Abstract
The ActA protein of Listeria monocytogenes is an essential virulence factor and is required for intracellular bacterial motility and cell-to-cell spread. plcB, cotranscribed with actA, encodes a broad-specificity phospholipase C that contributes to lysis of host cell vacuoles and cell-to-cell spread. Construction of a transcriptional fusion between actA-plcB and the green fluorescent protein gene of Aequorea victoria has facilitated the detailed examination of patterns of actA/plcB expression within infected tissue culture cells. actA/plcB expression began approximately 30 min postinfection and was dependent upon entry of L. monocytogenes into the host cytosol. L. monocytogenes Δhly mutants, which are unable to escape from host cell vacuoles, did not express actA/plcB at detectable levels within infected tissue culture cells; however, complementation of the hly defect allowed entry of the bacteria into the host cytoplasm and subsequent actA/plcB expression. These results emphasize the ability of L. monocytogenes to sense the different host cell compartment environments encountered during the course of infection and to regulate virulence gene expression in response.
Listeria monocytogenes is a facultative intracellular bacterial pathogen responsible for serious disease in immunocompromised patients and pregnant women (19, 42). L. monocytogenes enters host cells and escapes from membrane-bound vacuoles into the cytoplasm, where it begins to replicate (34, 53). Shortly after entry into the cytosol, the bacteria begin to move and spread to adjacent cells by using a host actin polymerization-based motility (11, 34, 41, 51, 53). Several gene products that are important for intracellular bacterial growth and cell-to-cell spread have been identified and described (reviewed in reference 22, 36, 43, and 46). These gene products include the hemolysin listeriolysin O, encoded by hly, which is required for escape of L. monocytogenes from host cell vacuoles; ActA, which is essential for actin polymerization-based bacterial motility and cell-to-cell spread; and a broad-range phospholipase C (PC-PLC) encoded by plcB that enhances escape from primary and secondary vacuoles (30, 45).
L. monocytogenes is similar to several other intracellular bacterial pathogens, such as Salmonella typhimurium (1, 5, 20, 29) and Legionella pneumophila (2, 3, 4, 49), in that it possesses gene products that are preferentially expressed within host cells or tissues. Gene products that facilitate escape of L. monocytogenes from primary vacuoles (listeriolysin O and plcA-encoded phosphatidylinositol-specific phospholipase C) are expressed when bacteria are grown in standard broth culture. However, gene products that contribute to intracellular spread of L. monocytogenes, such as those encoded by mpl, actA, and plcB, are expressed at low-to-undetectable levels by bacteria grown in standard broth culture, and expression increases following entry of L. monocytogenes into the host cell cytoplasm. As an example, immunoprecipitation experiments using antibodies directed against ActA have indicated that the protein becomes one of the most abundant bacterial surface proteins expressed by intracytoplasmic L. monocytogenes (7). L. monocytogenes strains that express high levels of actA (as well as hly and plcB) after growth in rich broth at 37°C have been described (for a review, see reference 46), but a study of a wide panel of L. monocytogenes isolates has suggested that these hypersecreting strains are variants or mutants (39). Indeed, Ripio et al. (38) have recently demonstrated that several L. monocytogenes strains that express elevated levels of virulence factors all carry a point mutation within prfA that converts the gene product to an “activated” state. Additional L. monocytogenes gene products preferentially expressed within infected mammalian cells have been described (25). These experiments and others suggest that L. monocytogenes is capable of sensing the different host cell environments it encounters during the course of infection and responding with the regulated expression of specific virulence gene products.
We are interested in understanding how L. monocytogenes regulates gene expression within infected host cells. As an initial step, it seemed important to better define the patterns of L. monocytogenes gene expression within infected host cells. This study describes the use of the green fluorescent protein (GFP) reporter gene system from Aequorea victoria to monitor L. monocytogenes intracellular gene expression. The use of GFP as a reporter has several advantages over other reporter gene systems in that it requires no cofactors and can be used in examination of fixed samples (10). GFP has been used to successfully monitor the expression of Mycobacterium smegmatis and Mycobacterium bovis BCG promoters within macrophages (14), as well as in the isolation of S. typhimurium genes preferentially expressed following bacterium-host association (54). We report here the successful use of GFP to monitor the timing and patterns of L. monocytogenes actA and plcB expression within different host cell compartments of infected tissue culture cells. actA/plcB expression was evident within 30 min to 1 h postinfection and was dependent upon the ability of L. monocytogenes to reach the host cytosol. actA/plcB did not appear to be expressed by bacteria located within host cell vacuoles. Our results indicate that GFP functions as a useful reporter system for the monitoring of the timing of L. monocytogenes intracellular gene expression and for determination of cell compartment expression patterns within infected host cells.
MATERIALS AND METHODS
Bacterial strains, growth media, and plasmids.
The bacterial strains used in this study are listed in Table 1. L. monocytogenes 10403S (serotype 1/2b) is resistant to streptomycin, and its 50% lethal dose for mice is 2 × 104 CFU (18). L. monocytogenes was stored at −70°C in brain heart infusion broth (BHI; Difco Laboratories, Detroit, Mich.) containing 20% glycerol. Escherichia coli HB101 or DH5α was used as the host strain for recombinant plasmids. All E. coli strains were grown in Luria-Bertani broth (12). Antibiotics were used at the following concentrations: carbenicillin, 50 μg/ml; chloramphenicol, 10 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Straina | Relevant characteristics(s) | Source or reference |
|---|---|---|
| 10403S | Wild type | 18 |
| NF-L357 | actA-gfp-plcB fusion | This work |
| DP-L2161 | Δhly | 24 |
| NF-L404 | Δhly actA-gfp-plcB fusion | This work |
| NF-L411 | NF-L404 + hlyb | This work |
| NF-L497 | Δhly + Pspac-gfpc | This work |
All strains were derived from 10403S.
hly provided by plasmid pDP906 (24).
IPTG-inducible gfp plasmid pNF496 (this work).
The thermosensitive shuttle vector plasmid pKSV7 has been described previously (47). Plasmid pSPAC, a shuttle vector containing the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter (56), and plasmid pRAY1, containing the A. victoria GFP gene (32), were gifts from David Dubnau. Plasmid pDP906, containing the hly gene, has already been described (24).
Construction of actA-gfp-plcB transcriptional gene fusion mutants.
Primers GFP-1 (5′-GCTCTAGAAGGAGGAAAAATATGAGTAAAGGAGAAGAAC-3′) and GFP-2A (5′-AACTGCAGCTATTTGTATAGTTCATCC-3′) were designed to amplify gfp coding sequences from plasmid pRAY1 by PCR (21) and to introduce a gram-positive ribosome binding site derived from SD1 of ermC (13) (underlined sequence of GFP-1) upstream of gfp. The PCR-amplified product was digested with XbaI and PstI and subcloned into pKSV7 (47) to yield pNF320. Primers ActA-1 (5′-GCGAATTCGAGTTGAACGGGAGAGGC-3′) and ActA-2 (5′-GCTCTAGAGTGTTTTTAATTATTTTTTC-3′) were used in conjunction with L. monocytogenes genomic DNA to PCR amplify an approximately 760-bp product containing C-terminal sequences of actA. The actA PCR product was digested with EcoRI and XbaI and subcloned upstream of gfp in plasmid pNF320 to yield plasmid pNF326. Primers PlcB-1 (5′ GCCTGCAGCGAAAGAAAAAGTGAGGT 3′) and PlcB-2 (5′ GCAAGCTTCGGGTAGTCCGCTTTCGC 3′) were used in conjunction with L. monocytogenes genomic DNA to PCR amplify an approximately 740-bp fragment containing plcB upstream and N-terminal coding sequences. This PCR product was digested with PstI and HindIII and subcloned downstream of gfp in plasmid pNF326 to yield plasmid pNF333. The pNF333 plasmid thus contains a transcriptional fusion of gfp to the actA gene of L. monocytogenes, as well as flanking L. monocytogenes chromosomal regions for introduction of the actA-gfp-plcB fusion into the L. monocytogenes chromosome via homologous recombination.
Construction of plasmid pSPAC-GFP.
Primers GFP-1 and GFP-2A (described above) were used to PCR amplify gfp from plasmid pRAY1. The purified gfp PCR product was digested with XbaI and PstI and subcloned into plasmid pSPAC to yield plasmid pSPAC-GFP.
Transfer of actA-gfp-plcB transcriptional fusions to the L. monocytogenes chromosome.
Plasmid pNF333 was introduced into L. monocytogenes 10403S and DP-L2161 (24) by electroporation (35), and transformants were isolated by growth at 30°C on BHI agar containing chloramphenicol. L. monocytogenes colonies containing the actA-gfp-plcB transcriptional fusion in single copy on the bacterial chromosome were isolated by a procedure previously described (9, 17) and designated NF-L357 (actA-gfp-plcB in strain 10403S) and NF-L404 (actA-gfp-plcB in strain DP-L2161). Southern analysis (40) was used to confirm the existence of the actA-gfp-plcB fusion in the correct location and in single copy within the L. monocytogenes chromosome.
Transformation of L. monocytogenes with pSPAC-GFP and pDP906 plasmids.
Plasmid pSPAC-GFP was introduced into L. monocytogenes 10403S and DP-L2161, and plasmid pDP906 was introduced into L. monocytogenes NF-L404 by electroporation (35), and transformants were isolated by growth at 37°C on BHI agar containing chloramphenicol.
Plaque formation in L2 cells.
Plaque assays were carried out as previously described by Sun et al. (48). Plaque size was measured as described by Camilli et al. (9).
Intracellular growth assays.
The cell lines used in these studies were the J774 mouse macrophage-like cell line and the potoroo kidney epithelial cell line PtK2; both were maintained as previously described (7, 51). Intracellular growth in J774 cells was monitored as described by Portnoy et al. (37). PtK2 cells were grown as previously described (51) on acid-washed glass coverslips and infected with approximately 2 × 107 CFU of L. monocytogenes 10403S or mutant strains for 60 min (multiplicity of infection, ∼10 CFU/cell). In experiments designed to examine the intracellular growth and GFP fluorescence of Δhly L. monocytogenes strains, infections of PtK2 cells were carried out with approximately 2 × 109 CFU of bacteria. This high concentration of bacteria resulted in the infection of nearly every PtK2 cell with one to two bacteria and increased the number of intracellular bacteria that could be visualized in each microscopic field. Following infection, cell monolayers were washed three times with 37°C phosphate-buffered saline (PBS), and then 5 ml of prewarmed medium containing 50-μg/ml gentamycin was added. At the indicated time points, coverslips were removed and processed for either bright-field or fluorescence microscopy. For bright-field microscopy, coverslips were stained with Diff-Quik (VWR Scientific, Chicago, Ill.) and mounted in Permount mounting medium (Fisher Scientific, Philadelphia, Pa.). For fluorescence microscopy, the cells were fixed by placing a drop of 3.7% formaldehyde in PBS on each coverslip and incubating it at room temperature for 5 min. The coverslips were then rinsed in PBS and mounted in Permafluor mounting medium (Immunon, Pittsburgh, Pa.) and allowed to set overnight. For experiments using rhodamine phalloidin, following formaldehyde fixation of cells, 1 drop of 0.1% Triton was placed on each of the coverslips, which were then incubated for 3 to 5 min at room temperature. Coverslips were washed by being dipped in PBS, and then 100 μl of 300 mM rhodamine phalloidin in 1-mg/ml bovine serum albumin in PBS was added to each coverslip. After 20 min at room temperature, the coverslips were washed in PBS, drained, and mounted in Permafluor. L. monocytogenes containing the actA-gfp-plcB reporter gene fusion were visualized in at least 20 separate microscopic fields for each data group. Bacteria were first visualized in a single field by using the fluorescein isothiocyanate (FITC) filter set (see below) and then scored for the presence of F-actin by switching to the rhodamine filter set. The data shown represents at least three independent experiments.
To quantitate percentages of fluorescent bacteria versus the total numbers of intracellular bacteria, PtK2 cells were infected with L. monocytogenes actA-gfp-plcB as described above and coverslips were removed at 30 min, 1 h, 1.5 h, and 2 h postinfection and fixed in PBS–3.2% formaldehyde. Coverslips were washed with PBS, and then 1 drop of PTB (0.1% Triton, 1-mg/ml bovine serum albumin in PBS) with a 1:320 dilution of Listeria O rabbit antiserum (Difco, Detroit, Mich.) was added and the coverslips were incubated at room temperature for 30 min. After washing, 1 drop of tetramethylrhodamine-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, Oreg.) at 5 μg/ml in PTB was added and the coverslips were incubated at room temperature for an additional 30 min. Coverslips were then washed in PTB and mounted in Permafluor. The relative percentage of intracellular versus total bacteria for each time point was determined by removing additional coverslips from each dish of infected cells at each time point and following the procedure described above with the omission of 0.1% Triton. Bacteria were counted in at least 50 different microscopic fields for each time point, and the number of intracellular L. monocytogenes bacteria (total number of bacteria times the percentage of intracellular bacteria) that were positive for GFP fluorescence was scored.
IPTG-mediated induction of gfp expression in PtK2 cells infected with NF-L497.
PtK2 cells were grown as previously described (51) on acid-washed glass coverslips and infected with approximately 109 CFU of L. monocytogenes NF-L497 for 60 min. Monolayers were washed three times with 37°C PBS, and then 5 ml of prewarmed medium containing 50-μg/ml gentamicin was added. After an additional 60 min, IPTG was added to a final concentration of 2 mM. At the indicated time points, coverslips were removed and processed for either bright-field or fluorescence microscopy.
Fluorescence microscopy and imaging.
Cells were observed by using a Zeiss Axiophot fluorescence microscope coupled to either a Diagnostic Instruments SPOT Digital Camera or a DAGE/MTI SIT camera. The filter sets used were Zeiss filter set no. 487915 for monitoring of rhodamine fluorescence and Zeiss FITC filter set no. 487910 with the substitution of an Omega short-pass emission filter (530 nm ± 20 nm, used to reduce rhodamine bleedthrough) for monitoring of GFP fluorescence.
RESULTS
Construction of L. monocytogenes chromosomal actA-gfp-plcB transcriptional fusion mutants.
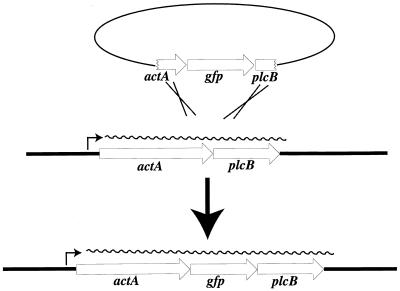
The PCR was used to amplify the A. victoria gfp gene (10) with the addition of a gram-positive ribosome binding site. A transcriptional fusion between actA and gfp was constructed in plasmid pKSV7 (47) and introduced into the L. monocytogenes chromosome via homologous recombination as described in Materials and Methods (Fig. 1). Two L. monocytogenes strains were used as recipients of the actA-gfp fusion: 10403S, the wild type strain, to generate NF-L357, and DP-L2161, a strain derived from 10403S that contains an hly deletion (24), to generate NF-L404. NF-L357 and NF-L404 each contain a single copy of a promoterless gfp gene located between actA and plcB in the L. monocytogenes chromosome.
FIG. 1.
Construction of an actA-gfp-plcB transcriptional gene fusion in L. monocytogenes. Expression of actA and plcB is transcriptionally coupled. A promoterless copy of gfp was inserted between the actA and plcB coding regions and then introduced into the L. monocytogenes chromosome via homologous recombination.
The actA-gfp-plcB transcriptional fusion does not affect intracellular bacterial growth or cell-to-cell spread.
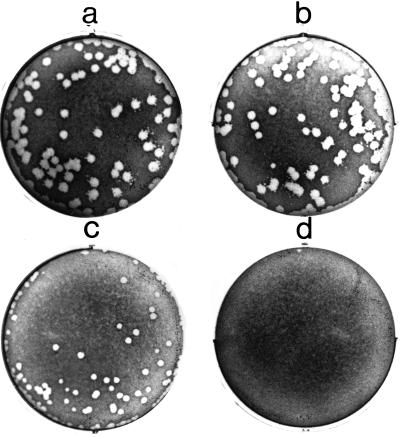
The capacity of L. monocytogenes to escape from a vacuole, replicate intracellularly, and spread to adjacent cells can be measured by the ability of the bacteria to form plaques in monolayers of mouse L cells. L. monocytogenes mutants lacking functional ActA or PC-PLC can be identified by their inability to form wild-type size plaques (26, 45, 55). We examined the plaque-forming ability of each of the L. monocytogenes actA-gfp-plcB fusion strains (Fig. 2). NF-L357 (Fig. 2a), which contains the actA-gfp-plcB chromosomal fusion in a wild-type background, formed plaques of the same approximate size and frequency as the parent strain (Fig. 2b). NF-L404, which contains a deletion within hly and does not escape from host cell vacuoles, did not form visible plaques in L2 cell monolayers (Fig. 2d). Complementation of NF-L404 by the introduction of plasmid-encoded hly (NF-L411) restored the ability of this strain to form plaques; however, the plaques were smaller than those formed by wild-type L. monocytogenes (Fig. 2c). The inability of hly on a plasmid to completely restore plaque size may result from the fact that the hly promoter is present in multiple copies and results in titration of PrfA, the transcriptional activator required for expression of several L. monocytogenes virulence genes (9, 24).
FIG. 2.
Plaque assay of intracellular growth and cell-to-cell spread of L. monocytogenes strains containing actA-gfp-plcB fusions. Confluent monolayers of mouse L2 fibroblast cells were infected with the indicated L. monocytogenes strain and stained after 3 days with neutral red. Panels: a, wild-type 10403S; b, NF-L357; c, NF-L411; d, NF-L404.
Intracellular growth of the actA-gfp-plcB fusion strains was also examined in the mouse macrophage-like cell line J774 and in the potoroo kidney epithelial cell line PtK2, as well as in mouse bone marrow-derived macrophages (23). Normal growth and spread were observed for NF-L357, whereas the NF-404 Δhly actA-gfp-plcB strain failed to replicate intracellularly. The results of the plaque assay and tissue culture infection experiments indicate that the introduction of gfp into the L. monocytogenes chromosome between actA and plcB produced no discernible effect on intracellular growth or cell-to-cell spread of the bacteria.
Characterization of intracellular L. monocytogenes actA-gfp-plcB expression by fluorescence in tissue culture cell lines.
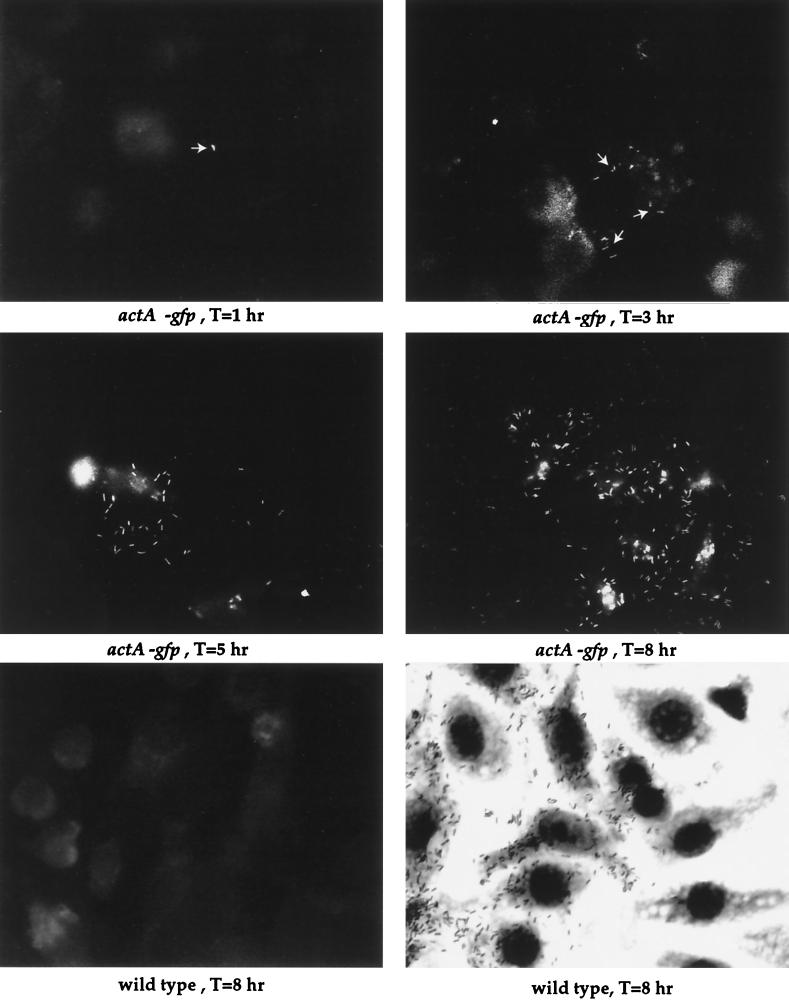
L. monocytogenes mutants containing the actA-gfp-plcB chromosomal fusion were examined by fluorescence microscopy following infection of mouse macrophage-like cell line J774 (Fig. 3). Fluorescent bacteria of strain NF-L357 were detectable between 30 min and 1 h postinfection. The numbers of fluorescent bacteria and the levels of fluorescence observed increased as the bacteria multiplied and spread to adjacent cells (Fig. 3, middle panels). No bacterial fluorescence was detected in cells infected with the 10403S parent strain at any time point postinfection (Fig. 3, bottom panel).
FIG. 3.
Intracellular fluorescence of NF-L357 in mouse macrophage-like cell line J774. Monolayers of J774 cells were grown on glass coverslips and infected with either NF-L357 or the wild-type 10403S strain as indicated. Coverslips were removed at the indicated time points and fixed prior to fluorescence microscopy. T, time postinfection.
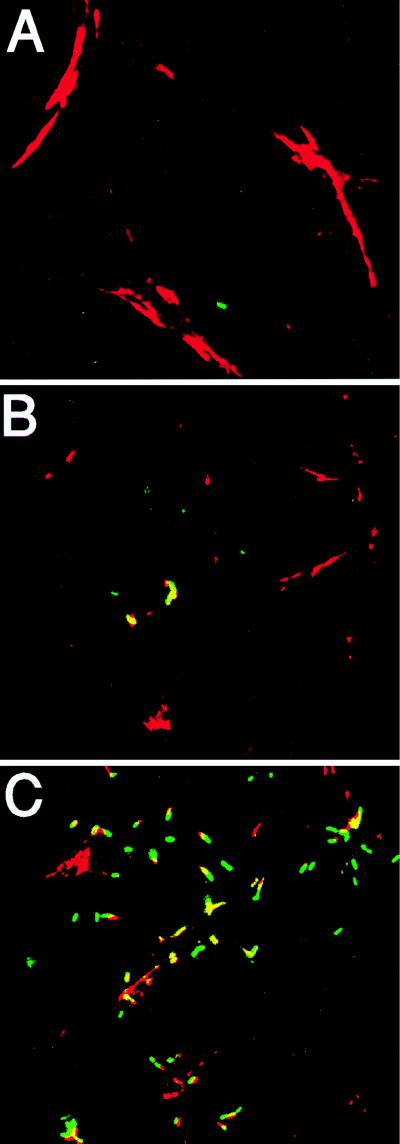
We also investigated infections of PtK2 cells, a potoroo kidney epithelial cell line that is relatively flat. Fluorescent bacteria were more easily visualized in this cell line (in comparison to J774 cells) with less background contributed from surrounding planes of focus. Fluorescent bacteria were initially detected beginning at approximately 30 min to 1 h postinfection. At the earliest time point examined, 30 min postinfection, approximately 80% of the intracellular NF-L357 bacteria were expressing actA-gfp, and by 2 h, at least 95% of the intracellular NF-L357 bacteria were scored as GFP positive (23). To obtain a functional correlation of actA expression with the expression patterns detected via actA-dependent GFP fluorescence, infected cells were permeabilized and stained with rhodamine phalloidin to allow detection of F-actin filaments accumulating around intracytoplasmic bacteria (Fig. 4 and Table 2). With a multiplicity of infection of approximately 10 CFU per cell, at 1.5 h postinfection L. monocytogenes bacteria expressing GFP are visible but detection of bacteria requires examination of large numbers of infected cells. Approximately 80% of the bacteria detected are expressing GFP in the absence of F-actin staining. The number of bacteria that express GFP and colocalize with F-actin increases steadily, such that at 2.5 h postinfection, over 70% colocalize, and at 4 h postinfection, greater than 90% of the bacteria colocalize with F-actin as detected by fluorescence staining. At 6 h postinfection, bacteria could be clearly seen either surrounded by actin filaments or followed by actin filament comet tails (Fig. 4C). These results indicated that actA expression, as detected by GFP fluorescence, preceded the accumulation of F-actin around intracytoplasmic bacteria. actA/gfp/plcB expression patterns therefore closely correlated with the functional appearance of ActA protein.
FIG. 4.
Intracellular fluorescence and F-actin staining of L. monocytogenes-infected PtK2 cells. Monolayers of PtK2 cells were grown on coverslips and infected with the NF-L357 actA-gfp-plcB fusion strain. Coverslips were removed at 3 h postinfection (A), 3 h postinfection (B) (an independent field adjacent to that shown in panel A), and 6 h postinfection (C) and treated with rhodamine phalloidin to identify F-actin. The yellow areas indicate overlapping fluorescence of GFP (green fluorescence) and F-actin (red fluorescence).
TABLE 2.
Quantitation of GFP expression versus F-actin staining of L. monocytogenes actA-gfp-plcB mutants following infection of PtK2 cells
| Expt no. and time (h) post-infection | No. of GFP-positive bacteriaa | No. of GFP- and F-actin- positive bacteriab | % GFP+ and:
|
|
|---|---|---|---|---|
| F-actin− | F-actin+ | |||
| 1 | ||||
| 1.5 | 26 | 2 | 92 | 8 |
| 2.0 | 34 | 8 | 76 | 24 |
| 2.5 | 61 | 50 | 18 | 82 |
| 4.0 | 65 | 63 | 3 | 97 |
| 2 | ||||
| 1.5 | 15 | 3 | 80 | 20 |
| 2.0 | 50 | 17 | 66 | 34 |
| 2.5 | 59 | 42 | 29 | 71 |
| 4.0 | 60 | 53 | 12 | 88 |
| 3 | ||||
| 1.5 | 63 | 17 | 73 | 27 |
| 2.0 | 62 | 27 | 56 | 44 |
| 2.5 | 57 | 38 | 34 | 66 |
| 4.0 | 55 | 51 | 7 | 93 |
| Mean ± SDc | ||||
| 1.5 | 82 ± 7 | 18 ± 7 | ||
| 2.0 | 66 ± 7 | 34 ± 7 | ||
| 2.5 | 27 ± 6 | 73 ± 6 | ||
| 4.0 | 7 ± 3 | 93 ± 3 | ||
Number of intracellular L. monocytogenes bacteria that scored positive for GFP expression following examination of infected cells by fluorescence microscopy. Numbers are taken from at least 20 different microscopic fields equivalent in size.
Number of intracellular L. monocytogenes bacteria that scored positive for GFP expression and F-actin accumulation following examination of cells by fluorescence microscopy. Numbers are taken from at least 20 different microscopic fields equivalent in size.
Mean values derived from three individual experiments are shown. Similar results were obtained from two additional independent experiments (data not shown).
L. monocytogenes actA/plcB expression is influenced by intracellular compartment location.
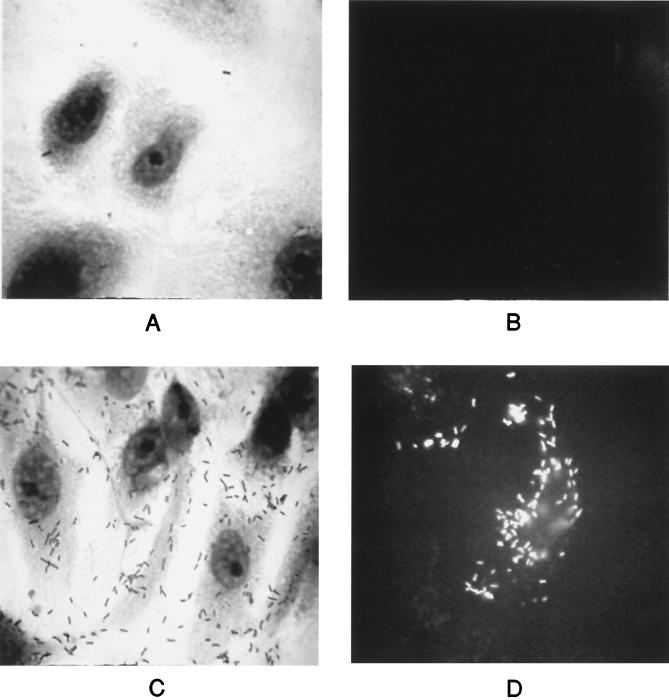
L. monocytogenes encounters several different host cell compartment environments during the course of infection, and these environments are thought to influence bacterial gene expression. As mentioned previously, immunoprecipitation experiments have demonstrated that ActA becomes one of the dominant surface proteins expressed by L. monocytogenes in the cell cytoplasm (7). Dietrich et al. (15) have recently reported that the actA promoter is preferentially activated in the cytosol of the infected host cell; however, it has not been definitively shown whether actA transcription occurs while L. monocytogenes resides within host cell phagosomes or if expression is absolutely dependent upon entry of the bacteria into the cytoplasm. L. monocytogenes NF-L404, which contains the actA-gfp-plcB transcriptional fusion in a Δhly background (24), was used to examine actA/plcB expression by bacteria trapped within host primary vacuoles. When PtK2 cells were infected with the L. monocytogenes Δhly actA-gfp-plcB fusion mutant, no fluorescence was detected at any time point postinfection (Fig. 5B). However, when the hly defect of this mutant was complemented by the introduction of plasmid-encoded hly, bacteria were able to escape from host vacuoles and multiply within the cytoplasm, where actA/gfp/plcB expression was readily detectable (Fig. 5D). These results indicate that actA/plcB expression does not occur while bacteria are located within host cell primary vacuoles (or occurs at low levels not detectable by this assay) and suggest that expression is triggered when the bacteria encounter a specific target host cell environment such as the cytosol.
FIG. 5.
L. monocytogenes actA/plcB is not expressed in primary host cell vacuoles. PtK2 cells were infected with NF-L404 (Δhly actA-gfp; A and B) or NF-L411 (NF-L404 plus plasmid-encoded hly; C and D). Infected cells were fixed and stained (A and C) or examined for fluorescence (B and D) at 8 h postinfection. At least 50 microscopic fields were examined per experiment; one of three experiments with similar results is shown.
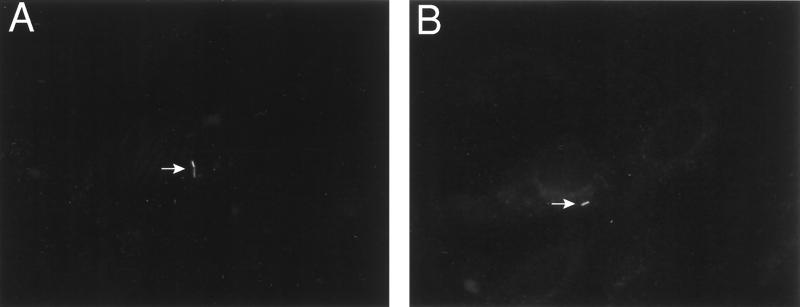
To verify that gene expression occurring in bacteria located within primary vacuoles would be detectable in our system using GFP as a reporter, L. monocytogenes Δhly mutants were transformed with plasmid pNF496, which contains gfp under the control of the IPTG-inducible Pspac promoter (56), to generate NF-L497. PtK2 cells were infected for 1 h with NF-L497 in the absence of IPTG induction. Infected cells were then washed, and gentamicin was added to kill any remaining extracellular bacteria. Following 1 h of gentamicin treatment, IPTG was added at a final concentration of 2 mM. Under these conditions, gfp expression should be induced only in the viable L. monocytogenes bacteria that were located within host cell vacuoles and that were thereby protected from gentamicin exposure. After 4 h of IPTG induction, the PtK2 cells were fixed and examined for fluorescent bacteria. Fluorescent bacteria located within host cell vacuoles were easily detectable (Fig. 6A and B), although it should be noted that these bacteria represented a relatively small percentage (approximately 10%) of the intracellular L. monocytogenes. The small numbers of fluorescent bacteria detected may be due in part to the requirement for IPTG to diffuse into L. monocytogenes-containing vacuoles at sufficient concentrations for induction of gene expression. These experiments demonstrate that it was therefore possible to observe GFP-mediated fluorescence from individual bacteria located within host vacuoles provided that gfp expression was occurring.
FIG. 6.
Induced expression of gfp in L. monocytogenes located within primary host cell vacuoles. NF-L497, a Δhly strain that contains a plasmid encoding an IPTG-inducible copy of gfp, was used in infection assays of PtK2 cells. At 1 h postinfection, monolayers were washed and treated with gentamicin to kill extracellular bacteria. At 2 h postinfection, 2 mM IPTG was added to induce gfp expression. Cells were fixed and examined for fluorescent bacteria at 4 h postinfection. White arrows identify fluorescent bacteria located within host cell vacuoles. At least 50 microscopic fields were examined per experiment; one of two experiments with similar results is shown.
DISCUSSION
It has been possible to define in detail the patterns of L. monocytogenes actA-plcB expression within infected host cells through the construction of transcriptional gene fusions with the gfp gene of A. victoria. Observation of infected tissue culture cells by fluorescence microscopy allowed determination of both the timing of actA-plcB expression and the intracellular location at which expression occurred. Such information should prove useful in gaining a better understanding of how L. monocytogenes responds to the different host cell compartment environments it encounters during the course of infection.
ActA and PC-PLC are critical components of L. monocytogenes pathogenesis. L. monocytogenes mutants that contain insertions or in-frame deletions within actA invade host cells and multiply within the cytoplasm but do not spread to adjacent cells and are 1,000-fold less virulent than wild-type bacteria in a mouse model of infection (7, 16, 26). ActA-dependent actin polymerization by L. monocytogenes has been examined in many cell types (7, 11, 16, 26–28, 51–53), as well as in cell extracts (50). Examination of the precise function and sites of action of PC-PLC has been more difficult. plcB deletion mutants are approximately 10-fold less virulent in mouse models of infection (45). PC-PLC is secreted as an inactive proenzyme; the activated form has recently been shown to localize within Lamp1-positive vacuoles thought to be formed during bacterial cell-to-cell spread (31). Our results suggest that activation of actA/plcB expression occurs shortly after L. monocytogenes gains access to the host cell cytosol. Timing of actA-plcB expression as monitored by GFP fluorescence correlated well with the detection of actin filaments surrounding the bacteria in our studies and agreed with previous actin filament data obtained in other laboratories (11, 26–28, 53). Preliminary data obtained at early time points postinfection suggests that actA/plcB expression may occur more rapidly in phagocytic cells, such as J774 cells, than in nonprofessional phagocytic cells such as PtK2 cells (23). The delay in actA/plcB expression observed in PtK2 cells may reflect the time needed for the bacteria to actively invade the host cell; alternatively, the timing of bacterial escape from the host cell vacuoles may vary between cell types.
By utilizing previously constructed and characterized L. monocytogenes Δhly mutants (24), it was possible to demonstrate that actA/plcB does not appear to be expressed by bacteria located within host cell vacuoles. Fluorescence was observed for Δhly mutants containing plasmid-encoded gfp under the control of an inducible promoter, indicating that fluorescence occurring within a single bacterium located in a vacuole could be detected. The IPTG-inducible Pspac promoter (56) is not a strong promoter in L. monocytogenes, thus, we believe that GFP fluorescence provides a relatively sensitive indicator of gene expression. However, previous experiments have demonstrated that PC-PLC is capable of mediating lysis of the primary vacuole in Henle 407 human epithelial cells (30), indicating that plcB expression does, in fact, occur in this environment. L. monocytogenes infections of Henle 407 cells are unusual in that bacterial lysis of the primary vacuole does not require the action of hly-encoded listeriolysin O (30, 37). It is unclear whether the primary vacuole of Henle 407 cells presents a unique environment in which actA/plcB expression occurs or whether Henle 407 primary vacuolar membranes are more sensitive to the action of the low levels of PC-PLC that are produced. No fluorescence of the Δhly actA-gfp-plcB strain was detected in PtK2 cells up to 5 h postinfection, suggesting that if low levels of expression were occurring, there was no accumulation of GFP. Complementation of the hly defect in the actA-gfp-plcB background resulted in brightly fluorescent bacteria following L. monocytogenes entry into the cytosol. These data strongly suggest that L. monocytogenes intracellular gene expression is dependent upon the bacteria encountering and sensing the appropriate host cell compartment environment and serve to illustrate the level and degree of intracellular gene regulation used by the bacteria during the course of infection.
ActA is reportedly one of the most abundant bacterial surface proteins expressed by L. monocytogenes in the host cell cytoplasm (6, 7). Several conditions have been reported to induce actA-plcB expression in extracellularly grown cultures of L. monocytogenes (6, 39, 44); however, the levels of NF-L357 fluorescence observed under any of these conditions appeared to be much lower than those of intracellular bacteria (23). Moors et al. (33) have recently described a lacZ/cat reporter gene system that can be used to monitor extracellular and intracellular L. monocytogenes gene expression, and their results indicate that actA is highly (226-fold) induced in infected host cells. We are in the process of devising similar ways to accurately quantitate differences observed in levels of actA/plcB expression, and our results thus far indicate that extracellular conditions have not yet been defined that result in levels of actA/plcB expression equivalent to those observed for intracellular bacteria (23).
The use of GFP to monitor L. monocytogenes intracellular gene expression presents several advantages over previous approaches. It provides a sensitive system that allows the monitoring of expression patterns of individual bacteria within an infected cell. No addition of cofactors is required; thus, gene expression that occurs within membrane-bound host cell compartments can be monitored without the complications of substrate diffusion. gfp fusions present in single copies on the bacterial chromosome eliminate potential artifacts resulting from multicopy plasmid-based experiments. Finally, no discernible effect on bacterial growth or viability was observed for L. monocytogenes gfp fusion mutants; thus, the patterns of gene expression observed are probably more reflective of the natural environment than are expression patterns obtained by using mutants with decreased viability (8).
Our results indicate that GFP provides a useful system for monitoring of the patterns of L. monocytogenes intracellular gene expression. We are in the process of constructing additional fusions of gfp to other L. monocytogenes genes important for intracellular growth and/or cell-to-cell spread, and it is hoped that examination of these fusion mutants will provide a more detailed understanding of host cell compartment influences on L. monocytogenes virulence gene regulation.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI41816 from the National Institutes of Health. N.E.F. thanks the Public Health Research Institute (New York, N.Y.) for initial support and encouragement.
We thank David Dubnau and Daniel Portnoy for gifts of plasmids and bacterial strains and for many helpful discussions. We thank Vojo Deretic for helpful suggestions regarding the preparation of GFP samples for fluorescence microscopy.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 5.Baumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohne J, Sokolovic Z, Goebel W. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol Microbiol. 1994;11:1141–1150. doi: 10.1111/j.1365-2958.1994.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 7.Brundage R A, Smith G A, Camilli A, Theriot J A, Portnoy D A. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc Natl Acad Sci USA. 1993;90:11890–11894. doi: 10.1073/pnas.90.24.11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubert A, Kestler H, Gotz M, Bockmann R, Goebel W. The Listeria monocytogenes iap gene as an indicator gene for the study of PrfA-dependent regulation. Mol Gen Genet. 1997;256:54–62. doi: 10.1007/s004380050545. [DOI] [PubMed] [Google Scholar]
- 9.Camilli A, Tilney L G, Portnoy D A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 11.Dabiri G A, Sanger J M, Portnoy D A, Southwick F S. Listeria monocytogenes moves rapidly through the host cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Denoya C D, Bechhofer D H, Dubnau D. Translational autoregulation of ermC 23S rRNA methyltransferase expression in Bacillus subtilis. J Bacteriol. 1986;168:1133–1141. doi: 10.1128/jb.168.3.1133-1141.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich G, Bubert A, Gentschev I, Sokolovic Z, Simm A, Catic A, Kaufmann S H E, Hess J, Szalay A A, Goebel W. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat Biotechnol. 1998;16:181–185. doi: 10.1038/nbt0298-181. [DOI] [PubMed] [Google Scholar]
- 16.Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wachter M, Wuenscher M, Chakraborty T. A novel bacterial gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag N E, Portnoy D A. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 18.Freitag N E, Rong L, Portnoy D A. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect Immun. 1993;61:2537–2544. doi: 10.1128/iai.61.6.2537-2544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M L, Killinger A H. Listeria monocytogenes and listeric infections. Bacteriol Rev. 1966;30:309–382. doi: 10.1128/br.30.2.309-382.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 21.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 22.Ireton K, Cossart P. Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes. Annu Rev Genet. 1997;31:113–138. doi: 10.1146/annurev.genet.31.1.113. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, K. E., and N. E. Freitag. Unpublished data.
- 24.Jones S, Portnoy D A. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect Immun. 1994;62:5608–5613. doi: 10.1128/iai.62.12.5608-5613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klarsfeld A, Goossens P L, Cossart P. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol Microbiol. 1994;13:585–597. doi: 10.1111/j.1365-2958.1994.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 26.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 27.Kocks C, Hellio R, Gounon P, Ohayon H, Cossart P. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J Cell Sci. 1993;105:699–710. doi: 10.1242/jcs.105.3.699. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn M, Prevost M C, Mounier J, Sansonetti P J. A nonvirulent mutant of Listeria monocytogenes does not move intracellularly but still induces polymerization of actin. Infect Immun. 1990;58:3477–3486. doi: 10.1128/iai.58.11.3477-3486.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 30.Marquis H, Doshi V, Portnoy D A. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquis H, Goldfine H, Portnoy D A. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J Cell Biol. 1997;137:1381–1392. doi: 10.1083/jcb.137.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall J, Molloy R, Moss G W, Howe J R, Hughes T W. The jellyfish green fluorescent protein: a new tool for studying ion channel expression and function. Neuron. 1995;14:211–215. doi: 10.1016/0896-6273(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 33.Moors M A, Levitt B, Youngman P, Portnoy D A. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect Immun. 1999;67:131–139. doi: 10.1128/iai.67.1.131-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mounier J, Ryter A, Coquis-Rondon M, Sansonetti P J. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect Immun. 1990;58:1048–1058. doi: 10.1128/iai.58.4.1048-1058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S F, Stewart G S A B. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 36.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portnoy D A, Jacks P S, Hinrichs D J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripio M-T, Dominguez-Bernal G, Lara M, Suarez M, Vazquez-Boland J-A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol. 1997;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripio M T, Dominguez-Bernal G, Suarez M, Brehm K, Berche P, Vazquez-Boland J A. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res Microbiol. 1996;147:371–384. doi: 10.1016/0923-2508(96)84712-7. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger J M, Mittal B, Southwick F S, Sanger J W. Analysis of intracellular motility and actin polymerization induced by Listeria monocytogenes in PtK2 cells. J Cell Biol. 1990;111:415A. [Google Scholar]
- 42.Seeliger H P R. Listeriosis—history and actual developments. Infection. 1988;16:81–85. doi: 10.1007/BF01639726. [DOI] [PubMed] [Google Scholar]
- 43.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 44.Shen H, Slifka M K, Matloubian M, Jensen E R, Ahmed R, Miller J F. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc Natl Acad Sci USA. 1995;92:3987–3991. doi: 10.1073/pnas.92.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith G A, Portnoy D A. How the Listeria monocytogenes ActA protein converts actin polymerization into a motile force. Trends Microbiol. 1997;5:272–276. doi: 10.1016/S0966-842X(97)01048-2. [DOI] [PubMed] [Google Scholar]
- 47.Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 48.Sun A N, Camilli A, Portnoy D A. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun. 1990;58:3770–3778. doi: 10.1128/iai.58.11.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Susa M, Hacker J, Marre R. De novo synthesis of Legionella pneumophila antigens during intracellular growth in phagocytic cells. Infect Immun. 1996;64:1679–1684. doi: 10.1128/iai.64.5.1679-1684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theriot J, Rosenblatt J, Portnoy D A, Goldschmidt-Clermont P J, Mitchison T. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 51.Theriot J A, Mitchison T J, Tilney L G, Portnoy D A. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 52.Tilney L G, Connelly P S, Portnoy D A. The nucleation of actin filaments by the bacterial intracellular pathogen, Listeria monocytogenes. J Cell Biol. 1990;111:2979–2988. doi: 10.1083/jcb.111.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tilney L G, Portnoy D A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 55.Vazquez-Boland J, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]