Abstract
Haemophilus influenzae requires heme for growth and can utilize hemoglobin and hemoglobin-haptoglobin as heme sources. We previously identified two hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA and HgpB, in H. influenzae HI689. Insertional mutation of hgpA and hgpB, either singly or together, did not abrogate the ability to utilize or bind either hemoglobin or the hemoglobin-haptoglobin complex. A hemoglobin affinity purification method was used to isolate a protein of approximately 120 kDa from the hgpA hgpB double mutant. We have cloned and sequenced the gene encoding this third hemoglobin/hemoglobin-haptoglobin binding protein and designate it hgpC. Insertional mutation of hgpC did not affect the ability of the strain to utilize either hemoglobin or hemoglobin-haptoglobin. An hgpA hgpB hgpC triple mutant constructed by insertional mutagenesis showed a reduced ability to use the hemoglobin-haptoglobin complex but was unaltered in the ability to use hemoglobin. A second class of mutants was constructed in which the entire structural gene of each of the three proteins was deleted. The hgpA hgpB hgpC complete-deletion triple mutant was unable to utilize the hemoglobin-haptoglobin complex and showed a reduced ability to use hemoglobin. We have identified three hemoglobin/hemoglobin-haptoglobin-binding proteins in Haemophilus influenzae. Any one of the three proteins is sufficient to support growth with hemoglobin-haptoglobin as the heme source, and expression of at least one of the three is essential for hemoglobin-haptoglobin utilization. Although the three proteins play a role in hemoglobin utilization, an additional hemoglobin acquisition mechanism(s) exists.
The human-specific pathogen Haemophilus influenzae causes a range of human infections in both adults and children, including otitis media, meningitis, epiglottitis, and pneumonia (10, 24). Since H. influenzae lacks most enzymes of the heme biosynthetic pathway, it has an absolute growth requirement for protoporphyrin IX (PPIX), the immediate precursor of heme (3). In vivo there is essentially no free PPIX and heme is intracellular, in the form of hemoglobin or heme-containing enzymes. Hemoglobin released by erythrocytes is avidly bound by the serum protein haptoglobin, and the complex is rapidly cleared by hepatocytes. Heme released by the breakdown of free hemoglobin will be bound by either of the serum proteins hemopexin or human serum albumin, and the complex(es) will be cleared from the circulation. Thus, heme is largely unavailable to invading microorganisms (1, 11). Hemoglobin and the hemoglobin-haptoglobin, heme-hemopexin, and heme-albumin complexes are all utilized by H. influenzae as heme sources in vitro (22). H. influenzae binds hemoglobin and the hemoglobin-haptoglobin complex at the cell surface, and we have previously identified two genes in H. influenzae type b strain HI689, hgpA and hgpB, which encode proteins that bind both hemoglobin and the hemoglobin-haptoglobin complex (5, 8, 9, 18). A distinctive feature of these genes is a length of CCAA nucleotide repeating units directly following the sequence encoding a putative leader peptide sequence (8, 18). Mutation of either hgpA or hgpB alone does not affect the ability of HI689 to use either hemoglobin or the hemoglobin-haptoglobin complex. However, a double mutation of hgpA and hgpB resulted in a reduced ability of the mutant strain to use the hemoglobin-haptoglobin complex (18). Both the single mutants and the double mutant retained the ability to bind hemoglobin and the hemoglobin-haptoglobin complex (18). These data indicate the presence of additional genes involved in the acquisition of heme from hemoglobin and hemoglobin-haptoglobin. Affinity isolation of hemoglobin-binding proteins from the hgpA hgpB double mutant resulted in the isolation of an approximately 120-kDa protein, which was shown by sequencing of internal peptide fragments to be a homologue of the putative gene product of the strain RdKW20 open reading frames (ORFs) HI0635 or HI0712 (4, 18). Similar to hgpA and hgpB, the ORFs HI0635 and HI0712 possess lengths of CCAA nucleotide repeats near the N-terminal of the coding sequence (4). Rd KW20 possesses two other putative genes containing lengths of CCAA repeats designated ORFs HI0661 and HI1566 (4). Homology at the amino acid level between the putative products of HI0635, HI0661, HI0712, and HI1566 and HgpA and HgpB suggests that these proteins have similar functions (7, 8, 18). Southern analysis with a (CCAA)6 oligonucleotide probe reveals three CCAA-containing regions in the chromosome of strain HI689 (15, 18). Subsequent analysis with probes designed to be specific for the structural region of the CCAA-containing genes or ORFs showed that the third CCAA-containing region in HI689 was a homologue of the ORF HI0712 (15).
The object of this study was to define the potential role of the HI0712 homologue in H. influenzae HI689 in the acquisition of heme from hemoglobin and/or the hemoglobin-haptoglobin complex and to elucidate the potential interaction(s) between HgpA, HgpB, and the HI0712 homologue.
MATERIALS AND METHODS
Bacterial strain growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains of H. influenzae were routinely maintained on brain heart infusion (BHI) agar (Difco, Detroit, Mich.) supplemented with 10 μg of both heme and β-NAD per ml (supplemented BHI; sBHI). For experiments in heme-replete media, H. influenzae was grown at 37°C in sBHI broth. Heme-restricted growth of H. influenzae took place in BHI supplemented with 10 μg of β-NAD per ml and 0.1 μg of heme per ml (heme-restricted BHI [hrBHI]), and heme-depleted growth took place in BHI supplemented with only 10 μg of β-NAD (heme-depleted BHI [hdBHI]). H. influenzae were made competent using the MII medium of Spenser and Herriott (21). Recombinant H. influenzae was grown on sBHI supplemented with antibiotics as indicated. Escherichia coli strains were maintained on Luria-Bertani medium supplemented with antibiotics as indicated.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| H. influenzae | ||
| HI689 | Type b | 17 |
| HI1704 | Type b, hgpAΔBglII Rbr | 8 |
| HI1705 | Type b, hgpBΔBclI Tcr | 18 |
| HI1706 | Type b, hgpC::SPEC Spr | This study |
| HI1707 | Type b, hgpAΔBglII hgpBΔBclI Rbr Tcr | 18 |
| HI1708 | Type b, hgpAΔBglII hgpC::SPEC Rbr Spr | This study |
| HI1709 | Type b, hgpBΔBclI hgpC::SPEC Tcr Spr | This study |
| HI1710 | Type b, hgpAΔBglII hgpBΔBclI hgpC::SPEC Rbr Tcr Spr | This study |
| HI1711 | Type b, ΔhgpA Rbr | This study |
| HI1712 | Type b, ΔhgpB Tcr | This study |
| HI1713 | Type b, ΔhgpC Spr | This study |
| HI1714 | Type b, ΔhgpA ΔhgpB Rbr Tcr | This study |
| HI1715 | Type b, ΔhgpA ΔhgpC Rbr Spr | This study |
| HI1716 | Type b, ΔhgpB ΔhgpC Tcr Spr | This study |
| HI1717 | Type b, ΔhgpA ΔhgpB ΔhgpC Rbr Tcr Spr | This study |
| E. coli | ||
| TOP10 | Used for cloning in pCR2.1-TOPO | Invitrogen |
| INVαF′ | Used for cloning in pUC19N and pUC19N-derived plasmids | Invitrogen |
| Plasmids | ||
| pCR2.1-TOPO | Plac, lacZα, Kanr Ampr, ColE1 origin, F1 origin, T7 promoter | Invitrogen |
| pUC19N | pUC19 with a NotI linker added at the HindIII end of the polylinker | 23 |
| pUCTSTE | pUC19 containing a 2.2-kbp Rbr cassette | 20 |
| pGESYII | pACYC177 containing a 2.8-kbp Tcr cassette | 18 |
| pSPECR | pCR-Blunt containing a 1.2-kbp Spr cassette | 25 |
| pHGPC | pCR2.1-TOPO, carrying a 5.5-kbp PCR product from H. influenzae HI689 which includes hgpC | This study |
| pHGPCSPEC | pHGPC with insertion of a PstI-digested 1.2-kbp Spr marker at a NsiI site internal to hgpC | This study |
| pQM | pCRII carrying a 3.2-kbp PCR product corresponding to hgpB | 18 |
| pA1A4 | pUC19N containing a PCR product of ∼1,200 bp corresponding to a region upstream of hgpA and PCR product of ∼800 bp corresponding to a region downstream of hgpA with a unique BamHI site between the two inserts | This study |
| pA1A4TSTE | pA1A4 with the 2.2-kbp Rbr marker from pUCTSTE inserted at the unique BamHI site | This study |
| pB1B4 | pUC19N containing a PCR product of ∼900 bp corresponding to a region upstream of hgpB and a PCR product of ∼900 bp corresponding to a region downstream of hgpB with a unique BamHI site between the two inserts | This study |
| pB1B4GESY | pB1B4 with the 2.8-kbp Tcr marker from pGESYII inserted at the unique BamHI site | This study |
| pC1C4 | pUC19N containing a PCR product of ∼1,100 bp corresponding to a region upstream of hgpC and a PCR product of ∼1,150 bp corresponding to a region downstream of hgpC with a unique BamHI site between the two inserts | This study |
| pC1C4SPEC | pC1C4 with the 1.2-kbp Spr marker from pSPECR inserted at the unique BamHI site | This study |
Rbr, ribostamycin resistance (15 μg/ml in H. influenzae); Tcr, tetracycline resistance (3 μg/ml in H. influenzae); Spr, spectinomycin resistance (150 μg/ml in H. influenzae); Kanr kanamycin resistance (50 μg/ml in E. coli); Ampr, ampicillin resistance (50 μg/ml in E. coli).
DNA isolation.
Bacterial genomic DNA was isolated with the DNA Now reagent (Biogentex, Seabrook, Tex). Plasmid DNA was isolated with plasmid kits (Qiagen, Chatsworth, Calif.) as directed by the manufacturer. DNA concentrations were assessed spectrophotometrically with a UV-1201S spectrophotometer and a DNA/protein program pack (Shimadzu, Kyoto, Japan).
Cloning of the HI0712 homologue from strain HI689.
A pair of primers, HGPCSPEC1 and HGPCSPEC4 (Table 2), were designed for use in the PCR based on the RdKW20 genomic sequence. This primer pair was designed to amplify the entire coding sequence of the HI0712 homologue from strain HI689 plus approximately 1,100 bp upstream and 1,150 bp downstream of the ORF. Reactions were performed in a 50-μl volume with 100 ng of H. influenzae HI689 chromosomal DNA as template, and the mixtures contained 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 10 pmol of each primer, and 2 U of Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.). PCR was carried out for 30 cycles, with each cycle consisting of denaturation at 95°C for 1 min, annealing at 64°C for 1 min, and primer extension at 72°C for 6 min, with one final extension of 10 min. An approximately 5.5-kbp amplicon was directly ligated into the TA cloning vector pCR2.1-TOPO (Invitrogen, San Diego, Calif.). The ligated vector was transformed into E. coli TOP10 competent cells (Invitrogen), and recombinants were selected on Luria-Bertani agar containing 50 μg of ampicillin per ml. A plasmid of the correct construct was identified and designated pHGPC.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| HGPATSTE1 | 5′-CCGCCGGAATTCGATCCGAACACAACTATCCC-3′ |
| HGPATSTE2 | 5′-CGCCGCGGTACCTAACGGAATAGGCAAGCACG-3′ |
| HGPATSTE3 | 5′-CCGCCGTCTAGAGTTGTAAAAATAAAATACG-3′ |
| HGPATSTE4 | 5′-TTGAAAGCGGCCGCCAAATTATGTTCATAAAGC-3′ |
| HGPBGESY1A | 5′-CCGGCCGAGCTCGAGGAACAATCGACAGGG-3′ |
| HGPBGESY2A | 5′-ATAATAGGATCCCAGGATTAGGCATTAACCG-3′ |
| HGPBGESY3 | 5′-AACGGCGGATCCCACCCTACATTAAGTACGC-3′ |
| HGPBGESY4 | 5′-TTGAAAGCGGCCGCGTTATCTAAAAAGCTACTGC-3′ |
| HGPCSPEC1 | 5′-ATAATAGAATTCGAAACTTGGTTACCTCACGCC-5′ |
| HGPCSPEC2A | 5′-CCTTTAGGATCCGCTATGTAAAAATAATCCGC-5′ |
| HGPCSPEC3 | 5′-GTCTTAGGATCCGAAAATCATTTTTGACCGC-3′ |
| HGPCSPEC4 | 5′-GCGGGTAAGCTTGAAACTGGTACATCAATCGG-3′ |
The insert of pHGPC was sequenced by automated sequencing (ABI model 373A) at the Recombinant DNA/Protein Resource Facility, Oklahoma State University, Stillwater, Okla.
Construction of an insertion mutation in hgpC.
The coding sequence of hgpC contains a single NsiI site at nucleotide 2408. Since NsiI produces cohesive ends compatible with those produced by PstI, the NsiI site was used for insertion of the PstI-excised spectinomycin resistance cassette from pSPECR (25). Plasmid pHGPC was linearized with NsiI and, following gel purification, was ligated with the 1.2-kbp spectinomycin resistance marker excised from pSPECR by using PstI. A plasmid of the correct construct was identified and designated pHGPCSPEC. Competent H. influenzae HI689 was transformed with pHGPCSPEC, and recombinants were selected on sBHI with 200 μg of spectinomycin per ml. Plasmid pHGPCSPEC was similarly transformed into the previously described hgpA single mutant (HI1705, previously designated HI689hgpAΔBglII), the hgpB single mutant (HI1706, previously designated HI689hgpBΔBclI), and the hgpA hgpB double mutant (HI1708, previously designated HI689hgpAΔBglII hgpBΔBclI) (18) to yield corresponding double and triple mutants. Appropriate chromosomal rearrangements were confirmed by Southern blot analyses.
Construction of complete deletions of hgpA, hgpB, and hgpC.
Complete deletions of hgpA, hgpB, and hgpC and replacement of the gene with an antibiotic resistance cassette were performed as described below. The method as it pertains specifically to deletion of hgpA is outlined in Fig. 1; it was essentially the same for deletion of hgpB and hgpC.
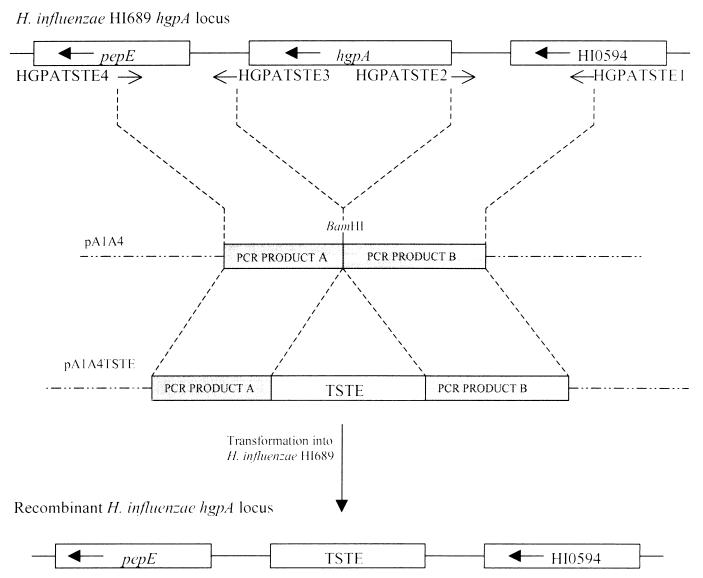
FIG. 1.
Construction of an H. influenzae HI689 mutant with a complete deletion of hgpA. Two PCR products, one encompassing a region upstream of hgpA and the second a region downstream of hgpA, were amplified with the primer pairs HGPATSTE1 plus HGPATSTE2 and HGPATSTE3 plus HGPATSTE4, respectively (Table 2). The first primer pair added EcoRI and KpnI sites to the ends of the PCR product, and the second primer pair added XbaI and NotI sites, to facilitate directional cloning. PCR products were initially cloned into the TA cloning vector pCR2.1-TOPO and then, utilizing the added restriction sites, were sequentially subcloned into pUC19N to yield pA1A4. The 2.2-kbp ribostamycin resistance marker (TSTE) was inserted at the single BamHI site between the cloned PCR products in pA1A4, to yield the mutagenic plasmid construct pA1A4TSTE. The construct pA1A4TSTE was used to transform H. influenzae HI689 to ribostamycin resistance. The wild-type hgpA locus is shown, and the direction of gene transcription is indicated. HI0594 refers to an ORF with no assigned function identified in the RdKW20 genome-sequencing project (4). The positions and direction of primers used in the PCR are indicated. The recombinant H. influenzae locus is shown where replacement of hgpA by the antibiotic resistance cassette TSTE has been achieved.
To delete hgpA, four primers were designed based on the nucleotide sequence of the regions flanking the gene (8, 9) for use in the PCR (Table 2; Fig. 1) with HI689 chromosomal DNA as template. Primer pair HGPATSTE1 and HGPATSTE2 amplified a product upstream of the gene (the PCR as above, except that annealing took place at 56°C and the extension time was 1 min). The second primer pair, HGPATSTE3 and HGPATSTE4, was designed to amplify a product downstream of hgpA (annealing at 42°C). The PCR products were cloned into pUC19N to yield pA1A4. Plasmid pA1A4 comprised upstream and downstream sequences of hgpA abutting each other, with a small portion of the pUC19N polylinker, notably the BamHI site, remaining in the middle. The residual BamHI site was used for insertion of the aminoglycoside resistance-encoding TSTE marker (20) to yield pA1A4TSTE. Competent H. influenzae HI689 was transformed with pA1A4TSTE, and recombinants were selected on sBHI with 15 μg of ribostamycin per ml.
For hgpB, the primers were designed based on the sequences flanking the ORF designated HI0661 in the RdKW20 genome (4). Separate PCRs were performed with primer pairs HGPBGESY1A plus HGPBGESY2A (annealing at 56°C) and HGPBGESY3 plus HGPBGESY4 (annealing at 53°C) with HI689 chromosomal DNA as the template. The first primer pair added SacI and BamHI sites to the PCR product, and the second added BamHI and NotI sites. The added sites were used to sequentially subclone the inserts into pUC19N to yield pB1B4. pB1B4 was subsequently linearized with BamHI, and the BamHI-excised tetracycline resistance marker from pGESYII was inserted to yield pB1B4TET. Competent H. influenzae HI689 was transformed with pB1B4TET and selected on sBHI with 3 μg of tetracycline per ml.
For hgpC, two PCRs were performed with primer pairs HGPCSPEC1 plus HGPCSPEC2A (annealing at 56°C) and HGPCSPEC3 plus HGPCSPEC4 (annealing at 54°C). The first primer pair added EcoRI and BamHI sites to the PCR product, and the second primer pair added BamHI and HindIII sites. The added sites were used to sequentially subclone the PCR products into pUC19N to yield pC1C4. pC1C4 was linearized with BamHI, and the BamHI-excised spectinomycin resistance marker from pSPECR was inserted to yield pC1C4SPEC. Competent H. influenzae HI689 was transformed with pC1C4SPEC and selected on sBHI with 150 μg of spectinomycin per ml.
The corresponding double and triple complete deletion mutants were constructed by sequential transformations followed by selection on appropriate antibiotics. Chromosomal integration was confirmed by Southern analyses with the coding region of hgpB and the antibiotic resistance marker cassettes as probes. Additional mutant characterization was achieved by amplifying the mutated locus from each mutant strain and sequencing across the deletion junctions. The primer pairs HGPATSTE1 plus HGPATSTE4, HGPBGESY1A plus HGPBGESY4, and HGPASPEC1 plus HGPCSPEC4 were used to amplify the mutated hgpA, hgpB, and hgpC loci, respectively. The PCR products were cloned and sequenced.
Southern blot and DNA hybridization.
DNA was digested with restriction enzymes as directed by the manufacturers, separated on agarose gels (0.5 or 0.8% [wt/vol] agarose) in TBE buffer (0.045 M Tris-borate, 0.001 M EDTA), and transferred to Magnagraph nylon membranes (MSI, Westbrook, Mass.) by the method of Southern as described by Sambrook et al. (19).
The enhanced chemiluminescence (ECL) 3′-oligolabeling system (Amersham) was used as directed by the manufacturer to label the 3′ end of the oligonucleotides. The ECL random-prime labeling kit (Amersham) was used as directed by the manufacturer to label DNA for use as probes. Labeled oligonucleotides or DNA were used to probe Southern blots. Following stringency washing, hybridization was detected with the ECL nucleic acid detection reagents (Amersham) as directed by the manufacturer. Blots were subsequently exposed to X-ray film (Fuji Photo Film Co., Tokyo, Japan).
Growth studies with H. influenzae.
H. influenzae strains were grown overnight in hrBHI broth and then subcultured at a 10% inoculum into hdBHI broth. After incubation for 3 h, the cultures were standardized to the same optical density at 605 nm and used to inoculate fresh hdBHI medium (0.5% inoculum). The cultures were supplemented with human hemoglobin at the specified concentrations or the human hemoglobin-haptoglobin complex (1 μg of hemoglobin equivalent per ml). Hemoglobin-haptoglobin complex was made by mixing hemoglobin with haptoglobin in a ratio of 1:2 (wt/wt) at room temperature for 30 min. Growth to stationary phase was monitored with a Shimadzu UV-1201S spectrophotometer and by measurement of the optical density at 605 nm.
In some growth studies bacteria were passaged through hemoglobin-haptoglobin prior to initiation of the growth study. Passage was performed as follows. An overnight culture of bacteria in sBHI was subcultured at a 0.1% inoculum into 5 ml of hdBHI broth. Following growth overnight to induce heme restriction, the bacteria were harvested by centrifugation and resuspended in 2 ml of hdBHI broth supplemented with human hemoglobin-haptoglobin complex (1 μg/ml hemoglobin equivalent) and incubated overnight. The overnight culture was subsequently used to initiate growth studies as described above.
Dot blot assay.
Binding of hemoglobin to H. influenzae was determined by the dot blot assay with biotinylated human hemoglobin as previously described (8). For some assays, the organisms were preincubated with trypsin (1 mg/ml) as previously described (16).
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence of the insert of pHGPC is AF094574.
RESULTS
Cloning and sequencing of a DNA fragment homologous to HI0712.
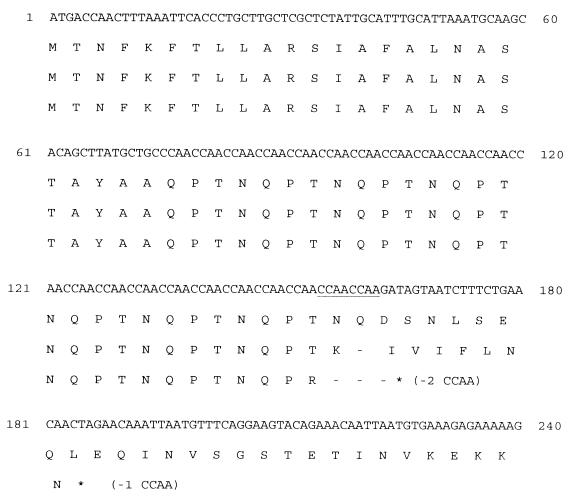
We previously cloned two hemoglobin/hemoglobin-haptoglobin-binding protein genes (hgpA and hgpB) from H. influenzae HI689 (8, 18). Insertional mutation of these genes, either alone or together, did not abrogate the ability of the mutant to bind or utilize either heme source (18). Affinity isolation of hemoglobin-binding proteins from the double mutant identified a third potential hemoglobin-binding protein as a homologue of either of the RdKW20 ORFs designated HI0635 and HI0712 (18). Southern analyses demonstrated that strain HI689 contained a gene homologous to HI0712 but not one homologous to HI0635 (15). Primers were designed based on the RdKW20 genomic sequence to amplify the homologue of HI0712 in strain HI689 and the flanking DNA. The primers amplified a product of the expected size from strain HI689, and the product was cloned into pCR2.1-TOPO, to yield pHGPC. Automated sequencing of the insert of pHGPC revealed an insert of 5,489 bp. The proposed gene includes a length of CCAA nucleotide repeats immediately following the sequence encoding a putative leader peptide (Fig. 2). The previously identified hemoglobin/hemoglobin-haptoglobin-binding protein genes, hgpA and hgpB, also contain these lengths of CCAA repeats (8, 18). Variation in the length of these CCAA repeating units can occur in H. influenzae (9) and presumably leads to variable expression of the encoded proteins. The insert of pHGPC contains a length of 21 CCAA repeating units resulting in a stop codon immediately downstream of the CCAA repeats. However, a hypothetical alteration in the length of the CCAA repeats by the addition of one unit would lead to expression of a mature protein (Fig. 2). The predicted protein consists of 1,011 amino acids preceded by a 23-residue leader peptide. The molecular mass of the mature protein was calculated to be 120,060 Da. The predicted protein was highly homologous to other bacterial iron and heme uptake-related proteins, particularly over the seven regions considered indicative of a TonB-dependent protein (12), including an identifiable “TonB box” starting at amino acid 40 of the mature protein and having the amino acid sequence EQINVSGSTETIN (12, 14). The predicted protein was highly homologous to the products of the RdKW20 ORFs designated HI0712 (90% similarity and 88% identity) and HI0635 (87% similarity and 84% identity). Homology to HgpA (57% similarity and 50% identity) and HgpB (59% similarity and 51% identity) was also significant. In addition, a hemoglobin-binding protein was isolated from an H. influenzae HI689 double mutant mutated in hgpA and hgpB and the amino acid sequence was obtained for three internal peptide fragments (18). The sequenced fragments from the previously purified protein have 100% amino acid sequence identity to the corresponding regions of the predicted protein identified here.
FIG. 2.
Nucleotide sequence of the portion of hgpC encoding the N-terminal region of the protein. The nucleotide sequence as shown contains 22 CCAA repeats; one CCAA unit has been added to the sequence obtained from the clone pHGPC to bring the gene into frame. The introduction of stop codons following the theoretical removal of one or two CCAA repeat units is shown. The underlined CCAA repeats are those that have been removed.
Based on homology of the gene product at the amino acid level to HgpA and HgpB and the previously reported isolation of a protein corresponding to this gene by affinity isolation with hemoglobin as the primary ligand (18), we designated the gene hgpC.
Construction of a mutant with an insertion mutation in hgpC.
To investigate the potential role of hgpC in acquisition of hemoglobin and/or hemoglobin-haptoglobin, an insertion mutation in hgpC was constructed. The NsiI site internal to hgpC was used to insert the spectinomycin marker from pSPECR into plasmid pHGPC to generate pHGPCSPEC. Plasmid pHGPCSPEC was used to transform HI689 to spectinomycin resistance. One spectinomycin-resistant HI689 colony was selected for further investigation. Southern hybridization with an HI0712-specific oligonucleotide probe (HI0712PROBE) and the labeled spectinomycin resistance marker confirmed that a single insertion of the spectinomycin marker had occurred at the correct site (data not shown). The mutant strain was designated HI1706. Plasmid pHGPCSPEC was used to transform the previously described hgpA and hgpB insertion-deletion mutants and the corresponding hgpA hgpB double mutant (18) to spectinomycin resistance. Southern analyses confirmed that the anticipated recombination events had occurred (data not shown).
Characterization of single, double, and triple mutants.
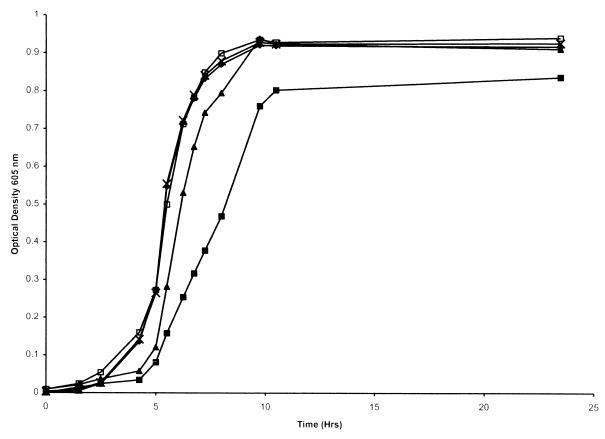
To determine the effect of insertional mutation of hgpC alone or in combination with insertion-deletion mutations of hgpA and hgpB, growth studies were performed. Previous studies demonstrated that insertion-deletion mutations in hgpA and hgpB alone had no effect on the utilization of either hemoglobin or hemoglobin-haptoglobin (18). An hgpA hgpB double mutant was also unaffected in the ability to utilize hemoglobin, although it initially demonstrated a reduced ability to use the hemoglobin-haptoglobin complex. However, passage of the double mutant through hemoglobin-haptoglobin before performing the growth studies restored growth to levels shown by the wild-type strain (18).
The single hgpC insertion mutant (HI1706) and the hgpB hgpC double mutant (HI1709) grew essentially the same as the wild type when hemoglobin-haptoglobin was the sole heme source (Fig. 3). However, the hgpA hgpC double mutant (HI1708) grew significantly slower and to a lower final density than the wild-type strain in hemoglobin-haptoglobin (Fig. 3). Following passage of the hgpA hgpC double mutant through hemoglobin-haptoglobin, growth with hemoglobin-haptoglobin as the sole heme source was restored to levels seen with the wild-type strain (Fig. 3). Growth of the hgpA hgpC double mutant was similar to growth of the hgpA hgpB double mutant (18). We have hypothesized that on initial isolation of the double-mutant strain, the majority of the population contains a copy of the remaining locus (either hgpB or hgpC) which is out of frame across the CCAA repeats. Since the remaining gene is essential for growth in media with hemoglobin-haptoglobin as the sole heme source, only the small portion of organisms which have an in-frame copy of the gene would grow, resulting in detection of apparently slower growth of the mutant compared to the wild-type strain. Passage through hemoglobin-haptoglobin would select for a population having an in-frame copy of the remaining gene; the growth characteristics of this population are similar to those of the wild-type strain. The hgpB hgpC double mutant does not exhibit the same pattern. This may result from hgpA being the gene that is predominantly in frame in the wild-type strain; thus, the hgpB hgpC double mutant would approximate the genotype of the wild-type strain and no significant growth defect would be noted. Since no information is available on the in-frame gene complement of wild-type HI689, this explanation for the growth characteristics of the hgpB hgpC double insertion mutant remains unproven.
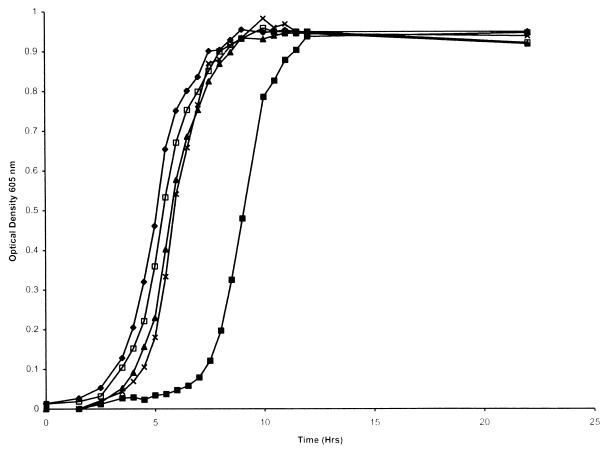
FIG. 3.
Growth of wild-type H. influenzae and insertional mutants in hdBHI with hemoglobin-haptoglobin as the heme source. Results for H. influenzae HI689 (×), the hgpC insertion mutant HI1706 (⧫), the hgpB hgpC double mutant HI1709 (▴), the hgpA hgpC double mutant HI1708 (■), and the hgpA hgpC double mutant HI1708 following passage through hemoglobin-haptoglobin (□) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
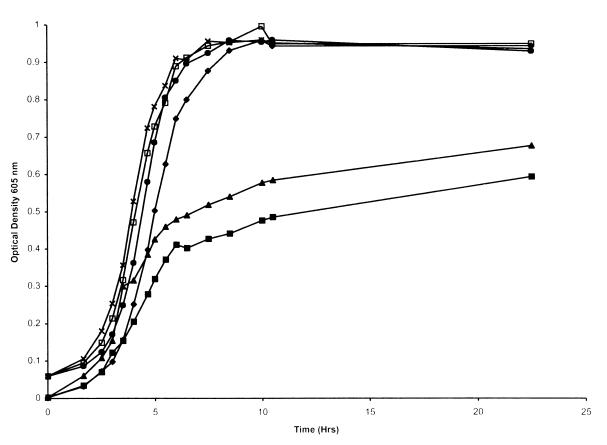
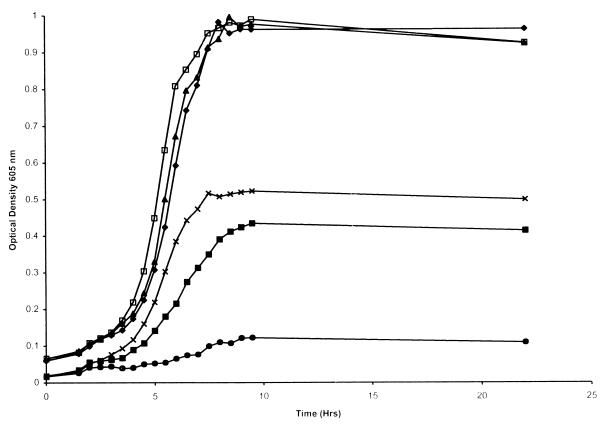
Growth studies with the hgpA hgpB hgpC triple-insertion mutant (H1710) showed that utilization of hemoglobin-haptoglobin was reduced compared to that by the wild type (Fig. 4). This defect was specific for hemoglobin-haptoglobin rather than being a general growth defect, since growth of the triple mutant in sBHI was identical to that of the wildtype strain (Fig. 4). Residual utilization of the complex by the triple mutant might result from an additional utilization mechanism. Alternatively, the portions of hgpA, hgpB, and hgpC remaining in the triple mutant constructed by insertional mutagenesis may retain some utilization capacity. The insertional mutagenesis protocol in each case left the “TonB box” intact. The TonB box is a short amino acid sequence which mediates the interaction between TonB and TonB-dependent receptors (14) and would presumably be essential for the correct function of these proteins. Passage of the triple insertion mutant through hemoglobin-haptoglobin resulted in an increased ability of the mutant to utilize the complex (Fig. 4). These data may reflect the second possibility, since passage through hemoglobin-haptoglobin would presumably result in a population with at least one hgp gene in frame in every organism. The wild-type strain similarly grows slightly better following passage through hemoglobin-haptoglobin than does the unpassaged wild-type strain (18).
FIG. 4.
Growth of wild-type H. influenzae and the triple-insertion mutant in sBHI or in hdBHI with hemoglobin-haptoglobin as the heme source. Results for H. influenzae HI689 in sBHI (●) and in hdBHI with hemoglobin-haptoglobin (⧫), the hgpA hgpB hgpC triple mutant HI1710 in sBHI (×) and in hdBHI with hemoglobin-haptoglobin (■), and the hgpA hgpB hgpC triple mutant HI1710, following passage through hemoglobin-haptoglobin, in sBHI (□) and in hdBHI with hemoglobin-haptoglobin (▴) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
None of the mutants discussed above were altered in the ability to utilize hemoglobin (1 μg/ml) as a source of heme (data not shown). The continued utilization of hemoglobin by the triple-insertion mutant, in particular, could indicate that HgpA, HgpB, and HgpC, although designated hemoglobin/hemoglobin-haptoglobin-binding proteins, in fact play no role in the utilization of hemoglobin and that their isolation in a hemoglobin affinity purification protocol (8, 18) may be an artifact resulting from their actual role in hemoglobin-haptoglobin binding. Alternatively, portions of the proteins remaining in the triple mutant could be sufficient for hemoglobin utilization, or there may be an additional hemoglobin utilization mechanism(s). We have previously shown that HgpA binds both hemoglobin and the hemoglobin-haptoglobin complex when expressed in E. coli (8, 9). Maciver et al. cloned a gene encoding a homologue of HgpA from the H. influenzae nontypeable strain TN106, which they designated hhuA (13). Based on homology and chromosomal locus, hgpA and hhuA are alleles of the same gene in different strains (9). Since hemoglobin-haptoglobin binding by HhuA was demonstrated in E. coli by using a fusion protein lacking approximately the first 150 amino acids of the mature protein (13), it is possible that the N-terminal portion of the protein is essential for hemoglobin binding.
Construction of mutants with complete deletion mutations of hgpA, hgpB, and hgpC.
To further investigate the role(s) of hgpA, hgpB, and hgpC in acquisition of hemoglobin and/or hemoglobin-haptoglobin, mutants with complete deletion mutations of each of the three genes were constructed. The strategy for mutant construction is outlined in Fig. 1 and resulted in complete deletion of each structural gene and replacement with an antibiotic resistance cassette. Southern hybridization with the coding sequence of hgpB as a probe (18) confirmed that deletion of the genes had occurred as proposed (Fig. 5). The labeled hgpB hybridized with three bands in an EcoRI digest of HI689 chromosomal DNA (data not shown). Each of the single mutants exhibited the loss of one band corresponding to the deletion of hgpA, hgpB, or hgpC as appropriate (Fig. 5, lanes A to C). Hybridization of the labeled probe was detected with only one band in each of the double mutants, and no hybridization with DNA from the triple mutant was detected (lanes D to G). Chromosomal DNA from each of the mutants was also probed with each of the labeled antibiotic resistance cassettes to confirm insertion of the marker as appropriate (data not shown). The mutants were additionally characterized by sequencing across the deletion junctions and the flanking regions. Sequence data from each of the single mutants and the triple mutant indicated that the flanking regions used for recombination were intact and that each gene had been deleted as anticipated (data not shown).
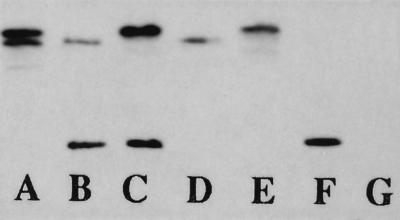
FIG. 5.
Southern analysis of H. influenzae HI689 and complete deletion derivatives with a universal probe for the three hgp genes. Lanes: A, H. influenzae hgpA mutant HI1711 chromosomal DNA EcoRI digest; B, H. influenzae hgpB mutant HI1712 chromosomal DNA EcoRI digest; C, H. influenzae hgpC mutant HI1713 chromosomal DNA EcoRI digest; D, H. influenzae hgpA hgpB mutant HI1714 chromosomal DNA EcoRI digest; E, H. influenzae hgpA hgpC mutant HI1715 chromosomal DNA EcoRI digest; F, H. influenzae hgpB hgpC mutant HI1716 chromosomal DNA EcoRI digest; G, H. influenzae hgpA hgpB hgpC mutant HI1717 chromosomal DNA EcoRI digest. The hybridizing bands in the H. influenzae digests correspond, from top to bottom, to hgpB, hgpC, and hgpA, respectively.
Characterization of the complete-deletion mutants.
Growth studies were performed to characterize the effect of complete deletion of hgpA, hgpB, and hgpC on utilization of hemoglobin-haptoglobin and hemoglobin.
The hgpB and hgpC single-deletion mutants grew comparably to the wild-type strain in hemoglobin-haptoglobin. Although the hgpA mutant was significantly delayed in entering the log phase, it grew at the same rate and to the same final density as the wild type did (Fig. 6). Passage of the hgpA single mutant through hemoglobin-haptoglobin restored growth to levels similar to those of the wild type (Fig. 6). Similarly, the hgpB hgpC double mutant grew as well as the wild-type strain while the hgpA hgpC double mutant was delayed in entering the log phase but grew at the same rate and to the same final density in the period examined (Fig. 7). The hgpA hgpB double mutant grew more slowly and to a lower final density than the wild type did (Fig. 7). Passage of the double mutants through hemoglobin-haptoglobin restored growth to levels comparable to those of the wild type (data not shown). The mutant in which all three genes had been deleted did not grow significantly with hemoglobin-haptoglobin as the sole heme source (Fig. 8). When the triple-deletion mutant was taken from late in the growth study and used in subsequent growth curve determinations, there was no change in the growth characteristics (data not shown). These data demonstrate that all three of the genes under investigation mediate hemoglobin-haptoglobin utilization and that any single gene is sufficient for utilization of this heme source.
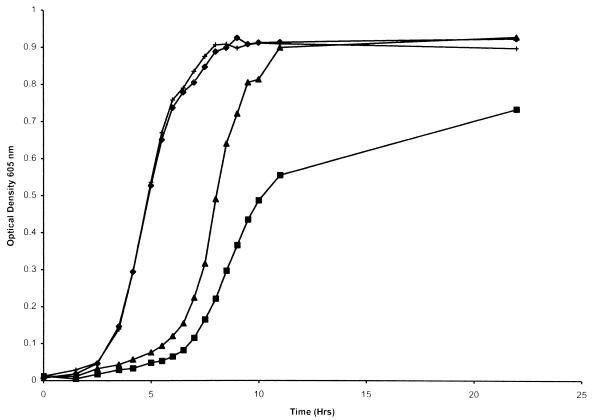
FIG. 6.
Growth of wild-type H. influenzae and the single-deletion mutants in hdBHI with hemoglobin-haptoglobin as the heme source. Results for H. influenzae HI689 (⧫), the hgpA deletion mutant HI1711 (■), the hgpA deletion mutant HI1711 following passage through hemoglobin-haptoglobin (□), the hgpB deletion mutant HI1712 (▴), and the hgpC deletion mutant HI1713 (×) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
FIG. 7.
Growth of H. influenzae in hdBHI with hemoglobin-haptoglobin as the heme source. Results for H. influenzae HI689 (⧫), the hgpA hgpB double-deletion mutant HI1714 (■), the hgpA hgpC double-deletion mutant HI1715 (▴), and the hgpB hgpC double-deletion mutant HI1716 (+) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
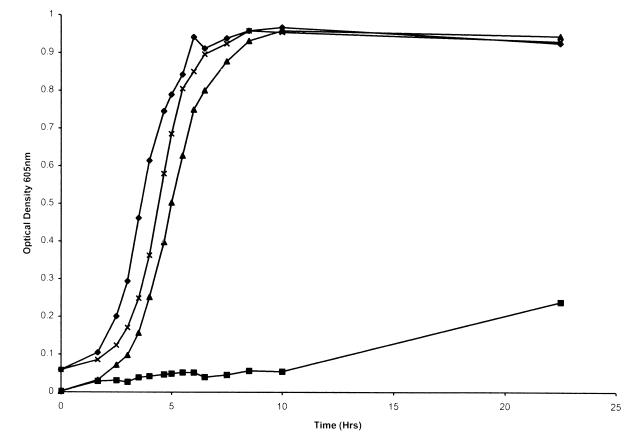
FIG. 8.
Growth of H. influenzae in sBHI or in hdBHI with hemoglobin-haptoglobin as the heme source. Results for H. influenzae HI689 in sBHI (×) and in hdBHI with hemoglobin-haptoglobin (▴) and the hgpA hgpB hgpC triple-deletion mutant HI1717 in sBHI (⧫) and in hdBHI with hemoglobin-haptoglobin (■) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
All of the deletion mutants grew as well as the wild type when hemoglobin at 1 μg/ml was present as the sole heme source (data not shown). Because each of the three proteins (HgpA, HgpB, and HgpC) binds hemoglobin either in dot blot binding assays or by purification in a hemoglobin affinity purification protocol (8, 18), they may play a role in hemoglobin utilization. To further investigate this question, we performed growth studies with concentrations of hemoglobin which were limiting for growth of the wild-type strain. When grown with concentrations of hemoglobin in the range of 31.25 to 125 ng/ml, the wild-type strain grew significantly better than the hgpA hgpB hgpC complete-deletion mutant did (Fig. 9). None of the single-deletion mutants (HI1711 to HI1713) or the hgpB hgpC double mutant (HI1716) were altered in the ability to utilize hemoglobin at 100 ng/ml compared to the wild-type strain. The hgpA hgpB and hgpA hgpC double-deletion mutants (HI1714 and HI1715) showed a slightly reduced ability to utilize hemoglobin at 100 ng/ml compared to the wild-type strain in some experiments, although in other experiments there was no obvious difference (data not shown). Due to the minor and inconsistent growth differences in these latter cases, it is not possible to determine whether the growth defect is genuine or whether such a growth defect would be corrected by passage through the heme source as for hemoglobin-haptoglobin utilization.
FIG. 9.
Growth of H. influenzae in sBHI or in hdBHI with hemoglobin (100 ng/ml) as the heme source. Results for H. influenzae HI689 in sBHI (⧫) and in hdBHI with hemoglobin (■), the hgpA hgpB hgpC triple-insertion mutant HI1710 in sBHI (▴) and in hdBHI with hemoglobin (×), and the hgpA hgpB hgpC triple-deletion mutant HI1717 in sBHI (□) and in hdBHI with hemoglobin (●) are shown. Each growth condition was used on at least two separate occasions in triplicate, and a representative curve is shown for each.
Since the difference in hemoglobin utilization became apparent only at limiting concentrations of hemoglobin, growth studies with the triple-insertion mutant (HI1710) were repeated with 100 ng of hemoglobin per ml. HI1710 consistently appeared to utilize hemoglobin better than the wild-type strain (Fig. 9).
Since deletion of hgpA, hgpB, and hgpC did not completely abolish the ability to utilize hemoglobin, hemoglobin-binding dot-blot studies were performed to determine whether the mutant retained the ability to bind hemoglobin. The hgpA hgpB hgpC triple-deletion mutant bound hemoglobin as well as the wild-type strain did (data not shown). To determine whether the hemoglobin binding of both the wild-type strain and the triple-deletion mutant was mediated through a surface-exposed protein, dot blots were performed with bacteria which had been preincubated with trypsin. Pretreatment with this proteolytic enzyme abolished hemoglobin binding by both the wild-type and triple-deletion mutant (HI1717) strains (data not shown). The organisms pretreated with trypsin remained viable, as determined by subsequent subculture on sBHI agar. These data indicate that hemoglobin binding in both the wild-type and triple-deletion mutant strains is mediated through a surface-exposed protein.
DISCUSSION
We have previously cloned two genes, hgpA and hgpB, encoding hemoglobin and hemoglobin-haptoglobin-binding proteins from H. influenzae HI689 (8, 18). A strain containing insertional mutations in both hgpA and hgpB grew less well than the wild-type strain when hemoglobin-haptoglobin complex was used as the heme source (18). However, the defect in the double mutant was reversed by passage of the mutant strain through media with hemoglobin-haptoglobin as the sole heme source (18). After selective passage, the double mutant also yielded an increased quantity of an approximately 120-kDa protein, compared to the unpassaged mutant strain, in a hemoglobin affinity protocol (18). The affinity purified protein was shown by amino acid sequencing of internal peptides and Southern hybridization analyses to be a homologue of the RdKW20 ORF HI0712, and was subsequently termed hgpC (15, 18). A common feature of hgpA, hgpB, and hgpC is a length of CCAA nucleotide repeats immediately following the sequence encoding the leader peptide. These CCAA nucleotides are believed to be involved in regulation of expression of the genes through a slipped-strand mispairing mechanism analagous to phase variation of the H. influenzae lipooligosaccharide through variation in length of a CAAT repeat region (6). We have recently demonstrated that alteration in the CCAA region of hgpA occurs in H. influenzae and is associated with alteration in gene expression (unpublished data). Based on the slip-strand hypothesis, we proposed that the initial hgpA hgpB double-mutant isolate possessed a copy of hgpC that was out of frame in most of the population. If hgpC were essential for growth of the double mutant when hemoglobin-haptoglobin was the sole heme source, passage through this heme source would result in selection of a population where hgpC was in frame, thus correcting the growth defect. Phase variation of H. influenzae hemoglobin- and hemoglobin-haptoglobin-binding proteins may provide a mechanism for evasion of the host immune system or may lead to expression of different proteins with different affinities for specific heme sources in different host microenvironments. Phase variation of the Neisseria gonorrhoeae hemoglobin utilization operon, hpuA and hpuB, has been demonstrated and is mediated through alterations in a length of guanine residues within the hpuA coding sequence (2).
This report demonstrates that H. influenzae type b strain HI689 possesses three highly homologous hemoglobin- and hemoglobin-haptoglobin-binding proteins. To define the function(s) of the three genes, two series of mutants were constructed. One series of mutants contained insertion mutations with the N-terminal region of each gene intact, and the second series contained complete deletions of the structural genes. Growth studies revealed different growth characteristics for the two sets of mutants.
The triple insertion mutant (HI1710) demonstrated a reduced ability to use hemoglobin-haptoglobin, while the triple deletion mutant (HI1717) was unable to utilize this heme source. It is possible that the residual portion(s) of one or more of the genes is sufficient to allow utilization of hemoglobin-haptoglobin, although utilization is not as efficient as when an entire gene is present. One of the CCAA-containing genes (ORF HI1566) of strain RdKW20 contains an internal stop codon, unrelated to the CCAA length; based on our findings, the truncated protein may retain function (4). Whether the residual portion of each of the genes alone is sufficient to allow the utilization of hemoglobin-haptoglobin is unclear, and construction of further mutants involving combinations of the insertion and deletion mutations is necessary to clarify this point. However, it is clear that each of the complete genes alone is sufficient to support growth in hemoglobin-haptoglobin since each of the deletion mutants (HI1714 to HI1716) grows as well as the wild-type strain in this heme source. An alternative explanation for the growth characteristic differences between the two classes of triple mutant would be polar effects due to the deletion mutations. This explanation is unlikely, however, since none of the single-deletion mutants (HI1711 to HI1713) or the double-deletion mutants (HI1714 to HI1716) has a growth deficiency.
In the case of growth in hemoglobin, the triple-deletion mutant (HI1717) exhibited a growth defect only when the hemoglobin concentration was reduced to levels which were limiting for growth of the wild-type strain. These data indicate that additional hemoglobin utilization mechanisms exist in H. influenzae. To determine whether these additional hemoglobin utilization mechanisms involved cell surface binding of hemoglobin, dot blot assays were performed. The triple mutant (HI1717) bound hemoglobin as well as the wild-type strain did. Binding of hemoglobin to both the wild-type and mutant strains was abolished by preincubation of the organisms with trypsin, indicating that binding is mediated via a surface-exposed protein moiety. No candidate hemoglobin-binding protein was identified by the hemoglobin affinity purification method with the triple-deletion mutant (data not shown). These data may indicate weak affinity between the additional putative hemoglobin-binding protein and its ligand. We have been unable to reduce the stringency of the washes used in our protocol without compromising the specificity of the purification.
The triple-insertion mutant (HI1710) appeared to grow slightly better than the wild-type strain in limiting concentrations of hemoglobin; however, the relevance of this observation is not clear.
In conclusion we have identified three hemoglobin/hemoglobin-haptoglobin-binding proteins (HgpA, HgpB, and HgpC) in H. influenzae HI689. Expression of any one of the three proteins is sufficient for utilization of hemoglobin-haptoglobin, while expression of at least one is essential for utilization of this heme source. The three proteins are not essential for hemoglobin utilization, and an additional surface-exposed hemoglobin binding protein(s) apparently exists in H. influenzae. Further studies will identify the additional hemoglobin-binding protein(s) and determine the hemoglobin utilization functional regions of HgpA, HgpB, and HgpC.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI29611 from the National Institute of Allergy and Infectious Diseases to T.L.S. and by health research contract HN5-055 from the Oklahoma Center for the Advancement of Science and Technology to D.J.M. We also acknowledge the support of the Children’s Medical Research Institute.
REFERENCES
- 1.Bezkorovainy A. Iron proteins. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological and clinical aspects. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 27–68. [Google Scholar]
- 2.Chen C J, Elkins C, Sparling P F. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans N M, Smith D D, Wicken A J. Haemin and nicotinamide adenine dinucleotide requirements of Haemophilus influenzae and Haemophilus parainfluenzae. J Med Microbiol. 1974;7:359–365. doi: 10.1099/00222615-7-3-359. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback R C, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Frangipane M E, Morton D J, Wooten J A, Pozsgay J M, Stull T L. Binding of human hemoglobin by Haemophilus influenzae. FEMS Microbiol Lett. 1994;118:243–248. doi: 10.1111/j.1574-6968.1994.tb06835.x. [DOI] [PubMed] [Google Scholar]
- 6.High N J, Jennings M P, Moxon E R. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol Microbiol. 1996;20:165–174. doi: 10.1111/j.1365-2958.1996.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 7.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H, Ren Z, Whitby P W, Morton D J, Stull T L. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin binding protein of Haemophilus influenzae. Microbiology. 1999;145:905–914. doi: 10.1099/13500872-145-4-905. [DOI] [PubMed] [Google Scholar]
- 10.Klein J O. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr Infect Dis J. 1997;16:S5–S8. doi: 10.1097/00006454-199702001-00002. [DOI] [PubMed] [Google Scholar]
- 11.Lee B C. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 12.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 13.Maciver I, Latimer J L, Liem H H, Muller-Eberhard U, Hrkal Z, Hansen E J. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect Immun. 1996;64:3703–3712. doi: 10.1128/iai.64.9.3703-3712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 15.Morton D J, Stull T L. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Haemophilus influenzae: species distribution of a family of genes containing CCAA nucleotide repeating units, abstr. B-224; p. 67. [Google Scholar]
- 16.Morton D J, Williams P. Siderophore-independent acquisition of transferrin-bound iron by Haemophilus influenzae type b. J Gen Microbiol. 1990;136:927–933. doi: 10.1099/00221287-136-5-927. [DOI] [PubMed] [Google Scholar]
- 17.Musser J M, Barenkamp S J, Granoff D M, Selander R K. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–191. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Z, Jin H, Morton D J, Stull T L. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook T, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sharetzsky C, Edlind T D, LiPuma J J, Stull T L. A novel approach to insertional mutagenesis of Haemophilus influenzae. J Bacteriol. 1991;173:1561–1564. doi: 10.1128/jb.173.4.1561-1564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spenser H T, Herriott R M. Development of competence in Haemophilus influenzae. J Bacteriol. 1965;90:911–920. doi: 10.1128/jb.90.4.911-920.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stull T L. Protein sources of heme for Haemophilus influenzae. Infect Immun. 1987;55:148–153. doi: 10.1128/iai.55.1.148-153.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tartof K D, Hobbs C A. New cloning vectors and techniques for easy and rapid restriction mapping. Gene. 1988;67:169–182. doi: 10.1016/0378-1119(88)90394-0. [DOI] [PubMed] [Google Scholar]
- 24.Turk D C. The pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 25.Whitby P W, Morton D J, Stull T L. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol Lett. 1998;158:57–60. doi: 10.1111/j.1574-6968.1998.tb12800.x. [DOI] [PubMed] [Google Scholar]