Abstract
Two new genes, mrpH and mrpJ, were identified downstream of mrpG in the mrp gene cluster encoding mannose-resistant Proteus-like (MR/P) fimbriae of uropathogenic Proteus mirabilis. Since the predicted MrpH has 30% amino acid sequence identity to PapG, the Galα(1-4)Gal-binding adhesin of Escherichia coli P fimbriae, we hypothesized that mrpH encodes the functional MR/P hemagglutinin. MR/P fimbriae, expressed in E. coli DH5α, conferred on bacteria both the ability to cause mannose-resistant hemagglutination and the ability to aggregate to form pellicles on the broth surface. Both a ΔmrpH mutant expressed in E. coli DH5α and an isogenic mrpH::aphA mutant of P. mirabilis were unable to produce normal MR/P fimbriae efficiently, suggesting that MrpH was involved in fimbrial assembly. Amino acid residue substitution of the N-terminal cysteine residues (C66S and C128S) of MrpH abolished the receptor-binding activity (hemagglutinating ability) of MrpH but allowed normal fimbrial assembly, supporting the notion that MrpH was the functional MR/P hemagglutinin. Immunogold electron microscopy of P. mirabilis HI4320 revealed that MrpH was located at the tip of MR/P fimbriae, also consistent with its role in receptor binding. The isogenic mrpH::aphA mutant of HI4320 was less able to colonize the urine, bladder, and kidneys in a mouse model of ascending urinary tract infection (P < 0.01), and therefore MR/P fimbriae contribute significantly to bacterial colonization in mice. While there are similarities between P. mirabilis MR/P and E. coli P fimbriae, there are more notable differences: (i) synthesis of the MrpH adhesin is required to initiate fimbrial assembly, (ii) MR/P fimbriae confer an aggregation phenotype, (iii) site-directed mutation of specific residues can abolish receptor binding but allows fimbrial assembly, and (iv) mutation of the adhesin gene abolishes virulence in a mouse model of ascending urinary tract infection.
Proteus mirabilis, commonly associated with complicated urinary tract infections (UTIs), expresses several types of fimbrial structures that promote attachment to and colonization of host mucosal surfaces (23, 24). One of these, mannose-resistant Proteus-like (MR/P) fimbria, a surface structure responsible for mannose-resistant hemagglutination (MRHA), has been shown to contribute significantly to the development of experimental UTIs. First, the majority of P. mirabilis strains isolated from patients with acute pyelonephritis express MR/P fimbriae as a single hemagglutinin type (25). Second, MR/P fimbriae are expressed in vivo and elicit a strong immune response in experimental UTIs (5). Third, an isogenic mrpA (which encodes the major structural subunit of MR/P fimbriae) mutant colonizes the urine, bladder, and kidneys of experimentally infected CBA mice in significantly smaller numbers than the wild-type strain does (3). Finally, our recent studies on the expression of MR/P fimbriae at the transcriptional level show that the invertible element which regulates transcription in a manner similar to Escherichia coli type 1 fimbria is >98% turned on in vivo (in the urine, bladder, and kidneys of infected mice) versus at most 50% in vitro (static culture) (27). Collectively, these observations imply a critical role for this adhesin in the development of UTIs.
To understand the mechanism by which the MR/P fimbria contributes to the development of UTIs, studies were carried out to define the gene that encodes the MR/P fimbrial adhesin. Sequence analysis of the structural and accessory genes previously identified (mrpA, mrpB, mrpC, mrpD, mrpE, mrpF, and mrpG) showed that most of the proteins predicted by these genes had homology to those of E. coli P fimbria and Serratia marcescens Smf fimbria, with the exception of MrpG, which showed no significant homology to any known fimbrial proteins (4). Interestingly, none of the predicted MR/P fimbrial proteins have any sequence homology to any known adhesins. Mutagenesis studies on the five predicted pilin-encoding genes (mrpA, mrpB, mrpE, mrpF, and mrpG) were unable to define any of them as encoding a functional fimbrial adhesin (references 3, 19, and 20 and unpublished data). The absence of a chaperone-binding domain at the C terminus of MrpG finally led us to question whether the previously determined 3′ end of the mrp operon represents the true end of the gene cluster. Newly generated sequence diverged from the old sequence in the middle of mrpG and predicted not only a new C terminus for MrpG with a consensus chaperone-binding domain but also another two open reading frames downstream, designated mrpH and mrpJ. mrpH predicted a protein of 29.2 kDa that has 30% amino acid sequence identity to PapG and 35% identity to SmfG, the fimbrial adhesins of P fimbria and Smf fimbria, respectively (21, 22). In this study, we tested the hypothesis that MrpH was the functional MR/P hemagglutinin.
While there are similarities to E. coli P fimbriae, there are more notable differences: (i) synthesis of the MrpH adhesin is required to initiate fimbrial assembly, (ii) MR/P fimbriae confer an aggregation phenotype, (iii) site-directed mutation of specific residues can abolish receptor binding but allow fimbrial assembly, and (iv) mutation of the adhesin gene abolishes virulence in a mouse model of ascending UTI.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. mirabilis HI4320 (urease-positive, hemolytic, and positive for MR/P, PMF, and ATF fimbriae), isolated from the urine of an elderly, long-term-catheterized woman with significant bacteriuria (≥105 CFU/ml) (26), has been used extensively by our group for virulence studies (summarized in reference 23). E. coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used as the host strain for transformation of plasmids other than suicide vector pCVD442 and its derivatives. E. coli DH5αλpir was used for the cloning with the pRK2-derived suicide vector pCVD442 (8).
Construction of an isogenic mrpH::aphA mutant of HI4320.
A 1.8-kb StyI-PvuII fragment of the mrp gene cluster that contains part of mrpG, mrpH, and part of mrpJ was cloned into pBluescript (Stratagene, La Jolla, Calif.). A kanamycin resistance (encoded by aphA) cassette (the 1.3-kb BamHI fragment of pUC4κ from Pharmacia Biotech Inc., Piscataway, N.J.) was inserted into the BamHI site within mrpH, so that it was flanked with approximately 1.0 kb of mrp sequence on its 5′ side and 0.8 kb on its 3′ side. This disrupted mrpH, along with its flanking homologous sequence, was cloned into pCVD442 (8), a pRK2-derived suicide vector, and electroporated into HI4320. About 500 kanamycin-resistant transformants were obtained on Luria-Bertani agar plates containing kanamycin (50 μg/ml). Of these, 400 were picked and passaged on to Luria-Bertani agar plates containing ampicillin (100 μg/ml) to screen for ampicillin resistance. Of 400 transformants, 19 were ampicillin susceptible and considered possible mrpH::aphA mutants. Four of the nineteen ampicillin-susceptible, kanamycin-resistant transformants (mrpH1011, mrpH1022, mrpH1023, and mrpH1100) were confirmed by Southern analysis as mrpH::aphA mutants.
Nucleotide sequencing.
Sequencing was performed by the dideoxy chain termination method with double-stranded DNA as the template. Reactions were run on a model 373A DNA sequencer (Applied Biosystems, Foster City, Calif.).
Southern blot analysis.
Chromosomal DNA was digested with either PvuII or PvuII plus PvuI, electrophoresed on a 0.8% agarose gel, and transferred to a membrane (QIABRANE Nylon Plus; Qiagen Inc., Chatsworth, Calif.). Probe labeling, hybridization, and signal detection were carried out with the enhanced chemiluminescence direct nucleic acid-labeling and detection system (Amersham Life Science, Little Chalfont, England) as specified by the manufacturer.
Expression of MR/P fimbriae in E. coli DH5α and construction of a ΔmrpH mutant.
The 9.2-kb AflIII-PstI fragment of the mrp gene cluster, including the phase-variable mrp promoter fixed in the “on” position (27), structural genes mrpA to mrpH, and a regulatory gene, mrpJ, was cloned into the AflIII-PstI site of pBluescript. This construct, designated pXL4206, was digested with PvuII to release a 1.2-kb fragment containing the majority of the 3′ end of mrpJ and its downstream sequences and then self-ligated to yield pXL9301. Since the deletion of mrpJ does not affect the production of normal MR/P fimbriae and is irrelevant to this study, constructs pXL9301 and pXL4206 are both referred to as wild type for MR/P fimbrial production throughout this report. Construct pXL4206 was digested with EcoRI to release a 1.8-kb fragment that contains the majority of mrpH and its downstream sequences and then self-religated to form pXL4401 (referred to as ΔmrpH). To complement this mutant, an 888-bp fragment containing the complete mrpH and its ribosomal binding site was PCR amplified and cloned into the EcoRV site of pBluescript, resulting in construct pXL5802. The “on” version of the mrp promoter (the promoter driving transcription of mrpA) was cut from pMRP-ON (27) by AflIII-EcoRI digestion and cloned into NcoI-EcoRI-digested pACYC184, generating the construct pON-184. E. coli DH5α containing both pXL4401 (ΔmrpH) and pON-184 (vector) is referred to as ΔmrpH + vector. To place it under the control of the mrp promoter, mrpH was cut out of pXL5802 by SstI-HincII digestion and ligated with SstI-XmnI-digested pON-184, resulting in construct pXL8906. E. coli DH5α containing both pXL4401 (ΔmrpH) and pXL8906 (mrpH) is referred to as ΔmrpH + mrpH (see Table 1 for a summary of the plasmid constructs).
TABLE 1.
Summary of mutagenesis studies in E. coli DH5α
| Construct | mrpH | Fimbrial productiona | MRHAb | BAc |
|---|---|---|---|---|
| pXL4206 or pXL9301 | Wild type | + | +++ | +++ |
| pXL4401 | Deletion (ΔmrpH) | − | − | − |
| pXL4401 and pXL8906d | Wild type | + | +++ | +++ |
| pLX1005 | C66S | + | + | + |
| pLX1105 | C128S | + | − | + |
| pLX1203 | C66S plus C128S | + | − | + |
| pLX1305 | C66 plus C128 | + | +++ | +++ |
+, normal fimbrial production; −, less than 1% of normal fimbrial production.
+++, positive reactions in the first three or more bacterial dilutions; +, positive only in the first dilution; −, negative from the first dilution.
+++, strong pellicle formation on the broth surface, which breaks into visible bacterial aggregates upon mild vortex; +, weak pellicle formation on the broth surface, which disperses into the broth upon mild vortex; −, no pellicle formation or any kind of BA.
E. coli DH5α(pXL8906) is negative for fimbrial production, MRHA, and BA.
Amino acid residue substitution (C66S and C128S) of MrpH.
Site-directed mutagenesis by overlap extension PCR (11) was used to amplify mrpH and replace the codon 66 TGT (encoding cysteine) or the codon 128 TGC (encoding cysteine) by TCT (encoding serine). The PCR-amplified mutant mrpH fragments were cloned into pCR-Blunt (Invitrogen Corp., San Diego, Calif.) and sequenced to confirm the mutations (data not shown). The SnaBI- and KpnI-digested mutant mrpH fragment was used to replace the wild-type SnaBI-KpnI fragment of pXL9301 to generate either pLX1005 (C66S) or pLX1105 (C128S). The 1.9-kb SmaI-EcoRI fragment (containing codon 66 but not codon 128) of pLX1105 was replaced by that of pLX1005 to generate pLX1203 (C66S and C128S), and the 1.9-kb SmaI-EcoRI fragment of pLX1005 was replaced by that of pLX1105 to reconstitute the wild-type version, designated pLX1305 (referred to as C66 and C128 to be distinguished from the original wild type).
MRHA assay.
Bacteria were collected by centrifugation (12,000 × g at 25°C for 1 min) after being grown under conditions optimal for MR/P fimbria production; for P. mirabilis strains, bacteria were passaged statically three times for 48 h each in Luria-Bertani broth at 37°C; for E. coli strains, bacteria were grown statically at 37°C for 72 h. Cell pellets were suspended in phosphate-buffered saline (PBS) to approximately 109 CFU/ml. A series of twofold dilutions of bacterial suspension was mixed with an equal volume of 3% (vol/vol) chicken erythrocytes (suspended in 0.85% saline containing 50 mM mannose) in a round-bottom 96-well microtiter plate. The plate was incubated at room temperature for 30 min to allow erythrocytes to settle to the bottom of the well. Nonagglutinated erythrocytes form a tight button, whereas agglutinated erythrocytes form a diffuse mat.
Isolation of MR/P fimbriae.
Bacteria grown under optimal conditions for MR/P fimbrial production, as described above, were collected by centrifugation and resuspended in 10 mM Tris-HCl, (pH 7.2). MR/P fimbriae were sheared from the cell surface by blending and purified by differential centrifugation as described previously (20). Partially purified fimbrial preparations were obtained by following the procedure for fimbrial isolation, excluding the CsCl gradient centrifugation step.
Expression and purification of MBP fusions.
A DNA fragment encoding the mature MrpH (lacking its N-terminal 22-amino-acid putative signal peptide) was PCR amplified and cloned into the EcoRI and HindIII sites of pMAL-C2 (New England Biolabs Inc., Beverly, Mass.) to express MrpH as a C-terminal fusion to maltose-binding protein (MBP). Also, a DNA fragment encoding the mature MrpA (lacking its N-terminal 23-amino-acid putative signal peptide) was PCR amplified and cloned into the EcoRI and HindIII sites of pMAL-C2 to express MrpA as a C-terminal fusion to MBP. Expression and purification of MBP-MrpH and MBP-MrpA fusion proteins were carried out as described by Ausubel et al. (2).
Preparation of antisera against MrpH or MrpA.
The purified MBP-MrpH (100 μg) and MBP-MrpA (100 μg) were each emulsified in Freund’s complete adjuvant and subcutaneously injected into separate New Zealand White rabbits. At 4 weeks after the primary immunization, animals were given booster injections of 100 μg of protein emulsified in Freund’s incomplete adjuvant. Blood samples taken during week 6 were assayed for reaction with antigen by Western blotting. A second booster injection of 100 μg of protein emulsified in Freund’s incomplete adjuvant was given to each rabbit during week 7. Sera were collected 2 weeks after the second booster.
The isopropyl-β-d-thiogalactopyranoside (IPTG)-induced cell lysate (about 10 mg protein) of E. coli DH5α transformed with plasmid pMAL-C2 was coupled to an AminoLink Plus column (Pierce Inc., Rockford, Ill.) as specified by the manufacturer. Polyclonal antisera from rabbits were passed through the column to remove antibodies against MBP as well as the antibodies reacting with E. coli DH5α proteins. Antibodies bound to the column were eluted through a cycle of pH change as described by Harlow and Lane (10). The procedure was repeated several times until antisera did not react on a Western blot with the induced cell lysate of E. coli DH5α(pMAL-C2).
Western blot analysis.
Partially purified fimbrial preparations were denatured in sodium dodecyl sulfate (SDS)-gel sample buffer (100°C for 30 min), electrophoresed on a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp., Bedford, Mass.). The blot was incubated with rabbit polyclonal antiserum against MrpA and then with goat anti-rabbit immunoglobulin G (IgG) alkaline phosphatase conjugate and then developed with BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate toluidinium/nitroblue tetrazolium) as a chromagenic substrate for alkaline phosphatase (1).
Immunogold EM.
Immunogold labeling was performed by a modification of the method of Faulk and Taylor (9). Bacteria were grown under conditions optimal for MR/P fimbrial production (see above). A drop of bacterial culture was placed on a Formvar-coated grid and processed as described previously (20). The grids were incubated at 37°C for 30 min each with a 1:100 dilution of rabbit antiserum against MrpH followed by a 1:25 dilution of goat anti-rabbit IgG (heavy plus light chains) conjugated to 30-nm-diameter gold beads (AuroProbe EM GAR G30; Amersham Life Science) and were then incubated at 37°C for 30 min each with a 1:100 dilution of rabbit antiserum against MR/P fimbriae (20) or rabbit antiserum against MrpA followed by a 1:25 dilution of protein G conjugated with 5-nm-diameter gold beads (AuroProbe EM protein GG5; Amersham Life Science). Between the incubations, the grids were washed three times with PBS–1% bovine serum albumin. At the end, they were washed three times with PBS–1% bovine serum albumin and three times with distilled water, negatively stained with 1% sodium phosphotungstate (pH 6.8), and examined by transmission electron microscopy (EM) with a JEM-1200EX II electron microscope (JEOL Ltd., Tokyo, Japan).
CBA mouse model of ascending UTI.
CBA mice were transurethrally challenged with 106 to 107 CFU of bacteria per mouse by a method described previously (20). After 7 days, the mice were sacrificed and bacteria recovered from the urine, bladder, and kidneys were enumerated on nonswarming agar plates (6) containing appropriate antibiotics. The range of detection in this assay is 102 to 109 CFU/ml of urine or CFU/g of tissue. Values of ≤102 were set to 102, and values of ≥109 were set to 109.
Nucleotide sequence accession number.
The nucleotide sequences of mrpG, mrpH, and mrpJ have been deposited in GenBank under accession no. Z32686.
RESULTS
Sequence analysis of MrpG, MrpH, and MrpJ.
The newly acquired 3′ sequence of mrpG revealed that the entire gene was 549 nucleotides and encoded a protein of 182 amino acid residues. MrpG has 48% amino acid sequence identity to SmfF and 29% identity to PapK. This is consistent with our previous result that MrpG is essential for fimbrial assembly and is located near the fimbrial tip but does not represent the adhesin itself (20). A similar role was assigned for PapK in the biogenesis of P fimbriae in E. coli (12).
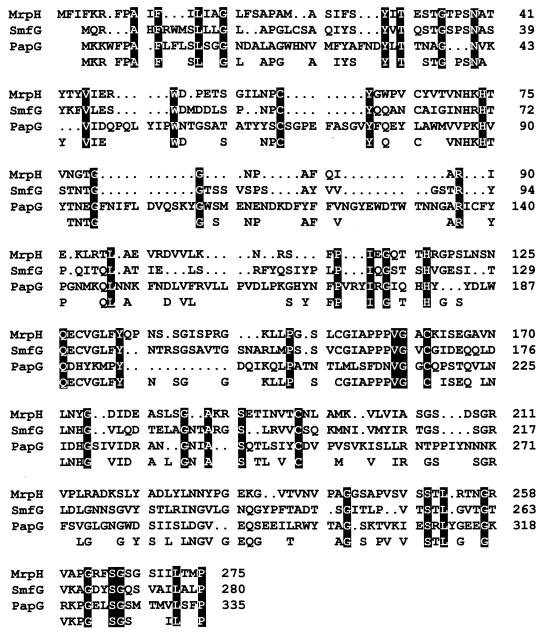
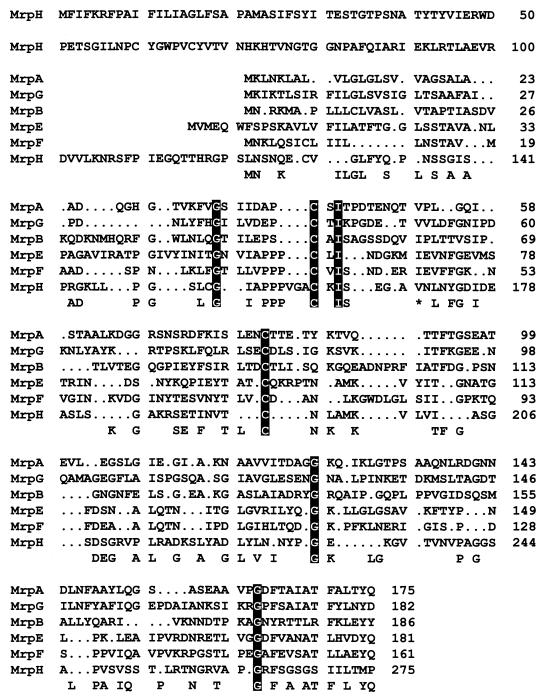
The mrpH gene is 828 nucleotides and is predicted to encode a polypeptide of 275 amino acid residues (Fig. 1). The predicted amino acid sequence of MrpH has 30% identity to PapG and 35% identity to SmfG (see Fig. 3), both of which were demonstrated to be fimbrial adhesins (21, 22). Protein sequence alignment of the six putative pilins of MR/P fimbriae (Fig. 2) shows that all of the pilins except MrpH were similar in size and had amino acid sequence identity throughout their entire predicted amino acid sequence, especially at the C-terminal chaperone-binding sequence (Gly and Tyr residues are amino acids 14 and 2 from the C terminus, respectively). Aligned with the other putative MR/P pilins, MrpH has a distinctive N terminus, perhaps a putative region for receptor-binding activity (Fig. 2). The C-terminal chaperone-binding domain of MrpH is unique in that it does not retain the conserved Tyr residue at the penultimate position. Also, MrpH has a Pro residue at the last position, a feature that has been conserved in many fimbrial adhesins from different species including PapG, PrsG, SmfG, Fim2, and Fim3 (Fig. 3) (16). Based on the sequence homology, we proposed that MrpH was the functional MR/P hemagglutinin.
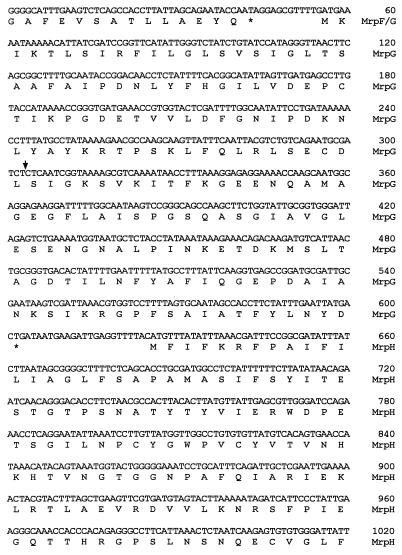
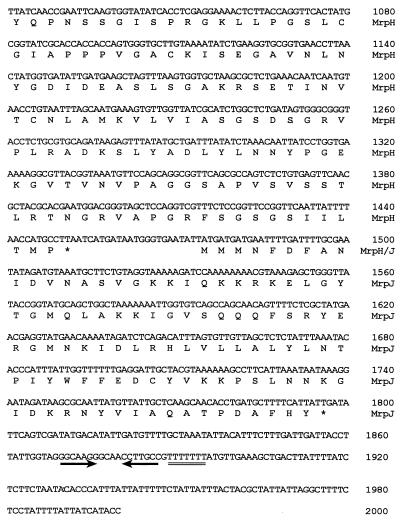
FIG. 1.
Newly acquired 3′-end sequence of the mrp gene cluster. Predicted polypeptides are translated below the nucleotide sequence. Letters representing amino acid residues are aligned with the first nucleotide of their corresponding codons. Asterisks, aligned with the first nucleotide of the stop codons, mark the end of polypeptides. Gene product designations are given in the right margin. Numbers in the right margin refer to the nucleotide sequence. The downward arrow indicates where the new sequence begins. The pair of inverted arrows and the double line underlie the putative rho-independent terminator.
FIG. 3.
Alignment of the amino acid sequence of putative MR/P adhesin with two other adhesins, SmfG of S. marcescens and PapG of E. coli. The amino acid sequences of MrpH, SmfG, and PapG were aligned by using the GCG Pileup software. Gaps (.) were introduced to obtain maximal fit. The amino acid residues that are conserved in two or more of the proteins are noted at the bottom of the alignment. Amino acid residues that are conserved in all three are highlighted in black boxes.
FIG. 2.
Alignment of amino acid sequences of putative pilins of the MR/P fimbria. The amino acid sequences of the six putative pilins of the MR/P fimbria were aligned by using the GCG Pileup software. Gaps (.) were introduced to obtain maximal fit. The amino acid residues that are conserved in three or more of the pilins are noted at the bottom of the alignment. The amino acid residues that are conserved in all six pilins are highlighted in black boxes. ∗, the valine residue and the isoleucine residue are both conserved in three of the six pilins. Aligned with the other putative MR/P pilins, MrpH is the only one that contains a distinctive N-terminal domain that could provide a possible basis for the receptor-binding activity.
Downstream of mrpH, there is an open reading frame of 324 nucleotides, designated mrpJ, which predicts a 107-amino-acid protein with a pI of 10.02 (Fig. 1). A BLAST search indicated that the predicted MrpJ might play a role in DNA binding and represent a transcriptional regulator. It is a unique feature for this mrp fimbrial gene cluster to have a transcriptional regulator gene following the structural genes. At 72 bp downstream of the stop codon of mrpJ, there is a predicted stem-loop structure followed by a run of seven T’s, suggestive of a rho-independent terminator and marking the end of the mrp gene cluster (Fig. 1). mrpJ is not discussed further in this report.
Insertional mutagenesis studies with P. mirabilis.
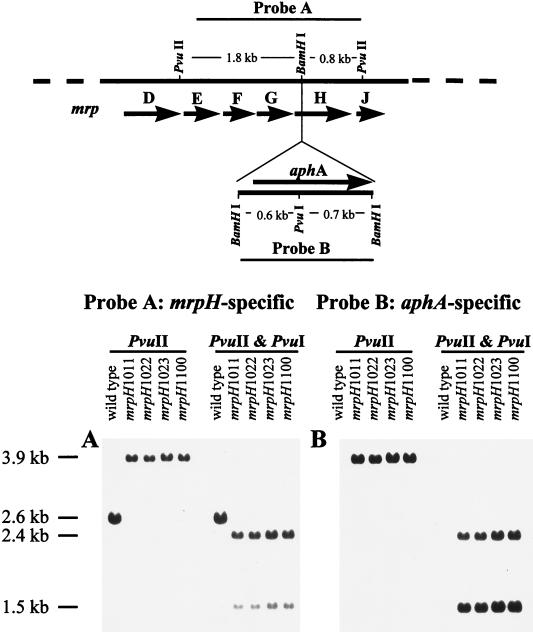
To test our hypothesis that MrpH is the functional adhesin of MR/P fimbriae, insertional mutagenesis was carried out to disrupt mrpH in the clinical isolate of P. mirabilis, strain HI4320, from which the mrp gene cluster was originally isolated. A kanamycin resistance (encoded by aphA) cassette was introduced into the BamHI site within mrpH on the chromosome through allelic exchanges (see Materials and Methods). The occurrence of the insertional mutation was verified by Southern blotting (Fig. 4). When probed with the mrpH-specific sequence (blot A), the 2.6-kb PvuII fragment reacted in the wild-type strain but was shifted to 3.9 kb in the mrpH::aphA mutants, suggestive of a 1.3-kb insertion (the size of the aphA insert). When probed with the aphA-specific sequence (blot B), the 3.9-kb band in the mutants hybridized with the probe, suggesting that the 1.3-kb insertion represents the kanamycin resistance cassette. The mutation was further confirmed by PvuII-PvuI double digestion. Since the kanamycin resistance cassette insertion also introduced a PvuI site into this region, PvuI cleaved the 3.9-kb fragment of the mrpH::aphA mutants into two fragments with predicted sizes of 2.4 and 1.5 kb. It was concluded from the Southern blotting results that the mrpH was disrupted by a kanamycin resistance cassette in these mrpH::aphA mutants.
FIG. 4.
Southern blot analysis of mrpH::aphA mutants. The predicted restriction map of the wild-type strain and mrpH::aphA mutant is illustrated in the top panel. The arrows represent predicted open reading frames within the defined region. The sizes of the bands that hybridized with the probes in the Southern blots are indicated along the left side.
Immunogold EM studies showed that the mrpH::aphA mutants were not capable of producing a normal complement of MR/P fimbriae. Indeed, less than 1% of the bacteria were MR/P fimbriated in the mrpH::aphA mutants, in contrast to about 50% in the wild type. The few mutant bacteria that did produce MR/P fimbriae carried for fewer of these fimbriae per bacterium than did the wild-type strain and the fimbriae produced by the mutant strains were often shorter and amorphous (data not shown). In fact, mutations in any other pilin-encoding genes (mrpA, mrpB, mrpE, mrpF, and mrpG) led to less MR/P fimbriation of HI4320 (references 3, 19, and 20 and unpublished data). However, even though the mrpH::aphA mutants were defective in producing MR/P fimbriae, they were still positive for MRHA (data not shown), suggesting that the MR/P fimbria may not be the only mannose-resistant hemagglutinin produced by the wild-type P. mirabilis HI4320.
Mutagenesis studies of mrpH in E. coli.
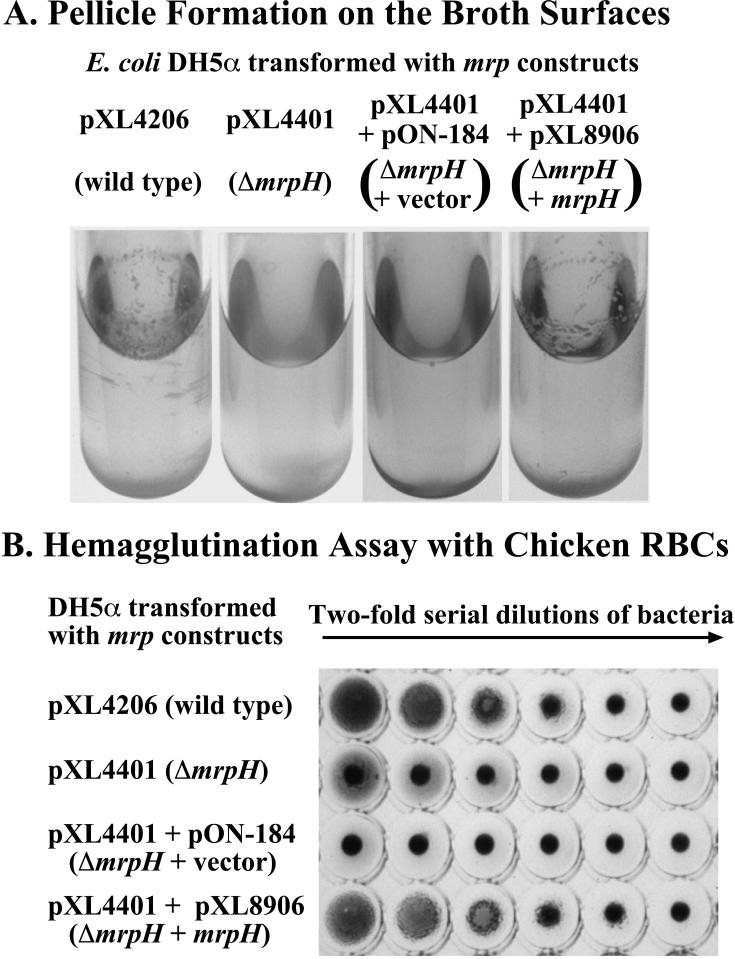
To avoid the complication of other mannose-resistant hemagglutinins produced by P. mirabilis HI4320 or the complication of native phase variation of the MR/P fimbriae (27), the structural genes of mrp gene cluster (mrpA to mrpH), together with the mrp promoter, were cloned into pBluescript (see Materials and Methods for details of all constructs) and electroporated into E. coli DH5α. Since mrpI, the gene encoding the putative recombinase that is responsible for the switch of the promoter region (27), was not included in this construct, the promoter was fixed in the “on” position. The constitutive expression of MR/P fimbriae conferred on bacteria not only the ability to cause MRHA but also the ability to aggregate (bacterial aggregation [BA]) in liquid broth and form a pellicle on the broth surface (Fig. 5). A simple deletion of mrpH (ΔmrpH) led to the loss of both abilities, which were restored by addition of mrpH on a compatible vector in trans, suggesting that mrpH may encode the aggregative mannose-resistant hemagglutinin of MR/P fimbria.
FIG. 5.
Bacterial aggregation and MRHA patterns of the ΔmrpH mutant expressed in E. coli. (A) Pictures of 72-h static cultures of E. coli DH5α containing various constructs grown in 5 ml of Luria-Bertani broth at 37°C. The tubes were handled carefully to avoid disrupting the pellicle. (B) MRHA assay of the bacterial cultures in panel A were performed as described in Materials and Methods.
It is known that mutations in minor pilins sometimes interfere with overall fimbrial assembly, as shown previously for the mrpG mutant (20). Immunogold electron micrographs revealed that the ΔmrpH mutant produced for fewer and much shorter MR/P fimbriae than did the wild-type strain (data not shown). The severity of this defect was clearly shown by the Western blot (Fig. 6). Partially purified fimbrial preparations (see Materials and Methods) of the wild type, the ΔmrpH mutant, and the complemented mutant (ΔmrpH + mrpH) were subjected to Western blotting with rabbit antiserum against MrpA. It was shown that the amount of MrpA assembled into fimbriae was undetectable in the ΔmrpH mutant (Fig. 6), suggesting that deletion of mrpH does indeed interfere with the assembly of MR/P fimbriae (such a phenomenon is not observed in papG mutants of E. coli P fimbrial genes). Therefore, it could not be concluded from this study that MrpH was the functional MR/P adhesin. Given the strong homology of MrpH to known adhesins, however, we still believed that it was the functional adhesin of MR/P fimbriae. The question of how to elucidate the involvement of MrpH in fimbrial assembly and its activity in receptor binding still remained.
FIG. 6.
Western blot of partially purified fimbrial preparations of various E. coli DH5α strains. Partially purified fimbrial preparations (see Materials and Methods for details) were separated on an SDS–12.5% polyacrylamide gel and subjected to Western blot analysis with polyclonal rabbit antiserum raised against MrpA.
Amino acid substitution of MrpH.
Given the significant homology of MrpH to PapG and SmfG, we believe that the mrpH gene encodes the adhesin for MR/P fimbria. The deficiency of the mrpH::aphA mutant in making MR/P fimbriae may simply due to a requirement for incorporation of MrpH into the fimbrial structure. MrpH, like its homolog PapG, may reside at the tip of the MR/P fimbria and may therefore be the first pilin to be translocated through the predicted usher MrpC. In an attempt to elucidate the receptor-binding activity of MrpH and its involvement in fimbrial assembly, we constructed amino acid substitutions in MrpH that abolished its receptor-binding activity but did not interfere with the fimbrial assembly process. As reported by Carnoy and Moseley (7), the cysteine residues in the N-terminal receptor-binding domain of Dr family adhesins form disulfide bonds that are crucial for receptor-binding activity. Intramolecular disulfide bond formation was detected in PapG as well (17). MrpH, compared with the other pilins (MrpA, MrpB, MrpE, MrpF, and MrpG), has a distinctive N-terminal domain that contains four cysteine residues (C60, C66, C128, and C152). Using overlap extension PCR, we replaced C66, C128, or C66 plus C128 by serine residues. The C66S, C128S, and C66S-plus-C128S substitution mutants and C66 plus C128 (the reconstituted wild type) were constructed as described in Materials and Methods. E. coli DH5α strains expressing the mutant MrpH (C66S, C128S, and C66S plus C128S) have dramatic decreases in MRHA and BA (summarized in Table 1). However, unlike the ΔmrpH mutant, these constructs were still capable of producing MR/P fimbriae at approximately the same level as the wild-type strain was (Fig. 6); this was confirmed by immunogold EM (data not shown).
Immunogold EM demonstrates that MrpH is located at the MR/P fimbrial tip.
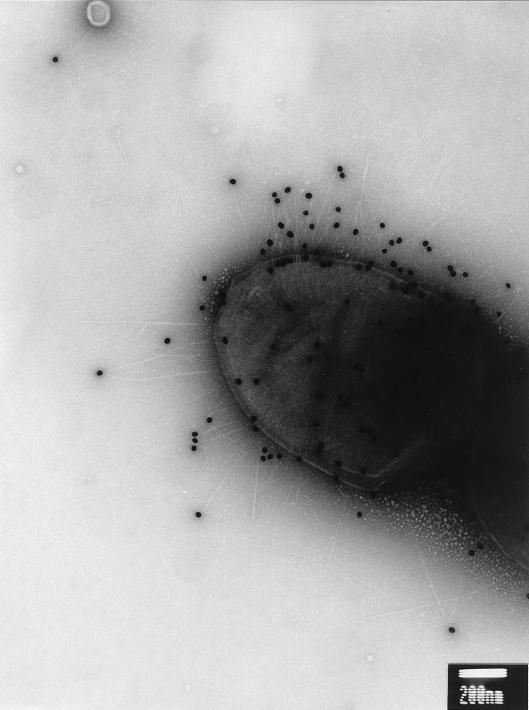
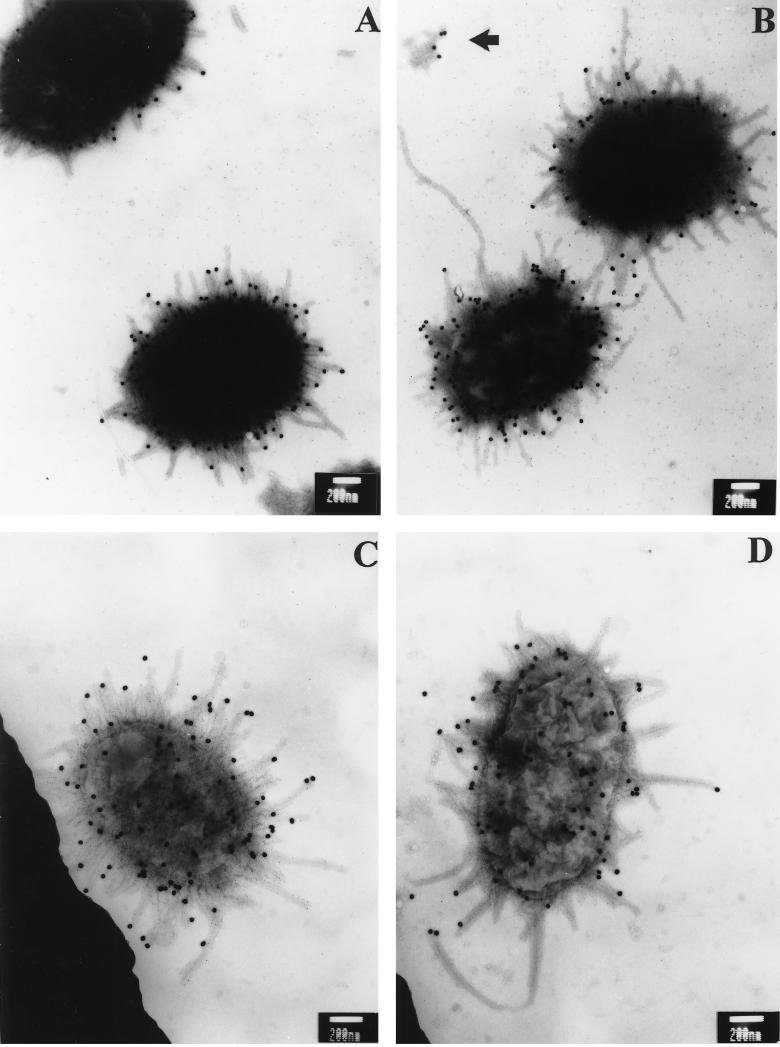
To examine whether MrpH is located at fimbrial tip like its homolog PapG, P. mirabilis HI4320, passaged under optimal conditions for fimbrial production, was subjected to immunogold labelling (see Materials and Methods). MrpH was labelled with 30-nm-diameter gold particles by using the primary rabbit antibodies against MrpH and the gold particle-coupled secondary antibodies against rabbit IgG (Fig. 7). To distinguish them from the other fimbriae produced by HI4320, MR/P fimbriae were labelled with 5-nm-diameter gold particles by using the primary rabbit antibodies against MR/P fimbriae (Fig. 8A and B) or MrpA (Fig. 8C and D) and the gold particle-coupled protein G. The electron micrographs clearly indicate that MrpH is located at the tip of the MR/P fimbria, a common feature for many fimbrial adhesins including PapG (21). It was noted that some MR/P fimbriae labelled with the 5-nm-diameter gold particles were not labelled with the 30-nm-diameter gold particle at the tip, indicating the absence of MrpH. This could be due to the mechanical shearing of the tips, since previous studies demonstrated that MR/P fimbriae are very fragile. The electron micrograph in Fig. 8B shows MR/P fimbria tips which may have been sheared off during preparation.
FIG. 7.
Immunogold electron micrograph of P. mirabilis HI4320. P. mirabilis HI4320 grown under conditions optimal for fimbrial production (see Materials and Methods) was reacted first with antiserum raised against MrpH and then with a secondary antibody (goat anti-rabbit IgG) conjugated to 30-nm-diameter gold particles. Bar, 200 nm. It is clearly shown that MrpH, targeted by the 30-nm-diameter gold particle, is located at the tip of bacterial fimbriae.
FIG. 8.
Immunogold electron micrographs of P. mirabilis HI4320. P. mirabilis HI4320 was reacted first with rabbit antiserum raised against MrpH and then with a secondary antibody (goat anti-rabbit IgG) conjugated to 30-nm-diameter gold particles. (A and B) Bacteria were then reacted with antiserum raised against MR/P fimbriae followed by a secondary antibody (recombinant protein G) conjugated to 5-nm-diameter gold particles. (C and D) Bacteria were then reacted with antiserum raised against MrpA followed by a secondary antibody (recombinant protein G) conjugated to 5-nm-diameter gold particles. Bars, 200 nm. The antiserum raised against MR/P fimbriae, as well as the antiserum raised against MrpA, labeled the shafts of the MR/P fimbriae. While the antiserum raised against MR/P fimbriae resulted in more extensive labeling, the antiserum raised against MrpA gave a cleaner background. The 5-nm-diameter gold particles that labeled the shafts of MR/P fimbriae are difficult to resolve but make the MR/P fimbriae appear thicker. The arrow in panel B indicates MR/P fimbria tips that may have been sheared off during preparation.
Analysis of virulence in a mouse model of ascending UTI.
As mentioned above, P. mirabilis HI4320 may produce other types of mannose-resistant hemagglutinin besides MR/P fimbria; therefore the importance of the MR/P fimbria to virulence was questioned. Can the role of the MR/P fimbria be fulfilled by the other mannose-resistant hemagglutinins? The contribution of MR/P fimbria to colonization and virulence in UTI was reassessed in a CBA mouse model of ascending UTI established previously (3).
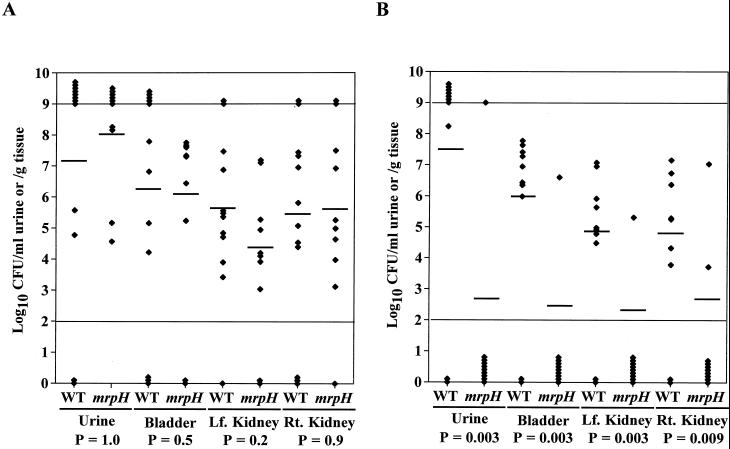
In an independent-challenge experiment, a group of 12 CBA mice were challenged with 4.8 × 106 CFU of the wild-type HI4320 per mouse and another group of 10 CBA mice were challenged with 5.5 × 106 CFU of the mrpH::aphA mutant per mouse. After 7 days, the mice were sacrificed and bacteria recovered from the urine, bladder, and kidneys were enumerated on nonswarming plates. Bacteria recovered from mice infected with the mrpH::aphA mutant were enumerated on both nonswarming plates and nonswarming plates containing kanamycin (50 μg/ml). No detectable kanamycin-sensitive revertants were found, indicating that the mutation is stable at least during the 7-day infection period. The geometric mean values of the bacterial counts were as follows: urine, 7.19 (wild type) versus 8.02 (mutant) log10 CFU/ml (P = 1.0); bladder, 6.25 (wild type) versus 6.09 (mutant) log10 CFU/g (P = 0.5); left kidney, 5.63 (wild type) versus 4.38 (mutant) log10 CFU/g (P = 0.2); and right kidney, 5.46 (wild type) versus 5.64 (mutant) log10 CFU/g (P = 0.9) (Fig. 9A). The data showed that the mrpH::aphA mutant colonized the urinary tracts of CBA mice just as well as the wild-type strain did.
FIG. 9.
Assessment of the virulence of the mrpH::aphA mutant of HI4320 in the CBA mouse model of ascending UTIs. (A) Independent-challenge experiment. A group of 12 mice and a group of 10 mice were transurethrally challenged with 4.8 × 106 CFU of the wild-type strain per mouse and 5.5 × 106 CFU of the mrpH::aphA mutant per mouse, respectively. (B) Cochallenge experiment. A group of 10 mice were transurethrally challenged with a roughly 1:1 mixture of the wild-type strain and the mrpH::aphA mutant (7.9 × 106 CFU of the wild-type strain and 1.2 × 107 CFU of the mrpH::aphA mutant per each mouse). After 7 days, the mice were sacrificed and the quantitative bacterial counts in the urine, bladders, and kidneys were calculated (see Materials and Methods). Each diamond represents the CFU per milliliter of urine or CFU per gram of tissue from an individual mouse. Horizontal bars represent the geometric means of the colony counts. The range of detection in this assay is 102 to 109 CFU/ml of urine or CFU/g of tissue. Values of ≤102 were set to 102, and values of ≥109 were set to 109. P values (bottom) were derived by using an unpaired, one-tailed Mann-Whitney test. Lf., left; Rt., right. WT, wild-type strain; mrpH, mrpH::aphA mutant.
A dramatically different outcome was observed in a cochallenge experiment. A group of 10 CBA mice were challenged with a roughly 1:1 mixture of the wild-type HI4320 and its isogenic mrpH::aphA mutant (7.9 × 106 CFU of the wild type and 1.2 × 107 CFU of the mutant were inoculated into the bladder of each mouse). After 7 days, the mice were sacrificed and bacteria recovered from the urine, bladder, and kidneys were enumerated on both nonswarming plates, where both the wild-type HI4320 and the mrpH::aphA mutant would grow, and nonswarming plates containing kanamycin (50 μg/ml), where only the mrpH::aphA mutant would grow. The geometric mean values of bacterial count were as follows: urine, 7.52 (wild type) versus 2.70 (mutant) log10 CFU/ml (P = 0.003); bladder, 5.98 (wild type) versus 2.46 (mutant) log10 CFU/g (P = 0.003); left kidney, 4.87 (wild type) versus 2.33 (mutant) log10 CFU/g (P = 0.003); and right kidney, 4.82 (wild type) versus 2.67 (mutant) log10 CFU/g (P = 0.009) (Fig. 9B). The mrpH::aphA mutant was shown to be significantly less competitive than the wild-type strain in the ability to colonize the lower and upper urinary tracts of mice.
DISCUSSION
The newly acquired sequence of the 3′ end of the mrp gene cluster revealed two new open reading frames downstream of mrpG, designated mrpH and mrpJ. In this study, we tested the hypothesis that MrpH was the functional MR/P hemagglutinin. First, sequence analysis showed that MrpH has 30% amino acid sequence identity to PapG and 35% identity to SmfG, the fimbrial adhesins of P fimbria and Smf fimbria, respectively (21, 22). Second, the amino acid sequence alignment of putative MR/P pilins, MrpA, MrpB, MrpE, MrpF, MrpG, and MrpH, showed that MrpH is the only polypeptide that has a distinctive N-terminal domain that could provide a possible basis for the receptor-binding activity. Third, amino acid substitutions of two cysteine residues within this N-terminal domain of MrpH with serine residues abolished its receptor-binding activity. Similar observations were reported for Dr family adhesins (7). Finally, immunogold electron micrographs showed that MrpH is located at the fimbrial tip, a feature shared by many fimbrial adhesins including PapG. However, despite all the evidence suggesting that MrpH is the functional adhesin of MR/P fimbria, direct evidence awaits further studies on the receptor of MR/P fimbriae or, more specifically, MrpH.
Since the defect of the ΔmrpH mutant in making MR/P fimbriae in E. coli could be complemented by mrpH in trans, it is unlikely to be caused by any polar effect resulting from the deletion rather than the deletion of mrpH itself. This suggests that unlike the adhesin of other fimbriae including E. coli type 1 fimbria and P fimbria (15, 21), MrpH is necessary for normal fimbrial biogenesis. Since mrpH is predicted to encode a pilin that is transported into the periplasm and incorporated into fimbriae, it is unlikely that MrpH will elicit any regulatory effect on the transcription of the mrp gene cluster. The absence of degraded fimbrial debris in the immunogold electron micrographs of ΔmrpH mutant (data not shown) undermines the argument that MrpH simply stabilizes a completed fimbrial structure (i.e., without MrpH, the MR/P fimbriae may be quickly degraded). Rather, we believe that the involvement of MrpH in fimbrial assembly may be related to its unique C-terminal chaperone-binding domain, as described above. As proposed by Hultgren’s group (12–14), the defined fimbrial assembly process relies on the molecular chaperone-usher system; the differential affinities of the various pilins for the periplasmic chaperone and the outer membrane usher appear to influence the order of pilins incorporated into fimbriae. The unique C terminus of MrpH may lead to a unique interaction between the chaperone MrpD and itself, and the unique MrpD-MrpH complex may simply trigger the opening of a gate in the MrpC usher, thereby initiating the fimbrial assembly process. Since the MrpH adhesin is required for fimbrial biogenesis, the assembly of MR/P fimbriae better supports the chaperone-usher model (12–14), in which the sequential export of pilins would be blocked in the absence of the adhesin.
The mrpH::aphA mutant of P. mirabilis HI4320 and the ΔmrpH mutant of E. coli DH5α have similarities in their defective fimbrial production: much shorter fimbriae and fewer fimbriae per bacterium. However, there is a major difference between them. For the mrpH::aphA mutant of HI4320, the percentage of MR/P fimbriated bacteria drops dramatically, from about 50% in the wild type to less than 1% in the mutant. For the ΔmrpH mutant of E. coli DH5α, since the mrp promoter was fixed at the “on” position, the percentage of bacteria with at least one MR/P fimbria was the same as that for the wild type, i.e., >98%. It seemed that either the blockade on the fimbrial assembly in the absence of MrpH was less severe in E. coli DH5α than in P. mirabilis HI4320, which probably was simply due to the high-copy-number vector used to carry the mrp gene cluster in E. coli, or the mutation in mrpH feedback to the regulatory components, which was absent in E. coli (e.g., MrpI), shut down the expression of MR/P fimbria in P. mirabilis.
Unlike many other adhesins including PapG, FimH, and Dr adhesins, MrpH, as well as SmfG, has four (instead of two) cysteine residues at its N-terminal receptor-binding site, potentially forming two (instead of one) disulfide bonds. Replacement of the MrpH N-terminal cysteine residues by serine residues (C66S, C128S, and C66S plus C128S) destroyed the receptor-binding activity of MR/P without interfering with fimbrial assembly. Since the absence of any MR/P pilins leads to abnormal fimbrial biogenesis (references 3, 19, and 20 and unpublished data), the normal fimbrial production of these cysteine substitution mutants implies the presence of all MR/P pilins. It indicated that cysteine substitutions in MrpH did not hinder the export or function of another MR/P pilin, itself the adhesin, but, rather, destroyed the receptor-binding activity residing in MrpH itself. The data provide strong evidence that MrpH is the functional adhesin of the MR/P fimbria. Also, it indicated that the N-terminal cysteine residues of MrpH are crucial to its receptor-binding activity, which is consistent with the findings of other researchers (7). However, it is inconclusive from this study whether there are only one or two disulfide bonds and exactly how these disulfide bonds are formed between the four cysteine residues.
That the isogenic mrpH::aphA mutant of P. mirabilis HI4320 was unable to produce MR/P fimbriae but was positive for MRHA uncovered the existence of another mannose resistant hemagglutinin in this clinical isolate. Previous studies showed that mutations in mrpB (19) or mrpG (20) abolished MRHA, suggesting that MR/P fimbria was the only mannose resistant hemagglutinin produced by HI4320. Therefore, the mrpH::aphA mutant differs from the mrpB::aphA and mrpG::aphA mutants in that it may stimulate the expression of an otherwise silent mannose resistant hemagglutinin in strain HI4320. The independent-challenge experiment showed that unlike the mrpG::aphA mutant, which showed 102- to 104-fold reduction in colonization of the mouse urinary tract (20), the mrpH::aphA mutant was able to colonize the mouse urinary tract at levels similar to the wild-type strain. Thus, the alternate mannose-resistant hemagglutinin produced by the mrpH::aphA mutant may be able to substitute for MR/P fimbriae and help the mutant to colonize the mouse urinary tract. However, in the cochallenge experiment, the mrpH::aphA mutant was unable to outcompete the wild-type strain and colonization of mouse urinary tract by the mutant was very poor. This indicated that the affinity of any alternate mannose-resistant hemagglutinin produced by the mrpH::aphA mutant for the mucosal surfaces of the urinary tract may not be as high as that of the MR/P fimbria.
The fact that most of the chromosomal mutations constructed with the kanamycin resistance cassette can be complemented with a single gene in trans suggests that insertion of this cassette has little polar effect. Preliminary experiments on mrpJ show that it does not affect MR/P fimbrial biogenesis in vitro. An in vitro competition experiment showed no detectable growth advantage of the wild type over the mutant strain in either minimal A medium or Luria broth. Still, these in vitro controls cannot completely rule out the possible outgrowth of the wild-type strain over the mutant in vivo, the possible polar effect on mrpJ, or the possible regulatory function of MrpJ in vivo, all of which may contribute to the loss of virulence in vivo.
Given the strong association of the MR/P fimbria with pyelonephritogenic P. mirabilis strains (25), the selective pressure for the mrp gene cluster to be turned on in vivo, and the tip localization and adhesive property of MrpH, we propose that MrpH represents a promising vaccine candidate. Antibodies against MrpH could not only opsonize bacteria for subsequent clearance by the host immune system but also prevent bacterial attachment to mucosal surfaces, the first step in colonization. Studies by Langermann et al. (18) showed that FimH adhesin-based systemic vaccination prevented type 1-piliated E. coli colonization in mice, supporting the use of adhesins in vaccination.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant DK47920 from the National Institutes of Health.
We thank David J. McGee for helpful discussions and a critical review of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 10.8.1–10.8.17. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 16.6.1–16.6.14. [Google Scholar]
- 3.Bahrani F K, Massad G, Lockatell C V, Johnson D E, Russell R G, Warren J W, Mobley H L T. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1994;62:3363–3371. doi: 10.1128/iai.62.8.3363-3371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahrani F K, Mobley H L T. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J Bacteriol. 1994;176:3412–3419. doi: 10.1128/jb.176.11.3412-3419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahrani F K, Johnson D, Robbins D, Mobley H L T. Proteus mirabilis flagella and MR/P fimbriae: isolation, purification, N-terminal analysis, and antibody response following experimental urinary tract infection. Infect Immun. 1991;59:3574–3580. doi: 10.1128/iai.59.10.3574-3580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belas R, Erskine D, Flaherty D. Transposon mutagenesis in Proteus mirabilis. J Bacteriol. 1991;173:6289–6293. doi: 10.1128/jb.173.19.6289-6293.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnoy C, Moseley S L. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 8.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulk W P, Taylor C M. An immunocolloid method for the electron microscope. Immunochemistry. 1971;8:1081–1083. doi: 10.1016/0019-2791(71)90496-4. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 313–315. [Google Scholar]
- 11.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme III J W, Normark S. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 13.Hultgren S J, Normark S. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 14.Jones C H, Jacob-Dubuisson F, Dodson K, Kuehn M, Slonim L, Striker R, Hultgren S J. Adhesin presentation in bacteria requires molecular chaperones and ushers. Infect Immun. 1992;60:4445–4451. doi: 10.1128/iai.60.11.4445-4451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 16.Kuehn M J, Ogg D J, Kihlberg J, Slonim L N, Flemmer K, Bergfors T, Hultgren S J. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 17.Kuehn M J, Normark S, Hultgren S J. Immunoglobulin-like PapD chaperon caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc Natl Acad Sci USA. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Mobley H L T. MrpB functions as the terminator for assembly of P. mirabilis mannose-resistant Proteus-like fimbriae. Infect Immun. 1998;66:1759–1763. doi: 10.1128/iai.66.4.1759-1763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Zhao H, Geymonat L, Bahrani F K, Johnson D E, Mobley H L T. Proteus mirabilis mannose-resistant Proteus-like fimbriae: MrpG is located at the fimbrial tip and is required for fimbrial assembly. Infect Immun. 1997;65:1327–1334. doi: 10.1128/iai.65.4.1327-1334.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg F, Lund B, Johansson L, Normark S. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature. 1987;328:84–87. doi: 10.1038/328084a0. [DOI] [PubMed] [Google Scholar]
- 22.Mizunoe Y, Matsumoto T, Amato K, Sekiguchi M, Kumazawa J. Identification and nucleotide sequence of the gene determining the adhesion capacity of Serratia marcescens. J Bacteriol. 1991;173:3257–3260. doi: 10.1128/jb.173.10.3257-3260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mobley H L T, Belas R. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 1995;3:280–284. doi: 10.1016/s0966-842x(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 24.Mobley H L T, Island M D, Massad G. Virulence determinants of uropathogenic Escherichia coli and Proteus mirabilis. Kidney Int. 1994;46:S129–S136. [PubMed] [Google Scholar]
- 25.Mobley H L T, Chippendale G R. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J Infect Dis. 1990;161:525–630. doi: 10.1093/infdis/161.3.525. [DOI] [PubMed] [Google Scholar]
- 26.Mobley H L T, Warren J W. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Li X, Johnson D E, Blomfield I, Mobley H L T. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol Microbiol. 1997;23:1009–1019. doi: 10.1046/j.1365-2958.1997.2791645.x. [DOI] [PubMed] [Google Scholar]