Significance
Understanding the mechanisms of spatial heterogeneity in tuberculosis (TB) transmission may enhance the effectiveness of spatially targeted interventions. In this work, we integrated whole-genome sequences of 2,712 Mycobacterial tuberculosis isolates and geographic coordinates of the patients’ residences in a large urban catchment area in Lima, Peru, to clarify if local transmission is the primary driver of high rates of TB disease in spatially distinct areas. We find that spatially specific TB incidence does not correlate closely to the frequency of local transmission. This suggests that understanding the causes that lead to spatially distinct TB hotspots is necessary for spatially targeting interventions.
Keywords: tuberculosis, genomic epidemiology, transmission, spatial analysis, whole-genome sequence
Abstract
Spatially targeted interventions may be effective alternatives to individual or population-based prevention strategies against tuberculosis (TB). However, their efficacy may depend on the mechanisms that lead to geographically constrained hotspots. Local TB incidence may reflect high levels of local transmission; conversely, they may point to frequent travel of community members to high-risk areas. We used whole-genome sequencing to explore patterns of TB incidence and transmission in Lima, Peru. Between 2009 and 2012, we recruited incident pulmonary TB patients and their household contacts, whom we followed for the occurrence of TB disease. We used whole-genome sequences of 2,712 Mycobacterial tuberculosis isolates from 2,440 patients to estimate pariwise genomic distances and compared these to the spatial distance between patients’ residences. Genomic distances increased rapidly as spatial distances increased and remained high beyond 2 km of separation. Next, we divided the study catchment area into 1 × 1 km grid-cell surface units and used household spatial coordinates to locate each TB patient to a specific cell. We estimated cell-specific transmission by calculating the proportion of patients in each cell with a pairwise genomic distance of 10 or fewer single-nucleotide polymorphisms. We found that cell-specific TB incidence and local transmission varied widely but that cell-specific TB incidence did not correlate closely with our estimates of local transmission (Cohen’s k = 0.27). These findings indicate that an understanding of the spatial heterogeneity in the relative proportion of TB due to local transmission may help guide the implementation of spatially targeted interventions.
With over 9.9 million cases and 1.2 million deaths estimated in 2020, tuberculosis (TB) is a major global health threat (1). While many tools have been developed for TB control, mathematical models suggest that TB cannot be eliminated without significant improvements in optimizing these existing tools by targeting them to the people who would most benefit from them (2, 3). While current World Health Organization (WHO) guidelines emphasize targeting them to high-risk individuals like people living with HIV, others have noted that much transmission occurs outside these defined risk groups and that a broader targeting strategy is needed (4).
Existing tools for TB control include active case finding, preventive therapy for latent TB infection, improved access to diagnosis and care, and bacillus Calmette-Guérin vaccination and/or revaccination (2, 3). Although recent innovations like short-course preventive therapy make expanded use of these interventions more feasible (5), it is not practical to implement these labor- and cost-intensive interventions across entire national populations. In many communities, TB tends to cluster geographically into high-incidence hotspots, and the targeting of interventions to spatially defined hotspots has been proposed as an effective alternative to individual or population-based interventions (4, 6–9). Although hotspots are usually defined by local TB incidence, it is not always clear if high rates of local TB transmission correspond to high rates of TB disease occurrence. Because most people who are infected with Mycobacterial tuberculosis (MTB) never develop TB disease, much TB transmission goes undetected and high local rates of disease may reflect spatial heterogeneity in the risk factors that lead to TB progression, rather than pinpointing areas where TB transmission is ongoing (4).
Understanding the causes of the spatially heterogeneous transmission may help inform efforts to develop effective spatially targeted TB control interventions. Here, we measure local TB transmission by examining the pairwise genomic distances between TB patients’ isolates as a function of spatial distance to identify transmission hotspots, and we compare these areas to high-incidence hotspots in the same regions.
Materials and Methods
Study Setting and Design.
We conducted a prospective cohort study of household TB transmission in Lima, Peru. The study was implemented in a defined catchment area of metropolitan Lima, consisting of 20 districts and including ∼3.3 million residents living in urban areas and periurban, informal shantytown settlements (10). In brief, between September 2009 and August 2012, we recruited consecutively diagnosed incident pulmonary TB patients 15 y of age or older as index TB patients. We confirmed their microbiological status by sputum smear microscopy and/or mycobacterial culture. Within 2 wk of patient enrollment in the study, we visited their households and invited their household contacts (HHCs) to participate in a longitudinal study of infection and disease occurrence. We obtained clinical and demographic data from index patients and HHCs, including age, body mass index, gender, education, type of housing, TB symptoms, bacillus Calmette-Guerin vaccination history, and comorbidities such as HIV and diabetes. The geographic coordinates of participating households were collected by a global positioning system.
We also examined TB registries at the participating health clinics to ensure we captured all incident TB cases during the 12-mo follow-up time. In addition to baseline samples, we collected sputum from all study patients, both index patients and HHCs who developed TB disease during follow-up. These were evaluated by sputum smear microscopy, mycobacterial culture and drug susceptibility testing. Where clinically indicated, patients underwent further microbiological tests up to 4 y after the initial diagnosis of TB disease.
Whole-Genome Sequencing and Genetic Distance.
We obtained whole-genome sequences (WGSs) from a subset of MTB isolates with Illumina HiSeq in paired-end mode with a read length of 100 to 150 base pairs and mean coverage of at least 50-fold. We mapped the paired-end raw sequencing data to the H37Rv reference genome using the BWA-MEM (Burroughs Wheeler Aligner-Maximal Exact Match) algorithm (11) and used SAMtools and Pilon to identify the single-nucleotide polymorphisms (SNPs) and the insertions and deletions using a coverage-based approach (12, 13). We assigned a variant call as missing if the valid depth of coverage at a specific variant was less than 12 reads, if the mean read mapping quality at the site did not reach 10, or if none of the alternative alleles accounted for at least 85% of the valid coverage. We retained SNPs in calculating the genetic distance of pairwise isolates. We excluded the variants in the PE/PPE gene family, as the error rate of short-read sequencing methodology is high in highly repetitive regions (14). We also excluded the isolates with evidence of mixed infection using the barcode method (15). We determined the genetic distance between two isolates using the number of SNPs that differ between the two isolates.
Pathogen Genetic Relatedness as a Function of Geographic Distance.
For all possible pairs of TB patients, we calculated the geographic distance between their households using an interface between R and the Open Source Routing Machine, a publicly available tool that analyzes OpenStreetMap road network data, to obtain the shortest motor vehicle travel time between points in both minutes and distance in kilometers (16). We estimated the genetic distance between all pairs of MTB isolates collected at the time of diagnosis and evaluated this distance by physical distance by levels of geographical proximity. For various levels (SNP difference cutoffs = 1, 5, and 10) of genetic relatedness, we compared the odds that a pair of isolates was closely related for various levels of geographical proximity to the odds of close genetic relatedness for paired isolates from individuals whose residential households were separated by 10 min or more of travel time.
Heterogeneity of TB Incidence and Local Transmission.
We divided Lima into areas using the 1 × 1 km WorldPop grid-cell surface areas and used household geographic coordinates to locate each TB patient in a specific cell (17). We considered only cells in which we had enrolled at least 10 TB patients. We estimated TB incidence for each cell by combining information on the number of TB patients, cell population estimates obtained from the WorldPop database, and patients’ study participation duration. In each cell, we used the proportion of clustered TB patients as a proxy of local transmission. To derive the proportion of clustered TB patients in each cell, we counted the number of TB patients who had a pairwise genomic distance of ≤10 with any other TB patient in that cell divided by the total number of TB study patients in that cell. We used the weighted Cohen’s kappa statistic test to assess the agreement between the quintiles of the two metrics (18). These analyses were conducted using ArcGIS 10.8, RStudio, and SAS 9.4 (SAS Institute).
Drug Resistance and Transmission Dynamics.
We repeated our analyses in the subgroup of patients with phenotypic drug-resistant TB (resistant to at least one of the following drugs: isoniazid, rifampicin, rifabutin, ethambutol, pyrazinamide, streptomycin, linezolid, moxifloxacin, amikacin, kanamycin, capreomycin, and ethionamide) to evaluate the impact of drug resistance on the transmission dynamics (10).
Temporal Variation of Spatial TB Burden.
To assess whether the spatial distribution of TB had changed since the time we had collected our data, we evaluated temporal variation in TB notification data from 11 districts of Lima Ciudad, one-third of our original study area, in 2015 and 2019. We ranked districts based on their annual TB burdens and calculated weighted Cohen’s kappa statistic test to determine the consistency of these rankings over time.
Ethical Considerations.
Before study participation, all study participants provided voluntary, written informed consent. The Harvard School of Public Health and Peru’s Research Ethics Committee of the NIH provided institutional review board approval.
Results
Sample Size Description.
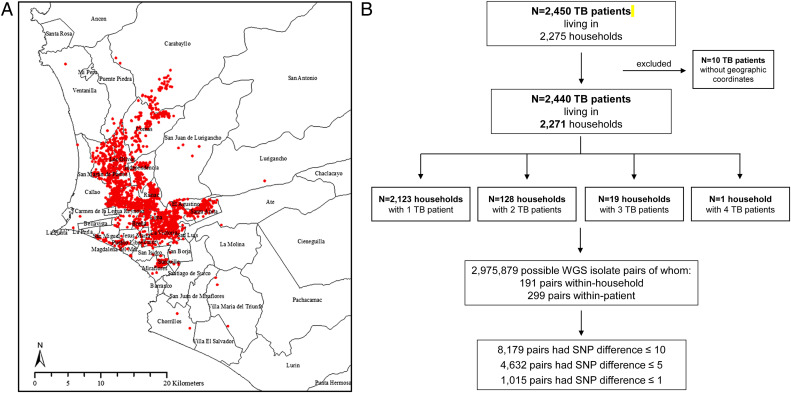
Among the 3,851 culture-positive TB patients, 2,916 isolates underwent WGS. After excluding 202 isolates with evidence of mixed infection and poor raw-read quality, we included 2,714 isolates from 2,440 TB patients living in 2,271 households, geographically located as shown in Fig. 1A. The number of TB patients by household is reported in Fig. 1B. We assessed 2,975,879 sequence pairs, 299 of which were repeat isolates from individual study participants and 191 of which were from distinct individuals within households. Among individuals with multiple isolates, 212 contributed two high-quality isolates, 27 had three isolates, and 1 had four isolates.
Fig. 1.
Spatial distribution and description of the TB cohort recruited between 2009 and 2012 in Lima, Peru. (A) Red dots indicate locations of the places of residency of TB patients in Lima districts. The districts of Lima are labeled. (B) Study flowchart.
Genetic Relatedness and Geographic Distance.
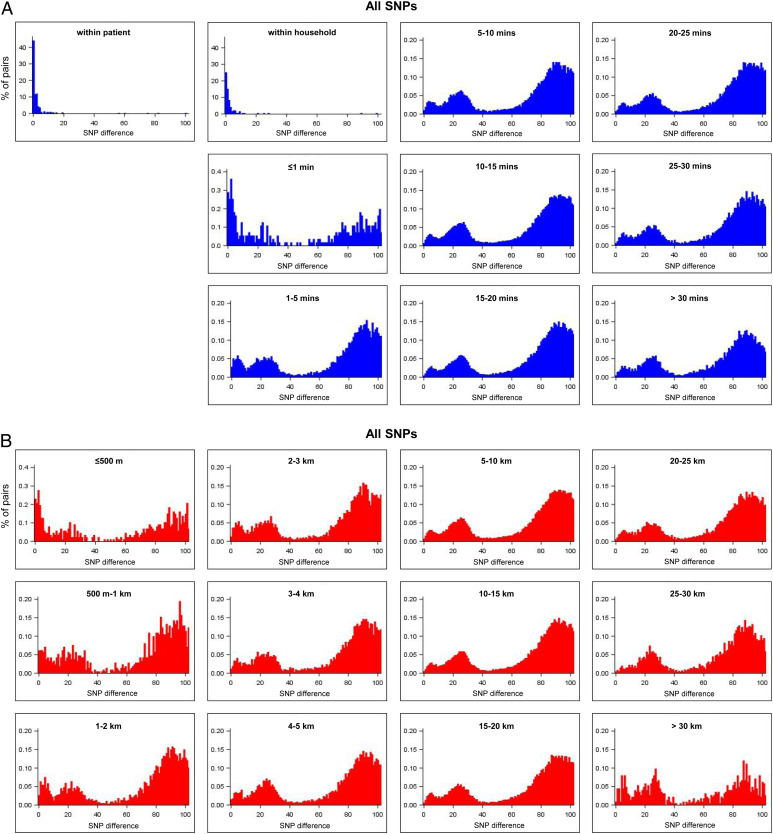
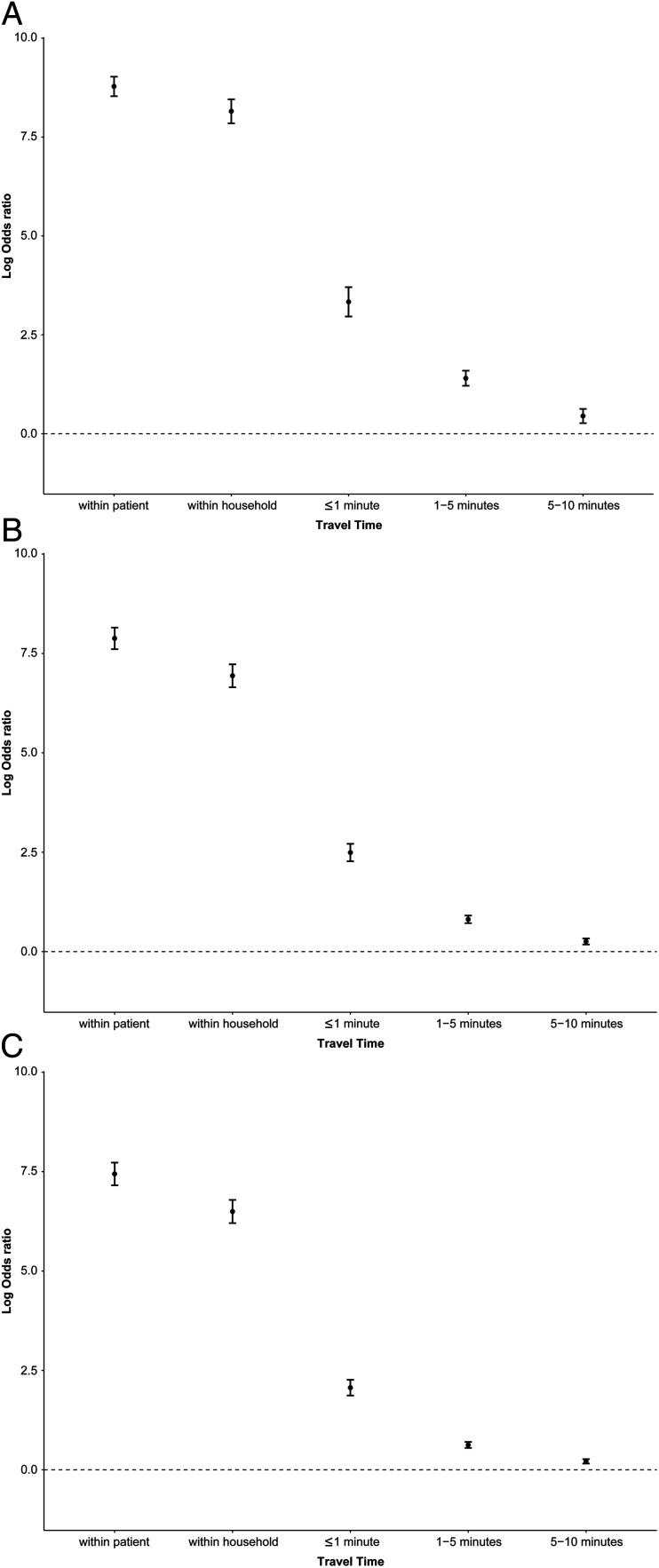
Fig. 2 A and B show that genomic distances were bimodally distributed across geographical distances, suggesting the presence of distinct closely related and more distantly related TB isolates. Genomic distances were smaller for within-household pairs and increased quickly with increasing geographical distances, suggesting that local transmission is restricted to nearby households of index patients (Fig. 2). The log odds of close genetic relatedness fell rapidly with increasing travel time between individuals’ places of residence (Fig. 3 and SI Appendix, Tables S1 and S2).
Fig. 2.
Distributions of pairwise-SNP differences stratified by geographical proximity (A) using travel time in minutes (zoom between 0 and 100 SNP differences) and (B) using distances in kilometers (zoom between 0 and 100 SNP differences).
Fig. 3.
Associations between pairwise geographical proximity (travel time between the places of residence) and SNP differences using pairs located 10 min or more apart as the reference category, measuring at a log odds ratio scale. (A) ≤1 SNP difference vs. >1 SNP difference, (B) ≤5 SNP difference vs. >5 SNP difference, and (C) ≤10 SNP difference vs. >10 SNP difference. Error bars are 95% CIs.
Genomically Linked Isolates.
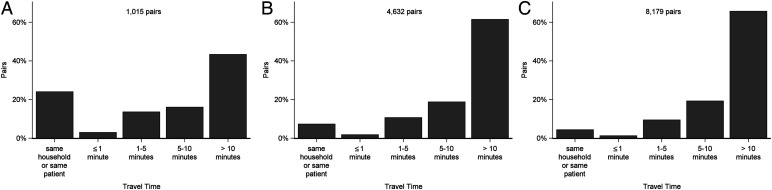
We chose a conservative genomic difference threshold of one SNP to identify the pairs most likely to be linked by direct transmission, of which we found 1,015 (0.03%) pairs among 783 patients. Among the 299 paired samples from the same individuals, 167 (55.8%) pairs were at or below the threshold, and among the 191 within-household pairs, 77 (40.3%) pairs were at or below the threshold. Among the genomically linked pairs, 43.3% were located more than 10 min apart, including some that were located at opposite ends of the city (Fig. 4A and SI Appendix, Table S1A). When we relaxed the thresholds, the proportion of the genomically linked pairs with more than 10 min apart increased (Fig. 4 B and C and SI Appendix, Table S1 B and C).
Fig. 4.
Histograms by travel time for pairs with (A) ≤1 SNP difference, (B) ≤5 SNP difference, and (C) ≤10 SNP difference.
TB Incidence and Local Transmission.
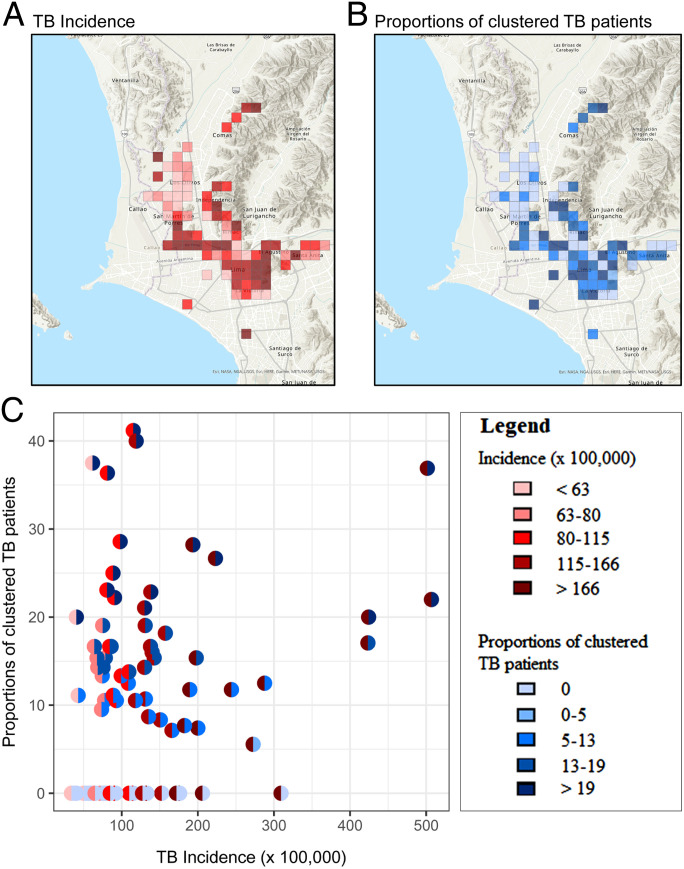
Among the 245 1 × 1 km cells in which Lima was divided, 190 cells contained at least a pair of TB patients and 87 cells at least 10 TB patients. We focused on these 87 cells, in which the median proportion of patients sharing a closely related isolate was 10.5% (Inter-quartile range: 0 to 16.7%), with a range of 0 to 41.2%. We observed no clustered TB patients for 39% of the cells. The median TB incidence was 91.4 cases per 100,000 people per year (IQR: 68.5 to 142.7), and TB incidence ranged from 33.9 to 506.9 cases per 100,000 people. We found only slight agreement between the TB incidence and the proportion of clustered TB patients (weighted Cohen’s kappa = 0.27; 95% confidence intervals [CIs]: 0.13 to 0.42) (Fig. 5).
Fig. 5.
Comparison of TB incidence and proportion of clustered TB patients for 1 × 1 km grids of Lima. (A) Map of the TB incidence by grids, in which the color of the grids represents the magnitude of the metric, with the lightest colors corresponding to the lowest TB incidence. (B) Map of the proportion of clustered TB patients by grids, with the lightest colors corresponding to the lowest proportions. (C) Each dot in the plot represents a grid. The left-half dot is colored based on the magnitude of the TB incidence and the right-half dot is colored based on the magnitude of the correspondent proportion of clustered TB patients.
Drug Resistance and Transmission Dynamics.
When we repeated our analyses in the subgroup of patients with drug-resistant TB, our results were almost identical to those that included all TB patients (SI Appendix, Figs. S1 to S3).
Temporal Variation of Spatial TB Burden.
We found substantial agreement (weighted Cohen’s k = 0.61; 95% CIs: 0.45 to 0.75) in the ranked TB burden in 11 districts in Lima Ciudad between 2015 and 2019, indicating that there had been little temporal variation in the spatial distribution of the TB burden in our study area.
Discussion
In this study, we combined spatial and genomic analysis of consecutively diagnosed TB patients to describe a spatially heterogeneous transmission pattern in a large catchment area in Lima, Peru. We observed that MTB WGSs from participants who had repeated isolates over time had few differences and that most sequence pairs from those living in the same household were nearly identical. While we observed that TB patients living in the same immediate vicinity were most likely to share a molecular link, most transmission events occurred between people residing more than a 5-min drive apart. While there was little variation in the spatial distribution of TB burden in Lima over time, we noted significant spatial variation in both TB incidence and local transmission as measured by weighted Cohen’s kappa in our study area.
While many previous studies have shown that geographically proximate TB patients are more likely to fall within a genotypic cluster, only one previous study has described the genetic relatedness of clinical MTB isolates as a function of geographic distance. In Shanghai, China, Yang et al. (19) found that for every additional kilometer separating the residences of any two patients, the odds of genomic clustering (defined as sharing 10 or fewer SNPs) decreased by 10%. In our study, we observed that the genetic link quickly dropped with spatial distance; even among pairs residing only 1-min driving distance apart, the proportion of similar strains declined sharply. Although they did not genotype MTB isolates, McAllister et al. (20) reported that the incidence of TB among HHCs and neighboring households in Bandung, Indonesia, was comparable and about twice that of randomly selected neighborhoods. Moonan et al. (21) evaluated a neighbor-based strategy that expanded TB contact investigation to nearest and next-nearest neighbors of an index TB patient in Botswana; this approach identified 146% more TB patients than an approach that implemented TB contact investigation to the index case's home residence would have. These findings suggest that both HHCs and residents of households in close proximity to an index TB patient may be high-risk groups that would benefit from contact investigation.
Our finding that a substantial proportion of transmission occurred between people who were not in close geographical proximity is also consistent with the results of two previous studies from China. A study from Shanghai found that the median pairwise geographic distances within each genomic cluster ranged from 6.5 to 10.1 km (19). In Shenzhen, Jiang et al. (22) found that 70% of the genomic clusters included patients residing in different districts. WHO guidelines recommend active screening for high-risk groups of people, such as HIV-infected individuals, as well as those exposed to TB at home (23). However, our results and those findings cited above suggest that this strategy will miss a substantial proportion of transmission that takes place outside the household. On top of screening high-risk groups, an alternative cost-effective strategy could be implementing active screening within geographically restricted areas (4).
Although several previous studies marked wide spatial heterogeneity of TB incidence or prevalence and described the local geographical transmission hotspots (24–26), none have formally reported the spatial heterogeneity of local transmission conditional on the same underlying TB incidence. In our study, we found low agreement of a spatially heterogeneous pattern between TB incidence and local transmission. This finding indicates that high TB incidence rates do not always correspond to high rates of local TB transmission. There are several possible explanations for the finding that an area of high TB incidence has a low proportion of clustered TB. First, people living in a low-transmission area may be frequent visitors to other areas with a high risk of transmission (4). Second, geospatial TB clustering may result from disease progression after infection in people with shared geospatially clustered risk factors (such as HIV, malnutrition, or exposure to biomass fuel use). Because only a small proportion of people who are infected with TB progress to disease, the clustering of risk factors for disease progression is likely to play a stronger role in the spatial patterns of TB incidence than it would for other infections (4). Our results demonstrate that the degree to which local transmission is responsible for high TB incidence hotspots may vary greatly.
We found that a substantial proportion of transmission events for this airborne pathogen occurred between people residing at a distance of more than 1 km. These results are in contrast to those from several previous studies that have used geolocated genotype and serotype data to study the spatiotemporal transmission of the vectorborne infection dengue fever. For example, Salje et al. (27) used dengue serotype data from Bangkok, Thailand, to evaluate the spatiotemporal distribution of disease risk at a 0.5-km geological scale over 5-y periods and found that localized transmission occurred at a scale of less than 1 km. Salje et al. (28) later combined dengue-geolocated WGSs and serotype data to show that more than 60% of cases living less than 200 m apart belonged to the same transmission chain. In a similar study, Villabona-Arenas et al. (29) used serotype data to reconstruct dengue transmission chains in Porto Alegre, Brazil, where they found that the majority of infections were transmitted by short-distanced human movement and that cluster size, spatial diameter, and duration were smaller in areas with more intense control interventions. Because TB transmission does not require a vector but is transmitted through infectious aerosols, its transmission patterns are likely to correspond to host mobility patterns, rather than the local distribution of nonhuman vectors (30). These studies demonstrate the potential for integrated spatiotemporal and pathogen genetic data to shed light on the transmission dynamics of infectious diseases (30).
We note some limitations of our study. First, because children often are unable to produce sputum, children diagnosed with TB disease were excluded from our cohort of index patients. Furthermore, we have only been able to obtain WGSs for a subset of the isolates that were cultured. It is thus very likely that we have missed transmission events. Second, our geographical data included only the locations where patients resided and did not consider their patterns of mobility within the city. Third, we may have missed some TB patients residing in our study area because they may have sought care at private clinics or health centers outside our study districts. Fourth, although we observed a low temporal variation of TB burden in Lima, we cannot rule out the possibility that the data we presented may not reflect the current situation. These four factors may have led us to underestimate transmission but should not change the spatially heterogeneous transmission pattern.
We observed that a portion of transmissions occurred between households with close proximity, as patients living in close proximity are more likely to have a molecular link. This finding suggests that expanding contact investigations to the neighborhood of an index TB patient could potentially help detect TB early or reduce TB transmission. Furthermore, we showed that high rates of TB incidence often do not correspond to high rates of local TB transmission. This finding indicates that an understanding of spatially heterogeneous transmission patterns is essential to evaluate the impact of active screening interventions within geographically restricted areas.
Supplementary Material
Acknowledgments
The work reported in this publication was supported by the NIH and National Institute of Allergy and Infectious Diseases grants U01AI057786, U19AI076217, U19AI109755 (Center for Excellence in Translational Research), and U19AI111224 (Tuberculosis Research Unit). The content is solely the responsibility of the authors and does not necessarily represent the official views of the founders.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207022119/-/DCSupplemental.
Data, Materials, and Software Availability
Anonymized MTB genomic distances and participant metadata are available in the SI Appendix. Some study data are available. All other study data are included in the article and/or SI Appendix.
References
- 1.World Health Organization, Global Tuberculosis Report (2021). www.who.int/publications/digital/global-tuberculosis-report-2021. Accessed 22 July, 2022.
- 2.Matteelli A., et al. , Tuberculosis elimination: Where are we now? Eur. Respir. Rev. 27, 180035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dye C., Glaziou P., Floyd K., Raviglione M., Prospects for tuberculosis elimination. Annu. Rev. Public Health 34, 271–286 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Cudahy P. G. T., et al. , Spatially targeted screening to reduce tuberculosis transmission in high-incidence settings. Lancet Infect. Dis. 19, e89–e95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sterling T. R., et al. ; TB Trials Consortium PREVENT TB Study Team, Three months of rifapentine and isoniazid for latent tuberculosis infection. N. Engl. J. Med. 365, 2155–2166 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Khundi M., et al. , Effectiveness of spatially targeted interventions for control of HIV, tuberculosis, leprosy and malaria: A systematic review. BMJ Open 11, e044715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaweno D., Trauer J. M., Denholm J. T., McBryde E. S., The role of geospatial hotspots in the spatial spread of tuberculosis in rural Ethiopia: A mathematical model. R. Soc. Open Sci. 5, 180887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaweno D., et al. , Methods used in the spatial analysis of tuberculosis epidemiology: A systematic review. BMC Med. 16, 193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alene K. A., Viney K., McBryde E. S., Clements A. C. A., Spatial patterns of multidrug resistant tuberculosis and relationships to socio-economic, demographic and household factors in northwest Ethiopia. PLoS One 12, e0171800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becerra M. C., et al. , Transmissibility and potential for disease progression of drug resistant Mycobacterium tuberculosis: Prospective cohort study. BMJ 367, l5894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker B. J., et al. , Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comas I., et al. , Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coll F., et al. , A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5, 4812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luxen D., Vetter C., “Real-time routing with OpenStreetMap data” in GIS: Proceedings of the ACM International Symposium on Advances in Geographic Information Systems (2011), pp. 513–516.
- 17.WorldPop, Open spatial demographic data and research. www.worldpop.org. Accessed 9 March 2022.
- 18.Tang W., Hu J., Zhang H., Wu P., He H., Kappa coefficient: a popular measure of rater agreement. Shanghai Jingshen Yixue 27, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C., et al. , Internal migration and transmission dynamics of tuberculosis in Shanghai, China: An epidemiological, spatial, genomic analysis. Lancet Infect. Dis. 18, 788–795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAllister S., et al. , Feasibility of two active case finding approaches for detection of tuberculosis in Bandung City, Indonesia. Public Health Action 7, 206–211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moonan P. K., et al. , A neighbor-based approach to identify tuberculosis exposure, the Kopanyo study. Emerg. Infect. Dis. 26, 1010–1013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Q., et al. , Citywide transmission of multidrug-resistant tuberculosis under China’s rapid urbanization: A retrospective population-based genomic spatial epidemiological study. Clin. Infect. Dis. 71, 142–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization, WHO consolidated guidelines on tuberculosis: Module 2: screening – systematic screening for tuberculosis disease. https://www.who.int/publications/i/item/9789240022676. Accessed 22 July 2022. [PubMed]
- 24.Yazdani-Charati J., Siamian H., Kazemnejad A., Mohammad V., Spatial clustering of tuberculosis incidence in the North of Iran. Glob. J. Health Sci. 6, 288–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S., et al. , Spatial dynamics of TB within a highly urbanised Asian metropolis using point patterns. Sci. Rep. 7, 36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verver S., et al. , Proportion of tuberculosis transmission that takes place in households in a high-incidence area. Lancet 363, 212–214 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Salje H., et al. , Revealing the microscale spatial signature of dengue transmission and immunity in an urban population. Proc. Natl. Acad. Sci. U.S.A. 109, 9535–9538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salje H., Cummings D. A. T., Lessler J., Estimating infectious disease transmission distances using the overall distribution of cases. Epidemics 17, 10–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villabona-Arenas C. J., et al. , Epidemiological dynamics of an urban Dengue 4 outbreak in São Paulo, Brazil. PeerJ 4, e1892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown T. S., Robinson D. A., Buckee C. O., Mathema B., Connecting the dots: Understanding how human mobility shapes TB epidemics. Trends Microbiol. 10.1016/j.tim.2022.04.005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized MTB genomic distances and participant metadata are available in the SI Appendix. Some study data are available. All other study data are included in the article and/or SI Appendix.