Abstract
Coronaviruses of the genera Gammacoronavirus and Deltacoronavirus are globally widespread and circulate primarily in wild and domestic birds. Prior studies have established frequently occurring crossover events from avian to mammalian reservoirs. However, there is limited understanding of the diversity and geographical distribution of coronaviruses among birds. In this study, the surveillance of coronaviruses in birds in Russia during 2020 revealed the presence of coronaviruses in 12% of samples from birds. Targeted NGS approach was used for the evaluation of genetic diversity based on RdRp gene. While gammacoronviruses were found in both wild birds and poultry, deltacoronaviruses were found in wild birds only and represent the first detections for Russia. A number of cases with the simultaneous detection of gamma- and deltacoronaviruses in one bird was reported. The results of this study highlight the importance of further research concerning the spread and diversity of coronaviruses among birds within and migrating throughout the territory of Russia across the globe.
Subject terms: Virology, Viral reservoirs, Next-generation sequencing
Introduction
Coronaviruses (CoVs) are an important subject of research considering they often serve as infectious agents that are capable of causing disease in human and animal hosts. Susceptible taxa include a wide range of mammals, as well as various species of wild and domestic birds. Currently there is a very limited understanding of the diversity of coronaviruses, their transmission mechanisms and the role of individual species in disparate populations. Coronaviruses are common name of representatives of family Coronaviridae, suborder Cornidovirineae, order Nidovirales. According to the International Committee on Taxonomy of Viruses (ICTV), the family Coronaviridae is classified into three subfamilies, one of which is subfamily Orthocoronavirinae and includes four genera of viruses. The Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus are of particular importance for agriculture and public health. Of the four genera of CoVs, the representatives of genera Alphacoronavirus and Betacoronavirus are considered to circulate most commonly among mammals, while birds serve as main hosts of viruses of genera Gammacoronavirus and Deltacoronavirus1–4.
The genus Gammacoronavirus is classified into three subgenera, two of which, subgenus Brangacovirus and subgenus Igacovirus, were identified only in birds. The subgenus Cegacovirus is represented by Beluga whale coronavirus SW1, which was isolated from a marine mammal5. The well-known and widespread representatives of the genus Gammacoronavirus are the Avian coronavirus (AvCov) and Avian coronavirus 9203 (AvCov9203), subgenus Igacovirus. These two species represent a group of viruses that includes all infectious bronchitus virus (IBV) genotypes as well as other genetically similar viruses. IBV is the causative agent of avian infectious bronchitis in chickens, while other viruses of this group cause disease in turkeys, guinea fowls and other species. These viruses are ubiquitous in agricultural settings, and are especially widespread in regions where commercial poultry farming. AvCovs cause significant economic damage due to the infection associated with serious respiratory, genitourinary and digestive problems in poultry6,7. In addition, there are many reports of the isolation of gammacoronaviruses (gammaCoVs) from other bird species, indicating the ability of the virus to infect a wider range of hosts. Thus, studies conducted in Brazil and China have shown the isolation of IBV from wild peacocks and Rock pigeons (Columba livia)8,9. In another European study, similar viruses have been isolated from the droppings of healthy wild ducks and Whooper swans (Cygnus cygnus). These findings suggest interspecies transmission of the IBV viruses among domestic and wild birds, and therefore possible spread over considerable distances by migrating birds10. A recent update of the ICTV (2019) includes Duck Coronavirus 2714 (DCoV) ratified species in the subgenus Igacovirus, which represent a large group of gammaCoVs commonly found in wild birds. There are reports that viruses from this group are able to cross interspecies barriers and infect poultry species commonly including domestic ducks, and chickens in some rare cases. Thus, there is a considerable diversity of gammaCoVs in domestic and wild birds that presents a substantial global challenge for the poultry industry concerning the surveillance of diverse viruses. In addition, gammaCoVs serve as a significant challenge when implementing strategic actions for the prevention and mitigation of poultry gammaCoVs outbreaks.
The genus Deltacoronavirus is classified into three subgenera—Andecovirus, Buldecovirus and Herdecovirus, which include seven ratified species, six of which represent groups of viruses that were found only in birds. Only one deltaCoV species Coronavirus HKU15, subgenus Buldecovirus, represents a group of viruses that were detected in birds and in mammalian species, including pigs, an Asian leopard cat (Prionailurus bengalensis) and a Chinese ferret badger (Melogale moschata). The Coronavirus HKU15 isolated from pigs and other viruses of subgenus Buldecovirus have been shown to have similar genetic characteristics (PDCoV HKU15 is more than 96% similar to SpCoV HKU17 in 7 aminoacid sequences of ICTV domains, which are used for species demarcation) and structure, indicating possible transmission of the virus from birds to mammals11–14. It was subsequently shown by phylogenetic analysis that porcine deltacoronaviruses (deltaCoVs) are capable of interspecies transmission13,15. Furthermore, a recent study describes cases of human infections with porcine deltacoronavirus (PDCoV) after probable contact with domestic animals16.
In general, studies on gammaCoVs and deltaCoVs show that wild birds play an important role in their circulation. Coronaviruses have been detected in many orders of wild birds, such as ducks, geese and swans (Anseriformes), storks (Ciconiiformes), shorebirds and gulls (Charadriiformes), pigeons (Columbiformes), chickens, turkeys, quails (Galliformes), songbirds (Passeriformes), pelicans (Pelecaniformes) and parrots (Psittaciformes)15,17. In general, the percentage of virus carriers depends on the order and sometimes the type of bird. For example, in one of the studies conducted in Sweden, it was found that the ducks were the most prominent hosts, carrying 39% of the gammaCoVs and deltaCoVs isolated from free-ranging birds. One the other hand, the overall proportion of carriers was 18.7% among all the studied wild birds17. Studies conducted in Korea demonstrated a lower percentage of virus carriers (0.95%) among wild birds, but, as in the previous study in Sweden, wild ducks were the dominant carriers. Overall, most studies indicate that the highest percentage of positive samples was obtained from birds of the order Anseriformes17,18.
At this time however, there are still many important, open questions regarding the full range of avian host species, the seasonal and geographic distribution of viral circulation and the genetic diversity of coronaviruses in birds. Limited numbers of studies are available that reveal the as-yet uncharted complexities in the ecology and evolution of CoVs in birds15. In particular, in Russian Federation, there are only a few reported studies of poultry CoVs, and these studies investigated the IBV group.
Bochkov et al., 2006 presented a comprehensive genetic characterisation of AvCoVs isolated from 1998 to 2002 during a surveillance study that sampled more than 250 poultry farms19. The authors revealed the circulation of several different IBV genotypes originating from the AvCoV group, predominated by the Massachusetts genotype (including H120, H52 vaccine lines). The predominance of viruses from this genotype among poultry subsequently persisted until at least 2010 as was shown by another surveillance study of IBV, which is described in the publication by Ovchinnikova et al., 2011. The publication presents the results of molecular biological characterisation of AvCoVs isolated in Russia, as well as Ukraine and Kazakhstan in the period from 2007 to 201020.
In effect, the surveillance and study of CoVs ecology and evolution contributes to the implementation of control measures in poultry agriculture21. Continued characterization of these viruses are also important for the prognosis of potential zoonotic spread of the viruses among mammalian species and humans. The aim of the work was to broaden the evaluation of ecology and diversity of CoVs in Russia. In this study we report the results of countrywide surveillance and the characterization of the genetic diversity of coronaviruses that were isolated from among wild birds and poultry in Russia in 2020.
Materials and methods
Ethics statements
The identification of bird species and their sampling was carried out by zoologists of regional laboratories of Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing (Rospotrebnadzor). Capture and sampling of birds were carried out in accordance with Russian legislation and under bioethics protocol № Vector/04-04.2018 issued by BioEthics Committee at FBRI SRC VB Vector Rosbotrebnadzor.
Samples collection
Samples from wild birds and poultry were collected during the National Avian Viruses Surveillance Program, which supports sample collection from 48 regions of Russia. Samples were collected from the territories of 14 regions of the western part of Russia (Stavropol region, Astrakhan region, Rostov region, Republic of North Ossetia-Alania, Belgorod region, Kostroma region), Siberia (Novosibirsk region, Kurgan region, Tyumen region, Yamalo-Nenets Autonomous Okrug) and the Russian Far East (Kamchatka Krai, Primorsky Krai, Sakhalin region, Magadan region). Samples from birds included cloacal swabs, feces and internal organ fragments. The Copan Universal Transport Medium (UTM-RT) System (Copan Diagnostics Inc., USA) was used for sample collection. Samples (including cloacal swabs, droppings or internal organ fragments) were collected individually from 584 birds and used for the study. Swabs and organ homogenates were prepared as previously described22. Organ fragments were homogenized using Qiagen Tissue Liser LT (Qiagen, Germany). Swabs and organ homogenates were vortexed vigorously for 7–10 s and then centrifugated for 5 min at 12,000×g at 4 °C.
One-step RT-PCR and nested PCR
Viral RNA was isolated from the collected samples using the RIBO-sorb or RIBO-prep kit (Interlabservice, Russia) according to the manufacturer’s instructions. With purpose of diagnostics of coronavirus, we used the published “modified PAN coronavirus PCR” and the test system developed in our laboratory using nested PCR18,23. All samples were analyzed using both methods. Modified PAN coronavirus PCR (one-step RT-PCR) was performed as described in Chamings et al. 18. Degenerate primers for the modified PAN coronavirus PCR (AC-CoV-F: GGTTGGGATTATCCWAARTGTG, AC-CoV-R: TGYTGTGARCAAAAYTCRTG) target the conserved region of the avian coronavirus polymerase gene (RdRp), producing a 602 bp amplicon. This test system allows for the detection of coronaviruses of all four genera (Alpha-, Beta-, Gamma- and Deltacoronaviruses). One step RT-PCR was performed using kits manufactured by Biolabmix LLC (Russia). These kits contain recombinant M-MuLV reverse transcriptase and DNA-dependent DNA polymerase in one enzyme mix. The reaction profile was as described in Chamings et al.18. Briefly, the reaction was performed at 50 °C for 20 min, 92 °C for 5 min, followed by 45 cycles of 93 °C for 30 s, with an annealing temperature for 30 s, 68 °C for 1 min, final 68 °C step for 3 min and held at 4 °C. The touchdown annealing temperature started at 60 °C for 3 cycles, then decreased by 2 °C every 3 cycles until 48 °C, which was used for the final 28 reaction cycles18. A test system was developed using nested PCR, which allows for detection with significantly increased sensitivity. DNA obtained from one-step RT-PCR using a modified PAN coronavirus PCR was used to perform nested PCR. The nested PCR test system included degenerate nested PCR primers for the detection of Gamma- and Deltacoronaviruses. The primers were made for the RdRp gene region, the coordinates of the nested primers were F: 14,203–14,224 and R: 14,607–14,629 and are given relative to the reference genome of the infectious bronchitis virus strain IBV D274 (GenBank ID: MH021175.1). The sequences of the primers were: Vec_CoVgd1 F2: CWAARTGTGAYAGRKCHATGCC, Vec_CoVgd1 R2: CCRTCRTCAGAMARDATCATNAR. The size of the amplicon with primers was 422 bp. For nested PCR, the BioMaster LR HS-PCR (2×) kit manufactured by Biolabmix (Russia) was used. This kit contains a mixture of HS-Taq DNA and Pfu DNA polymerases to ensure high fidelity amplification. The reaction profile was 94 °C for 3 min, 95 °C for 15 s, 50 °C for 30 s, 68 °C for 1 min for 40 cycles, then 68 °C for 7 min, held at 4 °C. The strain of chicken infectious bronchitis virus IBV D274 (GenBank ID: MH021175.1) was used as a positive control in all PCR performances. Purified water was used as a negative control in all PCR runs.
Sequencing
The sequencing of coronavirus amplicons of the RdRp gene was performed using Illumina MiSeq. Library was made using a v3 reagent kit (Illumina, USA). Sequences were assembled by aligning reads with known references using bwa-0.7.15 software24. Obtained sequences were deposited in the GenBank database (NCBI) (Supplementary Table S1). Amplicon coverage is shown in Supplementary Table S1.
Phylogenetic analysis
Phylogenetic trees were constructed using partial sequences of RdRp gene obtained in this study and reference sequences of avian coronaviruses from NCBI GenBank representing all relevant clades. Multiple sequence alignment was performed using the MUSCLE alignment option of MEGA X software. Phylogenetic analysis was performed using the neighbour joining method with 1000 bootstrap replications using MEGA X software (http://www.megasoftware.net/).
Results
Detection of coronaviruses in poultry and wild birds in Russia
A total of 584 avian samples, including 393 from wild birds and 191 from domestic birds were collected and examined during surveillance within the territory of the Russian Federation in July–December 2020 (Table 1). Samples from wild birds were collected from 8 regions and samples from poultry were collected from 9 regions of Russia (Table 1). The sampled wild birds belonged to 31 species of 3 orders (Table 2). Most of the bird species belonged to the Anseriformes and Charadriiformes orders, which were represented in the study by 16 and 13 species, respectively. The order Passeriformes was represented by 2 species of wild birds. Wild bird samples were from Charadriiformes (n = 200, 13 species), Anseriformes (n = 180, 16 species) and Passeriformes (n = 13, 2 species). One-step RT-PCR analysis revealed that 70 of 584 samples (12,0%) were positive for coronavirus: 56 samples from wild birds (14.2% among wild birds) and 14 samples from poultry (7.3% among poultry). Coronaviruses were identified from 20 species of birds, which belonged to 4 orders. The wild bird, in 25 cases of coronavirus RNA detection belonged to the order Anseriformes (9 bird species), 28 cases—to the order Charadriiformes (8 bird species) and 3 cases—to the order Passeriformes (2 bird species). Coronavirus RNA was detected in fourteen chickens, order Galliformes (1 species) (Table 2).
Table 1.
Results of the study of samples from wild and domestic birds.
| No. | Region of samples collection | Number of samples | Number of positive samples | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GammaCoV | DeltaCoV | Mixed gammaCoV + deltaCoV variants | Untyped | ||||||||
| Wild* | Dom.* | Wild | Dom | Wild | Dom | Wild | Dom | Wild | Dom | ||
| 1. | Novosibirsk region | 26 | – | 3 | – | 1 | – | – | – | 6 | – |
| 2. | Yamalo-Nenets Autonomous Okrug | 175 | – | 8 | – | 3 | – | 2 | – | 17 | – |
| 3. | Kamchatka Krai | 125 | – | – | – | 3 | – | 4 | – | 7 | – |
| 4. | Primorsky Krai | 6 | – | 1 | – | – | – | – | – | – | – |
| 5. | Sakhalin region | 27 | 60 | – | 7 | – | – | – | – | – | – |
| 6. | Stavropol region | 8 | 10 | 1 | – | – | – | – | – | – | – |
| 7. | Astrakhan region | – | 20 | – | 1 | – | – | – | – | – | – |
| 8. | Magadan region | 23 | 60 | – | – | – | – | – | – | – | – |
| 9. | Kurgan region | – | 15 | – | – | – | – | – | – | – | – |
| 10. | Tyumen region | – | 2 | – | – | – | – | – | – | – | – |
| 11. | Kostroma region | – | 10 | – | – | – | – | – | – | – | – |
| 12. | Rostov region | – | 4 | – | – | – | – | – | – | – | – |
| 13. | Republic of North Ossetia-Alania | 3 | – | – | – | – | – | – | – | – | |
| 14. | Belgorod region | – | 10 | – | 2 | – | – | – | – | – | 4 |
| Total | 393 | 191 | 13 | 10 | 7 | – | 6 | – | 30 | 4 | |
*Wild—samples from wild birds; dom.—samples from poultry.
Table 2.
Overview of the bird species and prevalence of coronaviruses.
| No. | Order | Species | No. of samples | No. of positive samples | Sequenced (MiSeq) | No. of viruses detected | Detected mixed variants in one sample* | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild birds | GammaCoV | DeltaCoV | ||||||||
| 1. | Anseriformes | Common teal (Anas crecca) | 41 | 2 | 1 | 1 | ||||
| 2. | Eurasian wigeon (Mareca penelope) | 30 | 3 | 2 | 1 | 1 | ||||
| 3. | Mallard (Anas platyrhynchos) | 28 | 3 | 2 | 3 | 2 GammaCoVs | ||||
| 4. | Gadwall (Mareca strepera) | 16 | 4 | 2 | 1 | 3 | GammaCoV + 2 DeltaCoVs | |||
| 5. | Tufted duck (Aythya fuligula) | 16 | 3 | 1 | 1 | |||||
| 6. | Northern pintail (Anas acuta) | 12 | 4 | 1 | 1 | |||||
| 7. | Common merganser (Mergus merganser) | 8 | 1 | 1 | 1 | |||||
| 8. | Greater scaup (Aythya marila) | 7 | ||||||||
| 9. | Common scoter (Melanitta nigra) | 7 | 4 | 1 | 2 | 2 GammaCoVs | ||||
| 10. | Greater white-fronted goose (Anser albifrons) | 3 | ||||||||
| 11. | Mute swan (Cygnus olor) | 3 | ||||||||
| 12. | Northern shoveler (Spatula clypeata) | 3 | 1 | 1 | 1 | |||||
| 13. | Velvet scoter (Melanitta fusca) | 2 | ||||||||
| 14. | Smew (Mergellus albellus) | 2 | ||||||||
| 15. | Common pochard (Aythya ferina) | 1 | ||||||||
| 16. | Bean goose (Anser fabalis) | 1 | ||||||||
| 17. | Charadriiformes | Dunlin (Calidris alpina) | 40 | 9 | 4 | 2 | 7 | 2 DeltaCoVs; GammaCoV + 2 DeltaCoVs; GammaCoV + 2 DeltaCoVs | ||
| 18. | Siberian gull (Larus fuscus heuglini) | 34 | 4 | 1 | 1 | 1 | GammaCoV + DeltaCoV | |||
| 19. | Slaty-backed gull (Larus schistisagus) | 30 | 1 | 1 | 1 | |||||
| 20. | Black-headed gull (Chroicocephalus ridibundus) | 28 | 2 | 1 | 1 | |||||
| 21. | Common gull (Larus canus) | 21 | ||||||||
| 22. | Herring gull (Larus argentatus) | 13 | 5 | 1 | 2 | 2 GammaCoVs | ||||
| 23. | Red-necked stint (Calidris ruficollis) | 9 | 4 | 2 | 2 | 3 | Gamma + 2 Delta; Gamma + Delta | |||
| 24. | Common tern (Sterna hirundo) | 8 | ||||||||
| 25. | Pomarine jaeger (Stercorarius pomarinus) | 7 | 2 | 1 | 1 | |||||
| 26. | Common sandpiper (Actitis hypoleucos) | 5 | 1 | 1 | 1 | |||||
| 27. | Eurasian woodcock (Scolopax rusticola) | 3 | ||||||||
| 28. | Great knot (Calidris tenuirostris) | 1 | ||||||||
| 29. | Broad-billed sandpiper (Calidris falcinellus) | 1 | ||||||||
| 30. | Passeriformes | Hooded crow (Corvus cornix) | 7 | 1 | 1 | 1 | ||||
| 31. | Rook (Corvus frugilegus) | 6 | 2 | 1 | 2 | 2 GammaCoVs | ||||
| Poultry | ||||||||||
| 32. | Galliformes | Chicken (Gallus gallus domesticus) | 134 | 14 | 10 | 11 | 2 GammaCoVs | |||
| 33. | Turkey (Meleagris gallopavo domesticus) | 10 | ||||||||
| 34. | Anseriformes | Domestic duck (Anas platyrhynchos domesticus) | 33 | |||||||
| 35. | Domestic goose (Anser anser domesticus) | 14 | ||||||||
| 4 orders | 36 species | 584 | 70 | 36 | 34 | 18 | ||||
*“GammaCoV + 2 DeltaCoVs” means that one gammacoronavirus and two different deltacoronaviruses were detected in one avian sample; “2 Gamma” means that two different gammacoronaviruses were detected in one avian sample etc.
Of the total number of samples from individual birds positive for coronaviruses, molecular genetic analysis was carried out for 36 samples (26 samples from wild birds, 10—from poultry). These samples were from all studied geographic locations and avian species. We performed partial sequencing of a conservative region for all coronavirus genera, specifically that of the RdRp gene (around 400 bp) using NGS. The nucleotide sequences of 51 viruses were deposited in NCBI GenBank, and their accession numbers are listed in Supplementary Table S1. The sequence length of the RdRp gene of one virus was not sufficient to deposit in GenBank, and it was only used for genotyping.
The 52 coronaviruses, which were identified, included 34 gammaCoVs (23 in wild bird, 11 in poultry) and 18 deltaCoVs (Table 2, Supplementary Table S1). Thirteen deltaCoVs were detected among birds of the order Charadriiformes and 4 deltaCoVs were detected among the birds of the order Anseriformes. The use of NGS made it possible to identify the presence of multiple coronaviruses in one positive sample. The simultaneous presence of genetic material of different coronaviruses in a single sample was detected from 12 avian hosts. The simultaneous presence of both gamma- and deltacoronaviruses isolated from a single bird host was detected in 6 cases in wild birds, representing 23% of all wild bird cases. (Table 2) (23% of all sequenced wild birds).
Evolutionary relationships were determined by phylogenetic analysis of the obtained nucleotide sequences, together with reference sequences. This phylogenetic analysis included sequences of the same region of the RdRp gene of the ratified species and of other relevant reference coronaviruses from GenBank.
Phylogenetic analysis of gammacoronaviruses in poultry and wild birds
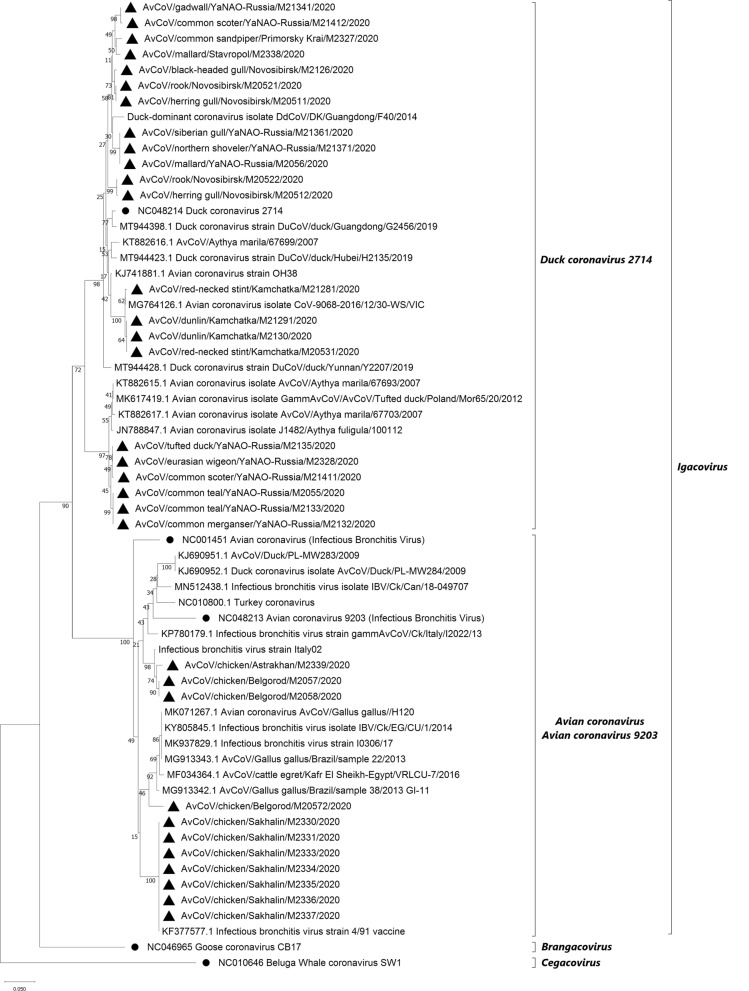
The phylogenetic analysis of the gammaCoVs identified in this study showed that viruses belong to the subgenus Igacovirus and were categorized into two groups—the AvCov/AvCov9203 group and the Duck coronavirus 2714 (DCoV 2714) group (Fig. 1).
Figure 1.
The phylogenetic analysis of the 360 base-pair region of the RdRp gene of select members of gammacoronaviruses by neighbour joining method using 1000 bootstraps. New viruses isolated within Russia are marked with black triangles. Ratified species (ICTV) are marked with black circles. The scale bar refers to a phylogenetic distance of 0.05 nucleotide substitutions per site.
All coronaviruses identified from poultry were grouped with the viruses of AvCov/AvCov9203 species. Partial sequences of the RdRp gene of gammaCoVs detected among chickens in the Astrakhan and Belgorod regions were classified into the group with infectious bronchitis virus strain Italy02 genotype and one virus grouped together with viruses of Massachusetts genotype (Mass genotype, strain AvCoV/Gallus gallus/H120). GammaCoVs detected in chickens in the Sakhalin region grouped with infectious bronchitis vaccine strain 4/91 (Fig. 1).
All gammaCoVs identified among wild birds in Russia belonged to the DCoV 2714 species group of subgenus Igacovirus (Fig. 1). The viruses belonging to the DCoV 2714 group identified in the study were further divided into two tentative subgroups. One subgroup included viruses that showed a high degree of identity with DCoV 2714 ratified species. The second tentative subgroup of viruses included CoVs detected in the Yamalo-Nenets Autonomous Okrug (YaNAO). This tentative subgroup includes only four viruses previously published in the literature and/or sequence repositories: AvCoV/Aythya marila/67693/2007, AvCoV/Aythya marila/67703/2007, GammaCoV/AvCoV/Tufted_duck/Poland/Mor65/20/2012, J1482/Aythya fuligula/100112 (Fig. 1). The nucleotide sequences of the RdRp gene fragment from the viruses within this subgroup were 91% or less similar to other viruses of the DCoV 2714 group (Supplementary Table S2).
Phylogenetic analysis of deltacoronaviruses in wild birds
The phylogenetic analysis based on the partial sequences from the RdRp gene of deltaCoVs identified in this study showed that the viruses belonged to the subgenus Buldecovirus (Fig. 2). One of the studied viruses belonged to the group represented by the ratified species Coronavirus HKU15 isolated from swine. Seven of the studied viruses tentatively grouped with the ratified species White-eye coronavirus HKU16, strain HKU16-6847 (Fig. 2). These 7 viruses were identified from the territory of the Kamchatka Krai, the Novosibirsk region, and the YaNAO, respectively, all belonged to the subgroup that includes previously identified three viruses. In addition, whole genome sequences have been obtained for these three viruses: Falcon coronavirus UAE-HKU27 988F (GenBank ID: LC364342.1), Pigeon coronavirus UAE-HKU29 (GenBank ID: LC364344.1), and Houbara coronavirus UAE-HKU28 285F (GenBank ID: LC364343.1). Ten viruses, whose circulation was detected from the territory of the Kamchatka Krai among birds of the order Charadriiformes, are grouped into a separate tentative subgroup of subgenus Buldecovirus, which includes a virus named Shorebird coronavirus isolate 85 (GenBank ID: JX548304.1). Nucleotide sequences of the RdRp gene fragment of these viruses share 80% or less sequence identity with the closest homologous ratified species and closest species, for which the complete genome is available (Supplementary Table S3).
Figure 2.
The phylogenetic analysis of a 357 base-pair region of the RdRp gene of select members of deltacoronaviruses by neighbour joining method using 1000 bootstraps. New viruses isolated in Russia are marked with black triangles. Ratified species (ICTV) are marked with black circles and other reference viruses with complete, published genome sequence is marked with empty circles. The scale bar refers to a phylogenetic distance of 0.05 nucleotide substitutions per site.
Discussion
Avian coronavirus surveillance in Russia
The reported coronavirus surveillance study among wild and domestic birds was conducted using samples that were collected from several regions that cover a large territory of Russia—from the western regions to the territories of the Russian Far East. The aforementioned surveillance of CoVs was performed for the first time in wild birds in Russia. Analysis of the samples showed that coronaviruses are circulating among wild and domestic birds across various geographic regions. In total, the percentage of avian hosts that were shedding virus in our studies was 12% for the studied birds in Russia (70 out of 584 birds sampled). The percentage of detection of coronaviruses among wild birds was 14.2%, and for poultry 7.3%. Such a high percentage of avian viral hosts that were actively shedding virus identified in our study is consistent with previously reported surveillance data from other countries. A study conducted in Sweden was found that among wild ducks, the percentage of virus carriers reached 18.7%17 and in Australia it was 15.3% among wild birds18. The diversity of birds (including one domestic species), among which coronaviruses were identified, was represented by four orders and 20 species. The orders Anseriformes and Charadriiformes contributed the greatest proportion of viral RNA to this study. Only gammaCoVs were detected among poultry, while both gammaCoVs and deltaCoVs were detected among wild birds. The detection of deltaCoVs in wild birds in Russia is reported for the first time, to our knowledge.
Ten of 18 deltaCoVs were isolated from birds of the order Charadriiformes. Among the confirmed samples with co-infecting avian CoVs, 5 of the 12 samples also belonged to the order Charadriiformes (Table 2).
No circulation of alphacoronaviruses or betacoronaviruses was detected among our sampled birds, which, together with previously reported studies from other countries, which also did not detect alphacoronaviruses and betacoronaviruses in birds4,15, may be a preliminary indication that birds do not currently spread the coronaviruses or that these viruses have not been detected in birds up-to-date. However further monitoring is required due to the ability of these viruses to cross species barriers and due to potential danger of these viruses to human hosts4,25.
Simultaneous presence of gammacoronaviruses and deltacoronaviruses from a single avian host
The coronavirus surveillance in wild and domestic birds within the territory of the Russian Federation in July–December 2020 revealed that a 12% of birds (70 of 584 sampled) carry a variety of deltaCoVs and gammaCoVs.
NGS analysis showed simultaneous presence of genetic material of different gammaCoVs and deltaCoVs from a single avian host in a 12 cases, six of which contained genetic material from viruses of both gammacoronavirus and deltacoronavirus genera. In most previous studies, the genetic characterization of CoVs from collected samples, i.e. swabs or organ homogenate, was performed using Sanger sequencing15,18,26. Unfortunately, Sanger sequencing does not allow for the characterization of two or more viruses in one from a single sample using PCR products. To overcome that limitation, in a study of coronaviruses in quail in Brazil, the cloning of PCR products from individual samples was used and obtained clones were sequenced. This protocol allowed for the detection of the viral RNA from Gammacoronavirus and Deltacoronavirus genera in one sample27. The limited number of publications detailing similar methods suggests that our study is among the first to demonstrate the simultaneous presence of different CoVs in the same bird. Our study demonstrated that the application of NGS is an effective and expedient way of conducting a more comprehensive characterization of surveillance samples, which may contain mixed populations of viruses and pseudo-viruses. The recently published and extensive genome analysis of Coronaviridae family representatives revealed the occurrence of recombination events. These recombination events often involve genomes of closely related viruses of the same subgenera, and in some rare cases, from different subgenera28,29. Although recombination events have not yet been detected among genomes of viruses belonging to different genera, genome analysis has suggested that such non-homologous recombination events may be possible. In addition, it has been demonstrated that genome recombination events may even occur among CoVs and highly dissimilar viruses such as toroviruses, gamma- and deltainfluenza viruses, reoviruses, rotaviruses, astroviruses and may even involve genetic material of infected hosts28. These findings highlight the need for more comprehensive surveillance of CoVs involving methods for evaluation of mixed populations of coronaviruses in various hosts.
Gammacoronaviruses in poultry
All gammaCoVs identified in this study in poultry belonged to two groups represented by viruses from three IBV genotypes: Massachusetts and Italy02 in the western part of Russia, and the 793/B genotype in Sakhalin region. These three genotypes belong to AvCov/AvCov9203 group of subgenus Igacovirus. Coronaviruses of the Massachusetts lineage (first reported in USA) and the 793/B (Europe origin) genotypes are not only found worldwide, but they are the most well-characterised and important IBV genotypes due to their agricultural significance21,30. Genotype 793/B emerged in the UK and France in the 1990s, and became prevalent in Europe and many other parts of the world31. Coronaviruses of Italy 02 genotype were prevalent in Europe a few years ago (in 2002–2004) but are now in decline according to surveillance reports31,32. The infectious bronchitis caused by IBV is a serious poultry disease that may include symptoms such as poor weight gains, coughing, sneezing, egg-laying disorders33 and it is associated with serious economic consequences among unvaccinated flocks21,34. Vaccination is used as an effective measure for the protection of poultry from avian infectious bronchitis. The avian infectious bronchitis vaccine based upon the Massachusetts genotype viruses are produced and used globally, e.g. based on H120 strain. In addition, vaccines based upon viruses of the 793/B genotype, e.g. vaccine strain 4/91, are available and used in many countries31,35.
In previously reported studies (1998–2002, 2007–2010) several IBV genotypes, including the ones genetically characterised from this study, were registered in poultry in Russia. The prevalence and geographical distribution of the detected genotypes varied in different time periods19,20. Data from these and our studies suggests some differences in the spatial distribution of gammaCoVs that are currently circulating in poultry in Russia. The practical importance of research on CoVs for poultry-based agriculture is very high, and such monitoring data is valuable for both risk assessment and the implementation of protective measures. Such protective measures include vaccine development and implementation of region appropriate vaccination strategies.
Gammacoronaviruses in wild birds
All gammaCoVs that were detected from wild birds in this study belonged to two subgroups in the DCoV 2714 group of viruses of subgenus Igacovirus. To our knowledge, this is the first report of detection of DCoV 2714 viruses in Russia. One subgroup of DCoV 2714 gammaCoVs was represented by ratified species Duck coronavirus 2714, which was previously identified in China in 201436. The second subgroup differed significantly from other group of the DCoV 2714 viruses. In this subgroup, only four viruses have been identified and subsequently published: two in Sweden, one from Poland, and one from Hong Kong17,26,37. Similar to this study, a published analysis of strains of this subgroup from Sweden showed 92% or less identity of the ORF1ab gene fragment with strains that were deposited in Genbank in 201617. These data tentatively indicate that these viruses may belong to a new subgroup of gammaCoVs, which are currently not represented by a ratified species.
According to previous studies, gammaCoVs may be transferred between wild birds and poultry. The occurrence of the virus transfer from poultry to wild birds was reported not only for wild type variants, but also for vaccine strains of IBV38,39. Overall, the circulation of a fairly wide variety of gammaCoVs both among wild and domestic birds was detected on the territory of the Russian Federation with potential importance for agriculture.
Deltacoronaviruses in wild birds
The circulation of CoVs that shared sequence similarity with the genus Deltacoronavirus was only detected only among wild birds in this study. This is consistent with the literature, which shows that all previously investigated deltaCoVs have been found in wild birds14,15. The deltaCoVs detected in Russia and reported upon in this study belonged to the subgenus Buldecovirus. Phylogenetic analysis showed that the viruses belonged into three separate groups of Buldecovirus subgenus. One group of deltaCoVs represented in Russia is in the same branch as ratified species White-eye coronavirus HKU16. These group included previously characterized viruses, which had their complete genome sequenced (Falcon coronavirus UAE-HKU27, Houbara coronavirus UAE-HKU28, Pigeon coronavirusUAE-HKU29). The analysis of the viruses showed that they are less than 90% similar in amino acid sequence to White-eye coronavirus HKU16 and other established viruses classified as deltaCoVs12. Ten viruses that were isolated from avian hosts in this study from the territory of the Kamchatka Krai are grouped into a separate branch, which differs from all established species (Supplementary Table S3). This group does not have previously characterized viruses from which whole genome sequences have been obtained. This group includes a previously reported virus named Shorebird coronavirus isolate 85 (GenBank ID: JX548304.1), isolated from the Ruddy turnstone (Arenaria interpres) in 2004, which was isolated in the United States. This virus has been described as a new deltaCoV40. The distinctiveness of the group is supported by phylogenetic analysis reported in previous studies26. However more genomic sequence for the viruses in this group is required for the designation of a new established species according to ICTV criteria. It should be noted that this group contains viruses that have been detected only among birds of order Charadriiformes, which includes the viruses identified during the course of this study in Russia, as well as viruses identified in other countries, including Poland, Australia and the USA15,18,26. One of the viruses identified in the current study belonged to the group that is represented by the ratified species Coronavirus HKU15 isolated from swine (PDCoV)13. This is the only group in genus Deltacoronavirus, which, in addition to avian viruses, contains viruses detected and in some cases, isolated from mammalian species (e.g. pigs, asian leopard cat, ferret badger) from many countries, including China, South Korea, Japan, Poland and the USA15,26. Analysis of the genomes of the viruses in the PDCoV group suggested a recent event of zoonotic transmission from birds to mammals13,15. Global spread of the viruses of this group in birds and various mammalian species, including a recent report of human infection demonstrate that these viruses may be concern for human health15,16.
Wild bird species involved in spreading coronaviruses
In this study, we examined 393 samples from wild birds belonging to 31 species. 380 samples were collected from birds of the Anseriformes and Charadriiformes orders and 13 samples were collected from birds of the Passeriformes order. The proportion of birds actively shedding viruses were the same (14%) for the birds of Anseriformes and Charadriiformes orders. GammaCoVs and deltaCoVs were detected from birds of both orders. These data suggest that birds of these orders play important role in the circulation of CoVs. These results are similar to previously reported studies that also showed important role of the birds of Anseriformes and Charadriiformes orders in avian CoVs ecology41,42.
In this study, the CoVs have been also detected among species such as Rook (Corvus frugilegus) and Hooded crow (Corvus cornix) from the order Passeriformes. It is worth noting that in this study, a virus from the species of Coronavirus HKU15, which have been isolated from both birds and mammals, was detected in a Hooded crow. In prior studies, PDCoVs were detected in sparrows (Passer domesticus), which belong to order Passeriformes15. The order Passeriformes is extremely important in the epidemiology and epizootiology of coronaviruses, since its representatives are often synanthropic species whose lifestyles are often associated with human activities and mayincludes close contacts with people and domestic animals15. The role of this taxonomic group in CoVs carriage and transmission need to be further evaluated.
Most bird species of the Anseriformes and Charadriiformes orders, among which we have reported upon the circulation of AvCoVs, undertake long-distance migration routes in Eurasia, twice per year43,44. For example, the Gadwall (Mareca strepera) migrates to the coastal regions of the Mediterranean and Black Seas, East Africa, the Caspian coast, Iran and India45. Eastern populations migrate to the east and southeast, reaching the territories of southeast Asia and the Japanese islands46. The nesting area of various populations of the Common teal (Anas crecca), a member of the Anatidae family, is extensive47. It inhabits much of western Europe and most of central Asia to northern Iran, northwestern Mongolia and Northeast China, and includes northern Japan and the western part of North America to the Great Lakes47. The spectacular migrations undertaken by these species suggest that the possibility exists for the intermingling of AvCoV viral RNA where these bird species are co-located43–46.
Conclusion
A comprehensive surveillance of various avian coronaviruses among wild and domestic birds was conducted in several regions of the Russian Federation in July–December 2020. This was the first study that included the monitoring of the circulation of CoVs in wild birds in Russia. The study revealed the broad circulation of various coronaviruses in Russia, which are shed and potentially transmitted by birds. This is also the the first study to report a considerable number6 of cases that simultaneously detected the shedding gammaCoVs and deltaCoVs from a single avian host. Viruses detected among wild and domestic birds belonged to groups of established species of gammaCoVs, i.e. AvCov/AvCov9203 group in poultry, DCoV 2714 group in wild birds, and deltaCoVs, which arecurrently designated by Coronavirus HKU15 and White-eye coronavirus HKU16. Genetic diversity of coronaviruses in wild birds was detected within genus Gammacoronavirus (DCoV 2714 group) and in genus Deltacoronavirus (HKU16 group). To our knowledge, this is the first report of the circulation of deltacoronaviruses among wild birds in Russia. No circulation of alphacoronaviruses or betacoronaviruses was detected among domestic or wild birds. Further monitoring is required due to the ability of these viruses to cross species barriers. Similarly to global trends, this study demonstrated the important role of wild birds of the Anseriformes and Charadriiformes orders in the circulation of gammaCoVs and deltaCoVs in Russia. Continuous surveillance of these viruses and further characterisation of avian coronaviruses will contribute to a better understanding of coronavirus spread and variability. This study underscores and provides critical data to the literature that could potentially be of great importance for epidemiological and epizootiological control within ecologically and agriculturally relevant species.
Supplementary Information
Acknowledgements
The reported study was funded by RFBR, project number 20-04-60051.
Author contributions
M.V.: Ideas; formulation of overarching research goals and aims, original draft preparation, supervision, D.A.: PCR analysis, N.G.S. experiments, bioinformatics analysis and data interpretation, writing- reviewing and editing, K.N.: NGS experiments, bioinformatics analysis and data interpretation, writing- reviewing and editing, M.M.: Sample collection and preparation, B.Y.: PCR analysis, B.M.: phylogenetic analysis, NGS experiments, G.V.: Sample collection, original draft reviewing and editing, L.S.: Sample collection, original draft reviewing and editing, G.A.: Sample collection and preparation, O.G.: Sample collection and preparation, S.S.: Sample collection and preparation, R.A.: conceptualisation the overall study and coordinated the investigation.
Data availability
All relevant data are within the paper and its Supporting Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-23925-z.
References
- 1.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alluwaimi AM, Alshubaith IH, Al-Ali AM, Abohelaika S. The coronaviruses of animals and birds: Their zoonosis, vaccines, and models for SARS-CoV and SARS-CoV2. Front. Vet. Sci. 2020 doi: 10.3389/fvets.2020.582287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF-W, To KK-W, Tse H, Jin D-Y, Yuen K-Y. Interspecies transmission and emergence of novel viruses: Lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islam A, Ferdous J, Islam S, Sayeed MA, Dutta Choudhury S, Saha O, et al. Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, domestic animals, and wildlife. Viruses. 2021;13(10):1908. doi: 10.3390/v13101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mihindukulasuriya KA, Wu G, St Leger J, Nordhausen RW, Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008;82(10):5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy JS. Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses: A review. Avian Pathol. 2000;29(3):207–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- 7.Ismail MM, Tang AY, Saif YM. Pathogenicity of turkey coronavirus in turkeys and chickens. Avian Dis. 2003;47(3):515–522. doi: 10.1637/5917. [DOI] [PubMed] [Google Scholar]
- 8.Felippe PAN, da Silva LHA, Santos MMAB, Spilki FR, Arns CW. Genetic diversity of avian infectious bronchitis virus isolated from domestic chicken flocks and coronaviruses from feral pigeons in Brazil between 2003 and 2009. Avian Dis. 2010;54(4):1191–1196. doi: 10.1637/9371-041510-Reg.1. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, Zhang G, Jiang J, Fu J, Ren T, Cao W, et al. A Massachusetts prototype like coronavirus isolated from wild peafowls is pathogenic to chickens. Virus Res. 2007;130(1):121–128. doi: 10.1016/j.virusres.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes LA, Savage C, Naylor C, Bennett M, Chantrey J, Jones R. Genetically diverse coronaviruses in wild bird populations of northern England. Emerg. Infect. Dis. 2009;15(7):1091–1094. doi: 10.3201/eid1507.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong BQ, Liu W, Fan XH, Vijaykrishna D, Tang XC, Gao F, et al. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81(13):6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SKP, Wong EYM, Tsang C-C, Ahmed SS, Au-Yeung RKH, Yuen K-Y, et al. Discovery and sequence analysis of four deltacoronaviruses from birds in the middle east reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J. Virol. 2018;92(15):e00265-18. doi: 10.1128/JVI.00265-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlasova AN, Kenney SP, Jung K, Wang Q, Saif LJ. Deltacoronavirus evolution and transmission: current scenario and evolutionary perspectives. Front. Vet. Sci. 2020;7:626785. doi: 10.3389/fvets.2020.626785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wille M, Holmes EC. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020;44(5):631–644. doi: 10.1093/femsre/fuaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature. 2021;600(7887):133–137. doi: 10.1038/s41586-021-04111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wille M, Muradrasoli S, Nilsson A, Järhult JD. High prevalence and putative lineage maintenance of avian coronaviruses in scandinavian waterfowl. PLoS ONE. 2016;11(3):e0150198. doi: 10.1371/journal.pone.0150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamings A, Nelson TM, Vibin J, Wille M, Klaassen M, Alexandersen S. Detection and characterisation of coronaviruses in migratory and non-migratory Australian wild birds. Sci. Rep. 2018;8(1):5980. doi: 10.1038/s41598-018-24407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochkov YA, Batchenko GV, Shcherbakova LO, Borisov AV, Drygin VV. Molecular epizootiology of avian infectious bronchitis in Russia. Avian Pathol. 2006;35(5):379–393. doi: 10.1080/03079450600921008. [DOI] [PubMed] [Google Scholar]
- 20.Ovchinnikova EV, Bochkov YA, Shcherbakova LO, Nikonova ZB, Zinyakov NG, Elatkin NP, et al. Molecular characterization of infectious bronchitis virus isolates from Russia and neighbouring countries: identification of intertypic recombination in the S1 gene. Avian Pathol. 2011;40(5):507–514. doi: 10.1080/03079457.2011.605782. [DOI] [PubMed] [Google Scholar]
- 21.Bande F, Arshad SS, Omar AR, Hair-Bejo M, Mahmuda A, Nair V. Global distributions and strain diversity of avian infectious bronchitis virus: A review. Anim. Health Res. Rev. 2017;18(1):70–83. doi: 10.1017/S1466252317000044. [DOI] [PubMed] [Google Scholar]
- 22.Spackman E, Killian ML. Avian Influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs BT—Animal Influenza Virus. In: Spackman E, editor. Springer. 2014. pp. 125–140. [DOI] [PubMed] [Google Scholar]
- 23.Vijgen L, Moës E, Keyaerts E, Li S, Van Ranst M. A pancoronavirus RT-PCR assay for detection of all known coronaviruses. Methods Mol. Biol. 2008;454:3–12. doi: 10.1007/978-1-59745-181-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. 2013;16:1303. [Google Scholar]
- 25.Nabi G, Wang Y, Lü L, Jiang C, Ahmad S, Wu Y, et al. Bats and birds as viral reservoirs: A physiological and ecological perspective. Sci. Total Environ. 2021;754:142372. doi: 10.1016/j.scitotenv.2020.142372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domańska-Blicharz, K., Miłek-Krupa, J., Pikuła, A. Diversity of coronaviruses in wild representatives of the aves class in Poland. Viruses. 13 (2021). [DOI] [PMC free article] [PubMed]
- 27.Torres CA, Hora AS, Tonietti PO, Taniwaki SA, Cecchinato M, Villarreal LYB, et al. Gammacoronavirus and deltacoronavirus in Quail. Avian Dis. 2016;60(3):656–661. doi: 10.1637/11412-032316-Reg.1. [DOI] [PubMed] [Google Scholar]
- 28.Nikolaidis M, Markoulatos P, Van de Peer Y, Oliver SG, Amoutzias GD. The neighborhood of the spike gene is a hotspot for modular intertypic homologous and nonhomologous recombination in coronavirus genomes. Mol. Biol. Evol. 2022;39(1):msab292. doi: 10.1093/molbev/msab292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amoutzias GD, Nikolaidis M, Tryfonopoulou E, Chlichlia K, Markoulatos P, Oliver SG. The remarkable evolutionary plasticity of coronaviruses by mutation and recombination: Insights for the COVID-19 pandemic and the future evolutionary paths of SARS-CoV-2. Viruses. 2022;14(1):78. doi: 10.3390/v14010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houta MH, Hassan KE, El-Sawah AA, Elkady MF, Kilany WH, Ali A, et al. The emergence, evolution and spread of infectious bronchitis virus genotype GI-23. Arch. Virol. 2021;166(1):9–26. doi: 10.1007/s00705-020-04920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackwood MW. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56(4):634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- 32.Cortés V, Sevilla-Navarro S, García C, Marín C, Catalá-Gregori P. Seroprevalence and prevalence of Infectious Bronchitis Virus in broilers, laying hens and broiler breeders in Spain. Poult. Sci. 2022;101(5):101760. doi: 10.1016/j.psj.2022.101760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ignjatović J, Sapats S. Avian infectious bronchitis virus. Rev. Sci. Tech. 2000;19(2):493–508. doi: 10.20506/rst.19.2.1228. [DOI] [PubMed] [Google Scholar]
- 34.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 35.Jordan B. Vaccination against infectious bronchitis virus: A continuous challenge. Vet. Microbiol. 2017;206:137–143. doi: 10.1016/j.vetmic.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang Q-Y, Wang K-C, Liu S, Hou G-Y, Jiang W-M, Wang S-C, et al. Genomic analysis and surveillance of the coronavirus dominant in ducks in China. PLoS ONE. 2015;10(6):e0129256. doi: 10.1371/journal.pone.0129256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu DKW, Leung CYH, Gilbert M, Joyner PH, Ng EM, Tse TM, et al. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85(23):12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34(6):439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 39.Rohaim MA, El Naggar RF, Helal AM, Hussein HA, Munir M. Reverse spillover of avian viral vaccine strains from domesticated poultry to wild birds. Vaccine. 2017;35(28):3523–3527. doi: 10.1016/j.vaccine.2017.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honkavuori KS, Briese T, Krauss S, Sanchez MD, Jain K, Hutchison SK, et al. Novel coronavirus and astrovirus in delaware bay shorebirds. PLoS ONE. 2014;9(4):e93395. doi: 10.1371/journal.pone.0093395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman MM, Talukder A, Chowdhury MMH, Talukder R, Akter R. Coronaviruses in wild birds—A potential and suitable vector for global distribution. Vet. Med. Sci. 2021;7(1):264–272. doi: 10.1002/vms3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wille M, Shi M, Klaassen M, Hurt AC, Holmes EC. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 2019;13(10):2603–2616. doi: 10.1038/s41396-019-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FAO. Wild Birds and Avian Influenza: An Introduction to Applied Field Research and Disease Sampling Techniques (eds. Whitworth, D., Newman, S. H., Mundkur, T. & Harris, P. (FAO Animal Production and Health Manual, No. 5, 2007). www.fao.org/avianflu.
- 44.Delany, S., Scott, D., Dodman, T., Stroud, D. The Wader Atlas: An Atlas of Wader Populations in Africa and Western Eurasia, Wetlands International, Wageningen, ISBN: 78-90-5882-047-1.
- 45.Scott, D. A. & Rose, P. M. Atlas of Anatidae Populations in Africa and Western Eurasia (Wetlands International Publication No. 41, Wetlands International, Wageningen, 1996).
- 46.Miyabayashi, Y. & Mundkur, T. Atlas of Key Sites for Anatidae in the East Asian Flyway (Wetlands International, 1999).
- 47.Veen, J., Yurlov, A. K., Delany, S. N., Mihantiev, A. I., Selivanova, M. A., Boere, G. C. An Atlas of Movements of Southwest Siberian Waterbirds. (Wetlands International, 2005). ISBN: 9789058829528.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.