Summary
Objective.
We examined whether post-traumatic epilepsy (PTE) is associated with measurable perturbations in gut microbiome.
Methods.
Adult Sprague-Dawley rats were subjected to Lateral Fluid Percussion Injury (LFPI). PTE was examined 7 months after LFPI, during a 4-week continuous video-EEG monitoring. 16S ribosomal ribonucleic acid gene sequencing was performed in fecal samples collected before LFPI/sham-LFPI and 1 week, 1 and 7 months thereafter. Longitudinal analyses of alpha diversity, beta diversity, and differential microbial abundance were performed. Short-chain fatty acids (SCFA) were measured in fecal samples collected before LFPI by Liquid Chromatography with Tandem Mass Spectrometry.
Results.
Alpha diversity changed over time in both LFPI and sham-LFPI subjects; no association was observed between alpha diversity and LFPI, the severity of post-LFPI neuromotor impairments, and PTE. LFPI produced significant changes in beta diversity and selective changes in microbial abundances associated with the severity of neuromotor impairments. No association between LFPI-dependent microbial perturbations and PTE was detected. PTE was associated with beta diversity irrespective of timepoint vis-à-vis LFPI, including at baseline. Preexistent fecal microbial abundances of four amplicon sequence variants belonging to the Lachnospiraceae family (three enriched and one depleted) predicted the risk of PTE with area under the curve (AUC) of 0.73. Global SCFA content was associated with the increased risk of PTE with AUC of 0.722, and with 2-Methylbutyric (depleted), valeric (depleted), isobutyric (enriched) and isovaleric (enriched) acids being most important factors (AUC of 0.717). When the analyses of baseline microbial and SCFA compositions were combined, AUC to predict PTE increased to 0.78.
Significance.
While LFPI produces no perturbations in the gut microbiome that are associated with PTE, the risk of PTE can be stratified based on preexistent microbial abundances and SCFA content.
Keywords: Fluid percussion injury, post-traumatic epilepsy, random forest classifier, microbiota, short-chain fatty acids
Introduction
Perturbations in gut microbiome have been well established after traumatic brain injury (TBI)1–9. Top-down, TBI impairs normal composition and function of gut microbiota, presumably through the dysregulation of autonomic nervous system10. Bottom-up, TBI-induced gut dysbiosis contributes to chronic neurological and neuropsychiatric sequela of brain trauma through the disruption of microbial products that are relevant to normal brain function7; 10. Examples of these products are molecules pertinent to inflammation, metabolism of gamma-aminobutyric acid, serotonin, dopamine, and short-chain fatty acids (SCFA)2; 11; 12. Potential outcomes include, but are not limited to impaired blood-brain barrier integrity, neurogenesis, microglia activation, myelination, and neuronal excitability11–14.
Studies examining the role of gut microbiome in brain disorders have been increasingly focusing on SCFA. SCFA are a major product of gut bacteria metabolism and are involved in blood-brain barrier permeability, innate immunity, histone acetylation, microglia function, and neuronal excitability13; 15–17. SCFA have been depleted in fecal samples after experimental TBI in mice, while their administration improved TBI-induced cognitive dysfunction2.
Post-traumatic epilepsy (PTE) is a common outcome of TBI18. There is a growing evidence that epilepsy is associated with altered composition of gut microbiome19–24. Studies involving microbiome transfer showed that microbiota taken from epilepsy-prone animals and transplanted in naïve subjects enhanced in the latter the susceptibility to epilepsy25; 26. Conversely, antiepileptic effects of ketogenic diet have been attributed to the modulation of gut microbiota27.
While the contribution of dysbiosis induced by precipitating insults (e.g. TBI) to brain disorders receives most attention, a less commonly discussed aspect is whether natural variations in gut microbial composition influence the vulnerability to a disease, including epilepsy23. Some evidence suggest that premorbid microbiome variations, when combined with other risk factors (e.g. genetic traits or dietary/nutritional specifics), may promote autoimmune disorders28, depression, anxiety29, Parkinson’s disease30 and epilepsy22.
Using a model of TBI-PTE continuum, lateral fluid percussion injury (LFPI) in the rat31–33, we investigated whether preexistent and/or TBI-induced alterations in microbial composition are associated with PTE. Further, considering the importance of SCFA in regulating epilepsy-relevant processes, we analyzed fecal content of SCFA vis-à-vis the susceptibility to PTE.
Methods
Subjects.
Female and male Sprague-Dawley rats (Charles River, Wilmington, MA), 50–60 days old, 350–450 g at the beginning of the study. The animals were singly housed at 20–25°C, 12 h normal light–dark cycle (Zeitgeber time [ZT] 0 = 7:00), with standard rodent chow (LabDiet 5001, LabDiet, St. Louis, MO, USA) and tap water ad libitum. The procedures were approved by the UCLA Animal Research Committee and by the Animal Care and Use Review Office of the United States Army Medical Research and Materiel Command.
Lateral Fluid Percussion Injury (LFPI) was performed as described31; 32. Details are in Supporting Information.
Neuromotor impairments.
In our studies31 such LFPI parameters as pressure pulse, apnea, the suppression of toe pinch and righting reflexes had no statistical association with gastrointestinal dysfunction following LFPI; conversely, the extent of neuromotor impairments one day after LFPI positively correlated with the disruption of intestinal barrier. For this study, we chose LFPI-induced neuromotor impairments, measured using composite neuromotor score31; 34, for examining the association between TBI and dysbiosis. Motor tasks included forelimb contraflexion, hindlimb flexion, lateral pulsion, and the ability to maintain position on the angled board (details are in the Supporting information). One day prior to LFPI, the animal was examined in all tasks, with the individual baseline score set at 28. One and seven days after LFPI, the tasks were repeated, with points deducted for each failure. Based on the conservative assessment of data distribution, rats with post-LFPI neuromotor score of ≤13 were defined as severely impaired, ≥14 and ≤24 as moderately impaired, and ≥25- as mildly or not impaired.
Fecal sample collection.
Fecal samples were collected from each animal one day before LFPI/sham LFPI, and one week, one and seven months thereafter. Between ZT3 and ZT4 the rat was placed in the autoclaved cage; 4–6 fecal boluses were collected immediately after defecation, frozen on dry ice, transferred into micro tubes (Rnase-, Dnase-, DNA-, pyrogen-, endotoxin-free), and stored at −80°C.
Monitoring of spontaneous seizures was performed independently in different rats by two groups - Mazarati and Staba. After the final sample collection, LFPI animals were randomized, with half of them transferred from the Mazarati’s to the Staba’s lab for seizure monitoring. For the analysis, data obtained by each group were combined. Seizure monitoring started 7 months after LFPI, after the last fecal sample collection, 1 week after surgery to implant recording electrodes, and continued for 4 weeks. Details are in Supporting Information. An electrographic seizure was defined as high-amplitude rhythmic discharges that represents a new pattern of activity (repetitive spikes, spike-and-wave discharges, and slow waves) that lasted ≥10 sec, showed evolution in pattern, frequency or field, and culminated in postictal depression35 (examples are in supporting information Fig. S1). The animal was considered to have epilepsy if during the 4-week observation period it presented with at least one electrographically identifiable seizure, notwithstanding its behavioral correlate.
16S ribosomal ribonucleic acid (rRNA) gene sequencing.
DNA was extracted from fecal samples using the ZymoBIOMICS DNA Microprep Kit (Zymo Research, Irvine, CA, USA) per the manufacturer’s protocol. The V4 hypervariable region of the 16S ribosomal RNA gene (515F/806R) was sequenced by 2×250 paired-end sequencing using an Illumina NovaSeq 6000 S prime flow cell36. The raw sequences were processed using the DADA2 pipeline in R to generate amplicon sequence variants (ASV) and taxonomy was assigned using the SILVA 138 database37, to provide fine taxonomic resolution that can differentiate bacterial species38. One sample with less than 35,000 sequences was excluded from the analysis. The sequence depth of the remaining samples ranged from 35,412 to 476,868, with a mean depth of 118,882.
Measurement of short-chain fatty acids (SCFA).
Fecal samples collected before LFPI, were analyzed for eight SCFA: acetic, propionic, isobutyric, butyric, 2-methylbutyric, isovaleric, valeric and hexanoic. Samples were analyzed at Metabolon Inc. (Morrisville, NC, USA) by means of Liquid Chromatography with Tandem Mass Spectrometry, using a proprietary Metabolon Methods TAM135: “LC-MS/MS Method for the Quantitation of SCFA (C2 to C6) in Feces”. Details are in Supporting Information.
Sample sizes.
Repeated microbiome analysis vis-à-vis neuromotor score was performed in 60 LFPI rats, vis-à-vis PTE- in 50 LFPI rats (18 with PTE and 32 without PTE), as well as in 21 sham-LFPI subjects. SCFA were analyzed in samples from 48 rats, collected before LFPI- 18 rats that would proceed to develop PTE, and 30 rats that would not develop PTE; samples from two rats of the “no PTE” category were randomly excluded due to procedural specifics. Further details are in Supporting Information.
Data analysis
Microbiota.
ASV count data was analyzed using the PhyloSeq package in R39. Alpha diversity (i.e. a measure of microbial diversity within a fecal sample of an individual rat) was assessed by calculating the Shannon index, which incorporates both microbial richness (i.e. how many ASVs were detected) and evenness (i.e. how evenly distributed the relative abundances of ASVs were in a sample) with data rarefied to 35,000 sequences/sample. Statistical significance was assessed using linear mixed effects models with subject as a random effect. Non-rarefied data was then filtered to remove ASVs present in less than 10% of samples; then, beta diversity (i.e. a measure of similarity/dissimilarity of microbial profiles between different groups of rats) analysis was performed using Bray-Curtis dissimilarity. Results were visualized by principal coordinates analysis. Repeated measures aware PERMANOVA was performed with 100,000 permutations to determine statistical significance of differences in beta diversity40; 41. Differential abundance testing was performed using non-rarefied 16S rRNA sequence data filtered to remove ASVs present in less than 25% of samples. The resulting filtered datasets were analyzed by mixed effects models implemented in Microbiome Multivariable Association with Linear Models (MaAsLin2) using log-transformed relative abundance data42. P-values for variables in the linear models were converted to q-values to correct for multiple hypothesis testing43. A random forests classifier to predict post-traumatic epilepsy development from baseline microbial abundances was created using the random Forest package in R with mtry=2 and 1001 trees44. Microbes were inputted into the algorithm if they were significantly associated with PTE in mixed effects MaAsLin2 models with q<0.25. An initial classifier was created then all features with an importance score greater than 2 in the preliminary classifier were used to construct a refined classifier with fewer features. The accuracy of the final random forests classifiers was assessed using 5-fold cross-validation with confidence intervals determined by bootstrapping in the pROC package in R45.
SCFA.
Statistical comparisons were done with the SAS 9.3 or JMP Pro 16.1.0 software (SAS Institute Inc., Cary, NC, USA). Comparisons of individual SCFAs between two groups (i.e. with and without impending PTE) were done with both t-test and Wilcoxon rank-sum test to account for both normal and abnormal distributions. Multiple logistic regression was used to determine the odds ratios (OR) for developing epilepsy for each SCFA and receiver operating characteristics curves (ROC), areas under the ROC (AUC), and 95% Wald confidence intervals (CI) values for individual or combined SCFAs were obtained. Significance was set at p<0.05.
Neuromotor score and seizures were analyzed using Prism 6 software (GraphPad, San Diego, CA); the applied statistical tests are indicated in Results.
Results.
Microbiome and LFPI
Analyses of alpha diversity, beta diversity and SCFA were performed both with and without the inclusion of sex as a covariate. We detected no discernable differences in microbial compositions between males and females, nor any of the outcome microbiome measures were affected by sex (not shown). Therefore, the respective results below are presented for females and males combined.
Time-dependent changes in microbial diversity and composition were analyzed using mixed-effects models, which included groups (LFPI and sham-LFPI), timepoint vis-à-vis craniotomy, and group:timepoint interaction, with individual animals as a random effect. There was a highly significant association of timepoint with Shannon index (p=0.0001; Fig. 1A). At the same time, for each timepoint, alpha diversity was not significantly different between subjects of LFPI and sham-LFPI groups (baseline p=0.3; 1 week p=0.11; 1 month p=0.64; 7 months p=0.87). The group:timepoint interaction was not significant (p=0.19), indicating that LFPI produced no specific changes to pre-existing alpha diversity profiles. Indeed, the distribution of alpha diversity between sham-LFPI and LFPI subjects remained unchanged over time (group p=0.003).
Fig. 1. Fecal microbial composition before and after LFPI and sham LFPI.

(A) Microbial alpha diversity as assessed by the Shannon index of richness and evenness before, 1 week, 1 month, and 7 months following craniotomy in feces from LFPI and sham-LFPI (denoted by “Sh” on the X-axis) rats. P-values were calculated using mixed effects linear models with group, timepoint, and group:timepoint interaction as fixed effects and rat identifier as a random effect. (B) Beta diversity analysis was performed using Bray-Curtis dissimilarity and visualized by principal coordinates (PC1 and PC2) analysis (PCoA). Each dot represents individual sample, differently colored for each timepoint on the left panel, and for each group on the right panel. Symbols in the right panel denote different timepoints. P-values were calculated by repeated measures aware PERMANOVA including group, timepoint, and group:timepoint interaction. (C) PCoA plots showing microbial composition at each of the four timepoints vis-à-vis craniotomy. P-values were calculated by analyzing data at each timepoint separately using PERMANOVA to assess group difference. (D) Phylum and (E) genus level composition in LFPI and sham-LFPI (denoted by S on the X-axis) groups at each timepoint. Shown are detected phyla (D) and the top 16 most abundant genera (E).
Beta diversity was also significantly associated with the timepoint (p<0.0001, Fig. 1B). Conversely, group type was not associated with microbial composition (p=0.99); however there was a significant group:timepoint interaction (p=0.005), indicating that while there were no pre-craniotomy differences between LFPI and sham-LFPI subjects, following craniotomy the microbiota in rats of LFPI and sham-LFPI groups diverged. When assessing each timepoint, the LFPI and sham-LFPI groups showed significant differences in beta diversity at 1 week (p=0.04), 1 month (p=0.006), and 7 months (p=0.01) but not at baseline (Fig. 1C). Relative abundances at phylum and genus levels matched the results of beta diversity analysis, showing clear differences between 1 and 7 months compared to baseline, and 1 week irrespective of the group type (Fig. 1D, E).
To examine whether specific bacteria were affected by LFPI, differential abundance testing was performed using mixed effects models implemented in MaAsLin2 (Fig. 2). Matching the beta diversity analysis, more differentially abundant microbes between LFPI and sham-LFPI groups appeared at 1 month and 7 months than at 1 week. At 1 week (Fig. 2A), three ASVs were significantly differentially abundant, including enrichment in LFPI rats of a Ruminoclostridium_9 ASV, depletion of a Pseudomonas ASV, and depletion of an unclassified Lachnospiraceae ASV (labeled as “1”) that was also depleted at 1 month (Fig. 2B). Three additional ASVs within the Lachnospiraceae family were also depleted at 1 month, as was a Lactobacillus ASV. Conversely, there was enrichment of Lactobacillus intestinalis, two Bacteroides ASVs (including B. uniformis), an abundant Clostridium_sensu_stricto_1 ASV, an unclassified Peptococcaceae ASV, and five ASVs within the Ruminococcaceae family. At 7 months (Fig. 2C), six ASVs were depleted in LFPI rats, all belonging to the Lachnospiraceae family similar to what was observed at 1 month. This was accompanied by enrichment of the same Peptococcaceae ASV enriched at 1 month, two ASVs within the Ruminococcaceae family, Parabacteroides goldsteinii, two Alistipes ASVs (A. ihumii and A. finegoldii), and a Streptococcus ASV.
Fig. 2. Differentially abundant fecal microbes between LFPI and sham-LFPI rats.
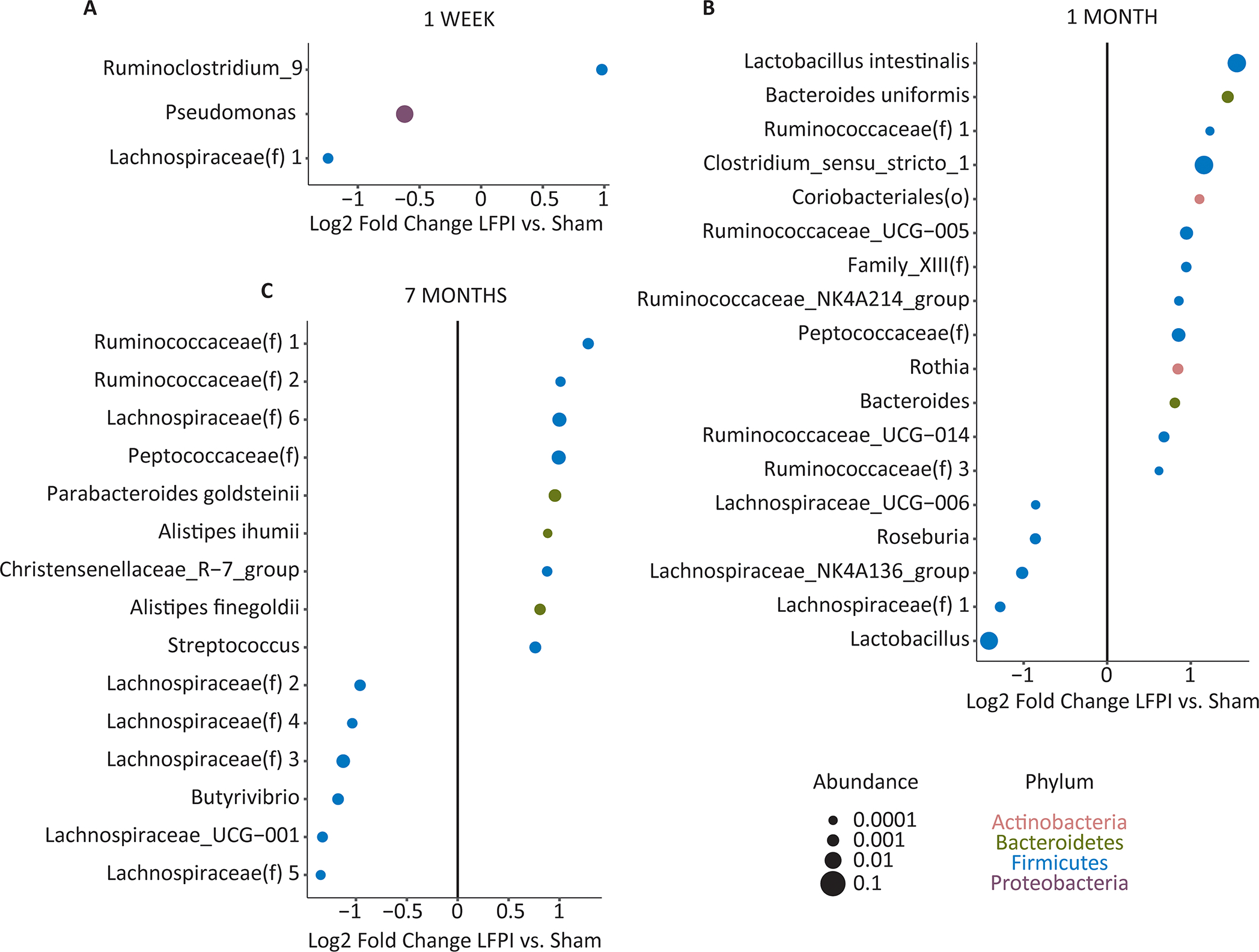
Microbes were analyzed at the level of amplicon sequence variants (ASVs), which were labeled according to the lowest level of taxonomic identification: species, genus, family [indicated by (f)], or order [indicated by (o)]. Multiple ASVs belonging to the same family were differentiated by arbitrary numbers starting from 1. ASVs with statistically significant association with LFPI (q<0.05) in a timepoint-dependent manner are shown for 1 week (A), 1 month (B), and 7 months (C). Effect size is shown as the log2 of the fold change. Dot sizes are proportional to mean ASV abundances and dot colors represents different phyla.
Microbiome and neuromotor impairments
Among the 60 LFPI rats, minimal/median/maximal (Min/Med/Max) neuromotor score 1 day after LFPI was 4/14.5/21 (95% confidence interval of median [CIMed] 13–15). Twenty-nine rats were severely impaired (Min/Med/Max score 4/12/13, 95% CIMed 10–13). Thirty-one rats fell were moderately impaired (Min/Med/Max score 14/16/21, 95% CIMed 15–18). None of the LFPI rats was mildly or not impaired. Neuromotor scores were significantly different between the two categories of the animals (Wilcoxon rank-sum, p<0.0001). Among 21 sham-LFPI rats, Min/Med/Max neuromotor score was 25/27/28, 95% CIMed 26–28. Neuromotor scores were significantly different between the groups of LFPI and sham-LFPI rats (Wilcoxon rank-sum p<0.0001), as well as between moderately impaired LFPI and sham-LFPI rats and severely impaired and sham-LFPI rats (Wilcoxon rank-sum p<0.0001). Female rats were more severely affected than males (Wilcoxon rank-sum, p=0.02). Seven days after LFPI, the animals recovered from neuromotor impairments (Min/Med/Max score was 26/27/28, 95% CIMed 27–28).
We compared alpha and beta diversities between the subgroups of moderately impaired (n=31) and severely impaired (n=29) LFPI rats. There were no associations between the extent of neuromotor impairments and either alpha diversity (p=0.39, Fig. 3A), or beta diversity (p=0.89, Fig. 3B). The group:timepoint effects were also insignificant (alpha diversity p=0.06; beta diversity p=0.25).
Fig. 3. Fecal microbial abundances and neuromotor impairments after LFPI.

(A) Microbial alpha diversity assessed by the Shannon index of richness and evenness. Mixed effects models were used with neuromotor impairments (moderate [“Mod” on X-axis] vs. severe [“Sev” on X-axis]), timepoint, and neuromotor impairment:timepoint interaction as fixed effects and rat identifier as a random effect. (B) Beta diversity analysis was performed using Bray-Curtis dissimilarity and visualized by principal coordinates analysis (PCoA). Each dot represents individual sample differently colored for moderate and severe impairment. Symbols denote different timepoints. P-values were calculated by repeated measures aware PERMANOVA including impairment, timepoint, and impairment:timepoint interaction. (C, D) Microbes that were differently abundant in moderately and severely impaired LFPI rats. Samples were analyzed together in mixed effects models implemented in MaAsLin2 with LFPI subgroups (moderately and severely impaired), timepoint, and subgroup:timepoint interaction as fixed effects and rat identifier as a random effect. ASVs that were significantly associated with neuromotor impairment (q<0.05) in a timepoint-dependent manner are shown for 1 week (C) and 7 months (D). Effect size is shown as the log2 of the fold change. Dot sizes are proportional to mean ASV abundances and dot colors represent different phyla. No ASVs were significant at q<0.05 at 1 month (not shown).
We further examined whether specific taxa might be related to the severity of post-LFPI neuromotor deficits. We performed differential abundance testing across all timepoints using mixed effects models to identify microbes that significantly differed between rats with severe and moderate neuromotor impairments at 1 week, 1 month, and 7 months. At 1 week (Fig. 3C) severely impaired rats, had significant depletion of six ASVs within the Lachnospiraceae and Ruminococcaceae families and enrichment of a single Ruminococcaceae ASV, as compared with moderately impaired subjects. There were no differentially abundant microbes at 1 month (not shown). At 7 months (Fig. 3D), severely impaired rats showed enrichment of a Streptococcus and Ruminococcaceae ASV and depletion of four ASVs within the Lachnospiraceae family. Of the differentially abundant microbes, the Streptococcus ASV had also been associated with LFPI, when compared with sham-LFPI subjects at the same timepoint.
Microbiome and PTE
During the 4-week EEG monitoring, spontaneous seizures were detected in 18 out of 50 LFPI rats (Mazarati- 11 out of 30, Staba- 7 out of 20). Individual animals displayed between 2 and 19 seizures during the observation period. No seizures were detected in sham-LFPI subjects. The proportions of females (5 out of 14) and males (13 out of 36) that developed PTE were statistically similar (Fisher exact test p=1.0). In rats that would proceed to develop PTE, Min/Med/Max neuromotor score 1 day after LFPI was 9/13/18 (95% CIMed l2–15). In rats, which would not develop PTE, Min/Med/Max neuromotor score was 8/15/21 (95% CIMed 13–16). The outcome measures were statistically similar between rats of the two categories (Wilcoxon rank-sum, p=0.31).
There were no significant differences in alpha diversity between rats that developed and did not develop PTE (p=0.07, Fig. 4A); the PTE:timepoint interaction was also not significant (p=0.61, Fig. 4A). Conversely, significant differences in beta diversity were observed between LFPI subjects that proceeded to develop and not to develop PTE (p=0.001, Fig. 4B); these differences was present irrespective of the timepoint vis-à-vis LFPI, including baseline (p=0.44). Prior to LFPI, rats that proceeded to develop PTE had enrichment of microbes within the Lachnospiraceae and Ruminococcaceae families and the depletion of Muribaculaceae family members, compared with LFPI rats that did not develop PTE (Fig. 4C). Of the PTE-associated taxa only Lachnospriaceae(f) one was differentially abundant after LFPI. Of note, this ASV was reduced at baseline in rats that would proceed to develop PTE and remained depleted following at 1 week and 1 month.
Fig. 4. Fecal microbial composition and PTE.
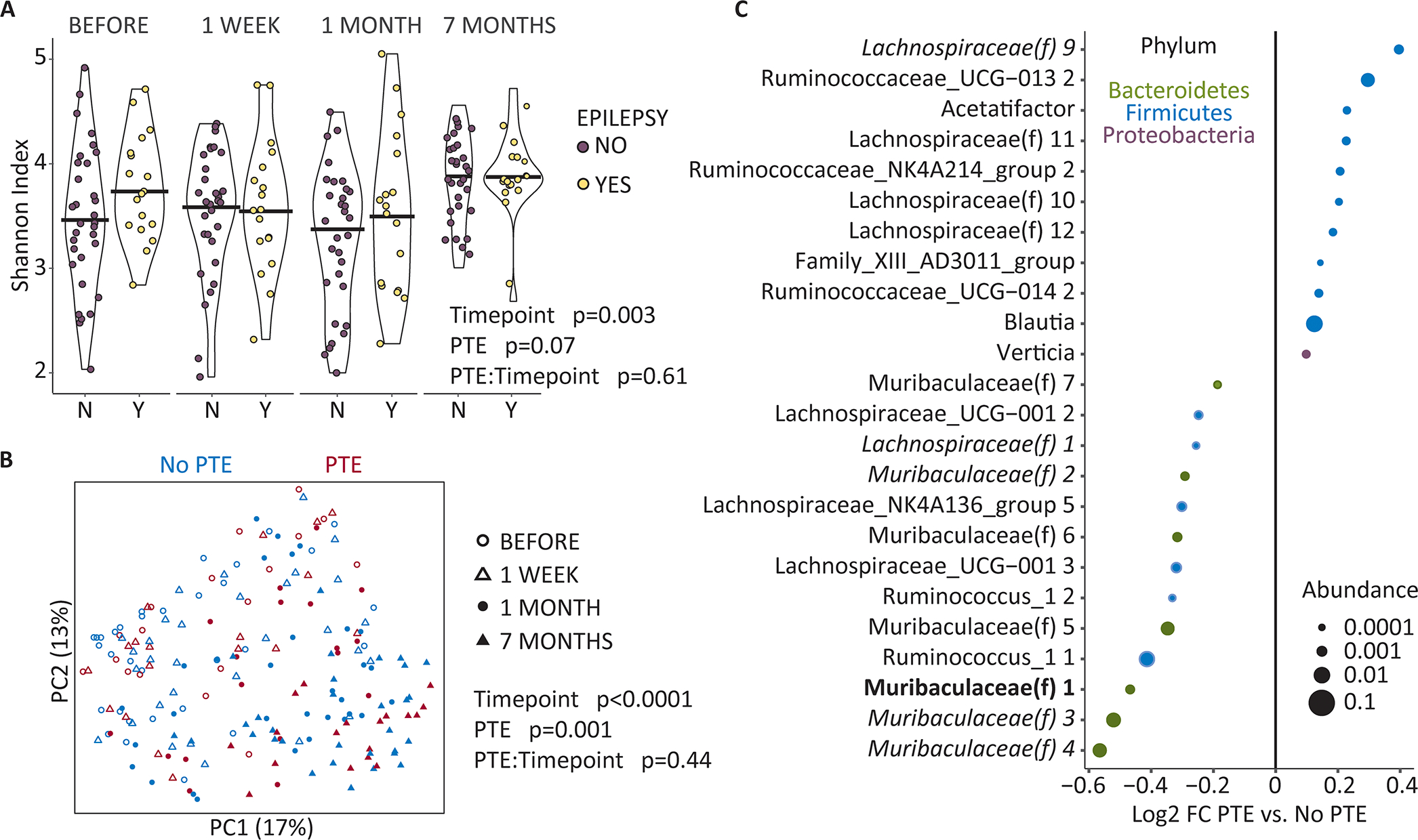
(A) Microbial alpha diversity was assessed by the Shannon index of richness and evenness. Mixed effects models were use with PTE status (EPILEPSY YES and NO), timepoint, and PTE status:timepoint interaction as fixed effects and rat identifier as a random effect. (B) Beta diversity analysis was performed using Bray-Curtis dissimilarity and visualized by principal coordinates (P1 and P2) analysis (PCoA). Each dot represents individual sample differently colored for the PTE status. Symbols denote different timepoints. P-values were calculated by repeated measures aware PERMANOVA including PTE status, timepoint, and PTE status:timepoint interaction. (C) Microbes that were differently abundant prior to LFPI, in rats that proceeded to develop and not develop PTE after LFPI. Samples were analyzed together in mixed effects models implemented in MaAsLin2 with PTE status and timepoint as fixed effects and rat identifier as a random effect. Interaction with time was not included as this was not significant in the beta diversity analysis. Shown are ASVs associated with PTE (q<0.25), with bold font indicating ASVs with q<0.05 and italics font indicating ASVs with q between 0.05 and 0.1. Effect size is shown as the log2 of the fold change. Dot sizes are proportional to mean ASV abundance and dot colors represent different phyla.
Preexistent fecal microbial composition and the risk of PTE
Considering the significant association of baseline microbial composition and specific taxa with PTE (Fig. 4C), we examined whether baseline microbial profiles could predict the development of epilepsy following LFPI. A random forests classifier was constructed from taxa that were differentially abundant before LFPI between rats that would and would not develop PTE. This classifier included four ASVs and demonstrated moderate accuracy for predicting PTE, with AUC of 0.73 in 5-fold cross-validation (Fig. 5A). All four ASVs belonged to the Lachnospiraceae family with two identified as belonging to the Blautia and Acetatifactor genera (Fig. 5B).
Fig. 5. Baseline fecal microbial abundances and SCFA and the risk of PTE.

(A) Receiver operating characteristics (ROC) curve for a random forest classifier predicting PTE from baseline abundances of 4 fecal microbes, selected from the 25 ASVs associated with epilepsy with q<0.25. Classifier performance was assessed by 5-fold cross-validation and is shown as the area under the ROC curve (AUC). The 95% confidence interval (shown as a blue area) was determined by bootstrapping. (B) Importance scores are shown for the four ASVs in the classifier shown in (A), representing their contribution to classifier accuracy. ASV color indicates whether the ASV was enriched (red) or depleted (green) prior to LFPI in rats that proceeded to develop PTE after LFPI. (C, D) ROC curves for PTE prevention derived from multiple logistic regression of 50 samples collected before LFPI. (C) When all 8 SCFAs are considered, AUC is 0.7222. (D) To avoid overfitting, we reduced the number of SCFA, and the best prediction (AUC=0.7167) was observed when we considered four SCFAs: 2-methylbutyrate (2Meth), isovaleric (Isov), isobutyric (Isobut), and valeric. Shown are ROCs for each individual SCFA and the 4 SCFAs combined (4 SCFA). (E) ROC curve for a random forest classifier predicting PTE from baseline abundances of four fecal microbes and four fecal SCFAs. (F) Importance scores are shown for the ASVs and SCFAs in the classifier shown in (E).
SCFA and the risk of PTE
Because the risk of PTE was associated with baseline microbial composition and abundances, we limited the examination of SCFA to fecal samples collected prior to LFPI, with PTE data applied for retrospective analysis. We found no association between any of the SCFAs and microbial composition or abundances (not shown), nor any differences between individual SCFA fecal content and impending PTE (t-test and Wilcoxon rank-sum; Fig. 6). Logistic regression showed that that risk of PTE increased with the depletion of 2-methylbutyric (OR=0.808, CI 0.668 – 0.979, p=0.029) and enrichment of isovaleric acids (OR=1.229, CI 1.004 – 1.503, p=0.045). Multiple logistic regression for PTE risk, considering all SCFAs yielded an AUC of 0.722 (CI 0.557 – 0.887; Fig. 5C). To avoid overfitting, we explored combinations of fewer SCFA and the smallest set yielding the best AUC (0.717) included 2-methylbutyric, isovaleric, isobutyric, and valeric acids (Fig. 5D). AUC of these SCFAs combined was better than the individual SCFA AUCs (0.5833 – 0.6343), suggesting possible SCFA interactions in predicting PTE. We further performed an analysis to identify correlations between PTE-associated microbes at baseline and SCFAs. We created a refined random forests classifier, which incorporated both the four ASVs and baseline levels of four SCFAs. This combined classifier had an AUC of 0.78 (Figure 5E), with the microbes and SCFAs having different importance scores (Fig. 5F).
Fig. 6. Short chain fatty acids (SCFA) in fecal samples collected prior to LFPI.

Data for each SCFA are presented for LFPI rats that proceeded to develop PTE (Yes on X-axis; n=18) and not to develop PTE (No on X-axis; n=30). No differences in individual SCFA content were found between the rats of two categories (Wilcoxon rank-sum and t-tests).
Discussion
Our major finding was that the risk of PTE was moderately associated with beta diversity and with selective microbial abundances, irrespective of the timepoint vis-à-vis LFPI, including at baseline. Baseline fecal composition of SCFAs was also moderately associated with the risk of PTE. The accuracy of predicting PTE based on preexistent microbiome profile was improved by integrating the identified ASVs and SCFAs.
Microbiome and LFPI
Alpha diversity, that is microbial diversity in an individual rat irrespective the microbial composition, was not affected by LFPI, the severity of neuromotor impairments and the presence of PTE. The observed time-dependent changes in alpha diversity reflected its natural variations when measured at random time points.
Beta diversity gauges the similarity/dissimilarity of microbial compositions between the rats of different groups and types. Beta diversity changed over time in both LFPI-and sham-LFPI subjects. At the same time, LFPI produced specific changes in microbial composition relative to the sham-LFPI, which were obvious shortly after the injury and persisted at 7 months. Since, on the one hand, neuromotor deficits lasted for <7 days after LFPI, and microbial changes were detectable even at 7 months, it was not likely that dysbiosis was directly due to neuromotor deficits. Of note, while we found no sex-specific associations between microbiome and PTE, we are not prepared to state this unequivocally, as the study was not powered for sex, and further increase in sex-targeted sample sizes may reveal such associations.
While many of identified changes in microbial composition were independent of the extent of post-LFPI neuromotor deficits, certain microbes were differently abundant in rats with severe vs. moderate impairments. The fact that these microbial perturbations occurred in LFPI rats with and without PTE alike, agreed with our earlier observations (also confirmed here), that the severity of post-LFPI neuromotor impairments had no bearing on the development of PTE31. The significance of these changes is outside the scope of this PTE-focused study; however, these findings merit further investigation, as they may provide insight in other chronic TBI sequela.
Sustained complex changes in gut microbial composition were reported in the weight drop46; 47 and in the controlled cortical impact (CCI) models in mice2; 4; 11, in CCI 48 and weight drop1 models in rats. Up to seven days after LFPI, changes in cecal microbiota were observed in rats6 and in fecal microbiota in mice8. Increased alpha-diversity and phylum-level changes in fecal samples were detected in people years after head injury5. Comparative analysis of different studies is complicated due to differences in species and strains, TBI paradigms, trauma severity, and analysis timepoints; overall assessments reveals little if any overlap, emphasizing an importance of standardized paradigms for determining which microbiota components are typically and critically associated with brain trauma9.
Microbiome and risk of PTE
When LFPI rats were retrospectively stratified vis-à-vis PTE, the animals with and without impending PTE had different microbial profiles even before LFPI, with the differences persisting thereafter. Hence, while PTE itself did not alter beta diversity, preexisting microbial composition informed the risk of epilepsy once TBI occurred. Multimodal involvement of gut microbes in regulating gut-brain axis and neuronal circuits relevant to epilepsy13; 19 make it impossible to draw definitive conclusions on mechanisms of this association. The answer is more likely to come from studies involving functional assessment by shotgun metagenomics and metabolomics, the latter of both fecal and blood samples to detect neuroactive microbial metabolites that enter the circulation and reach the brain. At the present stage, it would be reasonable to view our findings from a basic science perspective, whereby microbial profiling even before LFPI informs the risk of PTE. Following a thought expressed by Russo23, we may draw an analogy between our findings and genetic studies, which pursue the identification of genetic markers of susceptibility to PTE, that is such markers, which by their virtue (e.g. single nucleotide polymorphism) exist before TBI49. Certainly, an analysis performed soon after LFPI, would be more relevant for translational (e.g. diagnostic and therapeutic) purposes. Fundamentally, microbial profiles associated with impending PTE were universally present at different post-LFPI timepoints; this by itself lays a reasonable foundation for further translational studies.
One question is what “default states” of the identified microbial profiles are vis-à-vis PTE. One possibility is the default state is resistance to epilepsy; then microbial profile predicting PTE may be described as “proepileptic”. Alternatively, the default state is susceptibility to PTE; then, microbial profile predicting the absence of PTE may be described as “antiepileptic”. This question can be answered through fecal microbiome transfer, whereby “PTE-susceptible” recipients are transplanted with fecal samples from “PTE-resistant donors” and vice versa, followed by LFPI. Metagenomics and metabolomics may further help in identifying molecules and pathways that are involved in regulating susceptibility/resistance to PTE.
The involvement of microbiota in epilepsy has been documented in clinical observations (see19; 23; 24 for excellent reviews). Experimental confirmation comes from studies involving fecal microbiome transfer22; 25; 26, and as well as those establishing a pivotal role of microbiota in anti-seizure effects of ketogenic diet27.
Congruently with our findings, a study that used Wag-Rij rats, a genetic model of absence epilepsy, found that certain microbial profiles predated the development of seizures22. Even before the onset of epilepsy and at different timepoints thereafter, the most notable difference in fecal microbial composition between Wag-Rij rats and their normal Wistar counterparts was the increased Firmicutes:Bacteroidetes ratio. In our study, naive animals with impending PTE had reduced fecal abundance of specific bacteria in the Bacteroidetes phylum.
SCFA and risk of PTE
Global baseline fecal SCFA content was associated with the risk of PTE. Combination of three branched acids, 2-methylbutyric (depleted), isovaleric (enriched) and isobutyric (enriched), as well as a non-branched valeric (depleted) were most predictive of PTE risk. Branched SCFA are products of protein fermentation, which may result in additional metabolites that may damage the intestinal epithelium50, that latter suggested after LFPI31 and in genetic absence model22.
Global depletion of SCFAs was found in murine fecal samples up to one month after CCI, while exogenous SCFA administration improved CCI-induced cognitive deficits2. Treatment with butyric acid improved neurological dysfunction, brain edema, neurodegeneration, blood-brain barrier disruption in the weight drop model in mice51. Lower concentration of propionic and butyric acids was observed in fecal samples of Wag-Rij rats even before the emergence of absence epilepsy22. While our findings appear to be more subtle and complex, and are not well aligned with those reported by others (ostensibly for the reasons discussed for microbiota earlier), they do reflect an important role that is attributed to SCFAs in regulating brain functioning13; 15–17 and indicate that the analysis of SCFA in fecal samples may be useful for stratifying the risk of PTE (although SCFA involvement in epilepsy remains largely unknown, with data mostly limited to ketogenic diet23).
In conclusion, the analysis of natural variations in gut microbiome may inform the risk of PTE. This may be particularly useful when applied to individuals operating in such high-risk environments as war zone, hard-hat areas, and contact sports.
Supplementary Material
Key Points Box.
Lateral fluid percussion injury (LFPI) in rats modified beta diversity and selective microbial abundances in fecal samples
LFPI-induced dysbiosis was not associated with post-traumatic epilepsy (PTE)
The risk of PTE was moderately associated with beta diversity and with selective microbial abundances irrespective the time vis-à-vis LFPI
The risk of PTE was moderately associated with baseline fecal content of short-chain fatty acids (SCFA)
PTE risk stratification was improved by integrating baseline fecal microbial abundances and SCFA content
Acknowledgements
The work was supported by the Department of Defense Epilepsy Research Program grant W81XWH-19–1-0430 to AM; by Sudha Neelakantan and Venky Harinarayan Charitable Fund endowment to Epilepsy Research Laboratories to RS and AMM; partially by Veterans Administration grant CDA2 IK2CX001717 to JPJ; partially by National Institutes of Health grant U54NS100064 to ASG and RJS; partially by the Department of Defense Epilepsy Research Program grant W81XWH-18–1-0612 to ASG and AM.
Footnotes
Conflicts of interest
None of the authors has any conflict of interest to disclose.
Additional disclosure. ASG is Editor in Chief of Epilepsia Open, an Associate Editor of Neurobiology of Disease and has received royalties from Medlink and Morgan and Claypool publishers. These activities do not constitute conflict of interest with the reference to this article.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.Matharu D, Dhotre D, Balasubramanian N, Pawar N, Sagarkar S, Sakharkar A. Repeated mild traumatic brain injury affects microbial diversity in rat jejunum. J Biosci 2019;44. [PubMed] [Google Scholar]
- 2.Opeyemi OM, Rogers MB, Firek BA, Janesko-Feldman K, Vagni V, Mullett SJ, et al. Sustained Dysbiosis and Decreased Fecal Short-Chain Fatty Acids after Traumatic Brain Injury and Impact on Neurologic Outcome. J Neurotrauma 2021;38:2610–21. 10.1089/neu.2020.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pathare N, Sushilkumar S, Haley L, Jain S, Osier NN. The Impact of Traumatic Brain Injury on Microbiome Composition: A Systematic Review. Biol Res Nurs 2020;22:495–505. 10.1177/1099800420943961. [DOI] [PubMed] [Google Scholar]
- 4.Treangen TJ, Wagner J, Burns MP, Villapol S. Traumatic Brain Injury in Mice Induces Acute Bacterial Dysbiosis Within the Fecal Microbiome. Front Immunol 2018;9:2757. 10.3389/fimmu.2018.02757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban RJ, Pyles RB, Stewart CJ, Ajami N, Randolph KM, Durham WJ, et al. Altered Fecal Microbiome Years after Traumatic Brain Injury. J Neurotrauma 2020;37:1037–51. 10.1089/neu.2019.6688. [DOI] [PubMed] [Google Scholar]
- 6.Waligora-Dupriet AJ, Lafleur S, Charrueau C, Choisy C, Cynober L, Butel MJ, et al. Head injury profoundly affects gut microbiota homeostasis: Results of a pilot study. Nutrition 2018;45:104–07. 10.1016/j.nut.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Zhu CS, Grandhi R, Patterson TT, Nicholson SE. A Review of Traumatic Brain Injury and the Gut Microbiome: Insights into Novel Mechanisms of Secondary Brain Injury and Promising Targets for Neuroprotection. Brain Sci 2018;8. 10.3390/brainsci8060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You W, Zhu Y, Wei A, Du J, Wang Y, Zheng P, et al. Traumatic Brain Injury Induces Gastrointestinal Dysfunction and Dysbiosis of Gut Microbiota Accompanied by Alterations of Bile Acid Profile. J Neurotrauma 2021. 10.1089/neu.2020.7526. [DOI] [PubMed] [Google Scholar]
- 9.Hanscom M, Loane DJ, Shea-Donohue T. Brain-gut axis dysfunction in the pathogenesis of traumatic brain injury. J Clin Invest 2021;131. 10.1172/JCI143777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundman MH, Chen NK, Subbian V, Chou YH. The bidirectional gut-brain-microbiota axis as a potential nexus between traumatic brain injury, inflammation, and disease. Brain Behav Immun 2017;66:31–44. 10.1016/j.bbi.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Celorrio M, Abellanas MA, Rhodes J, Goodwin V, Moritz J, Vadivelu S, et al. Gut microbial dysbiosis after traumatic brain injury modulates the immune response and impairs neurogenesis. Acta Neuropathol Commun 2021;9:40. 10.1186/s40478-021-01137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George AK, Behera J, Homme RP, Tyagi N, Tyagi SC, Singh M. Rebuilding Microbiome for Mitigating Traumatic Brain Injury: Importance of Restructuring the Gut-Microbiome-Brain Axis. Mol Neurobiol 2021;58:3614–27. 10.1007/s12035-021-02357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jameson KG, Olson CA, Kazmi SA, Hsiao EY. Toward Understanding Microbiome-Neuronal Signaling. Mol Cell 2020;78:577–83. 10.1016/j.molcel.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Maiuolo J, Gliozzi M, Musolino V, Carresi C, Scarano F, Nucera S, et al. The Contribution of Gut Microbiota-Brain Axis in the Development of Brain Disorders. Front Neurosci 2021;15:616883. 10.3389/fnins.2021.616883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne) 2020;11:25. 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin CR, Osadchiy V, Kalani A, Mayer EA. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol 2018;6:133–48. 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol 2019;16:461–78. 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 18.Verellen RM, Cavazos JE. Post-traumatic epilepsy: an overview. Therapy 2010;7:527–31. 10.2217/THY.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum GR, Olson CA, Hsiao EY. Emerging roles for the intestinal microbiome in epilepsy. Neurobiol Dis 2020;135:104576. 10.1016/j.nbd.2019.104576. [DOI] [PubMed] [Google Scholar]
- 20.Peng A, Qiu X, Lai W, Li W, Zhang L, Zhu X, et al. Altered composition of the gut microbiome in patients with drug-resistant epilepsy. Epilepsy Res 2018;147:102–07. 10.1016/j.eplepsyres.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Lindefeldt M, Eng A, Darban H, Bjerkner A, Zetterstrom CK, Allander T, et al. The ketogenic diet influences taxonomic and functional composition of the gut microbiota in children with severe epilepsy. NPJ Biofilms Microbiomes 2019;5:5. 10.1038/s41522-018-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citraro R, Lembo F, De Caro C, Tallarico M, Coretti L, Iannone LF, et al. First evidence of altered microbiota and intestinal damage and their link to absence epilepsy in a genetic animal model, the WAG/Rij rat. Epilepsia 2021;62:529–41. 10.1111/epi.16813. [DOI] [PubMed] [Google Scholar]
- 23.Russo E The gut microbiota as a biomarker in epilepsy. Neurobiol Dis 2021;163:105598. 10.1016/j.nbd.2021.105598. [DOI] [PubMed] [Google Scholar]
- 24.Darch H, McCafferty CP. Gut microbiome effects on neuronal excitability & activity: Implications for epilepsy. Neurobiol Dis 2022;165:105629. 10.1016/j.nbd.2022.105629. [DOI] [PubMed] [Google Scholar]
- 25.Medel-Matus JS, Shin D, Dorfman E, Sankar R, Mazarati A. Facilitation of kindling epileptogenesis by chronic stress may be mediated by intestinal microbiome. Epilepsia Open 2018;3:290–94. 10.1002/epi4.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mengoni F, Salari V, Kosenkova I, Tsenov G, Donadelli M, Malerba G, et al. Gut microbiota modulates seizure susceptibility. Epilepsia 2021;62:e153–e57. 10.1111/epi.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 2018;174:497. 10.1016/j.cell.2018.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery TL, Kunstner A, Kennedy JJ, Fang Q, Asarian L, Culp-Hill R, et al. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc Natl Acad Sci U S A 2020;117:27516–27. 10.1073/pnas.2002817117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 2013;36:305–12. 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Heinzel S, Aho VTE, Suenkel U, von Thaler AK, Schulte C, Deuschle C, et al. Gut Microbiome Signatures of Risk and Prodromal Markers of Parkinson Disease. Ann Neurol 2021;90:E1–E12. 10.1002/ana.26128. [DOI] [PubMed] [Google Scholar]
- 31.Mazarati A, Medel-Matus JS, Shin D, Jacobs JP, Sankar R. Disruption of intestinal barrier and endotoxemia after traumatic brain injury: Implications for post-traumatic epilepsy. Epilepsia 2021;62:1472–81. 10.1111/epi.16909. [DOI] [PubMed] [Google Scholar]
- 32.Reid AY, Bragin A, Giza CC, Staba RJ, Engel J Jr. The progression of electrophysiologic abnormalities during epileptogenesis after experimental traumatic brain injury. Epilepsia 2016;57:1558–67. 10.1111/epi.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandel S, Gupta SK, Medhi B. Epileptogenesis following experimentally induced traumatic brain injury - a systematic review. Rev Neurosci 2016;27:329–46. 10.1515/revneuro-2015-0050. [DOI] [PubMed] [Google Scholar]
- 34.Kabadi SV, Hilton GD, Stoica BA, Zapple DN, Faden AI. Fluid-percussion-induced traumatic brain injury model in rats. Nat Protoc 2010;5:1552–63. 10.1038/nprot.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono T, Wagenaar J, Giorgi FS, Fabera P, Hanaya R, Jefferys J, et al. A companion to the preclinical common data elements and case report forms for rodent EEG studies. A report of the TASK3 EEG Working Group of the ILAE/AES Joint Translational Task Force. Epilepsia Open 2018;3:90–103. 10.1002/epi4.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs JP, Lin L, Goudarzi M, Ruegger P, McGovern DP, Fornace AJ Jr., et al. Microbial, metabolomic, and immunologic dynamics in a relapsing genetic mouse model of colitis induced by T-synthase deficiency. Gut Microbes 2017;8:1–16. 10.1080/19490976.2016.1257469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 2017;11:2639–43. 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. 10.1371/journal.pone.0061217.3632530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McArdle BH, Anderson MJ. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001;82:290–97. 10.1890/0012-9658(2001)082[0290:fmmtcd]2.0.co;2. [DOI] [Google Scholar]
- 41.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable Association Discovery in Population-scale Meta-omics Studies. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100:9440–5. 10.1073/pnas.1530509100.170937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breiman L Random Forests. Machine Learning 2001;45:5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 45.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma EL, Smith AD, Desai N, Cheung L, Hanscom M, Stoica BA, et al. Bidirectional brain-gut interactions and chronic pathological changes after traumatic brain injury in mice. Brain Behav Immun 2017;66:56–69. 10.1016/j.bbi.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angoa-Perez M, Zagorac B, Anneken JH, Briggs DI, Winters AD, Greenberg JM, et al. Repetitive, mild traumatic brain injury results in a progressive white matter pathology, cognitive deterioration, and a transient gut microbiota dysbiosis. Sci Rep 2020;10:8949. 10.1038/s41598-020-65972-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson SE, Watts LT, Burmeister DM, Merrill D, Scroggins S, Zou Y, et al. Moderate Traumatic Brain Injury Alters the Gastrointestinal Microbiome in a Time-Dependent Manner. Shock 2019;52:240–48. 10.1097/SHK.0000000000001211. [DOI] [PubMed] [Google Scholar]
- 49.Cotter D, Kelso A, Neligan A. Genetic biomarkers of posttraumatic epilepsy: A systematic review. Seizure 2017;46:53–58. 10.1016/j.seizure.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Rios-Covian D, Gonzalez S, Nogacka AM, Arboleya S, Salazar N, Gueimonde M, et al. An Overview on Fecal Branched Short-Chain Fatty Acids Along Human Life and as Related With Body Mass Index: Associated Dietary and Anthropometric Factors. Front Microbiol 2020;11:973. 10.3389/fmicb.2020.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Sun J, Du J, Wang F, Fang R, Yu C, et al. Clostridium butyricum exerts a neuroprotective effect in a mouse model of traumatic brain injury via the gut-brain axis. Neurogastroenterol Motil 2018;30:e13260. 10.1111/nmo.13260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


