Abstract
Synthetic green fluorescent protein (GFP) was used as a reporter to detect differential gene expression in the pathogenic fungus Cryptococcus neoformans. Promoters from the C. neoformans actin, GAL7, or mating-type alpha pheromone (MFα1) genes were fused to GFP, and the resulting reporter genes were used to assess gene expression in serotype A C. neoformans. Yeast cells containing an integrated pACT::GFP construct demonstrated that the actin promoter was expressed during vegetative growth on yeast extract-peptone-dextrose medium. In contrast, yeast cells containing the inducible GAL7::GFP or MFα1::GFP reporter genes expressed significant GFP activity only during growth on galactose medium or V-8 agar, respectively. These findings demonstrated that the GAL7 and MFα1 promoters from a serotype D C. neoformans strain function when introduced into a serotype A strain. Because the MFα1 promoter is induced by nutrient deprivation and the MATα locus containing the MFα1 gene has been linked with virulence, yeast cells containing the pMFα1::GFP reporter gene were analyzed for GFP expression in the central nervous system (CNS) of immunosuppressed rabbits. In fact, significant GFP expression from the MFα1::GFP reporter gene was detected after the first week of a CNS infection. These findings suggest that there are temporal, host-specific cues that regulate gene expression during infection and that the MFα1 gene is induced during the proliferative stage of a CNS infection. In conclusion, GFP can be used as an effective and sensitive reporter to monitor specific C. neoformans gene expression in vitro, and GFP reporter constructs can be used as an approach to identify a novel gene(s) or to characterize known genes whose expression is regulated during infection.
The number of invasive fungal infections has been increasing due to the growing number of immunocompromised patients worldwide. Cryptococcus neoformans is an encapsulated yeast that has become a significant human pathogen in individuals immunosuppressed by human immunodeficiency virus infection, malignancies, or organ transplants and in individuals receiving long-term treatment with corticosteroids. C. neoformans may also infect apparently healthy hosts. With these pathogenic features, C. neoformans has become a model yeast for the study of virulence factors of both primary and secondary fungal pathogens.
C. neoformans infection begins in the lung following the inhalation of yeasts or basidiospores and then spreads hematogenously to the brain, which results in life-threatening meningoencephalitis in high-risk individuals (5, 16). The pathogenesis of cryptococcosis is primarily influenced by three factors: (i) the status of the host defenses, (ii) the virulence of the C. neoformans strain, and (iii) the size of the inoculum. Numerous studies have documented the importance of host defenses and inoculum sizes by both experimental and clinical observations (8, 12, 15, 19, 25, 33). On the other hand, the importance of strain variation and the genetic basis of virulence have just begun to be explored. For instance, the study of the molecular pathogenesis of C. neoformans has recently been advanced by the introduction of new molecular tools and genetic analyses such as high-frequency transformation systems, site-directed gene disruption protocols, and genomic methods to capture differential gene expression at the site of infection (2, 9, 14, 27, 28, 38). These molecular strategies can now be used to identify the expression of specific genes associated with infection and then confirm their importance for virulence in animal models. The identification of these virulence genes and the genetic circuits which control expression will allow a better understanding of fungal pathogenesis in cryptococcosis and possibly allow researchers to exploit the concept that specific virulence genes might be used as novel targets for new antifungal drugs or vaccine development. Moreover, the development and adaptation of new technologies that allow the monitoring of the gene expression of C. neoformans in vivo will have an important impact on investigations of general fungal pathogenesis in the era of functional genomics (22).
To identify and characterize in vivo gene expression patterns for fungal pathogens, it will be particularly important to use relevant animal models. A series of excellent animal models has been developed for C. neoformans. For instance, the rabbit model of cryptococcal meningitis has been well established and shares many features with human cryptococcosis: (i) immunosuppression is required, (ii) cerebrospinal fluid (CSF) leukopenia develops, (iii) the infection is prolonged and eventually fatal, (iv) dissemination to multiple organs occurs, and (v) response to treatment regimens for cryptococcal meningitis parallels those in recent human trials. Furthermore, CSF can be continuously sampled throughout the infection and thus provides a “biological window” during studies of host-regulated gene expression (21, 23, 24, 26).
The green fluorescent protein (GFP) from the jellyfish Aequorea victoriae has been developed (1) and expressed as a reporter in a variety of heterologous systems, including Escherichia coli, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, mammals, and plants (1, 6, 30, 42). Cormack et al. have isolated a synthetic GFP (yEGFP3) that generates much more fluorescence than wild-type GFP in the fungi S. cerevisiae and Candida albicans (3). In this study, we used this synthetic GFP as a reporter to analyze in vitro and in vivo specific gene expression of C. neoformans. This study illustrates how promoter fusions can be used to monitor regulated gene expressions in C. neoformans during host infection. We also demonstrate that the expression of genes such as the mating-type alpha pheromone (MFα1) gene are regulated by the length and/or stage of infection. Therefore, it will be important to serially follow C. neoformans cells and their genetic expression in order to understand gene expressions which are regulated during infection; the rabbit model of cryptococcal meningitis allows continuous yeast cell sampling from the site of infection and is thus ideally suited for these studies.
MATERIALS AND METHODS
Strains and media.
C. neoformans M001, an ade2 auxotroph of H99, was used as the recipient of biolistic transformation. The pYGFP3 plasmid, containing the synthetic GFP, made by Cormack et al. (3) and the C. albicans CAI4, containing the aldehyde dehydrogenase::GFP expression plasmid (ADH1-yEGFP3), were gifts from Aaron P. Mitchell. C. albicans A39 and serotype A C. neoformans H99 were used as negative control strains. C. albicans strains and C. neoformans H99 and M001 were routinely grown on enriched medium (yeast extract-peptone-dextrose [YEPD]). V-8 starvation medium contained 5% V-8 vegetable juice (Campbell’s Soup Co.), 0.5 g of KH2PO4 per liter, and 4% agar and was adjusted to pH 7.2 with KOH before autoclaving. C. neoformans isolates transformed with pACT::GFP/ADE2 and pMFα1::GFP/ADE2 were selected on synthetic medium containing 6.7 g of yeast nitrogen base without amino acids (YNB w/o) per liter, 1.3 g of amino acid mix lacking adenine per liter, 180 g of sorbitol per liter, 20 g of glucose per liter, and 20 g of agar per liter. C. neoformans isolates transformed with pGAL7::GFP/ADE2 were selected on synthetic medium containing 6.7 g of YNB w/o per liter, 1.3 g of amino acid mix lacking adenine per liter, 180 g of sorbitol per liter, 180 g of sorbitol per liter, 20 g of galactose per liter, and 20 g of agar per liter. YNB-glucose and YNB-galactose media contained 6.7 g of YNB w/o per liter, 1.3 g of amino acid mix lacking adenine per liter, 20 g of agar per liter, and 20 g of glucose or galactose per liter, respectively.
Construction of plasmids to examine gene expression in C. neoformans.
Three cryptococcal promoters from the following genes were used: the actin gene isolated from serotype A strain H99 (4), the GAL7 gene (40), and an MFα1 gene from serotype D strain JEC21 (18). The GAL7 promoter was isolated from the plasmid pAUG-MF, originally cloned by Wickes and Edman (40), with PCR using two primers containing HindIII restriction sites (in bold and underlined): 6G, 5′-GAC CAA GCT TGT GGA AAG AAG CAG GTC TTG TCGA-3′, and 6H, 5′-GGC TAA GCT TTC TCA AGA GGG GAT TGA GCG CTGA-3′. PCR conditions were 95°C for 5 min (1 cycle); 93°C for 50 s, 50°C for 50 s, and 72°C for 80 s (25 cycles); and 72°C for 2 min (1 cycle). This amplification strategy produced a 585-bp fragment which was digested with HindIII and inserted into the HindIII site of pYGFP3. The C. neoformans ADE2 gene from strain B3501 was then inserted downstream from the GFP gene into an EcoRI site to yield plasmid pGAL7::GFP/ADE2 (Fig. 1A).
FIG. 1.
Construction of GAL7::GFP/ADE2, ACT::GFP/ADE2, and MFα1::GFP/ADE2 expression plasmids and the corresponding nucleotide sequences of the junctions.
The second fusion construct, pACT::GFP/ADE2, was engineered by cloning a HindIII-restricted, partially filled-in, and EcoRI-restricted 738-bp GFP fragment from pYGFP3 into an XbaI-restricted, partially filled-in, and EcoRI-restricted site of the expression plasmid pACT::lacZ/ADE2 (37) from which the 5.7-kb lacZ fragment had been deleted (Fig. 1B).
The third fusion construct, pMFα1::GFP/ADE2, was generated by cloning a HindIII- and EcoRI-restricted and blunt-ended 738-bp GFP fragment into a SalI restricted and filled-in site located at the 3′ promoter region of a putative pheromone gene, MFα1, using the pΔMFα1 plasmid. Briefly, the pΔMFα1 plasmid was made in two steps. First, the 2.1-kb fragment from the MATα locus of C. neoformans serotype D, strain JEC21, was generated by PCR with genomic DNA as the template, primer 1 (5′-TCG ACT ATC TAG AAA GCT TGG ATG TGA ATG CTAAA-3′), and primer 4 (5′-AGT TAA AGC AGT TTA TAG TGCA-3′). This fragment was cloned into pBluescript SK and the resulting plasmid was named pMFα1. Then, fragment A was generated by PCR with pMFα1 as the template and primers 1 and 2 (5′-CCGT AGA GTCGAC GGC AGT ATT GTA ACTGG-3′), which contains a SalI site (bold and underlined). Fragment B was generated by PCR with pMFα-1 as the template and the primers 4 and 3 (5′-CTGCC GTCGAC TCT ACG GTA GAC CCA ACG TCC CCT CTGC-3′), which also contains a SalI site (bold and underlined). Fragments A and B were combined and used as the template for PCR overlapping with primers 1 and 4, generating fragment C, which contains a new SalI site at the 3′ end of the MFα-1 promoter and the deletion of 114 bp of the open reading frame. This fragment was cloned and sequenced to make sure that no mutations were introduced by PCR manipulations, and the resulting plasmid was named pΔMFα1. Then, the HindIII- and EcoRI-restricted and blunt-ended 738-bp GFP fragment was cloned into the SalI-restricted and filled-in site of the pΔMFα-1 plasmid, generating the pMFα1::GFP plasmid. Finally, the ADE2 gene was inserted into an EcoRI site downstream from the pheromone gene, generating the pMFα1::GFP/ADE2 construct (Fig. 1C).
Nucleotide sequencing.
Sequencing was performed by the dideoxy chain termination method (35) with Sequenase, version 2.0 (Amersham Life Science, Cleveland, Ohio).
Transformation.
The three constructs were transformed into C. neoformans M001 by biolistic delivery of DNA following the protocol described by Toffaletti et al. (38). Adenine prototrophic transformants were selected on synthetic medium (1 M sorbitol) lacking adenine at 30°C, as described above. Adenine transformants were subcultured onto selective medium (YNB-glucose or YNB-galactose) and then passaged twice on YEPD agar. Stable adenine transformants, selected by the retention of a white colony color phenotype, were stored at 4°C.
Analysis of transformants.
Genomic DNA was isolated from each transformant as follows: yeast cells from a 10-ml mid- to late-log-phase YEPD broth culture were pelleted, transferred to a 2-ml screw-cap tube, and washed once in 1.5 ml of sterile distilled water. Cells were resuspended in 0.5 ml of TENTS (10 mM Tris [pH 7.5], 1 mM EDTA [pH 8.0], 100 mM NaCl, 2% Triton X-100, 1% sodium dodecyl sulfate) with a toothpick. Five milligrams of glass beads (diameter, 0.5 mm) and 0.5 ml of phenol-chloroform were added, and samples were vortexed for 2 min and centrifuged for 10 min in a microcentrifuge. The aqueous phase was transferred to a fresh tube, and DNA was precipitated by the addition of 2 volumes of 100% ethanol and incubated at −20°C for 10 min. DNA was pelleted, resuspended in 0.5 ml Tris-EDTA (pH 8.0) containing 10 μg of RNAse A per ml, and incubated at 37°C for 20 min. DNA was extracted once with phenol-chloroform, reprecipitated, washed with 70% ethanol, resuspended in 100 μl of Tris-EDTA, and stored at −20°C.
The integration of the fusion constructs was analyzed by Southern blot analysis (34). Briefly, 1 μg of genomic DNA, either undigested or digested with appropriate restriction enzymes, was electrophoresed in a 0.7% agarose gel, transferred to a nitrocellulose membrane, and probed with fragments carrying the GFP gene and the respective cryptococcal promoter. These DNA fragments carrying the GFP and the GAL7, actin, or pheromone promoters were labeled with [32P]dCTP (New England Nuclear) by using a random primer labeling kit (Gibco-BRL).
In vitro promoter expression.
Three stable transformants, each carrying the GFP gene fused to either the actin (Cn-ACT::GFP), GAL7 (Cn-GAL7::GFP), or pheromone (Cn-MFα1::GFP) promoter and integrated into the genome, were examined for the ability to express GFP when grown on enriched or selective medium. A Cn-ACT::GFP transformant was inoculated onto YEPD agar, and a Cn-MFα1::GFP transformant was inoculated onto both YEPD and V-8 agars. A Cn-GAL7::GFP transformant was inoculated onto both YNB-galactose and YNB-glucose agars. Yeast cells were incubated for 3 days at 30°C and assessed for GFP expression by fluorescent microscopy and flow cytometry. Wild-type H99, propagated on YEPD, was used as a negative C. neoformans control strain. C. albicans A39 and CAI4 carrying the ADH1-yEGFP3 expression plasmid were grown on YEPD and used as negative and positive candida control strains, respectively.
In vivo promoter expression.
A Cn-MFα1::GFP transformant was also assessed for the ability to detect the expression of GFP and thus measure the induction of MFα1 in the subarachnoid space of immunosuppressed rabbits. Both the wild-type H99 and the Cn-MFα1::GFP transformants were grown in YEPD broth for 48 h at 30°C. The cells were pelleted, washed once in 0.015 M phosphate-buffered saline (PBS), and resuspended in PBS at a concentration of 3.3 × 108 cells/ml. Approximately 108 viable yeast cells of each C. neoformans strain in a volume of 0.3 ml were inoculated intracisternally into two New Zealand White male rabbits that had received an intramuscular injection of cortisone acetate at 7.5 mg/kg (Merck Sharpe and Dohme, West Point, Pa.) 1 day earlier and then received daily injections for 22 days. Expression of GFP was monitored during the infection by withdrawing 0.5 ml of CSF from the infected rabbits at 6, 9, 16, and 22 days after inoculation and assessing the CSF yeast cells for fluorescence by epimicroscopy and flow cytometry. This experiment was repeated with a second set of rabbits. Moreover, two independent transformants containing fewer integrated copies of Cn-MFα1::GFP at different locations were also inoculated separately into rabbits and monitored for detection of fluorescence. C. neoformans H99 was used as a negative control.
Fluorescent microscopy and flow cytometry.
GFP expression was assessed in vitro and in vivo by fluorescent microscopy and flow cytometry. Yeast cells from a single colony (in vitro) and CSF (in vivo) were washed twice in 1 ml of sterile distilled water and resuspended in 0.5 ml of PBS. Microscopic analysis was performed with an Olympus BH2-RFCA epifluorescence microscope with a 420- to 490-nm excitation filter, a 500-nm dichroic filter, and a 515-nm emission filter. Images were recorded on Ektachrome color slide film (ASA 400; Kodak, Rochester, N.Y.).
Fluorescence-activated cell sorter (FACS) analysis was performed with a FACScan (Becton Dickinson Immunocytometry Systems). Analysis of the data was performed by the CellQUEST program (version 3.1f) and statistical analysis was performed with Kolmogorov-Smirnov statistic analysis, where the Kolmogorov-Smirnov statistic (D) is the index of similarity for two curves: if D is 0, the curves are identical; if D is 1, the curves are completely different (43).
RESULTS
We sought to determine the potential of GFP as a reporter to monitor gene expression in C. neoformans. The enhanced GFP probe of Cormack et al. was fused downstream of three C. neoformans promoters: actin (Cn-ACT::GFP), GAL (Cn-GAL7::GFP), or pheromone (Cn-MFα1::GFP). The resulting GFP gene fusion plasmids were introduced into C. neoformans by biolistic transformation of M001 and selected for adenine prototrophy.
Selection of stable transformants and Southern analysis.
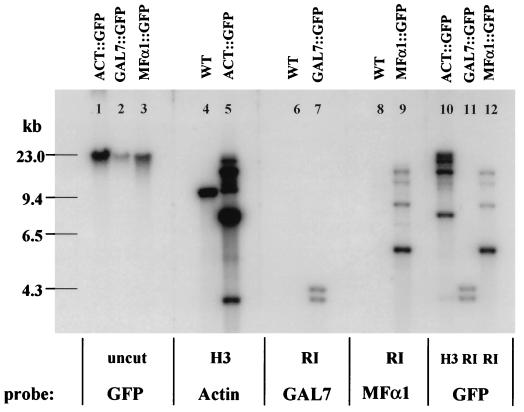
Thirty to forty transformants for each DNA construct were randomly selected and screened for mitotic stability. Stable transformants were necessary for analysis of gene expression in C. neoformans, and such transformants result from random integration of the transforming DNA into the genomic DNA of the recipient M001 strain. Southern analysis of the undigested genomic DNA isolated from multiple stable transformants revealed only high-molecular-weight DNA hybridizing to each of the respective promoter or GFP probes, and there was no evidence of extrachromosomal DNA in any of the transformants selected. Thus, the stable transformants carried only integrated copies of the transforming DNA (Fig. 2). Genomic DNA from stable transformants carrying ACT::GFP/ADE2, GAL7::GFP/ADE2, and MFα1::GFP/ADE2 constructs was digested with HindIII, EcoRI, and EcoRI, respectively, transferred to a nitrocellulose membrane and probed separately with regions of the GFP gene and promoters. Figure 2 shows the transformants containing intact copies of each promoter-GFP fusion gene which were selected for further analysis of GFP expression. These transformed strains were designated Cn-ACT::GFP, Cn-GAL7::GFP, and Cn-MFα1::GFP.
FIG. 2.
Southern analysis of Cn-ACT::GFP, Cn-GAL7::GFP, and Cn-MFα1::GFP genomic DNA with GFP and the actin, GAL7, and MFα1 promoters as probes. The Cn-ACT::GFP, Cn-GAL7::GFP, and Cn-MFα1::GFP transformants contain intact copies of each promoter-GFP fusion gene. H3, HindIII; RI, EcoRI; WT, wild type.
Expression of GFP in vitro.
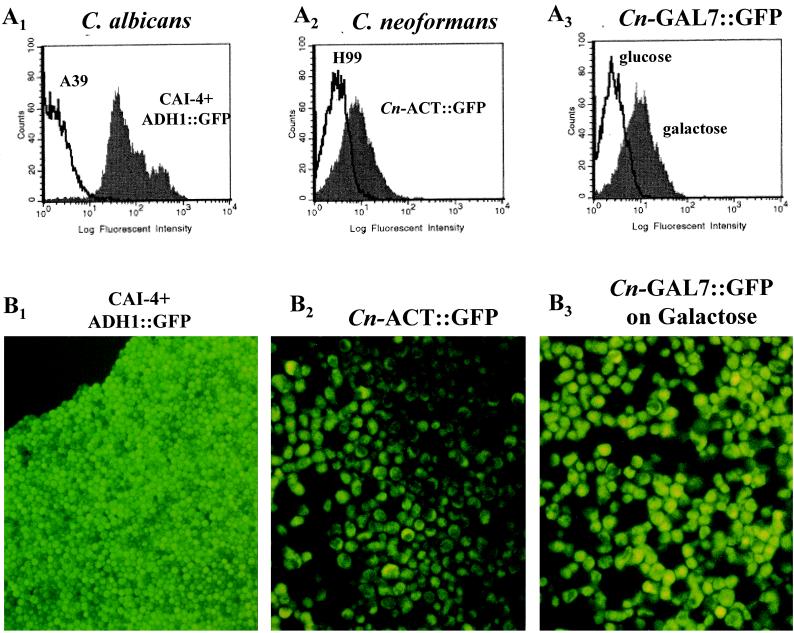
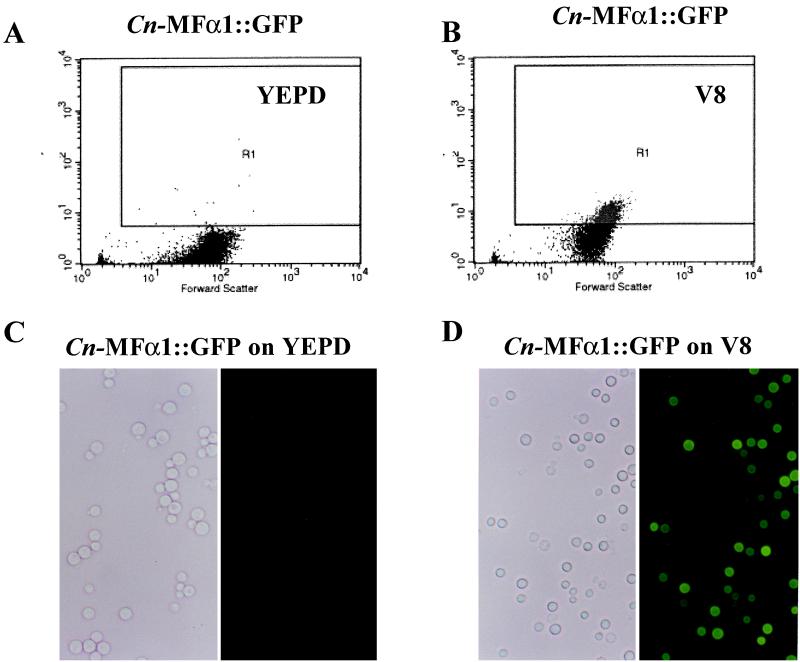
As a positive control for GFP expression, C. albicans CA14, in which the synthetic GFP is fused to the constitutive candida ADH1 promoter (5), was assessed for fluorescent activity. When cultured on YEPD agar, this transformant expressed intense fluorescent activity compared to C. albicans A39 (D = 0.99, P = 0.0001) (Fig. 3A1 and B1). We next assessed the ability of the three cryptococcal promoters (actin, GAL7, and MFα1) to drive GFP expression in each transformant. As expected, the Cn-ACT::GFP strain (actin promoter) expressed fluorescent activity when grown on a nonselective medium such as YEPD agar for 3 days, whereas the control parental strain H99 did not (D = 0.60, P = 0.001) (Fig. 3A2 and B2). This constitutive expression of a C. neoformans actin promoter at a stable environmental temperature confirms the results of Toffaletti and Perfect for actin gene expression in C. neoformans (37). On the other hand, GFP expression was regulated in the Cn-GAL7::GFP strain, which contained the galactose-inducible promoter. Significant fluorescent activity was detected in the Cn-GAL7::GFP strain grown on galactose but not on glucose media (D = 0.72, P = 0.001) (Fig. 3A3 and B3). This finding is consistent with the induction of the GAL7 gene by galactose and its repression by glucose as previously reported by Wickes and Edman (40). Furthermore, no detectable fluorescent activity was observed in the Cn-MFα1::GFP strain when it was propagated on YEPD agar (Fig. 4A and C). However, fluorescent activity from the Cn-MFα1::GFP reporter gene was detected in 23% of the yeast cells when the cells were grown on V-8 mating media for 3 days (D = 0.23, P = 0.01) (Fig. 4B and D). This finding supports the induction of the MFα1 gene as a pheromone responding to signals in the V-8 agar for the mating process.
FIG. 3.
(A) FACS analysis of transformed yeast cells grown on various media. (A1) C. albicans CAI4 carrying ADH1-yEGFP3 expression plasmid and the control C. albicans A39 strain, after growth on YEPD; (A2) Cn-ACT::GFP transformant and the control C. neoformans H99 strain, after growth on YEPD; (A3) Cn-GAL7::GFP transformant after growth on YNB-glucose and YNB-galactose. Each histogram represents 104 events. (B) Epifluorescent microscopy of GFP transformants. (B1) C. albicans CAI4 carrying ADH1-yEGFP3 expression plasmid after growth on YEPD; (B2) Cn-ACT::GFP strain after growth on YEPD; (B3) Cn-GAL7::GFP strain after growth on YNB-galactose.
FIG. 4.
FACS analysis and corresponding phase-contrast and epifluorescent microscopy of Cn-MFα1::GFP transformant grown in vitro. (A and B) FACS analysis of a Cn-MFα1::GFP strain after growth on YEPD (A) and V-8 (B). Log fluorescent intensity is plotted on the y axes. Each dot plot represents 104 events. Fluorescent activity is shown in gate R1. (C and D) Epifluorescent microscopy of a Cn-MFα1::GFP strain after growth on YEPD (C) and V-8 (D).
Expression of GFP in vivo.
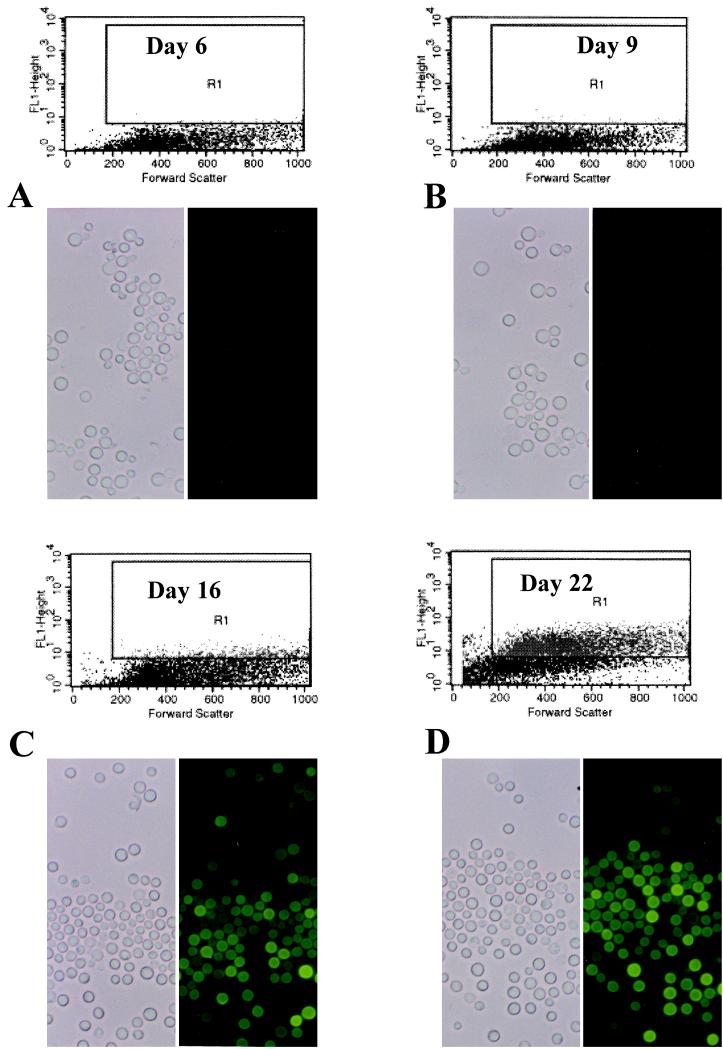
Since the Cn-MFα1::GFP strain appears to be appropriately regulated in vitro by environmental cues (i.e., “off” on YEPD and “on” on V-8 medium), we were able to explore another hypothesis. We tested whether the MFα1 promoter is activated in vivo in response to possible nutrient starvation signals which may naturally occur in vivo. The Cn-MFα1::GFP strain was inoculated intracisternally into cortisone-treated rabbits, and the presence of cell fluorescent activity was monitored throughout a 22-day period of infection observation. No fluorescent activity was detected in yeast cells obtained from the CSF on day 6 or 9 of infection (Fig. 5A and B). However, significant fluorescent activity was detected in 15% of the yeast cells after 16 days of infection, and by day 22 of infection 60% of yeast cells expressed fluorescent activity (Fig. 5C and D), and moreover, the overall intensity of fluorescence increased in the cells over the course of the infection. The intensities of fluorescent activities detected in yeast cells on days 6 and 22 of the infection were compared by statistical analysis and found to be statistically different (D = 0.76, P = 0.001). Wild-type H99 cells were used as a control and showed no fluorescent activity when examined at the same time points and compared to Cn-MFα1::GFP cells. Moreover, the Cn-MFα1::GFP yeast cells produced no fluorescence when removed from CSF at all time points, including days 16 and 22, and then regrown on YEPD or Sabouraud agar. To ensure that this induction was not related to a unique genomic position of the construct, two separate independent transformants with fewer copies of the construct at different locations were tested in vivo with separate rabbits. Both transformants followed the original strain with detection of fluorescence found only between days 14 and 21 of infection. By day 21, 30% of cells were fluorescent, compared to <5% at day 14 (data not shown). These observations indicate that the MFα1 promoter is induced by signals in the CSF during the proliferative stage of infection in the rabbit model of cryptococcal meningitis.
FIG. 5.
FACS analysis and corresponding phase-contrast and epifluorescent microscopy of Cn-MFα1::GFP from CSF. Yeast cells were isolated from the CSF on days 6 (A), 9 (B), 16 (C), and 22 (D) of infection. Log fluorescent intensity is plotted on the y axes. Each dot plot represents 104 events. Fluorescent activity is shown in gate R1.
DISCUSSION
With the completion of the S. cerevisiae genome project and progress being made with other microbial genomes, attention is now focused on functional genomic approaches. Multiple molecular tools to screen large numbers of genes for differential expression have been developed (11, 13, 20, 36, 44). The ability to monitor and identify gene expression patterns will provide insights into how microbial pathogens respond to the host environment. For instance, in a genomic screen of gene expressions Wodicka et al. employed high-density oligonucleotide arrays on glass chips and found that when S. cerevisiae is grown on rich or minimal media, only 10% of all mRNAs differ appreciably in expression and less than 3% of mRNAs differ more than fivefold in expression level (41). It is clear from these studies that fungi alter their gene expression in response to environmental cues and that identification of these regulated genes is both possible and foreseeable. In fact, De Bernardis et al. recently examined the expression of C. albicans aspartyl protease genes (SAP1 and SAP2) in vivo during an experimental candida vaginal infection of rats (7). For fungi like C. neoformans, for which the molecular biological databases are less fully developed, other techniques will be required to discover genes regulated during infection. For instance, both differential hybridization and differential display reverse transcription-PCR have been used to screen for regulated genes during C. neoformans infection (29, 31). Specific C. neoformans gene expression in the CSF has already been reported for one gene, CnLAC1, by reverse transcription-PCR (32), and another gene, COX1, has been identified by differential hybridization due to its expression at this CNS site of infection (29).
Three promoters (for the GAL7, actin, and MFα1 genes) fused with the synthetic reporter GFP gene were successfully constructed and confirmed by sequencing of the fusion junctions. Using flow cytometry, we found both in vitro and in vivo expression of GFP driven by these regulated C. neoformans promoters. Although the induced C. neoformans fluorescence was not as intense as the fluorescence for C. albicans CAI4 containing ADH1-yEGFP3 (for which it was originally optimized because of its unique codon usage), the GFP fluorescence from this construct was more than adequate for the detection of differential promoter expression in C. neoformans. Although 100% of the cells were not equally fluorescent (a phenomenon which is also seen in C. albicans [Fig. 3A1 and B1]) with both microscopy and flow cytometry, it was easy to distinguish the induction of the promoter construct in a strain from the baseline fluorescence of the uninduced strain. This study demonstrates that the synthetic GFP developed by Cormack et al. (3) can be used effectively as a reporter gene for monitoring gene expressions both in vivo and in vitro for this serotype A strain (H99). Future studies could attempt to further optimize GFP expression for C. neoformans.
Serotypes A and D are phylogenetically classified within the same variety (Cryptococcus neoformans var. neoformans), but further studies may actually determine that they are separated by millions of years of evolution. For instance, there are slight differences in their ribosomal DNA sequences, differences between 3 and 7% exist in their allelic sequences, and different karyotype patterns are observed. However, the ADE2 gene from a serotype D strain has been previously expressed in a serotype A strain (38). In this study, we confirm that these two serotypes can recognize and use promoters from each other. Since heterologous promoters from conserved genes of other basidiomycetes do not function well in C. neoformans (unpublished data), our observations with promoters from one serotype being recognized by another serotype suggest a functional evolutionary closeness between these two serotypes compared to other basidiomycetes.
Although it has been shown through analysis of congenic isolates which differed at the mating locus (12) that the MATα locus contributes to the virulence of C. neoformans in mice, this is the first study to specifically suggest the possibility that a putative pheromone gene within this locus might be directly implicated in the pathogenesis of C. neoformans. Expression of the MFα1 gene, which has been detected only during the mating process (18), is induced during growth on nutritionally depleted media, such as V-8 agar. We hypothesized that the low-nitrogen and -carbohydrate conditions of the subarachnoid space might contain a nutritional signal(s) similar to that of minimal media that results in the induction of MFα1 expression. This hypothesis may be correct, but the temporal gene activation during infection might support the presence of other inducible factors. For instance, the MFα1 promoter is activated during the proliferative stage of infection within the subarachnoid space and not during the early induction or exposure phase of infection. These findings suggest that the MFα1 promoter may be controlled or regulated by a central regulatory circuit that responds to either specific nutrient deprivation during infection (such as the changing of glucose or protein concentrations in CSF) or specific host signals (such as cytokines or chemokines). It is unlikely that delayed MFα1 induction is related to the aging of the yeast cells in vivo because the full expression of GFP by this strain is observed within 3 to 5 days after the strain is placed on V-8 agar. Its regulation also appears to be specific for the environmental site, since CSF yeasts returned to in vitro growth on complete media have the MFα1 promoter again repressed. Moreover, the specific in vivo MFα1 promoter induction is supported by the similar findings of three different and independent transformants.
It is important to recognize that these studies provide only an association of the regulated expression of MFα1 with infection. For instance, this up-regulation of the MFα1 promoter might be part of a global regulatory mechanism(s) for the stress response and growth of yeast under certain nutritional exposures both in vitro and in vivo. However, to prove whether MFα1 is directly related to the virulence composite of C. neoformans or simply part of an environmental response will require making a null mutant of MFα1 and testing the site-directed mutant’s effect on virulence in animal models.
Finally, the ability to use GFP as a reporter in C. neoformans suggests a number of interesting applications for studies of pathogenesis. For instance, the construction of heterologous fusion constructs comprising GFP fused to the promoters of genes that are preferentially expressed at a certain site of infection will be beneficial in identifying and timing the transcriptional regulation of these genes during infection, as we did with MFα1 in this study. GFP can also be used to detect unique gene regulations dependent on specific host infection sites. For example, yeast cells can migrate from the lung to the central nervous system during infection, and genes that are specifically induced in the lung but not in the central nervous system can be identified by this approach. Another strategy is to use GFP to find promoter sequences in C. neoformans that are induced or repressed during infection. By cloning small, random, genomic fragments (500 to 1,500 bp) upstream of the GFP gene, transforming these fragments into C. neoformans, and infecting rabbits with these transformants, it is feasible to identify promoters that are differentially expressed during infection. Viable yeast cells can then be specifically recovered by a FACS, and the promoters rescued from these cells can be used as probes to clone infection-regulated genes. These types of promoter-trap strategies used in conjunction with in vivo expression technology have been useful for detecting regulated promoters in single cells during bacterial infection. In fact, under in vivo conditions, GFP may be a more sensitive indicator of gene regulation in yeast than the original in vivo expression technology strategies which rely on both adenine complementation and the survival of the infecting organism (15).
ACKNOWLEDGMENTS
We are grateful to Wiley A. Schell for assistance with epifluorescence microscopy analysis and to Mary Ann Howard for assistance in manuscript preparation.
This work was supported by Public Health Service grants AI28388, AI41937, and AI-94-014 from the National Institute of Allergy and Infectious Diseases and as part of the Veterans Administration Research Center on AIDS and Human Immunodeficiency Virus Infection and the Duke University Mycology Research Unit.
REFERENCES
- 1.Chalfie M, Yu T, Guskirchen G, Ward W W, Parsher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack B P, Bertram G, Egerton M, Gow N A R, Falkow S, Brown A J P. Yeast-enhanced green fluorescent protein (yGFP): a reporter of gene expression in Candida albicans. Microbiology. 1992;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 4.Cox G M, Dykstra C, Rude T H, Perfect J R. Cryptococcus neoformans actin gene: characterization and its use as a phylogenetic marker. J Med Vet Mycol. 1995;33:261–266. doi: 10.1080/02681219580000521. [DOI] [PubMed] [Google Scholar]
- 5.Cox G M, Perfect J R. Fungal infections. Curr Opin Infect Dis. 1993;6:422–426. [Google Scholar]
- 6.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 7.De Bernardis F, Cassone A, Sturtevant J, Calderone R. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect Immun. 1995;63:1887–1892. doi: 10.1128/iai.63.5.1887-1892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dismukes W E. Cryptococcal meningitis in patients with AIDS. J Infect Dis. 1988;157:624–628. doi: 10.1093/infdis/157.4.624. [DOI] [PubMed] [Google Scholar]
- 9.Edman J C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoy J F, Murphy J W, Miller G G. T cell response to soluble cryptococcal antigens after recovery from cryptococcal infection. J Infect Dis. 1989;159:116–119. doi: 10.1093/infdis/159.1.116. [DOI] [PubMed] [Google Scholar]
- 11.Ivanova N B, Belyavsky A. Identification of differentially expressed gene by restriction endonuclease-based gene expression fingerprinting. Nucleic Acids Res. 1995;23:2954–2958. doi: 10.1093/nar/23.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang P, Pardee A B. Differential display of eucaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 14.Lodge J K, Jackson-Machelski E, Toffaletti D L, Perfect J R, Gordon J I. Targeted gene replacement demonstrates that myristoyl CoA:protein N-myristoyl transferase is essential for the viability of Cryptococcus neoformans. Proc Natl Acad Sci USA. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 16.Miller G P G. The immunology of cryptococcal disease. Semin Respir Infect. 1986;1:45–52. [PubMed] [Google Scholar]
- 17.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy J W. Influence of cryptococcal antigens on cell-mediated immunity. Rev Infect Dis. 1988;10:S432–S435. doi: 10.1093/cid/10.supplement_2.s432. [DOI] [PubMed] [Google Scholar]
- 20.Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- 21.Perfect J R. Fluconazole therapy for experimental cryptococcosis and candidiasis in the rabbit. Rev Infect Dis. 1990;12(Suppl. 3):299–302. doi: 10.1093/clinids/12.supplement_3.s299. [DOI] [PubMed] [Google Scholar]
- 22.Perfect J R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agents Chemother. 1996;40:1577–1583. doi: 10.1128/aac.40.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perfect J R. Differential gene display in Cryptococcus neoformans. Presented at the 98th General Meeting of the American Society for Microbiology. 1998. Atlanta, Ga. [Google Scholar]
- 24.Perfect J R, Durack D T. Treatment of experimental cryptococcal meningitis with amphotericin B, 5-fluorocytosine and ketoconazole. J Infect Dis. 1982;146:429–435. doi: 10.1093/infdis/146.3.429. [DOI] [PubMed] [Google Scholar]
- 25.Perfect J R, Durack D T, Gallis H A. Cryptococcemia. Medicine. 1983;62:98–109. doi: 10.1097/00005792-198303000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Perfect J R, Lang S D R, Durack D T. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 27.Perfect J R, Rude T H, Penning L M, Johnston S A. Cryptococcus neoformans TRP1 gene by complementation in Saccharomyces cerevisiae. Gene. 1992;122:213–217. doi: 10.1016/0378-1119(92)90053-r. [DOI] [PubMed] [Google Scholar]
- 28.Perfect J R, Toffaletti D L, Rude T H. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perfect, J. R., B. Wong, Y. Chang, K. J. Kwon-Chung, and P. R. Williamson.Cryptococcus neoformans: virulence and host defenses. J. Med. Vet. Mycol., in press. [PubMed]
- 30.Plautz J D, Day R N, Dailey G M, Welsh S B, Hall J C, Halpain S, Kay S A. Green fluorescent protein and its derivatives as versatile markers for gene expression in living Drosophila melanogaster, plant and mammalian cells. Gene. 1996;173:83–87. doi: 10.1016/0378-1119(95)00700-8. [DOI] [PubMed] [Google Scholar]
- 31.Rude T H, Perfect J R. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. C. neoformans genes regulated in the CSF during meningitis, abstr. F-45; p. 267. [Google Scholar]
- 32.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salkowski C A, Bartizal K F, Balish M J, Balish E. Colonization and pathogenesis of Cryptococcus neoformans in gnotobiotic mice. Infect Immun. 1987;55:2000–2005. doi: 10.1128/iai.55.9.2000-2005.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schena M, Shalon D, Davis R W, Brown P O. Qualitative monitoring of gene expression patterns with a complementary DNA microassay. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 37.Toffaletti D L, Perfect J R. Study of Cryptococcus neoformans actin gene regulation with a beta-galactosidase-actin fusion. J Med Vet Mycol. 1997;35:313–320. [PubMed] [Google Scholar]
- 38.Toffaletti D L, Rude T H, Johnston S A, Durack D T, Perfect J R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 40.Wickes B L, Edman J C. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 41.Wodica L, Dong H, Mittmann M, Ho M H, Lockart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 42.Yeh E, Gustafson K, Boulianne G L. Green fluorescent protein as a vital marker and reporter gene expression in Drosophila. Proc Natl Acad Sci USA. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young I T. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- 44.Zhao N, Hashida H, Takahashi N, Misumi Y, Sakaki Y. High-density cDNA filter analysis: a novel approach for large-scale quantitative analysis of gene expression. Gene. 1995;156:207–213. doi: 10.1016/0378-1119(95)00023-y. [DOI] [PubMed] [Google Scholar]