Abstract
Background
Nonalcoholic fatty liver disease (NAFLD) is a metabolic disease, the incidence of which increases annually. Shugan Xiaozhi (SGXZ) decoction, a composite traditional Chinese medicinal prescription, has been demonstrated to exert a therapeutic effect on NAFLD. In this study, the potential bioactive ingredients and mechanism of SGXZ decoction against NAFLD were explored via network pharmacology, molecular docking, and molecular dynamics simulation.
Methods
Compounds in SGXZ decoction were identified and collected from the literature, and the corresponding targets were predicted through the Similarity Ensemble Approach database. Potential targets related to NAFLD were searched on DisGeNET and GeneCards databases. The compound–target–disease and protein-protein interaction (PPI) networks were constructed to recognize key compounds and targets. Functional enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed on the targets. Molecular docking was used to further screen the potent active compounds in SGXZ. Finally, molecular dynamics (MD) simulation was applied to verify and validate the binding between the most potent compound and targets.
Results
A total of 31 active compounds and 220 corresponding targets in SGXZ decoction were collected. Moreover, 1,544 targets of NAFLD were obtained, of which 78 targets intersected with the targets of SGXZ decoction. Key compounds and targets were recognized through the compound–target–disease and PPI network. Multiple biological pathways were annotated, including PI3K-Akt, MAPK, insulin resistance, HIF-1, and tryptophan metabolism. Molecular docking showed that gallic acid, chlorogenic acid and isochlorogenic acid A could combine with the key targets. Molecular dynamics simulations suggested that isochlorogenic acid A might potentially bind directly with RELA, IL-6, VEGFA, and MMP9 in the regulation of PI3K–Akt signaling pathway.
Conclusion
This study investigated the active substances and key targets of SGXZ decoction in the regulation of multiple-pathways based on network pharmacology and computational approaches, providing a theoretical basis for further pharmacological research into the potential mechanism of SGXZ in NAFLD.
Keywords: Shugan Xiaozhi decoction, Nonalcoholic fatty liver disease (NAFLD), Network pharmacology, Molecular docking, Molecular dynamics simulation, Signaling pathway
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a progressive condition ranging from simple steatosis (NAFL) to nonalcoholic steatohepatitis (NASH), hepatic fibrosis, cirrhosis, and even hepatocellular carcinoma (HCC) (Challa et al., 2019; Raza et al., 2021). NAFLD is a metabolic disease representing the hepatic manifestation of a systemic metabolic disorder (Tilg & Effenberger, 2020) and has been demonstrated to be associated with obesity-related disorders and diabetes (Liu et al., 2020c; Stefan, Häring & Cusi, 2019). With the epidemic of obesity and metabolic-related comorbidities, the global incidence of NAFLD is estimated to be 25% and continues to rise (Mundi et al., 2020). NAFLD is one of the leading courses of chronic hepatic disease, affecting approximately 1.7 billion individuals worldwide (Zhou et al., 2020a). Moreover, NAFLD is one of the most common indications for liver transplantation and NAFLD-related HCC, which can be induced by steatosis-related lipotoxicity, is now the fastest growing reason for liver transplantation in the United States (Younossi et al., 2019; Younossi et al., 2016; Ioannou, 2021). Lifestyle modification including a healthy diet and increased physical activity, is recommended as the first-line treatment in NAFLD management (Jeznach-Steinhagen et al., 2019). Nevertheless, its effectiveness is limited in NAFLD patients due to their low readiness to change lifestyle, especially in terms of increasing physical activity (Centis et al., 2013). Another challenge for the lifestyle intervention has been the occurrence of weight regain (Kantartzis et al., 2009; Wadden et al., 2014).
At present, there are no US Food and Drug Administration- or European Medicines Agency–approved pharmacological therapies for the treatment of NAFLD (Chapman & Lynch, 2020; Cusi et al., 2022; Konerman, Jones & Harrison, 2018). Insulin sensitizers (rosiglitazone, pioglitazone and metformin), antioxidants (vitamin E), anti-inflammatory, and lipid-lowering drugs (atorvastatin and simvastatin) have been used for the treatment of NAFLD (Ahsan et al., 2020; Takahashi et al., 2015). However, the usage of rosiglitazone could increase the risk of cardiovascular events (Nissen & Wolski, 2007), while the long-term use of vitamin E (Klein et al., 2011; Miller 3rd et al., 2005) has been reported to increase the risk of prostate cancer and might increase all-cause mortality. Moreover, adverse outcomes of pioglitazone (Liao et al., 2017; Lincoff et al., 2007), atorvastatin (Pal et al., 2015) and simvastatin (Abdoli, Azarmi & Eghbal, 2014) including weight gain, edema, heart failure, cytotoxicity, and hepatotoxicity, should not be neglected. In recent years, dual peroxisome proliferator-activated receptor (PPAR) α/ γ agonists have appealed to global attention as promising new treatment options for metabolic syndrome, including diabetic complications and NAFLD (Cheng et al., 2019). Saroglitazar is a novel dual PPAR α/ γ agonist acting as a regulator of lipids and glucose metabolism (Sarkar et al., 2021). India approved saroglitazar in 2020 for the treatment of type-2 diabetes mellitus and NASH (Sarkar et al., 2021). However, a phase 2 trial conducted in the United States indicated that the role of saroglitazar in a particular subset of patients with elevated alanine aminotransferase without histological endpoints cannot be recommended (Kumar, Kulkarni & Jagdish, 2021; Shuja, Eqbal & Rehman, 2021). Therefore, there is an urgent need to develop pharmacological strategies for the treatment of NAFLD.
The complex pathophysiological mechanism of NAFLD reduces the efficacy of the application of a single agent. Traditional Chinese medicine (TCM) has been commonly used to treat hepatic disease and metabolic disorders–related to obesity and type 2 diabetes mellitus (Fu et al., 2021; Li, Zhang & Li, 2020a; Zhang et al., 2021). The therapeutic effect of TCM on NAFLD mainly involves holistic regulations including lipid metabolism modulation, anti-oxidative stress, anti-inflammation, and gut microbiota modulation (Dai et al., 2021; Liang et al., 2019; Ma, Zhou & Li, 2017). Shugan Xiaozhi (SGXZ) decoction is a composite traditional Chinese prescription composed of 12 herbs, including pleuri Radix, Paeoniae radix alba (stir-baked), Aurantii Fructus Immaturus, Glycyrrhizae Radixet Rhizoma, Artemisiae Scopariae Herba, Gardeniae Fructus, Poria, Alismatis Rhizoma, Crataegi Fructus, Cassiae Semen, Nelumbinis Folium, and Pumex in a ratio of 2: 1: 3: 1: 6: 2: 4: 6: 6: 6: 6: 6. Previous clinical studies have demonstrated that SGXZ could partially protect and restore the liver functions in NASH patients, which was associated with a significant reduction in aminotransferases, total cholesterol and triacylglycerol, and reparation of the intestinal mucosal barrier (Tang et al., 2018; Zhai et al., 2014). Mechanically, in a rat model, SGXZ decoction exerts the hepatoprotective effect through regulating fatty acid β-oxidation, relieving intestinal microecological disorder, and repairing the intestinal mucosal barrier (Xing et al., 2016; Zhai et al., 2021). However, the therapeutic mechanisms of SGXZ decoction in the treatment of NAFLD have not been fully clarified, and the active ingredients and key targets of SGXZ remain to be further investigated.
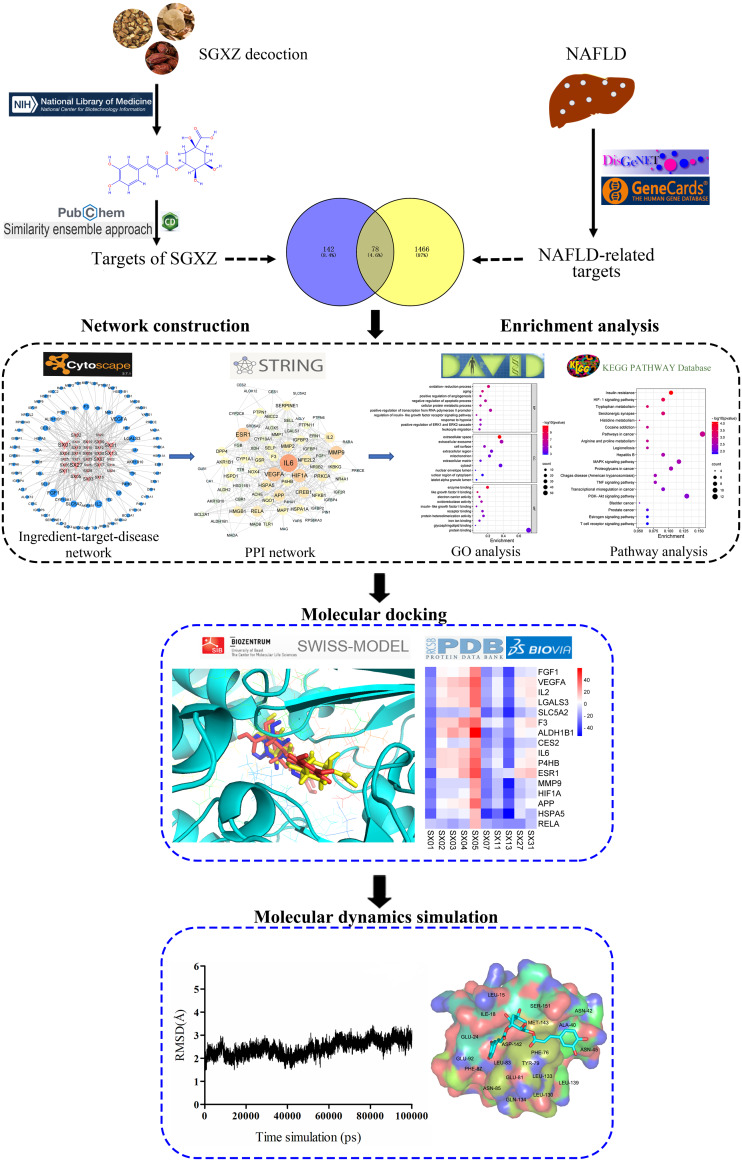
Based on the integration of system biology, bioinformatics and pharmacology (Lai et al., 2020; Liu & Sun, 2012; Zhang et al., 2019), network pharmacology is currently used to explore the potential pharmacological effects and underlying mechanisms of a drug on a disease (Guo et al., 2019; Zhang et al., 2020). Network pharmacology concentrates on the elucidation of complex biological relationships among the drugs, targets, pathways and diseases from a systemic and holistic perspective (Wang et al., 2021). The wholeness, relevance and systematic nature of network pharmacology are in line with the concept and treatment theory of TCM, providing a novel method and powerful tool to decipher the therapeutic mechanisms of TCM (Zhou et al., 2020b). Molecular docking is a computational method based on the analysis of the binding pose and affinity between small molecule and macromolecular target, which is widely utilized to predict and identify potentially active compounds (Álvarez-Carretero et al., 2018; Jiao et al., 2021). In addition, molecular dynamics (MD) simulations have been used widely to provide a full consideration of the flexibility of protein targets and obtain accurate binding modes and stability of compounds through Newtonian mechanics (Liu et al., 2018). In our study, network pharmacology combined with molecular docking and MD simulations was applied to explore the potential pharmacological and molecular mechanisms involved in the treatment of NAFLD by SGXZ decoction. The overall flowchart of this study is described in Fig. 1.
Figure 1. Schematic diagram of the whole study design.
Material and Methods
Collection of compounds and the putative targets of SGXZ decoction
The active compounds in SGXZ decoction were identified and collected from the literature. Then, 2D or 3D conformations of the compounds were downloaded either from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) or through sketching in ChemDraw software. Moreover, their simplified molecular input line entry system (SMILS) numbers (Table S1) were also acquired from PubChem or were converted by ChemDraw. To predict the putative gene target of SGXZ decoction, the SMILES of each chemical compound was submitted to Similarity Ensemble Approach (SEA) database (https://sea.bkslab.org/), which relates proteins based on the set-wise chemical similarity among their ligands (Keiser et al., 2007). The predicted protein targets of SGXZ decoction are listed in Table S2.
Identification of potential targets of NAFLD
“Nonalcoholic Fatty Liver Disease” or “Non-alcoholic Fatty Liver Disease” were used as the keywords to predict the potential NAFLD-related gene targets using GeneCards (https://www.genecards.org/) and DisGeNET (https://www.disgenet.org/) disease database. DisGeNET is a discovery platform containing one of the largest publicly available collections of genes associated with human diseases (Piñero et al., 2020). Based on all annotated and predicted human genes, GeneCards provides comprehensive, user-friendly information via a searchable, integrative database (Stelzer et al., 2016). After removing the repetitive targets from the two databases, the NAFLD-related protein targets were retained for further study. The detailed information on the targets is provided in Table S3.
Construction of compound-target-disease network
SGXZ decoction–target network was first constructed based on the compounds and their predicted targets of SGXZ using Cytoscape software (Version 3.7.1). Venny diagram 2.1 version (https://bioinfogp.cnb.csic.es/tools/venny/) was used to intersect the protein targets between the predicted targets of SGXZ decoction and the potential targets of NAFLD. Then, the compound–target–disease network was established based on the intersected common targets and their corresponding compounds. The degree value of nodes in the compound–target–disease network was analyzed with the NetworkAnalyzer plug-in in Cytoscape software. The degree indicates the total number of nodes connected to this node in the network, and nodes whose degree value is higher play a more essential regulatory role in the network. The compounds and targets that contributed significantly to the construction of the compound–target–disease network were recognized and selected as key active compounds and targets according to their higher degree value compared with the average degree value.
Construction of protein–protein interaction network
PPI network can elucidate the interaction relationship between targets, which further helps to identify the key nodes and targets in the network. The intersected common targets were submitted to the STRING database (https://string-db.org/; version 11.5) (Szklarczyk et al., 2019). The species was limited to “Homo sapiens”, and the confidence of interaction score was set at ≥ 0.400 as default, which ensured a reasonable interaction between targets. Results with tabular text output (TSV) format of the PPI network in STRING were exported and then visualized through Cytoscape software. The nodes with the highest degree value were considered the key targets that mediated the interaction of the network and participated in the treatment of NAFLD by SGXZ decoction.
Enrichment analysis
Functional annotation of the common targets in the PPI network was analyzed through the “Database for Annotation, Visualization, and Integrated Discovery” (DAVID) database (https://david.ncifcrf.gov/) to identify enriched biological themes of Gene Ontology (GO) and visualize genes on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps. DAVID can provide a comprehensive set of functional annotation tools for investigators to understand the biological meaning behind a large list of genes and is widely used in bioinformatics (Huang Da, Sherman & Lempicki, 2009a; Huang Da, Sherman & Lempicki, 2009b). The gene list of the common target proteins was uploaded to DAVID, and the species was limited to “Homo sapiens”. The functional annotation of biological processes (BPs), molecular functions (MCs), cellular components (CCs), and KEGG pathways with a P value <0.05 was then performed for further analysis. Accordingly, the target–pathway network was subsequently constructed using Cytoscape software.
Homology modeling and molecular docking
The 3D conformational SDF format of the selected key chemical compounds was downloaded from PubChem. The crystal structures of the key protein targets were retrieved in the RCSB Protein Data Bank database (PDB, https://www.rcsb.org/). For targets without released crystal structures, homology modeling was utilized to establish the structure based on the protein template. The construction was completed on the SWISS-MODEL website (https://swissmodel.expasy.org/), which is a fully automated protein structure homology–modeling server (Waterhouse et al., 2018). Then, the quality of the model structures was evaluated via the “Ramachandran plot” on the SAVES v6.0 website (https://saves.mbi.ucla.edu/). The target protein with neither crystal structure nor the template was excluded in molecular docking. Before docking, the compounds were charged with CHARMm force field and minimized for optimization in Discovery Studio 2020 (DS) software. The target proteins were also loaded in DS, and water and other redundant chains were first deleted. Then, excess structures of the protein were removed, incomplete amino acid residues were supplemented, and hydrogens were added to optimize the conformation. CDOCKER module in DS was applied to calculate the binding energy between the key active ingredients and key target proteins. CDOCKER is a grid-based molecular docking method based on CHARMm force field, which can produce high-precision docking results. For docking parameters of CDOCKER, the size of the docking box was set to 20 Å× 20 Å× 20 Å, “Top Hits” was set at 10; and “Pose Cluster Radius” was set at 0.5. Moreover, the docking site of each target protein was obtained from previous research or the active cavities detected in DS (Table S4). Only one top hit with the best docking pose of each complex was reported and saved for further analysis.
MD simulations
MD simulations using the Amber16 software package were performed on the complexes of the most potentially active compound and target proteins. Gaussian 16 and antechamber program were employed to prepare the force-filed parameters for the compound, and the ff14sb filed force parameters were applied for the proteins. The compound and protein were then loaded into the leap module, and the system was neutralized by automatically adding hydrogen atoms and antagonist ions. All the systems were immersed in the TIP3P water model with periodic boundary conditions and were first minimized through 2,000 steps of the steepest descent and 2,000 steps of conjugate gradients with the constraining of all atoms of protein and compound. Next, only the heavy atoms of the proteins were confined to minimize the water and the side chains by steepest descent minimization of 5,000 steps followed by a conjugate gradient minimization of 5,000 steps. Finally, the entire system was set with no constraints for the optimization with the same step as the last stage. The system was then heated slowly to 310 K and was balanced under nvt ensemble for 20 ps. Then, 100 ns simulations were performed, and the trajectory was saved every 500 ps. The CPPTRAJ module was exploited for the root mean square deviation (RMSD) analysis and the calculation of binding free energy of active compound and protein was carried out using molecular mechanics/generalized Born surface area (MM/GBSA) method by the MMPBSA.py program in AMBER 16.
Results
Active compounds of SGXZ decoction and the corresponding targets
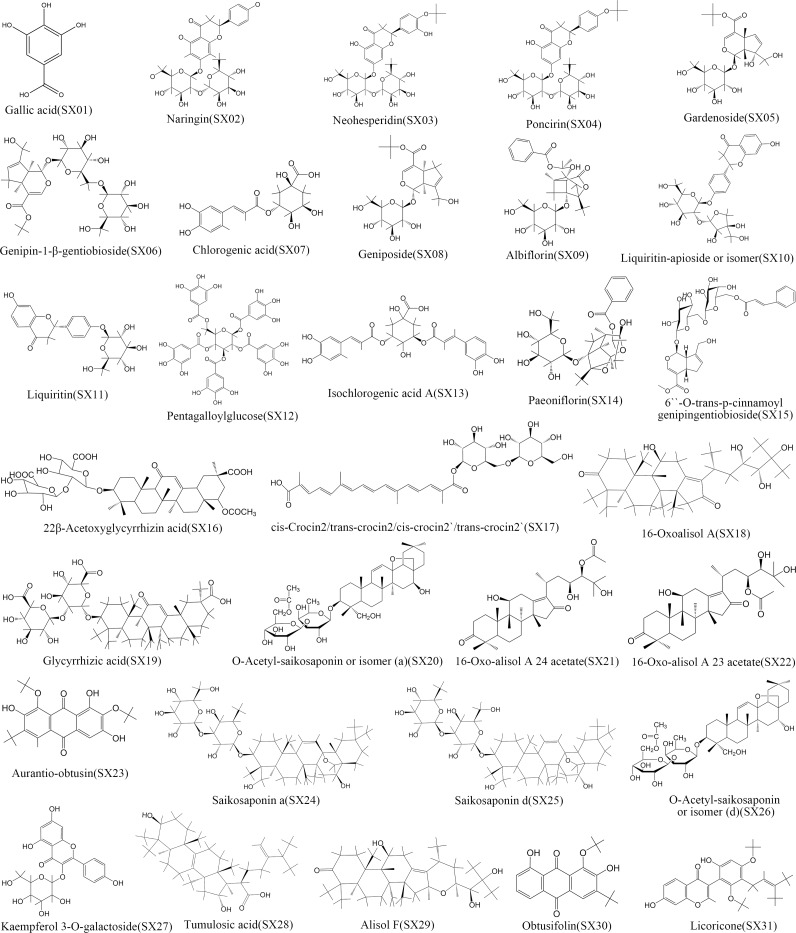
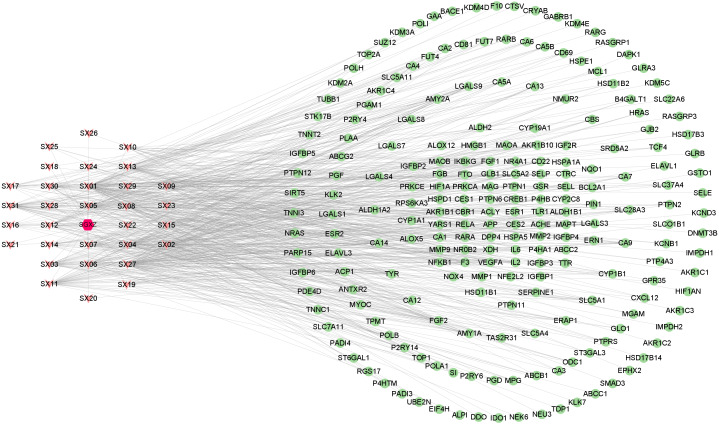
A total of 31 active compounds of SGXZ decoction (SX01 to SX31) were identified and collected from the previous study (Xing et al., 2016) with 2D chemical structures displayed in Fig. 2. Totally, 220 targets of SGXZ decoction were screened after removing duplicates. The interaction of compounds in SGXZ decoction with their corresponding targets was constructed (Fig. 3). The interaction network comprised 252 nodes and 686 edges with 31 compounds and 220 corresponding targets, revealing the multitarget effect of SGXZ decoction.
Figure 2. 2D structures of the 31 identified compounds in SGXZ decoction.
Figure 3. SGXZ decoction–targets interaction network.
Red hexagon, pink triangle, and green circle represent SGXZ decoction, compounds, and targets, respectively.
Potential targets of NAFLD and the compound–target–disease interaction
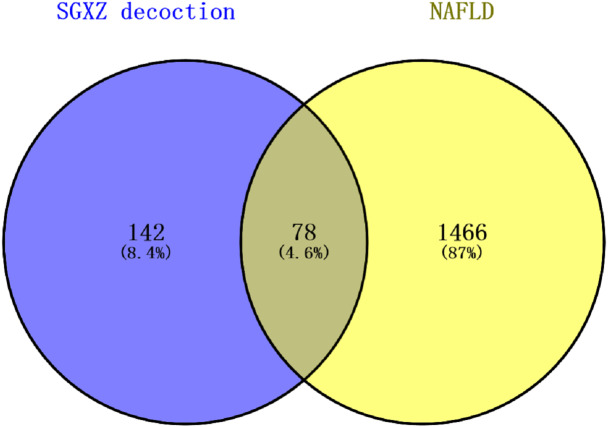
Potential targets of NAFLD were independently screened in DisGeNET and GeneCards databases. After the overlapped targets were deleted, 1544 potential protein targets of NAFLD from the two databases were retained. The Venny diagram showed that there were 78 common targets between SGXZ and NAFLD, which represented the potential targets of SGXZ in the treatment of NAFLD (Fig. 4).
Figure 4. Overlapped targets between SGXZ decoction and NAFLD.
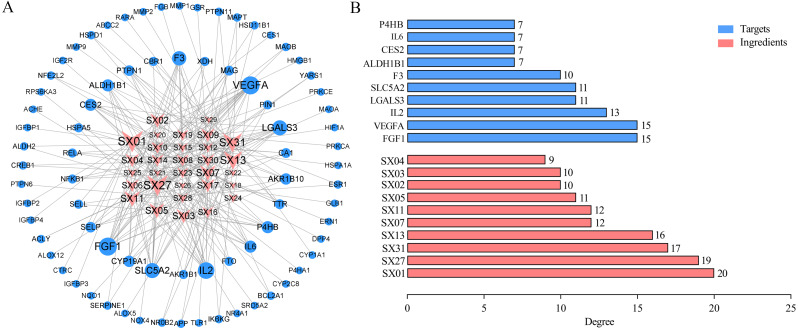
To further examine the direct regulatory effect of SGXZ decoction in treating NAFLD, the network of compounds with common intersection targets was constructed (Fig. 5A), which consisted of 109 nodes (31 compounds and 78 common targets) and 232 edges, suggesting multiple components involved with multiple targets of SGXZ decoction in the treatment of NAFLD. In addition, the top 10 compounds and common protein targets were obtained (Fig. 5B), and the detailed information is listed in Tables 1 and 2. SX01 (gallic acid) possessed the highest degree value (degree = 20), followed by SX27 (kaempferol-3-O-galactoside, degree = 19), SX31 (licoricone, degree = 17), SX13 (isochlorogenic acid A, degree = 16), SX07 (chlorogenic acid, degree = 12), SX11 (liquiritin, degree = 12), SX05 (gardenoside, degree = 11), SX02 (naringin, degree = 10), SX03 (neohesperidin, degree = 10), and SX04 (poncirin, degree = 9). These active compounds were recognized in SGXZ decoction to potentially exert the pharmacologic effect on NAFLD. Moreover, as listed in Table 2, the top 10 targets with the highest degree value included FGF1 (degree = 15), VEGFA (degree = 15), IL-2 (degree = 13), LGALS3 (degree = 11), SLC5A2 (degree = 11), F3 (degree = 10), ALDH1B1 (degree = 7), CES2 (degree = 7), IL-6 (degree = 7), and P4HB (degree = 7), which could be potentially acted on by the 10 key active compounds in SGXZ decoction.
Figure 5. Compound–target–disease interaction network.
(A) Interaction network of the compound–target–disease. Size of nodes corresponds to the value of the degree. SGXZ decoction chemical compounds and targets are colored in pink and blue, respectively; (B) Top 10 putative compounds and targets with the highest degree value in the compound–target–disease network.
Table 1. Key active compounds of SGXZ decoction in treatment of NAFLD.
| Number | Ingredient | Molecular formula | Pubchem_CID | Degree |
|---|---|---|---|---|
| SX01 | Gallic acid | C7H6O5 | 370 | 20 |
| SX02 | Naringin | C27H32O14 | 442428 | 10 |
| SX03 | Neohesperidin | C28H34O15 | 442439 | 10 |
| SX04 | Poncirin | C28H34O14 | 442456 | 9 |
| SX05 | Gardenoside | C17H24O11 | 24721095 | 11 |
| SX07 | Chlorogenic acid | C16H18O9 | 1794427 | 12 |
| SX11 | Liquiritin | C21H22O9 | 503737 | 12 |
| SX13 | Isochlorogenic acid A | C25H24O12 | 6474310 | 16 |
| SX27 | Kaempferol-3-O-galactoside | C21H20O11 | 5282149 | 19 |
| SX31 | Licoricone | C22H22O6 | 5319013 | 17 |
Table 2. Key targets of SGXZ decoction in the treatment of NAFLD.
| UniprotKB | Target | Protein name | Degree |
|---|---|---|---|
| P05230 | FGF1 | Fibroblast growth factor 1 | 15 |
| P15692 | VEGFA | Vascular endothelial growth factor A | 15 |
| P60568 | IL-2 | Interleukin-2 | 13 |
| P17931 | LGALS3 | Galectin-3 | 11 |
| P31639 | SLC5A2 | Sodium/glucose cotransporter 2 | 11 |
| P13726 | F3 | Tissue factor | 10 |
| P30837 | ALDH1B1 | Aldehyde dehydrogenase X, mitochondrial | 7 |
| O00748 | CES2 | Cocaine esterase | 7 |
| P05231 | IL-6 | Interleukin-6 | 7 |
| P07237 | P4HB | Protein disulfide-isomerase | 7 |
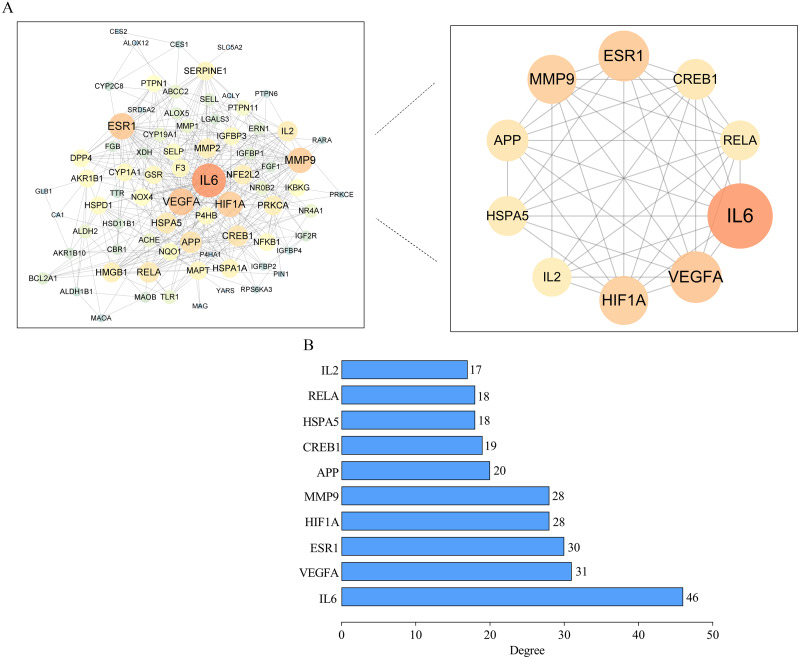
Outcome of the PPI network
The PPI network was constructed from the common 78 targets (Fig. 6). Due to the lack of any interactions with other targets, chymotrypsin-C (CTRC) and alpha-ketoglutarate-dependent dioxygenase (FTO) were removed from the network. Therefore, the network contained 76 nodes and 376 edges, with an average degree value of 9.64. The target proteins with higher degree value than average degree value were IL-6, VEGFA, estrogen receptor (ESR1), hypoxia-inducible factor 1-alpha (HIF1A), matrix metalloproteinase-9 (MMP9), amyloid-beta precursor protein (APP), cAMP-responsive element binding protein 1 (CREB1), heat-shock protein family A member 5 (HSPA5), transcription factor p65 (RELA), and IL-2. The 10 targets were indispensable for constructing the PPI network. IL-6 is a proinflammatory factors and is associated with metabolic disorders (Giraldez et al., 2021). Increased IL-6 level promotes hepatic insulin resistance and impairs lipid metabolism (Akbari & Hassan-Zadeh, 2018; Singh et al., 2021). Similarly, upregulated IL-2 potentially leads to increased insulin resistance and several other metabolic inflammatory markers in the obese population (Kochumon et al., 2020). VEGFA is a major proangiogenic cytokine that regulates angiogenesis. Increased VEGFA accelerates angiogenesis and therefore drives hepatic inflammation and fibrosis in NAFLD (Lefere, Devisscher & Geerts, 2020; Surapaneni, Vishnu Priya & Mallika, 2015). Upregulated HIF1A expression in the liver leads to aggravated steatosis by suppression of fatty acid (FFA) β-oxidation and by induction of FFA uptake and inflammatory factors (Holzner & Murray, 2021; Wu et al., 2019). RELA is a pivotal transcription factor that regulates inflammatory molecules (Gasparini & Feldmann, 2012). FGF1 exerts a protective role in a series of metabolic disorders (Nies et al., 2015). IL-6, IL-2, FGF1 and VEGFA are located at the upstream of the PI3K–Akt pathway and can stimulate the pathway, and RELA can be modulated by the stimulation of PI3K–Akt. MMP9 has a vital role in modulating and degrading gelatins, collagens and other ECM compounds (D’Amico et al., 2010; Lachowski et al., 2019). A decreased level of MMP9 is associated with more advanced fibrosis and serum liver injury indexes in NAFLD patients (Goyale et al., 2021). GRP78 (HSPA5) is a chaperone heat shock protein playing the central role in maintaining ER proteostasis under excessive stress (Pobre, Poet & Hendershot, 2019). LGALS3 is an essential regulator of insulin resistance, fibrosis and inflammation cytokines including TNF- α, IL-6 and IL-1β, and has been shown to participate in glucose intolerance and lipid metabolism disorders (Li et al., 2020b; Yu et al., 2021). CREB1 has been found to promote insulin resistance (Yoon et al., 2021) and ESR1 is associated with adiposity and mitochondrial metabolism (Zhou et al., 2020c). These targets were considered the putative key targets of SGXZ decoction for the treatment of NAFLD.
Figure 6. Protein–protein interaction (PPI) network.
(A) PPI network of potential targets of SGXZ decoction for treating NAFLD. The size of the node with the shade of the color indicates the corresponding degree value; (B) The identified top 10 putative targets with degree value higher than the average according to the PPI network.
GO and KEGG enrichment analysis
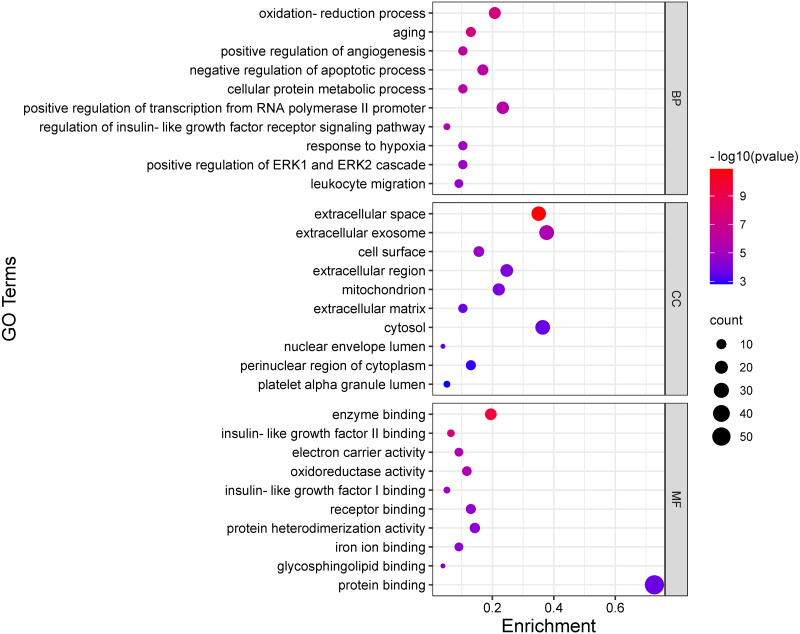
The GO functionally annotated targets in the PPI network involved three main aspects, including molecular function (MF), cellular component (CC), and biological process (BP). In total, 185 GO entries, including 45 of MF, 25 of CC, and 115 of BP, were acquired based on the P value (P < 0.05). As for BP, the targets were mainly concentrated on the oxidation–reduction process, positive regulation of angiogenesis, negative regulation of the apoptotic process, regulation of insulin-like factor receptor signaling pathway, and so on. As for CC, the targets were mainly responsible for the extracellular space, extracellular exosome, cell surface, extracellular region, mitochondrion, extracellular matrix, and so on. As for MF, the intersection targets were distributed in enzyme binding, insulin-like growth factor binding, electron carrier activity, oxidoreductase activity, receptor binding, protein heterodimerization activity, iron ion binding, glycosphingolipid binding, and protein binding. The bubble plot of the most significant enriched GO terms is shown in Fig. 7.
Figure 7. Representative GO enrichment analysis based on the targets in the PPI network.
Bubble plot of the top 10 terms of BP, CC, and MF. The bigger the dot, the more genes are enriched in the term, while the redder color indicates the smaller P value.
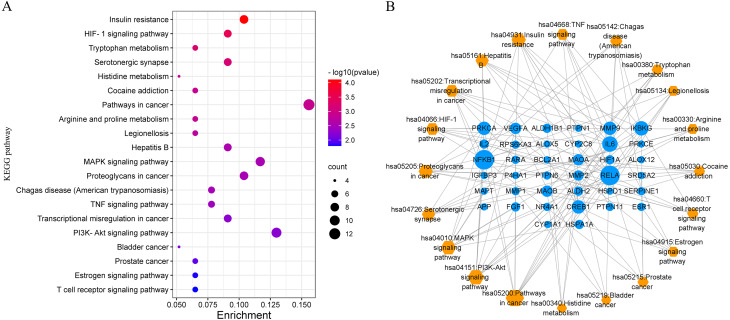
Forty-three annotated pathways were also obtained based on the targets in the PPI network, and the top 20 pathways with the smallest significance value are shown in Fig. 8A and Table 3. The bubble plot of the pathways suggested a concentration of insulin resistance, amino acid metabolism, cancer-related pathways, and inflammation-related pathways including MAPK signaling pathway, HIF-1 signaling pathway, TNF signaling pathway, and PI3K–Akt pathway. Moreover, the common targets also concentrated on the endocrine biological processes such as the estrone signaling pathway and immunological pathway of T cell receptors. To reveal the network interaction of the pathways and the involved targets, the target–pathway network was constructed and analyzed, which consisted of 58 nodes and 130 edges (Fig. 8B), indicating a complex interaction among them. The largest node size was for the pathways in cancer, followed by PI3K–Akt, MAPK signaling pathway, proteoglycan in cancer, and HIF-1 pathway, suggesting their significant role in the treatment of NAFLD.
Figure 8. Results of KEGG pathway enrichment analysis based on the targets in the PPI network.
(A) The bubble plot of the top 20 pathway enrichment analysis; (B) target–pathway interaction network suggests the underlying mechanisms of SGXZ decoction for the treatment of NAFLD. Blue circle nodes indicate the targets and orange diamonds represent the pathways. The gray edges link the interaction between the targets and pathways, and the node size is proportional to the degree value.
Table 3. Annotation of KEGG pathways and the involved potential targets.
| KEGG ID | Term | Count | P value | Targets |
|---|---|---|---|---|
| hsa04931 | Insulin resistance | 8 | 7.75E−05 | RPS6KA3, PTPN1, IL-6, CREB1, PRKCE, PTPN11, RELA, NFKB1 |
| hsa04066 | HIF-1 signaling pathway | 7 | 3.17E−04 | IL-6, SERPINE1, PRKCA, HIF1A, RELA, NFKB1, VEGFA |
| hsa00380 | Tryptophan metabolism | 5 | 5.77E−04 | MAOB, ALDH2, ALDH1B1, MAOA, CYP1A1 |
| hsa04726 | Serotonergic synapse | 7 | 6.91E−04 | APP, CYP2C8, MAOB, MAOA, ALOX5, PRKCA, ALOX12 |
| hsa00340 | Histidine metabolism | 4 | 0.001191 | MAOB, ALDH2, ALDH1B1, MAOA |
| hsa05030 | Cocaine addiction | 5 | 0.001253 | CREB1, MAOB, MAOA, RELA, NFKB1 |
| hsa05200 | Pathways in cancer | 12 | 0.001302 | IL-6, MMP1, MMP2, RARA, PRKCA, IKBKG, FGF1, HIF1A, MMP9, RELA, NFKB1, VEGFA |
| hsa00330 | Arginine and proline metabolism | 5 | 0.001352 | MAOB, P4HA1, ALDH2, ALDH1B1, MAOA |
| hsa05134 | Legionellosis | 5 | 0.001803 | IL-6, RELA, NFKB1, HSPD1, HSPA1A |
| hsa05161 | Hepatitis B | 7 | 0.002742 | IL-6, CREB1, PRKCA, IKBKG, MMP9, RELA, NFKB1 |
| hsa04010 | MAPK signaling pathway | 9 | 0.002986 | RPS6KA3, NR4A1, PRKCA, IKBKG, MAPT, FGF1, RELA, NFKB1, HSPA1A |
| hsa05205 | Proteoglycans in cancer | 8 | 0.003139 | MMP2, PTPN6, PTPN11, PRKCA, HIF1A, ESR1, MMP9, VEGFA |
| hsa05142 | Chagas disease (American trypanosomiasis) | 6 | 0.003296 | IL-6, SERPINE1, IKBKG, RELA, NFKB1, IL-2 |
| hsa04668 | TNF signaling pathway | 6 | 0.003726 | IL-6, CREB1, IKBKG, MMP9, RELA, NFKB1 |
| hsa05202 | Transcriptional misregulation in cancer | 7 | 0.005495 | IL-6, BCL2A1, IGFBP3, RARA, MMP9, RELA, NFKB1 |
| hsa04151 | PI3K-Akt signaling pathway | 10 | 0.005885 | NR4A1, IL-6, CREB1, PRKCA, IKBKG, FGF1, RELA, NFKB1, IL-2, VEGFA |
| hsa05219 | Bladder cancer | 4 | 0.007229 | MMP1, MMP2, MMP9, VEGFA |
| hsa05215 | Prostate cancer | 5 | 0.010389 | CREB1, SRD5A2, IKBKG, RELA, NFKB1 |
| hsa04915 | Estrogen signaling pathway | 5 | 0.015491 | CREB1, MMP2, ESR1, MMP9, HSPA1A |
| hsa04660 | T cell receptor signaling pathway | 5 | 0.016021 | PTPN6, IKBKG, RELA, NFKB1, IL-2 |
Verification of the binding between the key compounds and targets
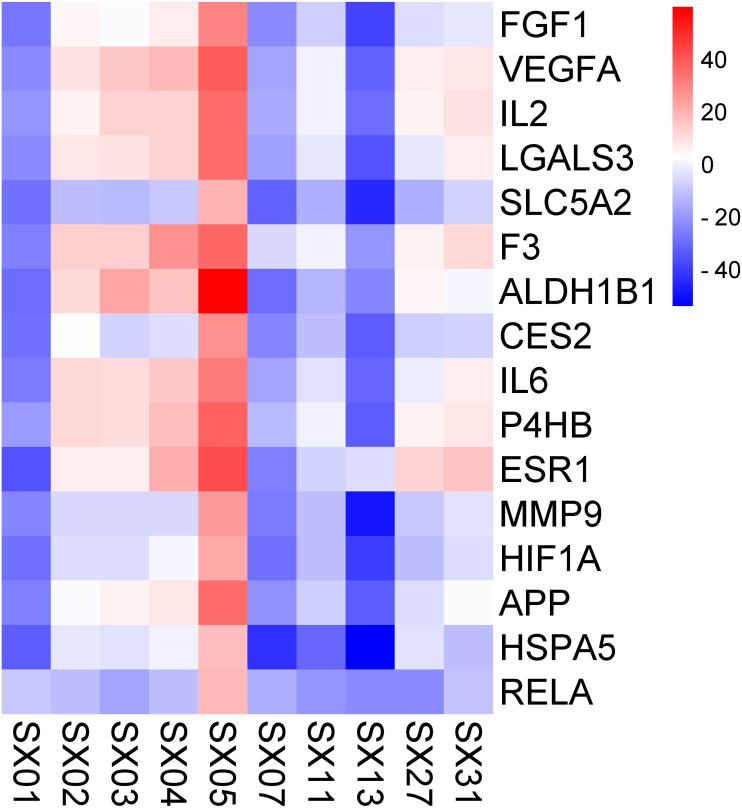
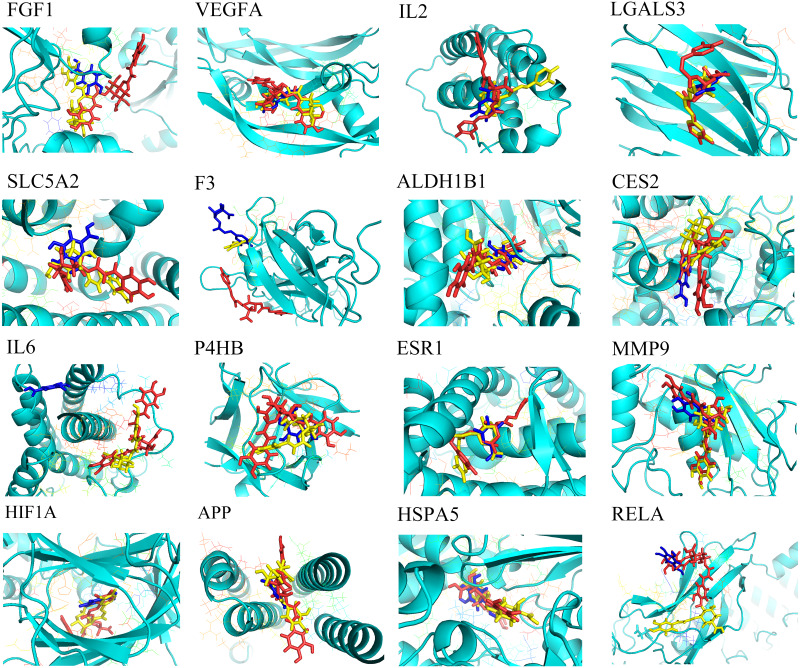
Molecular docking was employed to verify the binding between the key compounds and the key targets, revealing the potential therapeutic effect of SGXZ decoction acting on these targets. Crystal structures of the targets were downloaded from the PDB database, except for SLC5A2, CES2, and ALDH1B1 for their lack of 3D structures. Therefore, homology modeling was used to build the structures of SLC5A2, CES2, and ALDH1B1 (Song et al., 2021; Wang et al., 2019). The structural assessment of ALDH1B1 is provided in Fig. S1, which showed the good quality of the modeled structure. However, the crystal structure of CREB1 could not be obtained either from the PDB database or through the homology modeling since there were no templates of good quality. Besides, three duplicated key targets between the compound–target–disease network and the PPI network were removed. Thus, a total of 16 key targets were docked with 10 key compounds. After docking, 160 pairs of the compound–target complexes were retained according to their best binding affinity, and the heat map of their docking energy is shown in Fig. 9. Moreover, the binding energy of each pair of compound–target complex is provided in Table S5. Binding energy with a negative value indicated that the ligand molecule was able to combine with the receptor target proteins. The lower and more negative binding energy suggested a better binding affinity of active compounds with target proteins. Among the 10 compounds, SX01 (gallic acid), SX07 (chlorogenic acid), and SX13 (isochlorogenic acid A) could combine with all of the targets. The binding energy between the three compounds and targets and the residues that produce hydrogen bonds shared by all three active compounds are listed in Table 4. As shown, SX13 could bind with all of the key targets with the lowest binding energy, followed by SX07 and SX01. The binding location between the best poses of the three compounds and targets is displayed in Fig. 10. The results of molecular docking revealed that SX01, SX07, and especially SX13 might be the potentially active ingredients of SGXZ decoction for treating NAFLD through targeting these proteins.
Figure 9. Heat map of the docking energy (kcal/mol) between the key active ingredients and the key targets.
The bluer the color, the lower the binding energy and the stronger the binding ability.
Table 4. Binding energy between key active compounds and targets (kcal/mol).
| Target proteins | Ingredients | Shared Hydrogen Bond Interactions (Amino acid) | ||
|---|---|---|---|---|
| SX01 | SX07 | SX13 | ||
| FGF1 | −27.07 | −22.06 | −38.21 | LYS112, LYS113, ASN18, LYS128, LYS118 |
| VEGFA | −22.19 | −16.77 | −31.33 | N/A |
| IL-2 | −19.90 | −15.25 | −29.85 | LYS35 |
| LGALS3 | −22.81 | −17.71 | −34.94 | ASN174, GLU184 |
| SLC5A2 | −27.92 | −31.68 | −44.10 | SER77, ASP294, GLN295 |
| F3 | −24.94 | −5.43 | −19.86 | LYS65, LYS46 |
| ALDH1B1 | −29.62 | −28.87 | −24.23 | CYS302, CYS303 |
| CES2 | −27.76 | −23.35 | −32.97 | SER228 |
| IL-6 | −26.37 | −17.42 | −30.38 | N/A |
| P4HB | −19.07 | −12.82 | −33.19 | TRP396 |
| ESR1 | −35.57 | −24.86 | −3.86 | GLU353, LEU346 |
| MMP9 | −23.77 | −25.52 | −48.88 | HIS230, ALA191 |
| HIF1A | −28.66 | −28.22 | −39.94 | THR196, HIS199 |
| APP | −25.31 | −21.32 | −32.41 | LYS447, ASP444, HIS439 |
| HSPA5 | −33.25 | −43.27 | −53.91 | GLY364, SER365, LYS296, GLU293, ARG297 |
| RELA | −9.12 | −15.06 | −23.01 | N/A |
| Average | −25.21 | −21.85 | −32.57 | |
Notes.
none hydrogen bond interaction residues shared by SX01, SX07 and SX13 with the protein.
Figure 10. Combination between SX01, SX07, SX13, and targets.
Target proteins are displayed as a cyan cartoon, while SX01 (gallic acid), SX07 (chlorogenic acid), and SX13 (isochlorogenic acid A) are displayed on sticks and colored in blue, yellow, and pink, respectively.
RMSD analysis and binding free energy
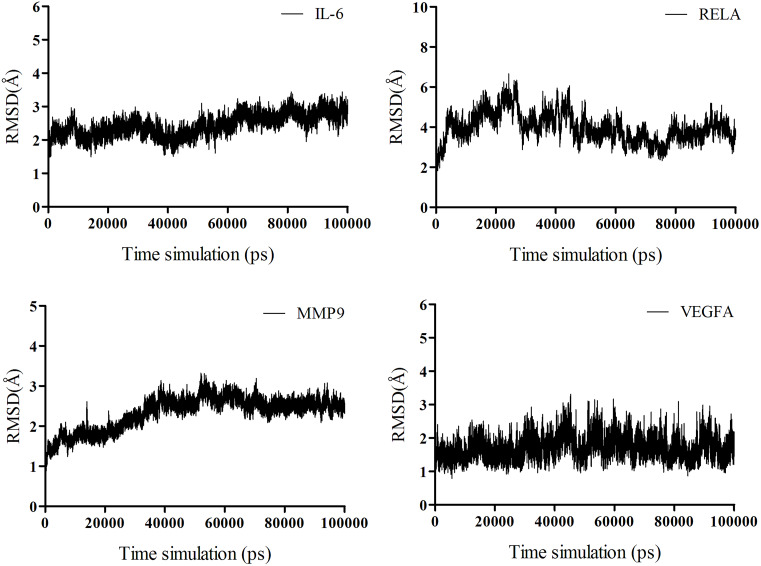
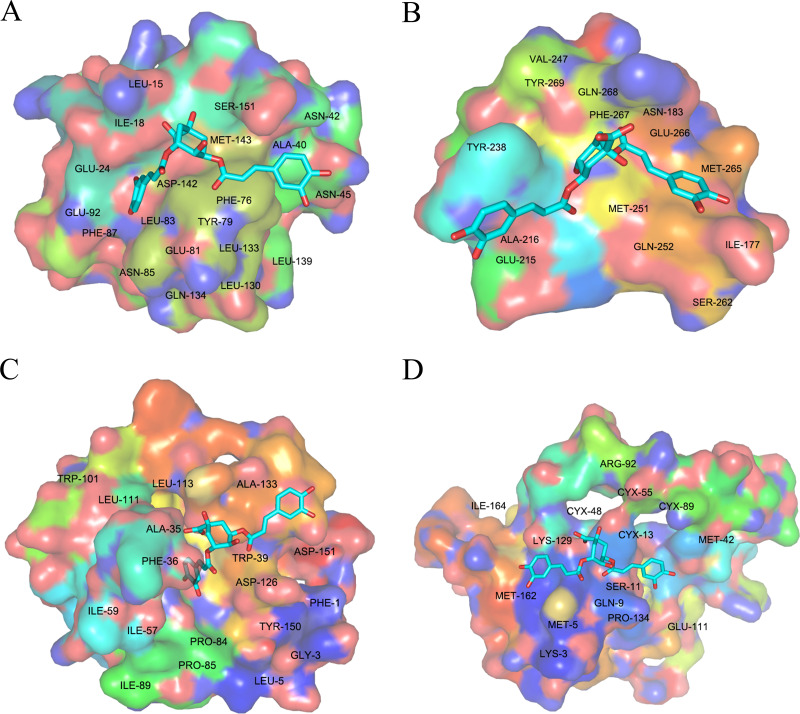
MD simulations were performed to validate the binding ability between the most potentially active compound and the key target proteins after molecular docking. SX13 was selected for MD simulations due to its most negative average docking energy compared to SX01 and SX07, which indicated a putative stronger binding ability of SX13. The proteins targets RELA, IL-6, MMP9, and VEGFA were chosen for their significant role both in the PPI network and the target–pathway interaction network. The equilibrium of all four systems was evaluated by RMSD, which represents the relatively stable combination of protein and compound. As shown in Fig. 11, the complex of VEGFA and SX13 maintained stability throughout the simulation, while the complexes IL-6/SX13 and MMP9/SX13 stabilized after 70 ns. As for the complex RELA/SX13, the conformation showed a slight larger fluctuation in the early stage and achieved the equilibrium at the last 20 ns simulations. As RELA is a macromolecular polymer consisting of more than 500 amino acids, it is reasonable that the complex of RELA and SX13 showed larger fluctuation than the other three complexes. Therefore, the MD trajectories between 80 and 90 ns simulations were selected for the calculations of binding free energy. The binding free energy and its components, including Van der Waals potential, electrostatic interaction, non-polar solvation free energy, and polar solvation free energy, which were calculated by MM/GBSA of the four systems, are listed in Table 5. The MM/GBSA results showed that the average binding free energy between SX13 and RELA, IL-6, MMP9, and VEGFA was −23.06 ± 1.69 kcal/mol, −21.93 ± 1.07 kcal/mol, −21.04 ± 1.31 kcal/mol, and −17.28 ± 1.37 kcal/mol, respectively. The second repeated simulation further strengthened the bind between SX13 and these target proteins (Fig. S2 and Table S6). The results demonstrated that SX13 could potentially combine directly with these key protein targets. The detailed binding of the protein with the compound is displayed in Fig. 12.
Figure 11. RMSD plot of the backbone atoms for isochlorogenic acid A with IL-6, RELA, MMP9, and VEGFA.
Table 5. Binding free energy and energy components calculated by MM/GBSA (kcal/mol).
| RELA | IL-6 | VEGFA | MMP9 | |
|---|---|---|---|---|
| ΔE vdW | −34.79 ± 2.30 | −27.05 ± 1.25 | −30.54 ± 1.02 | −33.50 ± 1.30 |
| ΔE elec | −11.59 ± 6.18 | −38.13 ± 1.85 | −11.26 ± 3.36 | −26.58 ± 4.06 |
| ΔG GB | 27.40 ± 5.81 | 47.48 ± 2.14 | 28.87 ± 3.42 | 43.96 ± 3.82 |
| ΔG SA | −4.09 ± 0.22 | −4.23 ± 0.19 | −4.35 ± 0.15 | −4.92 ± 0.18 |
| ΔG bind | −23.06 ± 1.69 | −21.93 ± 1.07 | −17.28 ± 1.37 | −21.04 ± 1.31 |
Notes.
- ΔEvdW
- van der Waals energy
- ΔEelec
- electrostatic energy
- ΔGGB
- electrostatic contribution to the solvation
- ΔGSA
- non-polar contribution to the solvation
- ΔGbind
- binding free energy
Figure 12. Conformational diagrams of the RELA–SX13 (A), IL-6–SX13 (B), MMP9–SX13 (C), and VEGFA–SX13 interactions (D).
Discussion
In the present study, the compounds of SGXZ decoction with corresponding targets were first identified, and the overlapping targets between SGXZ decoction and NAFLD were considered the main targets these compounds acted on. Ten compounds with their important contributions to the compound–target–disease network were selected as the key bioactive ingredients for the treatment of NAFLD. Target proteins, including VEGFA, FGF1, IL-2, LGALS3, SLC5A2, and IL-6, were excessively expressed in the compound–target–disease network and were selected as key targets. In addition, 10 targets with the highest degree in the PPI network were also recognized as key targets acted on by SGXZ decoction. The binding between the key compounds and the key targets was verified by molecular docking, which showed that gallic acid (SX01), chlorogenic acid (SX07), and isochlorogenic acid A (SX13) could bind with all of the key targets. Among them, isochlorogenic acid A showed the lowest binding energy, indicating its potential pharmacological activity. A previous study has suggested that isochlorogenic acid A possesses hepatoprotective properties and anti-hepatitis B effects through suppressing oxidation (Hao et al., 2012). Moreover, it has been indicated that isochlorogenic acid A exerts a protective effect on liver fibrosis by inhibiting inflammation via the HMGB1/TLR4/NF-κB signaling pathway (Liu et al., 2020b). Gallic acid, a simple polyphenol, has been reported to reduce lipid accumulation related to β-oxidation and ketogenesis (Chao et al., 2020). Specifically, the hepatoprotective effect of gallic acid attributed to the repression of inflammatory signaling pathways including nuclear factor-κB (NF-κB)/TNF-α/IL-6 and ROS/NF-κB/TNF-α in NAFLD rats (Fanaei et al., 2021a; Fanaei et al., 2021b). Chlorogenic acid, a natural polyphenol extracted from Artemisiae Scopariae (Yin-Chen in Chinese) (Cai et al., 2020) and Lonicera japonica (Jin-Yin-Hua) (Liu et al., 2020a), could ameliorate HFD-induced hepatic steatosis and inflammation via inhibition of TNF-α and IL-6 in the liver, which was associated with the regulation of gut microbiota and an increase in glucagon-like peptide-1 secretion (Shi et al., 2021; Zamani-Garmsiri et al., 2021).
After that, GO functional enrichment analysis revealed that the targets in the PPI network mainly involved oxidation, positive regulation of angiogenesis, metabolic process, hypoxia, extracellular matrix and insulin-like factor binding, and other biological processes. In addition, KEGG enrichment analysis indicated that the action pathways mainly included insulin resistance, amino acid metabolism, cancer-related pathways, inflammation-related pathways, estrone signaling pathway, and T cell receptor pathway. The target–pathway interaction network suggested that SGXZ decoction might perform a therapeutic role via multiple targets and multiple pathways. Through literature retrieval, signaling pathways including PI3K–Akt, MAPK, insulin resistance, HIF-1, tryptophan metabolism, and estrogen signaling pathway were recognized as core pathways involved in the treatment of NAFLD. The PI3K–Akt signaling pathway, the second largest node size in the target–pathway, is associated with inflammation, which is the driving force for the development and evolution of NAFLD (Byun et al., 2021; Sutti & Albano, 2020). Previous research has provided evidence that the regulation of PI3K–Akt signaling mediators could improve hepatocyte damage, hepatic gluconeogenesis, and lipid disorder (Chen, Liu & Peng, 2019; Liao et al., 2018; Yang et al., 2019). Moreover, inhibiting apoptosis and promoting autophagy via the reactive oxygen species (ROS)/MAPK pathway significantly decreased total cholesterol and triglyceride levels of both plasma and liver in HFD-fed mice (Wu et al., 2021). In addition, increased ROS formation, triglyceride, and lipid accumulation in hepatocytes led to excessive oxidative stress and inflammatory responses, which further produced a hypoxic microenvironment (He et al., 2021; Lei et al., 2021). HIF-1 signaling pathway acts as the key response to hypoxia. Loss of HIF-1α (HIF1A) activates oxidative stress and promotes lipid deposition and secretion of proinflammatory factors IL-6 and TNF-α, suggesting its regulatory role in the progression of NAFLD (Arai, Tanaka & Goda, 2018; He et al., 2021). Indeed, increased FFAs and lipogenesis in hepatocytes, the release of proinflammatory cytokines from adipose tissue such as IL-6 and TNF-α, together with altered gut microbiota give rise to insulin resistance, which predisposes to the development of NAFLD and progression to NASH (Buzzetti, Pinzani & Tsochatzis, 2016; Khan et al., 2019; Lim et al., 2010). Tryptophan is an amino acid that can give rise to indoles by the bacterial enzyme trytophanase A in intestinal epithelial cells. Indoles promote intestinal barrier function and could translocate from the intestines to the liver to modulate hepatic lipid metabolism and inflammation to protect against NAFLD (Ding et al., 2019). In general, the abovementioned core pathways and their interaction correlated to NAFLD might be acted on by SGXZ decoction in the treatment of NAFLD.
Combining the PPI network with the target–pathway network revealed the significant roles of proteins RELA, IL-6, VEGFA, and MMP9. Besides, the four targets played an important regulatory role in the PI3K–Akt signaling pathway according to the KEGG reference pathway map. To further validate the binding between the four targets and the most latent compound isochlorogenic acid A after molecular docking, MD simulations were employed due to their more accurate calculation of binding ability between ligand molecule and protein receptor. The binding free energy of isochlorogenic acid A with RELA, IL-6, MMP9, and VEGFA was −23.06 ± 1.69 kcal/mol, −21.93 ± 1.07 kcal/mol, −21.04 ± 1.31 kcal/mol, and −17.28 ± 1.37 kcal/mol, respectively. This suggests that the proteins could be potentially targeted by isochlorogenic acid A to exert a pharmacological action. RELA, also known as p65, is one of the five members of the NF-κB family that is regulated by its upstream regulator PI3K/Akt. It is a pivotal transcription factor regulating inflammatory molecules (Gasparini & Feldmann, 2012). Inhibition of NF-κB signaling alleviates hepatic lipid accumulation and hepatic inflammation in NAFLD (Daniel et al., 2021; Yang et al., 2021). IL-6 has inflammatory properties and is associated with metabolic disorders (Giraldez et al., 2021). Numerous studies have highlighted that increased IL-6 level promotes hepatic insulin resistance (Akbari & Hassan-Zadeh, 2018) and impairs lipid metabolism (Singh et al., 2021). IL-6 deficiency ameliorates the hepatic inflammation and injury in NASH mice fed methionine and choline–deficient diet (Mas et al., 2009a). VEGFA is a major proangiogenic cytokine that regulates angiogenesis. Increased VEGFA has been reported in the liver of animal models and serum of NASH patients (Lefere, Devisscher & Geerts, 2020; Surapaneni, Vishnu Priya & Mallika, 2015), which accelerates angiogenesis and therefore drives hepatic inflammation and fibrosis in NAFLD (Lefere, Devisscher & Geerts, 2020). The onset of hepatic inflammation causes fibrogenesis in NAFLD, which is manifested by deposited ECM proteins, including collagens, elastin, and fibronectin (Mas et al., 2009b; Munsterman et al., 2018). MMP9 performs a vital role in modulating and degrading gelatins, collagens, and other ECM compounds (D’Amico et al., 2010; Lachowski et al., 2019). A decreased MMP9 level is associated with more advanced fibrosis and serum liver injury indexes (AST, GGT) in NAFLD patients, while increased MMP9 activity could precede the clearance of the fibrotic matrix (Goyale et al., 2021; Trojanek et al., 2020).
In light of the above, we assume that SGXZ decoction might comprehensively regulate inflammation, lipid deposition, insulin resistance, and fibrosis in NAFLD by targeting multiple pathways and targets.
Conclusions
In this study, network pharmacology combined with molecular docking and MD simulations was performed to investigate the putative active ingredients, novel targets, and pivotal signaling pathways of SGXZ decoction in the treatment of NAFLD. It was found that chlorogenic acid, gallic acid, and especially isochlorogenic acid A might exert the pharmacological effect through the PI3K–Akt signaling pathway; this regulation was associated with RELA, IL-6, VEGFA, MMP9, and other protein targets. We further found that the MAPK signaling pathway, insulin resistance, HIF-1 signaling pathway, tryptophan metabolism, estrogen signaling pathway, and other pathways play a role in the regulation of NAFLD. This work provided insight into the therapeutic strategies of NAFLD and evidence for future research. Nonetheless, additional in vitro and in vivo studies should be carried out to corroborate the identified potent active substances, key targets and pivotal pathways.
Supplemental Information
Funding Statement
This work is financially supported by the Shenzhen Science and Technology Planning Project (grant no. JCYJ20190812161605538), the Shenzhen Key Laboratory (ZDSYS20210623092000002), the Guangdong Natural Science Foundation (grant no. 2018A030313181), and Shenzhen Traditional Chinese Medicine Hospital “3030 Program” Chinese Medicine Clinical Project (G3030202115). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Rong Yang conceived and designed the experiments, performed the experiments, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Huili Yang performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the article, and approved the final draft.
Dansheng Jiang performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Linyi Xu performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Lian Feng performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yufeng Xing conceived and designed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.
References
- Abdoli, Azarmi & Eghbal (2014).Abdoli N, Azarmi Y, Eghbal MA. Protective effects of N-acetylcysteine against the statins cytotoxicity in freshly isolated rat hepatocytes. Advanced Pharmaceutical Bulletin. 2014;4:249–254. doi: 10.5681/apb.2014.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan et al. (2020).Ahsan F, Oliveri F, Goud HK, Mehkari Z, Mohammed L, Javed M, Althwanay A, Rutkofsky IH. Pleiotropic effects of statins in the light of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Cureus. 2020;12:e10446. doi: 10.7759/cureus.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari & Hassan-Zadeh (2018).Akbari M, Hassan-Zadeh V. IL-6 signalling pathways and the development of type 2 diabetes. Inflammopharmacology. 2018;26:685–698. doi: 10.1007/s10787-018-0458-0. [DOI] [PubMed] [Google Scholar]
- Arai, Tanaka & Goda (2018).Arai T, Tanaka M, Goda N. HIF-1-dependent lipin1 induction prevents excessive lipid accumulation in choline-deficient diet-induced fatty liver. Scientific Reports. 2018;8:14230. doi: 10.1038/s41598-018-32586-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzetti, Pinzani & Tsochatzis (2016).Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Byun et al. (2021).Byun KA, Oh S, Son M, Park CH, Son KH, Byun K. Dieckol decreases caloric intake and attenuates nonalcoholic fatty liver disease and hepatic lymphatic vessel dysfunction in high-fat-diet-fed mice. Marine Drugs. 2021;19(9):495. doi: 10.3390/md19090495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2020).Cai Y, Zheng Q, Sun R, Wu J, Li X, Liu R. Recent progress in the study of Artemisiae Scopariae Herba (Yin Chen), a promising medicinal herb for liver diseases. Biomedicine & Pharmacotherapy. 2020;130:110513. doi: 10.1016/j.biopha.2020.110513. [DOI] [PubMed] [Google Scholar]
- Álvarez-Carretero et al. (2018).Álvarez-Carretero S, Pavlopoulou N, Adams J, Gilsenan J, Tabernero L. VSpipe, an integrated resource for virtual screening and hit selection: applications to protein tyrosine phospahatase inhibition. Molecules. 2018;23(2):353. doi: 10.3390/molecules23020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centis et al. (2013).Centis E, Moscatiello S, Bugianesi E, Bellentani S, Fracanzani AL, Calugi S, Petta S, Dalle Grave R, Marchesini G. Stage of change and motivation to healthier lifestyle in non-alcoholic fatty liver disease. Journal of Hepatology. 2013;58:771–777. doi: 10.1016/j.jhep.2012.11.031. [DOI] [PubMed] [Google Scholar]
- Challa et al. (2019).Challa TD, Wueest S, Lucchini FC, Dedual M, Modica S, Borsigova M, Wolfrum C, Blüher M, Konrad D. Liver ASK1 protects from non-alcoholic fatty liver disease and fibrosis. EMBO Molecular Medicine. 2019;11:e10124. doi: 10.15252/emmm.201810124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao et al. (2020).Chao J, Cheng HY, Chang ML, Huang SS, Liao JW, Cheng YC, Peng WH, Pao LH. Gallic acid ameliorated impaired lipid homeostasis in a mouse model of high-fat diet-and streptozotocin-induced NAFLD and diabetes through improvement of β-oxidation and ketogenesis. Frontiers in Pharmacology. 2020;11:606759. doi: 10.3389/fphar.2020.606759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman & Lynch (2020).Chapman RW, Lynch KD. Obeticholic acid-a new therapy in PBC and NASH. British Medical Bulletin. 2020;133:95–104. doi: 10.1093/bmb/ldaa006. [DOI] [PubMed] [Google Scholar]
- Chen, Liu & Peng (2019).Chen SH, Liu XN, Peng Y. MicroRNA-351 eases insulin resistance and liver gluconeogenesis via the PI3K/AKT pathway by inhibiting FLOT2 in mice of gestational diabetes mellitus. Journal of Cellular and Molecular Medicine. 2019;23:5895–5906. doi: 10.1111/jcmm.14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng et al. (2019).Cheng HS, Tan WR, Low ZS, Marvalim C, Lee JYH, Tan NS. Exploration and development of PPAR modulators in health and disease: an update of clinical evidence. International Journal of Molecular Sciences. 2019;20(20):5055. doi: 10.3390/ijms20205055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusi et al. (2022).Cusi K, Isaacs S, Barb D, Basu R, Caprio S, Garvey WT, Kashyap S, Mechanick JI, Mouzaki M, Nadolsky K, Rinella ME, Vos MB, Younossi Z. American association of clinical endocrinology clinical practice guideline for the diagnosis and management of nonalcoholic fatty liver disease in primary care and endocrinology clinical settings: co-sponsored by the american association for the Study of Liver Diseases (AASLD) Endocrine Practice. 2022;28:528–562. doi: 10.1016/j.eprac.2022.03.010. [DOI] [PubMed] [Google Scholar]
- Dai et al. (2021).Dai X, Feng J, Chen Y, Huang S, Shi X, Liu X, Sun Y. Traditional Chinese medicine in nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Chinese Medical Journal. 2021;16:68. doi: 10.1186/s13020-021-00469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico et al. (2010).D’Amico F, Consolo M, Amoroso A, Skarmoutsou E, Mauceri B, Stivala F, Malaponte G, Bertino G, Neri S, Mazzarino MC. Liver immunolocalization and plasma levels of MMP-9 in non-alcoholic steatohepatitis (NASH) and hepatitis C infection. Acta Histochemica. 2010;112:474–481. doi: 10.1016/j.acthis.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Daniel et al. (2021).Daniel PV, Dogra S, Rawat P, Choubey A, Khan AS, Rajak S, Kamthan M, Mondal P. NF-κB p65 regulates hepatic lipogenesis by promoting nuclear entry of ChREBP in response to a high carbohydrate diet. Journal of Biological Chemistry. 2021;296:100714. doi: 10.1016/j.jbc.2021.100714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding et al. (2019).Ding Y, Yanagi K, Cheng C, Alaniz RC, Lee K, Jayaraman A. Interactions between gut microbiota and non-alcoholic liver disease: the role of microbiota-derived metabolites. Pharmacological Research. 2019;141:521–529. doi: 10.1016/j.phrs.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanaei et al. (2021a).Fanaei H, Mard SA, Sarkaki A, Goudarzi G, Khorsandi L. Gallic acid protects the liver against NAFLD induced by dust exposure and high-fat diet through inhibiting oxidative stress and repressing the inflammatory signaling pathways NF-k β/TNF-α/IL-6 in Wistar rats. Avicenna Journal of Phytomedicine. 2021a;11:527–540. doi: 10.22038/ajp.2021.17835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanaei et al. (2021b).Fanaei H, Mard SA, Sarkaki A, Goudarzi G, Khorsandi L. Gallic acid treats dust-induced NAFLD in rats by improving the liver’s anti-oxidant capacity and inhibiting ROS/NF κβ/TNF α inflammatory pathway. Iranian Journal of Basic Medical Sciences. 2021b;24:240–247. doi: 10.22038/ijbms.2021.51036.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu et al. (2021).Fu K, Wang C, Ma C, Zhou H, Li Y. The potential application of Chinese medicine in liver diseases: a new opportunity. Frontiers in Pharmacology. 2021;12:771459. doi: 10.3389/fphar.2021.771459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini & Feldmann (2012).Gasparini C, Feldmann M. NF-κB as a target for modulating inflammatory responses. Current Pharmaceutical Design. 2012;18:5735–5745. doi: 10.2174/138161212803530763. [DOI] [PubMed] [Google Scholar]
- Giraldez et al. (2021).Giraldez MD, Carneros D, Garbers C, Rose-John S, Bustos M. New insights into IL-6 family cytokines in metabolism, hepatology and gastroenterology. Nature Reviews Gastroenterology & Hepatology. 2021;18:787–803. doi: 10.1038/s41575-021-00473-x. [DOI] [PubMed] [Google Scholar]
- Goyale et al. (2021).Goyale A, Jain A, Smith C, Papatheodoridi M, Misas MG, Roccarina D, Prat LI, Mikhailidis DP, Nair D, Tsochatzis E. Assessment of non-alcoholic fatty liver disease (NAFLD) severity with novel serum-based markers: a pilot study. PLOS ONE. 2021;16:e0260313. doi: 10.1371/journal.pone.0260313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2019).Guo W, Huang J, Wang N, Tan HY, Cheung F, Chen F, Feng Y. Integrating network pharmacology and pharmacological evaluation for deciphering the action mechanism of herbal formula zuojin pill in suppressing hepatocellular carcinoma. Frontiers in Pharmacology. 2019;10:1185. doi: 10.3389/fphar.2019.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao et al. (2012).Hao BJ, Wu YH, Wang JG, Hu SQ, Keil DJ, Hu HJ, Lou JD, Zhao Y. Hepatoprotective and antiviral properties of isochlorogenic acid A from Laggera alata against hepatitis B virus infection. Journal of Ethnopharmacology. 2012;144:190–194. doi: 10.1016/j.jep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- He et al. (2021).He Y, Yang W, Gan L, Liu S, Ni Q, Bi Y, Han T, Liu Q, Chen H, Hu Y, Long Y, Yang L. Silencing HIF-1α aggravates non-alcoholic fatty liver disease in vitro through inhibiting PPAR-α/ANGPTL4 singling pathway. Clinical Gastroenterology and Hepatology. 2021;44:355–365. doi: 10.1016/j.gastrohep.2020.09.014. [DOI] [PubMed] [Google Scholar]
- Holzner & Murray (2021).Holzner LMW, Murray AJ. Hypoxia-inducible factors as key players in the pathogenesis of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Frontiers in Medicine. 2021;8:753268. doi: 10.3389/fmed.2021.753268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Da, Sherman & Lempicki (2009a).Huang Da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Da, Sherman & Lempicki (2009b).Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ioannou (2021).Ioannou GN. Epidemiology and risk-stratification of NAFLD-associated HCC. Journal of Hepatology. 2021;75:1476–1484. doi: 10.1016/j.jhep.2021.08.012. [DOI] [PubMed] [Google Scholar]
- Jeznach-Steinhagen et al. (2019).Jeznach-Steinhagen A, Ostrowska J, Czerwonogrodzka-Senczyna A, Boniecka I, Shahnazaryan U, Kuryłowicz A. Dietary and pharmacological treatment of nonalcoholic fatty liver disease. Medicina. 2019;55(5):166. doi: 10.3390/medicina55050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao et al. (2021).Jiao X, Jin X, Ma Y, Yang Y, Li J, Liang L, Liu R, Li Z. A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Computational Biology and Chemistry. 2021;90:107402. doi: 10.1016/j.compbiolchem.2020.107402. [DOI] [PubMed] [Google Scholar]
- Kantartzis et al. (2009).Kantartzis K, Thamer C, Peter A, Machann J, Schick F, Schraml C, Königsrainer A, Königsrainer I, Kröber S, Niess A, Fritsche A, Häring HU, Stefan N. High cardiorespiratory fitness is an independent predictor of the reduction in liver fat during a lifestyle intervention in non-alcoholic fatty liver disease. Gut. 2009;58:1281–1288. doi: 10.1136/gut.2008.151977. [DOI] [PubMed] [Google Scholar]
- Keiser et al. (2007).Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nature Biotechnology. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- Khan et al. (2019).Khan RS, Bril F, Cusi K, Newsome PN. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatology. 2019;70:711–724. doi: 10.1002/hep.30429. [DOI] [PubMed] [Google Scholar]
- Klein et al. (2011).Klein EA, Thompson Jr IM, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM, Karp DD, Lieber MM, Walther PJ, Klotz L, Parsons JK, Chin JL, Darke AK, Lippman SM, Goodman GE, Meyskens Jr FL, Baker LH. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2011;306:1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochumon et al. (2020).Kochumon S, Al Madhoun A, Al-Rashed F, Thomas R, Sindhu S, Al-Ozairi E, Al-Mulla F, Ahmad R. Elevated adipose tissue associated IL-2 expression in obesity correlates with metabolic inflammation and insulin resistance. Scientific Reports. 2020;10:16364. doi: 10.1038/s41598-020-73347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konerman, Jones & Harrison (2018).Konerman MA, Jones JC, Harrison SA. Pharmacotherapy for NASH: current and emerging. Journal of Hepatology. 2018;68:362–375. doi: 10.1016/j.jhep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Kumar, Kulkarni & Jagdish (2021).Kumar K, Kulkarni A, Jagdish RK. Letter to the Editor: saroglitazar for treatment of NAFLD and NASH. Hepatology. 2021;74:3559–3560. doi: 10.1002/hep.32094. [DOI] [PubMed] [Google Scholar]
- Lachowski et al. (2019).Lachowski D, Cortes E, Rice A, Pinato D, Rombouts K, Hernandez ADelRio. Matrix stiffness modulates the activity of MMP-9 and TIMP-1 in hepatic stellate cells to perpetuate fibrosis. Scientific Reports. 2019;9:7299. doi: 10.1038/s41598-019-43759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai et al. (2020).Lai X, Wang X, Hu Y, Su S, Li W, Li S. Editorial: network pharmacology and traditional medicine. Frontiers in Pharmacology. 2020;11:1194. doi: 10.3389/fphar.2020.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefere, Devisscher & Geerts (2020).Lefere S, Devisscher L, Geerts A. Angiogenesis in the progression of non-alcoholic fatty liver disease. Acta Gastro-Enterologica Belgica. 2020;83:301–307. [PubMed] [Google Scholar]
- Lei et al. (2021).Lei L, Ei Mourabit H, Housset C, Cadoret A, Lemoinne S. Role of Angiogenesis in the Pathogenesis of NAFLD. Journal of Clinical Medicine. 2021;10:1338. doi: 10.3390/jcm10071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Zhang & Li (2020a).Li C, Zhang H, Li X. The mechanism of traditional Chinese medicine for the treatment of obesity. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy. 2020a;13:3371–3381. doi: 10.2147/dmso.S274534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2020b).Li YS, Li XT, Yu LG, Wang L, Shi ZY, Guo XL. Roles of galectin-3 in metabolic disorders and tumor cell metabolism. International Journal of Biological Macromolecules. 2020b;142:463–473. doi: 10.1016/j.ijbiomac.2019.09.118. [DOI] [PubMed] [Google Scholar]
- Liang et al. (2019).Liang Y, Lin C, Huang S, Xu Y. Traditional Chinese medicine and intestinal microbiota: a complementary and integrative health approach to ameliorate obesity-related diseases. Holistic Nursing Practice. 2019;33:259–265. doi: 10.1097/hnp.0000000000000311. [DOI] [PubMed] [Google Scholar]
- Liao et al. (2017).Liao HW, Saver JL, Wu YL, Chen TH, Lee M, Ovbiagele B. Pioglitazone and cardiovascular outcomes in patients with insulin resistance, pre-diabetes and type 2 diabetes: a systematic review and meta-analysis. BMJ Open. 2017;7:e013927. doi: 10.1136/bmjopen-2016-013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao et al. (2018).Liao X, Song L, Zhang L, Wang H, Tong Q, Xu J, Yang G, Yang S, Zheng H. LAMP3 regulates hepatic lipid metabolism through activating PI3K/Akt pathway. Molecular and Cellular Endocrinology. 2018;470:160–167. doi: 10.1016/j.mce.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Lim et al. (2010).Lim JS, Mietus-Snyder M, Valente A, Schwarz JM, Lustig RH. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nature Reviews Gastroenterology & Hepatology. 2010;7:251–264. doi: 10.1038/nrgastro.2010.41. [DOI] [PubMed] [Google Scholar]
- Lincoff et al. (2007).Lincoff AM, Wolski K, Nicholls SJ, Nissen SE. Pioglitazone and risk of cardiovascular events in patients with type 2 diabetes mellitus: a meta-analysis of randomized trials. JAMA. 2007;298:1180–1188. doi: 10.1001/jama.298.10.1180. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2020a).Liu T, Yang J, Liu S, Zhao Y, Zhou J, Jin Y, Huang L, Yuan Y. Regulation of chlorogenic acid, flavonoid, and iridoid biosynthesis by histone H3K4 and H3K9 methylation in Lonicera japonica. Molecular Biology Reports. 2020a;47:9301–9311. doi: 10.1007/s11033-020-05990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2020b).Liu X, Huang K, Zhang RJ, Mei D, Zhang B. Isochlorogenic acid A attenuates the progression of liver fibrosis through regulating HMGB1/TLR4/NF-κB signaling pathway. Frontiers in Pharmacology. 2020b;11:582. doi: 10.3389/fphar.2020.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu X, Shi D, Zhou S, Liu H, Liu H, Yao X. Molecular dynamics simulations and novel drug discovery. Expert Opinion on Drug Discovery. 2018;13:23–37. doi: 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
- Liu & Sun (2012).Liu ZH, Sun XB. Network pharmacology: new opportunity for the modernization of traditional Chinese medicine. Yao Xue Xue Bao. 2012;47:696–703. [PubMed] [Google Scholar]
- Liu et al. (2020c).Liu Z, Zhang Y, Graham S, Wang X, Cai D, Huang M, Pique-Regi R, Dong XC, Chen YE, Willer C, Liu W. Causal relationships between NAFLD, T2D and obesity have implications for disease subphenotyping. Journal of Hepatology. 2020c;73:263–276. doi: 10.1016/j.jhep.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Zhou & Li (2017).Ma J, Zhou Q, Li H. Gut microbiota and nonalcoholic fatty liver disease: insights on mechanisms and therapy. Nutrients. 2017;9(10):1124. doi: 10.3390/nu9101124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas et al. (2009a).Mas E, Danjoux M, Garcia V, Carpentier S, Ségui B, Levade T. IL-6 deficiency attenuates murine diet-induced non-alcoholic steatohepatitis. PLOS ONE. 2009a;4:e7929. doi: 10.1371/journal.pone.0007929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas et al. (2009b).Mas VR, Fisher RA, Archer KJ, Maluf DG. Proteomics and liver fibrosis: identifying markers of fibrogenesis. Expert Review of Proteomics. 2009b;6:421–431. doi: 10.1586/epr.09.59. [DOI] [PubMed] [Google Scholar]
- Miller 3rd et al. (2005).Miller 3rd ER, Pastor-Barriuso R, Dalal D, Riemersma RA, Appel LJ, Guallar E. Meta-analysis: high-dosage vitamin E supplementation may increase all-cause mortality. Annals of Internal Medicine. 2005;142:37–46. doi: 10.7326/0003-4819-142-1-200501040-00110. [DOI] [PubMed] [Google Scholar]
- Mundi et al. (2020).Mundi MS, Velapati S, Patel J, Kellogg TA, Abu Dayyeh BK, Hurt RT. Evolution of NAFLD and its management. Nutrition in Clinical Practice. 2020;35:72–84. doi: 10.1002/ncp.10449. [DOI] [PubMed] [Google Scholar]
- Munsterman et al. (2018).Munsterman ID, Kendall TJ, Khelil N, Popa M, Lomme R, Drenth JPH, Tjwa E. Extracellular matrix components indicate remodelling activity in different fibrosis stages of human non-alcoholic fatty liver disease. Histopathology. 2018;73:612–621. doi: 10.1111/his.13665. [DOI] [PubMed] [Google Scholar]
- Nies et al. (2015).Nies VJ, Sancar G, Liu W, Van Zutphen T, Struik D, Yu RT, Atkins AR, Evans RM, Jonker JW, Downes MR. Fibroblast growth factor signaling in metabolic regulation. Frontiers in Endocrinology. 2015;6:193. doi: 10.3389/fendo.2015.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen & Wolski (2007).Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England Journal of Medicine. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- Pal et al. (2015).Pal S, Ghosh M, Ghosh S, Bhattacharyya S, Sil PC. Atorvastatin induced hepatic oxidative stress and apoptotic damage via MAPKs, mitochondria, calpain and caspase12 dependent pathways. Food and Chemical Toxicology. 2015;83:36–47. doi: 10.1016/j.fct.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Piñero et al. (2020).Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobre, Poet & Hendershot (2019).Pobre KFR, Poet GJ, Hendershot LM. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. Journal of Biological Chemistry. 2019;294:2098–2108. doi: 10.1074/jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza et al. (2021).Raza S, Rajak S, Upadhyay A, Tewari A, Anthony Sinha R. Current treatment paradigms and emerging therapies for NAFLD/NASH. Frontiers in Bioscience-Landmark. 2021;26:206–237. doi: 10.2741/4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar et al. (2021).Sarkar S, Kumari D, Gupta SK, Sharma V, Mukhi S, Kamboj P, Sasibhushan V, Rai RK, Jatavallabhula SL, Mahajan D, Kumar Y, Kumar A, Dikshit M. Saroglitazar and Hepano treatment offers protection against high fat high fructose diet induced obesity, insulin resistance and steatosis by modulating various class of hepatic and circulating lipids. Biomedicine & Pharmacotherapy. 2021;144:112357. doi: 10.1016/j.biopha.2021.112357. [DOI] [PubMed] [Google Scholar]
- Shi et al. (2021).Shi A, Li T, Zheng Y, Song Y, Wang H, Wang N, Dong L, Shi H. Chlorogenic acid improves NAFLD by regulating gut microbiota and GLP-1. Frontiers in Pharmacology. 2021;12:693048. doi: 10.3389/fphar.2021.693048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuja, Eqbal & Rehman (2021).Shuja SH, Eqbal F, Rehman H. Saroglitazar—a potential therapeutic option in treating NASH? Drug Design, Development and Therapy. 2021;15:4227–4228. doi: 10.2147/dddt.S341223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh et al. (2021).Singh MK, Jayarajan R, Varshney S, Upadrasta S, Singh A, Yadav R, Scaria V, Sengupta S, Shanmugam D, Shalimar O, Sivasubbu S, Gandotra S, Sachidanandan C. Chronic systemic exposure to IL6 leads to deregulation of glycolysis and fat accumulation in the zebrafish liver. Biochimica et Biophysica Acta—Molecular and Cell Biology of Lipids. 2021;1866:158905. doi: 10.1016/j.bbalip.2021.158905. [DOI] [PubMed] [Google Scholar]
- Song et al. (2021).Song YQ, He RJ, Pu D, Guan XQ, Shi JH, Li YG, Hou J, Jia SN, Qin WW, Fang SQ, Ge GB. Discovery and characterization of the biflavones from ginkgo biloba as highly specific and potent inhibitors against human carboxylesterase 2. Frontiers in Pharmacology. 2021;12:655659. doi: 10.3389/fphar.2021.655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan, Häring & Cusi (2019).Stefan N, Häring HU, Cusi K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies. The Lancet Diabetes & Endocrinology. 2019;7:313–324. doi: 10.1016/s2213-8587(18)30154-2. [DOI] [PubMed] [Google Scholar]
- Stelzer et al. (2016).Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54:1.30.31–31.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Surapaneni, Vishnu Priya & Mallika (2015).Surapaneni KM, Vishnu Priya V, Mallika J. Effect of pioglitazone, quercetin, and hydroxy citric acid on vascular endothelial growth factor messenger RNA (VEGF mRNA) expression in experimentally induced nonalcoholic steatohepatitis (NASH) The Turkish Journal of Medical Sciences. 2015;45:542–546. doi: 10.3906/sag-1404-136. [DOI] [PubMed] [Google Scholar]
- Sutti & Albano (2020).Sutti S, Albano E. Adaptive immunity: an emerging player in the progression of NAFLD. Nature Reviews Gastroenterology & Hepatology. 2020;17:81–92. doi: 10.1038/s41575-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk et al. (2019).Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi et al. (2015).Takahashi Y, Sugimoto K, Inui H, Fukusato T. Current pharmacological therapies for nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World Journal of Gastroenterology. 2015;21:3777–3785. doi: 10.3748/wjg.v21.i13.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2018).Tang HH, Wei CS, Zheng YJ, Qiu M, Xing YF, Tong GD. Evaluating the clinical effect of modified shugan xiaozhi decoction in treating nonalcoholic fatty liver disease based on fibroscan technique. Journal of Traditional Chinese Medicine. 2018;59:594–598. doi: 10.13288/j.11-2166/r.2018.07.014. [DOI] [Google Scholar]
- Tilg & Effenberger (2020).Tilg H, Effenberger M. From NAFLD to MAFLD: when pathophysiology succeeds. Nature Reviews Gastroenterology & Hepatology. 2020;17:387–388. doi: 10.1038/s41575-020-0316-6. [DOI] [PubMed] [Google Scholar]
- Trojanek et al. (2020).Trojanek JB, Michałkiewicz J, Grzywa-Czuba R, Jańczyk W, Gackowska L, Kubiszewska I, Helmin-Basa A, Wierzbicka-Rucińska A, Szalecki M, Socha P. Expression of matrix metalloproteinases and their tissue inhibitors in peripheral blood leukocytes and plasma of children with nonalcoholic fatty liver disease. Mediators of Inflammation. 2020;2020:8327945. doi: 10.1155/2020/8327945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadden et al. (2014).Wadden JPBTA, Blackburn GL, Bolin P, Brancati FL, Bray GA, Clark JM, Coday M, Dutton GR, Egan C, Evans M, Foreyt JP, Sengardi SG, Gregg EW, Hazuda HP, Hill JO, Horton ES, Hubbard VS, Jakicic JM, Jeffery RW, Johnson KC, Kahn SE, Kitabchi AE, Knowler WC, Lewis CE, Maschak-Carey BJ, Montez MG, Montgomery B, Nathan DM, Nelson J, Patricio J, Peters A, Pi-Sunyer FX, Pownall H, Rickman AD, Vitolins M, Walkup MP, West DS, Williamson D, Wing RR, Wyatt H, Yanovski SZ. Eight-year weight losses with an intensive lifestyle intervention: the look AHEAD study. Obesity. 2014;22:5–13. doi: 10.1002/oby.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang L, Liu M, Yin F, Wang Y, Li X, Wu Y, Ye C, Liu J. Trilobatin, a novel SGLT1/2 inhibitor, selectively induces the proliferation of human hepatoblastoma cells. Molecules. 2019;24:3390. doi: 10.3390/molecules24183390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2021).Wang X, Wang ZY, Zheng JH, Li S. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chinese Journal of Natural Medicines. 2021;19:1–11. doi: 10.1016/s1875-5364(21)60001-8. [DOI] [PubMed] [Google Scholar]
- Waterhouse et al. (2018).Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2021).Wu D, Liu Z, Wang Y, Zhang Q, Li J, Zhong P, Xie Z, Ji A, Li Y. Epigallocatechin-3-gallate alleviates high-fat diet-induced nonalcoholic fatty liver disease via inhibition of apoptosis and promotion of autophagy through the ROS/MAPK signaling pathway. Oxidative Medicine and Cellular Longevity. 2021;2021:5599997. doi: 10.1155/2021/5599997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2019).Wu W, Li W, Wei J, Wang C, Yao Y, Zhu W, He W, Zhou W, Liu J. Chronic intermittent hypoxia accelerates liver fibrosis in rats with combined hypoxia and nonalcoholic steatohepatitis via angiogenesis rather than endoplasmic reticulum stress. Acta Biochimica et Biophysica Sinica. 2019;51:159–167. doi: 10.1093/abbs/gmy169. [DOI] [PubMed] [Google Scholar]
- Xing et al. (2016).Xing YF, Zhang Z, Fu WJ, Zhou DQ, Yuen AC, Mok DK, Chan CO, Tong GD. Shugan Xiaozhi decoction attenuates nonalcoholic steatohepatitis by enhancing PPAR α and L-FABP expressions in high-fat-fed rats. Evidence-Based Complementary and Alternative Medicine. 2016;2016:7870189. doi: 10.1155/2016/7870189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2019).Yang P, Liang Y, Luo Y, Li Z, Wen Y, Shen J, Li R, Zheng H, Gu HF, Xia N. Liraglutide ameliorates nonalcoholic fatty liver disease in diabetic mice via the IRS2/PI3K/Akt signaling pathway. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy. 2019;12:1013–1021. doi: 10.2147/dmso.S206867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2021).Yang XD, Chen Z, Ye L, Chen J, Yang YY. Esculin protects against methionine choline-deficient diet-induced non-alcoholic steatohepatitis by regulating the Sirt1/NF-κB p65 pathway. Pharmaceutical Biology. 2021;59:922–932. doi: 10.1080/13880209.2021.1945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon et al. (2021).Yoon YS, Liu W, Van de Velde S, Matsumura S, Wiater E, Huang L, Montminy M. Activation of the adipocyte CREB/CRTC pathway in obesity. Communications Biology. 2021;4:1214. doi: 10.1038/s42003-021-02735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi et al. (2016).Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, Racila A, Hunt S, Beckerman R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64:1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- Younossi et al. (2019).Younossi Z, Stepanova M, Ong JP, Jacobson IM, Bugianesi E, Duseja A, Eguchi Y, Wong VW, Negro F, Yilmaz Y, Romero-Gomez M, George J, Ahmed A, Wong R, Younossi I, Ziayee M, Afendy A. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clinical Gastroenterology and Hepatology. 2019;17:748–755.e743. doi: 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2021).Yu H, Yang F, Zhong W, Jiang X, Zhang F, Ji X, Xue M, Qiu Y, Yu J, Hu X, Chen J, Bao Z. Secretory Galectin-3 promotes hepatic steatosis via regulation of the PPAR γ/CD36 signaling pathway. Cell Signal. 2021;84:110043. doi: 10.1016/j.cellsig.2021.110043. [DOI] [PubMed] [Google Scholar]
- Zamani-Garmsiri et al. (2021).Zamani-Garmsiri F, Ghasempour G, Aliabadi M, Hashemnia SMR, Emamgholipour S, Meshkani R. Combination of metformin and chlorogenic acid attenuates hepatic steatosis and inflammation in high-fat diet fed mice. IUBMB Life. 2021;73:252–263. doi: 10.1002/iub.2424. [DOI] [PubMed] [Google Scholar]
- Zhai et al. (2014).Zhai FF, Sun H, Su XX, Xing YF. Clinical efficacy of Shugan Xiaozhi granules in treatment of nonalcoholic steatohepatitis. Chinese Journal of Integrated Traditional and Western Medicine on Liver Diseases. 2014;24:78–79+82. [Google Scholar]
- Zhai et al. (2021).Zhai FF, Wang YQ, Feng L, Xing YF. Study on the enterogenic mechanism of Shugan Xiaozhi decoction in treating NAFLD by regulating apoB48. Chinese Journal of Integrated Traditional and Western Medicine on Liver Diseases. 2021;31:721–725. [Google Scholar]
- Zhang et al. (2021).Zhang HY, Tian JX, Lian FM, Li M, Liu WK, Zhen Z, Liao JQ, Tong XL. Therapeutic mechanisms of traditional Chinese medicine to improve metabolic diseases via the gut microbiota. Biomedicine & Pharmacotherapy. 2021;133:110857. doi: 10.1016/j.biopha.2020.110857. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2020).Zhang M, Yuan Y, Zhou W, Qin Y, Xu K, Men J, Lin M. Network pharmacology analysis of Chaihu Lizhong Tang treating non-alcoholic fatty liver disease. Computational Biology and Chemistry. 2020;86:107248. doi: 10.1016/j.compbiolchem.2020.107248. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Frontiers in Pharmacology. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2020a).Zhou J, Zhou F, Wang W, Zhang XJ, Ji YX, Zhang P, She ZG, Zhu L, Cai J, Li H. Epidemiological features of NAFLD from 1999 to 2018 in China. Hepatology. 2020a;71:1851–1864. doi: 10.1002/hep.31150. [DOI] [PubMed] [Google Scholar]
- Zhou et al. (2020b).Zhou Z, Chen B, Chen S, Lin M, Chen Y, Jin S, Chen W, Zhang Y. Applications of network pharmacology in traditional Chinese medicine research. Evidence-Based Complementary and Alternative Medicine. 2020b;2020:1646905. doi: 10.1155/2020/1646905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2020c).Zhou Z, Moore TM, Drew BG, Ribas V, Wanagat J, Civelek M, Segawa M, Wolf DM, Norheim F, Seldin MM, Strumwasser AR, Whitney KA, Lester E, Reddish BR, Vergnes L, Reue K, Rajbhandari P, Tontonoz P, Lee J, Mahata SK, Hewitt SC, Shirihai O, Gastonbury C, Small KS, Laakso M, Jensen J, Lee S, Drevon CA, Korach KS, Lusis AJ, Hevener AL. Estrogen receptor α controls metabolism in white and brown adipocytes by regulating Polg1 and mitochondrial remodeling. Science Translational Medicine. 2020c;12(555):eaax8096. doi: 10.1126/scitranslmed.aax8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.