There is no consensus on the origin of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). We recently called for an independent inquiry (2). Garry has commented (3), as quoted, and we respond briefly here.
“FCSs are common in coronaviruses, and present in representatives of four out of five betacoronavirus subgenuses.”
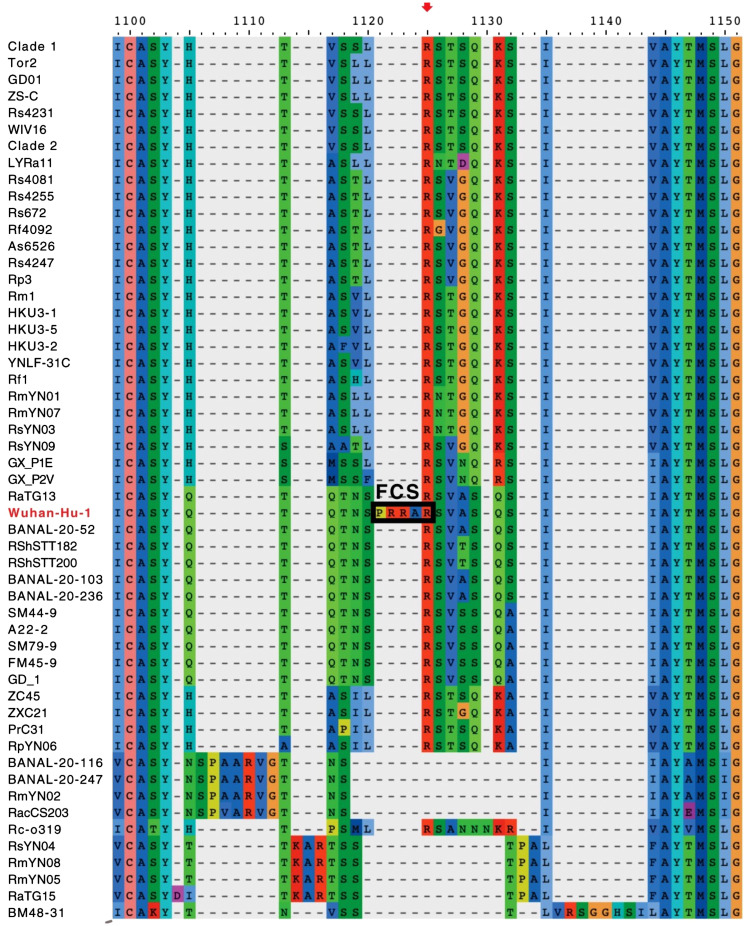
We stated that the furin cleavage site (FCS) in SARS-CoV-2 is unique among sarbecoviruses (ref. 2 and Fig. 1), although FCSs are common in other betacoronaviruses (2).
Fig. 1.
An alignment of the amino acid sequences of coronavirus spike proteins in the region of the S1/S2 junction, illustrating the sequence of SARS-CoV-2 (Wuhan-Hu-1) and some of its closest relatives. The FCS is indicated (PRRAR'SVAS), and furin cuts the spike protein between R and S, as indicated by the red arrowhead. Adapted from Chan and Zhan (4).
“The highly variable nature of the S1/S2 junction is easily ascertained by inspecting a precise alignment of sarbecovirus Spikes.”
Garry’s limited alignment of seven Spike protein sequences (3) is far from precise. RacCS203, for example, lacks the arginine, serine residues (RS) present in the other viruses. A comprehensive alignment (4) (Fig. 1) highlights the unusual FCS of SARS-CoV-2.
“Placing the insertion out of frame would be an ‘unusual and needlessly complex feat of genetic engineering.’”
The conclusion that the insertion is “out of frame” rests on knowing the “template” (proximal ancestor) for the insertion (whether in a laboratory or by natural evolution), which is not known here. In any case, an “out-of-frame” insertion, for example, to engineer a restriction site, is neither unusual nor complex.
“The immediate proximal ancestor of SARS-CoV-2 … first evolved in an intermediate host.”
This is complete conjecture. No intermediate host has been identified (5).
“Two related lineages—lineage A and lineage B—first infected humans via the wildlife trade at the Huanan Market in Wuhan.”
This is complete conjecture. No infected animal harboring lineage A or B was identified at the Huanan Market (6).
“Four extra amino acids (PRRA), not eight, were added…. There was an insertion of 12 nucleotides into the Spike gene.”
We stated clearly (2) that 12 nucleotides were inserted, and that the insertion of RRA completes a sequence identical to that in hENaCα.
“Except for one codon (cgu that encodes arginine 685), each of the codons for RRARSVAS is different in human ENaC and SARS-CoV-2.”
Exactly. If the insert arose naturally, these codons would be similar. They are not. If engineered, any codons could be selected for R682–A684.
“Harrison and Sachs make a serious accusation against scientists at University of North Carolina (UNC) and Wuhan Institute of Virology (WIV).”
No accusation was made (2). EcoHealth Alliance (EHA)–WIV–UNC have had a strong interest in protease cleavage in enhancing coronavirus infectivity (7), as expressed in the DEFUSE proposal (8) to insert FCS into novel viruses (2).
“UNC or WIV researchers would have had to possess the direct SARS-CoV-2 progenitor….”
We know little about unreported viruses available to EHA–WIV–UNC (2). Concerns about these should be clarified by release of databases, laboratory notebooks, and electronic communications (2).
“Harrison and Sachs allege that scientists … conspired to suppress theories of a laboratory origin of SARS-CoV-2.”
We did not use “allege,” “suppress,” or “conspire” (2). We believe that EHA, NIH, and others have not been transparent (2, 9), and that the origin of the virus remains unresolved (1).
Footnotes
Competing interest statement: J.D.S. is Chair of the Lancet Commission on COVID-19. The authors declare no competing financial or scientific interests.
References
- 1.Sachs J. D., et al. , The Lancet Commission on lessons for the future from the COVID-19 pandemic. Lancet 400, 1224–1280 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison N. L., Sachs J. D., A call for an independent inquiry into the origin of the SARS-CoV-2 virus. Proc. Natl. Acad. Sci. U.S.A. 119, e2202769119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garry R. F., SARS-CoV-2 furin cleavage site was not engineered. Proc. Natl. Acad. Sci. U.S.A. 119, e2211107119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan Y. A., Zhan S. H., The emergence of the spike furin cleavage site in SARS-CoV-2. Mol. Biol. Evol. 39, msab327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He W. T., et al. , Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 185, 1117–1129.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao G., et al. , Surveillance of SARS-CoV-2 in the environment and animal samples of the Huanan Seafood Market. Research Square [Preprint] (2022). 10.21203/rs.3.rs-1370392/v1 (Accessed 3 October 2022). [DOI]
- 7.Menachery V. D., et al. , Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 94, e01774-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lerner S., Hvistendahl M., New details emerge about coronavirus research at Chinese lab. The Intercept, 6 September 2021. https://theintercept.com/2021/09/06/new-details-emerge-about-coronavirus-research-at-chinese-lab/. Accessed 22 February 2022.
- 9.Kopp E., Timeline: The proximal origin of SARS-CoV-2. https://usrtk.org/biohazards/timeline-the-proximal-origin-of-sars-cov-2. Accessed 3 October 2022.