Abstract
C2H2-type zinc finger proteins (C2H2-ZFPs) play a key role in various plant biological processes and responses to environmental stresses. In Arabidopsis thaliana, C2H2-ZFP members with two zinc finger domains have been well-characterized in response to abiotic stresses. To date, the functions of these genes in strawberries are still uncharacterized. Here, 126 C2H2-ZFPs in cultivated strawberry were firstly identified using the recently sequenced Fragaria × ananassa genome. Among these C2H2-ZFPs, 46 members containing two zinc finger domains in cultivated strawberry were further identified as the C1-2i subclass. These genes were unevenly distributed on 21 chromosomes and classified into five groups according to the phylogenetic relationship, with similar physicochemical properties and motif compositions in the same group. Analyses of conserved domains and gene structures indicated the evolutionary conservation of the C1-2i subclass. A Ka/Ks analysis indicated that the C1-2i members were subjected to purifying selection during evolution. Furthermore, FaZAT10, a typical C2H2-ZFP, was isolated. FaZAT10 was expressed the highest in roots, and it was induced by drought, salt, low-temperature, ABA, and MeJA treatments. It was localized in the nucleus and showed no transactivation activity in yeast cells. Overall, these results provide useful information for enriching the analysis of the ZFPs gene family in strawberry, and they provide support for revealing the mechanism of FaZAT10 in the regulatory network of abiotic stress.
Keywords: strawberry, C2H2-ZFPs, abiotic stress, ZAT10
1. Introduction
Various abiotic and biotic stresses affect plant physiology and growth [1]. Severe drought occurred in the northern hemisphere in 2022, and the Yangtze River basin experienced the longest drought since records began in the 1960s. Strawberry (Fragaria × ananassa) is widely cultivated all over the world. It plays an important role due to its richness in minerals, vitamins, flavonoids, and other nutrients [2]. Additionally, it is desirable due to its bright color, enjoyable taste, short growth cycle, and high economic benefits [3]. However, it is easily affected by unfavorable environments, such as drought, high salinity, and extreme temperature, during production. These abiotic stresses eventually lead to reductions in strawberry yield and quality, and they are limiting factors for strawberry production [4,5].
To combat the detrimental effects of stressors, plants have evolved effective defense mechanisms by influencing their tolerance potential through integrating molecular- and cellular-level responses [6]. Typically, abiotic stress induces the production of excess reactive oxygen species (ROS) and causes oxidative damage to plants. After receiving ROS signals, plants activate both enzymatic and non-enzymatic scavengers, such as SODs, CATs, APXs, and anthocyanins, to scavenge excess ROS [7,8,9]. In addition, transcription factors (TFs), such as MYB, bHLH, WRKY, bZIP, NAC, AP2/ERF, and ZFP, are also key components of the plant stress response mechanism. Among them, the TFs that contain a zinc finger domain are widely involved in abiotic stress responses [10,11,12,13,14,15,16].
Zinc finger proteins (ZFPs) are one of the largest families in plants, and they play essential roles in many cellular functions, including transcriptional activation and inhibition, RNA binding, apoptosis regulation, and protein interaction [17]. ZFP TFs are divided into nine subfamilies, namely, C2H2, C2HC, C2HC5, C3H, C3HC4, C4HC3, Cys4, C6, and C8, according to the sequence and number of cysteine (Cys) and histidine (His) residues in the ZFP structure [17,18]. In Arabidopsis thaliana, there are 176 C2H2 zinc finger proteins (C2H2-ZFPs) divided into three groups (A, B, and C), and the C group is further classified into C1, C2, and C3 subsets. The C1 subset is one of the evolutionarily youngest families, comprising 64 members whose biological functions are related to developmental processes and stress responses [19]. The C1 subset can be subdivided into five subclasses, namely, C1-1i, C1-2i, C1-3i, C1-4i, and C1-5i (Ni indicates N zinc fingers), with 33, 20, 8, 2, and 1 members, respectively [18,19]. Among these subclasses, the C1-2i subclass has been the most extensively studied, and it has been demonstrated to be involved in plant development and stress responses; for example, AZF1/2/3, AtZAT6, AtZAT18, and AtZAT12 of A. thaliana have been found to be involved in water, salt, drought, and strong light stresses [20,21,22,23].
ZAT10, formerly known as STZ (salt tolerance zinc finger), belongs to a typical C2H2-ZFP, consisting of two C2H2-type zinc fingers CX2-4CX3FX5LX2HX3-5H (X: any amino acid; number: number of amino acids), and it contains two conserved Cys and His residues and an EAR (L/FDLNL/F(x)P) motif at its C-terminus [24,25,26]. Previous studies have reported that the constitutive expression of ZAT10 enhances tolerance to salt, heat, and osmotic stresses in A. thaliana. Interestingly, the knockout and RNAi mutants of AtZAT10 have also been found to be more tolerant to osmotic and salt stresses [24]. It has been demonstrated that the EAR motif of ZAT10 can function as a transcriptional repressor [27,28,29]; inhibiting the expression level of ZAT10 by RNAi or knockout mutation will lead to enhanced tolerance if this motif is directly involved in the suppression of stress [24]. These results suggest that ZAT10 may have different regulatory mechanisms in response to abiotic stress in A. thaliana.
At present, most of the studies on C2H2-ZFPs in abiotic stress are still focused on model plants, such as A. thaliana. In cultivated strawberries, the family members of C2H2-ZFPs and their regulatory mechanisms in response to abiotic stress are largely unknown. In the present study, C2H2-ZFPs in cultivated and wild strawberries were identified based on genomic data. Among these C2H2-ZFPs, the phylogenetic relationship, protein physicochemical properties, gene structure, conserved domains, promoter cis-acting elements, and gene collinearity of the C1-2i subclass were further analyzed. Then, FaZAT10 from the C1-2i subclass was isolated. The expression pattern, subcellular localization, and transcriptional activity of FaZAT10, as well as its expression pattern under different abiotic stresses and hormone treatments, were further analyzed. These results improve the characterization of strawberry C2H2-ZFP family members and provide a research basis for the further exploration of how FaZAT10 plays a role in strawberry abiotic regulatory networks.
2. Results
2.1. Identification and Chromosomal Localization of Strawberry C2H2-ZFP C1-2i Subclass
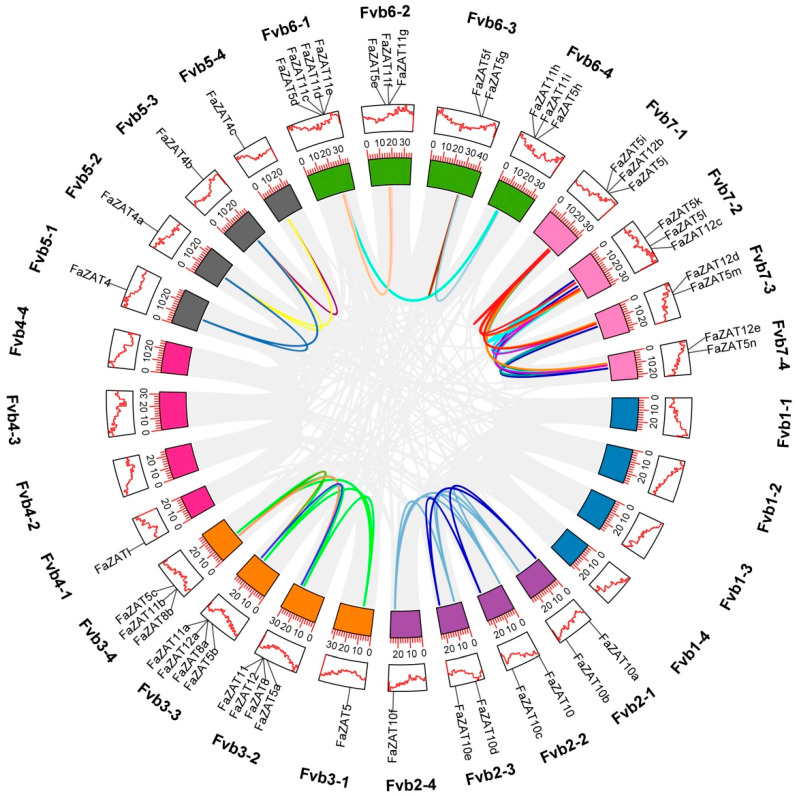
A total of 126 and 41 C2H2-ZFP candidate genes were identified in cultivated and wild strawberries, respectively. Among these, the following subclasses were identified: the 5i subclass (3 FaZATs and 1 FvZAT); the 4i subclass (11 FaZATs and 2 FvZATs); the 3i subclass (29 FaZATs and 8 FvZATs); the 2i subclass (46 FaZATs and 13 FvZATs); and the 1i subclass (37 FaZATs and 17 FvZATs). In the following work, the C1-2i subclass in cultivated strawberry was further studied. The nomenclature of the C1-2i subclass in cultivated strawberry was based on the homologous genes of diploid wild strawberry in NCBI, and the same alleles were numbered according to their position on chromosomes 1-7 (Figure 1 and Table S1).
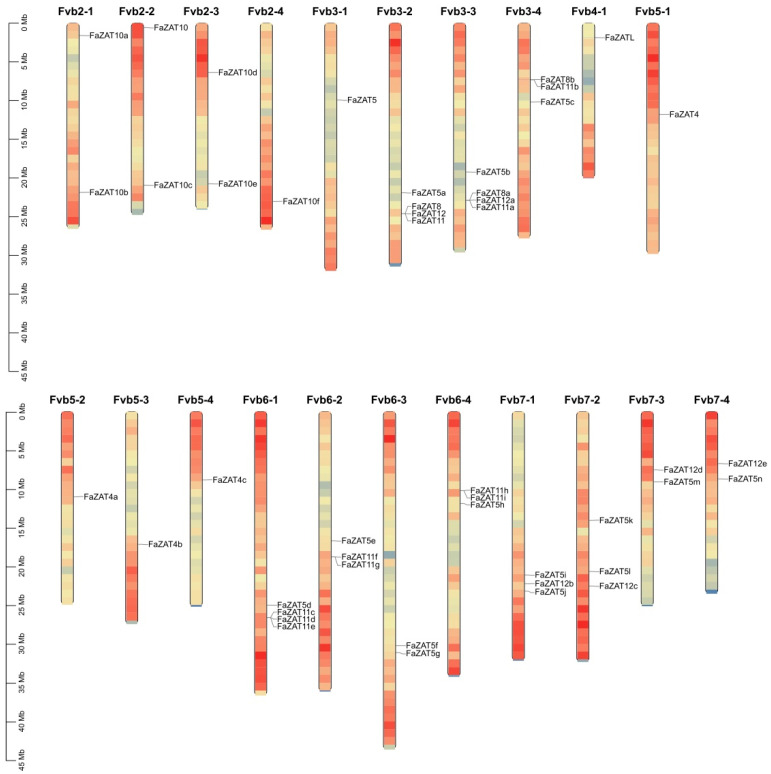
Figure 1.
Chromosomal localization of cultivated strawberry C2H2-ZFP C1-2i subclass.
The C1-2i subclass in cultivated strawberry was unevenly distributed on 21 chromosomes. The density of the genes was the highest on chromosomes Fvb3-2, Fvb3-3, and Fvb6-1, with four genes each. Only one gene was found on Fvb2-4, Fvb3-1, Fvb4-1, Fvb5-1, Fvb5-2, Fvb5-3, and Fvb5-4. Most of the FaZAT genes were in the regions at both ends of the chromosome (Figure 1).
2.2. Characterization of the C2H2-ZFP C1-2i Subclass
In the C1-2i subclass, the proteins ranged from 158 to 383 amino acids in length. The theoretical isoelectric point (pI) was between 6.39 and 10.14. The pIs of most members were greater than seven, indicating that they contained more basic amino acids. In addition, the molecular weights of all C1-2i proteins ranged from 17,378 to 42,826 Da. The predicted results of subcellular localization indicated that they were all located in the nucleus. The GRAVY values ranged from -0.895 to -0.198, revealing that strawberry C1-2i proteins were hydrophilic proteins. The instability index ranged from 39.08 to 75.27 (Table 1).
Table 1.
The physical and chemical properties of C2H2-ZFP C1-2i proteins in F. × ananassa.
| Gene Name | Amino Acid/aa | ORF/bp | Molecular Weight | pI | Instability Index | GRAVY | Location | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| FaZAT8b | 176 | 10,622 | 18,848 | 9.61 | 55.77 | −0.445 | 7,282,074–7,292,696 | Nucleus |
| FaZAT8 | 176 | 997 | 18,944 | 9.60 | 59.49 | −0.475 | 24,596,211–24,597,208 | Nucleus |
| FaZAT8a | 229 | 1115 | 24,872 | 9.70 | 53.08 | −0.264 | 22,862,614–22,863,729 | Nucleus |
| FaZAT12 | 173 | 1955 | 19,019 | 9.16 | 59.78 | −0.447 | 24,604,928–24,606,883 | Nucleus |
| FaZAT12a | 173 | 1300 | 18,957 | 9.59 | 59.83 | −0.482 | 22,874,029–228,75,359 | Nucleus |
| FaZAT11b | 158 | 476 | 17,378 | 9.69 | 67.74 | −0.645 | 7,323,096–7,323,572 | Nucleus |
| FaZAT11 | 160 | 482 | 17,496 | 9.90 | 62.24 | −0.599 | 24,610,951–24,611,433 | Nucleus |
| FaZAT11a | 160 | 482 | 17,586 | 10.14 | 60.52 | −0.555 | 22,888,095–22,888,577 | Nucleus |
| FaZAT12d | 221 | 1094 | 24,382 | 9.38 | 62.40 | −0.325 | 7,514,144–7,515,238 | Nucleus |
| FaZAT12c | 221 | 1086 | 24,400 | 9.38 | 63.82 | −0.357 | 22,530,332–22,531,418 | Nucleus |
| FaZAT12b | 190 | 1036 | 20,593 | 9.10 | 60.63 | −0.385 | 22,223,399–22,224,435 | Nucleus |
| FaZAT12e | 190 | 572 | 20,529 | 9.17 | 64.01 | −0.452 | 6,714,293–6,714,865 | Nucleus |
| FaZAT11c | 175 | 752 | 19,303 | 9.55 | 39.08 | −0.341 | 26,593,340–26,594,092 | Nucleus |
| FaZAT11i | 230 | 692 | 25,364 | 9.60 | 40.86 | −0.198 | 10,202,617–10,203,309 | Nucleus |
| FaZAT11f | 175 | 885 | 19,376 | 9.44 | 47.34 | −0.430 | 18,762,037–18,762,922 | Nucleus |
| FaZAT11g | 160 | 482 | 17,605 | 9.56 | 44..84 | −0.389 | 18,771,968–18,772,450 | Nucleus |
| FaZAT11d | 162 | 488 | 18,229 | 9.90 | 42.02 | −0.451 | 26,596,604–26,597,092 | Nucleus |
| FaZAT11e | 173 | 966 | 19,051 | 9.51 | 44.56 | −0.378 | 26,601,748–26,602,714 | Nucleus |
| FaZAT11h | 175 | 824 | 19,730 | 9.83 | 42.85 | −0.494 | 10,199,732–10,200,556 | Nucleus |
| FaZAT5l | 380 | 1142 | 41,776 | 6.96 | 73.60 | −0.889 | 20,647,253–20,648,395 | Nucleus |
| FaZAT5k | 383 | 2148 | 42,049 | 6.96 | 73.11 | −0.895 | 14,054,752–14,056,900 | Nucleus |
| FaZAT5i | 379 | 1139 | 41,510 | 6.81 | 70.02 | −0.868 | 21,096,734–21,097,873 | Nucleus |
| FaZAT5j | 380 | 2310 | 41,659 | 6.81 | 70.30 | −0.876 | 23,197,453–23,199,763 | Nucleus |
| FaZAT5n | 383 | 2944 | 42,340 | 6.94 | 71.63 | −0.930 | 8,687,658–8,690,602 | Nucleus |
| FaZAT5m | 380 | 1142 | 41,895 | 6.83 | 73.37 | −0.938 | 9,080,140–9,081,282 | Nucleus |
| FaZAT5g | 318 | 1041 | 34,736 | 6.60 | 66.74 | −0.634 | 31,122,729–31,123,770 | Nucleus |
| FaZAT5f | 324 | 1449 | 35,525 | 6.60 | 66.78 | −0.608 | 30,224,033–30,225,482 | Nucleus |
| FaZAT5e | 328 | 1126 | 36,077 | 6.64 | 69.83 | −0.612 | 16,681,442–16,682,568 | Nucleus |
| FaZAT5d | 330 | 1750 | 36,307 | 6.39 | 68.39 | −0.624 | 24,991,197–24,992,947 | Nucleus |
| FaZAT5h | 329 | 1626 | 36,126 | 6.42 | 75.27 | −0.595 | 11,865,273–11,866,899 | Nucleus |
| FaZAT5c | 324 | 1465 | 35,394 | 7.11 | 70.08 | −0.712 | 10,177,077–10,178,542 | Nucleus |
| FaZAT5a | 328 | 1609 | 35,634 | 7.11 | 67.31 | −0.642 | 21,919,033–21,920,642 | Nucleus |
| FaZAT5 | 323 | 1734 | 35,198 | 7.06 | 65.80 | −0.627 | 9,931,220–9,932,954 | Nucleus |
| FaZAT5b | 326 | 1876 | 35,598 | 7.38 | 66.85 | −0.644 | 19,227,512–19,229,388 | Nucleus |
| FaZAT10 | 269 | 1366 | 28,458 | 8.50 | 58.90 | −0.565 | 574,359–575,725 | Nucleus |
| FaZAT10b | 267 | 3084 | 28,341 | 8.50 | 57.18 | −0.600 | 21,865,334–21,868,418 | Nucleus |
| FaZAT10f | 262 | 2951 | 27,764 | 8.50 | 55.29 | −0.598 | 23,032,727–23,035,678 | Nucleus |
| FaZAT10d | 263 | 1590 | 28,206 | 8.95 | 56.46 | −0.622 | 6,379,625–6,381,,215 | Nucleus |
| FaZAT10e | 237 | 2343 | 25,184 | 8.45 | 73.44 | −0.557 | 20,755,846–20,758,189 | Nucleus |
| FaZAT10a | 237 | 1693 | 25,258 | 8.65 | 73.10 | −0.516 | 1,590,173–1,591,866 | Nucleus |
| FaZAT10c | 237 | 2571 | 25,317 | 8.15 | 70.18 | −0.578 | 20,926,689–20,929,260 | Nucleus |
| FaZAT4 | 323 | 1797 | 36,131 | 6.39 | 60.22 | −0.834 | 11,788,298–11,790,095 | Nucleus |
| FaZAT4a | 323 | 1655 | 36,131 | 6.39 | 60.22 | −0.834 | 11,007,603–11,009,258 | Nucleus |
| FaZAT4b | 323 | 1595 | 36,127 | 6.57 | 58.62 | −0.820 | 17,138,548–17,140,143 | Nucleus |
| FaZAT4c | 323 | 1862 | 36,213 | 6.44 | 59.36 | −0.831 | 8,825,171–8,827,033 | Nucleus |
| FaZATl | 383 | 7559 | 42,826 | 6.46 | 66.57 | −0.786 | 1,860,056–1,867,615 | Nucleus |
2.3. Phylogenetic Relationship and Sequence Alignment of the C2H2-ZFP C1-2i Subclass
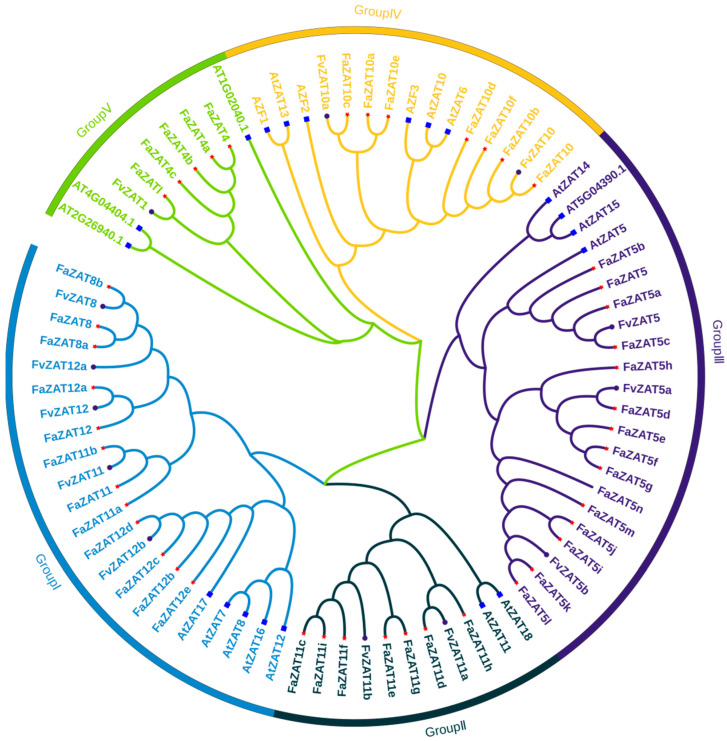
To further analyze the evolutionary relationship of the C1-2i subclass in cultivated strawberry, CLUSTALW was used to perform a multiple sequence alignment of 46 genes of the 2i subclass in cultivated strawberry and 13 in wild strawberry. Then, 20 C1-2i genes from A. thaliana [19] were introduced to construct a phylogenetic tree, as shown in Figure 2. The 46 C1-2i subclass members in cultivated strawberry and the 13 C1-2i subclass members in wild strawberry were mainly distributed in five groups: Group I (12 FaZATs and 5 FvZATs), Group II (7 FaZATs and 2 FvZATs), Group III (15 FaZATs and 3 FvZATs), Group IV (7 FaZATs and 2 FvZATs), and Group V (5 FaZATs and 1 FvZAT). The results of the multiple sequence alignment showed that most members contained a conserved motif, QALGGH, in the zinc finger helix (Figure S1), which is essential for the DNA-binding activity of C2H2-ZFPs [16].
Figure 2.
Unrooted phylogenetic tree of cultivated strawberry, wild strawberry, and A. thaliana C1-2i subclass members. The red asterisks represent cultivated strawberry, the purple circles represent wild strawberry, and the blue squares represent A. thaliana. The phylogenetic tree was developed on MEGA (version 7.0) using the neighborhood-joining phylogenetic method analysis. Both the bootstrap test and the approximate likelihood ratio test were set to 1000 times.
2.4. Gene Structure and Conserved Domain Analysis
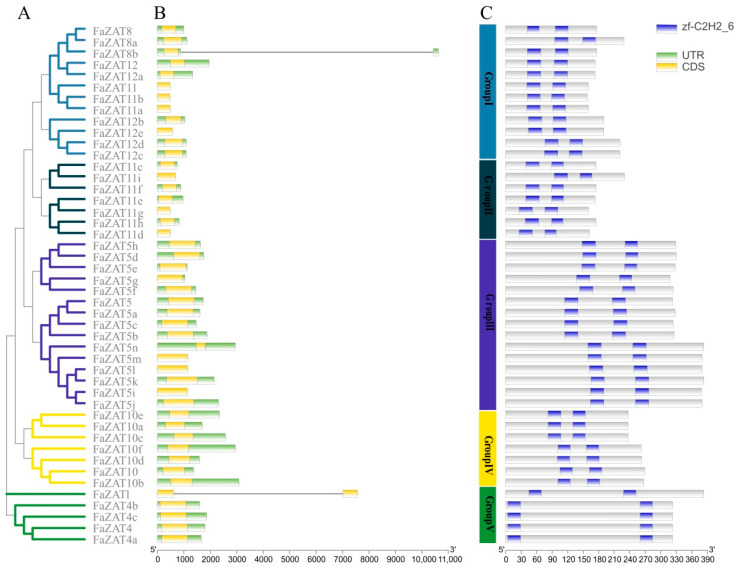
The gene structure of the C1-2i subclass changed irregularly; most genes contained two introns and one exon. In particular, FaZAT8b and FaZAT contained a 10,622 bp-long and 7559 bp-long open reading frame (ORF), respectively (Figure 3B and Table 1). Furthermore, three highly conserved motifs were analyzed on the MEME online website, including 29 conserved amino acid residues in Motif 1, 41 in Motif 2, and 21 in Motif 3. A sequence analysis showed that Motif 1 and Motif 2 were C2H2 domains and that Motif 3 was an EAR motif at the C-terminal, which is consistent with C1-2i member characteristics, indicating that these genes were evolutionarily conserved (Figure S3).
Figure 3.
The evolutionary tree (A), gene structures (B), conserved domains (C), and conserved motifs of the cultivated strawberry C1-2i subclass. Blue rectangles are conserved C2H2 domains, yellow rectangles are UTRs, and green rectangles are CDS. The phylogenetic tree was developed on MEGA (version 7.0) using the neighborhood-joining phylogenetic method analysis. Both the bootstrap test and the approximate likelihood ratio test were set to 1000 times.
2.5. Promoter Analysis of the C2H2-ZFP C1-2i Subclass Members in Strawberry
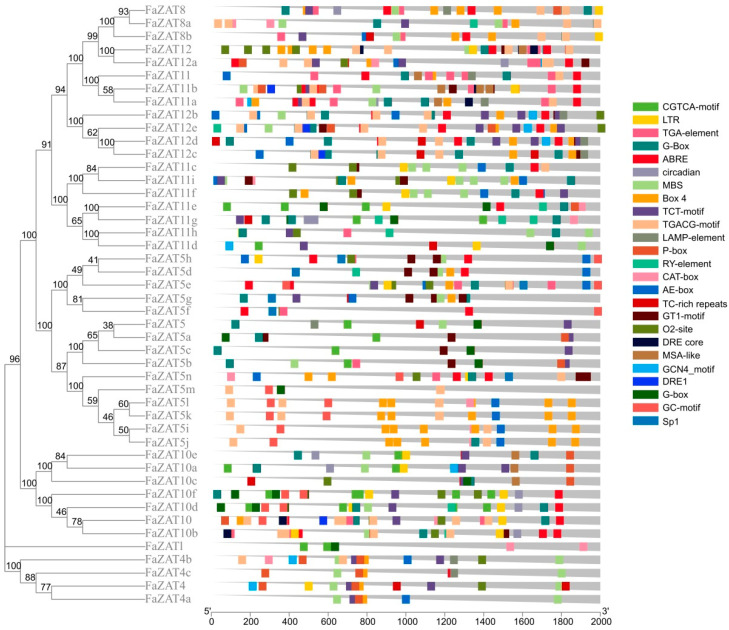
The cis-acting regulatory elements in the 2000 bp promoter region were predicted using the PlantCARE website. The results show that a total of 62 types of cis-regulatory elements were identified, except for the core promoter elements of a higher plant TATA-box and the universal promoter enhancement element CAAT-box, as well as unannotated elements. Among them, light responses were the most abundant, with 438 existing, followed by 182 MeJA-responsive elements and 124 ABA-responsive elements. Meanwhile, 15 elements related to plant development and 6 related to stress response were analyzed (Figure 4 and Table S2). In addition, MYB-related and MYC-related elements were also found to be abundant in the promoter region of the C1-2i subclass members in cultivated strawberry.
Figure 4.
Distribution of cis-acting elements in the promoter of C1-2i subclass members in cultivated strawberry. The phylogenetic tree was developed on MEGA (version 7.0) using the neighborhood-joining phylogenetic method analysis. Both the bootstrap test and the approximate likelihood ratio test were set to 1000 times.
2.6. Synteny Analysis of the C2H2-ZFP C1-2i Subclass in A. thaliana, F. vesca, and F. × ananassa
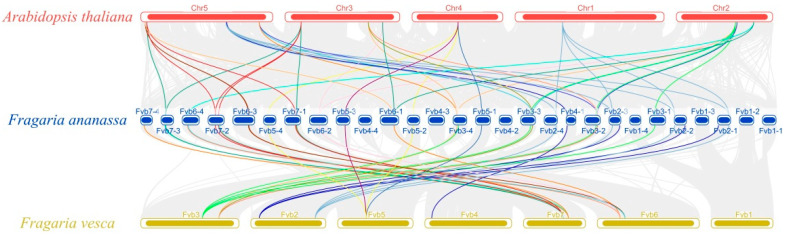
There were 57 syntenic gene blocks of the C1-2i genes among the cultivated strawberry chromosomes, and 61 FaZAT gene pairs were confirmed as collinear pairs (Table S3). To further explore the evolution of the C1-2i gene, interspecific synteny comparisons between F. × ananassa, F. vesca, and A. thaliana were also made. There were 91,807 and 42,389 collinear genes observed between F. × ananassa and F. vesca and between F. × ananassa and A. thaliana, respectively, with ratios of 64.45% and 31.17%. The interspecies collinearity analysis indicated that there may have been chromosomal and gene amplification events during the evolution of the two strawberries (Figure 5 and Figure 6).
Figure 5.
Synteny analysis of C2H2-ZFP C1-2i subclass in cultivated strawberry. Chromosomes are distinguished by different colors, the gray curve is the collinear gene region within the genome, and the colored curve is the collinear gene pair of the C2H2-ZFP C1-2i subclass.
Figure 6.
Synteny analysis of C2H2-ZFP C1-2i subclass in A. thaliana, F. × ananassa, and F. vesca genomes. The colored rounded rectangles represent the chromosomes of the three plants. The gray curves are the collinear gene regions within the genomes of the three species, and the colored curves represent the gene pairs that are collinear with the C2H2-ZFP C1-2i subclass.
The ratio of nonsynonymous substitution rates (denoted as Ka) to synonymous substitution rates (denoted as Ks) was used to assess the selective pressure on duplicate gene pairs within species. Ka/Ks >1 indicated positive selection, and Ka/Ks <1 indicated purifying selection [30]. The driving force of C1-2i gene pairs on replication was calculated using the Ka/Ks ratio. The Ka/Ks values of the C1-2i gene pairs were less than one, suggesting that these genes were influenced by purifying selection during evolution (Table S4).
2.7. Transcriptome Analysis of Strawberry C2H2-ZFP C1-2i Subclass under Salt Stress
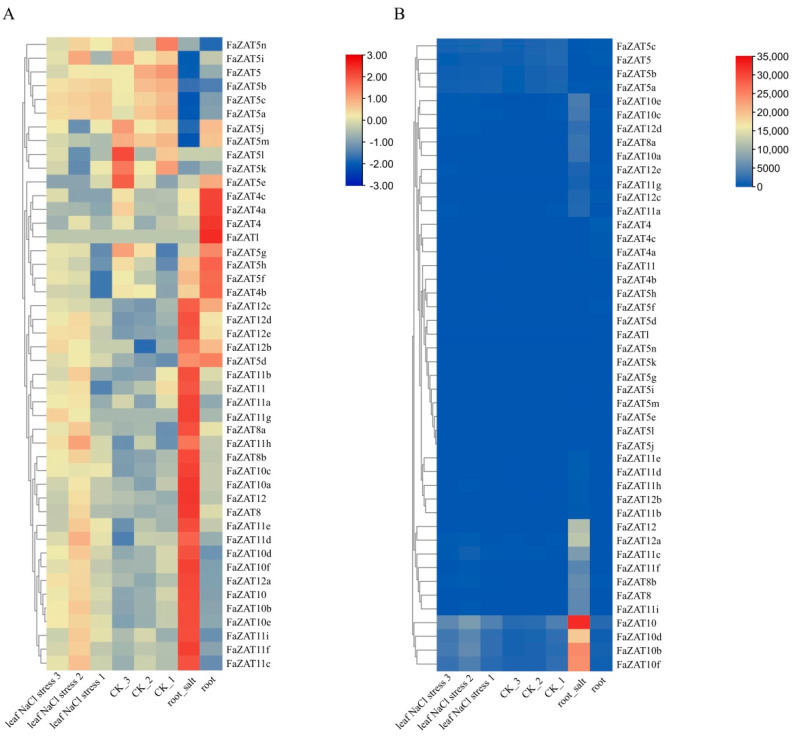
To better understand the response of FaZATs to stress at the transcriptional level, transcriptome data were used to detect the transcript abundance of FaZATs under salt stress. The results show that, under salt stress, FaZAT10, 10a, 10b, 10c, 10d, 10e, 10f, FaZAT12, 12a, 12c, 12d, FaZAT8, 8a, 8b, FaZAT11a, 11b,11c, 11f, and 11i were highly expressed in roots compared to the control (Figure 7A). In particular, the transcriptome data showed that FaZAT10 exhibited the highest expression in roots under salt stress (Figure 7B and Table S5).
Figure 7.
Transcript abundance of the C2H2-ZFP C1-2i subclass under salt stress. NaCl stress: 100 mM NaCl for 12 h; numbers indicate three biological replicates. The heatmap represents log2 value of FPKM. (A) The FPKM of each row was normalized. (B) The FPKM was not normalized.
2.8. Tissue-Specific Expression Profile of FaZAT10
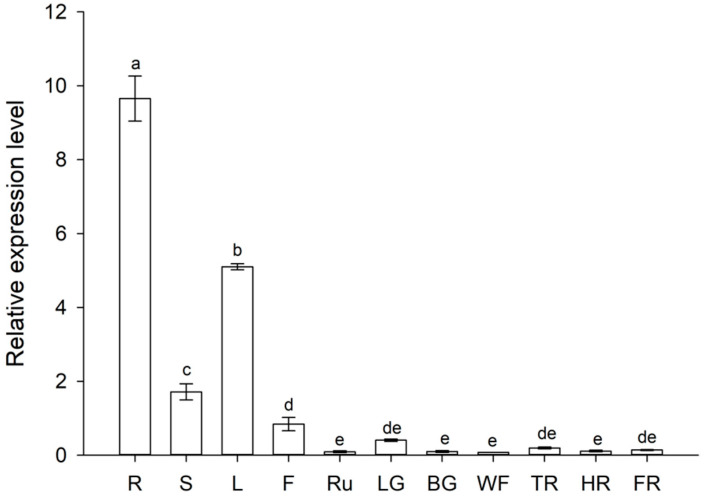
Based on the transcriptome data, FaZAT10 was cloned and studied in the following work. The results of the qRT-PCR analysis suggested that FaZAT10 was highly expressed in roots, followed by leaves and stems, and the expression level was the lowest in runners. In general, the transcript abundance of FaZAT10 was very low in different fruit developmental stages, and it had a higher expression level at the little green stage than at the other fruit developmental stages (Figure 8).
Figure 8.
Relative expressions of FaZAT10 in different tissues and developmental stages of fruits. R: root; S: stem; L: leaf; F: flower; Ru: runner; LG: little green; BG: big green; WF: white fruit; TR: turn red; HR: half red; FR: full red. Data contain three biological replicates, error bars represent standard error of the mean, and different letters are represented by the least significant difference test (LSD), p < 0.05.
2.9. The Subcellular Localization and Transcriptional Activity Analysis of FaZAT10
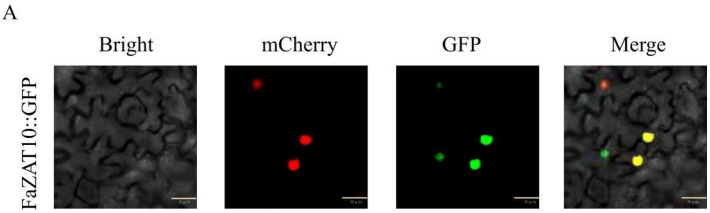
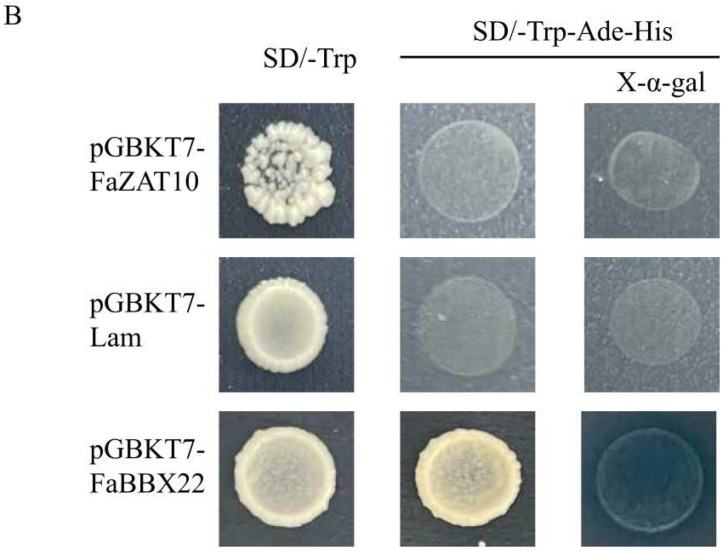
The GFP fluorescence signal of the FaZAT10::GFP fusion protein was located in the nucleus, which indicates that FaZAT10 is a nucleus-localized protein (Figure 9A). In addition, the result indicates that the yeast strains that transformed with pGBKT7-FaZAT10 (validation group) and pGBKT7-Lam (a negative control) could not grow normally on SD/-Trp/-His/-Ade/X-α-gal and SD/-Trp/-His/-Ade media. The positive group pGBKT7-FaBBX22 [31] grew well on SD/-Trp/-His/-Ade/X-α-gal medium and turned blue, which indicates that FaZAT10 has no trans-acting ability in yeast cells (Figure 9B).
Figure 9.
Transcription factor characterization of FaZAT10. (A) Subcellular localization of FaZAT10 protein. Bars, 30 µm. (B) Transcriptional activity analysis of FaZAT10 in yeast cells. pGBKT7-Lam is a negative control, and pGBKT7-FaBBX22 is a positive control.
2.10. FaZAT10 Was Induced by Abiotic Stress
Several cis-elements related to abiotic stress were found on the cloned 1,952 bp of the FaZAT10 promoter sequence, such as the DRE core (stress-induced), ABRE (in response to abscisic acid), LTR (in response to low temperature), MBS (in response to drought), CGTCA motif, and TGACG motif (in response to methyl jasmonate). We hypothesized that FaZAT10 would respond to these abiotic stresses, and, thereupon, abiotic stress and hormone treatments were designed to detect the expression of FaZAT10 (Figure S2 and Table 2).
Table 2.
Cis-regulatory elements of FaZAT10 promoter in cultivated strawberry.
| Function | Type | Motif |
|---|---|---|
| Light-responsive | 4 | Box4, G-box, G-Box, chs-CMA1a, TCT-motif |
| Meristem expression | 1 | CAT-box |
| ABA response | 1 | ABRE |
| Adversity induction | 2 | DRE core, DRE1 |
| Low-temperature response | 1 | LTR |
| Auxin-responsive | 1 | TGA-element |
| Gibberellin-responsive | 1 | P-box |
| MeJA response | 2 | CGTCA motif, TGACG motif |
| Drought stress | 1 | MBS |
| Anoxic specific inducibility | 1 | GC motif |
| MYB-binding site | 2 | MYB, Myb-binding site |
| MYC-binding site | 1 | MYC |
| WRKYs response element | 1 | W-box |
| Seed-specific regulation | 1 | RY element |
| Circadian control | 1 | Circadian |
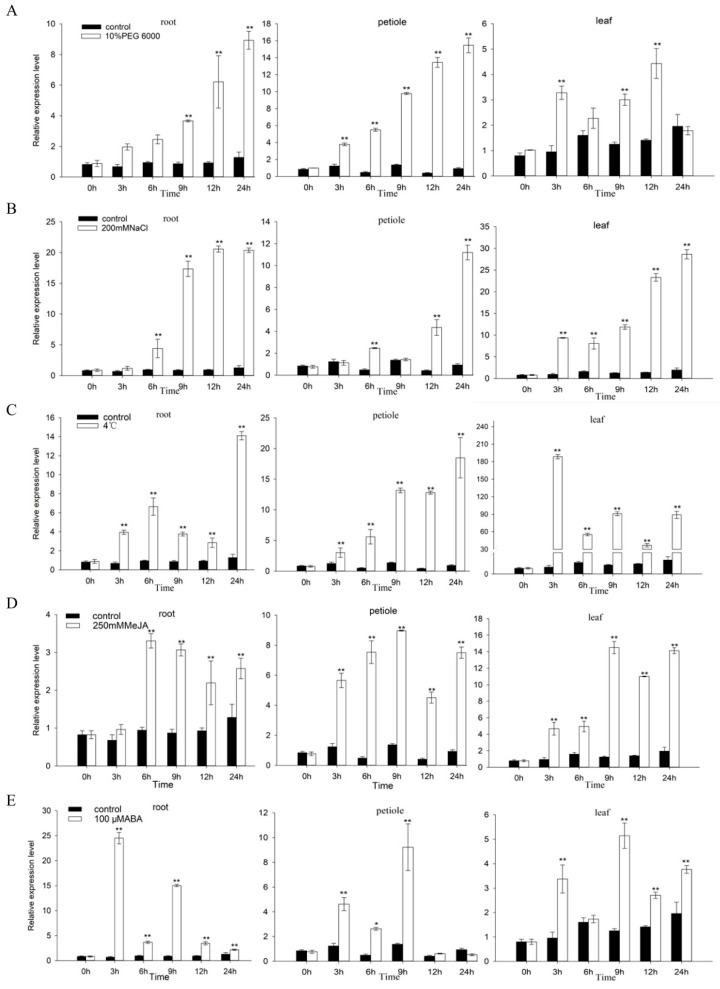
The result indicates that, under simulated drought stress, the expression of FaZAT10 increased continuously in roots and petioles, and it reached the highest level at 24 h. In leaves, the peak increased at 12 h and then gradually decreased. Under salt stress, the expression of FaZAT10 in the different tissues gradually increased along with the treatment time, and it reached the peak at 24 h, especially in roots. The expression pattern of FaZAT10 was quite different under low-temperature stress. It was significantly elevated in leaves after 3 h of stress, and then it decreased. The growth trend was similar in roots and petioles, and both reached the maximum at 24 h. Under the treatments of ABA and MeJA, FaZAT10 had no obvious tendency in roots, but the expressions of FaZAT10 in petioles and leaves both reached their peaks after 9 h of treatments, and then they decreased. Overall, these results suggest that FaZAT10 is more sensitive to salt, low-temperature, and drought stress treatments than to ABA and MeJA treatments.
3. Discussion
Transcription factors (TFs) play a central role in plant abiotic stress response networks. Currently, based on genome-wide analyses, different TF families have been identified in different plants. These TFs perform functions in various biological processes of plants, and some studies have shown that they are involved in plant abiotic stress responses [10,11,12,13,14,15,32,33]. C2H2-ZFPs are one of the best-studied TFs in plant abiotic stress responses, and 189, 109, and 321 C2H2-ZFPs have been identified in rice (Oryza sativa), poplar (Populus trichocarpa), and soybean (Glycine max), respectively [33,34,35,36,37]. Cultivated strawberry has a high economic value and is vulnerable to the negative effects of abiotic stress. Although A. thaliana provides an ideal model for studying abiotic stresses, no C2H2-ZFP family members have been reported in cultivated or wild strawberries to date. Accordingly, the whole-genome sequencing of the octoploid strawberry completed in 2019 and the re-annotation of the octoploid strawberry genome published in 2021 provided us with effective tools for a genome-wide analysis of the C2H2-ZFPs [38,39]. In this study, 126 and 41 C2H2-ZFPs in cultivated and wild strawberries were identified, and they contained one to five scattered zinc finger structures, with a maximum of five C2H2 zinc finger domains and a minimum of one. This identification is consistent with the zinc finger characteristics of subclass C1 reported in A. thaliana [17,19].
Knowing phylogenetic relationships among species is fundamental for many studies in biology. An accurate phylogenetic tree helps us to infer the origin of new genes and to understand morphological character evolution [40]. Our results show that genes in the same group are more similar to each other. They had similar but not identical protein physicochemical properties, conserved domains, and motifs, suggesting that they may have similar biological functions. For example, AtZAT6, AtZAT10, and AtZAT13 in the fourth group may be involved in the response to drought, salt, and low-temperature stress [19]. Moreover, in the analysis of the domains and motifs, we found that FaZAT4, 4a, 4b, and 4c were had characteristics consistent with the C2H2 domain, but their first zinc finger domain did not contain the conserved motif QALGGH and instead contained RALGGH (Figure S1). From the perspective of the zinc finger structure, the arbitrary amino acid X in the CX2-4CX3FX5LX2HX3-5H domain was significantly different. The presence of a highly conserved motif, QALLGGH, in the zinc finger helix is unique to plants, and the variable spacing between adjacent zinc fingers and each amino acid in the conserved sequence is thought to be important for DNA-binding activity, suggesting that this class of zinc finger proteins is involved in unique plant life processes [17,41,42]. Zinc finger helices containing this conserved motif are referred to as Q-type ZFPs, and those without any conserved motif are called C-type ZFPs [35]. This feature may affect the DNA-binding activity of these four members, which needs to be further demonstrated in experiments. We also found that the last amino acid in the first zinc finger structure of FaZAT11 is Q instead of H (Figure S1), but it conforms to other characteristics of zinc fingers, so FaZAT11 is still considered to be a C2H2-ZFP.
Plant cells have subcellular compartments, such as the nucleus, mitochondria, and chloroplast. The localization of plant proteins within cellular compartments was found to be essential in revealing their functions [43,44]. The prediction of the subcellular localization of the C1-2i members in cultivated strawberry showed that they were all localized to the nucleus. At the same time, FaZAT10 is also located in the nucleus. Previous studies in Arabidopsis have demonstrated that AtZAT10 functions as a transcriptional repressor due to the presence of the EAR domain at the C-terminus [28,29]. The EAR motif reduces the underlying transcriptional level of the reporter gene, and the transcriptional activation activity of other TFs [45]. The C-terminal inhibitory residues of AtERF4 are DLDLNL, and if a mutation occurs in this domain, the protein’s inhibitory function decreases or disappears [46]. The full-length FaZAT10 protein has no transactivation ability in yeast cells, and we conjectured that the EAR motif may be the reason why FaZAT10 does not have transactivation ability, as it may function as a transcriptional repressor in the abiotic stress signal transduction pathway. In cultivated strawberry, whether the DLNL sequence in the EAR motif of FaZAT10 is a necessary sequence for inhibitory activity needs further study.
The analysis of promoter regions is helpful to study the transcriptional regulation of TFs. The identification of plant promoters often involves the identification and characterization of genes expressed under specific tissue or physiological stress conditions [47]. Abiotic stress treatments were performed according to the cloned FaZAT10 promoter sequence. Our results indicate that FaZAT10 was continuously induced in roots and petioles; most pronounced in roots under salt and drought stresses; significantly induced in leaves after 3 h of stress at 4 °C; and significantly induced in petioles after 9 h of MeJA and ABA treatments (Figure 10). In Arabidopsis, the expression of AtZAT10 in leaves was induced by low temperatures, UV-B, oxidative stress, osmotic stress, and genotoxic stress, and it was strongly induced by low-temperature and salt stresses in roots [24]. It was also induced by a high amount of light, ABA, gibberellin, SA, and MeJA [47]. In apple, 10% PEG 6000, 150 mM NaCl, wounding, and 100 μM SA treatment induced the expression of MdZAT10 [48]. Our results are basically consistent with previous studies. The transcriptome data showed that only FaZAT10 was strongly expressed in the leaves and roots of all C1-2i members under salt stress, suggesting that FaZAT10 is more likely to play a key role in stress signal transduction. In Arabidopsis, Los2 binds to the promoter of ZAT10 in order to inhibit the expression of ZAT10, thereby attenuating the inhibitory effect of ZAT10 on RD29A and positively regulating the expression of RD29A, thus achieving the optimal response to cold environments [27]. The overexpression of AtZAT10 increased tolerance to salt and dehydration stresses and resulted in growth retardation in transgenic Arabidopsis thaliana [28]. In apple, the overexpression of MdZAT10 negatively regulates drought tolerance by regulating the expression of MdAPX2, increasing its sensitivity to PEG6000 treatment, and decreasing the ability to scavenge ROS [49]. In our study, FaZAT10 was more sensitive to salt, low-temperature, and drought stresses than hormone treatments, which indicates its potential function. However, the regulatory mechanism of FaZAT10 in response to abiotic stress remains to be further investigated.
Figure 10.
Expression patterns of FaZAT10 under different abiotic stresses. The relative expression levels of FaZAT10 under drought (A), salt stress (B), and low-temperature treatments (C) and MeJA (D) and ABA (E) hormone treatments. Data contain three biological replicates, error bars represent the standard error of the mean, and significant differences between samples were determined using Student’s t-test (**, p < 0.01; *, p < 0.05).
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of 2i Subclass in Strawberry
The gene sequences of the Arabidopsis C2H2 family were downloaded from TAIR (https://www.arabidopsis.org/) (accessed on 13 July 2022), and the Hidden Markov Model (HMM) profile of the conserved C2H2 domain (PF13912) was obtained from the Pfam database (http://pfam.xfam.org/) (accessed on 13 July 2022). To identify the ZAT genes in strawberries, wild (Fragaria_vesca_v4.0.a2) and cultivated (Fragaria x ananassa Camarosa Genome v1.0.a1) strawberry genome data were downloaded from GDR (https://www.rosaceae.org/) (accessed on 13 July 2022). Then, Simple HMM Search in Tbtools (Version 1.098752) [50] was used to pick out the candidate genes, and the redundant sequences were manually removed. Finally, all candidate genes were examined and analyzed using the SMART (http://smart.embl.de/#) (accessed on 13 July 2022) conserved domain prediction software Pfam (Pfam: Search Pfam (xfam.org)) (accessed on 13 July 2022), and NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (accessed on 13 July 2022) was used to ensure that they all contained the conserved C2H2 domain.
The ZAT proteins of Arabidopsis thaliana, the cultivated strawberries, and the wild strawberries were aligned using CLUSTALW, and a phylogenetic tree was developed on MEGA (version 7.0) using the neighborhood-joining phylogenetic method analysis. Both the bootstrap test and the approximate likelihood ratio test were set to 1000 times. The nomenclature of the ZAT gene in cultivated strawberry refers to the nomenclature of wild strawberry in NCBI. The same alleles were sequentially numbered according to their position on the chromosome, and the phylogenetic tree was embellished with iTOL (https://itol.embl.de/) (accessed on 15 July 2022).
4.2. Characteristic Analysis of the C1-2i Subclass
Online tools (https://web.expasy.org/protparam/) (accessed on 22 July 2022) were used to analyze the protein physicochemical properties of the cultivated strawberry 2i members: Plant-mPLoc (www.csbio.sjtu.edu.cn/cgi-bin/EukmPLoc2.cgi) (accessed on 22 July 2022) was used for subcellular localization prediction; a domain analysis was performed on the NCBI Conserved Domain Database (CDD: https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (accessed on 24 July 2022); and the motifs of cultivated strawberries were analyzed using the MEME Suite (https://meme-suite.org/meme/) (accessed on 24 July 2022). The physical gene location of the ZAT protein in cultivated strawberry was extracted from the cultivated strawberry genome annotation file.
The promoter sequence 2000 bp upstream of the ZAT start codon was extracted with Tbtools. The prediction of cis-acting elements on promoters was identified using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 26 July 2022) online website.
The nonsynonymous mutation rate (denoted as Ka), synonymous mutation rate (denoted as Ks), and the ratio of the nonsynonymous mutation rate to synonymous mutation rate (denoted as Ka/Ks) were calculated using Tbtools. Promoter, gene structure, conserved domains, chromosomal location, and collinearity analysis were all analyzed and visualized using Tbtools [50].
4.3. Plant Material and q-PCR Analysis
Octoploid cultivated strawberry ‘Benihoppe’ (Fragaria × ananassa Duch.) was asexually propagated using runners. The explant surface sterilization procedure was as follows: 70% alcohol soaking for 30 s, pouring out the alcohol, rinsing with sterile water two times, then soaking with 0.1% HgCl2 for 30 min, and rinsing with sterile water five times. Finally, the sterilized runners were inoculated into MS medium (0.5 mg/L 6-BA, 0.1 mg/L IBA), cultured for 45 days, and then transplanted to 1/2 MS medium. Plant material was cultured in a plant material culture room and a greenhouse under long-day conditions (16 h light, 5000 lx, 8 h dark, 22 °C). Plant material was grown in 1/2 MS medium before transplanting, and uniformly growing plants were transplanted in plastic pots with peat soil, perlite, and vermiculite (volume ratio = 4:1:1) and fertilized with regular irrigation.
Different tissues (roots, stems, functional leaves, flowers, ripe red fruits, and runners) of ‘Benihoppe’ were collected from the greenhouse for tissue-specific expression profiling; fruits at different developmental stages were collected for an expression profile analysis: small green (LG: little green, seven days after fruiting), big green (BG: big green, 15 days after fruiting), WF: white fruit, red (TR: turn red, 1/4 red), half red (HR: half red, 1/2 red), and ripe full red (FR: Full Red). Three biological replicates were established for all tissues, and all replicates were obtained from plants cultured under the same conditions. Each biological replicate contained at least six samples, with three or four samples taken from each plant. The total RNA of all samples was extracted using the modified CTAB method [51]; its integrity was checked using 1% agarose gel electrophoresis, and the concentration and quality were detected using a micronucleic acid protein analyzer. Reverse transcription was performed using a Primer Scripr™ RT Kit (TaKaRa, Dalian, China), with a gDNA removal Primer (TaKaRa, Dalian, China) per 1 µg of total RNA for PCR reactions. On the basis of the SYBR premixed ExTaqTM kit (Takara, Dalian, China), the 10 µL qRT-PCR system comprised 1µL cDNA (5 ng/µL), 0.4 µL forward/reverse primer (10 µM), 5 µL 2 × SYBR mix, and 3.2 µL ddH2O. qRT-PCR was performed on a CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA). Three technical replicates were used for each sample. The relative expression of genes was calculated using the 2−∆∆CT formula [52], and FaActin was used as a calibrated housekeeping gene. The primer sequences are listed in Table S6, Supplementary Materials. The RNA-seq-based expression levels of the FaZAT genes in strawberry were retrieved from the online transcriptomic data (SRA accessions: SRR19165835, SRR19165836, SRR19165837, SRR19165838, SRR19165841, SRR19165842, SRR8298771, SRR8298772).
4.4. Gene Clone and Sequence Analysis of FaZAT10
The full-length coding sequence (CDS) of FaZAT10 was cloned from an octoploid cultured strawberry ‘Benihoppe’, and primers were designed with reference to its homologue (FvZAT10, XM_004290912.2). The sequencing results were assembled by the software CLC Genomics Workbench (version 3.6.1) and compared with FvZAT10 using DNAMAN (version 8.0). The genome sequence of F.vesca_v2.0.a1 was used as the reference for promoter cloning, and the sequence of 2000 bp upstream of the initiation codon of FaZAT10 was cloned with the genomic DNA of cultivated strawberry as the template. The primer sequences are listed in Table S6.
4.5. Transcriptional Activation Activity Analysis of FaZAT10 Protein
The full-length CDS of FaZAT10 was inserted into the yeast expression vector pGBKT7 (BD) using the homologous recombination method. After confirming the positive vector, the transcriptional activity of the FaZAT10 protein was detected by transforming pGBKT7-Lam (a negative control), pGBKT7-FaBBX22 (a positive control) [31], and pGBKT7-FaZAT10 (validation group) into Y2H cold. After three days of culture at 28 °C, the growth of the yeast strains on different defective media (SD/ −Trp and SD/ −Trp-Ade-His /X-α-gal) was observed.
4.6. Abiotic Stress and Hormone Treatments
The octoploid cultivated strawberry ‘Benihoppe’ grown in plastic pots was pulled out, and the roots were washed with water and placed in a tissue culture bottle containing Hoagland nutrient solution (Hypo, HB8870-1) for seven days in a balanced culture, during which the nutrient solution was changed once. Low-temperature stress: the balanced octoploid strawberry ‘Benihoppe’ was put into the artificial climate box at 4°C, and the relative humidity was 75% in common Hoagland solution. Drought stress: Hoagland solution +10% PEG6000 simulated drought. Salt stress: Hoagland solution +200 Mm NaCl. Hormone treatment: 250 mM MeJA and 100 μM ABA were sprayed on the foliage with potted seedlings in common Hoagland solution. The control was potted seedlings in common Hoagland solution at 25 °C under long-day conditions. Roots, petioles, and leaves were sampled at 0 h, 3 h, 6 h, 9 h, 12 h, and 24 h. Each treatment contained at least six potted seedlings, with three biological replicates. Afterward, they were flash-frozen in liquid nitrogen at −80 °C for reservation.
4.7. Subcellular Localization
The amplified CDS of FaZAT10 and green fluorescent protein (GFP) was inserted into the plasmid vector (PYTSL-16) by homologous recombination, which was modified with pMDC83-35S and Psite-2NB [53]. The fusion expression of FaZAT10 and GFP (FaZAT10:GFP) was driven by the 35S promoter, and the nuclear localization maker for HY5-mCherry had the same 35S promoter drive [54]. The plasmid was transformed into Agrobacterium tumefaciens GV3101, and the two vectors were transiently co-expressed in tobacco epidermal cells [55]. All fluorescence signals of the samples were detected using a confocal laser scanning microscope system (FV3000 Olympus, Tokyo, Japan). The primers are listed in Table S6.
4.8. Statistical Analysis
All data were analyzed using IBM SPSS statistical software (version 23). Student’s t-test (**, p < 0.01; *, p < 0.05), one-way analysis of variance (ANOVA), Waller–Duncan test and multiple comparisons (p < 0.05) were carried out, and SigmaPlot (version 12.5) was used to plot the graphs. All of the experimental data are expressed as the mean ± standard error of the mean (SE).
5. Conclusions
In this study, for the first time, the C2H2-ZFPs in octoploid and diploid strawberry were identified. The C1-2i subclass members that contained two C2H2 domains were further systematically analyzed. These results contribute to our understanding of the evolution of the C1-2i subclass in cultivated strawberry. Meanwhile, FaZAT10 of the C1-2i subclass was cloned based on stress transcriptome data. Analyses of cis-acting elements in the promoter region and stress induction experiments revealed that FaZAT10 may play an important role in regulating stress signal transduction. Taken together, these results provide new information on the responses of C2H2-ZFPs to abiotic stress in strawberry. In addition, we preliminarily investigated the transcription factor properties of FaZAT10, and the specific molecular mechanism of FaZAT10 in abiotic stress needs to be further studied.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232113079/s1.
Author Contributions
Conceptualization, H.T. and H.L.; methodology, M.Y., H.L. and L.J.; performed the experiments, H.L. and M.Y.; software, Y.Y. and X.L. (Ximeng Lin); validation, N.Z., X.L. (Xiaoling Liu) and Y.L. (Yongqiang Liu); formal analysis, Y.Z. (Yong Zhang) and Y.L. (Yuanxiu Lin); investigation, Y.Z. (Yong Zhang) and Y.Z. (Yunting Zhang); resources, M.L., Y.W. and X.W.; data curation, Q.C. and Y.L. (Ya Luo); writing—original draft preparation, H.L.; writing—review and editing, H.T., Y.L. (Ya Luo) and Q.C.; project administration, H.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No.31872083).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Anzano A., Bonanomi G., Mazzoleni S., Lanzotti V. Plant metabolomics in biotic and abiotic stress: A critical overview. Phytochem. Rev. 2022;21:503–524. doi: 10.1007/s11101-021-09786-w. [DOI] [Google Scholar]
- 2.Giampieri F., Alvarez-Suarez J.M., Mazzoni L., Romandini S., Bompadre S., Diamanti J., Capocasa F., Mezzetti B., Quiles J.L., Ferreiro M.S., et al. The potential impact of strawberry on human health. Nat. Prod. Rep. 2013;27:448–455. doi: 10.1080/14786419.2012.706294. [DOI] [PubMed] [Google Scholar]
- 3.Kolle S.R., Shankarappa T.H., Rahimi B.A., Satish M.V. Review of Trends in Strawberry Research from 1960 to 2016. J. Agric. Food Inf. 2019;20:25–38. doi: 10.1080/10496505.2018.1507825. [DOI] [Google Scholar]
- 4.Kirnak H., Kaya C., Higgs D., Gercek S. A long-term experiment to study the role of mulches in the physiology and macro-nutrition of strawberry grown under water stress. Aust. J. Agric. Res. 2001;52:937–943. doi: 10.1071/AR01014. [DOI] [Google Scholar]
- 5.Nezhadahmadi A., Faruq G., Rashid K. The impact of drought stress on morphological and physiological parameters of three strawberry varieties in different growing conditions. Pak. J. Agric. Sci. 2015;52:79–92. [Google Scholar]
- 6.Brown J.G., Voth V. Salt damage to strawberries: Types of water, irrigation system, and soil condition found to influence salt accumulation in strawberry plantings. Calif. Agr. 1955;9:11–12. [Google Scholar]
- 7.Naing A.H., Kim C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021;172:1711–1723. doi: 10.1111/ppl.13373. [DOI] [PubMed] [Google Scholar]
- 8.Czarnocka W., Karpiński S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free. Radic. Biol. Med. 2018;122:4–20. doi: 10.1016/j.freeradbiomed.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury F.K., Rivero R.M., Blumwald E., Mittler R. Reactive oxygen species, abiotic stress and stresscombination. Plant J. 2017;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Niu Y., Zheng Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021;22:6125. doi: 10.3390/ijms22116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y.C., Zhang T.Y., Yu Y., Gou L.P., Yang J.T., Xu J., Pi E. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 2021;12:677611. doi: 10.3389/fpls.2021.677611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Pang S., Lu Z., Jin B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants. 2020;9:1515. doi: 10.3390/plants9111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi C.X., Yu Y.H., Dong C.H., Yang Y.X., Zhai Y.Q., Du F.P., Xia C., Ni Z.Y., Kong X.Y., Zhang L.C. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021;19:209–211. doi: 10.1111/pbi.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X.F., Kim M.Y., Ha J.M., Lee S.H. Overexpression of the Soybean NAC Gene GmNAC109 Increases Lateral Root Formation and Abiotic Stress Tolerance in Transgenic Arabidopsis Plants. Front. Plant Sci. 2019;10:1036. doi: 10.3389/fpls.2019.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin F.L., Zeng Y.L., Ji J.J., Wang P.G., Zhang Y.F., Li W.H. The Halophyte Halostachys caspica AP2/ERF Transcription Factor HcTOE3 Positively Regulates Freezing Tolerance in Arabidopsis. Front. Plant Sci. 2021;12:638788. doi: 10.3389/fpls.2021.638788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Ding Y.F., Cai C., Chen Z.X., Zhu C. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plant. 2019;165:690–700. doi: 10.1111/ppl.12728. [DOI] [PubMed] [Google Scholar]
- 17.Ciftci-Yilmaz S., Mittler R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008;65:1150–1160. doi: 10.1007/s00018-007-7473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie M., Sun J., Gong D., Kong Y. The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors. Genes. 2019;10:653. doi: 10.3390/genes10090653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Englbrecht C.C., Schoof H., Böhm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto H., Araki T., Meshi T., Iwabuchi M. Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene. 2000;248:23–32. doi: 10.1016/S0378-1119(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 21.Shi H.S., Wang X., Ye T.T., Chen F.F., Deng J., Yang P.F., Zhang Y.S., Chan Z.L. The Cysteine2/Histidine2-Type Transcription Factor ZINC FINGER OF ARABIDOPSIS THALIANA6 Modulates Biotic and Abiotic Stress Responses by Activating Salicylic Acid-Related Genes and C-REPEAT-BINDING FACTOR Genes in Arabidopsis. Plant Physiol. 2014;165:1367–1379. doi: 10.1104/pp.114.242404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin M.Z., Wang Y.P., Zhang L.H., Li J.Z., Quan W.L., Yang L., Wang Q.F., Chan Z.L. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017;68:2991–3005. doi: 10.1093/jxb/erx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida A., Kazuoka T., Torikai S., Kikuchi H., Oeda K. A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 2000;24:191–203. doi: 10.1046/j.1365-313x.2000.00864.x. [DOI] [PubMed] [Google Scholar]
- 24.Mittler R., Kim Y., Song L., Coutu J., Coutu A., Yilmaz S.C., Lee H., Stevenson B., Zhu J.K. Gain-andloss-of-functionmutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pabo C.O., Peisach E., Grant R.A. Design and Selection of Novel Cys2His2 Zinc Finger Proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 26.Lippuner V., Cyert M.S., Gasser C.S. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 1996;271:12859–12866. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- 27.Lee H., Guo Y., Ohta M., Xiong L.M., Stevenson B., Zhu J.K. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., Shinozaki K.Y. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold,and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta M., Matsui K., Hiratsu K., Shinshi H., Takagi M.O. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurst L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Ye Y., Wang Y., Jiang L., Yue M., Tang L., Jin M., Zhang Y., Lin Y., Tang H. B-Box Transcription Factor FaBBX22 Promotes Light-Induced Anthocyanin Accumulation in Strawberry (Fragaria × ananassa) Int. J. Mol. Sci. 2022;23:7757. doi: 10.3390/ijms23147757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan S.-A., Li M.-Z., Wang S.-M., Yin H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018;19:1634. doi: 10.3390/ijms19061634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo P., Jung J., Park C. Transcription factors: Improving abiotic stress tolerance in plants. In: Tuteja N., Gill S.S., Tiburcio A.F., Tuteja R., editors. Improving Crop Resistance to Abiotic Stress. John Wiley & Sons; Hoboken, NJ, USA: 2012. pp. 451–479. [DOI] [Google Scholar]
- 34.Liu Y., Khan A.R., Gan Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022;23:2730. doi: 10.3390/ijms23052730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal P., Arora R., Ray S., Singh A.K., Singh V.P., Takatsuji H., Kapoor S., Tyagi A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007;65:467–485. doi: 10.1007/s11103-007-9199-y. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q., Wang Z., Xu X., Zhang H., Li C. Genome-Wide Analysis of C2H2 Zinc-Finger Family Transcription Factors and Their Responses to Abiotic Stresses in Poplar (Populus trichocarpa) PLoS ONE. 2015;10:e0134753. doi: 10.1371/journal.pone.0134753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan S., Li X., Li R., Wang L., Zhang C., Chen L., Hao Q., Zhang X., Chen H., Shan Z., et al. Genome-Wide Identification and Classification of Soybean C2H2 Zinc Finger Proteins and Their Expression Analysis in Legume-Rhizobium Symbiosis. Front. Microbiol. 2018;9:126. doi: 10.3389/fmicb.2018.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edger P.P., Poorten T.J., VanBuren R., Hardigan M.A., Colle M., McKain M.R., Smith R.D., Teresi S.J., Nelson A.D.L., Wai C.M., et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019;51:541–547. doi: 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu T.J., Li M.Z., Liu Z.C., Ai X.Y., Li Y.P. Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Hort. Res. 2021;8:41. doi: 10.1038/s41438-021-00476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapli P., Yang Z.H., Telford M.J. Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 2020;21:428–444. doi: 10.1038/s41576-020-0233-0. [DOI] [PubMed] [Google Scholar]
- 41.Yu G.H., Jiang L.L., Ma X.F., Xu Z.S., Liu M.M., Shan S.G., Cheng X.G. A soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis. PLoS ONE. 2014;9:e109399. doi: 10.1371/journal.pone.0109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gourcilleau D., Lenne C., Armenise C., Moulia B., Julien J.L., Bronner G., Leblanc-Fournier N. Phylogenetic Study of Plant Q-type C2H2 Zinc Finger Proteins and Expression Analysis of Poplar Genes in Response to Osmotic, Cold and Mechanical Stresses. DNA Res. 2011;18:77–92. doi: 10.1093/dnares/dsr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glory E., Murphy R.F. Automated Subcellular Location Determination and High-Throughput Microscopy. Dev. Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Wattanapornprom W., Thammarongtham C., Hongsthong A., Lertampaiporn S. Ensemble of Multiple Classifiers for Multilabel Classification of Plant Protein Subcellular Localization. Life. 2021;11:293. doi: 10.3390/life11040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazan K. Negative regulation of defence and stress genes by EAR-motif containing repressors. Trends Plant Sci. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Han G.L., Wei X.C., Dong X.X., Wang C.F., Sui N., Guo J.R., Yuan F., Gong Z.Z., Li X.Z., Zhang Y. Arabidopsis ZINC FINGER PROTEIN1 Acts Downstream of GL2 to Repress Root Hair Initiation and Elongation by Directly Suppressing bHLH Genes. Plant Cell. 2020;32:206–225. doi: 10.1105/tpc.19.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porto M.S., Pinheiro M.P.N., Batista V.G.L., Santos R.C.D., Filho P.D.A.M., Lima L.M.D. Plant Promoters: An Approach of Structure and Function. Mol. Biotechnol. 2014;56:38–49. doi: 10.1007/s12033-013-9713-1. [DOI] [PubMed] [Google Scholar]
- 48.Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. Systemic and Intracellular Responses to Photooxidative Stress in Arabidopsis. Plant Cell. 2007;19:4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang K., Li C.Y., An J.P., Wang D.R., Wang X., Wang C.K., You C.X. The C2H2-type zinc finger transcription factor MdZAT10 negatively regulates drought tolerance in apple. Plant Physiol. Biochem. 2021;167:390–399. doi: 10.1016/j.plaphy.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Chen Q., Yu H.W., Wang X.R., Xie X.L., Yue X.Y., Tang H.R. An alternative cetyltrimethylammonium bromide-based protocol for RNA isolation from blackberry (Rubus L.) Genet. Mol. Res. 2012;11:1773–1782. doi: 10.4238/2012.June.29.10. [DOI] [PubMed] [Google Scholar]
- 52.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Ye Y., Liu Y., Li X., Wang G., Zhou Q., Chen Q., Li J., Wang X., Tang H. An Evolutionary Analysis of B-Box Transcription Factors in Strawberry Reveals the Role of FaBBx28c1 in the Regulation of Flowering Time. Int. J. Mol. Sci. 2021;22:11766. doi: 10.3390/ijms222111766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Xu P., Chen G., Wu J., Liu Z., Lian H. FvbHLH9 Functions as a Positive Regulator of Anthocyanin Biosynthesis by Forming a HY5-bHLH9 Transcription Complex in Strawberry Fruits. Plant Cell Physiol. 2020;61:826–837. doi: 10.1093/pcp/pcaa010. [DOI] [PubMed] [Google Scholar]
- 55.Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A. PGD Vectors: Versatile Tools for the Expression of Green and Red Fluorescent Protein Fusions in Agroinfiltrated Plant Leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313X.2002.01360.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.