Significance
The cell envelope of the extremophile bacterium Deinococcus radiodurans was studied by cryo-electron microscopy and described with unprecedented detail. In this bacterium, the outermost cell envelope layer, named surface layer, is characterized by a highly regular tiling of proteins extending their crystalline organization to the cell envelope layers below (until the inner membrane). The study shows three main protein complexes, with masses in the MDa range, regularly organized into an astonishing geometrical regularity. The observed organization contributes to protecting the cell against environmental stressors and maintaining an efficient permeation of environmental solutes.
Keywords: cryo-electron crystallography, cryo-electron tomography, S-layer, Type IV piliation system, SDBC
Abstract
Surface layers (S-layers) are highly ordered coats of proteins localized on the cell surface of many bacterial species. In these structures, one or more proteins form elementary units that self-assemble into a crystalline monolayer tiling the entire cell surface. Here, the cell envelope of the radiation-resistant bacterium Deinococcus radiodurans was studied by cryo-electron microscopy, finding the crystalline regularity of the S-layer extended into the layers below (outer membrane, periplasm, and inner membrane). The cell envelope appears to be highly packed and resulting from a three-dimensional crystalline distribution of protein complexes organized in close continuity yet allowing a certain degree of free space. The presented results suggest how S-layers, at least in some species, are mesoscale assemblies behaving as structural and functional scaffolds essential for the entire cell envelope.
Surface layers (S-layers) are ordered repetitions of proteinaceous units that tile the cell body of many prokaryotes (1–5). The periodical unit of S-layers consists of one or more proteins that, in some species, are further organized in multiprotein complexes (6–8). These coating structures are common in Bacteria and Archaea and have essential roles in providing cell rigidity, shape, adhesion, and resistance to extreme conditions (9–12). Despite their relevance in the cell envelope, detailed molecular and in situ studies on S-layers are still limited (13–15), leading to a distinct degree of uncertainty about their roles and functions. However, both the self-assembly properties of S-layer proteins (16) and the levels of energy consumption implied by their high expression and displacement suggest a refined specialization of these structures. Being at the forefront of the cell to the environment, S-layers represent one of the cell's compartments more exposed to selective pressure and, in extremophiles, cope with harsh conditions that are typically deviating from the canonical ones sustaining life (17). This fact ranks S-layers as one of the cell regions most subjected to specialization in bacteria.
One of the model organisms for S-layer studies is the radiation-resistant bacterium Deinococcus radiodurans. This bacterium is a ubiquitous coccus that occurs in a group of four cells to form tetrads (18) and is pink pigmented due to the presence of the strong-antioxidant xanthophyll deinoxanthin, a unique carotenoid of this organism (17, 19). Being known for its resistance to electromagnetic stress, such as high doses of ionizing radiation, this bacterium has been the subject of molecular studies to understand the DNA-repairing mechanisms as well as lipids and proteome damage protection (20, 21). Its S-layer has been investigated by several techniques, and its surface structure and porous organization were previously described at a low resolution (22–25). In the last decade, the research done on this S-layer provided important details on its organization, composition, and properties, eventually resulting in an overall vision with features somehow overlooked since then. In particular, this S-layer was found to be composed of several proteins further organized into different protein complexes (4, 26). Taken together, these studies provided multiple and indirect evidence suggesting the absence of a distinct separation between the S-layer and the underlying membranes (4, 8, 27–29). To shed a light on the S-layer extension and interaction with the rest of the cell envelope, here we explore the in situ three-dimensional (3D) organization of this S-layer. Cell-envelope patches were characterized by 3D cryo-electron crystallography (cryo-EC), which is a method used for the structural determination of micrometer- and nano-scale bidimensional crystals (30), compared and complemented by cryo-electron tomography (cryo-ET) and subsequent subtomogram averaging. Results describe an unexpectedly sophisticated crystalline regularity, with a 3D organization consisting of an ordered juxtaposition of three multiprotein complexes recently observed in their 2D organization (8) and here shown to extend into the entire cell-envelope thickness. The periodical organization of these complexes results in a highly packed cell-envelope structure that, despite the close continuity of its components, is still allowing a certain degree of free space in the whole cell-envelope thickness. While one of these complexes, indicated here as radial–dimeric complex, appears to be exclusive of the cell-envelope surface, involving the S-layer and the outer membrane (OM), the other two complexes, a type IV-like piliation system (T4P-like) and the S-layer deinoxanthin binding complex (SDBC), which were previously described as channels (8, 31), here are shown to span the layers below, representing the main sieving system in the cell. Such an organization forces the cell-envelope trafficking through the two main available ways, the T4P-like and the SDBC, opening for centralized and discrete trafficking across the cell envelope. The presented findings indicate S-layers as fundamental structures that “drive” the cell-envelope organization from the level of the cell body down to the molecular level according to a model where active and central functions of trafficking and protection are implied.
Results
A Characteristic Fourier Transform Pattern Indicates an Extended Crystalline Organization in the Cell Envelope of Deinococcus radiodurans.
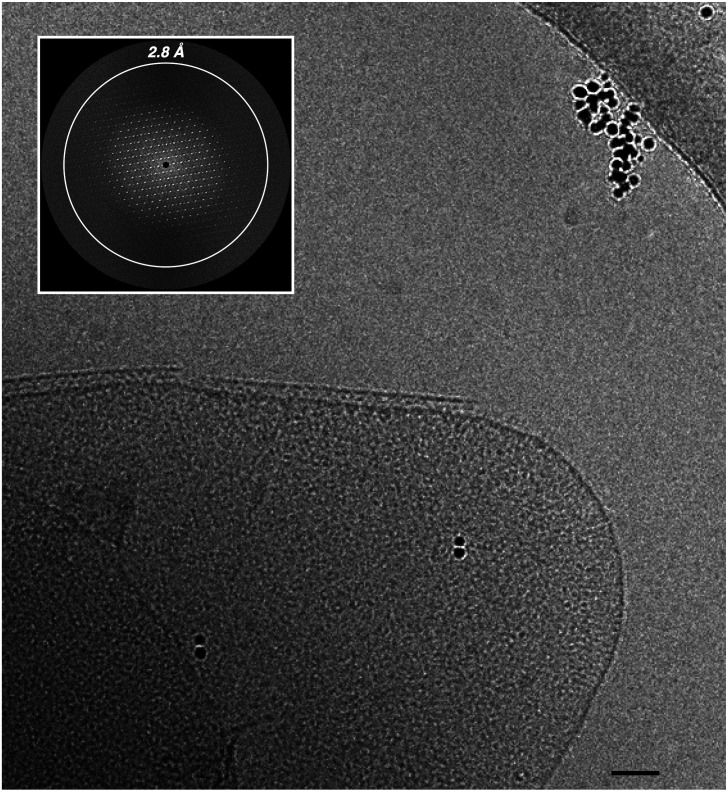
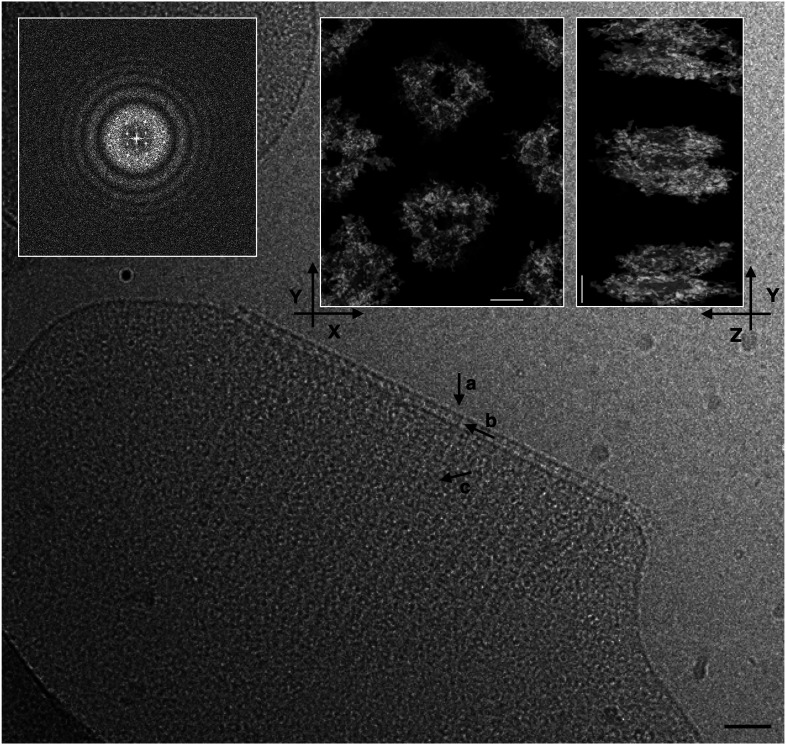
The general properties of radiation resistance and the abilities of its S-layer to shield the ultraviolet radiation (17, 32) make D. radiodurans an interesting candidate for studies on its cell envelope. Taking advantage of the S-layer crystallinity, the Fourier transforms of the cell envelope images were used as a discriminant for selecting the best patches to be subjected to cryo-electron microscopy (cryo-EM) studies by cryo-EC, cryo-ET, and subtomogram averaging. Accordingly, only integral cell-envelope patches (showing both membranes) with sharp spots extending to 2.8 Å in their Fourier transform (FT) were selected (Fig. 1). FTs analyses by independent processing and subsequent merging of 73 micrographs at different tilting angles of six different patches led to a 3D cryo-EM density map that allowed an indirect visualization in situ of the crystalline regions in the cell envelope (Fig. 2; EMD-14097). The phase residuals postprocessing indicated information up to 4 Å in plane and 10 Å along the z axis (Table 1), and interestingly, the reflections carried information not only of the S-layer, well known for its ∼70-Å-thick crystalline packing (23), but also of the underlying space, providing striking evidence of a crystallinity extended for ∼200 Å into the cell envelope (Fig. 2 and SI Appendix, Movie S1).
Fig. 1.
Continuous regularity of isolated cell-envelope patches. Cryo-EM micrographs of cell-envelope patches let glimpse a patterned regularity resulting from a crystalline organization. The crystalline nature of the sample can be demonstrated by Fourier transform analysis, which showed significant sampled Fourier coefficients beyond 2.8 Å resolution (Inset). The scale bar indicates 500 Å.
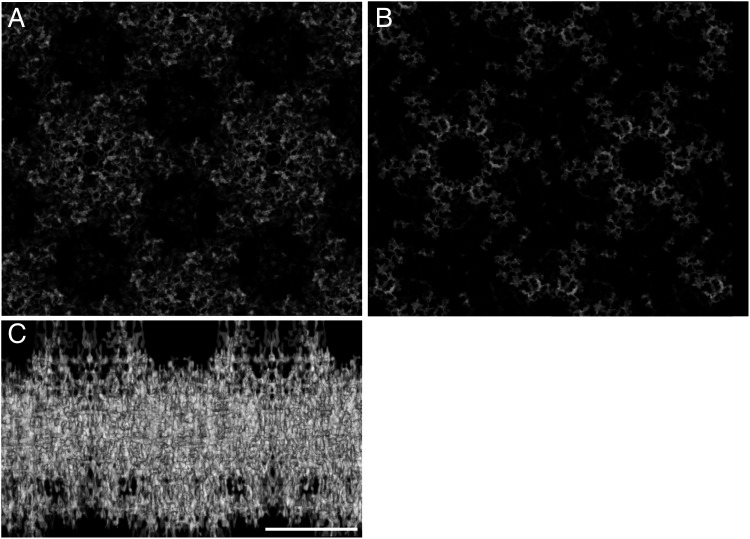
Fig. 2.
Projection map of the crystalline cell-envelope fraction. Map at 4 Å with imposed p6 symmetry (EMD-14097) visualized on the top view (A), bottom view (B), and side view (C). The map clearly shows the main structural constituents organized in individual units. In the side views, the thickness reaches ∼200 Å. The scale bar indicates 100 Å.
Table 1.
Electron crystallographic parameters and data
| Plane group symmetry | p6 |
| Unit cell dimensions | a (Å) = 190; b (Å) = 190; c (Å) = 200 γ (°) = 120 |
| No. of images | 73 with tilt angle distribution: [5 (0°); 6(+3°); 3(-3°); 4 (+6°); 5(-6°); 4(+9°);4(-9°); 4(+12°); 4(-12°); 2(+15°); 1(-15°); 2(+18°); 2(-18°); 2(+21°); 2(-21°); 2(+24°); 3(-24°); 2(+27°); 2(-27°); 2(+30°); 2(-30°); 1(+33°); 1(-36°); 2(+39°); 1(-39°); 1(+42°); 1(-42°); 2(+45°); 1(-45°)] |
| Defocus range (Å) | 39,500–123,738 |
| Magnification | 21,930x |
| IQ range used | 1–6 |
| Resolution cutoff in-plane (Å) | 4 |
| Resolution cutoff in-z (Å) | 10 |
| No. of observed reflections | 189,361 |
| No. of observed unique reflections | 49,905 |
| No. of possible unique reflections | 2,728 |
| No. of observed unique reflections with FOM>50% | 2,258 |
| Completeness counting reflections with FOM>50% | 82.80% |
| Redundancy (multiplicity) | 3.79 |
| Overall phase error for 1,000 spots (°) | 42.7 |
| Resolution (Å) | Unique reflections (N°)* | Phase residual (°)† |
|---|---|---|
| 12.7 | 102 | 24.9 |
| 8.9 | 104 | 41.2 |
| 7.3 | 101 | 41.4 |
| 6.3 | 102 | 41.3 |
| 5.7 | 104 | 47.9 |
| 5.2 | 99 | 43.9 |
| 4.8 | 104 | 47.1 |
| 4.5 | 103 | 45.8 |
| 4.2 | 96 | 52.3 |
| 4.0 | 85 | 41.1 |
FOM, figure of merit; IQ, intelligence quotient.
*With 1 ≤ IQ ≤ 6–4.0 Å.
†Amplitude-weighted, vectorial averaged phase residuals that show the phase deviation from theoretical 0°/180° (45° is random).
FT Analysis Reveals Three Different Protein Complexes Organized into a 3D Crystalline Packing.
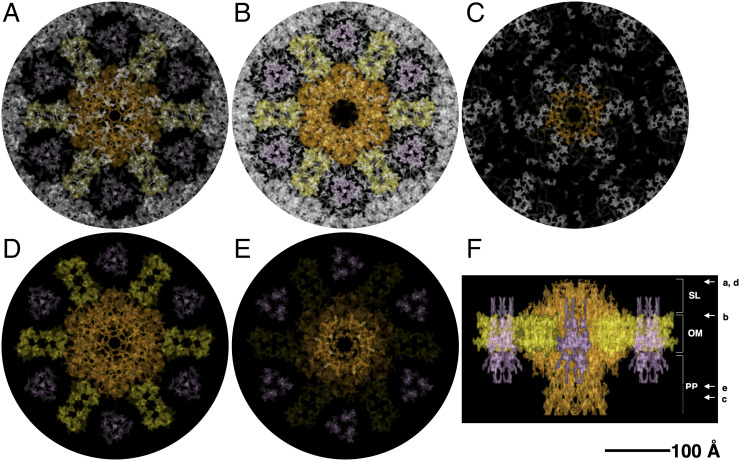
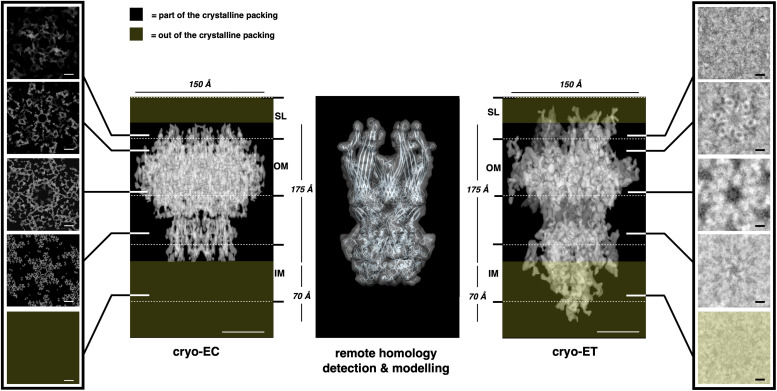
A consistent volume of the cell envelope appears to be occupied by three complexes. The larger complex shows an overall shape that resembles one of many secretion systems and fits well with the ∼1.1-MDa complex, called T4P-like (in orange; Fig. 3), previously identified. This complex was found to be abundant in this cell envelope and to have as a main component the protein DR_0774, a secretin pilQ commonly constituting the head region and interacting with the OM in many secretion systems (4). The T4P-like shows a height of 175 Å, a main hexagonal head with a side of 85 Å, and a diameter of 150 Å (Fig. 3 B and F), spanning the S-layer, the OM, and the periplasm (Fig. 3F) and possibly reaching the inner membrane (IM). On the top of the head, a multiple-spikes crown emerges and expands for 20 Å, defining the pore region (Fig. 3F). According to the p6 symmetry of this S-layer, each T4P-like is surrounded by six copies of a smaller complex, the SDBC, a multisubunit complex of which the main component, the protein DR_2577 (or SlpA), was extensively described and shown to be characteristic of these cell envelopes (8, 17, 29, 33). Recognizable by its triangular shape (with a side length of 90 Å and a thickness of ∼30 Å), this ∼0.9-MDa complex has the main body embedded in the OM and is located inside of a “cell-envelope case” (Fig. 3, in pink), as already observed in intermediate resolution studies on the surface of this cell envelope (8) and high-resolution studies (34; Protein Data Bank: 7ZGY). Differently from what was observed in purified SDBC samples (34), here from the main body depart three protrusions facing the external environment and the other three toward the periplasmic side (Fig. 3F). These protruding domains might be the loosely bound subunits of the SDBC that are not visible in the structure obtained by single-particle analysis (34) but that are found by mass spectrometry in the same samples (4, 29). Moreover, the possibility that specific domains, for example, the glomerular regions, might assume an alternative conformation in situ cannot be excluded. Finally, a third radial–dimeric complex of unknown identity, with a side of 70 Å, a width of 45 Å, and a height of 60 Å (Fig. 3, in yellow), was found to be localized in the S-layer and to span the underlying OM (Fig. 3F). By exclusion, we hypothesize that this third complex could be the DR_2508 protein, which is known as hexagonally packed intermediate (22, 23, 27) and is the only known protein of this S-layer for which a precise localization is still missing. These three individual units, the T4P-like, the SDBC, and the radial–dimeric complex, here represented in orange, pink, and yellow, respectively, were extracted from the map, providing a better view of their features and relative localization and their contribution to the different layers of the cell envelope (Fig. 3 D–F and SI Appendix, Movie S1).
Fig. 3.
Projection map with assigned densities. The specific occupancies for each complex are shown as top and side views with the T4P-like in orange, the radial–dimeric complex in yellow, and the SDBC in pink. For a better visualization, slices at the top-view level (A), at the outer-membrane level (B), and at the bottom level (C) of the map are shown. Colored complexes represent the extracted densities from the original map. Top (D), bottom (E), and side (F) views of the extracted complexes and their relative position are also shown. In F, on the right are indicated the S-layer (SL), the outer membrane (OM), and the periplasm (PP) thicknesses assigned using the T4P-like and the recent structure of the SDBC as a reference (34); the white arrows and the related letters (A–E) indicate the “view depth” of the images shown in the panels from A to E. The scale bar indicates 100 Å.
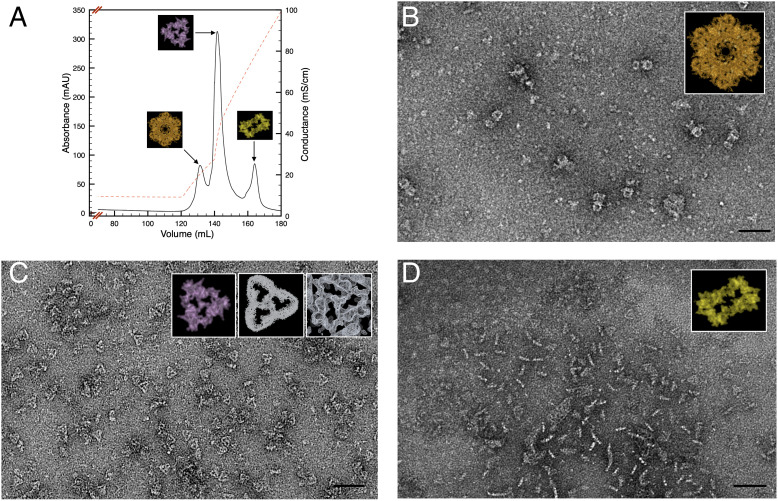
To corroborate these observations, the mild solubilization of the cell envelopes sample and its subsequent fractionation by anionic exchange chromatography confirmed the dominance of these three protein complexes (Fig. 4A), as shown by negative-stain EM (Fig. 4 B–D), here used to obtain a good contrast for low-concentrated samples. In particular, with respect to the pool of the second peak (Fig. 4A), the structure and identity of the particle were confirmed in a recently published report (34). The SDBC structure was compared here for dimensions and shapes (including the typical pore region) with both the full cryo-EM density map and the extracted particle (Fig. 4C, Insets). Similarly, the characterization of the T4P-like and radial–dimeric complexes will be reported elsewhere.
Fig. 4.
Chromatographic separation of the three cell-envelope complexes. The cell-envelope patches were mild solubilized and subsequently subjected to anionic exchange chromatography (A) resolving in three main protein complexes: the T4P-like complex (B), the SDBC (C), and the radial–dimeric complex (D). Insets show the particles as extracted from the whole cryo-EM density map; the Insets in C indicate the extracted particles (Left), the cryo-EM density map at atomic resolution as shown by Farci et al. (34) (Middle), and the particle visualized directly in the map (Right). The scale bar indicates 500 Å.
Cryo-ET and Subtomogram Averaging on Intact Cell Envelopes Display an Ordered Repetition of Channels.
Cryo-ET and related subtomogram averaging on the same data sets allowed an in situ direct analysis of intact cell-envelope fragments (Fig. 5; EMD-14095). At the tomographic level, raw movie slices (Fig. 5) and reconstructions (SI Appendix, Movie S2) showed a repetition of proteinaceous units as top and side views. Overall, these analyses confirmed the organization described by cryo-EC, showing a regular isoporous surface (interpore distance of 195 Å vs. 190 Å measured by 3D cryo-EC), with each pore corresponding to a T4P-like complex spanning the whole layers of the cell envelope (Fig. 5, Upper-Middle and Upper-Right Insets, respectively), hence permitting the in situ direct visualizations of the 3D regularity. A subsequent p6 symmetrization centered in the pore region allowed resolving better the T4P-like complex (8-Å best resolution; Fig. 6; SI Appendix, Fig. S1 for subtomogram averaging FSC; EMD-14096) and comparing it with the T4P-like structure extracted from the 3D cryo-EC map (Fig. 6). The subtomogram shows a dominance of the T4P-like occupancy, with each complex having a side of 85 Å, a diameter of ∼150 Å, and a total height of ∼260 Å (Fig. 6). Included in this structure are also noncrystalline regions (e.g., dynamic peripheral regions) that are missing in the model obtained by cryo-EC (Fig. 6, dark-yellow boxes). Noteworthy there is a 45-Å crown, which is more extended with respect to the one visible in the cryo-EC (∼20 Å), and a 70-Å-high conical protrusion in the bottom part of the complex involving the IM and the neighboring cytoplasmic region (Fig. 6). Furthermore, slicing along the complex's height allowed comparison of the patterning of symmetry at different levels, thus confirming the consistency of the two independent analyses (Fig. 6, left and right small boxes). As it can be envisaged by the differences in the T4P maps obtained by the two techniques, in subtomogram, averaging the missing wedge affected the final resolution. Similarly, being the cell-envelope patches mostly flat, as well as the SDBC and the radial–dimeric complexes, these could not be resolved by subtomogram averaging, explaining why they do not appear in the present analysis.
Fig. 5.
Tomography and related subtomogram averaging analysis. Slice of a raw tomographic movie and FT analysis (Upper-Left Inset) on a cell-envelope patch. A scale bar of 500 Å is shown. In the Inset, it is shown a typical subtomogram averaging of cell-envelope patch (EMD-14095) from the top view (Upper-Centered Inset) and side view (Upper-Right Inset). No symmetry was imposed. The three arrows on the main image indicate (a) the S-layer, (b) the regularity of the T4P-like complex in the membranes, and (c) the stripes created by the regularity of the complexes organized in the cell envelope. The scale bar indicates 50 Å.
Fig. 6.
Features comparison between T4P-like structure obtained by cryo-EC, remote homology detection, and subtomogram averaging. The main dimensions of the T4P-like complex obtained by 3D cryo-EC (Center Left; EMD-14097), remote homology detection (Center), and subtomogram averaging (Center Right; EMD-14096) are compared. The Left and Right boxes show comparable images of the slice through at equivalent levels, from Top to Bottom, for the T4P-like complex obtained by cryo-EC (left box) and by cryo-ET subtomogram averaging (right box), both computed imposing a p6 symmetry. The dark-yellow boxes indicate the noncrystalline regions that are missing in the model obtained by cryo-EC with respect to the model obtained by subtomogram averaging. The S-layer (SL), the outer membrane (OM), and the inner membrane (IM) thicknesses are indicated. Scale bars indicate 50 Å.
Finally, this analysis also allowed measurement of the thickness of the entire cell envelope and its regions. The whole thickness was found to be 299.5 ± 2.8 Å with a periplasm ∼99.3 ± 12.2 Å thick delimited by a compact “roof” represented by the S-layer and the OM known to be tightly interacting with each other (8, 34) and in here appearing as a single layer ∼98.9 ± 9.4 Å thick. Below, the cell envelope appears delimited by a “floor” consisting of an IM ∼69.8 ± 5.4 Å thick (SI Appendix, Fig. S2), representing the characteristic forefront of the cytosol. As shown in the previous paragraph, the samples from the first chromatographic peak are particles with shapes and dimensions resembling one of the secretion systems; hence, they could be ascribed to the T4P-like (Fig. 4B). To corroborate the consistency of this observation, we applied remote homology detection to the main gene of the T4P-like, the dr_0774, as previously reported (4). Between the possible models, all connected to different types of secretion systems, the protein invG from Salmonella typhimurium was selected to build a hexameric model of DR_0774 and then further convoluted to obtain a map at 10 Å, a resolution comparable with those of the maps obtained by the two cryo-EM techniques. This comparison showed a good degree of coherence between the structures obtained by cryo-EC and subtomogram averaging and the predicted structure obtained by remote homology detection, further confirming the obtained results (Fig. 6).
Discussion
We provide here the supramolecular description of the cell envelope in the radiation-resistant bacterium D. radiodurans. The combination of 3D cryo-EC and cryo-ET allowed us to exploit the full potential of the sample in terms of structural analysis. While cryo-ET and subtomogram averaging are deeply used methods for in situ studies in different subjects, the cryo-EC is a method applicable only in the case of micrometer- and nano-scale crystals, which here are naturally occurring in the sample. Therefore, the crystalline properties of this cell envelope were advantageous for performing a detailed characterization of the cell envelope, resulting in a well-preserved overall structure, and allowing an intermediate-resolution characterization (Figs. 1, 2, and 5).
We demonstrate that the well-known S-layers' order in this specific case is ascribable to three multiprotein complexes (Fig. 3). The most astonishing and remarkable result is that the S-layer regularity pervades the underlying layers (protein components of the OM, periplasm, and IM), imposing a defined order to the cell envelope (Fig. 3). The intricate structure resulting from a regular juxtaposition of protein complexes is expected to serve as an extended molecular sieving machine. This observation is unequivocally supported by its constituents, the T4P-like and the SDBC, already known for their gating properties (8, 31, 34). Accordingly, through this organization, the trafficking across the cell envelope has the potential to be finely regulated by the discrete distribution of its components. This organization, consisting of two types of gates isotropically distributed on the whole surface of the cell envelope, allows equalization of the solute exchange with the environment. The observation of this regularity appeared first by the FT of the cell-envelope patches and after was confirmed by the 3D structural analyses (Figs. 1 and 2). The assignment of the cell-envelope layers became straightforward when interpreting the results obtained here by cryo-EC and cryo-ET on the base of the structure at the atomic resolution of the SDBC (34). Having as reference this complex, of which the main part is a massive β-barrel structure embedded in the OM, it is possible to assign the S-layer, the OM, and consequently the periplasm in the 3D structure and the resulting model here shown (Figs. 2, 3, and 6).
Taken together, the present findings suggest that the S-layer and the underlying membranes occur according to an ordered organization at several levels. In fact, as a consequence of this cell-envelope topology, a continuous proteinaceous system can be envisaged from the range of individual cells down to single proteins (35). In such an organization, the crystalline unit cell represents a discrete structural unit, the repetition of which results in the association of six SDBC and six radial–dimeric complexes for each T4P-like complex (Fig. 3). According to this spatial displacement and the self-assembling properties of S-layer proteins (16), these three building blocks are expected to self-organize, forming a large functional structure with several hierarchic levels. If on one hand, this structural continuity provides extended compactness and statical isotropy at different levels, on the other hand, by this structure, the bacterium has an isotropic interaction with the surrounding environment by reaching an equally distributed exchange potential. Considering the well-known role of secretion systems and porins, such as the T4P-like (31, 36) and the SDCB (8, 37, 38), in exchanging across the cell envelope, this property is relevant not only for the dynamics associated with the trafficking but also with the specific pool of functions that might specialize a given S-layer. Moreover, it cannot be excluded that the T4P-like could be important for the biogenesis of the S-layer, allowing the deposition of the main S-layer proteins.
Notably, while the cell-envelope packing appears to be an intrinsic property due to the S-layer organization, the cryo-EC map indicates an extended free space in the periplasm and IM layers, where limited hindrances are needed for biological functions associated with proteins not involved in the crystalline packing (Fig. 3 C and F). This is particularly true for the periplasm, where trafficking processes and proteins not involved in the paracrystalline organization need to have enough space for the functional activities related to cell maintenance and homeostasis (e.g., osmotic and energetic balance).
At this stage, it is not given to know how much this organization is common among different S-layers. Considering that similar symmetries and surface organizations are also associated with other S-layers, this extended cell-envelope regularity could be a frequent feature for S-layers of isoporous type. These findings provide a breath for studying S-layers and understanding their active roles in the physiology of bacterial cells.
Materials and Methods
Experimental Model and Subject Details.
Cell lines.
Deinococcus radiodurans cultures (strain R1; American Type Culture Collection 13939; SI Appendix, Table S1) were cultivated in Tryptone Glucose Yeast extract broth (TGY) at 30 °C for 24 h, as described by Murray (39).
Method Details.
Cell envelope isolation and chromatography.
Cell envelopes were isolated at 4 °C in dim light as described in Farci et al. (8). Briefly, after harvesting by centrifugation (5,000g, 10 min, 4 °C), cells were resuspended in a 50 mM sodium phosphate buffer at pH 7.8 (buffer A) supplemented with deoxyribonuclease I (DNase I) (100 U, DNase I recombinant, RNase-free Roche) and disrupted using a French Pressure Cell (three cycles at 1,100 psi). The sample was centrifuged twice (2,000g, 10 min, 4 °C) to remove the debris, then the supernatant was centrifuged to collect cell-envelope fragments (48,000g, 10 min, 4 °C). The final pellet was resuspended in buffer A and digested with 100 μg/mL lysozyme (8 h, 30 °C, under shaking at 800 rpm) to remove surface polysaccharides. Digested cell-envelope fragments were centrifuged (48,000g, 10 min, 4 °C) and washed three times with buffer A by a serial sequence of centrifugation/resuspension steps. Final samples were used for the cryo-EC and cryo-ET.
Anionic exchange chromatography was performed according to Farci et al. (8). Briefly, cell-envelope fragments were resuspended (3–5 mg/mL total proteins) and solubilized for 30 min at room temperature with a final concentration of 1.1% of n-dodecyl-β-D-maltoside (β-DDM). After solubilization and centrifugation (48,000g, 10 min, 4 °C), the supernatant was subjected to anion-exchange chromatography (Hi-load HP column, Amersham). After five column volumes of washing with buffer B (50 mM sodium phosphate [pH 7.4]; 0.05% (wt/vol) β-DDM) at a flow rate of 0.5 mL/min, the elution was done with a linear gradient of 0–2.5M NaCl in buffer C (50 mM sodium phosphate [pH 7.4]; 2.5 M NaCl; 0.05% (wt/vol) β-DDM). All chromatography columns were subjected to the ReGenFix procedure (https://www.Regenfix.eu/) for regeneration and calibration prior to use.
EM.
Cryo-EM.
The grid preparation and data acquisition were done at Central European Institute of Technology, Brno, Czech Republic. Quantifoil R2/1.3 holey carbon grids (Quantifoil Micro Tools GmbH, Germany) were glow discharged prior to use. Cell-envelope samples were fast frozen into liquid ethane using a Vitrobot Mark IV (ThermoFisher, the Netherlands) plunge-freezing machine (4 °C, 100% humidity, blot force 2, blotting time 2 s) and placed in autogrid (FEI, Eindhoven, the Netherlands) prior to image acquisition. Grids were transferred to a Titan Krios transmission EM (TEM) (ThermoFisher, the Netherlands) operating at 300 kV and equipped with a Cs corrector (cs 2.7 mm), a Quantum GIF energy filter (slit width set to 20 eV), and a post-GIF K2 camera (Gatan, USA) at a magnification corresponding to a pixel size of 2.28 Å/px (∼21,000×). A dose-symmetric tilt scheme with a 3° increment (0, +3°, −3°, etc.; tilting range ± 60°) and a total dose of 2 e−/Å2 were used for tomography acquisition by SerialEM software (40). The defocus was set to vary between 2 µm and 5 µm.
Negative-staining EM.
The isolated protein complexes were negatively stained on glow-discharged copper grids (EMS200-Cu) covered with a 20-nm carbon film. The samples were applied at the grid in a volume of 3 µL, and the excess was removed after 1 min by capillarity through a delicate touching of the grid bar with a filter paper (Whatman No. 1) for 2 s. The grids were then stained for 1 min with 5 µL of 2% UranyLess TEM staining solution (Micro to Nano, the Netherlands), and the excess staining was removed by capillarity as in the previous step. Micrographs were acquired with a Tecnai F20 microscope (ThermoFisher, the Netherlands) operating at 200 kV, with an FEI Eagle 4K charge-coupled device camera, at a magnification of 53,000×.
Cryo-EC processing of cell-envelope fragments.
We collected 42 tomograms in movie mode (each tilting angle has a movie of eight frames), for which CTF (Contrast Transfer Function) and ice quality were inspected. A total of 40 tomograms were selected for further processing. CTFs and resolution were estimated by CTFFIND3 (41). For cryo-EC, eight-frame movies were drift corrected using MotionCor2 (42) on the Focus package (43). Micrographs, at different tilting angles (see Table 1 for tilt angle distribution) and from the six best-diffracting tomograms, were processed on the same package but this time in 2D crystallography mode according to the latest implementations of the 2dx package (44). By this procedure, a complete set of information was obtained for each micrograph (defocus, tilt geometry, lattice, and phase origin) and used to build a 2D projection map. Finally, data from the 73 best images were merged to obtain the final projection map by using the merge suite from Focus in 3D crystallography mode. The 3D model’s visualization and the fittings were done using the Chimera software (45) and Coot (46).
Cryo-ET and subtomogram averaging processing of cell-envelope fragments.
Tomograms were processed by the etomo software (40). Gold particles were used as fiducial markers, and only tomograms with minimal alignment errors were selected for the final reconstruction (40). For subtomogram averaging on 3D reconstructions, subtomogram boxes with a size of 300 × 300 × 300 voxels were manually selected from both in-plane and side views and finally extracted. Subtomogram alignment and averaging with missing-wedge compensation were performed with the Particle Estimation for Electron Tomography (PEET) software (47, 48) as follows. Subtomograms were initially rotationally aligned, assuming that translational shifts of subtomograms were approximately correct. Eulerian angles were determined by systematic search over some specified range of values, with the range and the coarseness of the search being reduced in successive iterations of the search (49). Absolute values of cross-correlation were used for alignment, potentially helping to prevent noise by reinforcing to match features in the ref. (47, 48). Finally, p6 symmetrization was performed using the scripts available for PEET. Isosurface visualization was accomplished using the Chimera software (45).
Remote Homology Detection and Modeling.
The sequence of the DR_0774 protein was taken from the Uniprot database, and its FASTA format was used on the Swiss-Model platform (https://swissmodel.expasy.org/; (50)) for remote homology detection, resulting in 50 templates and 12 models. Among them, we selected the DR_0774 model obtained from the protein invG from S. typhimurium (Uniprot entry A0A3Z1V7D5, PDB-6pee.1), which showed the highest sequence coverage with respect to DR_0774. Subsequently, this monomer was modeled into the maps obtained by cryo-EC and cryo-ET. This final model was convoluted into a map using the Chimera software and applying a resolution equal to 10 Å.
Quantification and Statistical Analysis.
Cell-envelope isolation for data analyses was performed on more than 20 independent preparations. Micrographs, tomograms, and maps are representative of at least three independent replicates. Electron crystallographic image statistics and main image parameters are indicated in Table 1. All attempts to reproduce the results here presented were successful.
Additional Resources.
No additional resources are available. All resources have been indicated in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This work was supported by the National Science Center (Poland) with the Sonata BIS 7 Program (2017) grant PRO-2017/26/E/NZ1/00344 and the Harmonia 10 Program (2018) grant PRO-2018/30/M/NZ1/00284 (both to D.P., D.F., and P.H.). We acknowledge Cryo-electron microscopy and tomography core facility Central European Institute of Technology Masaryk University of Czech Infrastructure for Integrative Structural Biology, Instruct-CZ Centre supported by MEYS CR (Ministry of Education, Youth and Sports of the Czech Republic) (LM2018127), and iNEXT-Discovery, project No. 871037, funded by the Horizon 2020 program of the European Commission. The authors are grateful to Dr. J. Nováček for the support at the cryo-EM core facility. D.F. kindly thanks Dr. J.M. Heumann (University of Colorado Boulder) for the support with the subtomogram averaging processing using PEET. While citing refs. scientifically relevant for this work, we actively worked to promote gender balance in our ref. list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209111119/-/DCSupplemental.
Data, Materials, and Software Availability
Materials, data, and associated protocols are available within the manuscript and/or supporting information, and the public repository Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/emdb/) with also the final 3D volumes and other relevant information about data acquisition and processing (accession codes EMD-14095 (51), EMD-14096 (52), EMD-14097 (53)). Data have been deposited on 24 December 2021 by D.F. and D.P. as authors.
References
- 1.Glaubert A. M., Sleytr U. B., Analysis of regular arrays of subunits on bacterial surfaces: Evidence for a dynamic process of assembly. J. Ultrastruct. Res. 50, 103–116 (1975). [DOI] [PubMed] [Google Scholar]
- 2.Bahl H., et al. , Molecular biology of S-layers. FEMS Microbiol. Rev. 20, 47–98 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Messner P., et al. , Biochemistry of S-layers. FEMS Microbiol. Rev. 20, 25–46 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Farci D., et al. , New features of the cell wall of the radio-resistant bacterium Deinococcus radiodurans. Biochim. Biophys. Acta 1838, 1978–1984 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Sleytr U. B., Schuster B., Egelseer E. M., Pum D., S-layers: Principles and applications. FEMS Microbiol. Rev. 38, 823–864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fagan R. P., Fairweather N. F., Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Farci D., et al. , On the S-layer of Thermus thermophilus and the assembling of its main protein SlpA. Biochim. Biophys. Acta Biomembr. 1860, 1554–1562 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Farci D., Kereïche S., Pangeni S., Haniewicz P., Bodrenko I.V., Ceccarelli M., Winterhalter M., Piano D.. Structural analysis of the architecture and in situ localization of the main S-layer complex in Deinococcus radiodurans. Structure. 29, 1279–1285.e3. (2021) [DOI] [PubMed] [Google Scholar]
- 9.Beveridge T. J., et al. , Functions of S-layers. FEMS Microbiol. Rev. 20, 99–149 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Pavkov-Keller T., Howorka S., Keller W., The structure of bacterial S-layer proteins. Prog. Mol. Biol. Transl. Sci. 103, 73–130 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Farci D., Slavov C., Piano D., Coexisting properties of thermostability and ultraviolet radiation resistance in the main S-layer complex of Deinococcus radiodurans. Photochem. Photobiol. Sci. 17, 81–88 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Kumar J., Ghosh P., Kumar A., Ultraviolet-B radiation stress-induced toxicity and alterations in proteome of Deinococcus radiodurans. Microb Physiol. 31, 1–15 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Bharat T. A. M., et al. , Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat. Microbiol. 2, 17059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambelli L., et al. , Architecture and modular assembly of Sulfolobus S-layers revealed by electron cryotomography. Proc. Natl. Acad. Sci. U.S.A. 116, 25278–25286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sexton D. L., Burgold S., Schertel A., Tocheva E. I., Super-resolution confocal cryo-CLEM with cryo-FIB milling for in situ imaging of Deinococcus radiodurans. Curr Res Struct Biol. 4, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pum D., Toca-Herrera J. L., Sleytr U. B., S-layer protein self-assembly. Int. J. Mol. Sci. 14, 2484–2501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farci D., Slavov C., Tramontano E., Piano D., The S-layer protein DR_2577 binds deinoxanthin and under desiccation conditions protects against UV-radiation in Deinococcus radiodurans. Front. Microbiol. 7, 155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eltsov M., Dubochet J., Fine structure of the Deinococcus radiodurans nucleoid revealed by cryoelectron microscopy of vitreous sections. J. Bacteriol. 187, 8047–8054 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian B., Hua Y., Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 18, 512–520 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Timmins J., Moe E., A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: Major findings, functional and mechanistic insight and challenges. Comput. Struct. Biotechnol. J. 14, 168–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krisko A., Radman M., Biology of extreme radiation resistance: The way of Deinococcus radiodurans. Cold Spring Harb. Perspect. Biol. 5, a012765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumeister W., et al. , The major cell envelope protein of Micrococcus radiodurans (R1). Structural and chemical characterization. Eur. J. Biochem. 125, 535–544 (1982). [DOI] [PubMed] [Google Scholar]
- 23.Baumeister W., et al. , Three-dimensional structure of the regular surface layer (HPI layer) of Deinococcus radiodurans. J. Mol. Biol. 187, 241–250 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Müller D. J., Baumeister W., Engel A., Conformational change of the hexagonally packed intermediate layer of Deinococcus radiodurans monitored by atomic force microscopy. J. Bacteriol. 178, 3025–3030 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister T. E., Pinhero P. J., In vivo atomic force microscopy of surface proteins on Deinococcus radiodurans. Langmuir 17, 2624–2628 (2001). [Google Scholar]
- 26.Farci D., et al. , Purification and characterization of DR_2577 (SlpA) a major S-layer protein from Deinococcus radiodurans. Front. Microbiol. 6, 414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothfuss H., Lara J. C., Schmid A. K., Lidstrom M. E., Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology (Reading) 152, 2779–2787 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Yu J., et al. , A tamB homolog is involved in maintenance of cell envelope integrity and stress resistance of Deinococcus radiodurans. Sci. Rep. 7, 45929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farci D., et al. , Structural insights into the main S-layer unit of Deinococcus radiodurans reveal a massive protein complex with porin-like features. J. Biol. Chem. 295, 4224–4236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renault L., et al. , Milestones in electron crystallography. J. Comput. Aided Mol. Des. 20, 519–527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold V. A. M., Salzer R., Averhoff B., Kühlbrandt W., Structure of a type IV pilus machinery in the open and closed state. eLife 4, e07380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly M. J., A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7, 237–245 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Farci D., Guadalupi G., Bierła K., Lobinski R., Piano D., The role of iron and copper on the oligomerization dynamics of DR_2577, the main S-Layer protein of Deinococcus radiodurans. Front. Microbiol. 10, 1450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farci D., et al. , The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions. J. Biol. Chem. 298, 102031 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodsell D. S., Olson A. J., Forli S., Art and science of the cellular mesoscale. Trends Biochem. Sci. 45, 472–483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Achouak W., Heulin T., Pagès J. M., Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199, 1–7 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Vergalli J., et al. , Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Murray R. G. E., The Family Deinococcaceae A. Balows, H. G. T., Dworkin H., Harder W., Schleifer K. H. (Eds.), The Prokaryotes (2nd ed.) (Springer, New York, 1992). pp. 3732–3744. [Google Scholar]
- 40.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Mindell J. A., Grigorieff N., Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biyani N., et al. , Focus: The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Gipson B., Zeng X., Zhang Z. Y., Stahlberg H., 2dx—User-friendly image processing for 2D crystals. J. Struct. Biol. 157, 64–72 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Cope J., Heumann J., Hoenger A., Cryo-electron tomography for structural characterization of macromolecular complexes. Curr. Protoc. Protein Sci. Chapter 17, Unit 17.13. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heumann J. M., Hoenger A., Mastronarde D. N., Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175, 288–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bostina M., et al. , Single particle cryoelectron tomography characterization of the structure and structural variability of poliovirus-receptor-membrane complex at 30 A resolution. J. Struct. Biol. 160, 200–210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farci D., Piano D., Subtomogram average of Deinococcus radiodurans’ cell envelope. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/search/EMD-14095. Deposited 24 December 2021.
- 52.Farci D., Piano D., Symmetrized subtomogram average of Deinococcus radiodurans’ cell envelope. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/search/EMD-14096. Deposited 24 December 2021.
- 53.Farci D., Piano D., 3D map of the crystalline organisation of Deinococcus radiodurans’ cell envelope. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/search/EMD-14097. Deposited 24 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials, data, and associated protocols are available within the manuscript and/or supporting information, and the public repository Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/emdb/) with also the final 3D volumes and other relevant information about data acquisition and processing (accession codes EMD-14095 (51), EMD-14096 (52), EMD-14097 (53)). Data have been deposited on 24 December 2021 by D.F. and D.P. as authors.