]), MW-2 (5′[555]-AGCATCGAGGAGCTGTGTA-3′[537]), and MCS-2 (5′[158]-TTGTCGTTGTCGCCTCAGA-3′[176]), described previously (39). Reaction products were resolved on 8 M urea–6.6% polyacrylamide gels (21) and visualized by autoradiography.
DtxR antigen analysis.
E. coli DH5α(pCMZ100) and SG22098(pTXZ184) reporter strains expressing wild-type or variant forms of DtxR from pWKS30 constructs were grown overnight in LB broth with aeration at 30°C. Samples containing 109 cells were centrifuged, and the pellets were resuspended in 125 μl of 1× sample buffer (21) and boiled for 5 min. Total cell samples were subjected to sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and Western blot analyses. A sample of polyclonal rabbit antiserum previously raised against a DtxR-MalE fusion protein (29) was adsorbed with an acetone powder prepared from E. coli DH5α (9) before being used to probe blots at a 1:10,000 dilution, followed by a secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce, Rockford, Ill.). The immobilized immune complexes bound to wild-type or mutant forms of DtxR were detected with a chromogenic substrate for the enzyme activity (Renaissance kit; DuPont NEN, Boston, Mass.). The amount of each immunoreactive DtxR variant was determined by scanning the Western blots and analyzing the digitized data by using imaging software from the National Institutes of Health (NIH Image 1.55), and the expression level for each variant was expressed as a percentage relative to wild-type DtxR, taken as 100%. Control Western blots with samples containing serial dilutions of cells expressing wild-type DtxR demonstrated that the amount of immunoreactive DtxR detected was proportional to the number of cells applied and that the signals obtained for the DtxR variants were in the linear range of the assay.
Measurement of β-galactosidase activity.
Because the toxO/P-lacZ translational fusion in pCMZ100 or pTXZ184 is negatively regulated by DtxR, intracellular levels of β-galactosidase activity decreased as DtxR repressor activity increased. β-Galactosidase activity levels were determined as previously described by using o-nitrophenyl-β-d-galactopyranoside as a substrate (12, 21). Levels of β-galactosidase activity were measured in Miller units.
RESULTS AND DISCUSSION
Effect of single site-directed mutations on DtxR activity.
Single substitutions of alanine were made for the anion-binding residues of site 1 (R80A, S126A, and N130A), the metal-binding residues of site 1 (H79A, E83A, and H98A), and the metal-binding residues of site 2 (C102A, E105A, and H106A). Based on the crystallographic finding that residue R80 of domain 2 of DtxR interacts with E20 of domain 1 (17, 20), a single alanine substitution for E20 was also constructed. In addition, selected double- and triple-substitution variants of DtxR were made. The phenotypic effects of these substitutions on the production and activity of DtxR in E. coli were investigated by using a previously described reporter gene system that is responsive to regulation by DtxR and iron (25, 26).
Two reporter strains were used for these analyses. Initially, E. coli DH5α containing the reporter plasmid pCMZ100, which carries a tox-lacZ gene fusion under the transcriptional control of the tox operator/promoter region, was used as the reporter strain to measure the phenotypic effects of various dtxR alleles expressed from a low-copy-number vector (pWKS30; ∼8 copies per cell [38]). In LB medium (high iron conditions), in the presence of the appropriate antibiotics to ensure stable maintenance of the plasmids, β-galactosidase activity was fully repressed by wild-type DtxR; the increased β-galactosidase activity of a strain containing a variant form of DtxR could reflect either decreased DtxR activity, a decreased amount of DtxR, or both. Quantitative analysis of DtxR by the scanning of Western blots and by image analysis showed that the DtxR variants were not expressed as much as wild-type DtxR in E. coli DH5α, although the variant proteins were usually present at more than 50% of the wild-type level (Tables 3 to 5). To minimize the effects of the decreased expression levels of the DtxR variants on the observed phenotypes, the constructs expressing each DtxR variant were transferred into the protease-deficient clpP mutant strain E. coli SG22098 containing the reporter plasmid pTXZ184, which carries a tox-lacZ gene fusion under the transcriptional control of the tox operator/promoter region and expresses tetracycline resistance (25). In this clpP mutant strain of E. coli, almost all of the DtxR variants were expressed at levels between 70 and 100% of the wild-type DtxR expression level (Tables 3 to 5).
TABLE 3.
Effects of single mutations within DtxR
| DtxR construct | DH5α(pCMZ100)
|
SG22098(pTXZ184)
|
||
|---|---|---|---|---|
| β-Galacto-sidase activitya,c | DtxR quanti-tationb | β-Galacto-sidase activitya,c | DtxR quanti-tationb | |
| Controls | ||||
| pWKS30 | 32 ± 2.5 | 0 | 42 ± 1.0 | 0 |
| dtxR-7 | <0.01 | 100 | 2.3 ± 1.0 | 100 |
| Site 1 metal-coordi-nating ligandsd | ||||
| H79A | 8.2 ± 0.9 | 78 | 4.3 ± 2.9 | 94 |
| E83A | 11 ± 0.5 | 67 | 10 ± 0.6 | 94 |
| H98A | 5.4 ± 1.8 | 82 | 2.9 ± 1.7 | 98 |
| Site 2 metal-coordi-nating ligandsd | ||||
| C102A | 35 ± 5.7 | 67 | 33 ± 0.1 | 69 |
| E105A | 27 ± 2.7 | 100 | 25 ± 6.5 | 86 |
| H106A | 32 ± 1.7 | 100 | 34 ± 5.8 | 92 |
| Site 1 anion-coordi-nating ligands | ||||
| R80A | 23 ± 4.9 | 60 | 20 ± 2.0 | 93 |
| S126A | 20 ± 2.1 | 54 | 14 ± 2.6 | 91 |
| N130A | 23 ± 2.6 | 49 | 19 ± 1.7 | 73 |
β-Galactosidase activities are given as Miller units.
Quantities of the DtxR variants are given as percentages of the expression of wild-type DtxR.
Assays were performed in triplicate with three separate cultures; mean values ± standard deviations are given.
These amino acid substitutions have been analyzed previously (3) but were included in this analysis as controls.
TABLE 5.
Effects of multiple mutations on DtxR activity
| DtxR construct | DH5α(pCMZ100)
|
SG22098(pTXZ184)
|
||
|---|---|---|---|---|
| β-Galacto-sidase activitya,c | DtxR quanti-tationb | β-Galacto-sidase activitya,c | DtxR quanti-tationb | |
| pWKS30 | 32 ± 2.5 | 0 | 42 ± 1.0 | 0 |
| dtxR-7 | <0.01 | 100 | 2.3 ± 1.0 | 100 |
| H79A/E83A | 31 ± 4.5 | 0 | 39 ± 6.6 | 0 |
| H79A/H98A | NDd | 0 | 36 ± 6.4 | 0 |
| H98A/E83A | 9.2 ± 2.6 | 64 | 1.6 ± 1.8 | 100 |
| N130A/S126A | 28 ± 4.6 | ND | 26 ± 3.6 | ND |
| R80A/H79A/H98A | 40 ± 2.4 | 58 | 33 ± 6.4 | 67 |
| H79A/E20A/R80A | 36 ± 3.2 | 56 | 28 ± 12.3 | 67 |
| R80A/H98A/E83A | 19 ± 2.5 | 67 | 11 ± 1.5 | 88 |
β-Galactosidase activities are given as Miller units.
Quantities of DtxR variants are given as percentages of the expression of wild-type DtxR.
Assays were performed in triplicate with three separate cultures; mean values ± standard deviations are given.
ND, not determined.
Levels of β-galactosidase activity were measured in both reporter systems for each of the DtxR variants with a single alanine substitution (Table 3). The amino acid substitutions within DtxR are designated by the one-letter code for the wild-type amino acid, its number in the sequence of the DtxR polypeptide, and the one-letter code for the substituting alanine (e.g., H79A for the variant with the alanine replacing histidine at residue 79). In the absence of dtxR (pWKS30 control), β-galactosidase was expressed maximally from the toxO/P-lacZ gene fusion. No β-galactosidase activity was detectable in the presence of wild-type dtxR, demonstrating the complete repression of expression from the tox operator/promoter by DtxR. The β-galactosidase activity levels measured for the variants expressed in either reporter strain were similar, and both data sets are shown in Table 3. These data demonstrated that the levels of β-galactosidase expression from the reporter plasmid were determined by the effects of the alanine substitutions on DtxR activity and that variations in the level of expression of DtxR protein did not determine the observed DtxR activity in these experiments.
Recent crystallographic studies of DtxR demonstrated that binding site 1 coordinates both a divalent cation and an anion (sulfate or phosphate) (17, 20), and sequence analysis revealed that the residues involved in anion coordination (Arg80, Ser126, and Asn130) are conserved among members of the DtxR family (4, 8, 14, 30), suggesting that the binding of the anion might be important for DtxR activity (17). We therefore examined the effects of substituting an alanine residue for each of the residues involved in anion binding at site 1 (Table 3). The alanine substitution variants designated R80A, S126A, and N130A each had dramatically decreased repressor activity, and the phenotypes of these variants were similar. It was reported previously that substitution of alanine for the metal-binding residues of site 2 (C102, E105, or H106) abolished DtxR repressor activity and that substitution of alanine for the metal-binding residues of site 1 had no effect (H79A and H98A) or a very slight effect (E83A) on DtxR activity under high-iron conditions (3). We confirmed that alanine substitutions for metal-binding residues at site 2 completely inactivated repressor activity, and it was striking that the phenotypes of variants with alanine substitutions for the anion-binding residues at site 1 were almost as dramatic as those of variants with substitutions for the metal-binding residues at site 2. The E83A, H79A, and H98A variants all had significantly impaired repressor activity in E. coli DH5α, but only the E83A variant had significantly decreased repressor activity in E. coli SG22098. The impaired activity of the E83A variant in E. coli DH5α was not caused by its proteolytic degradation, because the mutant phenotype persisted when it was present in E. coli SG22098 at 94% of the wild-type DtxR level. In contrast, proteolytic degradation did contribute to the decreased repressor activity of H79A and H98A in E. coli DH5α, since their repressor activities were indistinguishable from that of wild-type DtxR when they were present in E. coli SG22098 at 94 to 98% of the wild-type DtxR level. Our findings that alanine substitutions for the anion-binding residues R80, S126, and N130 cause dramatic inactivation of DtxR activity and that the alanine substitution for the metal-binding residue E83 causes moderate inactivation of DtxR activity demonstrate a significant role for the anion- and metal-binding ligands of site 1, as well as for the metal-binding ligands of site 2, in the biological activity of DtxR.
These data provide strong evidence that both sites 1 and 2 are necessary for the activity of DtxR and establish for the first time that the anion-binding ligands at site 1 are essential for DtxR activity. Single alanine substitutions for the metal-binding ligands at site 2 completely abolished DtxR activity, in confirmation of previous findings (3, 34). Substitution of alanine for the metal-binding ligand E83 of site 1 had a significant effect on DtxR activity in our reporter system under high-iron conditions that was greater than the effect reported previously for a similar DtxR E83A variant (3).
Comparison of the effects of substitutions for residues R80 and E20.
Recent crystallographic studies also demonstrated that R80 forms hydrogen bonds not only with the sulfate anion in site 1 but also with E20 in domain 1 of DtxR (Fig. 1). To determine whether the interaction between E20 and R80 is required for DtxR activity, the effects of alanine substitutions for E20 and R80 were tested both individually and in combination with alanine substitutions for other cation- or anion-binding residues in site 1 (Table 4). It was presumed that if an interaction between R80 and E20 is important for the function of the DtxR molecule, an E20A substitution might give the same results as an R80A substitution. This proved to be the case, as is indicated in Table 4. The effects of the single R80A and E20A substitutions on DtxR activity were indistinguishable and resulted in a dramatic decrease of repressor activity. It is important to note that random substitutions within the first domain of DtxR do not necessarily affect the activity of DtxR. In 1994, it was shown that several amino acid substitutions within domain 1, including T7I, R13C, E19K, T24I, T44I, and T67I, had little to no effect on DtxR activity (39).
TABLE 4.
Comparison of the R80A and E20A substitutions in DtxR, alone and in combination with each other or with substitutions for other site 1 ligands
| DtxR construct | DH5α(pCMZ100)
|
SG22098(pTXZ184)
|
||
|---|---|---|---|---|
| β-Galacto-sidase activitya,c | DtxR quanti-tationb | β-Galacto-sidase activitya,c | DtxR quanti-tationb | |
| Controls | ||||
| pWKS30 | 32 ± 2.5 | 0 | 42 ± 1.0 | 0 |
| dtxR-7 | <0.01 | 100 | 2.3 ± 1.0 | 100 |
| Substitutions for R80 and E20 | ||||
| R80A | 23 ± 4.9 | 60 | 20 ± 2.0 | 93 |
| E20A | 19 ± 3.5 | 53 | 21 ± 2.3 | 72 |
| R80A/E20A | 20 ± 3.5 | 82 | 21 ± 0.5 | 91 |
| Substitutions for R80 and other site 1 ligands | ||||
| R80A/H79A | 26 ± 4.3 | 80 | 30 ± 3.9 | 81 |
| R80A/E83A | 15 ± 2.5 | 81 | 8.5 ± 1.3 | 100 |
| R80A/H98A | 19 ± 1.5 | 81 | 18 ± 5.2 | 91 |
| R80A/S126A | 29 ± 2.6 | 74 | 33 ± 6.3 | 85 |
| R80A/N130A | 23 ± 1.7 | 71 | 21 ± 2.4 | 79 |
| Substitutions for E20 and site 1 ligands | ||||
| E20A/H79A | 30 ± 4.2 | 73 | 31 ± 1.5 | 55 |
| E20A/E83A | 24 ± 2.3 | 76 | 26 ± 2.5 | 92 |
| E20A/H98A | 22 ± 4.7 | 88 | 19 ± 1.6 | 100 |
| E20A/S126A | 30 ± 4.7 | 79 | 40 ± 0.3 | 96 |
| E20A/N130A | 23 ± 4.4 | 74 | 26 ± 4.4 | 90 |
β-Galactosidase activities are given as Miller units.
Quantities of the DtxR variants are given as percentages of the expression of wild-type DtxR.
Assays were performed in triplicate with three separate cultures; mean values ± standard deviations are given.
The multiple amino acid substitutions within DtxR are designated as indicated above for the single amino acid substitutions, with a slash separating the individual alanine substitutions (e.g., R80A/E20A for the variant with alanines replacing arginine at residue 80 and glutamic acid at residue 20). The combination of E20A and R80A substitutions within the same DtxR molecule had the same effect as either substitution alone and therefore did not display a cumulative effect (Table 4). The effects of the R80A and E20A substitutions on DtxR activity were also similar whether they were present alone or in combination with single alanine substitutions for other ligands of the anion/cation binding site, except for the R80A/E83A combination, which caused less inactivation of DtxR repressor activity than did the R80A substitution alone (Table 4). Taken together, these results indicate that the specific interaction between the R80 and E20 residues, originally detected by X-ray crystallography (20), is important for DtxR activity.
Effect of multiple substitutions within DtxR metal binding site 1.
Single amino acid substitutions of the metal-coordinating ligands of site 1 did not affect DtxR activity as dramatically as did substitutions of the amino acids involved in coordination of the anion. To further assess the relative requirements for the various residues involved in metal and anion binding at site 1, additional multiple substitutions were constructed and analyzed (Table 5) as described above. Interestingly, the effect of the H98A/E83A double substitution in strain DH5α was no greater than the effect of either mutation alone, and in strain SG22098 the H98A/E83A variant was similar to wild-type DtxR both in repressor activity and in the level of expression of the mutant protein. In striking contrast, both the H79A/E83A and H79A/H98A double-substitution variants had no DtxR activity and were not detectable as immunoreactive proteins in Western blots. Double substitutions for residues involved in the coordination of the anion at site 1 (N130A/S126A [Table 5] and R80A/S126A and R80A/N130A [Table 4]) had no greater effect than the substitutions for each residue individually. The triple substitutions R80A/H79A/H98A and H79A/E20A/R80A completely inhibited DtxR repressor activity and had greater effects than any of the single or double substitutions within the same residues, although the triple-substitution variants were produced in amounts nearly comparable to those of the single- and double-substitution variants (Table 5). In contrast, the R80A/H98A/E83A triple substitution had no greater effect on DtxR activity than did the R80A substitution alone (compare Tables 4 and 5). This was consistent with the weak effect of the H98A/E83A double substitution and the similarity in effects of the R80A/H98A and R80A/E83A double substitutions with that of the R80A substitution alone. These results further show the requirement of anion/cation binding site 1 for DtxR activity and suggest that various combinations of alanine substitutions for the ligands affect the activity of the DtxR molecule to various degrees.
In summary, the studies presented here show that binding sites 1 and 2 within the second domain are both important for the biological activity of DtxR. The present study demonstrated for the first time that residues R80, S126, and N130, which are involved in anion coordination at binding site 1, are essential for DtxR activity. In addition, our data provide strong evidence that the interaction between E20 in domain 1 and R80 in the anion/cation binding site 1 of domain 2, which is present both in apo-DtxR and in holo-DtxR, is an important structural element for DtxR activity.
ACKNOWLEDGMENTS
This research was supported in part by Public Health Service grants R01 AI14107 to R.K.H., 1 F32 AI10038-01 to J.G.-S., and RO1 CA65656 to W.G.J.H. and by a Schering Forschungsgesellschaft postdoctoral fellowship to E.P.
REFERENCES
- 1.Barksdale L, Arden S B. Persisting bacteriophage infections, lysogeny, and phage conversion. Annu Rev Microbiol. 1974;28:265–299. doi: 10.1146/annurev.mi.28.100174.001405. [DOI] [PubMed] [Google Scholar]
- 2.Boyd J, Oza M N, Murphy J R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding X, Zeng H, Schiering N, Ringe D, Murphy J R. Identification of the primary metal ion-activation sites of the diphtheria tox repressor by X-ray crystallography and site-directed mutational analysis. Nat Struct Biol. 1996;3:382–387. doi: 10.1038/nsb0496-382. [DOI] [PubMed] [Google Scholar]
- 4.Doukhan L, Predich M, Nair G, Dussurget O, Mandic-Mulec I, Cole S T, Smith D R, Smith I. Genomic organization of the mycobacterial sigma gene cluster. Gene. 1995;165:67–70. doi: 10.1016/0378-1119(95)00427-8. [DOI] [PubMed] [Google Scholar]
- 5.Edwards D C, Seamer P A. The uptake of iron by Corynebacterium diphtheriae growing in submerged medium. J Gen Microbiol. 1960;22:715–721. doi: 10.1099/00221287-22-3-705. [DOI] [PubMed] [Google Scholar]
- 6.Freeman V J. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 1951;61:675–688. doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman V J, Morse I U. Further observations on the change to virulence of bacteriophage-infected avirulent strains of Corynebacterium diphtheriae. J Bacteriol. 1952;63:407–414. doi: 10.1128/jb.63.3.407-414.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunter-Seeboth K, Schupp T. Cloning and sequence analysis of the Corynebacterium diphtheriae dtxR homologue from Streptomyces lividans and S. pilosus encoding a putative iron repressor protein. Gene. 1995;166:117–119. doi: 10.1016/0378-1119(95)00628-7. [DOI] [PubMed] [Google Scholar]
- 9.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 10.Lee J H, Wang T, Ault K, Liu J, Schmitt M P, Holmes R K. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:4273–4280. doi: 10.1128/iai.65.10.4273-4280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke A, Main E R. The relation of copper and iron to the production of toxin and enzyme action. J Infect Dis. 1931;48:419–435. [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 13.Mueller J H, Miller P A. Production of diphtheria toxin of high potency (100 LF) on a reproducible medium. J Immunol. 1941;40:21–32. [Google Scholar]
- 14.Oguiza J A, Tao X, Marcos A T, Martín J F, Murphy J R. Molecular cloning, DNA sequence analysis, and characterization of the Corynebacterium diphtheriae dtxR homolog from Brevibacterium lactofermentum. J Bacteriol. 1995;177:465–467. doi: 10.1128/jb.177.2.465-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pappenheimer A M., Jr Diphtheria toxin. Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 16.Pappenheimer A M, Jr, Johnson S J. Studies in diphtheria toxin production. I. The effect of iron and copper. Br J Exp Pathol. 1936;17:335–341. [Google Scholar]
- 17.Pohl E, Qiu X, Must L M, Holmes R K, Hol W G J. Comparison of high-resolution structures of the diphtheria toxin repressor in complex with cobalt and zinc at the cation-anion binding site. Protein Sci. 1997;6:1114–1118. doi: 10.1002/pro.5560060519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pohl E, Holmes R K, Hol W G J. Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor in the crystal structures of apo- and holo-DtxR. J Biol Chem. 1998;273:22420–22427. doi: 10.1074/jbc.273.35.22420. [DOI] [PubMed] [Google Scholar]
- 19.Qiu X, Verlinde C L M J, Zhang S, Schmitt M P, Holmes R K, Hol W G J. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure. 1995;3:87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu X, Pohl E, Holmes R K, Hol W G J. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry. 1996;35:12292–12302. doi: 10.1021/bi960861d. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiering N, Tao X, Zeng H, Murphy J R, Petsko G A, Ringe D. Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1995;92:9843–9850. doi: 10.1073/pnas.92.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt M P. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect Immun. 1997;65:4634–4641. doi: 10.1128/iai.65.11.4634-4641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt M P, Holmes R K. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt M P, Holmes R K. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect Immun. 1991;59:3903–3908. doi: 10.1128/iai.59.11.3903-3908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt M P, Holmes R K. Analysis of diphtheria toxin repressor-operator interactions and characterization of a mutant repressor with decreased binding activity for divalent metals. Mol Microbiol. 1993;9:173–181. doi: 10.1111/j.1365-2958.1993.tb01679.x. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt M P, Holmes R K. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J Bacteriol. 1994;176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt M P, Twiddy E M, Holmes R K. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci USA. 1992;89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt M P, Predich M, Doukhan L, Smith I, Holmes R K. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect Immun. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt M P, Talley B G, Holmes R K. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:5364–5367. doi: 10.1128/iai.65.12.5364-5367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai S-P, Kraft A E, Nootheti P, Holmes R K. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb Pathog. 1990;9:267–273. doi: 10.1016/0882-4010(90)90015-i. [DOI] [PubMed] [Google Scholar]
- 33.Tao X, Murphy J R. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J Biol Chem. 1992;267:21761–21764. [PubMed] [Google Scholar]
- 34.Tao X, Murphy J R. Cysteine-102 is positioned in the metal binding activation site of the Corynebacterium diphtheriae regulatory element DtxR. Proc Natl Acad Sci USA. 1993;90:8524–8528. doi: 10.1073/pnas.90.18.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao X, Nikolaus S, Zeng H, Ringe D, Murphy J R. Iron, DtxR and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 36.Tao X, Zeng H, Murphy J R. Transition metal ion activation of DNA binding by the diphtheria tox repressor requires the formation of stable homodimers. Proc Natl Acad Sci USA. 1995;92:6803–6807. doi: 10.1073/pnas.92.15.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida T, Gill D M, Pappenheimer A M., Jr Mutation in the structural gene for diphtheria toxin carried by temperate phage β. Nat New Biol. 1971;233:8–11. doi: 10.1038/newbio233008a0. [DOI] [PubMed] [Google Scholar]
- 38.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 39.Wang Z, Schmitt M P, Holmes R K. Characterization of mutations that inactivate the diphtheria toxin repressor gene (dtxR) Infect Immun. 1994;62:1600–1608. doi: 10.1128/iai.62.5.1600-1608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White A, Ding X, vanderSpek J C, Murphy J R, Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998;394:502–506. doi: 10.1038/28893. [DOI] [PubMed] [Google Scholar]