Abstract
A low-molecular-weight protein named NspA (neisserial surface protein A) was recently identified in the outer membrane of all Neisseria meningitidis strains tested. Antibodies directed against this protein were shown to protect mice against an experimental meningococcal infection. Hybridization experiments clearly demonstrated that the nspA gene was also present in the genomes of the 15 Neisseria gonorrhoeae strains tested. Cloning and sequencing of the nspA gene of N. gonorrhoeae B2 revealed an open reading frame of 525 nucleotides coding for a polypeptide of 174 amino acid residues, with a calculated molecular weight of 18,316 and a pI of 10.21. Comparison of the predicted amino acid sequence of the NspA polypeptides from the gonococcal strains B2 and FA1090, together with that of the meningococcal strain 608B, revealed an identity of 93%, suggesting that the NspA protein is highly conserved among pathogenic Neisseria strains. The level of identity rose to 98% when only the two gonococcal predicted NspA polypeptides were compared. To evaluate the level of antigenic conservation of the gonococcal NspA protein, monoclonal antibodies (MAbs) were generated. Four of the seven NspA-specific MAbs described in this report recognized their corresponding epitope in 100% of the 51 N. gonorrhoeae strains tested. Radioimmunobinding assays clearly indicated that the gonococcal NspA protein is exposed at the surface of intact cells.
Neisseria meningitidis and Neisseria gonorrhoeae are pathogenic Neisseria species. These species, which cause quite dissimilar diseases, are closely related, having more than 80% DNA genome homology and up to 98% sequence similarity for housekeeping genes (18, 40). This high degree of relatedness is reflected in their many common genetic, biochemical, and antigenic features. For example, it was shown that N. meningitidis produces proteins highly similar to the gonococcal PI (2, 12, 17, 21), PII (3, 22, 33), and PIII (6, 16) outer membrane (OM) proteins as well as the pilin protein (30, 34), the iron-repressible proteins (32), and the H.8 antigen (5, 9, 10, 16). The high levels of inter- and intrastrain antigenic variations of the OM components of N. gonorrhoeae appear to allow this organism to evade the host immune system and limit the capacity of those antigens to serve as vaccines (37). Identification of conserved antigens is of great interest, considering the high levels of heterogeneity and antigenic variations for the different gonococcal outer membrane components.
Martin et al. (28) recently reported the identification in the OM of N. meningitidis of a low-molecular-weight protein, which they named NspA (neisserial surface protein A). Using NspA-specific monoclonal antibodies (MAbs), they showed that this protein was antigenically highly conserved and accessible at the surface of intact bacterial cells of all N. meningitidis isolates tested. Two of these NspA-specific MAbs were shown to be bactericidal in vitro against several meningococcal isolates (27). Intraperitoneal injection of these bactericidal MAbs passively protected mice against a lethal meningococcal challenge. It was also demonstrated that the injection of recombinant NspA (rNspA) protein produced by Escherichia coli protected mice against experimental meningococcal infection (28).
In this study, gonococcal NspA-specific MAbs were generated to further investigate the antigenic conservation of the NspA protein. The gonococcal nspA gene was cloned and sequenced to obtain additional information about the molecular conservation of nspA genes among the two pathogenic Neisseria species.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A collection of 51 clinical and laboratory strains of N. gonorrhoeae and 8 strains of N. meningitidis was used in this study. Of the N. gonorrhoeae strains, seven were isolates from patients with disseminated gonococcal infections and were provided by P. Turgeon, St-Luc Hospital, Montreal, Canada. N. gonorrhoeae FA1090 (13) and MS11 (31) were kindly provided by A. Jerse, Uniformed Services University of the Health Sciences, Bethesda, Md. All other strains were obtained from the culture collection of the National Reference Center for Neisseria and from the Antimicrobial and Molecular Biology Division of the Laboratory Center for Disease Control, Ottawa, Canada. The N. gonorrhoeae strains were grown overnight on chocolate agar plates (Quelab Laboratories, Montreal, Canada) at 37°C in an atmosphere containing 8% CO2. The strains were stored at −70°C in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) containing 20% (vol/vol) glycerol (Sigma Chemical Co., St. Louis, Mo.). E. coli XL1-Blue MRF′ [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac (F′ proAB lacIq ZΔM15 Tn10 [TetrI]) (Stratagene, La Jolla, Calif.) and E. coli B strain BL21 [F− dcm ompT hsdS (rB− mB− gal] (Stratagene) were grown on Lennox Luria-Bertani agar or broth (Gibco BRL, Gaithersburg, Md.) at 37°C. Where appropriate, 100 μg of ampicillin (Sigma) per ml was added to the medium. The low-copy-number plasmid pWKS30 (41) and plasmid p629 (15) were used to clone the nspA gene as well as to produce the gonococcal rNspA protein.
Colony hybridization with an N. meningitidis nspA probe.
A DNA probe was prepared by PCR amplification of the nspA gene from N. meningitidis 608B (28) with oligonucleotide primers NC-01 (5′-ATG AAA AAA GCA CTT GCC ACA CTG-3′) and NC-18 (5′-TCA GAA TTT GAC GCG CAC GCC G-3′) synthetized on an ABI synthesizer (Applied Biosystems, Inc., Mississauga, Canada). The amplification reactions were performed in 50-μl reaction mixtures containing 1 mM each primer, 100 ng of template genomic DNA of N. meningitidis 608B, and 2 U of Taq polymerase (Pharmacia Biotech, Baie d’Urfé, Canada). The samples were overlaid with 50 μl of mineral oil and subjected to 25 cycles of amplification consisting of denaturation at 94°C for 1 min, annealing at 65°C for 1 min, and polymerization at 72°C for 1 min. The 525-bp amplification product was purified by electrophoresis on a low-melting-point agarose gel and labeled by random priming with the DIG DNA labeling and detection kit (Boehringer Mannheim, Laval, Canada.).
The colonies from each bacterial strain to be tested were dotted onto a positively charged nylon membrane (Amersham Life Science, Oakville, Canada), dried, and then treated as specified by the manufacturer. Prehybridizations and hybridizations were done at 42°C with solutions containing 50% (vol/vol) formamide according to the DIG kit user’s guide for filter hybridization (Boehringer Mannheim). The prehybridization solution also contained 100 μg of denatured herring sperm DNA (Gibco BRL) per ml to prevent nonspecific hybridization of the DNA probe. The stringency washes and detection steps with the chemiluminescent lumigen substrate were also done as described in the DIG kit user’s guide.
Cloning and sequencing of the gonococcal nspA gene.
A Southern hybridization (36) assay with a 32P-labeled DNA probe corresponding to a 2.75-kb ClaI fragment from the N. meningitidis genome with the nspA gene (28) was used for the identification of a 2.75-kb ClaI fragment containing the N. gonorrhoeae nspA gene. Briefly, chromosomal DNA from N. gonorrhoeae B2 was purified as described previously (26), digested with ClaI (Pharmacia Biotech), subjected to electrophoresis on an agarose gel, and transferred to a positively charged nylon membrane (Amersham Life Science). The N. meningitidis nspA probe was obtained by ClaI digestion of the plasmid pN2202 (28) and purification of the resulting 2.75-kb fragment with a QIAquick gel extraction kit (Qiagen Inc., Valencia, Calif.). The DNA probe was labeled with [α-32P]dCTP (ICN Pharmaceuticals, Inc., Montreal, Canada), using the T7 Quickprime Kit as specified by the manufacturer (Pharmacia Biotech), and then purified with the QIAquick nucleotide removal kit (Qiagen Inc.). The prehybridization, hybridization, and washes were performed as described previously (36). The membrane was exposed to a Kodak X-OMAT AR film (InterSciences, Markham, Canada) with an intensifying screen. The N. gonorrhoeae ClaI fragments ranging from 2.5 to 3 kb that reacted positively to the nspA probe were purified from an agarose gel and were ligated into the low-copy-number plasmid pWKS30, which had been previously digested with ClaI and dephosphorylated. The recombinant plasmids were then transformed into E. coli XL1-Blue MRF′ as described by Hanahan (20). Colony hybridization was performed with the N. meningitidis nspA gene as a probe for the identification of positive transformants (36). Finally, positively hybridizing colonies were transferred onto a nitrocellulose membrane (Amersham). The Me-7 MAb (27), which recognized the meningococcal NspA protein and is bactericidal for N. meningitidis, was used in a dot immunoassay (39) to identify clones producing the gonococcal NspA protein. The selected recombinant plasmid pWKS30 containing the gonococcal 2.75-kb insert was named pURV1. The nspA gene from N. gonorrhoeae was sequenced with the primers previously described for sequencing the meningococcal nspA gene (28). DNA sequencing was performed with the Taq Dye Deoxy Terminator cycle-sequencing kit and Applied Biosystems Inc. (Foster City, Calif.) automated sequencer model 373A as recommended by the manufacturer. A series of new primers were synthesized and used to obtain the sequence of the entire gene. Assembling and alignment of the sequences were accomplished by using the Sequencher program (Gene Codes Corp, Ann Arbor, Mich.) and Geneworks software (IntelliGenetics, Inc. Mountain View, Calif.). The gonococcal nspA gene from strain B2 was amplified directly from purified chromosomal DNA by PCR with primers OCRR-120 (5′-TTT GGA TCC TCA GAA TTT GAC GCG CAC-3′) and OCRR-119 (5′-GGC AGA TCT ATG AAA AAA GCA CTT GCC G-3′), which contained the BamHI and BglII restriction sites, respectively. The nspA PCR product of approximately 525 bp was ligated into plasmid p629 containing the λpL promoter controlled by a thermolabile λ repressor (15). The sequence of the nspA insert was verified as previously described, and the resulting recombinant plasmid was identified as pURV24.
Production and purification of the N. gonorrhoeae NspA protein.
Recombinant plasmids pURV1 and pURV24 were purified from E. coli XL1-Blue MRF′ transformants producing the rNspA protein by using the Plasmid Midi kit (Qiagen Inc.) and transformed into E. coli B strain BL21 by electroporation with the Gene Pulser II apparatus (Bio-Rad Laboratories). An overnight culture of E. coli BL21 transformed with plasmid pURV1 was inoculated in Lennox Luria-Bertani broth containing 100 μg of ampicillin per ml and was grown for 4 h at 37°C with agitation. Heat induction of expression of the nspA gene insert present on plasmid pURV24 was performed by the method described by George et al. (15). The bacterial cells were then removed from the cultures by centrifugation at 12,000 × g for 30 min at 4°C. The supernatant was filtered through a 0.22-μm-pore-size filter and concentrated with an ultrafiltration apparatus and a Diaflo ultrafiltration membrane YM 10 (Amicon Inc., Beverly, Mass.). The concentrated culture supernatant was subjected to 48% (wt/vol) ammonium sulfate precipitation before being fractionated by two successive chromatography steps with phenyl-Sepharose (Pharmacia) and cation-exchange Highload SP columns (Pharmacia). The purity of the rNspA protein was shown to be >85% as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with the discontinuous buffer system of Laemmli (24) and staining with Coomassie brilliant blue.
Preparation of OM extracts.
Lithium chloride extractions of the OM of N. gonorrhoeae B2, MS11, and 5776 and N. meningitidis 608B were performed as described previously (7). The protein concentrations were determined by the Lowry et al. method adapted to membrane fractions (25). Meningococcal and gonococcal OM preparations and rNspA proteins were resolved by SDS-PAGE with 14% (wt/vol) polyacrylamide gels, and immunoblot analyses were performed as previously described (28).
Immunization of mice and generation of MAbs against the gonococcal NspA protein.
BALB/c mice (Charles River Laboratories, Montreal, Canada) were injected subcutaneously three times at 3-week intervals with 20 μg of purified N. gonorrhoeae rNspA protein in phosphate-buffered saline (PBS) containing 0.2 mg of QuilA (Cedarlane Laboratories, Hornby, Canada) per ml. Serum samples were collected from each mouse via the periorbital sinus before each immunization and 2 weeks after the third injection. The sera were tested by enzyme-linked immunosorbent assay (ELISA) as described previously (19) with, as solid-phase antigens, gonococcal OM preparations (7.5 μg/ml), meningococcal OM preparations (7.5 μg/ml), and purified gonococcal rNspA protein (1 μg/ml). Three days before the fusion, the selected mouse was injected intravenously with 20 μg of gonococcal rNspA protein. Hybridoma cell lines were produced as previously described (19). Supernatants collected from wells containing growing clones were tested for specific antibody production by ELISA. A second screening was performed on positive clones by Western immunoblotting (28) with the same protein preparations. Selected positive hybridoma cell lines were cloned by limiting dilution, expanded, and frozen in liquid nitrogen. The class, subclass, and light-chain type of MAbs were determined by ELISA with commercially available reagents (Southern Biotechnologies Associates Inc., Birmingham, Ala.).
Detection of the NspA protein in N. gonorrhoeae strains by whole-cell ELISA.
To evaluate the reactivity of hybridoma supernatants against a panel of N. gonorrhoeae strains, whole-cell ELISA was used as previously described (1) with some modifications. Briefly, overnight gonococcal cultures were removed from chocolate agar plates, suspended in PBS, and spectrophotometrically adjusted to an optical density at 620 nm (OD620) of 0.6. Cell suspensions were aliquoted and frozen at −70°C until needed for the assay. The day before the assay, 0.1-ml portions of selected bacterial suspensions were dispensed into appropriate wells of a flat-bottom microtitration plate and allowed to evaporate to dryness at 37°C. The wells were washed three times with PBS containing 0.02% (vol/vol) Tween 20 (J. T. Baker, Phillipsburg, N.J.) and blocked with 3% (wt/vol) bovine serum albumin (Sigma) in PBS per well. The plates were incubated for 30 min at 37°C, and the blocking solution was discarded. A 0.1-ml volume of hybridoma culture supernatant containing the NspA-specific MAb to be tested was added to each well, and the plates were incubated for 1 h at 37°C and then washed three times. Alkaline phosphatase-conjugated AffiniPure Goat anti-mouse immunoglobulins (Jackson ImmunoResearch Laboratories, Mississauga, Canada) were diluted in PBS containing 0.5% (wt/vol) bovine serum albumin, and 0.1 ml of this solution was added to each well. The plates were incubated for 1 h at 37°C and washed three times with PBS containing 0.02% Tween 20, and 0.1 ml of p-nitrophenyl disodium phosphate solution in 10% diethanolamine (pH 9.6) was added to each well. Following incubation for 1 h at room temperature, the OD405 was read with a Dynatech MRX microplate reader. Controls included wells containing cell culture medium without antibody and wells containing culture supernatant with unrelated MAbs. Values were considered positive when the OD405 was greater than twice the reading obtained for control wells.
Radioimmunobinding assay.
Binding of NspA-specific antibodies to whole gonococci was measured by a radioimmunobinding assay as previously described (29). Gonococcal and meningococcal strains were incubated with culture supernatant without antibody or culture supernatant containing MAbs specific to Haemophilus influenzae as negative controls. Results are expressed as means and standard deviation of counts per minute (cpm) obtained from the different experiments. Background values were substracted from all the binding values.
Nucleotide sequences.
The nucleotide sequence of the gonococcal nspA gene described in this report has been assigned GenBank accession no. U52069. The nucleotide sequence of the N. meningitidis 608B gene is available from GenBank under the accession no. U52066. Contig 206 obtained from N. gonorrhoeae FA1090 is also available from the Gonococcal Genome Sequencing Project (B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer), University of Oklahoma.
RESULTS
Distribution of the nspA gene in N. gonorrhoeae.
A DNA probe derived from the N. meningitidis 608B nspA gene was used in colony hybridization assays to evaluate the distribution of the nspA gene among N. gonorrhoeae isolates. The nspA DNA probe was shown to hybridize with all 15 N. gonorrhoeae strains tested. These strains included eight serological reference strains and seven clinical isolates. As expected, the nspA probe also hybridized with the three N. meningitidis strains tested, which included one strain of each of serogroups A, B, and C. The nspA probe did not hybridize with any of the 19 nonneisserial bacterial strains tested: Alcaligenes faecalis (ATCC 8750), Bordetella pertussis (9340), Bordetella bronchiseptica, Citrobacter freundii (ATCC 2080), Edwardsiella tarda (ATCC 15947), Enterobacter cloacae (ATCC 23355), Enterobacter aerogenes (ATCC 13048), Escherichia coli, Flavobacterium odoratum, Haemophilus influenzae type b (Eagan strain), Klebsiella pneumoniae (ATCC 13883), Proteus rettgeri (ATCC 25932), Proteus vulgaris (ATCC 13315), Pseudomonas aeruginosa (ATCC 9027), Salmonella typhimurium (ATCC 14028), Serratia marcescens (ATCC 8100), Shigella flexneri (ATCC 12022), Shigella sonnei (ATCC 9290), and Xanthomonas maltophila. These strains were obtained from the American Type Culture Collection or from our own strain collection.
Molecular conservation of the gonococcal nspA gene.
As observed for N. meningitidis, the gonococcal nspA gene was shown by Southern hybridization to be on a 2.75-kb ClaI restriction endonuclease fragment on the gonococcal genome (data not shown). Genomic ClaI fragments of the corresponding size were then ligated into the ClaI-digested vector pWKS30. The recombinant plasmids were used to transform E. coli XL1-Blue MRF′. Nine positive transformants were identified by a colony hybridization assay with a DNA probe derived from the original meningococcal nspA gene isolated from strain 608B. Digestion of the nine recombinant DNA plasmids with ClaI revealed that three clones carried the expected 2.75-kb gonococcal insert. Dot immunoassay on bacterial colonies by using NspA-specific MAbs showed that the NspA protein was produced by the three clones. This result indicated that the nspA gene was transcribed under the control of a promoter located on the 2.75-kb insert. One clone was selected for further studies, and plasmid DNA from this clone, identified as pURV1, was purified and used to sequence the gonococcal nspA gene. The sequence analysis revealed the presence on the 2.75-kb insert of an open reading frame of 525 nucleotides. This open reading frame codes for a 174-amino-acid polypeptide with a molecular weight of 18,316 and a predicted isoelectric point of 10.21 (Fig. 1). Analysis of the sequence also indicated the presence of a putative leader sequence corresponding to amino acids 1 to 19 (M-K-K-A-L-A-A-L-I-A-L-A-L-P-A-A-A-L-A) that was previously shown to be cleaved in the mature meningococcal NspA protein (28). Comparison of the deduced amino acid sequence of the NspA protein with the sequences compiled in GenBank and Swissprot revealed, as first observed for the meningococcal NspA protein, the existence of weak homology to Neisseria opacity proteins. The homology between the NspA protein and the Opa proteins was found to be clustered in two particular regions, located at the carboxy terminus and between amino acid residues 126 and 144 of the NspA protein.
FIG. 1.
Predicted amino acid sequences of the NspA protein of N. gonorrhoeae B2, N. gonorrhoeae FA1090, and N. meningitidis 608B (28). Differences are indicated by the one-letter code, and identities are indicated by a period. Evaluation of the identities between these sequences indicated that these three proteins are highly similar, with a global identity of 93%. Sequence analysis revealed the presence of an additional amino acid residue (glutamine) at position 73 for the NspA protein from N. gonorrhoeae FA1090.
An alignment of the two NspA deduced amino acid sequences derived from N. gonorrhoeae B2 and FA1090 and the sequence from N. meningitidis 608B is shown in Fig. 1. The NspA sequence from strain FA1090 was obtained from the gonococcal genome-sequencing project, which is under way, and was found on contig 206. These three deduced polypeptide sequences are 93% identical. The level of identity increased to 98% when the two gonococcal sequences were compared, showing differences in only 3 positions out of 174 amino acid residues. In addition, the polypeptide from N. gonorrhoeae FA1090 has an insertion of one glutamine residue at position 73.
Antigenic conservation of the gonococcal NspA protein.
The mouse selected to generate the NspA-specific MAbs was immunized three times at 3-week interval with 20 μg of purified gonococcal rNspA protein mixed with 0.2 mg of Quil A per ml. This protein was immunogenic, since a high NspA-specific titer was recorded when the serum collected from this mouse was tested by ELISA against the rNspA protein. Screening of the hybridoma cell lines was performed by ELISA and immunoblotting with OM preparations from N. gonorrhoeae B2, N. meningitidis 608B, and the purified gonococcal rNspA protein. Seven NspA-specific hybridomas, secreting MAbs 11E9, 1D4, 13E5, 5D1, 14D8, 14D7, and 2F10, which strongly recognized the native as well as the recombinant gonococcal protein, were selected for subcloning by limiting dilution and further characterized. The class and subclass of each of these MAbs were determined and are presented in Table 1. Whole-cell ELISA were performed with a collection of 51 N. gonorrhoeae strains and 8 N. meningitidis strains to evaluate the specificity of the NspA-specific MAbs. Four of the seven MAbs, namely, 11E9, 1D4, 13E5, and 5D1, reacted with all 51 N. gonorrhoeae strains tested, while MAbs 14D8, 14D7, and 2F10 recognized 50, 14, and 8 gonococcal strains, respectively. All of these MAbs recognized at least N. meningitidis 608B. This particular strain was previously shown to produce large amounts of NspA protein in the OM. MAbs 5D1, 14D7, and 2F10 reacted with all eight N. meningitidis strains. MAb 5D1 reacted with all meningococcal and gonococcal strains tested.
TABLE 1.
Reactivity of the NspA-specific MAbs with different strains of N. gonorrhoeae and N. meningitidis
| MAb | Isotypea | No. of strains recognized/total no. testedb
|
|
|---|---|---|---|
| N. gonorrhoeae | N. meningitidisc | ||
| 11E9 | IgG1 | 51/51 | 1/8 |
| 1D4 | IgG2a | 51/51 | 1/8 |
| 13E5 | IgG1 | 51/51 | 3/8 |
| 5D1 | IgG2b | 51/51 | 8/8 |
| 14D8 | IgG1 | 50/51 | 3/8 |
| 14D7 | IgG1 | 14/51 | 8/8 |
| 2F10 | IgG1 | 8/51 | 8/8 |
| Me-7 | IgG2a | 9/51 | 8/8 |
IgG, immunoglobulin G.
Reactivity was considered positive when the ODs were greater than twice the reading obtained for control wells without antibody.
N. meningitidis strains tested included two strains of serogroup B and one strain each of serogroups D, X, Z, A, Y, and W-135.
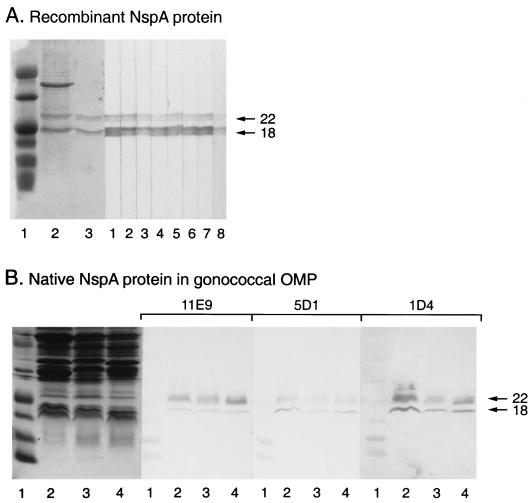
Immunoblot analysis clearly indicated that the protein produced in the culture supernatant of E. coli BL21 transformed with pURV1 was full-length and was reactive with the seven NspA-specific MAbs described in this report (Fig. 2A). As was the case for the meningococcal NspA protein, both the 22- and 18-kDa protein bands, which are known to be the two forms of the NspA protein that can be found after SDS-PAGE, were produced by the recombinant E. coli and were reactive with the NspA-specific MAbs. Figure 2B also shows the reactivity of MAbs 1D4, 5D1, and 11E9 with the native NspA protein present in OM preparations extracted from N. gonorrhoeae B2, MS11, and 5776. These MAbs also efficiently recognized in Western immunoblot analyses the two protein bands at 18 and 22 kDa, which are characteristic of the NspA protein after SDS-PAGE and were produced by E. coli BL21 transformed with pURV24, containing only the nspA gene insert (data not shown). This result clearly indicated that these MAbs recognized only the NspA protein and not other gonococcal proteins that might have been produced by plasmid pURV1.
FIG. 2.
Reactivity of the NspA-specific MAbs with recombinant and native NspA proteins. (A) SDS-PAGE migration profile of concentrated culture supernatant of recombinant E. coli BL21(pURV1) producing the gonococcal NspA protein (lane 2), and purified gonococcal rNspA protein (lane 3), and an immunoblot showing the reactivity of MAbs 11E9 (lane 1), 5D1 (lane 2), 1D4 (lane 3), 14D8 (lane 4), 13E5 (lane 5), 14D7 (lane 6), 2F10 (lane 7), and Me-7 (lane 8) with purified gonococcal recombinant NspA protein. (B) SDS-PAGE migration profile of OM preparations extracted from N. gonorrhoeae 5776 (lane 2), MS11 (lane 3), and B2 (lane 4), and an immunoblot showing the reactivity of MAb 11E9, 5D1, and 1D4 with these OM preparations. SDS-PAGE was performed with 14% polyacrylamide gels and the proteins were visualized by Coomassie blue staining. The first lane of each gel contained the prestained low-molecular-weight markers 43,000, 29,000, 18,400, 14,300, 6,200, 3,400, and 2,300 (Gibco BRL). The arrows indicate the locations of the NspA protein bands.
Surface exposure of the gonococcal NspA protein.
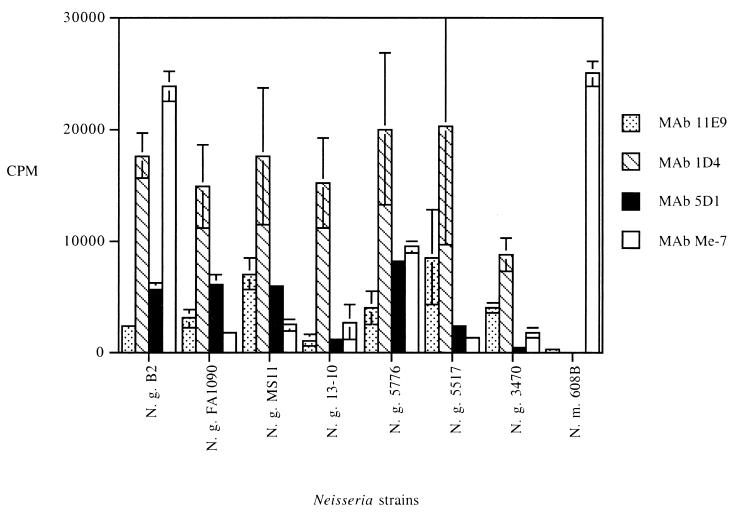
Radioimmunobinding assays were performed to determine whether the MAbs generated against the gonococcal NspA protein were able to recognize their specific epitopes at the surface of the gonococcus. MAbs specific to Haemophilus influenzae (P2-4 and 6B11) and hybridoma culture supernatant without antibody were used as negative controls. Preliminary assays indicated that six of the eight MAbs tested, namely, 14D7, 1D4, 5D1, 12E12, 2F10, and Me-7 (anti-NspA of N. meningitidis 608B), bound strongly to their epitope at the surface of the gonococcus, with cpm values between 6,340 ± 393 and 23,649 ± 585 (data not shown). In these experiments, the background values were 572 ± 26 cpm when hybridoma culture supernatant without antibody was used as a negative control and 1,087 ± 307 cpm when bacteria were incubated with MAb 6B11. MAbs 11E9, 1D4, 5D1, and Me-7 were selected to conduct attachment study on additional strains. This panel of strains contained three gonococcal laboratory strains, one strain isolated from a patient with localized infection (strain 13-10), three strains isolated from patients with disseminated infection (strains 5776, 5517, and 3470) and the serogroup B meningococcal strain 608B. As shown in Fig. 3, high cpm values (between 8,861 ± 1,473 and 20,326 ± 10,525) were recorded for MAb 1D4, while MAb 11E9 attached only weakly at the surface of intact N. gonorrhoeae cells with cpm values between 1,159 ± 529 and 8,611 ± 4,171. Values obtained for MAb 5D1 were intermediate with respect to the other two MAbs tested. Background values were recorded when MAbs 11E9 and 1D4 were tested against meningococcal strain 608B, indicating that their corresponding epitopes are not exposed at the surface of intact meningococcal cells. The binding values obtained with these MAbs varied from one gonococcal strain to another. MAb Me-7, which reacted in whole-cell ELISA with only 9 of 51 gonococcal strains, attached efficiently at the surface of meningococcal strain 608B, with a mean cpm value of 24,955 ± 1,057. This MAb also recognized its specific epitope at the surface of intact gonococcal cells from strains B2 and 5776 but not the other strains.
FIG. 3.
Binding properties of the NspA-specific MAbs to intact bacterial cells. Hybridoma culture supernatants were incubated with live intact N. gonorrhoeae (N. g.) or N. meningitidis (N. m.) cells. The results are expressed as means and standard deviations of cpm obtained from at least two different experiments. Background values were substracted from all the binding values.
DISCUSSION
We recently reported the identification of the NspA protein in the OM of N. meningitidis by using MAbs (27, 28). Some of these MAbs, such as Me-7, also reacted with certain gonococcal isolates, suggesting that a homologue of the meningococcal NspA protein could also be found in this closely related species.
Colony hybridization was performed with the nspA gene, which was originally cloned from the serogroup B meningococcal strain 608B as a probe to demonstrate the presence of an homologue of the nspA gene in the genome of N. gonorrhoeae. Strong hybridization signals were recorded with all 15 gonococcal and 3 meningococcal strains tested. This result indicated that the level of homology between the meningococcal nspA probe and a related sequence in the gonococcal genome was high enough to enable hybridization and detection. This was not surprising since most of the genes studied in either one of these bacterial species have a homologue in the other (38).
A strategy which yielded the first meningococcal nspA gene was adapted and successfully used to clone and sequence homologous gene from gonococcal strain B2. Interestingly, this nspA gene was found to be present on a 2.75-kb ClaI genomic DNA fragment, as was the case for the meningococci. This is in accordance with what was previously reported, which indicated that the localization on the chromosome of most of the genes that have been mapped so far in both species is the same and that many restriction sites are also conserved (14). Analysis of the data which were available from the gonococcal genome-sequencing project indicated that the nspA gene was also present in the genome of gonococcal strain FA1090. When the deduced amino acid sequences of the NspA proteins from both gonococcal strains were compared, they were determined to be 98% identical. They differed at only 3 of the 174 amino acid residues. The NspA polypeptide from N. gonorrhoeae FA1090 was also shown to possess an additional glutamine residue at position 73. The addition of one glutamine residue is not restricted to a particular gonococcal strain, since it was also found to be present in the NspA protein of a serogroup A meningococcal strain (unpublished data). The impact of this particular modification on the immunogenicity of the protein is presently under study. The level of molecular conservation of the NspA protein is similar to or even higher than what was reported for other gonococcal surface antigens. For example, pairwise comparison of two PIB porin protein sequences obtained from strain R10 (11) and MS11 (17) revealed that they were 95% identical, with only 17 differences in the 350 amino acid residues (8). However, the PI protein occurs in the gonococcal population as two mutually exclusive immunochemical classes of outer membrane proteins, called PIA and PIB, each composed of multiple serovars (23). The overall level of identity falls to only 63% when PIA and PIB were compared.
With 93% identity, the level of molecular conservation remained very high when the deduced amino acid sequences of the two gonococcal NspA proteins were compared to that of the meningococcal NspA protein. The presence of closely related analogous proteins in both pathogenic Neisseria species is well documented (38, 42). For example, comparison of the predicted amino acid sequences of the meningococcal class I (PorA) protein and the gonococcal PIB protein have revealed that a large proportion of residues are conserved between the two proteins. It was shown that 166 residues were identical and 84 were functionally equivalent. Comparable results were also obtained when the class I protein and the PIA protein were compared (2). The level of molecular conservation recorded for the NspA proteins produced by either pathogenic Neisseria species is very high compared to that for the other neisserial surface proteins. Indeed, higher levels of molecular conservation were reported only for proteins encoded by housekeeping genes such as recA, where a level of identity of 100% was reported (43).
Since the hybridization experiment suggested that the nspA gene was present in the genome of every gonococcal strains tested, we decided to generate additional NspA-specific MAbs to investigate the antigenic conservation of the gonococcal NspA protein. As expected, whole-cell ELISA results clearly showed that the NspA protein was produced by every gonococcal strain and that certain epitopes such as the ones recognized by MAbs 11E9, 1D4, 13E5, and 5D1 were highly conserved (Table 1). Interestingly, all these MAbs also recognized at least one of the eight meningococcal strains tested. In addition, MAb 5D1 was found to recognize every meningococcal and gonococcal strain tested. Similar findings were reported previously for another antigenically highly conserved outer membrane protein called Lip or H.8 (10, 44). Indeed, MAb directed against this protein efficiently recognized N. gonorrhoeae as well as N. meningitidis strains but did not react with most nonpathogenic neisserial species. For this reason, it was suggested that this protein might be involved in pathogenesis. Unfortunately, no protective immunity was induced by this particular protein (4).
It was postulated that the failure of anti-Lip antibodies to promote bactericidal killing or to confer passive protection could be related to limited exposure of the antigen on the bacterial cells (35). To evaluate the surface exposure of the gonococcal NspA protein, the attachment of the NspA-specific MAbs was evaluated by a radioimmunobinding assay. In contrast to what was observed for the Lip protein, the NspA-specific MAbs 1D4 and 5D1 efficiently recognized their corresponding epitopes at the surface of intact gonococcal cells, thus indicating that portions of the NspA protein are exposed and easily accessible to the antibodies. Other epitopes of the gonococcal NspA protein, such as the one recognized by MAb 11E9, seemed to be less accessible to the antibodies. The mouse used to generate the MAbs was immunized with purified gonococcal rNspA protein. The conformation of the purified rNspA protein would not be expected to correspond exactly to the spatial organization of the native gonococcal NspA protein when inserted in the OM. Interestingly, when tested by immunoblotting or whole-cell ELISA, these MAbs also reacted well with the NspA protein present in the OM of meningococcal strain 608B. However, radioimmunobinding results indicated that their specific epitopes were not accessible at the surface of intact meningococcal cells (Fig. 3). This latter result suggested that the insertion of the NspA protein is not exactly identical in these two species, thus directly affecting the attachment of the specific MAbs, or that other components present at the surface of intact meningococcal cells might mask these epitopes, thus preventing the binding of the MAbs. Other epitopes on the meningococcal NspA protein were shown to be easily accessible at the surface of intact meningococcal cells (unpublished data). In addition, MAb Me-7 was shown to be bactericidal against serologically distinct meningococcal strains, clearly confirming the notion that certain portions of the meningococcal NspA protein are exposed at the cell surface (27).
In conclusion, we have shown that the gonococcal NspA protein is closely related to its analogue, the meningococcal NspA protein. This protein is present in the OM of all gonococcal strains tested so far, where it is easily accessible to specific antibodies. We are presently studying the possible role of the NspA protein in the colonization of mucosal surfaces by the gonococcus.
ACKNOWLEDGMENT
This research was funded by a grant from Biochem Vaccines, Inc.
REFERENCES
- 1.Abdillahi H, Poolman J T. Whole-cell Elisa for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol Lett. 1987;48:367–371. [PubMed] [Google Scholar]
- 2.Barlow A K, Heckels J E, Clarke I N. The class 1 outer membrane protein of Neisseria meningitidis: gene sequence and structural and immunological similarities to gonococcal porins. Mol Microbiol. 1989;3:131–139. doi: 10.1111/j.1365-2958.1989.tb01802.x. [DOI] [PubMed] [Google Scholar]
- 3.Barritt D S, Schwalbe R S, Klapper D G, Cannon J G. Antigenic and structural differences among six proteins II expressed by a single strain of Neisseria gonorrhoeae. Infect Immun. 1987;55:2026–2031. doi: 10.1128/iai.55.9.2026-2031.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee A K, Moran E E, Zollinger W D. Antibodies to meningococcal H.8 (Lip) antigen fail to show bactericidal activity. Can J Microbiol. 1990;36:117–122. doi: 10.1139/m90-021. [DOI] [PubMed] [Google Scholar]
- 5.Black W J, Cannon J G. Cloning of the gene for the common pathogenic Neisseria H.8 antigen from Neisseria gonorrhoeae. Infect Immun. 1985;47:322–325. doi: 10.1128/iai.47.1.322-325.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake M S, Wetzler L M, Gotschlich E C, Rice P A. Protein III: structure, function and genetics. Clin Microbiol Rev. 1989;2(Suppl.):S60–S63. doi: 10.1128/cmr.2.suppl.s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodeur B R, Larose Y, Tsang P, Ashton F, Ryan A. Protection against infection with Neisseria meningitidis group B serotype 2b by passive immunization with serotype-specific monoclonal antibody. Infect Immun. 1985;50:510–516. doi: 10.1128/iai.50.2.510-516.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butt N J, Virji M, Vayreda F, Lambden P R, Heckels J E. Gonococcal outer-membrane protein PIB: comparative sequence analysis and localization of epitopes that are recognized by type-specific and cross-reacting antibodies. J Gen Microbiol. 1990;136:2165–2172. doi: 10.1099/00221287-136-11-2165. [DOI] [PubMed] [Google Scholar]
- 9.Cannon J G. Conserved lipoproteins of pathogenic Neisseria bearing the H.8 epitope: lipid-modified azurin and H.8 outer membrane protein. Clin Microbiol Rev. 1989;2(Suppl.):S1–S4. doi: 10.1128/cmr.2.suppl.s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon J G, Black W J, Nachamkin I, Stewart P. Monoclonal antibodies that recognizes an outer-membrane antigen common to the pathogenic Neisseria species but not to most nonpathogenic species. Infect Immun. 1984;43:994–999. doi: 10.1128/iai.43.3.994-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carbonetti N H, Simnad V I, Seifert H S, So M, Sparling F. Genetic of protein I of Neisseria gonorrhoeae: construction of hybrid porins. Proc Natl Acad Sci USA. 1988;85:6841–6845. doi: 10.1073/pnas.85.18.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbonetti N H, Sparling P F. Molecular cloning and characterization of the structural gene for protein I, the major outer membrane protein of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1987;84:9084–9088. doi: 10.1073/pnas.84.24.9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dempsey J F, Litaker W, Madhure A, Snodgrass T L, Cannon J G. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers including opa and pil genes. J Bacteriol. 1991;173:5476–5486. doi: 10.1128/jb.173.17.5476-5486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dempsey J F, Maurer M E, Cannon J G. Abstracts of the 10th International Pathogenic Neisseria Conference 1996. 1996. Chromosome organization in Neisseria gonorrhoeae and Neisseria meningitidis; p. 341. [Google Scholar]
- 15.George H J, Watson R J, Harbrecht D F, DeLorbe W J. A bacteriophage λ cI857 cassette controls λPL expression vectors at physiologic temperature. Bio/Technology. 1987;5:600–603. [Google Scholar]
- 16.Gotschlich E C, Blake M S, Koomey J M, Seiff M, Derman A. Cloning of the structural gene of three H.8 antigens and protein III of Neisseria gonorrhoeae. J Exp Med. 1986;164:868–881. doi: 10.1084/jem.164.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotschlich E C, Seiff M E, Blake M S, Koomey J M. Porin protein of Neisseria gonorrhoeae: cloning and gene structure. Proc Natl Acad Sci USA. 1987;84:8135–8139. doi: 10.1073/pnas.84.22.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guibourdenche M, Popoff M Y, Riou J Y. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea and “Neisseria polysaccharea.”. Ann Inst Pasteur. 1986;137B:177–185. doi: 10.1016/s0769-2609(86)80106-5. [DOI] [PubMed] [Google Scholar]
- 19.Hamel J, Brodeur B R, Larose Y, Tsang P S, Belmaaza A, Montplaisir S. A monoclonal antibody directed against a serotype-specific, outer-membrane protein of Haemophilus influenzae type b. J Med Microbiol. 1987;23:163–170. doi: 10.1099/00222615-23-2-163. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 21.Judd R C. Protein I: structure, function, and genetics. Clin Microbiol Rev. 1989;2(Suppl.):S41–S48. doi: 10.1128/cmr.2.suppl.s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawula T H, Aho E L, Barritt D S, Klapper D G, Cannon J G. Reversible phase variation of expression of Neisseria meningitidis class 5 outer membrane proteins and their relationship to gonococcal protein II. Infect Immun. 1988;56:380–386. doi: 10.1128/iai.56.2.380-386.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knapp J S, Tam M R, Nowinski R C, Holmes K K, Sandström E G. Serological classification of Neisseria gonorrhoeae with the use of monoclonal antibodies to gonococcal outer membrane protein I. J Infect Dis. 1984;150:44–48. doi: 10.1093/infdis/150.1.44. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Marmur J. A procedure for the isolation of ribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–216. [Google Scholar]
- 27.Martin D, Cadieux N, Hamel J, Brodeur B R. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Identification of a highly conserved outer membrane protein of Neisseria meningitidis, abstr. B-187; p. 187. [Google Scholar]
- 28.Martin D, Cadieux N, Hamel J, Brodeur B R. Highly conserved Neisseria meningitidis surface protein confers protection against experimental infection. J Exp Med. 1997;185:1173–1183. doi: 10.1084/jem.185.7.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin D, Peppler M S, Brodeur B R. Immunological characterization of the lipooligosaccharide B band of Bordetella pertussis. Infect Immun. 1992;60:2768–2725. doi: 10.1128/iai.60.7.2718-2725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer T F, Billyard E, Haas R, Storzbach S, So M. Pilus genes of Neisseria gonorrhoeae: chromosomal organization and DNA sequence. Proc Natl Acad Sci USA. 1984;8:6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer T F, Mlawer, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 32.Mietzner T A, Barnes R C, Jean Louis Y A, Shafer W A, Morse S A. Distribution of an antigenically related iron-regulated protein among the Neisseria species. Infect Immun. 1986;51:60–68. doi: 10.1128/iai.51.1.60-68.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newhall W J, Mail L B, Wilde III C E, Jones R B. Purification and antigenic relatedness of proteins II of Neisseria gonorrhoeae. Infect Immun. 1985;49:576–580. doi: 10.1128/iai.49.3.576-580.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potts W J, Saunders J R. Nucleotide sequence of the structural gene for class I pilin from Neisseria meningitidis: homologies with the pilE locus of Neisseria gonorrhoeae. Mol Microbiol. 1988;2:647–653. doi: 10.1111/j.1365-2958.1988.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 35.Robinson E N, McGee Z A, Buchanan T M, Blake M S, Hitchcock P J. Probing the surface of Neisseria gonorrhoeae: simultaneous localization of PI and H.8 antigens. Infect Immun. 1987;55:1190–1197. doi: 10.1128/iai.55.5.1190-1197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2rd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sparling P F, Elkins C, Wyrick P B, Cohen M S. Vaccine for bacterial sexually transmitted infections: a realistic goal? Proc Natl Acad Sci USA. 1994;91:2456–2463. doi: 10.1073/pnas.91.7.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez J A, de la Fuente L, Berron S, O’Rourke M, Smith N H, Zhou J, Spratt B G. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol. 1993;3:567–571. doi: 10.1016/0960-9822(93)90001-5. [DOI] [PubMed] [Google Scholar]
- 41.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene (Amsterdam) 1991;100:195–199. [PubMed] [Google Scholar]
- 42.West S E H, Clark V L. Genetic loci and linkage associations in Neisseria gonorrhoeae and Neisseria meningitidis. Clin Microbiol Rev. 1989;2(Suppl.):S92–S103. doi: 10.1128/cmr.2.suppl.s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Spratt B G. Sequence diversity within the argF, fbp and recA genes of natural isolates of Neisseria meningitidis: interspecies recombination within the argF gene. Mol Microbiol. 1992;6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]
- 44.Zollinger W D, Ray J S, Moran E E, Seid R. Identification by monoclonal antibody of an antigen common to the pathogenic Neisseria species. In: Schoolnik G K, editor. The pathogenic neisseriae. Washington, D.C: American Society for Microbiology; 1985. pp. 579–584. [Google Scholar]