Abstract
Upon entry into the host, Salmonella enterica strains are presumed to encounter an iron-restricted environment. Consequently, these bacteria have evolved a variety of often-redundant high-affinity acquisition systems to obtain iron in this restricted environment. We have identified an iron transport system that is encoded within the centisome 63 pathogenicity island of Salmonella typhimurium. The nucleotide composition of this locus is significantly different from that of the rest of this pathogenicity island, suggesting a different ancestry and a mosaic structure for this region of the S. typhimurium chromosome. This locus, designated sit, consists of four open reading frames which encode polypeptides with extensive homology to the yfe ABC iron transport system of Yersinia pestis, as well as other ABC transporters. The sitA gene encodes a putative periplasmic binding protein, sitB encodes an ATP-binding protein, and sitC and sitD encode two putative permeases (integral membrane proteins). This operon is capable of complementing the growth defect of the enterobactin-deficient Escherichia coli strain SAB11 in iron-restricted minimal medium. Transcription of the sit operon is repressed under iron-rich growth conditions in a fur-dependent manner. Introduction of a sitBCD deletion into wild-type S. typhimurium resulted in no apparent growth defect in either nutrient-rich or minimal medium and no measurable virulence phenotype. These results further support the existence of redundant iron uptake systems in S. enterica.
It has long been recognized that virulence factors of bacterial pathogens are often encoded in mobile genetic elements such as plasmids, transposons, or bacteriophages. More recently, it has become apparent that virulence factors are frequently found in discrete contiguous regions of the chromosome termed pathogenicity islands (33, 43, 54). Pathogenicity islands often have a G+C content that is significantly different from the overall G+C content of the chromosome of the host organism. This observation, coupled with the frequent presence of sequences resembling transposable elements in the boundaries of pathogenicity islands, has led to the notion that these regions constitute genetic information that may have been acquired horizontally from a heterologous microorganism, a process that most likely contributed significantly to the speciation of different bacteria. It is also a common occurrence that genes located within a pathogenicity island encode functionally related proteins.
Salmonella spp. are enteropathogenic bacteria that have sustained, longstanding associations with their vertebrate hosts. As a consequence, these bacteria display very sophisticated means to interact with host cells, resulting in the stimulation of a variety of host cellular responses (27). These responses ultimately allow these bacteria to gain access to host cells and survive within the host’s environment. The ability of Salmonella to stimulate host cellular responses is largely associated with a type III secretion system encoded within a pathogenicity island located at centisome 63 (26). This system directs the translocation of a number of bacterial proteins into the host cell, resulting in the stimulation of cellular responses such as membrane ruffling, activation of transcription factors, and, in some cells, programmed cell death (14, 35, 39). Ultimately, these responses allow Salmonella to initiate the processes that lead to the establishment of inflammatory diarrhea and the invasion of deeper tissues.
Upon entry into deeper tissues, Salmonella spp. encounter an iron-restricted environment. Consequently, similar to many other bacterial pathogens, Salmonella spp. have evolved a variety of high-affinity iron acquisition systems to obtain iron from this limiting environment. A number of iron uptake systems have been identified in Salmonella (7, 8, 10, 15, 22, 46, 47, 50, 51, 53). These include systems that make use of siderophores such as enterobactin or aerobactin to capture iron and specialized transport systems that mediate the uptake of the siderophore-iron(III) complexes. The activity of most of these specialized transport systems requires the function of the bacterial outer membrane protein TonB (42, 60). Another type of system identified in Salmonella is encoded by the feoAB locus and mediates the transport of iron(II) through the inner membrane (60). This system does not require siderophores, as iron(II) is soluble and therefore readily enters the periplasmic space by diffusion through the porins. Salmonella strains carrying mutations in known iron uptake systems are either minimally affected in virulence or not affected at all (10, 34, 40, 62). This is surprising, as Salmonella spp. are predicted to encounter iron-restricted environments in the course of their pathogenic cycle. The lack of strong phenotypes associated with mutations in iron uptake systems is therefore most likely due to the existence of several redundant systems that can mediate the uptake of this critical nutrient. Here, we report the identification and characterization of a novel iron uptake system encoded in the centisome 63 pathogenicity island of Salmonella typhimurium. This system belongs to the ABC family of transporters and complements the growth defect of an enterobactin-deficient mutant of Escherichia coli in iron-restricted medium.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. All S. typhimurium strains were derived from the wild-type strain SL1344 (40). Clinical isolates from different Salmonella enterica serovars, Shigella spp., and E. coli were from the laboratory collection. The sit operon deletion mutant strain SB801 was constructed by deleting an HpaI-to-NdeI DNA fragment (which encompasses the sitB, sitC, and sitD genes) and inserting a kanamycin resistance cassette lacking a transcription terminator (aphT) (29). The deletion construct was introduced into the chromosome by allelic exchange as previously described (41). The SL1344 derivative strain SB833, which carries the fur::Tn10 allele from strain JF2043 (30), was constructed by P22HTint-mediated transduction (58). A reporter strain carrying a fusion of sitB to the promoterless lacZ gene was constructed as follows. A promoterless lacZ cassette was cloned into the EcoRV site of sitB, and the gene fusion was cloned into the R6K-derived suicide vector pGP704 (37). The resulting plasmid, pSB1005, was subsequently integrated into the chromosome of wild-type S. typhimurium SL1344 or its fur::Tn10 derivative strain SB833 by conjugation and homologous recombination (41), yielding the reporter strains SB804 and SB835, respectively. The control reporter strain SB836 was constructed by moving the iroA::MudJ allele from strain JF2043 (30) into SB833 by P22HTint-mediated transduction. All E. coli and S. typhimurium strains were grown in Luria-Bertani medium at 37°C, and, when appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; streptomycin, 100 μg ml−1; and tetracycline, 10 μg ml−1. Iron-depleted conditions were created by using Curtiss’s minimal salts (5 g of NH4Cl, 1 g of NH4NO3, 3 g of Na2SO4, 9 g of K2HPO4, 3 g of KH2PO4, 96 mg of MgSO4 · 7H2O, and 40 mg of histidine, per liter of medium) supplemented with a 150 mM concentration of the iron chelator 2,2′-dipyridyl (Sigma, St. Louis, Mo.). Iron-replete media were supplemented with 40 μM FeSO4. Sodium citrate was added at a final concentration of 1 mM when required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Description, source and/or reference |
|---|---|---|
| E. coli strains | ||
| DH5αMCR | F−mcrA Δ(mrr-hsdRMS-mcrBC) | GIBCO BRL |
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Knr) λpir | 48 |
| BL21(DE3) | F−ompT hsdS | 59 |
| SAB11 | proA leu lacY1 Ent− | Enterobactin-deficient strain (5) |
| S. typhimurium strains | ||
| JF2043 | fur-1 zbf-5123::Tn10 iroA1::MudJ | 30 |
| SL1344 | Wild type | 40 |
| SB136 | invA | 29 |
| SB801 | ΔsitBCD::aphT | This study |
| SB804 | sitB::lacZ | SL1344 derivative with pSB1000 integrated in the chromosome |
| SB930 | sitB::lacZ | SL1344 derivative with pSB1005 integrated in the chromosome |
| SB833 | fur-1 | P22HTint[JF2043]→SL1344 |
| SB835 | fur-1 sitB::lacZ | SB833 derivative with pSB1005 integrated in the chromosome |
| SB836 | fur-1 iroA::MudJ | P22HTint[JF2043]→SB833 |
| Plasmids | ||
| pSB377 | R6K-derived replicon (41) | |
| pGP704 | 48 | |
| pSB857 | sitABCD | 36 |
| pSB992 | ΔsitBCD::aphT | pSB377 backbone |
| pSB998 | sitABCD | pSC101-derived replicon |
| pSB1000 | sitB::lacZ | pSB377 backbone |
| pSB1005 | sitB::lacZ | pGP704 backbone |
Recombinant DNA methods, DNA sequencing, and analyses.
Recombinant DNA procedures were carried out by standard protocols (55). Chromosome walking of the S. typhimurium chromosome was carried out as previously described (36). Nucleotide sequence determination was carried out with both strands of the template DNA by the dideoxy termination method. DNA and protein sequences were analyzed with the Genetics Computer Group (GCG) package from the University of Wisconsin (19). The Blastp program was used for searching protein sequence databases (GenBank, EMBL, and Swissprot) (3). PCR amplification of the sitA-flhA intergenic region was carried out by standard procedures with primers complementary to of the 3′ end of flhA (5′-TGTGGGCACTGGCTTTCATA-3′) and the 5′ end of sitA (5′-CGTGCGGGTTCGGTTTAC-3′). The predicted size of the amplified fragment based on the S. typhimurium nucleotide sequence is 646 bp.
Southern and dot blot hybridizations.
DNA samples were separated on a 1% agarose gel and transferred onto a Hybond-N nylon membrane (Amersham Life Science, Arlington Heights, Ill.). For dot blots, appropriate amounts of chromosomal DNA were spotted on Hybond-N nylon membranes. Southern and dot blot hybridizations were performed with an enhanced chemiluminescense-based kit (Amersham Life Science) according to the manufacturer’s instructions. Fluorescence-labeled probes were generated by using a random-primed nonradioactive labeling reaction (Amersham Life Science).
Tissue culture cell invasion, macrophage cytotoxicity, and mouse infection.
Bacterial internalization, macrophage cytotoxicity determination, and mouse infections were carried out as described elsewhere (13, 28).
β-Galactosidase assay.
β-Galactosidase activity was measured by the Miller assay as previously described (55).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in GenBank under accession number AF128999.
RESULTS
Identification of a putative iron transport system in the centisome 63 pathogenicity island of S. typhimurium.
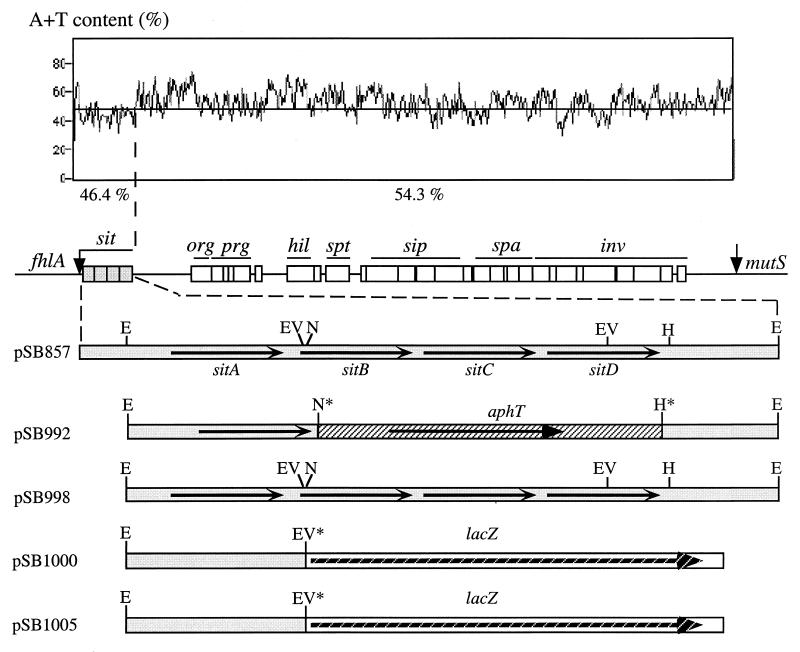
As part of our ongoing effort to characterize the centisome 63 pathogenicity island of S. typhimurium, we determined the nucleotide sequence of one of its border regions between the avrA and fhlA genes (36, 49). Plasmid pSB851, which harbors an insert that contains this entire region, was used as the source of DNA for nucleotide sequencing. This plasmid contains a segment of the centisome 63 pathogenicity island from wild-type S. typhimurium retrieved by chromosome walking (36). Four open reading frames (ORFs) apparently arranged in a single transcriptional unit were identified (Fig. 1). Putative ribosome-binding sites positioned at the appropriate distance from the initiation codons were found upstream of each of the four ORFs. A Blast search of the available databases revealed that the predicted polypeptides encoded by this operon exhibit extensive sequence similarity to ABC transport systems. The highest homologies are to the yfe ABC iron transport operon of Yersinia pestis (9), to the mnt manganese transport system of Synechocystis sp. strain 6803 (6), and to an ABC transporter of unknown function identified during sequencing of the Haemophilus influenzae genome (23). Significant similarity to a number of ABC transporters thought to mediate attachment of several gram-positive bacteria to host cells was also detected (45, 56). The arrangement of the S. typhimurium locus, which we have named sit, is characteristic of all binding-protein-dependent or ABC transport systems (11, 38) (Fig. 1). sitA encodes a 305-amino-acid polypeptide with closest similarity with the Y. pestis YfeA (9) and the cyanobacterium Synechocystis strain 6803 MntC (6) proteins (Fig. 2). YfeA and MntC are thought to function as periplasmic binding proteins in ABC transport systems involved in the transport of iron and manganese, respectively. In addition, SitA shows sequence similarity to EfaA and PsaA, which are putative adhesins from Enterococcus faecalis (45) and Streptococcus pneumoniae (56), respectively. The polypeptide encoded by sitB exhibits the signature motif (LSGGQKKRVFLARAI) of the ABC transporter family of proteins (25, 38). In addition, SitB displays a canonical nucleotide binding motif (GVNGSGKS), which is another characteristic feature of this protein family (17, 61). This region, generally referred to as Walker box A, is thought to form a flexible loop between a β-strand and an α-helix which interacts with one of the phosphate groups of the nucleotide. sitC and sitD encode polytopic integral membrane proteins which are predicted to function as permeases in this ABC system (25, 38, 57). Consistent with the homologies of the other polypeptides encoded in the sit locus, SitC and SitD are most closely related to the putative permeases of the Yersinia ABC iron transporter encoded by the yfe locus (data not shown).
FIG. 1.
A+T content and genetic organization of the sit locus encoded in the centisome 63 pathogenicity island of S. typhimurium. The region between orgA and fhlA was cloned by chromosomal walking, resulting in the plasmid pSB857. The nucleotide sequence of the region located immediately downstream of fhlA was determined. Four ORFs (sitA, sitB, sitC, and sitD) apparently arranged in a single transcriptional unit were identified. Abbreviations for restriction enzymes: E, EcoRI; EV, EcoRV; N, NdeII; H, HpaI. An asterisk indicates that the restriction site has been destroyed by cloning.
FIG. 2.
Sequence alignment of SitA with periplasmic binding proteins of binding-protein-dependent transport systems. Sequences were aligned by using the Pileup program of the GCG software package from the University of Wisconsin (19). Black boxes indicate identical amino acids, and shaded boxes indicate conservative substitutions.
Previous analysis of the nucleotide composition of the type III secretion genes encoded within the centisome 63 pathogenicity island has shown that the G+C content of this region is lower than the average for the S. typhimurium chromosome (29, 31, 32). This observation supports the hypothesis that this region of the Salmonella chromosome was acquired via horizontal gene transfer from a heterologous source. Analysis of the nucleotide composition of the sit operon shows a G+C content of 53.6%, which is significantly different from that of the rest of the centisome 63 pathogenicity island and is similar to the overall nucleotide composition of the S. typhimurium chromosome. In fact, there is a sharp transition in the nucleotide composition of the intergenic region that separates sitD, the last gene in the sit operon, and avrA, encoding a substrate of the type III secretion system (36). This observation suggests that the type III secretion system and the ABC transporter encoded by the sit operon have different ancestries and may have been acquired from different sources. Thus, the centisome 63 pathogenicity island may be a mosaic of at least two different regions with different ancestries and different functions.
Distribution of the sit operon.
To investigate the distribution of the sit operon among different strains of S. enterica, as well as other bacterial species, high-stringency dot blot DNA hybridization was performed with a 2-kb fragment encompassing a region between the 3′ end of sitD and the 5′ end of sitC as a probe. Chromosomal DNAs from 30 different S. enterica serovars, two pathogenic strains of E. coli, E. coli K-12, Shigella sonnei, and Shigella flexneri were used for the dot blotting. Under stringent conditions, the DNA probe hybridized strongly with all S. enterica isolates tested but did not hybridize with DNA samples from Yersinia and E. coli strains. A weak signal was detected in samples from Shigella spp. These results indicate that the sit operon is widely distributed among S. enterica serovars (Table 2). We also examined the location of the sit operon in representative serovars of S. enterica by PCR analysis with primers complementary to the sit locus and to the immediately adjacent gene fhlA, which constitutes one of the boundaries of SPI-1 (49). A fragment of ∼650 bp was obtained from all of the strains tested, which include isolates of S. typhimurium (serogroup B), Salmonella gallinarum (serogroup D), Salmonella pullorum (serogroup D1), Salmonella enteritidis (serogroup D1), Salmonella typhi (serogroup D1), Salmonella dublin (serogroup D), Salmonella nierstedten (serogroup C4), Salmonella thompson (serogroup C1), Salmonella duisburg (serogroup E1), and Salmonella choleraesuis (serogroup C1). These results indicate that the sit locus is located in the same region of the chromosome in most likely all serovars of S. enterica.
The S. typhimurium sit operon allows utilization of chelated iron by an enterobactin-deficient E. coli strain.
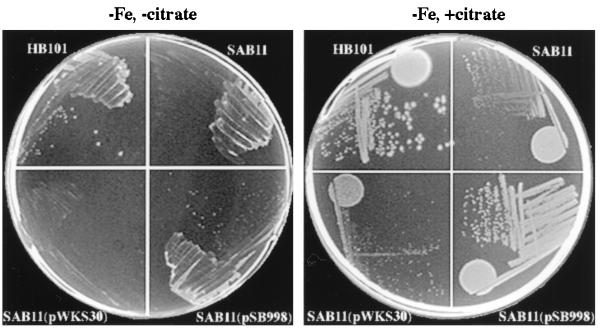
The close sequence similarity of the components of the ABC transporter encoded by the sit operon with similar systems involved in iron transport prompted us to test the possibility that this Salmonella system may be capable of transporting iron. To test this hypothesis, we introduced the plasmid pSB998, which contains the entire sit operon, into the enterobactin-deficient E. coli strain SAB11 (5, 9). This strain is incapable of growing in iron-limited media without the presence of exogenous siderophores (5, 9). As shown in Fig. 3, E. coli SAB11 expressing the S. typhimurium sit operon was able to grow in iron-deficient minimal medium. Growth was almost equivalent to that of the ent+ parent strain HB101 (average colony sizes after 48 h, 1.1 ± 0.05 mm for SAB11 and 2.1 ± 0.04 mm for HB101). In contrast, the same strain carrying the vector plasmid (pWKS30) alone failed to form visible colonies after 48 h of incubation at 37°C in the same medium, although it was able to grow in the presence of an exogenous siderophore such as citrate (Fig. 3). These results demonstrate that the sit operon encodes an ABC system that is capable of transporting chelated iron to allow growth of an enterobactin-deficient strain of E. coli and suggest that this system may perform an equivalent function in S. typhimurium.
FIG. 3.
Functional complementation of E. coli SAB11 by the S. typhimurium sit operon. The indicated strains were grown on Curtiss’s minimal agar plates in the presence or absence of citrate. Growth plates after 3 days of incubation at 37°C are shown. The enterobactin-deficient E. coli SAB11 carried either no plasmid, pSB998 (which contains the S. typhimurium sti operon), or the plasmid vector pWKS30. The enterobactin-proficient HB101 strain was included as positive control.
The expression of the sit operon is regulated by iron concentration.
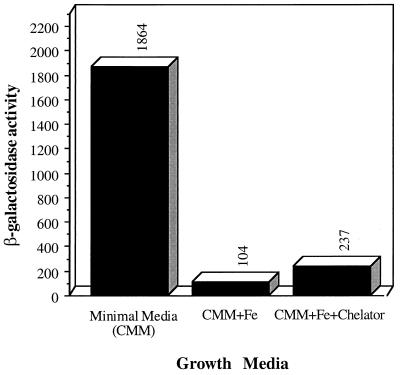
Binding-protein-dependent transport systems are often expressed only under certain conditions, such as with a specific nutrient limitation or in the presence of an appropriate substrate (16, 38). The sequence similarity of the predicted Sit proteins with components of iron transport systems, coupled to the ability of the sit genes to allow the utilization of chelated iron by an enterobactin-deficient strain of E. coli, prompted us to examine the effect of iron on the expression of the sit operon. To monitor sit gene expression, a transcriptional fusion of sitB to a promoterless β-galactosidase reporter gene was constructed and the resulting gene fusion was integrated into the chromosome of wild-type S. typhimurium by homologous recombination, resulting in strain SB804 (see Materials and Methods). The expression of the sit operon under iron-limiting and nonlimiting conditions was then monitored by measuring the levels of the β-galactosidase reporter enzyme. The expression of the sit operon was induced 18-fold when strain SB804 was grown under iron-limiting conditions (Fig. 4). The induction of sit gene expression was prevented by the addition Fe2+ but not by the addition of Ca2+. No induction was observed when strain SB804 was grown in Luria-Bertani medium, which is rich in iron. These results demonstrate that the expression of the sit operon is regulated at the transcriptional level by the iron concentration in the medium and further support the hypothesis that this operon encodes an iron transport system in S. typhimurium.
FIG. 4.
Expression of the sit operon is regulated by iron concentration. The expression of the sit operon in bacteria grown under iron-restricted or iron-sufficient conditions as indicated was monitored. CMM, Curtiss’s minimal medium. The iron chelator was 2,2′-dipyridyl (150 mM). β-Galactosidase activity is expressed in Miller units.
Effect of a fur mutation on the expression of the sit operon.
The expression of iron transport systems is negatively regulated by the transcriptional repressor Fur (16). When bound to iron, this protein is capable of binding to a consensus operator sequence located between the −10 and −35 promoter elements of iron-responsive genes, thereby repressing their transcription (18). We therefore examined the sequence upstream of the predicted ATG start codon of sitA for the presence of putative promoter elements and a Fur consensus binding site. Using the neural network algorithm (52), we identified putative −10 and −35 promoter elements in the region immediately upstream of sitA, the first gene in the sit operon (data not shown). Further analysis identified a 19-nucleotide sequence within the sit promoter region which resembles the Fur consensus binding site (Fig. 5).
FIG. 5.
Location of a putative fur box in the −10 to −35 region upstream of sitA. Alignment with the fur box sequences was done with the GCG Pileup program. Uppercase letters indicate consensus residues. de Lorenzo’s consensus sequence was previously published (18).
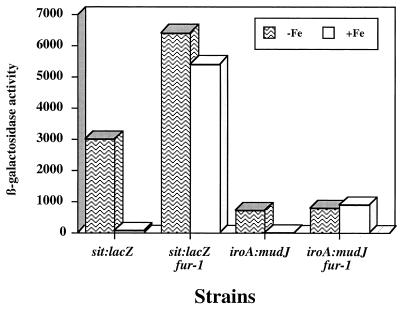
We then examined the effect of fur on sit expression. For this purpose, a sitB::lacZ gene fusion was introduced into the chromosome of an S. typhimurium fur null mutant strain, resulting in strain SB835. The expression of the sit operon in strain SB835 was then tested under both iron-limiting and nonlimiting conditions. As shown in Fig. 6, a mutation in fur completely abolished the repression of the sit operon in the presence of iron. iroA, a previously described iron-regulated gene (24), showed similar derepression in the fur background strain. This result demonstrates that the iron-dependent repression of the sit operon is mediated by Fur.
FIG. 6.
Role of fur in expression of the sit operon. The influence of a fur mutation on the expression of the sit operon in bacteria grown under iron-restricted or iron-rich conditions as indicated in Materials and Methods was monitored. The iroA reporter gene (24) was used as a control for fur-regulated genes. The values are from one experiment and are equivalent to the results obtained in several repetitions of this experiment. β-Galactosidase activity is expressed in Miller units.
Effect of sit mutations on phenotypes associated with the centisome 63 pathogenicity island.
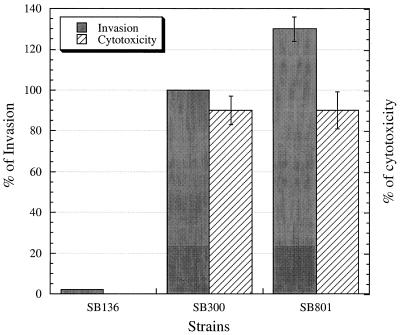
The association of an iron transport system with the centisome 63 pathogenicity island prompted us to investigate the role of this operon in the interaction of Salmonella with host cells. An S. typhimurium strain, SB801, carrying a deletion of the sitB, sitC, and sitD genes was constructed by allelic exchange as indicated in Materials and Methods. Strain SB801 was tested for its ability to enter cultured Henle-407 epithelial cells and for its ability to induce apoptosis in cultured macrophages. As shown in Fig. 7, the ability of the mutant strain to enter into Henle-407 cells and its toxic effect in macrophages were indistinguishable from those of the wild-type S. typhimurium strain SL1344. These results indicate that the iron transport system encoded by the sit operon is most likely not associated with these phenotypes. We then investigated the potential contribution of the sit operon to S. typhimurium pathogenesis by examining the virulence of a sitBCD deletion mutant strain in a mouse model of infection. No difference in the virulences of the wild-type and ΔsitBCD strains was observed after oral infection of susceptible BALB/c mice (data not shown). These results are consistent with previous reports indicating that iron uptake systems are functionally redundant in S. typhimurium and that single mutations most often translate into either a weak or no virulence phenotype (10, 34, 40, 62).
FIG. 7.
Effect of sit on S. typhimurium entry into cultured epithelial cells and macrophage cytotoxicity. The internalization levels, measured by the gentamicin protection assay, were standardized by considering the levels for the wild-type S. typhimurium strain SL1344 to be 100% (the actual values in this case were 17% ± 0.8%). Macrophage J774 cytotoxicity is presented as the percentage of cells exhibiting cytotoxicity after 30 min of infection and was determined as described previously (13). Fewer than 1% of macrophages infected with S. typhimurium SB136 exhibited cytotoxicity.
DISCUSSION
We have described here an ABC transporter that is encoded within the centisome 63 pathogenicity island of S. typhimurium. The nucleotide composition of the 5 kb of DNA that comprises this locus (54% G+C) is significantly different from the nucleotide composition of the rest of the pathogenicity island (44% G+C), which encodes a type III secretion system. Our observation suggests that this pathogenicity island may be the result of independent events that allowed the acquisition of genetic material from different sources. The mosaic structure of this region is also supported by the unrelated functions encoded in these two loci. Our hybridization studies, although not exhaustive, clearly indicate that the sit locus is widely conserved among different S. enterica serotypes. This observation suggests that the acquisition of the sit region of the centisome 63 pathogenicity island must have occurred early in the evolution of S. enterica.
The ABC transporter encoded by the sit operon is most closely related to a Y. pestis iron transport system encoded by the yfe locus (9). Several ABC iron transporters have been described for gram-negative bacteria (38). Most of these systems are involved in the transport of a siderophore-iron complex across the inner membrane. Examples of these systems in S. enterica are the enterobactin and aerobactin iron uptake systems (21). However, more recently another family of ABC transport systems that mediate the transport of iron from the periplasm to the cytosol in a siderophore-independent manner has been recognized. These systems are thought to utilize several outer membrane proteins as iron receptors to capture iron from the medium into the periplasmic space. The S. typhimurium sitABC system is functionally more closely related to this family of iron transporters, which includes, in addition to the Y. pestis yfeABC system, the Neisseria gonorrhoeae fbpABC, H. influenzae hitABC, and Serratia marcescens sfuABC systems (1, 2, 4, 9, 12). The ability of the S. typhimurium sit operon to allow the utilization of chelated iron by an enterobactin-deficient strain of E. coli when grown in iron-deficient minimal medium supports this hypothesis.
The expression of iron uptake systems is stringently regulated by the concentration of iron in the growth medium (16). In most cases, control of gene expression is exerted through the function of the iron-sensitive transcriptional repressor protein Fur. Consistent with its involvement in iron uptake, the expression of the sit operon was strongly influenced by the levels of iron in the growth medium. When growth was under iron-limiting conditions, expression of the sit operon increased 18-fold. This induction of sit gene expression was readily prevented by the addition of Fe2+ to the growth medium but not by the addition of Ca2+. Furthermore, repression by iron was completely abrogated by the introduction of a fur null mutation. These results demonstrate that the expression of the sit operon is regulated at the transcriptional level by the iron concentration in the medium, further supporting its involvement in iron uptake.
During the pathogenic cycle, S. enterica strains are thought to encounter iron-limited environments. It is therefore not surprising that these bacteria have evolved several iron uptake systems (8, 10, 15, 22, 46, 47, 50, 51, 53). Despite the expected importance of these systems for pathogenesis, the experimental demonstration of their involvement in Salmonella virulence has remained the subject of some controversy, as deficiency in any individual system has resulted in either a limited or no virulence phenotype (10, 34, 40, 62). Consistent with these results, the virulence of a strain carrying a deletion of the sit operon remained virtually indistinguishable from that of wild-type S. typhimurium. This is most likely a consequence of the redundancy of iron uptake systems in these bacteria rather than a reflection of the lack of importance of iron acquisition in Salmonella pathogenesis. Unambigous demonstration of the importance of iron in Salmonella virulence awaits the construction of a strain deficient in all described, and perhaps yet-to-be-discovered, iron uptake systems.
In summary, we have identified an ABC transporter encoded within the centisome 63 pathogenicity island of S. typhimurium. This transporter is most likely involved in iron uptake, since its expression is regulated by iron concentration and the Fur transcriptional repressor and it can confer the ability to grow in an iron-deficient minimal medium to an enterobactin-deficient strain of E. coli. Our results further support the existence of redundant iron uptake systems in S. enterica and provide evidence for a mosaic structure in the centisome 63 pathogenicity island.
TABLE .
1. Distribution of sitCD genes
| Hybridization signal | Organism | Serotype |
|---|---|---|
| Positive | S. enterica serovar agona | B |
| S. enterica serovar anatum | E1 | |
| S. enterica serovar arizona | ||
| S. enterica serovar bovis morbidificans | C2 | |
| S. enterica serovar braenderup | C1 | |
| S. enterica serovar Brandenburg | B | |
| S. enterica serovar bredeney | B | |
| S. enterica serovar choleraesuis | C1 | |
| S. enterica serovar dublin | D1 | |
| S. enterica serovar duisburg | B | |
| S. enterica serovar enteritidis | D1 | |
| S. enterica serovar gallinarum | D1 | |
| S. enterica serovar hadar | C2 | |
| S. enterica serovar heidelberg | B | |
| S. enterica serovar infantis | C1 | |
| S. enterica serovar java | B | |
| S. enterica serovar manhattan | C2 | |
| S. enterica serovar montevideo | C1 | |
| S. enterica serovar newport | C2 | |
| S. enterica serovar nienstaedten | C4 | |
| S. enterica serovar ohio | C1 | |
| S. enterica serovar othmarschen | C1 | |
| S. enterica serovar panama | D1 | |
| S. enterica serovar pullorum | D1 | |
| S. enterica serovar schwarzengrund | B | |
| S. enterica serovar tennessee | C1 | |
| S. enterica serovar thompson | C1 | |
| S. enterica serovar typhi | D1 | |
| S. enterica serovar typhimurium | B | |
| S. enterica serovar virchov | C1 | |
| Negative | S. sonnei | |
| S. flexneri | ||
| Enteropathogenic E. coli (strain E2348) | ||
| Enteroinvasive E. coli (strain EIC32) | ||
| E. coli K-12 |
ACKNOWLEDGMENTS
We thank John Foster for providing the fur-1 S. typhimurium mutant strain and Jorge Crosa for the enterobactin-deficient E. coli strain.
This work was supported by Public Health Service grant AI30492 from the National Institutes of Health. J.E.G. is an investigator of the American Heart Association.
REFERENCES
- 1.Adhikari P, Berish S A, Nowalk A J, Veraldi K L, Morse S A, Mietzner T A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J Bacteriol. 1996;178:2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari P, Kirby S D, Nowalk A J, Veraldi K L, Schryvers A B, Mietzner T A. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J Biol Chem. 1995;270:25142–25149. doi: 10.1074/jbc.270.42.25142. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Angerer A, Klupp B, Braun V. Iron transport systems of Serratia marcescens. J Bacteriol. 1992;174:1378–1387. doi: 10.1128/jb.174.4.1378-1387.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barghouthi S, Payne S M, Arceneaux J E, Byers B R. Cloning, mutagenesis, and nucleotide sequence of a siderophore biosynthetic gene (amoA) from Aeromonas hydrophila. J Bacteriol. 1991;173:5121–5128. doi: 10.1128/jb.173.16.5121-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartsevich V V, Pakrasi H B. Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 1995;14:1845–1853. doi: 10.1002/j.1460-2075.1995.tb07176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumler A J, Norris T L, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumler A J, Tsolis R M, van der Velden A W, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 9.Bearden S W, Staggs T M, Perry R D. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin W H, Jr, Turnbough C L, Jr, Posey B S, Briles D E. The ability of Salmonella typhimurium to produce the siderophore enterobactin is not a virulence factor in mouse typhoid. Infect Immun. 1985;50:392–397. doi: 10.1128/iai.50.2.392-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boos W, Lucht J M. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1175–1209. [Google Scholar]
- 12.Chen C Y, Berish S A, Morse S A, Mietzner T A. The ferric iron-binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol Microbiol. 1993;10:311–318. doi: 10.1111/j.1365-2958.1993.tb01957.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Zychlinsky A. Apoptosis induced by bacterial pathogens. Microb Pathog. 1994;17:203–212. doi: 10.1006/mpat.1994.1066. [DOI] [PubMed] [Google Scholar]
- 15.Colonna B, Nicoletti M, Visca P, Casalino M, Valenti P, Maimone F. Composite IS1 elements encoding hydroxamate-mediated iron uptake in FIme plasmids from epidemic Salmonella spp. J Bacteriol. 1985;162:307–316. doi: 10.1128/jb.162.1.307-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean M, Allikmets R. Evolution of ATP-binding cassette transporter genes. Curr Opin Genet Dev. 1995;5:779–785. doi: 10.1016/0959-437x(95)80011-s. [DOI] [PubMed] [Google Scholar]
- 18.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorman C J, Barr G C, Bhriain N N, Higgins C F. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988;170:2816–2826. doi: 10.1128/jb.170.6.2816-2826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earthart C. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1075–1090. [Google Scholar]
- 22.Ernst J F, Bennett R L, Rothfield L I. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J Bacteriol. 1978;135:928–934. doi: 10.1128/jb.135.3.928-934.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 24.Foster J W, Park Y K, Bang I S, Karem K, Betts H, Hall H K, Shaw E. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology. 1994;140:341–352. doi: 10.1099/13500872-140-2-341. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs R. Predicting protein function: a versatile tool for the Apple Macintosh. Comput Appl Biosci. 1994;10:171–178. doi: 10.1093/bioinformatics/10.2.171. [DOI] [PubMed] [Google Scholar]
- 26.Galán J E. Molecular bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 27.Galán J E, Bliska J B. Cross-talk between bacterial pathogens and their host cells. Annu Rev Cell Dev Biol. 1996;12:219–253. doi: 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- 28.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-del Portillo F, Foster J W, Finlay B B. Role of acid tolerance response genes in Salmonella typhimurium virulence. Infect Immun. 1993;61:4489–4492. doi: 10.1128/iai.61.10.4489-4492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginocchio C, Pace J, Galán J E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groisman E A, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groisman E A, Ochman H. Pathogenicity islands—bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 34.Hall H K, Foster J W. The role of Fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol. 1996;178:5683–5691. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galán J E. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 36.Hardt W-D, Galán J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 39.Hobbie S, Chen L M, Davis R, Galán J E. Involvement of the mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 40.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 41.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 42.Kingsley R, Rabsch W, Roberts M, Reissbrodt R, Williams P H. TonB-dependent iron supply in Salmonella by α-ketoacids and α-hydroxyacids. FEMS Microbiol Lett. 1996;140:65–70. doi: 10.1111/j.1574-6968.1996.tb08316.x. [DOI] [PubMed] [Google Scholar]
- 43.Lee C A. Pathogenicity islands and the evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 44.Liu S L, Ezaki T, Miura H, Matsui K, Yabuuchi E. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun. 1988;56:1967–1973. doi: 10.1128/iai.56.8.1967-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe A M, Lambert P A, Smith A W. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luckey M, Neilands J B. Iron transport in Salmonella typhimurium LT-2: prevention, by ferrichrome, of adsorption of bacteriophages ES18 and ES18.h1 to a common cell envelope receptor. J Bacteriol. 1976;127:1036–1037. doi: 10.1128/jb.127.2.1036-1037.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luckey M, Pollack J R, Wayne R, Ames B N, Neilands J B. Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J Bacteriol. 1972;111:731–738. doi: 10.1128/jb.111.3.731-738.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills D B, Bajaj V, Lee C A. A 40 kilobase chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 50.Pollack J R, Ames B N, Neilands J B. Iron transport in Salmonella typhimurium: mutants blocked in the biosynthesis of enterobactin. J Bacteriol. 1970;104:635–639. doi: 10.1128/jb.104.2.635-639.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabsch W, Paul P, Reissbrodt R. A new hydroxamate siderophore for iron supply of Salmonella. Acta Microbiol Hungarica. 1987;34:85–92. [PubMed] [Google Scholar]
- 52.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Presented at the Pacific Symposium on Biocomputing, Kona, Hawaii, 2 to 7 January 1996.
- 53.Reissbrodt R, Kingsley R, Rabsch W, Beer W, Roberts M, Williams P H. Iron-regulated excretion of α-keto acids by Salmonella typhimurium. J Bacteriol. 1997;179:4538–4544. doi: 10.1128/jb.179.14.4538-4544.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhierl B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 56.Sampson J S, O’Connor S P, Stinson A R, Tharpe J A, Russell H. Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect Immun. 1994;62:319–324. doi: 10.1128/iai.62.1.319-324.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saurin W, Koster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:74–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 59.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsolis R M, Baumler A J, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Mol Microbiol. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunit of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yancey R J, Breeding S A, Lankford C E. Enterochelin (enterobactin): virulence factor for Salmonella typhimurium. Infect Immun. 1979;24:174–180. doi: 10.1128/iai.24.1.174-180.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]