Significance
Vision starts when a photon isomerizes the 11-cis-retinylidene chromophore of rhodopsin to initiate phototransduction. The all-trans-retinylidene product is hydrolyzed and all-trans-retinal released, allowing rebinding of 11-cis-retinal to regenerate rhodopsin for sustained vision. Subsequently, all-trans-retinal is cleared, preventing aldehyde toxicity by reduction to all-trans-retinol. Defects in these metabolic processes could lead to severe retinopathies. Studying the biochemistry of key proteins responsible for these fundamental steps of vision in their native membrane environments has remained challenging. Using rapid quantitative chemical and analytical methods, we directly captured and quantified the kinetics and energetics of these essential biochemical processes occurring in native membranes. Our results shed light on the entire rhodopsin photocycle and chromophore regeneration and are broadly applicable to other retinylidene proteins.
Keywords: rhodopsin, chromophore, photoreceptors, retinoids, eye
Abstract
For sustained vision, photoactivated rhodopsin (Rho*) must undergo hydrolysis and release of all-trans-retinal, producing substrate for the visual cycle and apo-opsin available for regeneration with 11-cis-retinal. The kinetics of this hydrolysis has yet to be described for rhodopsin in its native membrane environment. We developed a method consisting of simultaneous denaturation and chromophore trapping by isopropanol/borohydride, followed by exhaustive protein digestion, complete extraction, and liquid chromatography–mass spectrometry. Using our method, we tracked Rho* hydrolysis, the subsequent formation of N-retinylidene-phosphatidylethanolamine (N-ret-PE) adducts with the released all-trans-retinal, and the reduction of all-trans-retinal to all-trans-retinol. We found that hydrolysis occurred faster in native membranes than in detergent micelles typically used to study membrane proteins. The activation energy of the hydrolysis in native membranes was determined to be 17.7 ± 2.4 kcal/mol. Our data support the interpretation that metarhodopsin II, the signaling state of rhodopsin, is the primary species undergoing hydrolysis and release of its all-trans-retinal. In the absence of NADPH, free all-trans-retinal reacts with phosphatidylethanolamine (PE), forming a substantial amount of N-ret-PE (∼40% of total all-trans-retinal at physiological pH), at a rate that is an order of magnitude faster than Rho* hydrolysis. However, N-ret-PE formation was highly attenuated by NADPH-dependent reduction of all-trans-retinal to all-trans-retinol. Neither N-ret-PE formation nor all-trans-retinal reduction affected the rate of hydrolysis of Rho*. Our study provides a comprehensive picture of the hydrolysis of Rho* and the release of all-trans-retinal and its reentry into the visual cycle, a process in which alteration can lead to severe retinopathies.
Rod photoreceptors possess a unique cellular structure, including the rod outer segment (ROS), composed of a long stack of lipid discs packed with rhodopsin (Rho) to optimize photon capture (1). This dense packing allows for the high sensitivity to light required for scotopic vision. The responsiveness of Rho to light is afforded by the photochemistry of its Schiff base (SB)–bound 11-cis-retinal (11cRAL) chromophore, whose cis-trans isomerization induces a series of protein conformational changes, leading to the photoactivated Rho (Rho*) state (2, 3). The binding site of 11cRAL has been meticulously explored by multiple groups and finally determined in 1982 to be Lys296 by using sodium borohydride (NaBH4) reduction to trap the retinylidene SB (RSB) in place, thereby preventing loss of chromophore to hydrolysis during proteolysis and chromatography (4).
Rho* generated after light exposure exists in an equilibrium of metarhodopsin states, of which metarhodopsin II (MII) greatly dominates and interacts with the G protein transducin to initiate phototransduction (5). Therefore, the cis-trans photoisomerization of retinal converts the native Rho ligand, which behaves as an inverse agonist, into a transient agonist that must be hydrolyzed from Rho* (6). Hydrolysis provides all-trans-retinal (atRAL) substrate for the visual cycle, where it is isomerized back to 11cRAL. The amine group of the Lys296 residue then reacts with 11cRAL to regenerate ground-state Rho. Thus, hydrolysis primes the photoreceptor for restoration of dark sensitivity (dark adaptation), which in humans takes up to 40 min for complete recovery of maximal light sensitivity (7). Notably, 11cRAL enters and atRAL exits the protein through different sites (8). The entry site does not bind atRAL and is highly selective for 11cRAL. The exit site solely allows atRAL to leave without reentry (8). Released atRAL is subsequently reduced to all-trans-retinol (atROL) catalyzed by NADPH-dependent retinol dehydrogenases (RDHs) (9).
To date, the crucial step of all-trans-RSB (atRSB) hydrolysis in the Rho photocycle has largely been studied using indirect spectrophotometric techniques that require the solubilization of Rho in detergent micelles. As such, these methods do not capture Rho biochemistry in the native membrane environment (10). The broad overlapping absorbance bands of Rho, its metastates after light exposure, and released atRAL present challenges in the study of Rho* atRSB hydrolysis using spectrophotometric techniques (11). The metastates differ in structure, including anti- versus syn-stereoisomerism and protonation state of the atRSB (12). Interestingly, atRSB protonation in Rho* behaves inversely with pH in the following manner: lower pH stabilizes the deprotonated atRSB, MII, whereas higher pH stabilizes the protonated atRSBs, MI and MIII (6, 13, 14). Among these photoproducts, MII is understood to be the signaling state of Rho and has also been observed to be most susceptible to atRSB hydrolysis (5, 15). However, the hydrolysis of SBs is facilitated via nucleophilic attack of water on a protonated SB-carbon rather than a deprotonated SB-carbon, as found in MII (16). Therefore, the mechanisms of the hydrolysis and the Rho intermediate most susceptible to hydrolysis require more investigation.
Another factor that complicates the study of atRSB hydrolysis with detergent-solubilized Rho is the loss of phospholipids, in particular phosphatidylethanolamine (PE), which can form adducts with the released atRAL and potentially affect the rate of the hydrolysis reaction (17). Moreover, these N-retinylidene-PE (N-ret-PE) adducts have documented roles as substrates for the visual cycle transport protein ABCA4, as well as in photic regeneration pathways (18). As such, a consideration of their formation and clearance is critical for understanding alternative chromophore regeneration pathways, as well as the pathophysiology of ABCA4-associated retinal diseases, such as Stargardt disease-1 (19). Furthermore, the reduction of atRAL by RDHs in the context of the retina, where it can form PE adducts, needs clarification.
In this study, the hydrolytic release of atRAL from Rho* within native bovine ROS membranes was monitored by direct quantification of the opsin-bound RSBs in their different cis-trans configurations after light exposure of Rho in ROS membranes and of the atRAL hydrolysis product. This was achieved by using simultaneous alcohol protein denaturation and NaBH4 reduction to trap RSBs as nonhydrolyzable retinyl secondary amine adducts. After proteolysis, these retinyl signatures and their cis-trans isomeric configurations were detected by liquid chromatography–tandem mass spectrometry (LC-MS/MS) and used to monitor opsin RSBs over time. Concurrently, both free and PE-bound atRAL were tracked in the alcohol-solubilized lipid fraction. The energetic landscape of Rho*-atRSB hydrolysis was further elucidated using quantum chemical methods.
Results
Proteolysis and LC-MS/MS Analysis of Nε-at-retinyl-peptide (bovine Rho 292–302).
Stereospecific detection of the trapped RSB within opsin has not been achieved to date because of the high hydrophobicity of the resulting retinyl peptides and the lability of the retinyl moiety. Therefore, RSB isomerization and hydrolysis could not be studied, except indirectly by spectrophotometric techniques. To establish a direct LC-MS/MS detection method, we first tested a synthetic peptide consisting of residues 292 to 302 of the bovine Rho sequence (bRhoPep) (SI Appendix, Fig. S1A). NaBH4 reduction trapped the atRAL-SB adducted to the ε amine group of Lys296 of bRhoPep, forming a nonhydrolyzable retinyl secondary amine adduct (SI Appendix, Fig. S1B). The LC-MS peak of this retinyl peptide showed the characteristic ultraviolet-visible (UV-Vis) spectrum of the retinyl moiety (λmax = 328 nm), which greatly enhanced the hydrophobicity of the peptide, as reflected in the increased chromatographic retention time from 5 to 31 min (SI Appendix, Fig. S1 C and D). MS/MS showed electrospray ionization (ESI)–source fragmentation of the Nε-retinyl-peptide analyte into a retinyl cation and a peptide ion, whose sequence was confirmed by collision-induced dissociation (CID) fragmentation to be that of the bRhoPep (SI Appendix, Fig. S1 E–G).
Because stereospecific MS detection of the whole Nε-retinyl-opsin protein is unfeasible, a proteolytic digestion of Nε-retinyl-opsin to Nε-retinyl-peptides would be necessary for detection and sequence determination by LC-MS/MS. This principle was tested by proteolysis of the Nε-retinyl-bRhoPep using proteinase K or the protease mixture pronase. The proteinase K digest yielded four unique Nε-retinyl-peptides, identified by MS/MS to be KretTS, KretT, AKret, and FAKret, each with a sequence containing Lys296 adducted to the retinyl moiety (SI Appendix, Fig. S2). Complete proteolysis of Nε-at-retinyl-bRhoPep down to Nε-at-retinyl-Lys by pronase at 40 °C yielded one prominent peak and two minor peaks, which were all identified to be Nε-retinyl-Lys (SI Appendix, Fig. S3). Based on the UV-Vis and MS spectra of each synthetic retinyl isomer standard of Nε-retinyl-Lys (SI Appendix, Fig. S4), the most prominent peak was determined to be the all-trans isomer, whereas the two minor peaks were determined to be the 9-cis and 13-cis isomers. These two minor isomers are known to form spontaneously by thermal isomerization of the all-trans isomer (20). Pronase was found to completely proteolyze Nε-retinyl-opsin at 10 °C, which minimized the thermal isomerization and retained the isomeric composition. The LC separation followed by UV-Vis and MS spectral identification of the four Nε-retinyl-Lys isomers (9-cis, 11-cis, 13-cis, and all trans) as standards (SI Appendix, Fig. S4) demonstrated the potential of this method for capturing the photochemistry of retinylidene proteins following trapping of the RSB by NaBH4 and proteolysis by pronase.
Proteolysis and LC-MS/MS Analysis of Nε-retinyl-opsin.
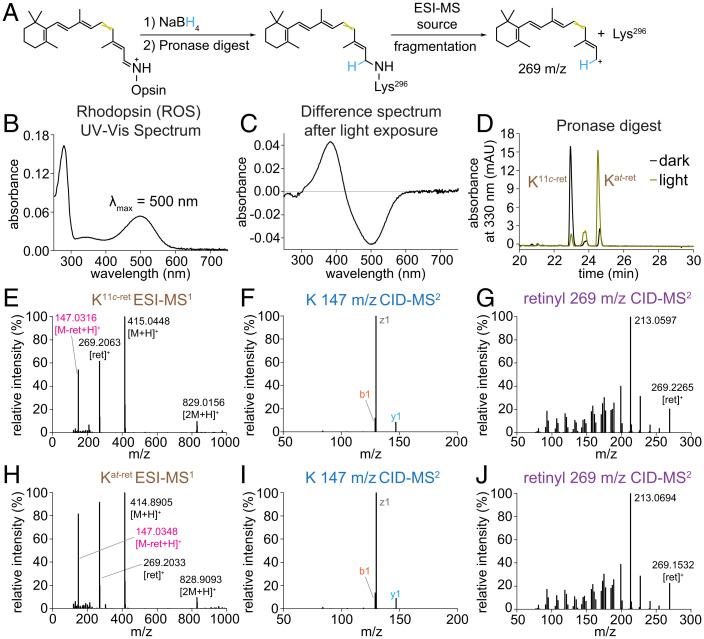
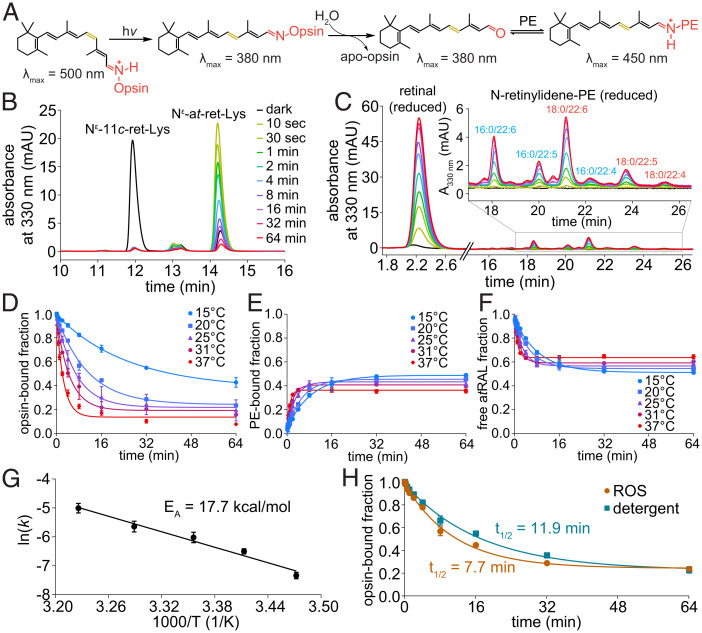
To study Rho biochemistry in native membranes, a saturated solution of NaBH4 in isopropanol (iPrOH) was added to ROS to rapidly denature the protein and, simultaneously, reduce the RSB, thus trapping the retinal moiety bound to opsin. Remarkably, this procedure can be used to study Rho either in native membranes (Fig. 1A) or purified in detergent solutions (SI Appendix, Fig. S5). Pronase converted a suspension of the denatured proteins into a clarified solution that could be analyzed by LC-MS/MS. Prior to processing the samples, we used UV-Vis spectroscopy to determine the photostate of Rho in ROS membranes. The 500-nm absorbance maximum of Rho declines after light exposure, producing a new maximum at 380 nm, which is characteristic of MII (Fig. 1 B and C). From the pronase digest, Rho yielded a dominant Nε-11-cis-retinyl-Lys peak, whereas Rho* yielded a dominant Nε-all-trans-retinyl-Lys peak, as confirmed by LC-MS/MS and comparison with our synthetic standards (Fig. 1 D–J).
Fig. 1.
LC-MS/MS analyses of pronase digests of Rho and Rho* from native membranes. (A) Schematic diagram of sample preparation and MS workflow. (B) UV-Vis spectrum of Rho from ROS membranes solubilized in DDM. (C) Difference absorbance spectrum of DDM-solubilized Rho versus Rho* in ROS membranes illuminated with 565-nm fiber light at 625 μW for 10 s at 4 °C. (D) Chromatographic separation of Nε-retinyl-Lys peaks from the pronase digests of Rho and Rho* in ROS membranes treated with NaBH4 in iPrOH. (E, H) ESI-MS spectrum showing characteristic cleavage of retinyl cation (269 m/z) from precursor Nε-retinyl-Lys analyte (415 m/z), producing a product Lys ion (147 m/z). (F, I) CID-MS/MS spectrum of the product Lys ion exhibiting a characteristic fragmentation pattern of Lys. (G, J) CID-MS/MS spectrum of retinyl cation demonstrating a complex fragmentation pattern with a characteristic dominant 213-m/z signal (39, 40). mAU, milli absorbance unit.
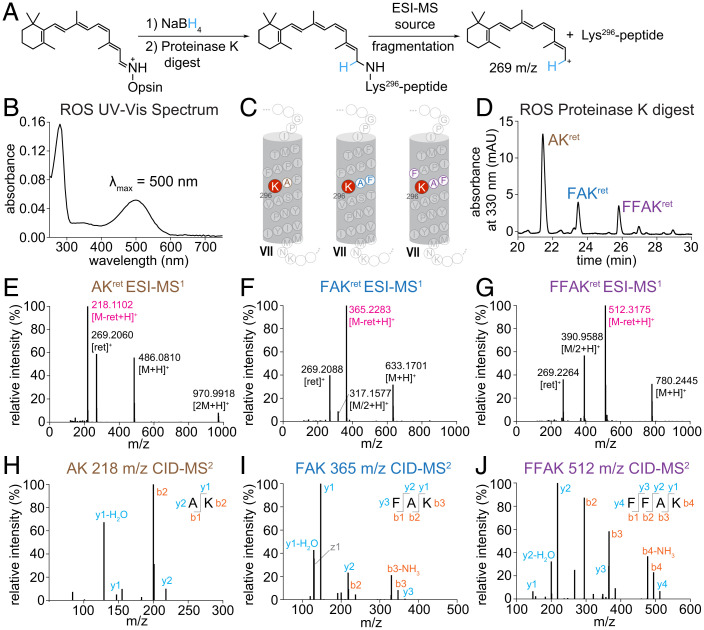
The Nε-retinyl-Lys peaks were confirmed to originate from Rho in ROS membranes by LC-MS/MS analysis of the Nε-retinyl-peptides generated from a proteinase K digestion of the ROS-protein precipitate. Proteinase K also clarified the suspension of protein precipitate into a solution of Nε-retinyl-peptides that were identified by LC-MS/MS to be similar to those from the proteinase K digest of Nε-retinyl-bRhoPep (Fig. 2). The proteinase K and pronase digests of ROS-protein precipitates from treatment of ROS membranes with sodium borodeuteride (NaBD4) in iPrOH generated similar results, showing the same Nε-retinyl analytes with an increase in mass corresponding to deuteration and source fragmentation producing the C15-D1-retinyl cation at 270 m/z (SI Appendix, Figs. S6 and S7). Our NaBD4 results align with the NaBH4 reduction of the RSB of Rho or Rho* and with the characteristic ESI-source fragmentation of the retinyl moiety.
Fig. 2.
LC-MS/MS analysis of proteinase K digest of Rho from native membranes. (A) Schematic diagram of sample preparation and MS workflow. (B) UV-Vis spectrum of Rho from ROS membranes solubilized in DDM. (C) Location of chromophore binding residue within helix VII of bovine Rho, with labeled Nε-retinyl-peptide fragments detected from the proteinase K digest. (D) Chromatographic separation of Nε-retinyl-peptides from proteinase K digestion of Rho treated with NaBH4 in iPrOH. (E–G) ESI-MS spectrum showing characteristic cleavage of retinyl cation (269 m/z) from precursor Nε-retinyl-peptide analyte, producing a product peptide ion. (H–J) MS/MS spectrum of CID fragmentation of the product peptide ion for sequence determination. mAU, milli absorbance unit.
Separation and Identification of N-ret-PEs.
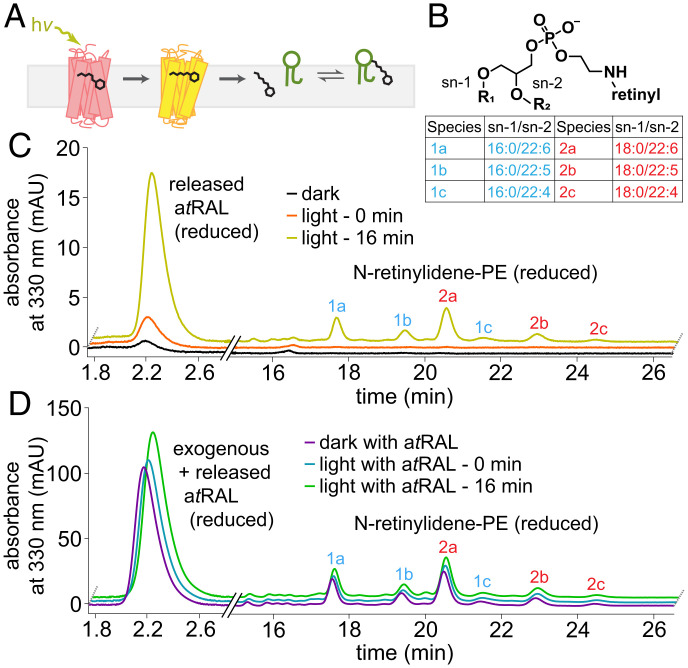
We expected that, concomitantly with the trapping of the bound atRAL moiety to Rho* by NaBH4, any N-ret-PEs formed by SB condensation of atRAL with PE after Rho* hydrolysis would also be captured by NaBH4 reduction as N-retinyl-PEs (Fig. 3A). Using LC-MS/MS and CID fragmentation of N-retinyl-PEs, we determined that the main fatty acyl chains in the sn-2 position of PE are docosahexaenoic acid (22:6), docosapentaenoic acid (22:5), and docosatetraenoic acid (22:4) (Fig. 3B), which are most abundant in ROS membranes (21) (SI Appendix, Fig. S8).
Fig. 3.
Formation of N-ret-PE in native membranes after light exposure. (A) Diagram of N-ret-PE formation after illumination of ROS membranes. (B) Chemical structure of N-ret-PE, with a table of major species found in the ROS membranes. (C) Chromatogram of atRAL released and resultant N-ret-PE in ROS at 0 and 16 min after illumination for 10 s with 565-nm fiber light at 625 μW. (D) Analogous experiment to that in C, but with exogenous atRAL added to the ROS membrane suspension prior to illumination. mAU, milli absorbance unit.
In the dark, ROS treated with NaBH4 in iPrOH had a small signal of atROL, the NaBH4-reduction product of atRAL, with no other discernible signals of retinyl species (Fig. 3C). A small increase in atROL was observed in ROS briefly exposed to light. Incubation in the dark for 16 min after brief exposure to light led to a large increase in the atROL signal, as well as the appearance of six major peaks with distinct relative peak height/area ratios identified by LC-MS/MS as the N-retinyl-PEs described above. The addition of exogenous atRAL to ROS membranes unexposed to light produced the same fingerprint of these six N-retinyl-PE peaks (Fig. 3D). Exposure to light followed by a 16-min incubation in the dark led to an increase in atROL and N-retinyl-PE signals corresponding to the contribution from NaBH4-reduced free and PE-bound atRAL from Rho* hydrolysis (Fig. 3D). Therefore, addition of NaBH4 in iPrOH to ROS membranes enabled continuous tracking of the atRAL following its hydrolytic release from Rho*, even in the presence of exogenous atRAL.
Kinetics of Rho* Hydrolysis and Subsequent N-ret-PE Formation.
Our trapping procedure enabled a detailed kinetic analysis of each step as it occurred in the native phospholipid bilayer (Fig. 4A). NaBH4 in iPrOH was added to ROS at different time points after illumination to trap the remaining Rho*-bound atRAL, the free atRAL, and subsequently formed N-ret-PEs. Snapshots of the progressive hydrolysis of Rho* to form free atRAL and the subsequent reaction of the atRAL with the lipid membrane are shown by chromatograms of the NaBH4-reduced opsin-bound RSBs over time in Fig. 4B and the NaBH4-reduced free and PE-bound atRAL over time in Fig. 4C. The decreasing Nε-all-trans-retinyl-Lys signal and increasing atROL and N-retinyl-PE signals over time reflect the time course of Rho*-atRSB hydrolysis and atRAL release (Fig. 4 B and C). The rates of hydrolysis followed pseudo first-order decay kinetics and increased with temperature (Fig. 4D).
Fig. 4.
Temperature dependence of Rho* hydrolysis and subsequent N-ret-PE formation. (A) Scheme of hydrolysis reaction following light exposure. (B) Chromatograms of pronase digests of Rho* protein pellets obtained from ROS membranes treated with NaBH4 in iPrOH after brief light exposure (565-nm fiber light at 625 μW for 10 s) and subsequent incubation in the dark for different periods of time at 37 °C. (C) Chromatograms of the iPrOH-solubilized lipid fractions from the corresponding samples from B for detection of NaBH4-reduced free and PE-bound atRAL from Rho* hydrolysis at 37 °C. (D) Rho* hydrolysis exhibited pseudo first-order decay kinetics at various temperatures. (E) The formation of N-ret-PEs at different temperatures from atRAL produced by Rho* hydrolysis followed pseudo first-order kinetics. (F) The proportion of the atRAL from Rho* hydrolysis that was unbound to PE tracked inversely to the proportion that formed PE adducts. (G) The Arrhenius plot of hydrolysis rate constants at different temperatures yielded an EA of 17.7 kcal/mol. (H) Rho* hydrolysis was faster in ROS membranes than in DDM detergent micelles at 20 °C. mAU, milli absorbance unit.
The atRAL released by hydrolysis of Rho* subsequently reacted reversibly with PE, reaching a steady state of ∼40% atRAL bound to PE as N-ret-PE (Fig. 4E). The kinetics of this N-ret-PE formation also followed pseudo first-order kinetics. The overall rate of N-ret-PE formation, starting from the hydrolysis of Rho* to the release of atRAL for SB condensation with PE, was found to increase with increased temperature; however, the steady-state level of N-ret-PE decreased with increased temperature (Fig. 4E). The proportion of atRAL released via Rho* hydrolysis and not bound to PE showed an inverse relationship with the formation of N-ret-PEs (Fig. 4F).
From the temperature dependence of the Rho* hydrolysis reaction, an Arrhenius plot (Fig. 4G) was generated, yielding an activation energy (EA) of 17.7 ± 2.4 kcal/mol. The rate of Rho* hydrolysis was also measured in n-dodecyl-β-D-maltoside (DDM) micelles to mimic conditions of mild detergent solutions that are typically used for spectrophotometric measurements of Rho. The rate of hydrolysis in DDM was found to be ∼50% slower than that in the ROS membranes (Fig. 4H).
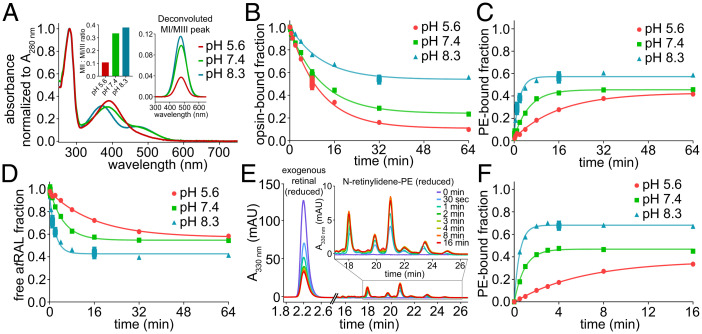
pH Dependency of Rho* Hydrolysis and N-ret-PE Formation.
The protonation state of the iminium nitrogen of Rho*-atRSB is known to behave inversely to changes in the external protic environment of Rho* (22). This behavior was observed by UV-Vis spectroscopy, showing that higher pH shifts the metarhodopsin equilibrium from MII, the deprotonated state, toward protonated states, namely MI and MIII (Fig. 5A). Rho*-atRSB hydrolysis occurred to greater extents with decreasing pH, where MII is favored (Fig. 5B). However, lower pH appeared to slow down the overall rate of N-ret-PE formation (Fig. 5C). The amount of free atRAL was inversely proportional to the amount of N-ret-PE (Fig. 5D). To isolate the reaction between atRAL and PE and obtain the rate of N-ret-PE formation independently, exogenous atRAL was added to ROS membranes and tracked by addition of NaBH4 in iPrOH at different time points. The representative chromatograms of the SB condensation reaction of atRAL with PE over time showed the decline in atRAL starting material with the formation of N-ret-PEs (Fig. 5E). The rate of N-ret-PE formation exhibited pH dependency, with a faster rate of reaction as pH increased (Fig. 5F). In summary, Rho*-atRSB hydrolysis was faster at lower pH, which favors MII, whereas N-ret-PE formation was slower at lower pH.
Fig. 5.
pH-dependent Rho* hydrolysis and N-ret-PE formation in native membranes. (A) UV-Vis spectrum of Rho after illumination with a 565-nm fiber light at 625 μW for 10 s and incubation for 12 h at 4 °C. Spectral analysis and deconvolution of the protonated retinylidene SB peak demonstrated inverse pH dependency, with a greater proportion of protonated retinylidene SB (MI/III) appearing with increasing pH. (B) Hydrolysis of atRAL from Rho* at 20 °C occurred to the greatest extent at lower pH in which the MI-MII equilibrium shifts toward MII. (C) The rate of N-ret-PE formation at 20 °C exhibited pH dependency and was fastest at high pH. (D) The proportion of Rho* hydrolysis–derived atRAL that did not form PE adducts tracked inversely to the proportion that formed PE adducts. (E) Chromatographic traces of N-ret-PE formation over time at pH 8.3 and 20 °C, after addition of exogenous atRAL to ROS membranes. (F) The mole fraction of N-ret-PE formed at 20 °C from exogenous atRAL over time exhibited pseudo first-order kinetics, with faster rates and higher steady state as pH increased. mAU, milli absorbance unit.
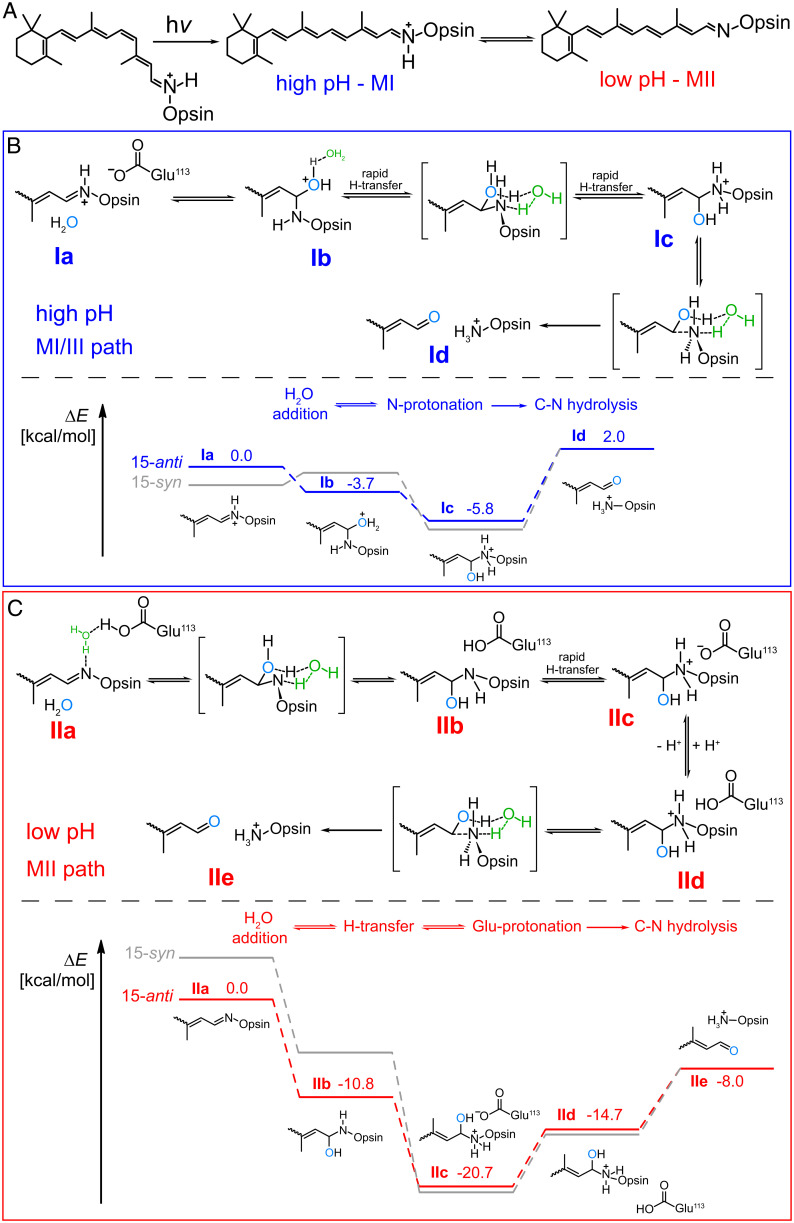
Computational Analysis of Rho*-atRSB Hydrolysis.
We explored the mechanistic basis for the pH dependency of Rho*-atRSB hydrolysis by computationally modeling the energy landscapes of atRSB hydrolysis for each metarhodopsin species. We examined the influence of Glu113 and Glu181 on the energy levels of each metarhodopsin species and subsequent intermediates of hydrolysis, considering the location of these Glu residues in the chromophore binding pocket, their influence on spectral characteristics of the retinylidene chromophore, and their role in forming the water and H-bonding network that surrounds the chromophore and spans the Rho molecule (23, 24). Upon photoactivation to MII, Glu113 and Glu181 shift from their position in Rho, translocating so that both surround and interact with the atRSB (23, 25). Plausibly, the protonation states of Glu113 and Glu181 could vary with pH and determine whether a charged or neutral carboxyl residue would exist near the atRSB in Rho*, which can exist in either a syn- or anti-configuration. In MI, the configuration of its protonated atRSB is 15-anti, whereas in MIII, its protonated atRSB is 15-syn (12, 26). The exact stereoisomerism of the deprotonated atRSB of MII is unclear, but it could loosely adopt either a 15-anti or 15-syn configuration (12, 26). For either the protonated or deprotonated form of the atRSB, the anti-configuration is still dominant over syn, because the anti-syn interconversion requires additional light or thermal energy (12, 26). We carried out quantum chemical calculations to map the energetic landscape of hydrolysis as influenced by the syn-anti configuration and protonation state of the atRSB, as well the protonation states of Glu113 and Glu181. Our calculations were performed on a hydrogen-saturated cluster model of the Rho active site derived from the current highest-resolution structure of a metarhodopsin species to date, 3PQR, proposed to correspond to MII, with a deprotonated anti-atRSB structure (SI Appendix, Fig. S9).
For protonated atRSB, favored at high pH, our initial calculations showed preference for the deprotonated states of both Glu113 and Glu181. Notably, the protonated syn-atRSB was found to be 3.3 kcal/mol lower in energy (more stable) than the anti-configuration. However, water addition is only exothermic for the transition of the protonated anti-atRSB to an oxonium intermediate (Fig. 6 B, Ib) (−3.7 kcal/mol), followed by exothermic proton transfer (−2.0 kcal/mol) between the oxonium and nitrogen, as facilitated by the polar protic environment of the pocket (Fig. 6). The final C-N bond cleavage is considerably endothermic such that the final products (Fig. 6 B, Id) were 2.0 kcal/mol higher in energy than reactants (Fig. 6 B, Ia). The hydrolysis of the protonated syn-atRSB was also found to be an overall endothermic process (Fig. 6). In contrast, hydrolysis of either the anti- or syn-isomers of the deprotonated atRSB was overall exothermic (Fig. 6). Under low pH conditions favoring the deprotonated atRSB, our calculations showed a preference for the deprotonated state of Glu181 and the protonated state of Glu113. The deprotonated anti-atRSB was markedly lower in energy than the corresponding syn-isomer by 4.4 kcal/mol. Water addition to either the anti- or syn-isomers of deprotonated atRSB was considerably exothermic, as was the proton transfer from Glu113 eventually to the nitrogen of the Lys296-carbinolamine intermediate, likely via water or the H-bonding network of the pocket (Fig. 6 C, IIb). At low pH, Glu113 becomes protonated (Fig. 6 C, IId). The reprotonation of Glu113 followed by the C-N bond cleavage are both endothermic; however, the overall energy change in the process is exothermic by ∼8.0 kcal/mol (Fig. 6 C, IIe), in strong contrast to the endothermic reaction at higher pH. Assuming linear relationships between effective activation barriers and overall change in energy, we conclude that changes in pH affect the protonation patterns of Glu113, Glu181 carboxyl moieties, and the Lys296-N-atom, which in turn have a decisive effect on the energetics of hydrolysis of the different metarhodopsin species and release of atRAL.
Fig. 6.
Energy landscape of hydrolysis of the atRSB adduct of different metarhodopsin species. (A) Rho photoisomerization and production of Rho* in pH-dependent equilibrium of metarhodopsin states. (B and C) Highlighted in blue (high pH, MI/III path) (B) and red (low pH, MII path) (C) are computationally determined energy landscapes for hydrolysis of both the 15-anti and 15-syn configurations of protonated and deprotonated atRSBs. The numbers reported are relative energies with respect to the initial states Ia or IIa. Energy levels for the 15-syn configuration are shown in gray for reference.
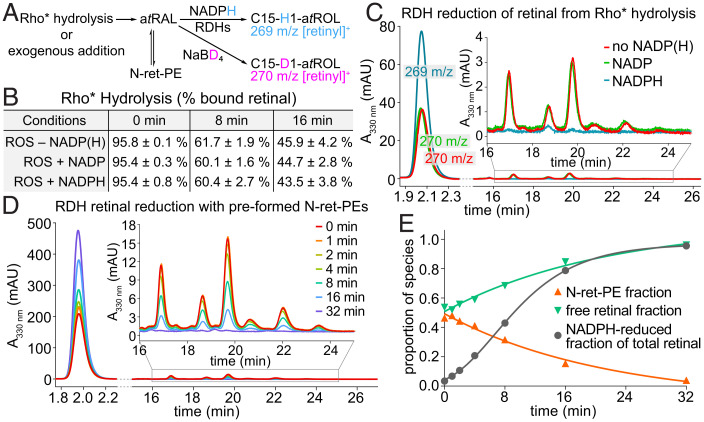
Reduction of atRAL in Native Membranes.
In the visual cycle, atRAL is reduced to atROL by RDHs (9). Our method using NaBD4 in place of NaBH4 allowed us to track this step of the visual cycle in ROS, thereby studying the fate of atRAL after its hydrolytic release from Rho* in the presence of the essential dinucleotide cofactor NADPH (Fig. 1A, SI Appendix, Fig. S10). We found that atRAL release from hydrolysis of Rho*-atRSB occurred to a similar extent in the presence or absence of NADPH (NADP was used as a control), suggesting that atRAL reduction by RDHs does not influence the biochemistry of Rho*-atRSB hydrolysis (Fig. 7B). However, nearly all of the released atRAL was reduced to atROL in the presence of NADPH at both 8 and 16 min postillumination (Fig. 7C). Notably, there were almost undetectable levels of N-ret-PEs in the presence of NADPH at both time points (Fig. 7C).
Fig. 7.
Reduction of atRAL by RDHs in native membranes. (A) Simplified diagram from SI Appendix, Fig. S10 depicting NaBD4-based monitoring of RDH activity by tracking NADPH-reduced vs. NaBD4-reduced retinal. (B) In ROS membranes, Rho* hydrolysis at pH 7.4 and 20 °C is unaffected by addition of dinucleotides, NADP, or NADPH. (C) At 16 min of Rho* hydrolysis, the released atRAL was reduced by RDHs in the presence of NADPH, substantially attenuating the formation of N-ret-PEs. (D) In the presence of both free atRAL and N-ret-PEs in equilibrium at pH 7.4 and 20 °C after addition of exogenous atRAL, RDH activity was initiated by addition of NADPH. The NADPH-reduced atRAL and NaBD4-reduced free and PE-bound atRAL were monitored over time by LC-MS. (E) In samples corresponding to those of D, the fraction of NADPH-reduced atRAL followed sigmoidal growth kinetics, with the concurrent reduction of the initially available free atRAL and the atRAL gradually supplied from the equilibrium with N-ret-PE, as demonstrated by the slow decline of RDH activity over the time course. mAU, milli absorbance unit.
To determine whether N-ret-PEs play a role in mediating the delivery of atRAL, NADPH was added to ROS-containing N-ret-PE, as prepared by addition of exogenous atRAL followed by a 16-min incubation to reach the steady state of N-ret-PE formation at pH 7.4. The atRAL from N-ret-PEs appeared not to be a direct source of substrate for the RDHs, because the proportion of N-ret-PEs was unaffected at the instant of NADPH addition (Fig. 7 D and E). Furthermore, the presence of NADPH did not accelerate the dissociation of N-ret-PEs as would have been observed if N-ret-PEs played an indispensable role in atRAL-substrate delivery to RDHs (Fig. 7 D and E). Within the first 4 min, N-ret-PE levels were reduced to a small extent, but nearly 25% of free atRAL was reduced by the RDHs (Fig. 7E). Therefore, the RDHs exhibited their activity on free atRAL in native membranes rather than on PE-bound atRAL. By 32 min, nearly all of the atRAL had been reduced, with most atRAL dissociated from PE (Fig. 7E).
Discussion
A critical step in the visual cycle is the release of the spent chromophore, atRAL, which is ultimately transformed back to 11cRAL. The steps immediately following retinal photoisomerization (i.e., hydrolytic release of atRAL, formation of N-ret-PE, and reduction of atRAL to atROL) have only been subjected to preliminary investigation because of technical issues, particularly in membranes. The problems, just to list a few, include the difficulty in isolating long hydrophobic peptides, heterogeneity of the digestion product from the transmembrane segments of Rho, complete decomposition of retinyl peptides by elimination of the retinyl carbocation moiety, and lack of quantitative preservation of the geometric isomers of the retinoids. Because of the importance of these fundamental biochemical processes in the physiology of mammalian vision, we focused on developing robust and direct methods to study them, thereby furthering our understanding of these biochemical reactions.
The highly robust proteases, proteinase K and pronase, were effective in the digestion of Nε-retinyl-opsin for subsequent MS analysis. Proteinase K unequivocally produced peptide profiles unique to Lys296, whereas pronase digested Nε-retinyl-opsin completely, yielding the definitive retinyl-Lys296 adduct. The complete proteolysis of iPrOH/NaBH4-treated Rho and Rho* by pronase and subsequent LC-MS/MS analyses enabled the determination of the cis-trans isomeric composition of the RSB. Thereby, Rho photochemistry was captured, with the observation of a dominant Nε-11-cis-retinyl-Lys from Rho and a dominant Nε-all-trans-retinyl-Lys from Rho*. Thus, our methods captured the 11-cis to all-trans photoisomerization characteristic of Rho and the precise Lys296 residue that formed visual pigment via SB condensation with 11cRAL chromophore, demonstrating the ability to directly follow the course of Rho photochemistry.
MS analyses of Nε-retinyl-peptides and Nε-retinyl-Lys revealed the tendency of the retinyl group to cleave as a retinyl cation during source fragmentation, likely because of the resonance stabilization of the retinyl cation and the formation of a stable peptide or Lys leaving group upon protonation of the ε-amine by formic acid in the mobile phase. With other sites of protonation, residual precursor molecules can also be readily detected, but with retinyl esters or retinol, the retinyl cation is the predominant species observed on ESI-MS full scan (27–30). The partial source fragmentation of the precursor molecule consistently yields a product retinyl cation and a product peptide or Lys ion, which can be further characterized by CID fragmentation for sequence determination. We expect that our method can be extended to other molecules that possess a retinyl moiety as a fluorophore or a tag, together with a variety of protonatable functional groups.
After characterizing the photochemistry of Rho, we tracked Rho* hydrolysis directly by monitoring the opsin-SB–bound and –released atRAL over time. The rate of hydrolysis was faster with Rho* in ROS membranes than in DDM detergent micelles, demonstrating how the nature of the lipid environment can alter molecular dynamics. Our measured EA of 17.7 kcal/mol for Rho* hydrolysis in ROS membranes at pH 7.4 is in agreement with early measurements by photocalorimetry, which produced values of 22.1 kcal/mol at pH 8.0 and 11.7 kcal/mol at pH 5.4 for Rho* in ROS membranes (31). The lower EA of hydrolysis at pH 5.4 also aligned with the results of our Rho* hydrolysis experiments with ROS, which showed hydrolysis occurring to a greater extent at lower pH, thereby supporting the longstanding notion that hydrolysis proceeds via the MII signaling state of Rho. Our computational results further support the favorability of hydrolysis proceeding via the deprotonated SB of MII over the protonated SB of MI or MIII. Our computational studies confirmed the key influence of the protonation states of the Glu113 and Glu181 residues on the energetic landscape of the reaction, with the hydrolysis process being exothermic only at lower pH.
The formation of N-ret-PE also displayed pH dependency, with faster rates and higher steady states as pH increased, likely because of a greater proportion of free PE available for nucleophilic attack on the C15 aldehyde carbon of atRAL to initiate the SB condensation reaction. Therefore, pH changes had opposite effects on the rate of Rho* hydrolysis versus the rate of N-ret-PE formation; at lower pH, there was more Rho* hydrolysis but less N-ret-PE formation. Furthermore, the overall rate of N-ret-PE formation from released atRAL was slower than the rate of Rho* hydrolysis at low pH (khyd = 0.001537 s−1 vs. kNretPE = 0.001062 s−1). Prior to our examination of the kinetics of N-ret-PE formation, we hypothesized that PE could potentially play a role in facilitating the hydrolysis of the atRSB of Rho* by serving as a proximal acceptor of the released atRAL. However, based on our pH studies of the Rho* hydrolysis and the N-ret-PE condensation reaction, the rate of hydrolysis did not appear to be influenced by the subsequent formation of N-ret-PE with the atRAL from Rho* hydrolysis. Instead, the hydrolysis of the atRSB relied on the protein dynamics of Rho, reflecting the dependency of the hydrolysis kinetics on pH-driven changes in the metarhodopsin equilibrium.
At physiological pH, the rate of N-ret-PE formation was an order of magnitude faster than the rate of Rho* hydrolysis (khyd = 0.001479 s−1 vs. kNretPE = 0.01819 s−1). The observations of N-ret-PE formation from exogenous or Rho*-derived atRAL and their faster rate of formation as compared with Rho* hydrolysis are significant to our understanding of the formation of toxic bisretinoids that are abundant with Stargardt disease and age-related macular degeneration (19, 32, 33). This notion is consistent with hydrolysis being rate limiting such that soon after atRAL is released, it can be captured by PE.
Furthermore, our results show that exogenous or Rho* hydrolysis–derived atRAL react reversibly with PE to reach a steady-state mole fraction of N-ret-PE of 0.4. Therefore, after intense light exposure to the retina, close to half of all the resultant atRAL released from Rho* has the propensity to form N-ret-PEs, which serve as the starting point to drive the formation of bisretinoids. Although bisretinoids were not observed under the conditions and timescale of our studies of Rho* hydrolysis, their formation could be facilitated by any number of factors, including delayed reduction (e.g., by intense illumination and depletion of NADPH), attenuated phagocytosis, and genetic alterations of pertinent visual cycle proteins and enzymes (34, 35). Longer durations of light exposure and incubation periods with released atRAL or accelerated supply of 11cRAL that is constitutively supplied by the retinal pigment epithelium could also lead to bisretinoid formation (36). However, N-ret-PE formation was recently shown to not always lead to a pathologic outcome but could very well serve as a nonclassical means of chromophore regeneration via production of 9-cis or 11-cis retinal from N-ret-PE photoisomerization (18).
Another physiological role that we hypothesized for N-ret-PEs was as a shuttle for atRAL delivery to RDHs at the membrane periphery. However, RDHs were found to exhibit activity selectively on free atRAL, with little effect on the proportion of atRAL sequestered as N-ret-PE adducts. Following Rho* hydrolysis, RDH activity led to conversion of nearly all of the released atRAL to atROL and thereby near complete attenuation of N-ret-PE formation. Such an observation highlights the significance of RDHs in the prevention of the formation and accumulation of N-ret-PEs after light exposure. Our method using NaBD4 therefore allowed us to capture outer-segment membrane-bound RDH activity and can be broadly used to study other redox biochemical reactions involving functional groups, such as aldehydes or ketones, that are susceptible to borohydride/deuteride chemistry. In short, transformation of atRAL via the visual cycle is essential for sustainability of our vision. These reactions are carried out in the membrane environment and involve very active aldehydes (atRAL and 11cRAL) that undergo several enzymatic and nonenzymatic transformations. Using rapid quantitative chemical and analytical methods, we are able now to follow each step quantitatively with preservation of stereospecificity.
Recently, in a collaboration with Chen et al. (37), we described a partially overlapping method that used state-of-the-art native MS to investigate the first steps of the visual cycle and phototransduction in ROS membranes. Native MS enables real-time monitoring of events that cause changes in molecular mass, such as SB formation and retinal release upon hydrolysis of Rho; formation of retinyl-phospholipid adducts; protein-protein, protein-ligand, and protein-lipid interactions; and so on. The method described in the current study requires simpler instrumentation, which is available in many more laboratories and institutions. In addition, the method in this report allows for nonvolatile buffers and salts, including NADPH, which is required for reduction of atRAL by RDHs. Furthermore, the isomeric composition of the retinylidene adduct of opsin could be determined, in addition to identifying the specific Lys residue where the SB adduct occurs. Thus, the current method nicely complements the capabilities of the native MS method for investigating the visual cycle.
In this study, we captured and quantified the kinetics and energetics of the critical steps of our vision. We demonstrate that the rate-limiting step is hydrolytic release of atRAL from Rho*, and this reaction is dependent on the presence of native lipids. This methodology should be applicable to investigate the photochemistry occurring with other retinylidene-opsins, including cone opsins, melanopsin, channel Rho, and nonvisual pigments, like RGR or peropsin. Furthermore, our method of capturing bound retinal chromophore could also translate to detecting whether chromophore analogs successfully form an SB linkage in the chromophore binding pocket of various opsin proteins.
Materials and Methods
Proteinase K Digestion of Rho from ROS Membranes.
ROS membranes from InVision Bioresources were prepared from fresh bovine retinas using sucrose gradient isolation as described previously and washed with hypotonic buffer (20 mM Hepes; pH 7.4) (38). In the dark room under dim red light, one part ROS membranes (2 mg/mL or 50 μM Rho) was treated with three parts saturated solution of NaBH4 granules (∼100 μM) in cold iPrOH for immediate reduction of the SB and isolation of Rho by precipitation. Cold ethanol (EtOH) can also be used in place of iPrOH. After centrifugation at 20,000 × g to pellet the protein precipitate, the pellet was washed with cold methanol (MeOH) followed by cold water. The protein pellet was then resuspended in proteinase K buffer (4 M urea, 100 mM bis-Tris propane [BTP; pH 7.8], 100 mM CaCl2). Subsequently, proteinase K (20 mg/mL) was added at ∼10× the weight of the Rho substrate. Then, the digestion mixture was incubated at room temperature for 24 h, with occasional mixing by brief gentle spinning on a vortex mixer. The digest was passed through a BioPureSPN C18 spin column for desalting. The column was washed with 20% acetonitrile (ACN) in water with 0.1% formic acid (FA), and peptides were eluted using 60% ACN. The eluted Nε-retinyl-peptide products were separated using a Dionex UHPLC with an XBridge C18 column and a 40-min gradient of 20 to 60% ACN in water with 0.1% FA at a flow rate of 0.3 mL/min. Nε-retinyl-peptide products were detected by UHPLC absorbance at 330 nm and identified by MS/MS with CID fragmentation using an LTQ XL spectrometer.
Pronase Digestion of Rho from ROS Membranes.
In the dark room under dim red light, one part ROS membranes (2 mg/mL or 50 μM Rho) was treated with three parts saturated solution of NaBH4 granules in cold iPrOH (∼100 μM) for immediate reduction of the SB and isolation of Rho by precipitation. After centrifugation at 20,000 × g to pellet the protein precipitate, the pellet was washed with cold MeOH followed by cold water. The protein pellet was then resuspended in pronase buffer (100 mM BTP [pH 7.8], 100 mM CaCl2). Subsequently, pronase (20 mg/mL in water) was added at ∼10× the weight of Rho substrate. Then, the digestion mixture was incubated at 8 to 10 °C for 24 h with gentle agitation using a shaker. The digest was passed through a BioPureSPN C18 spin column for desalting. The column was washed with 20% can in water, and digest products were eluted using 60% ACN. The eluted Nε-retinyl-Lys products were separated using a Dionex UHPLC with an XBridge C18 column and a 30-min gradient of 20 to 50% ACN in water with 0.1% FA at a flow rate of 0.3 mL/min. Nε-retinyl-Lys products were detected by UHPLC absorbance at 330 nm and identified by MS/MS with CID fragmentation using an LTQ XL spectrometer.
Separation and Identification of N-ret-PE Species from ROS.
A suspension of ROS membranes (2 mg/mL or 50 μM Rho) in 20 mM Hepes-buffered solution (at pH 5.6, 7.4, or 8.3) was either supplied with 2.5 molar equivalents of atRAL relative to the amount of Rho or illuminated to supply atRAL via Rho* hydrolysis. For illumination, a 565-nm fiber light set to an intensity of 625 μW was applied to ROS for 10 s; three parts of a saturated solution of NaBH4 (∼100 μM) in cold iPrOH was added to one part ROS membranes to solubilize lipids while immediately halting hydrolytic release of atRAL from Rho* and the progression of SB formation between atRAL and PE. After centrifugation at 20,000 × g to pellet the denatured membrane proteins, the solubilized lipids within the supernatant were analyzed by LC-MS/MS. N-retinyl-PEs were separated using an XBridge C18 column and a mobile phase consisting of water with 0.1% FA (solvent A) and MeOH with 0.1% FA (solvent B). The conditions were as follows: a 10-min gradient of 95 to 100% solvent B, followed by 20 min of isocratic 100% solvent B at a flow rate of 0.3 mL/min. The separated N-retinyl-PE products were detected by UHPLC absorbance at 330 nm and identified by MS/MS with CID fragmentation using an LTQ XL spectrometer.
Quantum Chemical and Kinetic Analyses.
Details of the quantum chemical and kinetic analyses of the Rho* hydrolysis can be found in the SI Appendix. Details of the analyses of N-ret-PE formation and RDH activity can also be found in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Gregory Tochtrop and Vladimir J. Kefalov and members of the P.D.K. and K.P. laboratories for helpful comments on this project. This research was supported in part by NIH Research Grant EY030873 (National Eye Institute) to K.P. and NIH Training Grants 1F30EY033659-01 and T32-GM08620 to J.D.H. The research was also supported by grants from the Department of Veterans Affairs (I01BX004939) and the NSF (CHE-2107713) to P.D.K. The authors acknowledge support to the Gavin Herbert Eye Institute at the University of California, Irvine from an unrestricted grant from Research to Prevent Blindness and from NIH core grant P30 EY034070. M.A.K. acknowledges funding from the European Union Horizon 2020 research and innovation program under Marie Skłodowska-Curie Grant Agreement 847413. This work has been published as part of an international cofinanced project funded by the program of the Polish Minister of Science and Higher Education entitled PMW in the years 2020 to 2024 (Agreement 5005/H2020-MSCA-COFUND/2019/2). A.K. acknowledges support from the National Science Centre, Poland (Grant 2020/39/B/ST4/01952).
Footnotes
Reviewers: T.S., The Rockefeller University; J.S., Columbia University Irving Medical Center.
Competing interest statement: K.P. is the Chief Scientific Officer of Polgenix, Inc., and consultant of Prime Medicine and Editas Medicine.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2213911119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Falk G., Fatt P., Distinctive properties of the lamellar and disk-edge structures of the rod outer segment. J. Ultrastruct. Res. 28, 41–60 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Yoshizawa T., Wald G., Pre-lumirhodopsin and the bleaching of visual pigments. Nature 197, 1279–1286 (1963). [DOI] [PubMed] [Google Scholar]
- 3.Wright W. E., Brown P. K., Wald G., Orientation of intermediates in the bleaching of shear-oriented rhodopsin. J. Gen. Physiol. 62, 509–522 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ovchinnikov YuA, Rhodopsin and bacteriorhodopsin: Structure-function relationships. FEBS Lett. 148, 179–191 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Emeis D., Kühn H., Reichert J., Hofmann K. P., Complex formation between metarhodopsin II and GTP-binding protein in bovine photoreceptor membranes leads to a shift of the photoproduct equilibrium. FEBS Lett. 143, 29–34 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K., G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht S., Haig C., Chase A. M., The influence of light adaptation on subsequent dark adaptation of the eye. J. Gen. Physiol. 20, 831–850 (1937). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schädel S. A., et al. , Ligand channeling within a G-protein-coupled receptor. The entry and exit of retinals in native opsin. J. Biol. Chem. 278, 24896–24903 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiser P. D., Palczewski K., Pathways and disease-causing alterations in visual chromophore production for vertebrate vision. J. Biol. Chem. 296, 100072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrens D. L., Khorana H. G., Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 270, 5073–5076 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Janz J. M., Farrens D. L., Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J. Biol. Chem. 279, 55886–55894 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Bartl F. J., Vogel R., Structural and functional properties of metarhodopsin III: Recent spectroscopic studies on deactivation pathways of rhodopsin. Phys. Chem. Chem. Phys. 9, 1648–1658 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Sakmar T. P., Structure of rhodopsin and the superfamily of seven-helical receptors: The same and not the same. Curr. Opin. Cell Biol. 14, 189–195 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Sakmar T. P., Menon S. T., Marin E. P., Awad E. S., Rhodopsin: Insights from recent structural studies. Annu. Rev. Biophys. Biomol. Struct. 31, 443–484 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Knowles A., Pepe I. M., Can metarhodopsin I activate rod outer segment phosphodiesterase? Cell Biophys. 13, 43–53 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Cordes E. H., Jencks W. P., The mechanism of hydrolysis of Schiff bases derived from aliphatic amines. J. Am. Chem. Soc. 85, 2843–2848 (2002). [Google Scholar]
- 17.Beharry S., Zhong M., Molday R. S., N-retinylidene-phosphatidylethanolamine is the preferred retinoid substrate for the photoreceptor-specific ABC transporter ABCA4 (ABCR). J. Biol. Chem. 279, 53972–53979 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Kaylor J. J., et al. , Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate. Nat. Commun. 8, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparrow J. R., Wu Y., Kim C. Y., Zhou J., Phospholipid meets all-trans-retinal: The making of RPE bisretinoids. J. Lipid Res. 51, 247–261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futterman S., Rollins M. H., The catalytic isomerization of all-trans-retinal to 9-cis-retinal and 13-cis-retinal. J. Biol. Chem. 248, 7773–7779 (1973). [PubMed] [Google Scholar]
- 21.van Deenen L. L., Chemistry of phospholipids in relation to biological membranes. Pure Appl. Chem. 25, 25–56 (1971). [DOI] [PubMed] [Google Scholar]
- 22.Matthews R. G., Hubbard R., Brown P. K., Wald G., Tautomeric forms of metarhodopsin. J. Gen. Physiol. 47, 215–240 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada T., et al. , Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 5982–5987 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R., Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lüdeke S., et al. , The role of Glu181 in the photoactivation of rhodopsin. J. Mol. Biol. 353, 345–356 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Vogel R., Siebert F., Zhang X. Y., Fan G., Sheves M., Formation of meta III during the decay of activated rhodopsin proceeds via Meta I and not via Meta II. Biochemistry 43, 9457–9466 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Van Breemen R. B., Huang C. R., High-performance liquid chromatography-electrospray mass spectrometry of retinoids. FASEB J. 10, 1098–1101 (1996). [DOI] [PubMed] [Google Scholar]
- 28.van Breemen R. B., et al. , Development of a method for quantitation of retinol and retinyl palmitate in human serum using high-performance liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry. J. Chromatogr. A 794, 245–251 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Wingerath T., Kirsch D., Spengler B., Kaufmann R., Stahl W., High-performance liquid chromatography and laser desorption/ionization mass spectrometry of retinyl esters. Anal. Chem. 69, 3855–3860 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Rocchi S., et al. , Quantitative profiling of retinyl esters in milk from different ruminant species by using high performance liquid chromatography-diode array detection-tandem mass spectrometry. Food Chem. 211, 455–464 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Cooper A., Rhodopsin photoenergetics: Lumirhodopsin and the complete energy profile. FEBS Lett. 123, 324–326 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Wu Y., Yanase E., Feng X., Siegel M. M., Sparrow J. R., Structural characterization of bisretinoid A2E photocleavage products and implications for age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 107, 7275–7280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sparrow J. R., et al. , The bisretinoids of retinal pigment epithelium. Prog. Retin. Eye Res. 31, 121–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Ueda K., Riera M., Kim H. J., Sparrow J. R., Bisretinoids mediate light sensitivity resulting in photoreceptor cell degeneration in mice lacking the receptor tyrosine kinase Mer. J. Biol. Chem. 293, 19400–19410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda A., Golczak M., Maeda T., Palczewski K., Limited roles of Rdh8, Rdh12, and Abca4 in all-trans-retinal clearance in mouse retina. Invest. Ophthalmol. Vis. Sci. 50, 5435–5443 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer N. P., Thompson D. A., Koutalos Y., Relative contributions of all-trans and 11-cis retinal to formation of lipofuscin and A2E accumulating in mouse retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 62, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S., et al. , Capturing a G-protein coupled receptor signalling cascade across a native membrane. Nature 604, 384–390 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papermaster D. S., Preparation of retinal rod outer segments. Methods Enzymol. 81, 48–52 (1982). [DOI] [PubMed] [Google Scholar]
- 39.McCaffery P., et al. , Retinoid quantification by HPLC/MS(n). J. Lipid Res. 43, 1143–1149 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Khaksari M., Mazzoleni L. R., Ruan C., Kennedy R. T., Minerick A. R., Data representing two separate LC-MS methods for detection and quantification of water-soluble and fat-soluble vitamins in tears and blood serum. Data Brief 11, 316–330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.