Abstract
Exoenzyme S (ExoS), an ADP-ribosylating enzyme produced by the opportunistic pathogen Pseudomonas aeruginosa, is directly translocated into eukaryotic cells by bacterial contact. Within the cell, ExoS ADP-ribosylates the cell signaling protein Ras and causes inhibition of DNA synthesis and alterations in cytoskeletal structure. To further understand the interrelationship of the different cellular effects of ExoS, functional analyses were performed on HT-29 epithelial cells after exposure to ExoS-producing P. aeruginosa 388 and the non-ExoS-producing strain 388ΔS. Two different mechanisms of morphological alteration were identified: (i) a more-transient and less-severe cell rounding caused by the non-ExoS-producing strain 388ΔS and (ii) a more-severe, long-term cell rounding caused by ExoS-producing strain 388. Long-term effects of ExoS on cell morphology occurred in conjunction with ExoS-mediated inhibition of DNA synthesis and the ADP-ribosylation of Ras. ExoS was also found to cause alterations in HT-29 cell function, leading to the loss of cell adhesion and microvillus effacement. Nonadherent ExoS-treated cells remained viable but had a high proportion of modified Ras. While microvillus effacement was detected in both 388- and 388ΔS-treated cells, effacement was more prevalent and rapid in cells exposed to strain 388. We conclude from these studies that ExoS can have multiple effects on epithelial cell function, with more severe cellular alterations associated with the enzymatic modification of Ras. The finding that ExoS had greater effects on cell growth and adherence than on cell viability suggests that ExoS may contribute to the P. aeruginosa infectious process by rendering cells nonfunctional.
Pseudomonas aeruginosa is an opportunistic pathogen which affects compromised hosts such as individuals with cystic fibrosis, neutropenia, leukemia, and extensive wounds or burns. A number of proteins produced by P. aeruginosa contribute to its virulence, including exotoxin A, elastase, and phospholipase C, each through characterized mechanisms of action. Less is known about the cellular effects of exoenzyme S (ExoS), an ADP-ribosylating enzyme known to contribute to P. aeruginosa virulence by causing increased tissue damage and bacterial dissemination (14). Difficulties in elucidating the cellular effects of ExoS relate to its type III mechanism of secretion from P. aeruginosa, which requires direct contact between the bacterium and the target cell for the translocation of ExoS (34).
ExoS was first identified in P. aeruginosa 388 culture supernatants as an aggregate of two proteins: 49-kDa ExoS and 53-kDa ExoT (20, 23). These immunologically cross-reactive proteins (14) were subsequently found to be encoded by separate, coregulated genes, with ExoT having 75% amino acid identity with ExoS, but only 0.2% of the catalytic activity (33). The integral relationship between ExoS function and the eukaryotic cell environment was first indicated by the requirement of a eukaryotic cofactor, 14-3-3 proteins, for ExoS ADP-ribosyltransferase (ADPRT) activity (4). In vitro analyses identified multiple, functionally diverse proteins in mammalian cell lysates as substrates for ExoS ADPRT activity. Preferred substrates include specific low-molecular-mass GTP-binding (LMMG) proteins in the Ras superfamily, including Ras (3, 5), and the cytoskeletal protein vimentin (2).
Recognition of the requirement of bacterial contact for the induction and delivery of ExoS into eukaryotic cells led to the development of model systems which allowed bacterial translocation of ExoS and yet the differentiation of the cellular effects of ExoS from those of other Pseudomonas factors. The model system adapted by our laboratory used the prototype P. aeruginosa ExoS-producing strain 388 to translocate ExoS, differentiating the effects of ExoS from those of other strain 388 factors in comparative studies with the isogenic non-ExoS-producing strain 388ΔS (25). This system allowed the identification of inhibitory effects of ExoS production on DNA synthesis and cellular viability, thus confirming a toxic effect of ExoS on cell function (25). Later studies with this system identified Ras as an in vivo substrate of ExoS and found the efficiency of ADP-ribosylation of Ras to directly correlate with the inhibition of cellular DNA synthesis, suggesting a cause-and-effect relationship (22). In a different model system with the Yersinia type III secretory apparatus to translocate recombinant ExoS, ExoS expression was found to cause disruption of the actin cytoskeleton (7). In these latter studies, although more severe cell rounding was detected upon the translocation of wild-type ExoS, cell rounding was also caused by a mutated form of ExoS having a >2,000-fold-reduced ADPRT activity, thus indicating that cytoskeletal alterations could occur independently of this enzymatic activity.
The multiple and diverse effects of ExoS on cell function can be explained, to some extent, by its multidomain structure, which includes an amino-terminal domain possessing aggregation properties and required for ExoS secretion and a carboxy-terminal domain having ADPRT enzymatic activity (17, 34). Further diversity in the cellular effects of ExoS may also relate to Ras functioning as an in vivo substrate of ExoS and the potential of Ras to play a pivotal role in multiple signal transduction pathways. The purpose of the studies described here was to further characterize and define the cellular effects of ExoS upon its translocation into human epithelial cells via the P. aeruginosa type III secretory process. The studies identified additional effects of ExoS on cell adherence and microvillus effacement and distinguished two mechanisms by which P. aeruginosa 388 can cause alterations in HT-29 cell morphology: an ExoS-dependent mechanism and an ExoS-independent mechanism. More-severe or long-term effects of ExoS on cell function consistently correlate with the ADP-ribosylation of Ras, suggesting a role of ExoS enzymatic activity in permanent alterations of ExoS in cell function.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. aeruginosa strains used in this study include the parental strain 388 (1); strain 388ΔexoS, which lacks ExoS production due to an allelic exchange of the majority of the structural gene with a tetracycline gene cartridge (19); strain 388popD::Tc*, which lacks production of the PopD translocation protein due to the insertion of the omega cartridge encoding tetracycline resistance (31); and strain exs1::Tn1, which has a Tn1 insertion in the pscC type III secretory gene (34). Strain 388ΔS complementation studies were performed by using a two-step cloning strategy to construct a gentamicin-resistant plasmid containing the exoS structural gene. First, a 1.7-kb PstI/BamHI fragment containing the exoS structural gene was isolated from plasmid pUCPexoS (21, 34) by using the HindIII site within the multiple cloning site of pUCPexoS and the BamHI site downstream of the exoS structural gene. The 1.7-kb HindIII/BamHI fragment was then cloned into the broad-host-range vector pCP13 (6) digested with HindIII and BamHI. Ligation products were transformed into Escherichia coli JM109, and the exoS-containing pCP13 vector (pCP13exoS) was confirmed by restriction analyses. Second, a 1.6-kb HindIII fragment containing the gentamicin resistance cartridge from pGmΩ1 (29) was cloned into the HindIII site of pCP13exoS to yield pCP13exoSGm. Plasmid DNA isolated from gentamicin-resistant E. coli transformants was confirmed by restriction analyses and then transformed by triparental mating (12) into strain 388ΔS. Gentamicin-resistant 388ΔS clones were selected and maintained by using 250 to 300 μg of gentamicin per ml.
Bacteria were grown and prepared for coculture with HT-29 cells as previously described (22, 25), except when eukaryotic cell induction of ExoS effects on cell function were assessed. In these experiments bacteria were grown in non-ExoS-inducing medium containing unchelated Trypticase soy broth, 1% glycerol, and 100 mM monosodium glutamate (TSB). ExoS ADPRT activity in bacterial culture supernatants was quantified as previously described (25) and reported as femtomoles of ADP-ribose transferred per minute per milliliter of culture supernatant. For coculture studies, bacteria were diluted to the indicated concentration in McCoy 5A tissue culture medium containing 0.6% bovine serum albumin (McCoy-BSA) (Gibco-BRL, Gaithersburg, Md.). These suspensions were subsequently plated to confirm the bacterial concentration and to determine the multiplicity of infection (MOI) in HT-29 cell coculture studies. MOIs of 10:1 to 100:1 bacterial versus eukaryotic cells were used in these studies, as specifically indicated, with this range previously found to be optimal for detection of effects of ExoS on cell function and Ras modification (22, 25).
HT-29 cell culture.
HT-29 cells, a human carcinoma cell line obtained from the American Type Culture Collection (ATCC), were cultured as specified by ATCC in McCoy 5A medium supplemented with 10% fetal bovine serum (McCoy-FBS) at 37°C in 5% CO2–95% air. Cells were split 1:6 and passaged as the culture reached confluence. For coculture with bacteria, cells were detached from the growth surface with 0.25% trypsin–1 mM EDTA (trypsin-EDTA) (Gibco-BRL), resuspended in McCoy-FBS, counted, diluted to the appropriate density in McCoy-FBS, and then seeded in 48- or 6-well plates (Costar, Cambridge, Mass.) and cultured for 48 h. Bacteria were cocultured with HT-29 cell monolayers for 1 to 4 h, and HT-29 cells were processed and assayed in cell function assays described below.
Examination of bacterial effects on HT-29 cell function.
In all experiments, control and experimental cells were identically treated, except that in controls, coculture medium did not contain bacteria.
(i) Quantification of DNA synthesis.
After the coculture period, bacteria were removed from HT-29 cell monolayers; monolayers were then washed with McCoy-FBS containing 200 μg of gentamicin/ml and 100 μg of ciprofloxacin/ml (McCoy-FBS-GC), pulsed in McCoy-FBS-GC, and analyzed for [3H]thymidine incorporation after 20 h, as previously described (25).
(ii) Immunoprecipitation, detection, and quantification of Ras proteins.
Ras was immunoprecipitated from cells after coculture with bacteria by using monoclonal Y13-259 Ras antibody (ATCC), rabbit anti-rat immunoglobulin G (Sigma), and protein A-Sepharose (Sigma), as previously described (22). Proteins were resolved on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels, and Ras immunoblots were developed by using pan-Ras OP22 antibody (Oncogene Research, Cambridge, Mass.) and enhanced chemiluminescence (Amersham).
(iii) Examination of cell morphology.
Long-term effects of strain 388 and mutant strains on cell morphology were assessed by phase-contrast microscopy in conjunction with DNA synthesis assays at the end of the 20-h pulse period. For video time-lapse microscopy, cells were grown and cultured with bacteria in 25-cm2 flasks. After the removal of bacteria and the addition of medium containing antibiotics, culture vessels were placed in an incubation chamber mounted on the stage of an adapted Zeiss ICM-405 inverted phase-contrast microscope. The temperature in the chamber was maintained at 37°C, and the atmosphere contained 5% CO2–95% air. Selected fields of ca. 100 cells were examined by using a 40× objective lens, and images were captured by using a Hitachi KP-M1U charge-coupled device camera. Recordings were made at a temporal ratio of 20:1 by using a time-lapse video recorder.
(iv) Analysis of cellular adherence.
After a 3-h coculture period, bacteria were removed, HT-29 cell monolayers were washed three times with McCoy-FBS-GC, and then cells were detached from the growth surface with trypsin-EDTA. Detached cells were resuspended in the original volume of McCoy-FBS-GC, replated, and allowed to grow for 20 h. At this time, nonadherent cells were recovered and saved and adherent cells were detached with trypsin-EDTA. Each cell population was assayed for cell number, viability, and necrosis by trypan blue exclusion, as previously described (25), and for DNA synthesis, Ras modification, and apoptosis.
(v) Analysis of apoptosis.
To quantify internucleosomal fragmentation, 106 HT-29 cells, treated as indicated, were lysed, and DNA fragmentation was analyzed in a Cell Death Detection enzyme-linked immunosorbent assay (ELISA; Boehringer Mannheim, Indianapolis, Ind.). The relative amount of apoptosis was determined by calculating an enrichment factor in which the absorbance of the sample (after subtracting the background level) was divided by the absorbance of the control.
Scanning electron microscopy.
Bacterial strains 388, 388ΔS, ΔPopD, and ΔPscC (108 CFU/ml) or no bacteria were cultured for 1, 2, or 3 h with HT-29 cell monolayers grown on 12-mm glass coverslips. Bacteria were removed, and cells were washed twice with McCoy-BSA, fixed in situ in 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), rinsed, postfixed in 1% osmium in 0.1 M cacodylate buffer, and then dehydrated in a graded series of ethanol mixtures and treated with hexamethyldisilane. After they were dried, coverslips were coated with gold and examined by using a JEOL JSEM-LV5410 scanning electron microscope.
RESULTS
Identification of ExoS-specific effects on HT-29 cell DNA synthesis and morphology.
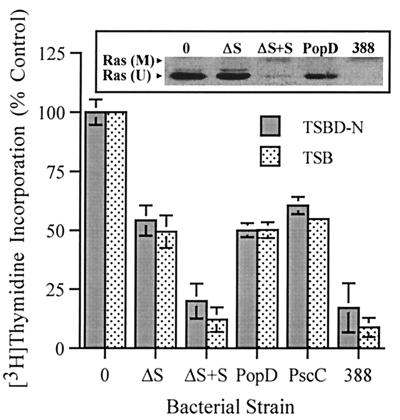
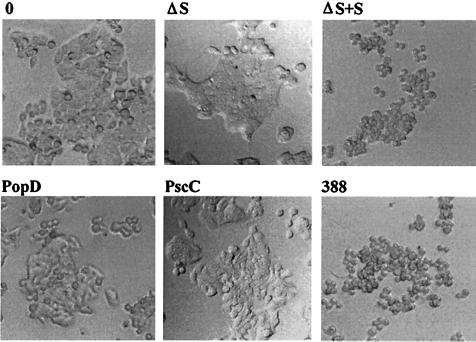
Identifying the effects of ExoS on cell function has been complicated by its type III mechanism of secretion and the requirement for bacterial contact in the delivery of ExoS into target cells. To differentiate the cellular effects of ExoS from those of other bacterial factors during coculture studies, P. aeruginosa strains having mutations in ExoS production or components of the type III secretory process were compared for their effects on DNA synthesis and cellular morphology. The specific strains examined included the following: (i) ExoS-producing strain 388; (ii) the non-ExoS-producing isogenic strain 388ΔS; (iii) strain 388ΔS complemented with a pCP13exoSGm plasmid containing the exoS structural gene (ΔS+S); (iv) strain 388popD::Tc*, which lacks expression of PopD, a homolog of Yersinia YopD involved in type III translocation (ΔPopD) (36); and (v) strain 388 exs1::Tn1, which lacks production of PscC, a homolog of the Yersinia YscC protein involved in type III secretion (ΔPscC) (34). The differential effect of each of the 388 strains on HT-29 epithelial cell function was examined relative to the induction or noninduction of ExoS production prior to the coculture period. High levels of ExoS production were confirmed in culture supernatants of strains 388 and ΔS+S grown in TSBD-N inducing medium prior to coculture with HT-29 cells (46.6 and 36.9 fmol of ADP-ribose transferred min−1 ml−1, respectively), but not in the respective strains grown in TSB noninducing medium (<0.3 fmol of ADP-ribose transferred min−1 ml−1). Regardless of the bacterial preculture conditions, the same pattern of DNA synthesis inhibition by the bacterial strains was detected, indicating that contact with HT-29 cells is sufficient for the induction of ExoS effects on DNA synthesis (Fig. 1). Strains 388 and ΔS+S were found to cause the greatest (and comparable) levels of inhibition of [3H]thymidine incorporation (∼80%), which correlated with an equally efficient shift in Ras mobility (Fig. 1, inset). Less inhibition of DNA synthesis was caused by strains 388ΔS, ΔPopD, and ΔPscC (∼50%), the comparable inhibition by these mutants reflecting an undefined bacterial factor not related to ExoS production nor the expression of the respective type III secretory products. The 30% greater inhibition of DNA synthesis and the modification of Ras by strains 388 and ΔS+S, when compared to that of 388ΔS, identifies an ExoS-specific effect. The lack of this 30% greater inhibition in the type III secretory mutants also confirms the requirement of the P. aeruginosa PopD translocation and PscC secretion proteins for ExoS effects on DNA synthesis. When HT-29 cell morphological alterations were examined 20 h after exposure to bacteria, at the end of the DNA pulse period, severe rounding was detected in HT-29 cells exposed to strains 388 and ΔS+S but not in HT-29 cells exposed to the other bacterial strains (Fig. 2). We conclude from these studies that ExoS production by strain 388 is directly associated with increased inhibition of DNA synthesis, Ras modification, and long-term alterations in cell morphology.
FIG. 1.
ExoS-specific effect on cell proliferation and Ras modification. DNA synthesis was examined in HT-29 cells, seeded at 105 cells/well, cultured for 48 h, and then cocultured with 107 CFU of 388, 388ΔS (ΔS), 388ΔS complemented with ExoS (ΔS+S), ΔPopD, or ΔPscC bacteria per ml (MOI of 10:1). Bacteria were either induced or noninduced for ExoS production in TSBD-N or TSB media, respectively, prior to coculture with HT-29 cells. After 4 h, bacteria were removed and replaced with medium containing [3H]thymidine and antibiotics to inhibit further bacterial growth, and DNA synthesis was assayed after 20 h. The means and standard deviations of assays performed in triplicate are indicated, and the results are expressed as percentages of non-bacterium-treated monolayer DNA synthesis. (Inset) To analyze for Ras modification, cells were cocultured in a manner identical to that described above, the culture supernatants were removed, the cells were lysed, and Ras was immunoprecipitated by using Ras monoclonal Y13-259 antibody. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Ras immunoblots were developed by using pan-Ras OP22 antibody and detected by using enhanced chemiluminescence. The mobility of unmodified Ras (U) and modified Ras (M) are indicated.
FIG. 2.
ExoS-specific effects on HT-29 cell morphology. HT-29 cells were cocultured with strain 388, 388ΔS (ΔS), ΔS+S, ΔPopD, or ΔPscC and examined for morphological alterations by phase-contrast microscopy after 20 h. Studies were performed in conjunction with the DNA synthesis assays shown in Fig. 1.
Examination of the temporal course of morphological changes in HT-29 cells caused by strain 388.
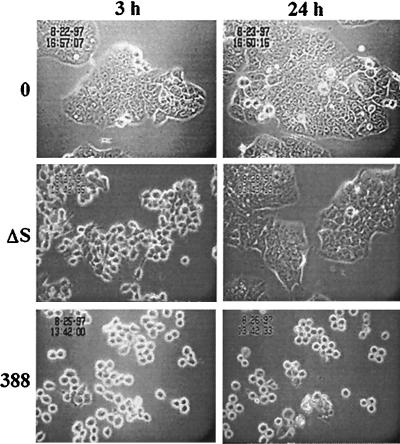
Further understanding of the effects of ExoS on HT-29 cell morphology was gained by using time-lapse microscopy. In these studies, HT-29 cells were cocultured with strain 388 or 388ΔS for 3 h, and then bacteria were removed and the cells were treated with antibiotics to inhibit further bacterial growth and monitored for morphological alterations over a 24-h period. Cell rounding was evident 3 h after exposure to both strains 388 and 388ΔS compared to non-bacterium-treated control cells, but the rounding caused by strain 388 was more severe (Fig. 3). The differential effects of strains 388 and 388ΔS on cell morphology became more evident after 24 h, at which time cells exposed to strain 388ΔS regained normal morphology, while those exposed to strain 388 remained rounded. These studies draw attention to different mechanisms by which strain 388 can cause morphological alterations. One mechanism, which appears less severe and more transient, is recognized in strain 388ΔS and occurs in the absence of enzymatically active ExoS. A second, which causes a long-term alteration in cellular morphology, occurs in the presence of enzymatically active ExoS.
FIG. 3.
Time-lapse video analysis of effects of strains 388 and 388ΔS on HT-29 cell morphology. HT-29 cells were seeded at 5 × 105 cells/ml in 25-cm2 tissue culture flasks and grown overnight to ∼30% confluency. Cells were then cocultured with 108 CFU of 388 or 388ΔS bacteria per ml (MOI of 20:1) or no bacteria (0) for 3 h. Bacteria were removed and replaced with medium containing antibiotics. A single field of each culture was videotaped for 24 h by using a Zeiss ICM-405 inverted-phase microscope equipped with a warm stage heater-recirculator device and maintained at 37°C in a 5% CO2–95% air atmosphere. Illustrative images obtained in a single field were captured and recorded at a temporal ratio of 20:1. Magnification, ×40.
Effects of ExoS production on cell adherence.
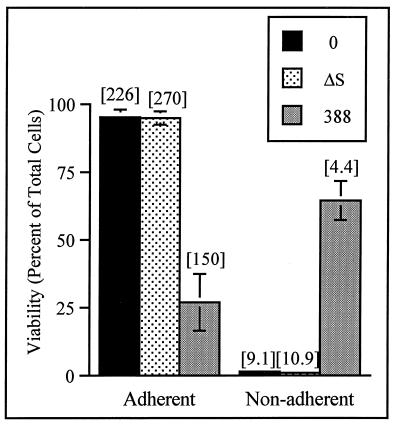
Functional consequences of differential effects of strains 388 and 388ΔS on cellular morphology were examined by using cell re-adherence assays. Since minimal loss of HT-29 cell adherence to cell matrix was observed in association with cell rounding after the 3-h coculture period with either strain 388 or strain 388ΔS, adherence was functionally evaluated in replating assays where HT-29 cells were passaged after a 3-h exposure to bacteria. Cell viability and the efficiency of re-adherence were then examined 20 h later. As shown in Fig. 4, more than 95% of HT-29 cells exposed to strain 388ΔS were able to re-adhere at 20 h. This compared to only 27% of the 388-treated cells being able to re-adhere, with 65% of the 388-treated cells remaining viable but nonadherent. DNA synthesis assays detected the relative rate of [3H]thymidine incorporation in cells exposed to strain 388 to be approximately twofold lower than in cells exposed to strain 388ΔS, regardless of the re-adherence. When the eventual fate of the 388-treated cells was assessed, the number of necrotic cells was two- to threefold higher after treatment with strain 388 compared to treatment with strain 388ΔS or with no bacteria, averaging 13.2 ± 3.6% at 24 h and 26.4 ± 6.6% at 48 h. A similar relative increase in the number of apoptotic cells was detected in 388-treated cells with an ELISA to quantify nucleosomal DNA fragments present in the cell lysates. Levels of apoptosis in cells exposed to strains 388 and 388ΔS were increased by factors of 7.4 ± 1.1 and 2.6 ± 1.1, respectively, compared to non-bacterium-treated control cells. A DNA ladder, characteristic of apoptosis, was detected in HT-29 cells exposed to strain 388 but not in cells exposed to strain 388ΔS or to no bacteria (not shown). While we cannot determine from our studies whether the observed decrease in cell viability after exposure to strain 388 was due to loss of adherence or a direct effect of ExoS, recent studies have found the ADPRT domain of ExoS to be cytotoxic when transiently expressed in CHO and Vero cells (26), implying that both mechanisms are possible. The finding, however, that cell viability was maintained in a high percentage of 388-treated cells 20 h after exposure to bacteria indicates that the immediate effect of ExoS on HT-29 cells was not on viability but on cell morphology, adherence, and DNA synthesis.
FIG. 4.
Effect of ExoS production on HT-29 cell re-adherence. HT-29 cells were cocultured with 108 CFU of bacteria per ml for 3 h (MOI of 30:1); bacteria were then removed, and cells were detached by trypsin-EDTA treatment and reseeded in culture wells. At 20 h, adherent and nonadherent cells were harvested separately, cell viability and numbers were assessed by trypan blue staining, and DNA synthesis was assayed as described in Fig. 1. Viability is expressed as the percentage of total adherent plus nonadherent cells, and the means and standard errors of three independent assays are shown. DNA synthesis, indicated in brackets above bars, is reported as the disintegrations per minute per 1,000 cells in the respective adherent or nonadherent populations. Results of one of three independent DNA assays are shown, and the mean of assays performed in triplicate is represented.
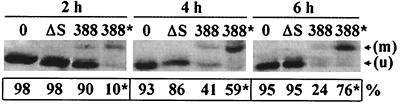
When alterations in cell adherence were related to Ras modification, Ras was found to be more efficiently modified in nonadherent than in adherent 388-treated cells. In time course studies comparing Ras modification and loss of adherence to increasing time of exposure to bacteria, Ras modification was found to precede the loss of re-adherence (Fig. 5). In the adherent 388-treated HT-29 cell population, levels of unmodified Ras decreased with increasing time, a finding which corresponded to an increased proportion of modified Ras in the nonadherent 388-treated HT-29 cells. These studies imply that the ADP-ribosylation of Ras by ExoS is coordinated with the loss of HT-29 cell matrix adherence.
FIG. 5.
Association of Ras modification with loss of adherence after exposure to ExoS-producing bacteria. HT-29 cells were cocultured with no bacteria (0) or with 108 CFU of strain 388 or strain 388ΔS (ΔS) bacteria per ml for 2, 4, or 6 h (MOI of 95:1). Bacteria were removed, and cells were reseeded in culture wells; adherent and nonadherent cells were then harvested after 20 h as described for Fig. 4. Ras was immunoprecipitated from cell lysates, and Ras modification was analyzed as described in Fig. 1 in adherent cells treated with no bacteria or with strain 388ΔS and in both adherent and nonadherent (indicated by an asterisk) cells treated with strain 388. The percentage of adherent or nonadherent viable cells in each population is indicated below the respective Ras modification patterns. The mobility of unmodified Ras (u) and modified Ras (m) is indicated.
Examination of HT-29 cellular alterations associated with ExoS production and type III secretory processes.
Scanning electron microscopy was used to compare the progression of cellular changes after a 1-, 2-, or 3-h exposure of HT-29 cells to strains 388, 388ΔS, ΔPopD, and ΔPscC. As shown in Fig. 6A and B, cell rounding was evident in HT-29 cells after a 1- or 3-h exposure to strain 388 and, in many instances, rounded cells showed a severe loss of cell surface microvilli. Bacteria appeared to remain securely attached to HT-29 cells in the absence of microvilli, and no cellular structures, such as pedestals, were evident underneath the bacteria. While cell rounding and loss of microvilli were also apparent in cells exposed to strain 388ΔS, both events occurred more slowly, with loss of microvilli detected only after 3 h and three- to fourfold less frequently than was observed with strain 388. Cell rounding was also detected in HT-29 cells after coculture with strains ΔPopD and ΔPscC; however, no notable loss of microvilli was observed during the 3-h exposure to bacteria. The finding that ExoS facilitates but is not required for cell rounding or effacement draws attention to the ability of non-ExoS-related bacterial factors to initiate alterations in cell structure and loss of microvilli. The inability of PopD and PscC mutant strains to cause effacement within the same time frame also implies the role of the type III secretory process in cellular alterations leading to the loss of HT-29 cell surface microvilli.
FIG. 6.
Scanning electron microscopy of HT-29 cells cocultured with ExoS-producing bacteria, non-ExoS-producing bacteria, or type III secretory mutant bacteria. Strains 388, 388ΔS, ΔPopD, and ΔPscC (MOI of ∼30:1) were cocultured for 1, 2, or 3 h with HT-29 monolayers grown on 12-mm-diameter glass coverslips. Cells were then washed, fixed in situ in 2% glutaraldehyde, postfixed in 1% osmium, dehydrated in a graded series of ethanol mixtures, and treated with hexamethyldisilane. After cells were dried, the coverslips were coated with gold and examined by using a JEOL JSEM-LV5410 scanning electron microscope. Images representative of indicated time points are shown, with each image at a ×5,000 magnification. (A and B) Cells treated with strain 388 show cell rounding and loss of cell surface microvilli after 1 and 3 h of exposure to bacteria. (C and D) Cells treated with strain 388ΔS show cell rounding after 1 and 3 h, with microvillus effacement not evident until 3 h. (E and F) Cells treated with strains ΔPopD and ΔPscC show cell rounding but no microvillus effacement after 3 h.
DISCUSSION
As an understanding of ExoS and its regulatory and secretory processes unfolds, it becomes increasingly evident that ExoS functions in a manner distinctly different from other known P. aeruginosa virulence factors. ExoS is part of a complex regulon that includes a type III secretory process and responds to eukaryotic cell contact. The contact-mediated translocation of ExoS into target cells, as with other type III secreted proteins, allows ExoS direct access to eukaryotic cell signaling processes. Current data support that ExoS can have multiple effects on cell function, which likely relates to independent, but coordinated, activities of the amino- and carboxy-terminal domains of ExoS, as well as to its enzymatic modification of cellular Ras, a protein integral to cell signaling through multiple pathways.
To gain a more precise understanding of the interrelationship of cellular processes involved in the effects of ExoS on cell function, the differential effects of the ExoS-producing strain 388 on HT-29 epithelial cell function were compared to those of 388 mutant strains lacking production of ExoS or defined components of the type III secretory system. A 30% greater inhibition of DNA synthesis was identified as being a direct result of the production and type III-mediated translocation of ExoS. This occurred in addition to an approximate 50% inhibition of HT-29 cell DNA synthesis caused by non-ExoS- or non-type III-related bacterial factors produced by strain 388 during the coculture period. While the bacterial factors responsible for the non-ExoS-related inhibition have not been specifically investigated, this inhibition appears to differ from that of ExoS in becoming more pronounced upon prolonged exposure to bacteria. Previous studies found the differential effects of ExoS production on DNA synthesis to be initiated relatively early in the coculture period, detectable after a 0.5-h exposure to bacteria, but becoming optimal within a 3- to 4-h exposure (22, 25). Other, more generalized effects on cell function, including inhibition of protein synthesis, decreased cell viability, as well as increased inhibition of DNA synthesis, become apparent upon prolonged coculture times or exposure to higher concentrations of bacteria. The complex and multiple effects of strain 388 on cell function reflect the multifactorial nature of P. aeruginosa virulence. The effects of ExoS on Ras modification and DNA synthesis, however, appear to precede other effects on cell function, implying a role in early stages of the P. aeruginosa infectious process.
Consistent with common or coordinated signaling events being involved in the effects of ExoS on DNA synthesis and cytoskeletal alterations, both events were initiated early in the coculture period, required an intact type III secretory apparatus, and were associated with the ADP-ribosylation of Ras. Time-lapse video studies revealed that strain 388 could cause alterations in cell morphology by an ExoS-independent mechanism, as well as by an ExoS-dependent mechanism. Long-term alterations in cell morphology required ExoS production, while short-term alterations in morphology were apparent in strain 388ΔS-treated HT-29 cells, and both appeared to be facilitated by an intact type III secretory process. Strain 388ΔS produces two known type III effector proteins, ExoT and ExoY, both of which have been found to cause cell rounding (30, 37). It is notable that the less severe, short-term cell rounding detected in HT-29 cells cocultured with strain 388ΔS resembled that of the less severe cell rounding observed by Frithz-Lindsten et al. (7) when the effects of wild-type ExoS and of the E381A enzymatic mutant form of ExoS on HeLa cells were compared. Although the effects of an ADPRT inactive form of ExoS were not directly examined in our studies, strain 388ΔS produces ExoT, which is highly homologous to ExoS, yet it has <0.2% of the catalytic activity. Thus, it is possible that ExoT, produced by strain 388ΔS, may be functioning in a manner similar to that of E381A mutant ExoS in causing the less-severe, short-term cell rounding. The amino-terminal domain of ExoS shows homology with the Yersinia virulence factor YopE (35), a Yersinia cytotoxin that disrupts cytoskeletal structure by an as-yet-undefined mechanism (28). Similarly, the amino-terminal domain of SptP, a virulence factor secreted by Salmonella typhimurium via the type III process, has been found to share homology with YopE and ExoS and cause a similar disruption of actin cytoskeleton structure (8). The seemingly comparable alteration in cell morphology caused by type III effector proteins, independent of other identified enzymatic activities, suggests that a transient alteration in host cytoskeletal structure may be a common feature of proteins translocated by the type III secretory process.
Functional consequences of differences in the effects of strains 388 and 388ΔS on HT-29 cell morphology became more evident in cellular adherence analyses. Although neither strain 388 nor strain 388ΔS caused significant loss of HT-29 cell adherence during the coculture period, when cells were passaged after exposure to bacteria and assayed for re-adherence, less than one-third of strain 388-treated cells were able to re-adhere to tissue culture wells. The ADP-ribosylation of Ras by ExoS appeared to precede and increase with the loss of re-adherence, suggesting coordination between loss of adherence and Ras modification. These studies indicate that long-term alterations in cell morphology caused by strain 388 can be functionally differentiated from the short-term alterations of strain 388ΔS, based on their association with the disruption of focal adhesion. Focal adhesion complexes play an integral role in the transduction of cellular signals, and the formation of stable focal adhesion complexes are linked to the actin cytoskeletal structure (38). The effects of ExoS on cell adherence and cytoskeletal structure could therefore both be linked to an interruption of actin polymerization. The previous finding that ExoS can exert less-severe effects on cell morphology in an ADPRT-independent manner (7), coupled with our observation that more-severe cytoskeletal alterations and loss of cell matrix adherence occur in association with Ras modification, is consistent with the multifunctional domain structure of ExoS, with the amino-terminal YopE homologous region of ExoS causing transient alterations in cytoskeletal structure that are coordinated with more-severe cytoskeletal alterations caused by the ADP-ribosylation of cellular proteins by the carboxy-terminal domain.
In previous studies, the modification of Ras by ExoS was found to correlate directly with inhibition of DNA synthesis (22). Although a large number of Ras-effector interactions have been identified (16), the best characterized of these, the activation of Raf by interaction with Ras, induces the transfer of a proliferative or differentiation signal to the nucleus. Recent in vitro and in vivo studies have found that the ADP-ribosylation of Ras by ExoS interferes with Raf activation (10, 32), thus providing a mechanism for the inhibitory effects of ExoS on DNA synthesis and PC12 neurite outgrowth (9, 25). A current understanding of cell signaling pathways finds that Ras can also affect cytoskeletal structure through indirect interactions with the Rho subfamily of LMMG proteins that regulate actin polymerization. Rho, Rac, and Cdc42 play distinct roles in actin rearrangement, forming stress fibers, lamellipodia, and filopodia, respectively. These three proteins also form an interactive network, with Cdc42 able to activate Rac, and Rac able to activate Rho, and all three can associate with integrin-based adhesion complexes (reviewed in references 11 and 13). Ras, via its direct interaction with phophatidylinositol 3-kinase (PI3K), can activate Rac and cause actin rearrangement (27). The guanine nucleotide exchange factor Sos has also been found to couple Ras with Rac in a PI3K-dependent manner (24). Thus, it is possible that effects of ExoS on cytoskeletal structure may result from the modification of Ras. An alternative explanation for the cytoskeletal effects of ExoS relates to the potential diverse substrate specificity of ExoS and the possibility that ExoS might directly modify or alter the function of proteins, such as Rac, Cdc42, Rho, or the intermediate filament vimentin, that affect cytoskeletal structure. Effects of ExoS on cell morphology and adherence would thus appear to be coordinated with Ras modification and the inhibition of DNA synthesis but mediated by the modification of proteins in alternative signal transduction pathways.
While many similarities are observed in eukaryotic cell functions altered by the type III secretory process of different bacteria, each bacterium appears to manipulate eukaryotic pathways in a slightly different manner which best enhances the survival of the specific organism. P. aeruginosa, like enteropathogenic E. coli (EPEC), is predominantly an extracellular organism, and both bacteria use type III secreted proteins to mediate their infectious processes. Electron microscopy studies revealed that contact between EPEC and host epithelial cells results in the effacement of microvilli from the surface of cells and the production of a densely packed cytoskeletal structure forming a pedestal beneath the bacterium (18). Microvilli effacement and pedestal formation were found to be associated with the accumulation of cellular actin and to require the EPEC type III secretory system (15). When the interaction of the ExoS-producing strain 388 with HT-29 cells was examined by scanning electron microscopy, microvillus effacement, similar to that observed for EPEC, was detected, often in conjunction with severe cell rounding. No pedestal-like structures, however, were evident underneath strain 388 P. aeruginosa. Some microvillus effacement was also apparent in HT-29 cells exposed to strain 388ΔS, indicating that the process can occur independently of ExoS production; however, effacement was facilitated by ExoS production. Scanning electron microscopy revealed minimal loss of microvilli in HT-29 cells treated with strain 388 bacteria having mutations in their type III secretory apparatus, indicating, as with EPEC, that the type III secretory system facilitates the P. aeruginosa effacement process. Thus, like other bacteria whose infectious process is mediated by the type III secretory system, P. aeruginosa appears to have acquired a slightly different mechanism of manipulating eukaryotic cell function, but alterations in actin cytoskeletal structure appear to be part of this mechanism.
The studies described here have increased the list of effects that ExoS can have on cell function, providing further insight into the complex effects of ExoS on eukaryotic cell signaling processes. Although the coordinated activities of both enzymatic and nonenzymatic portions of ExoS cause alterations in cell function, the direct association of the ADP-ribosylation of Ras with effects of ExoS on DNA synthesis, cell adherence, and long-term morphological alterations are consistent with ExoS ADPRT activity contributing to the more-severe alterations in cell function. While different signaling pathways may be involved in these cellular effects, a common signal leading to an interruption of cell function appears to be delivered to HT-29 epithelial cells. This becomes most apparent in functional analyses of HT-29 cell populations that lose adherence after exposure to ExoS-producing bacteria. These cells have a lower DNA proliferative index, lack functional focal adhesion complexes and cytoskeletal structures required for the integration of cell signals, and have a high proportion of ADP-ribosylated Ras, and yet the majority of cells remain viable after 48 h. This indicates that the immediate outcome of exposure to ExoS is not cell death but rather an inhibition of normal cellular processes required for growth and adherence. While the effects of ExoS on cell viability can be observed upon more-prolonged exposure to bacteria (25), we hypothesize that the initial role of ExoS in the P. aeruginosa infectious process is one of cellular inactivation. ExoS production, in combination with other P. aeruginosa virulence factors, then leads to the eventual death of the target cell via necrotic and apoptotic mechanisms. Relative to epithelial tissue, an interruption of cell growth and adherence would result in increased bacterial dissemination.
ACKNOWLEDGMENTS
We thank Dennis Ohman for his help in the construction of the pCP13exoSGm plasmid used in the 388ΔS complementation studies. We also thank Debra Hazen-Martin and Carol Moskos for their assistance in the scanning electron microscopy studies and Lisa Rucks for her assistance in the coculture studies.
This work was supported by NIH grant AI41694.
REFERENCES
- 1.Bjorn M J, Pavlovskis O R, Thompson M R, Iglewski B H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979;24:837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coburn J, Dillon S T, Iglewski B H, Gill D M. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coburn J, Gill D M. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn J, Kane A V, Feig L, Gill D M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 5.Coburn J, Wyatt R T, Iglewski B H, Gill D M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–9008. [PubMed] [Google Scholar]
- 6.Darzins A, Chakrabarty A M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, Galan J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 9.Ganesan A K, Frank D W, Misra R P, Schmidt G, Barbieri J T. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 10.Ganesan, A. K., T. S. Vincent, J. C. Olson, and J. T. Barbieri. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor catalyzed nucleotide exchange. Submitted for publication. [DOI] [PubMed]
- 11.Giancotti F G. Integrin signaling: specificity and control of cell survival and cell cycle progression. Curr Opin Cell Biol. 1997;9:691–700. doi: 10.1016/s0955-0674(97)80123-8. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 14.Iglewski B H. Pseudomonas toxins. In: Hardegree M C, Tu A T, editors. Handbook of toxins. Vol. 4. New York, N.Y: Marcel Dekker; 1988. pp. 249–265. [Google Scholar]
- 15.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 17.Knight D A, Finck-Barbancon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulich S M, Frank D W, Barbieri J T. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect Immun. 1995;63:1–8. doi: 10.1128/iai.63.1.1-8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulich S M, Frank D W, Barbieri J T. Purification and characterization of exoenzyme S from Pseudomonas aeruginosa 388. Infect Immun. 1993;61:307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 22.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicas T I, Iglewski B H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 25.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pederson K J, Barbieri J T. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol Microbiol. 1998;30:751–759. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 28.Rosqvist R, Forsberg A, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweizer H P. Small broad-host range gentamicin cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 30.Vallis A J, Finck-Barbancon V, Yahr T L, Frank D W. Biological effects of Pseudomonas type III-secreted proteins on CHO cells. Infect Immun. 1998;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallis A J, Yahr T L, Barbieri J T, Frank D W. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol., in press. [DOI] [PubMed]
- 33.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 35.Yahr T L, Hovey A K, Kulich S M, Frank D W. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yahr T L, Mende-Mueller L M, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yahr T L, Vallis A J, Hancock M K, Barbieri J T, Frank D W. ExoY, an adenylate cyclase secreted by Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada K M, Geiger B. Molecular interactions in cell adhesion complexes. Curr Opin Cell Biol. 1997;9:76–85. doi: 10.1016/s0955-0674(97)80155-x. [DOI] [PubMed] [Google Scholar]