Significance
To ensure proper cell differentiation during development requires, on the one hand, cell-intrinsic potentials and correct inductive signals, and on other hand needs to resist incorrect inductive signals, particularly in cases that different precursor cells are of comparable differentiation potentials and located in close proximity. The precursor cells of liver and anterior intestine represent a typical example, but little is known how the intestinal precursors resist liver-inductive signals. Herein, we elucidate mechanisms showing how the intestinal precursors resist the liver-inductive signal to avoid being misinduced to liver fate, in which the Cdx1b-Wnt inhibition cascade plays essential roles. This study provides an example of the mechanisms used by precursor cells to resist incorrect inductive signals and avoid wrong cell fate.
Keywords: intestine, liver, cell fate, Cdx, Wnt
Abstract
During coordinated development of two neighboring organs from the same germ layer, how precursors of one organ resist the inductive signals of the other to avoid being misinduced to wrong cell fate remains a general question in developmental biology. The liver and anterior intestinal precursors located in close proximity along the gut axis represent a typical example. Here we identify a zebrafish leberwurst (lbw) mutant with a unique hepatized intestine phenotype, exhibiting replacement of anterior intestinal cells by liver cells. lbw encodes the Cdx1b homeoprotein, which is specifically expressed in the intestine, and its precursor cells. Mechanistically, in the intestinal precursors, Cdx1b binds to genomic DNA at the regulatory region of secreted frizzled related protein 5 (sfrp5) to activate sfrp5 transcription. Sfrp5 blocks the mesoderm-derived, liver-inductive Wnt2bb signal, thus conferring intestinal precursor cells resistance to Wnt2bb. These results demonstrate that the intestinal precursors avoid being misinduced toward hepatic lineages through the activation of the Cdx1b-Sfrp5 cascade, implicating Cdx/Sfrp5 as a potential pharmacological target for the manipulation of intestinal-hepatic bifurcations, and shedding light on the general question of how precursor cells resist incorrect inductive signals during embryonic development.
Development of the digestive system initiates from the convergence of postgastrulational endoderm to form a nascent gut tube, which eventually produces the digestive tract and accessory organs. The precursors of foregut digestive organs—including liver, pancreas, and anterior intestine—arise from common endodermal progenitors and locate in close proximity at different spatial domains along the undifferentiated gut tube (1–3). For proper organogenesis and demarcation of organ boundaries, precursors of each foregut digestive organ need, on the one hand, to receive proper inductive signals, and on the other hand should develop intrinsic mechanisms to resist the inductive signals of other organs. The former aspect has been intensively investigated in past years in different organisms, including zebrafish, Xenopus, and mammals. Multiple transcriptional factors (4–8) and inductive signals (9–17) particularly derived from the mesoderm have been identified to positively or negatively regulate the specification of different foregut digestive organs. However, because of their close proximity on the undifferentiated gut tube, precursors of different foregut digestive organs might all be reached and could not be distinguished by the signaling molecules diffused from the mesoderm. The latter aspect, mechanisms of how precursors of a foregut digestive organ avoid being misinduced to wrong cell fate, still remain largely unknown.
The liver bud is induced to sprout from the undifferentiated foregut tube. Prior to liver specification, hepatic precursors need to establish the competence so as to respond to the mesoderm-derived liver-inductive signals (18, 19). However, expression patterns of key factors involved in this competence establishment, including members of forkhead box A (FoxA) and GATA families, as well as EpCAM, do not distinguish the precursors of liver and anterior intestine (6, 20–22). Therefore, anterior intestinal precursors must develop intrinsic mechanisms to resist the liver-inductive signals, but these mechanisms are poorly understood. This gap of knowledge is reflected by the lack of genetic mutations that mislead anterior intestinal precursors toward hepatic lineage and cause a hepatized intestine phenotype. The forward genetic screen covers the whole genome without bias, which becomes a powerful tool to obtain genetic mutants with unique phenotypes.
Results
The lbw Mutant Exhibits Hepatized Anterior Intestine.
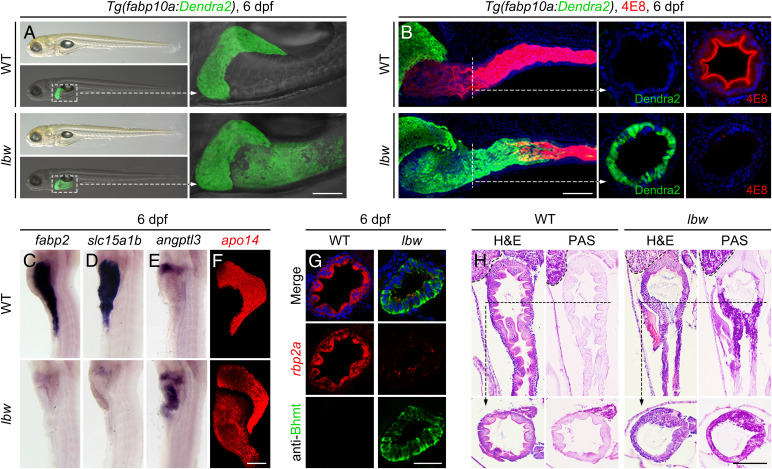
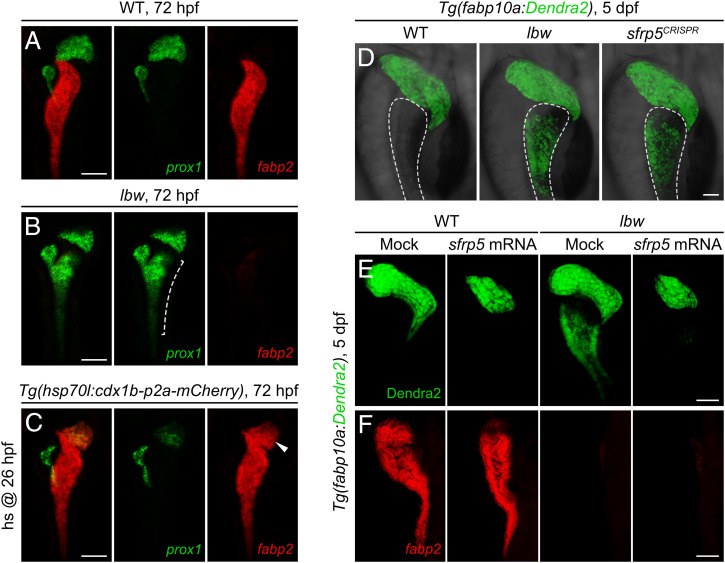
A zebrafish N-Ethyl-N-Nitrosourea (ENU) mutagenesis and forward genetic screen were conducted under the Tg(fabp10a:Dendra2)cq102 transgenic background, in which Dendra2 was driven by a hepatocyte-specific promoter fabp10a. To characterize mechanisms underlying the resistance of intestinal precursors to liver-inductive signals, we screened mutant phenotypes exhibiting abnormal expression of liver markers in the intestine. A recessive mutant named leberwurst (lbwcq93), which displayed ectopic expression of fabp10a:Dendra2 in the anterior intestine without other visible phenotypes at 6 d postfertilization (dpf), was identified (Fig. 1A). These ectopic Dendra2-expressing cells replaced the 4E8-labeled intestinal enterocytes in the lumen of the anterior intestine (Fig. 1B), displaying a posterior shift of the intestine–liver boundary. The anterior intestine in lbw lost the intestinal markers fabp2, slc15a1b, and rbp2a (Fig. 1 C, D, and G), while in the meantime it obtained functional hepatocyte markers angptl3, apo14, Bhmt, gc, and ttr (Fig. 1 E–G and SI Appendix, Fig. S1A). Sections, H&E staining, and periodic acid-Schiff (PAS) staining showed that the lbw intestinal epithelium lost the morphologies of intestinal folds, but gained the glycogen storage function of hepatocytes (Fig. 1H). This hepatized anterior intestine consistently maintained in lbw until it died from the inability to digest and absorb the ingested food at 12 dpf. These data demonstrate the replacement of intestinal cells by functional liver cells in the anterior intestine of lbw, showing hepatized anterior intestine and suggesting that the mutated gene encodes a key factor to resist the liver-inductive signals in the intestinal precursors.
Fig. 1.
The lbw mutant exhibits hepatized anterior intestine. (A) Ectopic expression of Dendra2 in the anterior intestine was observed in the lbw mutant under the Tg(fabp10a:Dendra2) transgenic background. The dashed frames are shown at high magnification. (B) The 4E8-labeled intestinal enterocytes (red) were replaced by the Dendra2-expressing cells (green) in the anterior intestine of lbw, exhibiting posterior shift of the intestine–liver boundary (WT, n = 34 of 35; lbw, n = 28 of 32). The dotted lines indicated the cross-section planes. (C–F) In contrast to the wild-type (WT), the intestinal markers fabp2 (C, WT, n = 28 of 28; lbw, n = 28 of 33) and slc15a1b (D, WT, n = 33 of 34; lbw, n = 25 of 28) were absent in lbw, whereas the functional hepatocyte markers angptl3 (E, WT, n = 23 of 25; lbw, n = 34 of 37), and apo14 (F, WT, n = 34 of 34; lbw, n = 36 of 40) became detectable in the anterior intestine of lbw. (G) The intestinal sections showed that the enterocyte marker rbp2a was nearly lost in the lbw intestine; instead, the mature hepatocyte marker Bhmt became expressed. (H) H&E and PAS staining showed that the lbw intestinal epithelium lost the morphologies of intestinal folds but gained the function of glycogen storage. The liver is outlined at the upper left corner, and the dotted lines indicate the cross-section planes. (Scale bars, 100 μm.) (A) Lateral views, anterior left; (B) lateral and sectional views; (C–F) lateral views, anterior up; (G and H) sectional views. n = number of embryos with indicated phenotype/total analyzed in each class.
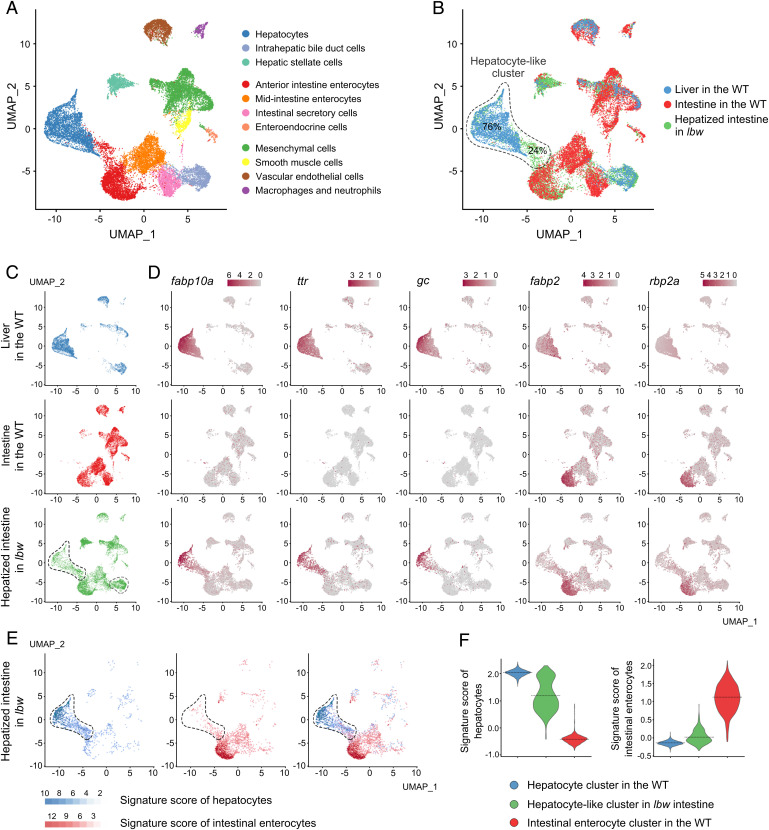
Recent advances in single-cell RNA-sequencing (scRNA-seq) provide powerful tools for characterizing cell identities during liver and intestine development based on molecular and genetic characteristics (23–25). To further characterize the liver cells in lbw intestine, we dissected lbw intestine at 6 dpf and carried out scRNA-seq to compare its cell population clusters with wild-type liver and intestine. A total of 21,548 cells from three samples (lbw intestine, wild-type liver, and intestine) with more than 6,000 cells per sample were analyzed. Approximately 20,000 mean reads, 2,100 unique molecular identifier (UMI) counts, and 550 median genes in each cell were captured. Uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction analyses identified a total of 11 clusters (Fig. 2A and SI Appendix, Fig. S1B). Among these 11 clusters, the dark blue and dark red clusters were characterized as hepatocyte and intestinal enterocyte populations, respectively, based on relative expression levels of well-known marker genes (SI Appendix, Fig. S1C and Dataset S1).
Fig. 2.
scRNA-seq identifies hepatic clusters in the anterior intestine of lbw. (A) The UMAP analysis identified a total of 11 clusters from three samples, including dissected lbw intestine, wild-type liver, and intestine. Different colors correspond to different cell clusters as annotated. (B and C) Of total cells in the hepatocyte-like cluster (dashed outlines), 76% from lbw intestine (green dots) were colocalized with the hepatocyte cluster from wild-type liver (blue dots). The merged UMAP (B) and three samples (C) are exhibited. Note that both the hepatocyte-like cluster and biliary-like cluster were identified in lbw intestine (C, dashed circles). (D) Gene-expression patterns showed expression of the hepatocyte markers fabp10a, ttr, and gc in the hepatocyte-like cluster from lbw intestine. (E and F) In the hepatocyte-like cluster (dashed outlines) from lbw intestine, the signature score of hepatocyte was significantly higher than the signature score of anterior intestinal enterocyte (E), which was validated in violin plots of signature scores (F). The dotted line indicates the mean of each violin. The corresponding score for each cell is calculated based on the expression dataset of top 50 genes in the wild-type hepatocytes and intestinal enterocytes.
In contrast to the wild-type intestine, two major extra cell clusters, hepatocyte-like cells and biliary-like cells, could be identified in the lbw intestine by color-coding the tissues on the UMAP (Fig. 2C, dashed frames). Of the total cells in the hepatocyte-like cell cluster, 76% were colocalized with wild-type hepatocytes, while the rest were localized between the wild-type hepatocyte and enterocyte clusters (Fig. 2 B and C), which might represent a hybrid state. Single-cell gene-expression patterns showed that the hepatocyte markers fabp10a, ttr, gc, bhmt, angptl3, and sePb were all expressed in the hepatocyte-like cluster, validating that the gene-expression profiles of these cells were similar to hepatocytes (Fig. 2D and SI Appendix, Fig. S1D). Furthermore, using the top 50 highly expressed genes in the wild-type hepatocytes and intestinal enterocytes (Dataset S1), we have calculated the signature score for each cell in the lbw intestine sample. UMAP feature (Fig. 2E) and violin plots (Fig. 2F) of signature scores showed that the cells in the hepatocyte-like cluster were closer to the hepatocyte identities. These data authenticate the presence of hepatocyte-like cells in the anterior intestine of lbw, suggesting that the mutation leads to misinduction of anterior intestinal precursors into hepatic lineages.
The lbw-Encoding Cdx1b Resists the Liver-Inductive Wnt2bb Signal.
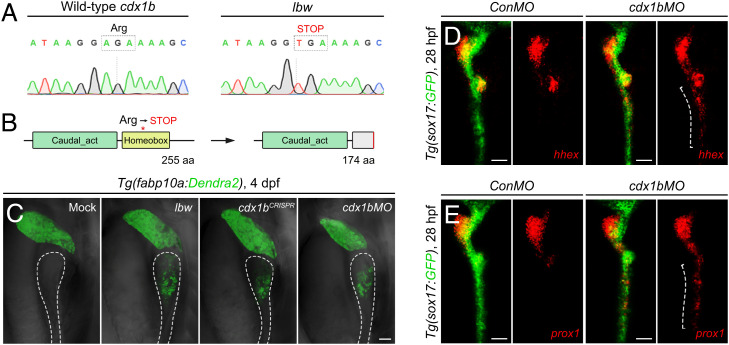
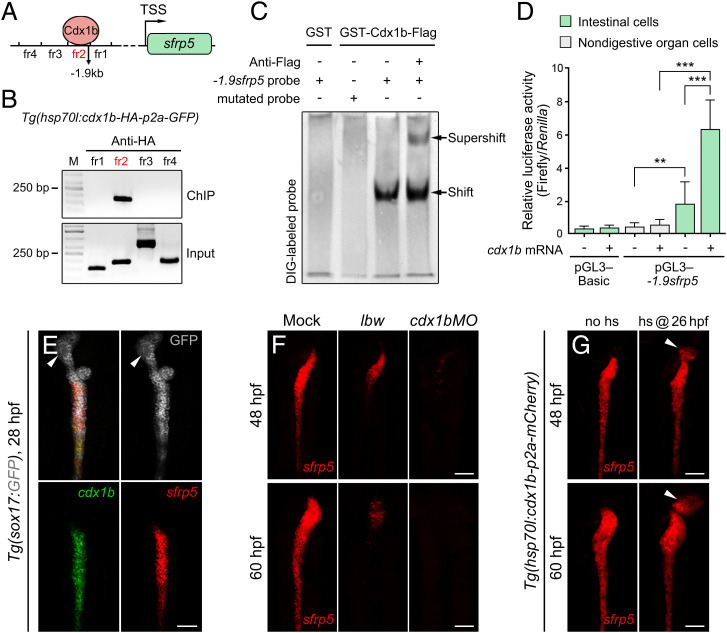
Using a standard set of simple sequence length polymorphism markers, we placed the lbw locus to chromosome 21 and positioned it within an interval of about 200 kb containing seven open reading frames, through fine-mapping (SI Appendix, Fig. S2A). Sequencing results showed that the lbw mutant allele contained an A→T mutation in the second exon of cdx1b gene (SI Appendix, Fig. S2 B and C), which led to the formation of a premature stop codon and the consequent truncation of the Cdx1b homeobox domain (Fig. 3 A and B). The nonsense-mediated mRNA decay (26) resulted in significant reductions of the cdx1b mRNA level in lbw homozygotes (SI Appendix, Fig. S2D). A cdx1bCRISPR,cq145 mutant generated by CRISPR/Cas9, the cdx1blbw/CRISPR double heterozygotes, and antisense morpholino oligonucleotides targeting the ATG region of cdx1b (cdx1bMO) (27), all phenocopied the hepatized anterior intestine of lbw (Fig. 3C and SI Appendix, Fig. S2 B–F), validating that the lbw phenotypes were caused by the dysfunction of Cdx1b. The Cdx1b dysfunction was ineffective to the swim bladder marker anxa5b (SI Appendix, Fig. S2G). The cdx1b mRNA is maternally contributed, and its zygotic expression became detectable exclusively in the nascent gut tube from 24 h postfertilization (hpf) and maintained in the intestine throughout development (27, 28). When translations of both maternal and zygotic cdx1b mRNAs were blocked by cdx1bMO, the earliest liver specification markers hhex and prox1 appeared in the intestinal domain at 28 hpf (Fig. 3 D and E), the stage when liver bud emerged. These results suggest Cdx1b as the key local factor in the anterior intestinal precursors to resist liver-inductive signals.
Fig. 3.
Loss of Cdx1b leads to misinduction of intestinal precursors to liver fate. (A and B) An A→T mutation in lbw led to the formation of a premature stop codon in the coding region of cdx1b gene (A), which disrupted of the Cdx1b homeobox domain (B). (C) The cdx1bCRISPR mutant (n = 58 to 67) and injection of cdx1bMO (n = 44 of 58) validated that the hepatized anterior intestine phenotype in lbw (n = 63 of 69) was caused by the dysfunction of Cdx1b. The dashed outlines mark the intestine. (D and E) Expressions of the earliest liver specification markers hhex (D, conMO, n = 58 of 62; cdx1bMO, n = 40 of 53) and prox1 (E, conMO, n = 51 of 54; cdx1bMO, n = 45 of 59) were detectable in the anterior intestinal domain (dashed brackets) of cdx1b morphants at 28 hpf. (Scale bars, 50 μm.) (C) Lateral views, anterior up; (D and E) dorsal views, anterior up. n = number of embryos with indicated phenotype/total analyzed in each class.
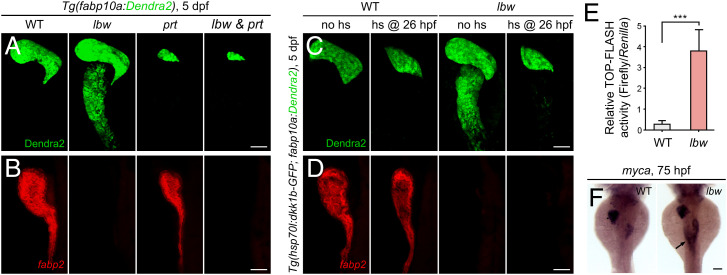
Zebrafish Cdx1b is required for the intestinal differentiation (28), but its roles and mechanisms in resisting the liver-inductive signals have previously been unappreciated. Because the mesoderm-derived Wnt2bb signal induces the initial specification of liver anlage from an undifferentiated gut tube (10) and EpCAM licenses the whole gut tube to respond to Wnt2bb (22), we checked whether the Wnt2bb signal is the antagonizing target of Cdx1b. The lbw/cdx1b and prt/wnt2bb double mutant exhibited strongly reduced liver and absence of fabp2, rescuing the hepatized anterior intestine but maintaining the defective intestinal differentiation as in the lbw single mutant (Fig. 4 A and B). Similarly, the hepatized anterior intestine but not loss of fabp2 in lbw was rescued by the heat-shock–induced overexpression of Dkk1b, an inhibitor of Wnt/β-catenin signaling (Fig. 4 C and D). These data showed that in the absence of the liver-inductive Wnt2bb signal, cdx1b mutation failed to cause hepatized anterior intestine, while still led to defects in intestinal differentiation. Both Wnt reporter activities in the sorted intestinal cells, as indicated by TOP-FLASH (Fig. 4E), and the expression of the Wnt downstream target gene myca in the anterior intestine were derepressed in lbw (Fig. 4F). All of these results demonstrate that independent of its roles in the intestinal differentiation, Cdx1b confers anterior intestinal precursors resistance to the liver-inductive Wnt2bb signal.
Fig. 4.
Cdx1b resists the liver-inductive Wnt2bb signal in the anterior intestine. (A and B) The hepatized anterior intestine was rescued in the lbw/cdx1b and prt/wnt2bb double mutant (A, WT, n = 29 of 30; lbw, n = 27 of 31; prt, n = 29 of 33; lbw & prt, n = 18 of 22), whereas the defective intestinal differentiation phenotype of lbw retained (B, WT, n = 38 of 38; lbw, n = 38 of 43; prt, n = 29 of 35; lbw & prt, n = 19 of 24). (C and D) The heat-shock (hs)–induced overexpression of Dkk1b rescued the hepatized anterior intestine (C, no hs in WT, n = 30 of 32; hs in WT, n = 34 of 46; no hs in lbw, n = 33 of 38; hs in lbw, n = 25 of 32), but not loss of fabp2 (D, no hs in WT, n = 32 of 35; hs in WT, n = 31 of 39; no hs in lbw, n = 25 of 28; hs in lbw, n = 28 of 35) in lbw. Note that overexpression of Dkk1b leads to small liver in both siblings and lbw. (E) TOP-FLASH reporter showed activation of Wnt signaling in the sorted lbw intestinal cells (n = 10). (F) In contrast to the wild-type (n = 37 of 40), expression of the Wnt downstream target gene myca became activated in the anterior intestine of lbw (arrow, n = 34 of 40). Data are expressed as mean ± SD, ***P < 0.001, Student's t test. hs @ 26 hpf, heat shock at 26 hpf. (Scale bars, 50 μm.) (A–D) Lateral views, anterior up; (F) dorsal views, anterior up. n = number of embryos with indicated phenotype/total analyzed in each class.
Cdx1b Activates sfrp5 Transcription to Block the Wnt2bb Signal.
To understand mechanisms underlying the resistance to Wnt2bb signaling, we analyzed whether the Cdx1b binding sequences were present within the promoter/enhancer regions of well-known Wnt inhibitors. A sequence in the promoter/enhancer region of sfrp5, 1.9 kb upstream of its transcriptional starting site (TSS), hereafter called −1.9sfrp5, was identified as a potential binding site of Cdx1b (Fig. 5A). Sfrp5 is a secreted protein that competes to the Frizzled receptor for binding to Wnt ligands, thus blocking Wnt signaling (29, 30). Chromatin immunoprecipitation (ChIP) from zebrafish embryos showed the association of Cdx1b-HA to the genomic DNA at the −1.9sfrp5 locus (Fig. 5B). Electrophoretic mobility shift assay (EMSA) indicated the direct binding of GST-Cdx1b recombinant protein to the −1.9sfrp5 DNA sequence (Fig. 5C). Activities of the luciferase reporter driven by the −1.9sfrp5 sequence were elevated by Cdx1b in the intestinal cells, but not in nondigestive organ cells (Fig. 5D). Expressions of sfrp5 and cdx1b in the gut tube appeared to be overlapped and posterior to the liver anlage during the emergence and growth of liver bud (Fig. 5E and SI Appendix, Fig. S3). Expression of sfrp5 in the intestine was significantly reduced in lbw, which became even more evident in the cdx1b morphant (Fig. 5F). In contrast to the inactivated sfrp5 expression and hepatized anterior intestine in lbw, the heat-shock–induced overexpression of cdx1b led to significantly reduced prox1 as well as ectopic expressions of sfrp5 and fabp2 in the liver (Figs. 5G and 6 A–C). A sfrp5CRISPR,cq95 mutant generated by CRISPR/Cas9 exhibited expression of the hepatocyte markers gc and ttr in the anterior intestine (SI Appendix, Fig. S4 A–D), which phenocopied the hepatized intestine of lbw (Fig. 6D). In contrast to the wild-type or single heterozygotes, the sfrp5CRISPR/cdx1blbw or sfrp5CRISPR/cdx1bCRISPR double heterozygotes exhibited reduced sfrp5 expression to some extent, but devoid of the hepatized intestine phenotype (SI Appendix, Fig. S4 E and F). Overexpression of sfrp5 led to small liver in both siblings and lbw, indicating the rescue of hepatized anterior intestine but not defective intestinal differentiation in lbw (Fig. 6 E and F). All of these results imply that Cdx1b may directly bind to the sfrp5 regulatory region and activates its transcription in the anterior intestinal precursors, thus blocking the Wnt2bb liver-inductive signal. Sfrp5 mediates the anti-Wnt2bb function, but not the intestinal differentiation function, of Cdx1b.
Fig. 5.
Cdx1b binds to genomic DNA at the sfrp5 regulatory locus to activate its transcription. (A and B) A genomic locus 1.9 kb upstream of the sfrp5 TSS was identified as the binding site of Cdx1b (A, fr2 marked in red), which was shown by ChIP analyses (B). fr represent the genomic DNA fragments upstream of the sfrp5 TSS. (C) The EMSAs showed the direct binding of GST-Cdx1b recombinant protein to the DIG-labeled −1.9sfrp5 DNA sequence in vitro. The binding specificity was controlled by the supershift with anti-Flag antibodies. (D) Activities of luciferase reporter driven by the −1.9sfrp5 DNA sequence in the sorted intestinal cells, but not in the nondigestive organ cells, were activated by cdx1b mRNA (n = 10). (E) The cdx1b and sfrp5 showed overlapped expression patterns in the undifferentiated gut tube at 28 hpf (n = 52 of 56). Note that the expressions are posterior to the liver bud (arrowheads). (F and G) In contrast to the controls (Mock at 48 hpf, n = 40 of 43; Mock at 60 hpf, n = 36 of 37), expression of sfrp5 in the intestine was significantly reduced in lbw and cdx1b morphant at 48 hpf and 60 hpf (F, lbw at 48 hpf, n = 30 of 34; lbw at 60 hpf, n = 34 of 37; cdx1bMO at 48 hpf, n = 48 of 62; cdx1bMO at 60 hpf, n = 50 of 65), whereas the heat-shock–induced cdx1b overexpression led to ectopic expression of sfrp5 in the liver bud (G, arrowheads, no hs, 48 hpf, n = 37 of 37; 60 hpf, n = 32 of 35; hs, 48 hpf, n = 46 of 55; 60 hpf, n = 44 of 52). Data are expressed as mean± SD, ***P < 0.001; **P < 0.01, Student’s t test. hs @ 26 hpf, heat shock at 26 hpf. (Scale bars, 50 μm.) (E) Dorsal views, anterior up; (F and G) ventral views, anterior up. n = number of embryos with indicated phenotype/total analyzed in each class.
Fig. 6.
Overexpression of Sfrp5 rescues the hepatized intestine but not defective intestinal differentiation of lbw. (A–C) In contrast to the wild-type control (A, n = 22 of 22), the lbw mutant exhibited loss of fabp2 and ectopic expression of prox1 in the anterior intestine (B, dashed bracket, n = 27 of 31). In contrast, the heat-shock–induced cdx1b overexpression resulted in significantly reduced prox1 and ectopic fabp2 expression in the liver (C, arrowhead, n = 42 of 48). (D) The sfrp5CRISPR mutant phenocopied the hepatized anterior intestine of lbw (WT, n = 33 of 34; lbw, n = 32 of 36; sfrp5CRISPR, n = 32 of 39). The dashed outlines mark the intestine. (E and F) Overexpression of Sfrp5 rescued the hepatized anterior intestine (E, Mock into WT, n = 32 of 34; sfrp5 mRNA into WT, n = 53 of 65; Mock into lbw, n = 30 of 34; sfrp5 mRNA into lbw, n = 30 of 38), but not defective intestinal differentiation, of lbw (F, Mock into WT, n = 36 of 36; sfrp5 mRNA into WT, n = 37 of 45; Mock into lbw, n = 28 of 32; sfrp5 mRNA into lbw, n = 26 of 35). (Scale bars, 50 μm.) (A–C) Ventral views, anterior up; (D–F) lateral views, anterior up. n = number of embryos with indicated phenotype/total analyzed in each class.
Fgf Signaling Acts Additively with Cdx1b to Resist Liver Fate.
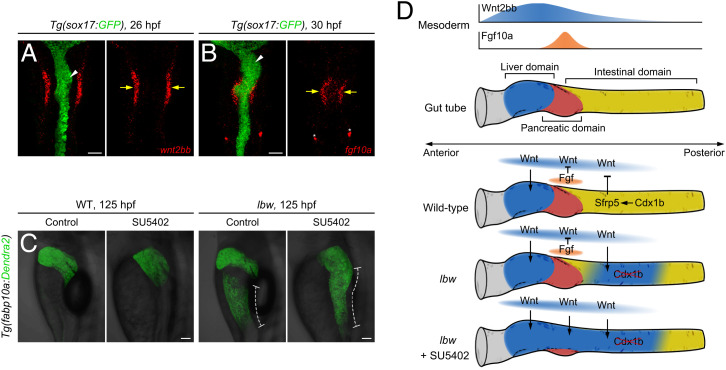
Note that the most anterior part of the intestine was not hepatized in lbw (Figs. 1H, 3C, and 6D), which might be caused by other factors acting in an additive fashion with Cdx1b to resist liver fate in this domain. Fgf10a has been reported to repress ectopic hepatocyte formation induced by Wnt overexpression (31). During the emergence of liver bud, wnt2bb was expressed in the lateral plate mesoderm flanking the foregut tube (Fig. 7A). While fgf10a was restrictively expressed in the mesenchyme surrounding the pancreatic and most anterior intestinal domain, more medial than wnt2bb (Fig. 7B). The wnt2bb and cdx1b are detectable at 24 hpf, which is earlier than the expression of fgf10a from 28 to 30 hpf (31, 32). Inhibition of Fgf signaling by SU5402 in lbw led to expansion of the hepatized tissue to the most anterior part of intestine, which did not occur in lbw or SU5402 treatment alone (Fig. 7C). Thus, in the most anterior part of intestine, Cdx1b and Fgf10a act in an additive fashion to resist liver fate. Taken together, the intestinal Cdx1b activates sfrp5 transcription, conferring anterior intestinal precursors resistance to the liver-inductive Wnt2bb signal, while Fgf10a acts additively with Cdx1b to resist liver fate in the most anterior part of intestine (Fig. 7D).
Fig. 7.
Inhibition of the Fgf signaling in lbw causes anterior expansion of hepatized intestine. (A) wnt2bb was expressed in the lateral plate mesoderm flanking the foregut tube (yellow arrows, n = 34 of 37). (B) fgf10a was expressed in the mesenchyme surrounding the pancreatic and most anterior intestinal domain, more medial than wnt2bb (yellow arrows, n = 55 of 61). The arrowheads indicate the liver bud. The asterisks indicate the lateral line neuromasts. (C) Inhibition of the Fgf signaling by SU5402 in lbw resulted in anterior expansion of hepatized intestine (dotted bars) to the most anterior part, but not SU5402 treatment or lbw alone (Control treatment in WT, n = 48 of 51; SU5402 treatment in WT, n = 41 of 56; Control treatment in lbw, n = 29 of 33; SU5402 treatment in lbw, n = 28 of 36). (D) Illustrations of mechanisms underlying the resistance of intestinal precursors to the liver-inductive Wnt signal. The intestinal Cdx1b activates sfrp5 transcription, conferring anterior intestinal precursors resistance to the liver-inductive Wnt2bb signal. Note that the repression of liver fate by Fgf10a is effective on the most anterior intestine, in which Cdx1b and Fgf10a act in an additive fashion. (A and B) Ventral views, anterior up; (C) lateral views, anterior up. n = number of embryos with indicated phenotype/total analyzed in each class.
Discussion
The liver and anterior intestinal precursors are in close proximity, and the liver anlage arises from the undifferentiated foregut tube. Key molecules involved in the intrinsic competence establishment for liver formation such as FoxA and GATA factors express in the precursors of both liver and anterior intestine (6, 20, 21). This expression pattern in the whole undifferentiated gut tube is also the case for EpCAM, the key molecule that licenses the responsive competence to the Wnt2bb signal (22). These studies implicate that before specification to the intestinal fate, anterior intestinal precursors are competent to respond to liver-inductive signals. Thus, intestinal precursors must develop mechanisms to resist liver-inductive signals. In this study, we have presented evidences on how intestinal precursors resist the liver-inductive Wnt signal to avoid being misinduced toward hepatic lineages.
In previous zebrafish studies, the Cdx1b-deficient larvae display hypoplastic intestines (27, 28). Here we show that lbw exhibits hepatized anterior intestine, suggesting previously unappreciated roles of Cdx1b in the resistance of liver-inductive signals. The knockout of mammalian Cdx2, the homolog of zebrafish cdx1b, leads to sporadic transformation of duodenal cells to pyloric stomach-like identities (33), implicating potential functions of Cdx2 in the duodenal precursors to resist the inductive signals of more anterior digestive organs. The zebrafish hhex mutant develops an intrahepatic intestinal tube due to derepression of Cdx1b and Pdx1 (34), suggesting that the liver precursors also need to resist the intestine-inductive signals, such as Cdx1b. These reports, together with this study, imply that the intrinsic transcriptional factors such as Cdx1b and Hhex not only promote cellular development autonomously, but also orchestrate organogenesis of the entire foregut endoderm.
Frizzled (Fzd) is a large family of proteins that serve as receptors in the Wnt signaling pathway (35). Fzd5 is required for early liver formation and interacts genetically with Wnt2 and Wnt2bb (36). Different thresholds of Wnt-Fzd7 signaling are essential to maintain foregut progenitors (37). The secreted frizzled-related proteins, such as Frzb1 and Sfrp2, are identified to express in the chicken intestinal tract (38). Sfrp5, previously reported in Xenopus, coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt signaling (29). In this study, Sfrp5 acts as the downstream effector of Cdx1b to resist the liver-inductive Wnt2bb signaling. The C-terminal of SFRPs contains segments of positive-charged residues that facilitate binding extracellular matrix (ECM) molecules with affinity (39). FGFs enhance the extracellular accumulation of SFRPs through cobinding the complexes of SFRP and ECM molecules (40, 41), which may explain why the Sfrp5 protein in the wild-type embryos cannot diffuse across the pancreatic domain to reach the liver domain.
Note that the ectopic expressions of sfrp5 and fabp2 induced by the cdx1b overexpression is restricted to the liver, but not other tissues (Figs. 5G and 6C). Additionally, Cdx1b fails to activate the luciferase reporter driven by the −1.9sfrp5 sequence in nondigestive organ cells (Fig. 5D). These data hypothesize that either cofactors of Cdx1b are specifically expressed in the liver and intestinal precursors, or only the liver and intestinal precursors maintain the competence to induce sfrp5 expression after 26 hpf. But the identities of cofactors or mechanisms underlying the competence maintenance need further investigations.
Materials and Methods
Zebrafish Strains.
The zebrafish facility and studies were approved by the Institutional Review Board of Southwest University (Chongqing, China). Zebrafish strains were maintained under standard laboratory conditions according to Institutional Animal Care and Use Committee protocols. Embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU, Sigma) from 24 hpf to prevent pigmentation. All experimental protocols were approved. All results involved in the study were acquired according to ethical standards.
Zebrafish of lbwcq93, cdx1bCRISPR,cq145, prts416 (10), sfrp5CRISPR,cq95 mutants, and Tg(fabp10a:Dendra2)cq102, Tg(sox17:GFP)s870 (13), Tg(hsp70l:cdx1b-HA-p2a-GFP)cq103, Tg(hsp70l:cdx1b-p2a-mCherry)cq104, and Tg(hsp70l:dkk1b-GFP)w32 (31) transgenic lines were established and used in this study.
scRNA-Seq.
scRNA-seq was performed using the 10X Chromium platform according to a published protocol (23, 25). The lbw intestine, wild-type liver, and intestine were dissected at 6 dpf and collected in 500 μL of DMEM-10%FBS (Gibco). The viability of cells (∼90%) was determined using Trypan blue staining. The libraries were constructed using Chromium Single Cell 3′ GEM v3.1 Reagent Kit and sequenced on an Illumina NovaSEq. 6000 with ∼30% sequencing saturation. Raw sequence reads were aligned to the zebrafish genome (GRCz11) using the 10X Genomics Cell Ranger 4.0. The output gene-expression matrices were further analyzed using the Seurat v3.2 package. UMAP and t-SNE were used for clustering analysis at a resolution of 0.7 to identify distinct clusters of cells and to visualize the clustering results. The Signature score was performed using the AddModuleScore function in Seurat with default parameters.
Positional Cloning of lbw.
Heterozygous lbw was outcrossed with the polymorphic line SJD to perform mapping as previously described (42); 357 mutant embryos were used for fine-mapping with simple sequence length polymorphism markers. The primer sequence of z-markers can be found at www.zfin.org/, and the following primers were designed to narrow down the determined interval further: L21m25 forward (5′-CAGTCATCAGGGCTGACAGCAG-3′), reverse (5′-GCATGATGGTCTACATCTGACAG-3′); L21m32 forward (5′-CTAACTGTTTGCCTTCAG-3′), reverse (5′-ATGTCCCCACTAGGTCAC-3′); L21m50 forward (5′-GAGGAGAGCAGAGCCATTTGTC-3′), reverse (5′-GTGTGTACGTGTGCATATTCACAG-3′). The lbw/cdx1b genotype was finally determined by sequencing the PCR fragment containing the mutated bases.
Generation of Zebrafish CRISPR Mutants.
The cdx1bCRISPR and sfrp5CRISPR mutants in which mutagenesis were targeted at exon 2 and exon 1, respectively, were generated by CRISPR/Cas9 as previously described (43, 44). The 20-nt guide RNA (gRNA) target sequences were used as follows: cdx1b exon 2 (5′-ACAGCCGCTACATCACAATA-3′); sfrp5 exon 1 (5′-GGACCACGAGACCATGCCTG-3′). A mixture including 300 pg of Cas9 mRNA and 80 pg of gRNA was microinjected into the one-cell–stage embryos under the AB genetic background. The DNA of embryos or tail fins was extracted through the alkaline lysis procedure. The genomic regions flanking the gRNA target sites were amplified and sequenced for validation. The validated embryos were raised to adults, then screened to identify founders. The identified founders were crossed back to the AB genetic background for two generations to obtain cdx1bCRISPR+/− or sfrp5CRISPR+/− stable lines.
mRNAs and Morpholinos.
The cDNAs of zebrafish cdx1b and sfrp5 were amplified using PfuUltra High-Fidelity DNA polymerase (Stratagene) and subcloned into pCS2(+) vector for mRNA preparation. Linearized plasmids were used to synthesize mRNAs by the mMESSAGE mMACHINE Transcription Kit (Ambion). cdx1b and sfrp5 mRNAs were injected at the one-cell stage with 150 to 200 pg per embryo. Antisense morpholino oligonucleotides targeting the ATG region of cdx1b (5′-CATTTTTTCTGGTGGCTCCAGTGC-3′) (27) were injected at the one-cell stage with 2 ng per embryo. The control morpholino (conMO) contained the same nucleotide sequences as cdx1bMO except for four mismatches: (5′-CAaTTTTTgTGGTGcCTCCAcTGC-3′).
Antibody Staining, In Situ Hybridization, and Histological Staining.
Zebrafish embryos were fixed overnight in 4% formaldehyde solution. Whole-mount in situ hybridization was performed using fabp2 (28), slc15a1b, angptl3 (45), gc, anxa5b (46), and myca (31) antisense probes. Fluorescent in situ hybridization (FISH) and the combination of FISH and antibody staining were performed as previously described (47–49) using apo14, ttr, rbp2a (50), hhex (51), prox1 (52), fabp2, cdx1b (27), sfrp5, wnt2bb (10), and fgf10a (31) antisense probes labeled with digoxigenin or fluorescein, and antidigoxigenin-POD (1:1,000; Roche), antifluorescein-POD (1:1,000; Roche), anti-GFP (1:1,000; Abcam), anti-Dendra2 (1:1,000; Evrogen) antibodies, and tyramide signal amplification and fluorescence detection system (TSA, PerkinElmer). Antibody staining on tissue sections was performed using anti-Bhmt (1:500; gift from the Peng Laboratory, Zhejiang University, Hangzhou, China) primary antibodies. Whole-mount antibody staining was performed using 4E8 (1:1,000; Abcam) primary antibody. The nuclei were stained with DNA fluorochrome DAPI (Sigma). The secondary antibodies used in the study included Alexa 488-conjugated donkey anti-rabbit IgG (1:1,000; Invitrogen), Alexa 568-conjugated donkey anti-mouse IgG (1:1,000; Invitrogen), and Alexa 647-conjugated donkey anti-goat IgG (1:1,000; Invitrogen).
For histological staining, paraffin sections were first dewaxed and rehydrated, then stained with H&E staining solution or 0.5% PAS reagent according to standard procedures.
Generation of Transgenic Lines and Heat Shock Induction.
To generate pBluescript-hsp70l:cdx1b-HA-p2a-GFP and pBluescript-hsp70l:cdx1b-p2a-mCherry plasmids, the cdx1b-HA-p2a-GFP and cdx1b-p2a-mCherry fragments were subcloned downstream of hsp70l promoter. Constructs flanked by the I-SceΙ restriction sites were coinjected with I-SceΙ (New England Biolabs) into zebrafish embryos of the AB genetic background at the one-cell stage for transgenesis. Primers for cloning cdx1b-HA: forward (5′-ATGTACGTGAGTTATCTCCTAGATA-3′), reverse (5′-AGCGTAGTCTGGGACGTCGTATGGGTAATACTCTTCTTTGATGGACATTGGC-3′).
The Tg(hsp70l:dkk1b-GFP), Tg(hsp70l:cdx1b-HA-p2a-GFP), or Tg(hsp70l:cdx1b-p2a-mCherry) transgenic embryos at 26 hpf were heat-shocked at 39 °C for 40 min, then incubated at 28.5 °C until the time point of analyses.
ChIP.
ChIP was performed as previously described (53). The Tg(hsp70l:cdx1b-HA-p2a-GFP) embryos were heat-shocked at 26 hpf, then collected and dissociated in the lysis buffer. The chromatin was sheared by sonication to an average fragment size of 150 to 300 bp. IP were performed using 4 µg of anti-HA (Roche) antibodies. ChIP samples and corresponding inputs were detected by PCR using 10 pairs of primers covering every 150 to 300 bp of the 2,500-bp genomic DNA fragment upstream of the sfrp5 TSS. A genomic DNA fragment at the -1.9sfrp5 locus was identified to be associated with Cdx1b-HA using the -1.9sfrp5 primers (5′-GGCTTATCTAGGTTAATTAGG-3′ and 5′-GTTTAACAGAGCAAGGACAT-3′).
EMSA.
EMSA was performed using the DIG Gel Shift Kit (Roche) according to the manufacturer’s instructions. In vitro transcribed and translated GST-Cdx1b-Flag was incubated with DIG-labeled −1.9sfrp5 probe containing the Cdx1b-binding site. For negative control and supershift experiments, DIG-labeled mutated probe and 1 µg of anti-Flag antibody (Sigma) were added to the reaction, respectively. The reaction mixtures were separated on a 5% native polyacrylamide gel and electrically transferred to a nylon membrane. The chemiluminescent signals were detected by alkaline phosphatase-conjugated anti-DIG antibodies (Roche) and CSPD (Roche).
TOP-FLASH and Luciferase Reporter Assay.
The 50 ng/μL TOP-FLASH or 40 ng/μL pGL3 –1.9sfrp5 luciferase reporter plasmids were injected into the embryos at the one-cell stage. The injected embryos were divided into siblings and lbw at 96 hpf based on phenotypes, and their intestinal tissues were isolated with tweezers. The cells in the intestine, controlled by nondigestive organ cells, were sorted and collected by flow cytometry, then incubated with 50 μL of passive lysis buffer for 30 min at room temperature. Luciferase activity was measured by GloMax 20/20 Luminometer (Promega) using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized by Renilla luciferase activities in embryos coinjected with the 5 ng/μL vectors of pRL-CMV.
Chemical Treatments.
The 1 mM stock of the Fgf receptor inhibitor SU5402 (Sigma-Aldrich) was prepared in 100% DMSO and diluted to 2 µM with egg water. The 0.2% DMSO solution in egg water was used as a control. The larvae were treated with SU5402 from 24 to 38 hpf, then incubated in egg water with PTU until the time point of analyses.
Quantification and Statistical Analysis.
The lbw homozygotes could not survive beyond 12 dpf, and the heterozygotes were raised to adulthood. More than nine pairs of fish were used to set one specific cross. For a cross between lbw heterozygous parents, more than 40 embryos or larvae were used for analyses in each treatment. The genetic ratio of lbw mutant conformed with the Mendelian inheritance, and genotyping preceded phenotyping in homozygotes and heterozygotes analyses. All experiments comparing treatment samples were performed using randomly assigned siblings without investigator blinding. Sample sizes were chosen according to the estimation of effect sizes. After at least two repeats, data were analyzed for statistical significance using the comparison of means, two-tailed Student’s t test and Fisher’s exact test with IBM SPSS Statistics v25. Variance for all groups of data were presented as ±SD. No data were excluded. A value of P < 0.05 was considered to be statistically significant. For all statistical comparisons: *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank D. Y. Stainier, D. Wang, and J. Peng for fish lines, plasmids, and antibodies. This work was supported by the National Natural Science Foundation of China (32192400, 32270859, and 31730060) and the National Key R&D Program of China (2021YFA0805000).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. K.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205110119/-/DCSupplemental.
Data, Materials, and Software Availability
All the scRNA-seq data in this study have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA857143) (54).
References
- 1.Field H. A., Ober E. A., Roeser T., Stainier D. Y., Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279–290 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Zaret K. S., Grompe M., Generation and regeneration of cells of the liver and pancreas. Science 322, 1490–1494 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., et al. , A single-cell-resolution fate map of endoderm reveals demarcation of pancreatic progenitors by cell cycle. Proc. Natl. Acad. Sci. U.S.A. 118, e2025793118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke Z., Oliver G., Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech. Dev. 118, 147–155 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Yoshitomi H., Zaret K. S., Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development 131, 807–817 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Lee C. S., Friedman J. R., Fulmer J. T., Kaestner K. H., The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bort R., Signore M., Tremblay K., Martinez Barbera J. P., Zaret K. S., Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 290, 44–56 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Gao N., et al. , Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 22, 3435–3448 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi J. M., Dunn N. R., Hogan B. L., Zaret K. S., Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 15, 1998–2009 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ober E. A., Verkade H., Field H. A., Stainier D. Y., Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Calmont A., et al. , An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev. Cell 11, 339–348 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Shin D., et al. , Bmp and Fgf signaling are essential for liver specification in zebrafish. Development 134, 2041–2050 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Chung W. S., Shin C. H., Stainier D. Y., Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev. Cell 15, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wandzioch E., Zaret K. S., Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science 324, 1707–1710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yimlamai D., et al. , Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens M. L., et al. , Genomic integration of Wnt/β-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs. Development 144, 1283–1295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nissim S., et al. , Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev. Cell 28, 423–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao R., Duncan S. A., Embryonic development of the liver. Hepatology 41, 956–967 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Burke Z. D., Thowfeequ S., Tosh D., Liver specification: A new role for Wnts in liver development. Curr. Biol. 16, R688–R690 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Ang S. L., et al. , The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 119, 1301–1315 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Holtzinger A., Evans T., Gata4 regulates the formation of multiple organs. Development 132, 4005–4014 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Lu H., Ma J., Yang Y., Shi W., Luo L., EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Farnsworth D. R., Saunders L. M., Miller A. C., A single-cell transcriptome atlas for zebrafish development. Dev. Biol. 459, 100–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capdevila C., et al. , Single-cell transcriptional profiling of the intestinal epithelium. Methods Mol. Biol. 2171, 129–153 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Cai P., et al. , Farnesoid X receptor is required for the redifferentiation of bipotential progenitor cells during biliary-mediated zebrafish liver regeneration. Hepatology 74, 3345–3361 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Chang Y. F., Imam J. S., Wilkinson M. F., The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Cheng P. Y., et al. , Zebrafish cdx1b regulates expression of downstream factors of Nodal signaling during early endoderm formation. Development 135, 941–952 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Flores M. V., et al. , Intestinal differentiation in zebrafish requires Cdx1b, a functional equivalent of mammalian Cdx2. Gastroenterology 135, 1665–1675 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Li Y., et al. , Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev. 22, 3050–3063 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satoh W., Matsuyama M., Takemura H., Aizawa S., Shimono A., Sfrp1, Sfrp2, and Sfrp5 regulate the Wnt/beta-catenin and the planar cell polarity pathways during early trunk formation in mouse. Genesis 46, 92–103 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Shin D., Lee Y., Poss K. D., Stainier D. Y., Restriction of hepatic competence by Fgf signaling. Development 138, 1339–1348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manfroid I., et al. , Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development 134, 4011–4021 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Grainger S., Savory J. G., Lohnes D., Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 339, 155–165 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Gao C., et al. , Zebrafish hhex-null mutant develops an intrahepatic intestinal tube due to de-repression of cdx1b and pdx1. J. Mol. Cell Biol. 11, 448–462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malbon C. C., Frizzleds: New members of the superfamily of G-protein-coupled receptors. Front. Biosci. 9, 1048–1058 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Poulain M., Ober E. A., Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development 138, 3557–3568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z., Rankin S. A., Zorn A. M., Different thresholds of Wnt-Frizzled 7 signaling coordinate proliferation, morphogenesis and fate of endoderm progenitor cells. Dev. Biol. 378, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride H. J., Fatke B., Fraser S. E., Wnt signaling components in the chicken intestinal tract. Dev. Biol. 256, 18–33 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Uren A., et al. , Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 275, 4374–4382 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Zhong X., et al. , Regulation of secreted Frizzled-related protein-1 by heparin. J. Biol. Chem. 282, 20523–20533 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Prochazkova M., et al. , FGF signaling refines Wnt gradients to regulate the patterning of taste papillae. Development 144, 2212–2221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahary N., et al. , The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 77, 305–329 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Chen J., et al. , Acute brain vascular regeneration occurs via lymphatic transdifferentiation. Dev. Cell 56, 3115–3127.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Xia Z., et al. , Metal transporter Slc30a1 controls pharyngeal neural crest differentiation via the zinc-Snai2-Jag1 cascade. MedComm (2020) 2, 778–797 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S. H., et al. , Angiopoietin-like 3 regulates hepatocyte proliferation and lipid metabolism in zebrafish. Biochem. Biophys. Res. Commun. 446, 1237–1242 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Teoh P. H., Shu-Chien A. C., Chan W. K., Pbx1 is essential for growth of zebrafish swim bladder. Dev. Dyn. 239, 865–874 (2010). [DOI] [PubMed] [Google Scholar]
- 47.He J., Mo D., Chen J., Luo L., Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nat. Protoc. 15, 3361–3379 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Liu C., et al. , Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 44, 1162–1176 (2016). [DOI] [PubMed] [Google Scholar]
- 49.He J., et al. , Mammalian target of rapamycin complex 1 signaling is required for the dedifferentiation from biliary cell to bipotential progenitor cell in zebrafish liver regeneration. Hepatology 70, 2092–2106 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Lickwar C. R., et al. , Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 15, e2002054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J., Lu H., Zou Q., Luo L., Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e8 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Chen J., et al. , Cerebrovascular injuries induce lymphatic invasion into brain parenchyma to guide vascular regeneration in zebrafish. Dev. Cell 49, 697–710.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Das P. M., Ramachandran K., vanWert J., Singal R., Chromatin immunoprecipitation assay. Biotechniques 37, 961–969 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Yang Y., et al. , Data for “Intestinal precursors avoid being misinduced to liver cells by activating Cdx-Wnt inhibition cascade.” NCBI SRA. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA857143. Accessed 25 October 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the scRNA-seq data in this study have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA857143) (54).