Abstract
The major diagnostic antigens of Histoplasma capsulatum are the H and M antigens, pluripotent glycoproteins that elicit both humoral and T-cell-mediated immune responses. These antigens may play a role in the pathogenesis of histoplasmosis. M antigen is considered immunodominant because antibodies against it are the first precipitins to arise in acute histoplasmosis and are commonly present during all phases of infection. The biological activity of monomolecular M antigen and its ability to elicit a protective immune response to H. capsulatum are largely unknown. A molecular approach was used to identify the biological nature of M antigen, including its purification from histoplasmin, partial digestion with proteinases, and reverse-phase high-performance liquid chromatography to separate the released peptides. The amino acid sequences of the purified peptides were obtained by Edman degradation, and using degenerate oligonucleotide primers for PCR, a 321-bp fragment of the gene encoding the M antigen was amplified from genomic H. capsulatum DNA. This fragment was used to screen an H. capsulatum genomic DNA library, leading to the isolation, cloning, and sequencing of the full-length gene. The M gene consists of 2,187-bp DNA encoding a protein of 80,719 Da, which has significant homology to catalases from Aspergillus fumigatus, Aspergillus niger, and Eimericella nidulans. A cDNA was generated by reverse transcription-PCR and cloned into the expression vector pQE40. The identity of the cloned, expressed protein was confirmed by Western blotting. The recombinant fusion protein was immunoreactive with monoclonal antibodies raised against M antigen, with polyclonal mouse anti-M antiserum, and with a serum sample from a patient with histoplasmosis. The gene encoding the major immunodominant M antigen of H. capsulatum is a presumptive catalase, and the recombinant protein retains serodiagnostic activity.
Histoplasmosis is a systemic fungal disease caused by Histoplasma capsulatum var. capsulatum, a dimorphic fungus which grows in the mycelial form at room temperature in the environment and in the yeast form at 37°C on enriched medium or in infected tissues. The significance of histoplasmosis results from its worldwide distribution associated with bird or bat excrement, its ability to mimic other serious diseases (23), and its recognition as a common and serious opportunistic infection in patients with AIDS (24).
The major diagnostic antigens of H. capsulatum are the H and M antigens, pluripotent glycoproteins that elicit both humoral and T-cell-mediated immune responses (9). These antigens may also play a role in the pathogenesis of histoplasmosis. M antigen is considered immunodominant because antibodies against it are the first precipitins to arise in acute histoplasmosis and are more commonly present during all phases of disease (11, 22). The biological activity of monomolecular M antigen and its ability to elicit a protective immune response to H. capsulatum are largely unknown because of difficulties in obtaining sufficient quantities of monomolecular M antigen by biochemical purification procedures. M glycoprotein has been proposed to be a catalase, with an activity that could protect yeast forms from oxidative fungicidal mechanisms, but definitive evidence to support this hypothesis awaits confirmation (8). Immunochemical analysis of M antigen indicated it is a glycoprotein with a molecular mass of ∼70 to 94 kDa (8, 17, 25, 26) and an isoelectric point of 4.7 (7). M antigen contains species-specific protein epitopes and glycosidic epitopes; the latter are N linked to the peptide core and probably belong to the high-mannose-type glycan (26). The peptide epitopes react with human antibodies and are not affected by N deglycosylation (27). Biochemically purified M antigen has been shown to evoke proliferation of T cells (9).
The diagnosis of histoplasmosis is based on the results of careful clinical evaluation and associated laboratory tests. Isolation of H. capsulatum from biological specimens provides a definitive diagnosis, but this fungus frequently fails to grow under artificial culture conditions. In such cases, serologic tests, such as complement fixation and immunodiffusion, are indicated for detection of antibodies against M antigen. Serodiagnosis has some limitations, including misleading positive results for patients with other diseases caused by microbes that cross-react with H. capsulatum. Much of this cross-reactivity is due to carbohydrate determinants (1a, 7, 11, 12, 16, 27), and they can be reduced after chromatographic purification of the M antigen (25). Even purified M antigen evokes cross-reactivity in more-sensitive assays, such as enzyme immunoassay or Western blotting, owing to covalent carbohydrate determinants that are cross-reactive with similar determinants present among genera of dimorphic fungal pathogens and in certain bacteria (27). The detection of antibodies against peptide epitopes within native or recombinant M antigen could improve the specificity and sensitivity of the serologic tests and provide an earlier and more specific diagnosis of acute histoplasmosis.
In this study, our objective was to identify the biological nature of M antigen as a step in the eventual development of a recombinant antigen. With monomolecular M antigen, it would then be possible to conduct epitope mapping, improve the immunodiagnosis of histoplasmosis, and evaluate the role of M antigen in eliciting delayed-type hypersensitivity or its in vitro correlates and in the pathogenesis of histoplasmosis. Therefore, the gene encoding M glycoprotein was cloned, expressed, and sequenced.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Yeast phase cells of H. capsulatum 6623 (ATCC 26320) were grown at 37°C in Pine’s liquid medium (15, 16) for 48 h to late log phase. Escherichia coli q358 was used as the host for the bacteriophage λGem11, and E. coli INV F′ (Invitrogen Co., Carlsbad, Calif.) was used as the recipient for the subcloning vector pBluescript SK(−) (Stratagene, La Jolla, Calif.).
Purification and amino acid sequence of M antigen.
M antigen was purified from histoplasmin by cation-exchange chromatography in columns of carboxymethyl-Sepharose CL-6B, as described previously (25). Samples of M antigen were subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and electrotransferred for 1 h at 400 mA to polyvinylidene difluoride membranes (Immobilon-P; Millipore Corp., Bedford, Mass.) in 25 mM Tris, 192 mM glycine, and methanol (20% [vol/vol]). The membrane was washed several times with 1 mM dithiothreitol (DTT), stained with Ponceau S, and destained with 10% aldehyde-free acetic acid–1 mM DTT. Several washings with 1 mM DTT were made to remove the acetic acid. The band was identified by its molecular mass, and its identity was confirmed by Western blotting. The band in the membrane corresponding to M antigen (200 pmol/protein band) was excised and submitted to Edman degradation without prior modification. To obtain the internal amino acid sequences, the band was digested in situ with lysyl endopeptidase (Boehringer Mannheim Corp., Indianapolis, Ind.), and peptides were purified by microbore reverse-phase high-performance liquid chromatography on reverse-phase C18 silica. All amino acid sequences were obtained with a model 477A protein sequencer or Procise (Perkin-Elmer/Applied Biosystems, Foster City, Calif.).
DNA isolation.
Yeast form cells grown in 50 ml of Pine’s broth were harvested by filtration on a 0.45-μm-pore-size membrane, washed three times with deionized H2O, and blotted to remove excess moisture. Yeast form cells were placed in a sterile mortar with 1 g of glass beads (0.5-mm diameter); liquid N2 was added, and the mixture was ground to a fine powder in a biological safety cabinet. Personal protective equipment was used, and other precautions were taken according to established biosafety guidelines (20). The triturate was resuspended in 20 ml of TE Buffer, pH 8.0 (10 mM Tris–1 mM EDTA), and DNA was extracted with phenol, ethanol (EtOH) precipitated, and then dried and redissolved in 0.05 M TE. The RNA was removed after digestion with RNase (final concentration, 10 μg/ml) (Boehringer Mannheim) at 37°C for 1 h, followed by proteinase K treatment (50 mg/ml) (Sigma Chemical Co., St. Louis, Mo.) for an additional 1 h at 37°C. The DNA was subjected again to phenol extraction, EtOH precipitated, and redissolved in TE (18).
Generation of M DNA probe by PCR.
H. capsulatum genomic DNA was used as a template for PCR amplification of a DNA fragment encoding an internal portion of the M protein. Degenerate oligonucleotide primers were designed based on the amino acid sequences of the two internal peptides (V22 and V18) of M antigen because of initial ambiguity in the NH2 terminal sequence (Table 1). The sense primer, M4F, 5′-AA(AG)AA(CT)CC(AGC)GA(CT)TT(CT)-3′, was a 17-mer with 48-fold degeneracy, and the antisense primer, M8R, 5′-TT(AGCT)CC(AGT)AT(AGCT)GT(AG)AA-3′, also was a 17-mer with 96-fold degeneracy. PCR was conducted in a total volume of 100 μl containing 100 ng of DNA as a template, 100 mM (each) deoxynucleoside triphosphate, 1 mM (each) oligonucleotide primer, and 10× PCR buffer containing 500 mM KCl, 100 mM Tris-HCl (pH 8.3), 25 mM MgCl2, and 2.5 U of Taq polymerase (Boehringer Mannheim). The amplification conditions consisted of denaturation at 95°C for 5 min followed by 35 cycles of the succeeding steps: denaturation at 95°C for 5 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min. A final elongation step was done at 72°C for 5 min. A 321-bp PCR product was subcloned into the pCRII vector with the TA cloning kit (Invitrogen, Inc., Carlsbad, Calif.) by procedures recommended by the vendor and sequenced with a dye-labeled terminator and automated sequencer (Perkin-Elmer/Applied Biosystems).
TABLE 1.
Amino acid sequences of the NH2 terminus and lysyl endopeptidase-digested fragments of M antigen
| Origin | Amino acid sequence |
|---|---|
| NH2 terminus | S D P T D Q F L |
| Internal sequences | |
| 2642-m1947/19 | D F I F R Q K I Q H F D H E R |
| 5070-m1941/20 | T L Q G R A G L V |
| V22-m1947/20 | A Q A L G G K N P D F H R Q D L |
| V21-m1947/12 | S G R Y P E |
| V16-m1941/21 | F D F D L L D P T K |
| V18-m1941/23 | I I P E E L V P F T P I G K |
Screening of a H. capsulatum genomic library.
The 321-bp amplicon was labeled with [α-32P]dCTP by High Prime DNA labeling mix (Boehringer Mannheim), purified by passage through a DEAE column (NACS Prepac Convertible, Bio-Rad Laboratories Life Technologies, Inc., Gaithersburg, Md.), and used for screening the genomic library, derived from DNA partially digested with Sau3AI and cloned into λGem11 via the XhoI half-site, kindly provided by Glenmore Shearer. An E. coli q358 bacterium infected with the genomic library was replica plated onto nitrocellulose membranes. The plaques were lysed and then heat fixed. The filters were hybridized with a 32P-labeled probe. Twelve positive colonies were picked and rescreened as large plaques. Two strongly positive plaques were purified and mapped by Southern analysis (18). These clones were digested with BamHI, and one fragment of 4.0 kb was obtained.
Gene sequence analysis.
The 4.0-kb fragment was subcloned into pBluescript II KS and sequenced by the strategy of “primer walking” by the dideoxy chain termination method. Oligonucleotides (22-mer) were synthesized on the basis of DNA sequence and applied to initiate the sequencing reaction. The clone was sequenced in both directions. To determine the sites of putative introns, 5 μg of RNA was reverse transcribed, with oligo(dT) to initiate the cDNA reaction. The first strand of cDNA was amplified with a sense primer located at the start site of the mature protein: the sequence of this primer was 5′-CGGAATTCTCCGACCCTACGGA-3′. The antisense primer was 5′-ACCAAGCTTCTATCCAACGGGAACCGA-3′. A 5′ EcoRI site (underlined) was added to the sense primer, and a HindIII site (underlined) was added to the antisense primer to facilitate cloning in pBluescript SK(−). PCR was performed for 35 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 2 min, with 5 U of Vent polymerase (New England Biolabs, Beverly, Mass.). The PCR product was digested with EcoRI and HindIII and cloned into pBluescript SK(−), restriction mapped, and sequenced in its entirety.
Southern hybridization.
Standard conditions for electrophoresis and Southern blotting were used (18). Hybridization was performed with digoxigenin (DIG)-labeled 321-bp probe at 42°C overnight. All the prehybridization, hybridization, and membrane-washing steps were performed with DIG Easy Hyb kit and DIG Wash and Block buffer set (Boehringer Mannheim) according to the manufacturer’s recommendations (1).
Expression of M gene.
cDNA was synthesized and used as a template for PCR. The sense primer was 5′-CGGAATTCAGATCTGACCCTACGGACCAG-3′, and the antisense primer was 5′-ACCAAGCTTCTAGCTTCTATCCAACGGGAA-3′. The cDNA encoding M antigen was amplified by PCR with these primers in the presence of 2 U of Vent polymerase (New England Biolabs). The conditions were 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min. Thirty-five cycles were performed. The amplified cDNA was digested with BglII and HindIII (Gibco BRL, Grand Island, N.Y.) and ligated into pQE40 (Qiagen, Valencia, Calif.) that had been digested with the same enzymes. The ligation reaction product was cloned into E. coli XL-1 Blue (Stratagene). Colonies of E. coli were examined for the presence of the proper plasmid by restriction mapping. One clone, pQE40M6, was randomly selected for expression. The recombinant protein was purified by affinity chromatography on a nickel-Sepharose column as described previously (4).
SDS-PAGE and Western blotting.
Native and recombinant M antigens were first dissociated at 100°C for 5 min in 0.125 M Tris-HCl buffer (pH 6.8) containing 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.025% bromophenol blue. SDS-PAGE was then conducted with 7.5% separatory and 4% stacking gels in an electrophoresis cell (Mini-Protean II; Bio-Rad Laboratories, Richmond, Calif.) operated at 10 mA of constant current for stacking and 20 mA for protein separation. The gel contents were electrotransferred to nitrocellulose membranes in a Mini Trans-Blot cell (Bio-Rad Laboratories) containing the transfer buffer, 25 mM Tris-HCl, 192 mM glycine, and methanol (20% [vol/vol], pH 8.3) operated at 400 mA for 1 h. Free binding sites in the membranes were blocked by incubation for 30 min in 5% nonfat milk in phosphate-buffered saline (PBS)–0.3% Tween 20 (pH 7.5) (PBS-T). The membrane was sliced vertically, and the strips were incubated for 60 min at room temperature with (i) polyclonal mouse anti-M serum and polyclonal mouse anti-H serum, a gift from Sandra L. Bragg, Centers for Disease Control and Prevention; (ii) ascitic fluids containing murine monoclonal antibodies (MAbs) EC2-EC7 and EC2-AH12, a pool of MAbs specific for the M antigen, and MAb CA1-CB4, which binds C antigen (12, 17); and (iii) a serum specimen from a patient with culture-confirmed histoplasmosis. A preimmune mouse serum and a serum sample from a healthy individual were used as negative controls. Both polyclonal antisera and the MAbs were diluted 1:100 to 1:1,000 in PBS-T containing 5% nonfat milk. After each strip was washed in PBS-T four times for 20 min each time, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G and goat anti-human immunoglobulin G (Bio-Rad Laboratories), optimally diluted 1:1,000 in PBS-T, were added and incubated as described above. The blot strips were washed again and incubated with a substrate solution of 0.5 mg of 3,3′-diaminobenzidine tetrahydrochloride (Bio-Rad Laboratories) per ml plus 5 μl of H2O2 (30% [vol/vol]) per 50 ml of PBS. After color development, the strips were rinsed exhaustively in deionized water.
Nucleotide sequence accession number.
The M gene sequence was deposited in GenBank, and its accession number is AF026268.
RESULTS
Amino acid sequencing of M antigen.
The amino acid sequences of the NH2 terminus and six internal peptides are shown in Table 1. The amino acid sequences of two internal peptides, V22 and V18, of the M protein (Table 1) showed 66 to 73% identity with sequences of catalases of Schizosaccharomyces pombe (gpD55675 YSPC-1) and Aspergillus niger (gpZ23138 ANCATRGNA-1).
Cloning and sequencing of the M gene.
The significant degree of homology of these two internal peptides to the catalases suggested a certain arrangement in the protein (Fig. 1). Considering their positions, two degenerate oligonucleotides were designed to amplify a fragment from genomic H. capsulatum DNA. A 321-bp PCR product was achieved by using HistoM4F and HistoM8R as primers and was confirmed by Southern blotting to represent a unique gene of H. capsulatum. Sequence analysis of this amplicon obtained by the dideoxy chain terminator method enclosed the two native internal peptides, confirming that the PCR product encoded a region of the M gene.
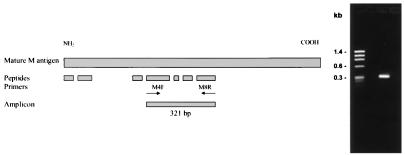
FIG. 1.
M gene cloning strategy. The M antigen peptide sequences are shown in their presumed positions, inferred from amino acid homology with fungal catalases. Primers M4F (sense primer) and M8R (antisense primer) were designed based on two internal peptides, V22-m1947/20 and V18-m1941/23, respectively (Table 1), and were used in PCR amplification of a 321-bp fragment of H. capsulatum genomic DNA.
The 321-bp PCR fragment was gel purified in 1% agarose and used to screen an H. capsulatum genomic DNA library to isolate the entire M gene. A BamHI genomic fragment of 4.0 kb carrying the M gene was isolated and characterized. This fragment was subcloned into pBluescript II KS and was sequenced in its entirety in both directions. Figure 2 shows the complete nucleotide sequence of the M gene and the deduced amino acid sequence, consisting of 729 residues with an estimated molecular mass of 80,719 Da.
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of M antigen of H. capsulatum. The TATA element, the CAAG motifs, the T+C-rich block, and the stop codon are in bold, italic, and shaded type. Base positions are shown on the left, and amino acid positions are shown on the right. Introns are indicated by lowercase letters. The leader peptide (1 to 40) is bold and shaded. The potential N-glycosylation sites are in italics, and the 321-bp fragment is underlined.
The coding region of the M gene is interrupted by five introns (Fig. 2), with the 5′ and 3′ extremities presenting the GT/AG consensus (14). The coding region of the M gene presented a 0.46 A/T ratio. The 5′ 565-bp flanking sequence of this gene exhibited regions similar to the promoter regions of other eukaryotic genes. A putative TATA element is at position 318, and a T+C-rich pyrimidine block is found downstream at position 365. The CAA motif is found twice upstream of the T+C block at positions 34 and 341. The 3′ region downstream from the M gene open reading frame contains a pentanucleotide (5′-AAATA-3′) at position 3138, 19 nucleotides downstream from the termination codon, similar to the polyadenylation consensus sequence described in other eukaryotes (14). This sequence is known to be involved in termination of transcription, processing, and addition of poly(A) at the 3′ terminus.
Protein structure.
Sequencing of the N terminus of the native protein showed that the first residue of the mature protein is a serine residue at position 614, giving a mature protein of 689 amino acids with a predicted mass of 76,491 Da. Therefore, the expected M gene has a leader peptide composed of 40 amino acids. The initiator methionine is followed by an arginine (R), a positively charged residue, and a core of 10 or more hydrophobic residues following the positively charged amino acid, features described as characteristics of signal peptides (10). The sequence also has a purine (adenine) in the −3 position, a prerequisite for an initiation codon (13). Five potential N glycosylation sites (NXT or NXS) were predicted (Fig. 2).
Comparison of the deduced amino acid sequence of M gene with known sequences.
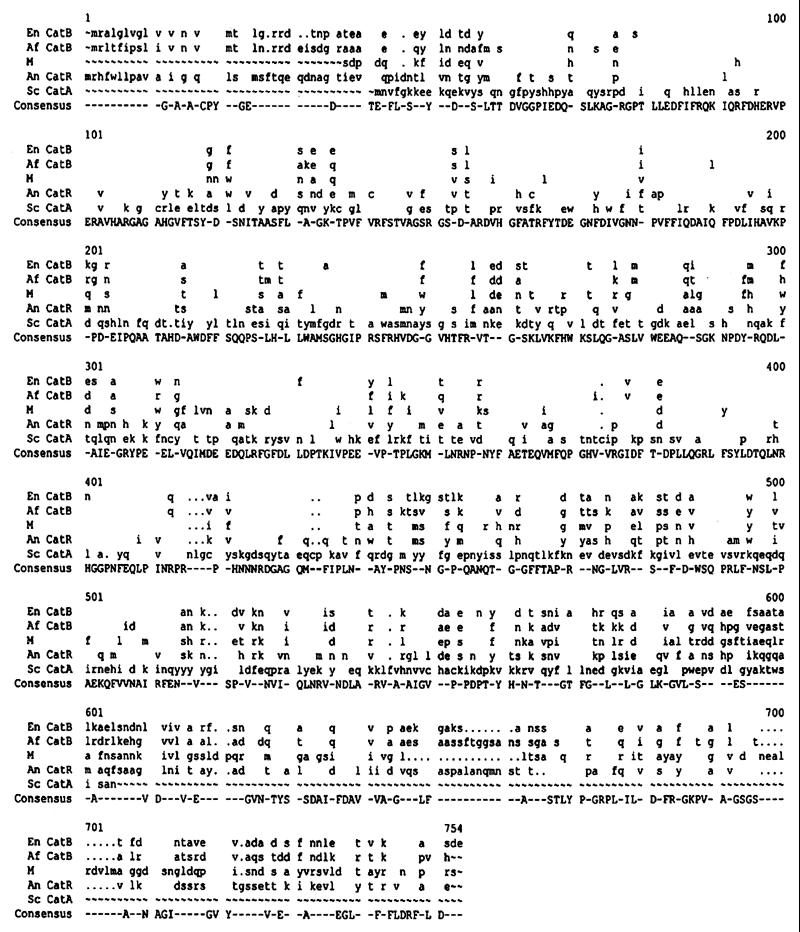
The earlier database results showing that two peptide sequences of M protein had 66 to 73% identity with sequences of catalases of S. pombe (gpD55675 YSPC-1) and A. niger (gpZ23138 ANCATRGNA-1) suggested that M antigen could be a catalase. Comparison of the complete deduced amino acid sequence of M with known fungal catalases from Aspergillus fumigatus, A. niger, Eimericella nidulans, and Saccharomyces cerevisiae demonstrated 61.2, 53.2, 60.4, and 21.7% similarity, respectively, at the amino acid level. Figure 3 shows the multiple alignment of the amino acid sequences encoded by the M gene and those of fungal catalases. The M sequence can be divided into parts with high and low homology, suggesting functional domains.
FIG. 3.
Comparison of the deduced amino acid sequence of M gene with those of other catalases from eukaryotes: A. fumigatus (Af), E. nidulans (En), A. niger (An), and S. cerevisiae (Sc). The GenBank accession numbers for the catalases are u87850, u80672, l15474, and x13028, respectively. Alignments were made with Genetics Computer Group, Inc. (Madison, Wis.) software. Blank spaces indicate identity with the consensus; ∼, gaps placed at the ends of sequences; dots indicate internal gaps; and dashes indicate no consensus at a plurality of 3.
Copy number of M gene.
Southern blotting of H. capsulatum genomic DNA digested with various restriction enzymes was probed with the 320-bp PCR product in order to evaluate the genomic organization of the M gene. A single hybridized band of 4.0 kb was seen with the BamHI-digested genomic DNA, which corresponded to the sizes of the λGem11-purified inserts. The hybridization profile obtained when genomic DNA was digested with BamHI and with other restriction endonucleases (data not shown) suggested that a single copy or very few copies of the M gene occur(s) in the genome.
Immunologic reactivity of recombinant M protein.
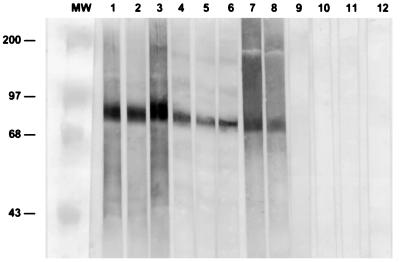
SDS-PAGE of recombinant M protein revealed a band with a molecular mass of ∼73 kDa. The identity of the cloned, expressed protein was confirmed by Western blotting probed with MAbs specific for M antigen (EC2-EC7 and EC2-AH12), with polyclonal mouse anti-M antiserum, and with a serum sample from a patient with culture-confirmed histoplasmosis. The recombinant fusion protein was immunoreactive with all anti-M antigen MAbs and with polyclonal anti-M antiserum samples but not with the MAbs and polyclonal serum samples used as controls (Fig. 4).
FIG. 4.
Western blot analysis of recombinant M antigen probed with MAbs EC2-EC7 and EC2-AH12 and M pool anti-M antigen (lanes 1, 2, and 3, respectively); with polyclonal mouse serum anti-M antigen at 1:100 (lane 4), 1:500 (lane 5), and 1:1,000 (lane 6); with a serum sample from a patient with a culture-confirmed case of histoplasmosis at 1:100 and 1:500 (lanes 7 and 8); and with negative controls: a preimmune mouse serum at 1:100 (lane 9), a healthy human serum sample at 1:100 (lane 10), polyclonal mouse serum anti-H antigen (lane 11), and MAb CA1-CB4 anti-C antigen (lane 12).
DISCUSSION
Despite the accumulated evidence of its importance in humoral immune responses, measured in clinical serologic tests (1a, 16, 27), the biological function of M antigen remains obscure, and its interaction with cells of the immune system is largely uncharacterized. The lack of availability of monomolecular M antigen has delayed efforts to determine whether it can induce protective immunity. Recombinant DNA technology was used to begin to address these issues by cloning, expressing, and characterizing the gene encoding M protein. Amino acid sequences from the N-terminal and internal peptides were determined, and the significant sequence similarity of two internal peptides of M protein with sequences of catalases of other fungi was observed. This sequence homology facilitated the design of degenerate primers to isolate a gene fragment by PCR, which then was used to screen an H. capsulatum genomic library. Several lines of evidence confirmed that the cloned cDNA and gene described in this research represent the authentic M antigen. The deduced amino acid sequence includes all the peptide sequences obtained from the native protein. The deduced mature M open reading frame encodes a polypeptide of 689 amino acids with a predicted mass of 76,491 Da, which is concordant with the molecular mass of the native glycoprotein described by immunochemical analysis of M antigen (8, 12). In addition, the recombinant M antigen, expressed as a fusion protein in E. coli, reacted strongly with MAbs against M antigen as well as with polyclonal mouse M antiserum and with an antiserum sample from a patient with histoplasmosis. No reactivity was observed with the anti-C antigen MAb (CA1-CB4) and with serum samples used as controls.
Sequence analysis of cDNA obtained by reverse transcription-PCR demonstrated that the gene encoding M protein has five introns, consistent with data already observed for genes that encode other H. capsulatum proteins (4).
A comparison of the deduced M protein sequence to the sequences of catalases from other fungi showed a high degree of homology to catalases from A. fumigatus, A. niger, and E. nidulans. Fungal catalases are very characteristic and are distinguished from those produced by other eukaryotic organisms. They are larger molecules than those from the other eukaryotes, being composed of four subunits with molecular masses ranging from 80,000 to 97,000 Da (6, 21). M antigen was previously suggested to be a catalase, based on indirect evidence obtained from an analysis by a panel of monoclonal anti-catalases (8). The high degree of homology between M protein and other fungal catalases, as well as the estimated molecular mass of the M protein, 80,719 Da, which is within the size range of fungal catalases, strongly implies that the biological nature of M antigen is a catalase.
Catalases play a protective role for aerobic microbes, neutralizing the toxic effects of hydrogen peroxide (H2O2) via its hydrolysis to O2 and H2O. Calderon and Shennan (3) have demonstrated that the resistance of Trichophyton quinckeanum and Trichophyton rubrum to H2O2-mediated killing is due to endogenous catalase. H. capsulatum is capable of replicating within polymorphonuclear cells and macrophages despite the generation of a respiratory burst (2, 5, 19). Therefore, M protein may aid in the intracellular survival of this fungus within polymorphonuclear cells by conferring resistance to oxidative fungicidal mechanisms.
The cloning and sequencing of the M gene and its expression as a recombinant protein open the possibility of mapping individual peptide epitopes to permit an analysis of B- and T-cell reactive sites within the protein. Additional knowledge gained in this area would enhance the understanding of the fungus-host interaction and would broach an exploration of the role of M antigen in the protective immune response. Moreover, these data may contribute to the development of antifungal therapy directed against fungal catalases. Finally, the recombinant protein and/or its selected peptide epitopes will be useful in the development of more sensitive and specific diagnostic tests. Studies are already in progress in our laboratory to explore this last possibility.
ACKNOWLEDGMENTS
The research of R. M. Zancopé-Oliveira at the Mycotic Diseases Branch, Centers for Disease Control and Prevention, was supported by a fellowship from the American Society for Microbiology, and the work was supported in part by grant Proc. 522249/94-4 from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. G. S. Deepe was supported by grants AI-34361, AI-42747, and HL-55949.
We thank José Mauro Peralta for review of the manuscript.
REFERENCES
- 1.Boehringer Mannheim Biochemicals. Genius system user’s guide for membrane hybridization. Indianapolis, Ind: Boehringer Mannheim Biochemicals; 1995. [Google Scholar]
- 1a.Bradley G, Pine L, Reeves M W, Moss C W. Purification, composition and serological characterization of histoplasmin H and M antigens. Infect Immun. 1974;9:870–880. doi: 10.1128/iai.9.5.870-880.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock W E, Wright S D. Role of the adherence-promoting receptors, CR3, LFA-1, and p150,95, in binding of Histoplasma capsulatum by human macrophages. J Exp Med. 1987;165:195–210. doi: 10.1084/jem.165.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderon R A, Shennan G I. Susceptibility of Trichophyton quinckeanum and Trichophyton rubrum to products of oxidative metabolism. Immunology. 1987;61:283–288. [PMC free article] [PubMed] [Google Scholar]
- 4.Deepe G S, Jr, Durose G G. Immunobiological activity of recombinant antigen from Histoplasma capsulatum. Infect Immun. 1995;63:3151–3157. doi: 10.1128/iai.63.8.3151-3157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissenberg L G, Goldman W E. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev. 1991;4:411–421. doi: 10.1128/cmr.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler T, Rey M W, Vaha-Vahe P, Power S D, Berka R M. The catR gene encoding catalase from Aspergillus niger: primary structure and elevated expression through increased gene copy number and use of a strong promoter. Mol Microbiol. 1993;9:989–998. doi: 10.1111/j.1365-2958.1993.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 7.Green J H, Pine L. Preparation of H and M antigens of Histoplasma capsulatum free of heterologous antigens. Curr Microbiol. 1985;12:209–216. [Google Scholar]
- 8.Hamilton A J, Bartholomew M A, Figueroa J, Fenelon L E, Hay R J. Evidence that the M antigen of Histoplasma capsulatum var. capsulatum is a catalase, which exhibits cross-reactivity with other dimorphic fungi. J Med Vet Mycol. 1990;28:479–485. [PubMed] [Google Scholar]
- 9.Harris J E, Deepe G S. Characterization of antigenic determinants in histoplasmin that stimulate Histoplasma capsulatum-reactive T cells in vitro. Infect Immun. 1988;56:2343–2349. doi: 10.1128/iai.56.9.2343-2349.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman L, Kovacs J A, Reiss E. Clinical immunomycology. In: Rose N R, de Macario E C, Folds J D, Lane H C, Nakamura R M, editors. Manual of clinical laboratory immunology. Washington, D.C: American Society for Microbiology; 1997. pp. 575–584. [Google Scholar]
- 12.Knowles J B. Immunochemical characterization of monoclonal antibodies to factors in histoplasmin. Ph.D. thesis. Chapel Hill: University of North Carolina; 1985. [Google Scholar]
- 13.Kozak M. Point mutation defines a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 14.Lewin B. Genes V. New York, N.Y: Oxford University Press Inc; 1994. pp. 127–159. [Google Scholar]
- 15.Pine L. Growth of Histoplasma capsulatum. VI. Maintenance of the mycelial phase. Appl Microbiol. 1970;19:413–420. doi: 10.1128/am.19.3.413-420.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pine L, Gross H, Bradley G, George J R, Gray S B, Moss C W. Procedures for the production and separation of H and M antigens in histoplasmin: chemical and serological properties of the isolated products. Mycopathology. 1977;61:131–141. doi: 10.1007/BF00468007. [DOI] [PubMed] [Google Scholar]
- 17.Reiss E, Knowles J B, Bragg S L, Kaufman L. Monoclonal antibodies against the M-protein and carbohydrate antigens of histoplasmin characterized by the enzyme-linked immunotransfer blot. Infect Immun. 1986;53:540–546. doi: 10.1128/iai.53.3.540-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Schaffner A, Davis C E, Schaffner T, Markert M, Douglas F, Braude A I. In vitro susceptibility of fungi to killing by neutrophil granulocytes discriminates between primary pathogenicity and opportunism. J Clin Investig. 1986;78:511–524. doi: 10.1172/JCI112603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. Department of Health and Human Services. Biosafety in microbiological and biochemical laboratories, 3rd ed. Health and Human Services publication (CDC) 93-8395. Washington, D.C: U.S. Government Printing Office; 1993. pp. 80–81. [Google Scholar]
- 21.Vainshtein B K, Melik-Adamyan W R, Barynin V V, Vagin A A, Grabenko A I, Borisov V V. Three-dimensional structure of catalase from Penicillium vitale at 2.0A resolution. J Mol Biol. 1986;188:63–72. doi: 10.1016/0022-2836(86)90479-1. [DOI] [PubMed] [Google Scholar]
- 22.Wheat L J, French M L V, Kohler R B, Zimmerman S E, Smith W R, Norton J A, Eitzen H E, Smith C D, Slama T G. The diagnostic laboratory tests for histoplasmosis. Ann Intern Med. 1982;97:680–685. doi: 10.7326/0003-4819-97-5-680. [DOI] [PubMed] [Google Scholar]
- 23.Wheat L J. Diagnosis and management of histoplasmosis. Eur J Clin Microbiol Infect Dis. 1989;8:480–490. doi: 10.1007/BF01964063. [DOI] [PubMed] [Google Scholar]
- 24.Wheat L J. Histoplasmosis in the acquired immunodeficiency syndrome. Curr Top Med Mycol. 1996;7:7–18. [PubMed] [Google Scholar]
- 25.Zancopé-Oliveira R M, Bragg S L, Hurst S F, Peralta J M, Reiss E. Evaluation of cation exchange chromatography for the isolation of M glycoprotein from histoplasmin. J Med Vet Mycol. 1993;31:29–41. [PubMed] [Google Scholar]
- 26.Zancopé-Oliveira R M, Bragg S L, Reiss E, Peralta J M. Immunochemical analysis of the H and M glycoproteins from Histoplasma capsulatum. Clin Diagn Lab Immunol. 1994;1:563–568. doi: 10.1128/cdli.1.5.563-568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zancopé-Oliveira R M, Bragg S L, Reiss E, Wanke B, Peralta J M. Effects of histoplasmin M antigen chemical and enzymatic deglycosylation on cross-reactivity in the enzyme linked immunoelectrotransfer blot method. Clin Diagn Lab Immunol. 1994;1:197–207. doi: 10.1128/cdli.1.4.390-393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]