Abstract
The protective antigen (PA) protein of anthrax toxin binds to a cellular receptor and is cleaved by cell surface furin to produce a 63-kDa fragment (PA63). The receptor-bound PA63 oligomerizes to a heptamer and acts to translocate the catalytic moieties of the toxin, lethal factor (LF) and edema factor (EF), from endosomes to the cytosol. In this report, we used nondenaturing gel electrophoresis to show that each PA63 subunit in the heptamer can bind one LF molecule. Studies using PA immobilized on a plastic surface showed that monomeric PA63 is also able to bind LF. The internalization of PA and LF by cells was studied with radiolabeled and biotinylated proteins. Uptake was relatively slow, with a half-time of 30 min. The number of moles of LF internalized was nearly equal to the number of moles of PA subunit internalized. The essential role of PA oligomerization in LF translocation was shown with PA protein cleaved at residues 313-314. The oligomers formed by these proteins during uptake into cells were not as stable when subjected to heat and detergent as were those formed by native PA. The results show that the structure of the toxin proteins and the kinetics of proteolytic activation, LF binding, and internalization are balanced in a way that allows each PA63 subunit to internalize an LF molecule. This set of proteins has evolved to achieve highly efficient internalization and membrane translocation of the catalytic components, LF and EF.
Protective antigen (PA), lethal factor (LF), and edema factor (EF) are three large proteins secreted by Bacillus anthracis which are collectively known as anthrax toxin (16, 17, 32). The PA component binds to a specific cell surface receptor (7) and causes internalization and translocation of LF and EF across the endosomal membrane to the cytosol (10, 15). EF and LF contain the catalytic domains of anthrax toxin. EF has adenylyl cyclase activity (15), and LF is a zinc-dependent metalloprotease (12) that is known to cleave at least two targets, mitogen-activated protein kinase kinases 1 and 2 (6, 33). The combination of PA and LF, known as lethal toxin, causes lysis of mouse macrophages within 120 min (8, 11). The PA protein has structurally distinct domains for performing the functions of receptor binding and translocation of the catalytic moieties across endosomal membranes (17, 25).
The mechanism by which individual toxin components interact to cause toxicity was recently reviewed (17). PA binds to a specific cell surface receptor (7) and is cleaved at the sequence RKKR167 by cellular proteases including furin (13, 21). The receptor-bound 63-kDa carboxyl-terminal fragment (PA63) has a site to which LF or EF binds (18). The receptor-bound PA63 complex with LF or EF is internalized by endocytosis to an acidic compartment. The transfer of LF or EF to the cytosol appears to occur through a channel formed by low-pH-induced insertion of PA63 into the membrane (34). The formation of ion-conductive channels by PA63 has been demonstrated in both artificial lipid membranes (5) and CHO cells (19). A soluble, oligomeric form of PA63 produced in vitro which may correspond to the membrane channel was demonstrated by sedimentation equilibrium (16) and gel electrophoresis (16, 30). Electron microscopy showed that the oligomer formed in vitro is predominantly a heptamer (20). In a recent study, residues of PA lining the lumen of the heptameric channel were identified (3). PA internalized by cells or exposed to low pH on the cell surface produces oligomers which survive heating in sodium dodecyl sulfate (SDS) (20).
Injection of lethal toxin produces symptoms in experimental animals that closely resemble those seen during B. anthracis infections, and B. anthracis strains lacking only LF have very low virulence (26). This and other evidence show that the anthrax lethal toxin is the major virulence factor. The potency of lethal toxin for animals and cells depends on the amount of LF delivered to the cytosol. We wished to delineate the interactions of LF and PA which determine the efficiency of the internalization process. Here, we report that both the monomeric and the oligomeric forms of PA63 bind LF in a 1:1 ratio. The potency of anthrax lethal toxin depends upon the amount of LF associated with the PA oligomer. In addition, we demonstrate that 50% of receptor-bound PA is internalized in 30 min.
MATERIALS AND METHODS
Reagents and general procedures.
Chemicals and resins were purchased from Pharmacia. 125I-Bolton-Hunter reagent (2,000 Ci/mmol) and 125I-streptavidin (20 mCi/mg) were purchased from Amersham Corp. N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP), maleimide-activated (EZ-Link) horseradish peroxidase (HRP), and modified avidin-coated plates (NeutrAvidin) were purchased from Pierce. RAW264.7 cells were obtained from the American Type Culture Collection. Trypsin and chymotrypsin were purchased from Sigma.
Preparation of PA and LF proteins.
For preparation of PA proteins, plasmid pYS5 (29) or derivatives carrying mutated PA genes were transformed into B. anthracis UM23C1-1 and grown in FA medium with 20 μg of neomycin per ml for 16 h at 37°C. PA G735C was constructed by overlap PCR; it retains full toxicity. PA proteins were purified as described previously (13). For some experiments, PA and LF were radiolabeled with 125I-Bolton-Hunter reagent.
LF truncated at residue 254 and extended with the sequence GGCGG was prepared as a fusion to glutathione S-transferase (GST) in the vector pGEX-KG according to methods previously described for LF fusion proteins (2). The resulting plasmid pNA86 was transformed into Escherichia coli SG12036 (kindly provided by Susan Gottesman, National Cancer Institute), which has mutations in the gal, lon, and sulA genes. Cultures were grown in super broth to an A600 of 0.8, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM. The culture was grown for an additional 4 h, and bacteria were pelleted at 4°C. The protein was purified on glutathione-Sepharose 4B resin (Pharmacia) and cleaved with thrombin. The resulting LF1–254-GGCGG protein was separated from GST by chromatography on a MonoQ column (Pharmacia) with an NaCl gradient in 10 mM Tricine, pH 8.
Biotinylation of PA.
PA G735C was biotinylated with 17 molar equivalents of biotin-HPDP (Pierce Chemicals) for 90 min at 23°C in 10 mM HEPES–25 mM NaCl, pH 7.4. Excess reagent was removed by four cycles of concentration and dilution in an ultrafiltration device (Centricon-30; Amicon, Inc.).
Purification of PA63 oligomer and PA63 oligomer containing biotin.
Native PA (600 μg) alone or mixed with biotinylated PA G735C (100 μg) was cleaved with 1 μg of trypsin per ml for 30 min at 25°C in 25 mM HEPES–1 mM CaCl2–0.5 mM EDTA, pH 7.4. Protease was inactivated by adding 10 μg of 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF; Boehringer Mannheim) per ml. The cleaved PA protein was run on a MonoQ column (Pharmacia) and eluted with an NaCl gradient in 10 mM BisTrisPropane, pH 9.0. The fraction containing the PA63 oligomer was pooled, and protein was assayed with bicinchoninic acid reagent (Pierce).
Coupling of LF1–254-GGCGG to maleimide-activated HRP.
Purified LF1–254-GGCGG was coupled to a maleimide-containing HRP. LF1–254-GGCGG (1.0 mg) was treated with 0.5 mM dithiothreitol (DTT) in 10 mM HEPES, pH 7.4, for 30 min to fully reduce the sulfhydryl residue of the cysteine. DTT was removed by gel filtration on Sephadex G-25 (PD-10 column; Pharmacia) in 100 mM Na phosphate–5 mM EDTA, pH 7.6. Maleimide-activated HRP (1.6 mg; Pierce Chemicals) was added and incubated for 12 h at 23°C. Any remaining maleimide residues were inactivated by adding DTT to 1 mM for 60 min at 23°C. DTT was removed by passing the protein through a PD-10 column.
Binding of LF to PA in solution.
Gradient Phast gels (4 to 15%; Pharmacia) were soaked in buffer (0.112 M acetic acid, 0.112 M Tris [pH 6.4], containing 2 mg of 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid [CHAPS] per ml) for 2 h, blotted to remove excess liquid, and then allowed to dry in air until they reached their original weight. Buffer strips were made from 2% isoelectric focusing agarose type VIII (Sigma)–0.88 M l-alanine–0.25 M Tris (pH 8.8), containing 2 mg of CHAPS per ml. Both PA and protease-treated PA were incubated with LF in a sample buffer of 50 mM 2-[N-cyclohexylamino]ethanesulfonic acid (CHES), pH 9.0, containing 2 mg of CHAPS per ml for 30 min at room temperature. Gels were run for 80 V · h with the Pharmacia Phast gel program recommended for nondenaturing gels. The gels were stained in Coomassie brilliant blue R-250, destained, and dried.
LF binding studies using PA immobilized on 96-well plates.
The ability of LF to bind to PA was measured with LF1–254-GGCGG–HRP and the biotinylated PA G735C immobilized on 96-well plates coated with a modified avidin (NeutrAvidin plates; Pierce Chemicals). All additions were made in 100-μl portions, and incubations were at 23°C. Varying concentrations of biotinylated PA G735C were added to the 96-well plates and incubated for 2.5 h. Unbound PA was removed by washing four times with 25 mM Tris (pH 7.6)–50 mM NaCl–0.05% Tween 20–1% bovine serum albumin (BSA) (buffer A). In some wells, the bound PA was treated for 25 min with 1 μg of trypsin per ml in 10 mM HEPES, pH 7.4. Trypsin was inactivated by adding 10 μg of soybean trypsin inhibitor per ml and washing three times with buffer A. LF1–254-GGCGG–HRP was added at 15 μg/ml and incubated for 90 min. The wells were washed four times with buffer A, and the amount of bound HRP was determined by adding 1 mg of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) in 100 mM Na citrate, pH 4.5, and recording the A405.
Cell culture and cytotoxicity assays.
RAW264.7 macrophages were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 0.45% glucose, 10% fetal bovine serum, and 2 mM glutamine. LF bound to PA63 oligomer in various ratios was assayed for toxicity in the macrophage lysis assay with 3-[4,5-dimethylthiazol-2-yl]2,5-diphenyltetrazolium bromide (MTT) to determine viability (28).
Binding and uptake of LF and PA by cells.
Binding of LF and PA to RAW264.7 cells was measured at 4 and 37°C in 12-well plates with radiolabeled proteins. LF and PA were iodinated with 125I-Bolton-Hunter reagent to specific activities of 1.86 × 106 and 1.6 × 106 dpm/μg, respectively, as determined by densitometry and subsequent counting of Coomassie blue-stained gels. Iodination with Bolton-Hunter reagent had no effect on the biological activity of PA or LF. For binding experiments at 4°C, cells were placed at 4°C for 15 min. The medium was replaced with 4°C minimum essential medium containing Earle’s salts without bicarbonate and supplemented with 1% BSA and 25 mM HEPES, pH 7.4, and containing 125I-PA at 0.1 μg/ml either alone or with 0.5 μg of 125I-LF per ml. The cells were incubated for 12 h and washed, and the cell-associated radioactivity was measured. The protocol for binding and uptake at 37°C was identical, except that DMEM containing 10% serum was used and uptake was allowed for 2 h.
Cellular internalization of biotinylated PA.
The internalization of PA was also measured by using biotin-labeled PA G735C and 125I-streptavidin as described by Pinet et al. (27). PA G735C was biotinylated, and a portion was cleaved with trypsin as described above. RAW264.7 cells in 24-well plates were incubated with 0.5 μg of biotinylated PA G735C per ml in DMEM with 0.45% glucose, 2 mM glutamine, 1% BSA, and 25 mM HEPES (medium A) for 5 h at 4°C. The cells were washed four times with cold medium A to remove unbound biotinylated PA G735C. Warm medium was added to cause PA internalization for various times. The internalization of PA was stopped by adding cold medium A containing 0.4 μCi of 125I-streptavidin per ml, and cells were incubated for 1 h at 4°C. The cells were washed with cold medium A, and radioactivity was determined. Nonspecific binding was determined by incubating the cells for 15 min in 50 mM glutathione–75 mM NaCl–10 mM EDTA, pH 7.2, in DMEM containing 1% BSA to remove surface-bound biotin prior to addition of 125I-streptavidin.
RESULTS AND DISCUSSION
Proteolytic cleavage of PA at the furin site, RKKR167, either in solution (16) or on the cell surface (13), leads to the exposure of a high-affinity binding site for LF and EF on the 63-kDa carboxyl-terminal fragment (PA63). Cleavage of PA also results in the formation of a defined oligomeric form of PA63 (16), which has been visualized by electron microscopy as a heptameric ring (20). We and others believe that this heptameric PA63 is a soluble form of the membrane-inserted channel through which LF and EF translocate to the cytosol (17, 20). Little is known about the stoichiometry or affinity of LF and EF binding to PA63. We sought to characterize the oligomerization of PA63, the binding of LF, and the role of these events in the internalization process that leads to toxicity. All the work reported here used LF rather than EF, because LF is more easily purified. However, previous work showed that the interactions of LF and EF with PA are qualitatively identical (16), so it follows that results described here for LF are likely to apply equally well to EF.
Interaction of LF with PA63 in solution.
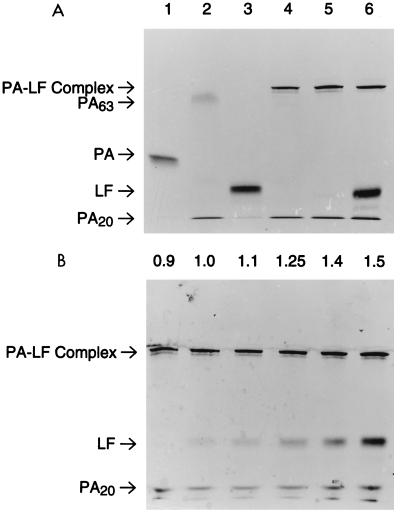
The interaction of PA63 and LF in solution was studied by gel electrophoresis under nondenaturing conditions. Previous work showed that the purified PA63 oligomer was most soluble at elevated pH in CHAPS detergent (16, 30). Because the heptameric PA63 is very large (calculated molecular weight [MW] = 440,000), gels with low polyacrylamide concentrations were used to obtain adequate electrophoretic mobility. An effective electrophoresis system was developed by modifying commercial gels prepared on plastic backing (Phast gels; 4 to 15% polyacrylamide gradient). These gels were soaked in a pH 8.8 buffer containing CHAPS. Agarose buffer strips containing an alanine solution that produces a stacking effect were prepared. In this electrophoresis system, native PA and LF have high mobility (Fig. 1A, lanes 1 and 3). PA was cleaved at the RKKR167 site with low concentrations of trypsin and then incubated at pH 9.0 with LF to allow complex formation. The trypsin-cleaved PA in the absence of LF formed a diffuse, slowly migrating band, suggesting that it had oligomerized either prior to or during electrophoresis (Fig. 1A, lane 2). Addition of LF caused this diffuse band to sharpen and migrate even more slowly (Fig. 1A, lanes 4 to 6), indicating that LF was binding to the PA63 complex. The change in mobility of LF due to binding to PA63 will be referred to here as a gel shift.
FIG. 1.
Binding of LF to PA in solution. Increasing amounts of LF were incubated with PA that had been nicked with trypsin for 30 min at room temperature. Samples were separated on 4 to 15% gradient gels (Phast) and stained with Coomassie blue. (A) Lane 1, 270 ng of native (unnicked) PA; lane 2, 270 ng of PA nicked with trypsin; lane 3, 270 ng of LF; lanes 4 to 6, 270 ng of trypsin-nicked PA and 135, 270, and 540 ng of LF, respectively. (B) Lanes 1 to 6, 270 ng of trypsin-nicked PA and increasing amounts of LF (243, 270, 297, 337, 378, and 405 ng, respectively). The mass ratio of LF to PA is indicated above each lane. The results are representative of the three analyses performed.
The stoichiometry of binding was estimated by adding increasing amounts of LF to a fixed amount of PA63. No free LF was observed when the amount of LF was one-half of or equal to the amount of PA (Fig. 1A, lanes 4 and 5), indicating that all the LF was associated with the slowly migrating PA63 band. When the amount of LF was increased to twice that of PA, half of the LF migrated as free LF (Fig. 1A, lane 6). This result suggests that the PA63 heptamer is able to bind approximately an equal weight of LF. The absence of any free LF when the mass ratio is 1:1 indicates that the affinity of PA63 for LF is very high.
A more precise measurement of the capacity of PA63 to bind LF was made by adding amounts of LF that were more nearly equal to the amount of PA63 (Fig. 1B). In this case, small amounts of free LF first became visible in lane 2, which contained 270 ng of LF and 270 ng of trypsin-nicked PA, for an LF/PA mass ratio of 1.0, as labeled in the figure. Because the MWs of LF and full-size PA are 90,200 and 82,700, respectively, this corresponds to a molar ratio of 0.9. As the amount of LF was increased further, the band of free LF increased proportionately, indicating that the slower-migrating PA63 oligomer was saturated with LF. These data suggest that the PA63 heptamer binds six to seven LF molecules. Symmetry considerations suggest that the actual number is seven.
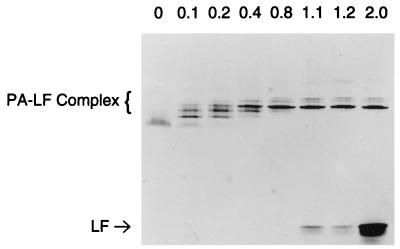
The fact that the oligomeric PA63 can bind an equal amount of LF shows that multiple molecules of LF are binding. Indeed, the data of Fig. 1 imply that a heptameric PA63 binds approximately seven LF molecules. It therefore appeared probable that intermediate species containing fewer LF molecules might exist at low relative concentrations of LF. Alternatively, if LF binding were highly cooperative, then intermediate species might not be detectable. To analyze PA63 complexes containing less than stoichiometric amounts of LF, we used purified PA63 because it produces sharper bands in the gel shift system. PA63 was obtained by chromatography of trypsin-nicked PA as described in Materials and Methods. Gel electrophoresis of mixtures containing limiting amounts of LF with purified PA63 is shown in Fig. 2. As in Fig. 1, saturating amounts of LF produce a band of free LF near the bottom of the gel (Fig. 2, lane “2.0”). Incubation of the PA63 oligomer with limiting amounts of LF caused formation of PA63-LF complexes with varying numbers of LF molecules. The intensities of the bands did not change in direct proportion to their mobilities, implying that individual LF molecules may not bind independently. The prominent band observed in all samples with LF/PA ratios of 0.4 to 2.0 (Fig. 2) may be a complex having equal numbers of PA63 and LF molecules, presumably seven of each. These lanes also contain two faint bands above the putative 7:7 complex. These bands may represent complexes containing denatured proteins which can interact but not pack into a compact structure. A more “open” structure of the same MW would have lower mobility. The possibility that these bands are a complex containing five to six LF molecules, and therefore are less tightly packed, is also possible, although one would expect such a complex to disappear at higher LF concentrations like those in the right-hand lanes.
FIG. 2.
Determination of multimeric forms of PA-LF complex. Purified PA63 (468 ng) was incubated with various amounts of LF for 30 min at room temperature. Samples were separated on 4 to 15% Phast gradient gels. Lanes 1 to 8, increasing amounts of LF (0, 47, 94, 187, 374, 515, 562, and 936 ng, respectively). The mass ratio of LF to PA is indicated above each lane. The results are representative of the three analyses performed.
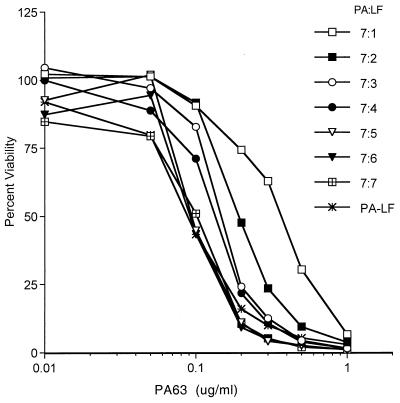
The data from the nondenaturing gels suggest that the PA63 oligomer is able to bind variable numbers of LF molecules up to a maximum of seven. However, the data do not indicate which of these PA-LF complexes is biologically active. It is possible that PA-LF complexes with any number of LF molecules will be active and that the potency may increase with increasing amounts of LF. The potency of PA63 oligomer associated with various amounts of LF was resolved in an another experiment in which the PA63 oligomer complexed with various amounts of LF was added to the RAW264.7 cells and incubated for 2.5 h at 37°C. The potency of PA63 oligomer in lysing RAW264.7 cells increased with increasing amounts of associated LF (Fig. 3). These results further confirmed the observation for Fig. 2 that PA oligomer can bind one or more LF molecules and also showed that the potency of the lethal toxin depends on the amount of LF bound to PA oligomer.
FIG. 3.
Potency of PA oligomer bound to various ratios of LF. Aliquots of purified PA63 at 10 μg/ml were incubated with various amounts of LF for 30 min at room temperature. The resulting PA63-LF complexes were serially diluted and added to the RAW264.7 cells and incubated for 2.5 h at 37°C in a CO2 incubator. The labels on the lines refer to the molar ratios of PA63 to LF during the preincubation. For the curve “PA-LF,” LF was added at 0.5 μg/ml to all wells and the indicated concentration of PA63 was added separately. Cell lysis was determined by MTT assay. The results are representative of the three analyses performed.
Role of PA63 oligomerization in binding of LF.
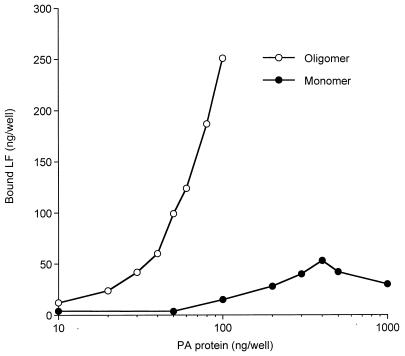
For LF to be delivered to the cytosol of target cells, it must bind to the receptor-bound, proteolytically activated form of PA, PA63. It is well established that proteolytic activation of PA is a prerequisite for membrane insertion, oligomerization, and LF binding. However, it is not known whether each of these three events can occur independently or whether these events must occur in a particular order. To begin answering these questions, we determined whether monomeric PA63 is able to bind LF. Simple measurements in solution cannot answer this question because trypsin- or furin-nicked PA oligomerizes rapidly. Therefore, we immobilized PA on a plastic surface so as to prevent its oligomerization after cleavage. This approach was aided by the availability of PA G735C, a fully toxic mutant (unpublished results). Because native PA contains no Cys residues, the single Cys residue added at the C terminus provides a unique sulfhydryl group through which PA can be immobilized in a known orientation. The carboxyl terminus of PA is involved in receptor binding (31) and is at the opposite end of the protein from the LF binding site. Therefore, modifications at the COOH terminus are not expected to have any effect on trypsin cleavage or LF binding. The single cysteine was reacted with a sulfhydryl-specific biotinylating reagent, and varying amounts of the resulting biotinylated PA were then bound to commercial plastic 96-well plates coated with a modified avidin and blocked to prevent nonspecific binding. The immobilized, biotin-PA G735C was treated with trypsin, and the ability to bind LF was measured with a chemical conjugate of the PA-binding region of LF with HRP (LF1–254GGCGG–HRP). The binding of the LF conjugate increased approximately linearly with the amount of PA used as a coating on the 96-well plates (Fig. 4, monomer), saturating at about 200 to 300 ng/well. Competition with native LF showed that >90% of the LF1–254GGCGG–HRP binding was specific. Binding in the absence of trypsin treatment was less than 20% of the specific, trypsin-dependent binding (data not shown). Although it might be argued that some of the immobilized PA63 could assemble into oligomers, we consider it unlikely that a significant fraction of the tethered PA63 molecules could contact each other in the precise orientation needed to form oligomers. Furthermore, oligomerization would be highly dependent on the concentration of surface-bound PA63, so that there would be an exponential increase in LF binding versus PA concentration, and this is not observed. The ability of the LF conjugate to bind to the immobilized PA protein strongly suggests that monomeric PA has high affinity for LF.
FIG. 4.
Binding of LF1–254-GGCGG–HRP to PA monomer or oligomer on enzyme-linked immunosorbent assay plates. PA G735C-biotin (0.01 to 1 μg/well; monomer) or PA63 made from a 6:1 mixture of PA and PA G735C-biotin (0.01 to 0.1 μg/well; oligomer) was applied as a coating to avidin-coated plates for 3 h at 23°C. Unbound PA was removed by washing four times with 25 mM Tris, pH 7.6, containing 1% BSA, 0.05% Tween 20, and 50 mM NaCl (buffer A). The PA G735C-biotin attached to the immobilized avidin (monomer) was treated with trypsin (100 μl, 1 μg/ml) in 10 mM HEPES, pH 7.4, for 25 min at 23°C. Trypsin was inactivated by adding soybean trypsin inhibitor (100 μl, 10 μg/ml) and again washed three times with buffer A. LF1–254-GGCGG–HRP (100 μl, 15 μg/ml) was added to all wells and incubated for 90 min at 23°C. The excess LF1–254-GGCGG–HRP was removed by washing the wells four times with buffer A. The color was developed with ABTS, and A405 was measured in a microplate reader. A405 values were corrected for the small activity (<10% of total) in controls containing excess native LF to compete with LF1–254-GGCGG–HRP. Parallel assays of known amounts of soluble HRP were used to construct a standard curve by which A405 values were converted to nanograms of LF. The experiment was performed three times with results similar to those shown.
To compare the amounts of LF that bind to PA monomer and heptamer, a similar experiment was performed with PA63 oligomer formed by mixing native PA and biotinylated PA G735C at a 6:1 ratio and purifying the oligomer after trypsin cleavage as described in Materials and Methods. In this case, each avidin site could bind one heptamer, so that there may be as much as sevenfold more PA63 bound than in the case where intact biotin-PA is added and then trypsin treated. Consistent with this view, the oligomer bound much more LF conjugate than did monomer (Fig. 4, oligomer). These results show that both monomeric and oligomeric forms of PA63 can bind LF and confirm the earlier results that the oligomeric PA63 can bind multiple LF molecules.
Stoichiometry of PA and LF binding to cells.
The results described above show that LF binds very tightly to PA63 and that oligomerization of PA63 is not required prior to LF binding. To determine whether similar events occur on the surface of cells, we measured the stoichiometry of LF and PA binding to the mouse macrophage cell line RAW264.7. Cells were incubated with 0.1 μg of 125I-PA per ml alone or in combination with 0.5 μg of 125I-LF per ml for 12 h at 4°C or 2 h at 37°C, and cell-associated radioactivity was measured (Table 1). The amounts of LF bound to the cells were estimated by subtracting the radioactivity in wells containing PA alone. At 4°C, PA binds, becomes nicked by cell surface furin (13, 21), and then binds LF. Prior studies show that a 12-h incubation at 4°C leads to nearly complete nicking of receptor-bound PA (30). If the binding interactions identified by the in vitro studies described above accurately reflect events on cells, it would be expected that LF would bind in amounts equal to that of PA. Equimolar binding of LF and PA was indeed observed (Table 1, 4°C data).
TABLE 1.
Binding of PA and LF to RAW264.7 cellsa
| Temp (°C) | ng of toxin/mg of cell protein
|
PA/LF ratio | |
|---|---|---|---|
| PA | LF | ||
| 4 | 2.25 ± 0.35 | 2.22 ± 0.19 | 1:0.98 |
| 37 | 9.15 ± 0.32 | 8.32 ± 0.27 | 1:0.91 |
RAW264.7 cells cultured in 12-well plates were incubated with 0.1 μg of 125I-PA per ml alone or in combination with 0.5 μg of 125I-LF per ml at 4°C for 12 h or at 37°C for 2 h. Cells were washed four times and solubilized in 0.1 M NaOH. The amount of LF bound to the cells was determined by subtracting the radioactivity in wells containing PA alone. Each value is the average of three individual wells and representative of a typical experiment performed three times. Nonspecific binding was determined by incubating the cells with nonradioactive PA or LF at 100-fold excess over labeled protein. The specific counts per minute per well and percentages of nonspecific radioactivity were as follows: 4°C, PA, 1,545 cpm (10.6%), and LF, 1,797 cpm (8.9%); 37°C, PA, 6,381 cpm (9.0%), and LF, 4,515 cpm (3.47%).
A similar analysis was done at 37°C, under conditions where endocytic uptake of both PA and LF occurs. The amounts of PA and LF associated with the cells after the 2-h incubation were fourfold greater than those at 4°C (Table 1), suggesting that an amount of receptor equal to that on the cell surface is internalized and replaced every 30 min. The molar amounts of LF bound and internalized were nearly equal to that of PA. These data suggest that nearly every molecule of PA binds and delivers a molecule of LF into endosomes. The rate of internalization was also measured by a more direct assay to be described below.
Role of PA63 oligomerization in LF translocation.
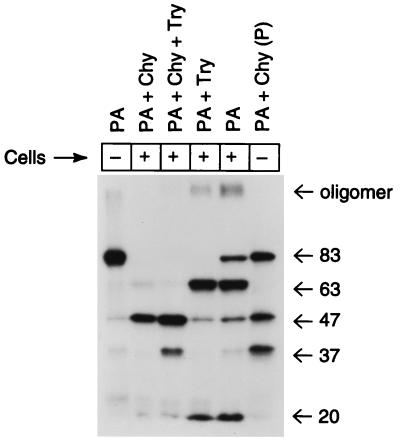
Although proteolytic cleavage of PA is essential to toxicity (29) and cleavage leads to oligomerization of PA63 (16, 20), it has not been proven that oligomerization is required for LF translocation and toxicity. To study the role of oligomerization in productive internalization, we performed experiments using chymotrypsin-nicked, radioiodinated PA. Chymotrypsin-nicked PA can bind to the cellular receptor, be activated by furin, and bind LF, but it is biologically inactive (16, 24). Although the gel shift assay showed that chymotrypsin-nicked PA could form oligomers in vitro (30), assays for oligomer formation on cells were not reported. The data in Fig. 5 show that chymotrypsin-nicked PA incubated with cells at 37°C did not produce SDS- and heat-resistant oligomers. Oligomer was present only in those cells incubated with native or trypsin-nicked PA. The inability of chymotrypsin-nicked PA to form stable oligomers and its lack of toxicity when combined with LF suggest that the stable oligomer formation seen in cells is an essential step in delivery of LF to the cytosol.
FIG. 5.
Oligomerization of PA on cells. RAW264.7 cells were cultured in 12-well tissue culture plates. 125I-PA was cleaved with trypsin, chymotrypsin, or both and then incubated with cells for 3 h at 37°C in DMEM containing 10% serum and 2 mM glutamine. Cells were washed four times with the same medium, solubilized in SDS sample buffer, heated for 5 min at 95°C, and separated on 10% polyacrylamide gels in SDS. Volumes containing 900 cpm were loaded in each lane, and the gel was exposed to X-ray film for 6 days. Lane 1, PA (no cell incubation); lane 2, chymotrypsin-nicked PA; lane 3, trypsin- and chymotrypsin-nicked PA; lane 4, trypsin-nicked PA; lane 5, PA; lane 6, PA partially nicked with chymotrypsin (no cell incubation; “P” denotes partial digest). The results shown are representative of the two analyses performed. Numbers at right show molecular mass in kilodaltons.
PA receptor internalization rate.
The rate and extent of internalization of the PA-LF complex are determined largely by the properties of the PA receptor. Cell surface receptors for several bacterial toxins have been identified and characterized (14, 23), but that for PA remains unidentified. The PA receptor appears to be a protein which is present at 2,000 to 50,000 copies per cell (7, 9, 16). Reported dissociation constants average 10−9 M. Because of the unique requirement that receptor-bound PA be cleaved before it can bind LF, the rate of PA internalization relative to proteolytic activation is a critical determinant of the efficiency of LF delivery to the cytosol. That is, if internalization of PA were rapid relative to proteolytic cleavage, then most PA molecules would be endocytosed before capturing an LF molecule.
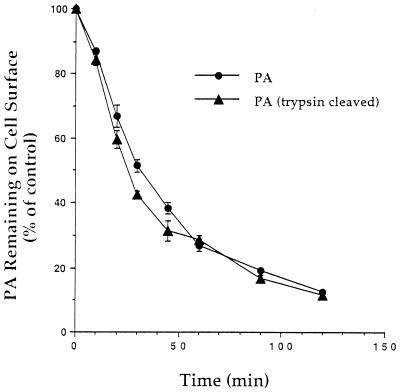
To investigate the rate of internalization of the PA receptor, we adopted a sensitive assay utilizing biotinylated PA G735C (27). Because the trafficking of some receptors is influenced by the properties of the ligand, it was also of interest to determine whether uncleaved PA and PA63 are internalized and trafficked differently. PA G735C was labeled with biotin-HPDP. Although the Cys substitution in this mutant is in the region implicated in receptor binding (31), the full toxicity of this mutant PA shows that receptor binding is unaffected. A portion of the biotin-labeled PA G735C was nicked with trypsin, and both proteins were incubated with cells. 125I-streptavidin was used to measure the amount of biotinylated PA G735C remaining on the cell surface after various periods of internalization. It was found that approximately half the PA entered cells in 30 to 40 min (Fig. 6). Trypsin nicking had no effect on the rate of internalization, suggesting that the receptor does not distinguish between various forms of PA. The rate of internalization found in this assay correlates well with that estimated from the data of Table 1. This rate of internalization (3% per min) is very slow compared to that for receptors for many protein ligands (25% per min) but is similar to the rate at which diphtheria toxin is internalized (1).
FIG. 6.
Internalization of PA. RAW264.7 cells in 24-well tissue culture plates were incubated for 5 h at 4°C in medium A (see Materials and Methods) containing 0.5 μg of biotinylated PA G735C per ml, either intact or trypsin nicked. The cells were washed and then incubated at 37°C for varying periods. Internalization of PA was stopped by adding cold medium containing 125I-streptavidin. Cells were washed to remove unbound 125I-streptavidin, and radioactivity was measured. Nonspecific binding was determined by first stripping surface biotin by incubation in glutathione prior to addition of 125I-streptavidin. The experiment was repeated three times with similar results.
Overview of toxin internalization.
The data presented above provide a basis for refining the description of the interaction of PA and LF with cells. As described above, the process of toxin action begins with PA binding to its receptor and being nicked by furin or another protease at Arg167. Release of the amino-terminal fragment removes steric hindrance or allows a conformational change that creates a high-affinity site for LF or EF. Nicking of PA appears to be quite slow, with a half-time of at least 30 min at 37°C (30). The low rate of nicking is not surprising if furin is the responsible enzyme, because only small amounts of furin are found on the cell surface compared to the amounts in the trans-Golgi region (22). It was previously assumed that PA internalization was relatively rapid and that many molecules of PA would therefore be endocytosed without being nicked. The new data presented here include evidence that PA is actually internalized quite slowly. The recognition that internalization, like nicking, is quite slow makes it evident that PA remains on the cell surface long enough to become fully nicked. This interpretation is consistent with, and indeed dictated by, the data in Table 1 showing that nearly equal amounts of PA and LF are internalized at 37°C. This can occur only if every PA molecule becomes nicked and then captures and internalizes an LF molecule.
The next step in toxin action is endocytosis of the PA63-LF complex. The data do not directly show whether the endocytosed LF-containing species is a monomeric or an oligomeric PA63. The demonstration that monomeric PA63 can bind LF shows that oligomerization does not necessarily precede endocytosis. Perhaps the species endocytosed is a mixture of monomeric and oligomeric PA63-LF species. In the endosome, acidification leads to membrane insertion of PA63 and LF translocation to the cytosol (10, 16). An unresolved question that our data do not address is whether membrane insertion and oligomerization must occur in a particular sequence. It can be noted that oligomerization does not appear to be a rate-limiting step, because dose-response curves do not show the high cooperativity characteristic of many multimeric systems.
Little is understood about the translocation of LF across the endosomal membrane. Two types of mechanisms can be distinguished. In one of these, insertion of the PA63 oligomer into the membrane would be the driving force which simultaneously inserts a molecule of LF into the membrane. This mechanism implies that the PA63 oligomer might deliver only a single LF molecule to the cytosol. The second type of mechanism would view a preformed PA63 channel as being competent to translocate multiple LF molecules. Studies with artificial lipid membranes containing PA63 show that translocation of organic cations is voltage dependent and that larger cations can be forced through by increasing the voltage (4). If LF behaves similarly to these organic cations and multiple molecules can be translocated, then a mechanism must exist to transfer LF from a site at which it is bound with very high affinity on each PA63 molecule to a site from which it can be forced into the channel lumen to initiate translocation. Direct measurements of the translocation efficiency have not been made, so it is not known whether the amount of LF translocated to the cytosol is more consistent with the first or the second of these mechanisms.
The anthrax toxin proteins constitute a unique membrane translocation system which is quite accessible to experimental study. The proteins are easily expressed and purified, and translocation can be measured in simple cytotoxicity assays. The PA63 channel appears able to translocate a number of different polypeptides (2). For these reasons, the anthrax toxin proteins provide an attractive model system of membrane transport, while also providing a basis for the development of reagents for cell biology and therapeutics.
ACKNOWLEDGMENTS
We thank J. K. Batra and Valery M. Gordon for helpful discussions, Maria Sandkvist and Valery M. Gordon for assistance with the manuscript, Naveen Arora for constructing the plasmid encoding GST–LF1–254-GGCGG, and Jerry M. Keith and S. K. Brahmachari for making this work possible.
Y.S. was partially supported by the Rockefeller Foundation.
REFERENCES
- 1.Almond B D, Eidels L. The cytoplasmic domain of the diphtheria toxin receptor (HB-EGF precursor) is not required for receptor-mediated endocytosis. J Biol Chem. 1994;269:26635–26641. [PubMed] [Google Scholar]
- 2.Arora N, Leppla S H. Fusions of anthrax toxin lethal factor with Shiga toxin and diphtheria toxin enzymatic domains are toxic to mammalian cells. Infect Immun. 1994;62:4955–4961. doi: 10.1128/iai.62.11.4955-4961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson E L, Huynh P D, Finkelstein A, Collier R J. Identification of residues lining the anthrax protective antigen channel. Biochemistry. 1998;37:3941–3948. doi: 10.1021/bi972657b. [DOI] [PubMed] [Google Scholar]
- 4.Blaustein R O, Finkelstein A. Voltage-dependent block of anthrax toxin channels in planar phospholipid bilayer membranes by symmetric tetraalkylammonium ions. Effects on macroscopic conductance. J Gen Physiol. 1990;96:905–919. doi: 10.1085/jgp.96.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaustein R O, Koehler T M, Collier R J, Finkelstein A. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc Natl Acad Sci USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland T D, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Vande Woude G F. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 7.Escuyer V, Collier R J. Anthrax protective antigen interacts with a specific receptor on the surface of CHO-K1 cells. Infect Immun. 1991;59:3381–3386. doi: 10.1128/iai.59.10.3381-3386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedlander A M. Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. J Biol Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 9.Friedlander A M, Raziuddin A. Anthrax protective antigen receptor: identification of a protective antigen binding protein by chemical cross-linking. In: Withholdt B, editor. Bacterial protein toxins. New York, N.Y: Gustav Fischer; 1992. pp. 365–366. [Google Scholar]
- 10.Gordon V M, Leppla S H, Hewlett E L. Inhibitors of receptor-mediated endocytosis block the entry of Bacillus anthracis adenylate cyclase toxin but not that of Bordetella pertussis adenylate cyclase toxin. Infect Immun. 1988;56:1066–1069. doi: 10.1128/iai.56.5.1066-1069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna P C, Acosta D, Collier R J. On the role of macrophages in anthrax. Proc Natl Acad Sci USA. 1993;90:10198–10201. doi: 10.1073/pnas.90.21.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimpel K R, Arora N, Leppla S H. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 13.Klimpel K R, Molloy S S, Thomas G, Leppla S H. Anthrax toxin protective antigen is activated by a cell-surface protease with the sequence specificity and catalytic properties of furin. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kounnas M Z, Morris R E, Thompson M R, FitzGerald D J, Strickland D K, Saelinger C B. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J Biol Chem. 1992;267:12420–12423. [PubMed] [Google Scholar]
- 15.Leppla S H. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leppla S H. The anthrax toxin complex. In: Alouf J E, Freer J H, editors. Sourcebook of bacterial protein toxins. London, United Kingdom: Academic Press; 1991. pp. 277–302. [Google Scholar]
- 17.Leppla S H. Anthrax toxins. Handb Nat Toxins. 1995;8:543–572. [Google Scholar]
- 18.Leppla S H, Friedlander A M, Cora E. Proteolytic activation of anthrax toxin bound to cellular receptors. In: Fehrenbach F, Alouf J E, Falmagne P, Goebel W, Jeljaszewicz J, Jurgen D, Rappuoli R, editors. Bacterial protein toxins. New York, N.Y: Gustav Fischer; 1988. pp. 111–112. [Google Scholar]
- 19.Milne J C, Collier R J. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol Microbiol. 1993;10:647–653. doi: 10.1111/j.1365-2958.1993.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 20.Milne J C, Furlong D, Hanna P C, Wall J S, Collier R J. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J Biol Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- 21.Molloy S S, Bresnahan P A, Leppla S H, Klimpel K R, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 22.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naglich J G, Metherall J E, Russell D W, Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 24.Novak J M, Stein M P, Little S F, Leppla S H, Friedlander A M. Functional characterization of protease-treated Bacillus anthracis protective antigen. J Biol Chem. 1992;267:17186–17193. [PubMed] [Google Scholar]
- 25.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 26.Pezard C, Berche P, Mock M. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinet V, Vergelli M, Martin R, Bakke O, Long E O. Antigen presentation mediated by recycling of surface HLA-DR molecules. Nature. 1995;375:603–606. doi: 10.1038/375603a0. [DOI] [PubMed] [Google Scholar]
- 28.Quinn C P, Singh Y, Klimpel K R, Leppla S H. Functional mapping of anthrax toxin lethal factor by in-frame insertion mutagenesis. J Biol Chem. 1991;266:20124–20130. [PubMed] [Google Scholar]
- 29.Singh Y, Chaudhary V K, Leppla S H. A deleted variant of Bacillus anthracis protective antigen is non-toxic and blocks anthrax toxin action in vivo. J Biol Chem. 1989;264:19103–19107. [PubMed] [Google Scholar]
- 30.Singh Y, Klimpel K R, Arora N, Sharma M, Leppla S H. The chymotrypsin-sensitive site, FFD315, in anthrax toxin protective antigen is required for translocation of lethal factor. J Biol Chem. 1994;269:29039–29046. [PubMed] [Google Scholar]
- 31.Singh Y, Klimpel K R, Quinn C P, Chaudhary V K, Leppla S H. The carboxyl-terminal end of protective antigen is required for receptor binding and anthrax toxin activity. J Biol Chem. 1991;266:15493–15497. [PubMed] [Google Scholar]
- 32.Smith H, Stanley J L. Purification of the third factor of anthrax toxin. J Gen Microbiol. 1962;29:517–521. doi: 10.1099/00221287-29-3-517. [DOI] [PubMed] [Google Scholar]
- 33.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem Biophys Res Commun. 1998;248:706–711. doi: 10.1006/bbrc.1998.9040. [DOI] [PubMed] [Google Scholar]
- 34.Wesche J, Elliott J L, Falnes P O, Olsnes S, Collier R J. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]