Abstract
Novel bio-therapeutic agents that harness the properties of small, non-coding nucleic acids hold great promise for clinical applications. These include antisense oligonucleotides that inhibit messenger RNAs, microRNAs (miRNAs), or long non-coding RNAs; positive effectors of the miRNA pathway (short interfering RNAs and miRNA mimics); or small RNAs that target proteins (i.e. aptamers). These new therapies also offer exciting opportunities for cardiovascular diseases and promise to move the field towards more precise approaches based on disease mechanisms. There have been substantial advances in developing chemical modifications to improve the in vivo pharmacological properties of antisense oligonucleotides and reduce their immunogenicity. Carrier methods (e.g. RNA conjugates, polymers, and lipoplexes) that enhance cellular uptake of RNA therapeutics and stability against degradation by intracellular nucleases are also transforming the field. A number of small non-coding RNA therapies for cardiovascular indications are now approved. Moreover, there is a large pipeline of therapies in clinical development and an even larger list of putative therapies emerging from pre-clinical studies. Progress in this area is reviewed herein along with the hurdles that need to be overcome to allow a broader clinical translation.
Keywords: Antisense oligonucleotides, Cardiovascular disorders, MicroRNAs, RNA therapeutics, Short interfering RNAs
Graphical Abstract
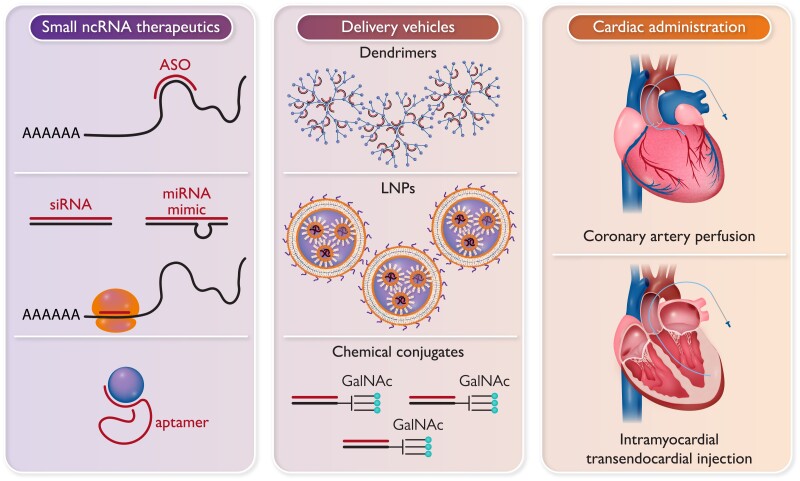
Graphical abstract.
Types of small non-coding RNA therapeutics (left panel); some of the methods for in vivo delivery (centre); and routes for administration to the heart (right). ASO, antisense oligonucleotides; GalNAc, N-acetylgalactosamine; LNPs, lipid nanoparticles; miRNAs, microRNAs; siRNA, short interfering RNAs.
Clinical need for new cardiovascular therapies
Cardiovascular disease (CVD) remains the leading cause of death globally despite major advances in prevention and invasive treatments. Symptomatic ischaemic heart disease (IHD), sudden death, and heart failure (HF) in survivors of acute myocardial infarction (MI) remain major problems. Indeed, much CVD, such as IHD, cardiomyopathy, HF, and peripheral vascular disease, is ultimately relatively intractable. Current therapies generally only delay disease progression. With few exceptions, they target a limited number of disease pathways identified decades ago. Furthermore, the overall approach is largely ‘one-size-fits-all’ with relatively little stratification, e.g. by disease stage or mechanism.
Conceptual progress in understanding the pathomechanisms of CVD, including appreciation of the role of non-coding RNAs (ncRNAs) in these processes, and technological advances, now permit the development of an innovative class of therapies based on small nucleic acids.
Here, we discuss the different types of ncRNA-based therapies (Graphical Abstract), emerging clinical and advanced pre-clinical data in CVD, and the challenges that need to be overcome to maximize the potential of this exciting new therapeutic modality.
Types of small, non-coding nucleic acid therapeutics
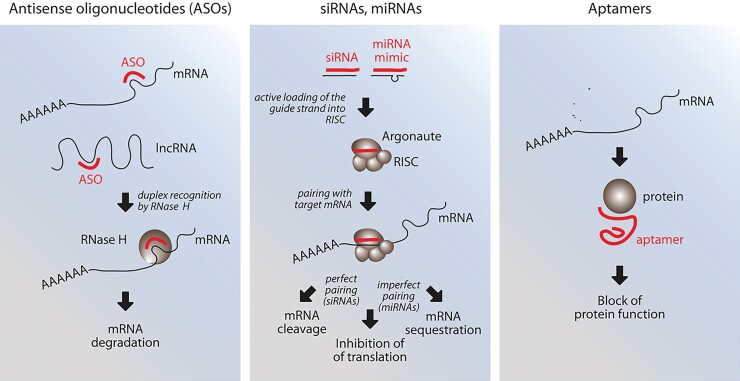
Small non-coding nucleic acid therapeutics that have reached clinical application fall into three categories (Figure 1): (i) short antisense oligonucleotides (ASOs) that pair with and inhibit messenger RNAs (mRNAs), microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs); (ii) positive effectors of the RNA interference (RNAi) pathway [small interfering RNAs (siRNAs), miRNAs]; and (iii) conformational small RNAs (aptamers) that target proteins. Other therapeutic ncRNAs (e.g. miRNA sponges), lncRNAs, and circRNAs, which have longer dimensions and are less advanced in clinical development, or CRISPR guide RNAs for gene editing are not discussed.
Figure 1.
Schematic representation of the three main classes of small, non-coding RNA therapeutics. Not shown here for simplicity is also the possibility of using antisense oligonucleotides to target circular RNAs (reviewed in Holdt et al.1). ASO, antisense oligonucleotide; lncRNA, long non-coding RNA; mRNA, messenger RNA; RISC, RNA-induced silencing complex.
Antisense oligonucleotides
Antisense oligonucleotides are usually 17–22 nt long and chemically modified to decrease degradation by endogenous nucleases and improve cellular uptake. Pairing with the antisense cellular RNA is followed by steric inhibition of target RNA function and/or induction of target RNA degradation.
To date, nine ASOs have been approved in Europe and the USA (Table 1). The first of these, fomivirsen (https://www.accessdata.fda.gov/drugsatfda_docs/nda/98/20961_vitravene.cfm), was a 21-mer ASO against cytomegalovirus (CMV) for the treatment of CMV retinitis; its commercialization ended in 2002 due to the development of more effective antivirals. Three of the approved ASOs have a specific cellular mRNA target [mipomersen against apolipoprotein B (ApoB), inotersen against transthyretin, and volanesorsen against ApoC3]. The remaining five ASOs are alternative splicing modulators of the dystrophin (eteplirsen, golodirsen, viltolarsen, and casimersen) or the SMN2 (nusinersen) pre-mRNAs. Recent reviews have discussed the development of these products.2,3 None of these ASOs target miRNAs, lncRNAs or circRNAs, but at least 10 other ASOs are in advanced Phase 2/3 clinical development,3 including molecules targeting miRNAs.
Table 1.
Antisense oligonucleotides and short interfering RNAs approved by the European Medicines Agency and/or the Food and Drug Administration and their main characteristics
| Product (commercial name; developer/manufacturer) | Length | Modifications | Vehicle | Route of administration | Indication | Target organ | Target gene and mechanism | Year of approval |
|---|---|---|---|---|---|---|---|---|
| Antisense oligonucleotides (ASOs) | ||||||||
| Fomivirsen (Vitravene; Isis Pharmaceuticals, Novartis) | 21-mer | PS | None | Intravitreal | CMV retinitis | Eye | CMV IE-2 mRNA | 1998 (FDA), 1999 (EMA); 2002 withdrawn |
| Mipomersen (Kynamro; Ionis Pharmaceuticals, Kastle Therapeutics) | 20-mer | PS, 2′-MOE, GapmeR | None | Subcutaneous | Familiar hypercholesterolaemia (FH) | Liver | Apolipoprotein B (ApoB) mRNA | 2013 (FDA); 2019 withdrawn |
| Nusinersen (Spinraza; Ionis Pharmaceuticals, Biogen) | 18-mer | PS, 2′-MOE | None | Intrathecal | Spinal muscular atrophy | Survival of motoneuron 2 (SMN2) pre-mRNA splicing (exon 7 inclusion) | 2017 (EMA), 2016 (FDA) | |
| Eteplirsen (Exondys 51, Sarepta Therapeutics) | 30-mer | PMO | None | Intravenous | Duchenne muscular dystrophy (DMD) | Skeletal muscle | Dystrophin pre-mRNA splicing (exon 51 skipping) | 2016 (FDA) |
| Inotersen (Tesgedi; Ionis Pharmaceuticals, Akcea Therapeutics) | 20-mer | PS, 2′-MOE, GapmeR | None | Subcutaneous | Hereditary transthyretin amyloidosis | Liver | Transthyretin (TTR) mRNA | 2018 (EMA), 2018 (FDA) |
| Golodirsen (Vyondys 53; Sarepta Therapeutics) | 25-mer | PMO | None | Intravenous | Duchenne muscular dystrophy (DMD) | Muscle | Dystrophin pre-mRNA splicing (exon 53 skipping) | 2019 (FDA) |
| Viltolarsen (Viltepso, NS Pharma) | 21-mer | PMO | None | Intravenous | Duchenne muscular dystrophy (DMD) | Muscle | Dystrophin pre-mRNA splicing (exon 53 skipping) | 2020 (FDA) 2020 (EMA) |
| Volanesorsen (Waylivra; Ionis Pharmaceuticals, Akcea Therapeutics) | 20-mer | PS, 2′-MOE, GapmeR | None | Subcutaneous | Familiar chylomicronaemia syndrome (FCS) | Liver | Apolipoprotein C3 (ApoC3I) mRNA | 2019 (EMA) |
| Casimersen (Amondys 45; Sarepta Therapeutics) | 22-mer | PMO | None | Intravenous | Duchenne muscular dystrophy (DMD) | Muscle | Dystrophin pre-mRNA splicing (exon 45 skipping) | 2021 (FDA) |
| Small interfering RNAs (siRNAs) | ||||||||
| Patisiran (Onpattro; Anylam Pharmaceuticals) | 21-nt ds | 2′-O-Me | SNALP LNP | Intravenous | Hereditary transthyretin amyloidosis | Liver | Transthyretin mRNA | 2018 (EMA), 2019 (FDA) |
| Givosiran (Givlaari; Anylam Pharmaceuticals) | 21-nt ds | PS, 2′-O-Me, 2′-F, GalNAc-conjugated | None | Subcutaneous | Acute hepatic porphyria (AHP) | Liver | Delta aminolevulinic acid synthase 1 mRNA | 2020 (EMA), 2019 (FDA) |
| Inclisiran (Leqvio; Novartis Pharmaceuticals) | 22-nt ds | PS, 2′-O-Me, 2′-F, GalNAc-conjugated | None | Subcutaneous | Primary hypercholesterolaemia or mixed dyslipidaemia | Liver | Proprotein convertase subtilisin/kexi)n type 9 (PCSK9) mRNA | 2020 (EMA) 2021 (FDA) |
| Lumasiran (Oxlumo; Anylam Pharmaceuticals) | 21-nt ds | PS, 2′-O-Me, 2′-F, GalNAc-conjugated | None | Subcutaneous | Primary hyperoxaluria type 1 (PH1) | Liver | Hydroxyacid oxidase-1 mRNA | 2020 (EMA), 2020 (FDA) |
ASO, antisense oligonucleotide; ds, double stranded; GalNAc, N-acetylgalactosamine; PMO, phosphoroamidate morpholino oligomer; PS, phosphorothioate modification; siRNA, short interfering RNA.
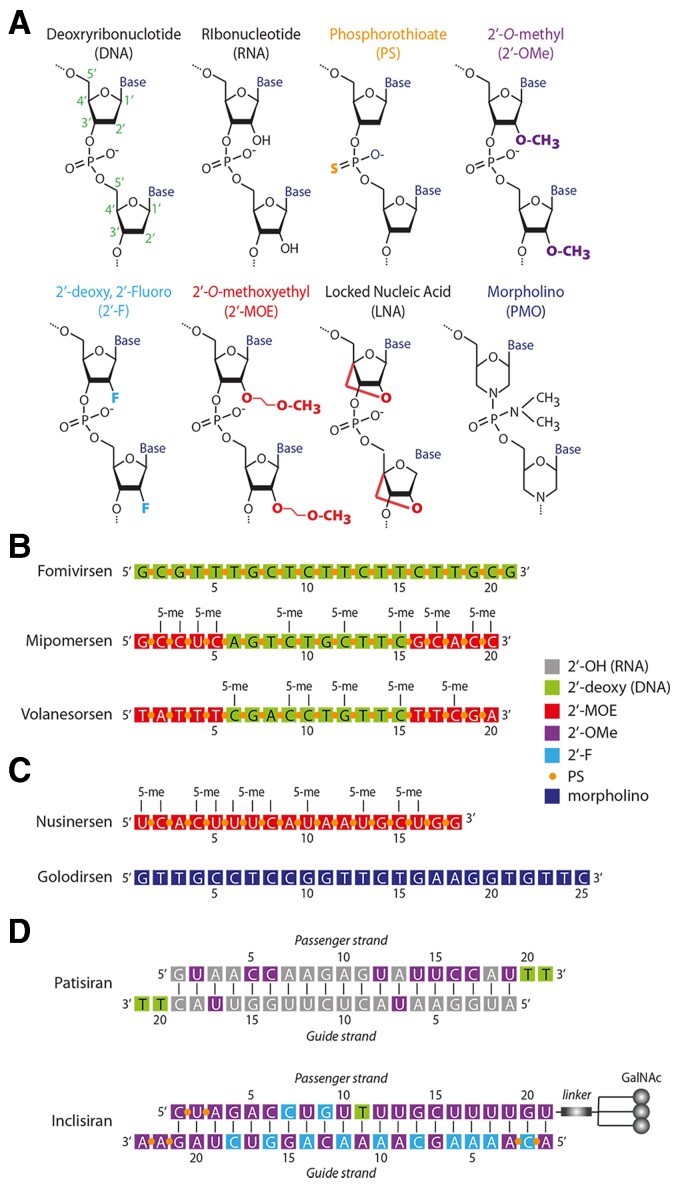
The original idea of therapeutic ASOs dates back three decades,4 with over 100 Phase 1 studies performed and 25% reaching Phase 2/3.5,6 Clinical success, however, was only rendered possible by the progressive identification of chemical modifications that improve stability and reduce immune recognition. Classic DNA ASOs undergo DNA:RNA pairing but are then relatively rapidly degraded by RNAse H. This problem can be circumvented by substituting non-bonding oxygen atoms in the phosphate groups of ASOs with sulphur atoms (phosphorothioates, PS) or introducing chemical modifications in the 2′ position of the sugar molecule of nucleotides (e.g. 2′-O-methyl-, 2′-O-methoxy-ethyl-, or 2′′-fluoro-nucleotides; 2′-OMe, 2′-MOE, or 2′-F, respectively; Figure 2A; reviewed in Lennox and Behlke7 and van Rooij and Olson8).
Figure 2.
Chemical modifications and structure of selected, clinically approved, non-coding RNAs. (A) Chemical modification of ribonucleotides. The abbreviations commonly used to indicate these modified nucleotides are indicated. (B) Chemical structure of three clinically approved antisense oligonucleotides. Mipomersen and volanesorsen have a typical GapmeR configuration, in which the first five nucleotides at both the 5′ and 3′ ends are modified, while the central region is formed of oligodeoxynucleotides to allow recruitment of RNAse H to the central target RNA:antisense oligonucleotide DNA duplex. (C) Chemical structure of the two splicing modulators nusinersen and golodirsen. The latter is formed of morpholino (phosphoroamidate morpholino oligomer) monomers, same as eteplirsen, vitolarsen and casimersen. (D) Chemical structure of two short interfering RNAs. Patisiran is formulated in a lipid nanoparticle, while, in the case of inclisiran, three GalNAc moieties are attached through a linker to the 3′ end of the siRNA passenger strand. Both are designed to target the liver.
An effective modification to increase stability of ASOs is the inclusion of RNA nucleotides in which the ribose moiety contains an extra bond between the 2′ oxygen and 4′ carbon (2′-O,4′-C-methylene bridge).9 These locked nucleic acid (LNA) nucleotides confer remarkable thermal stability against enzymatic degradation while maintaining base-pairing specificity.10 However, for mRNA and larger ncRNA targeting, substrate duplexes with either LNA or 2′-O-alkylated RNA nucleotides are unable to recruit RNAse H and thus inhibition can only occur through steric hindrance.11,12 This issue can be solved using a GapmeR approach, where a central continuous stretch of RNase H-recruiting nucleotides (e.g. PS DNA) is flanked by modified nucleotides. This arrangement improves ASO stability while still permitting RNAse H recruitment to the central DNA–RNA duplex for degradation. A gap of at least seven to eight DNA nucleotides is necessary for activation of RNAse H.13–15 The GapmeR design also permits the use of shorter ASOs (typically, 16-mers),14–16 with increased target knockdown efficacy, the possibility of targeting several miRNA family members sharing the same seed sequence and neighbouring regions, and most relevant, reduced innate immune activation.
The first small nucleic acid therapeutic to be approved (fomivirsen) was a DNA ASO in which all bonds were phosphorothiates. A significantly more effective GapmeR design is present in mipomersen (this drug, however, showed significant liver toxicity and was withdrawn), inotersen, and volanesorsen (Figure 2B).
The modulation of pre-mRNA splicing and the inhibition of miRNA processing do not require RNAse H-mediated duplex degradation. In the case of splicing, steric block ASOs selectively exclude or retain a specific exon (exon skipping and inclusion, respectively) by masking a splicing signal in the pre-mRNA or to affect RNA structure thus preventing access to cis splicing signals by the spliceosome (reviewed in Neil et al.17). Five steric block ASOs that modulate splicing have reached clinical approval. Nusinersen is an 18-mer ASO for spinal muscular atrophy formed of 2′-MOE nucleotides linked by PS bonds. Other approved ASOs are for Duchenne muscular dystrophy and consist of phosphorodiamidate morpholino oligomers in which the nitrogen bases are connected to a morpholino ring rather than to the deoxyribose ring and a phosphorodiamidic bond substitutes the natural phosphodiester bond (Figure 2A). A schematic representation of eteplirsen, one of these ASOs, alongside that of nusinersen, is shown in Figure 2C.
RNA interference therapeutics (microRNAs and small interfering RNAs)
Endogenous miRNAs are double-stranded RNA molecules of 19–23 nt with 2 nt overhangs at their 5′ ends, which pair with cellular mRNAs and block their function by inducing mRNA degradation or blocking translation18,19 (Figure 1). As pairing with the mRNA target only requires short regions of homology (in particular, a 7 nt seed sequence at position +2/+8 of the miRNA), each miRNA can target tens or hundreds of different transcripts. The delivery of double-stranded RNA molecules having the same structure as natural miRNAs but with perfect complementarity with a given target mRNA (siRNAs) intercepts and exploits the RNAi pathway to obtain more specific, single gene down-regulation.
The first clinically approved siRNA was patisiran in 2018, against the transthyretin mRNA for hereditary transthyretin amyloidosis (HTT-amyloid).20 This was followed by givosiran, an siRNA against the delta aminolevulinic acid synthase 1 mRNA for acute hepatic porphyria,21 inclisiran, an siRNA against PCSK9 for familiar hypercholesterolaemia,22 and lumasiran, an siRNA against the hydroxy acid oxidase 1 mRNA for primary hyperoxaluria Type 123 (Table 1). The structures of patisiran and inclisiran are shown in Figure 2D. As siRNAs need to interact with the cellular RISC complex to exert their function, they usually tolerate less chemical modification than ASOs, in particular in their guide strand (the one that pairs with the mRNA targets).
No miRNA has so far reached clinical approval, yet these molecules have significant therapeutic appeal for several reasons. First, endogenous miRNAs are naturally occurring molecules with a precise and physiological mechanism of action. Second, having evolved to target several different mRNAs, miRNAs can elicit complex yet defined phenotypes [e.g. cardiomyocyte (CM) proliferation24 or transdifferentiation25]. Third, the human miRNome has relatively low complexity (1973 hairpin precursor miRNAs, corresponding to 2675 human mature sequences annotated in miRBase v.22.126); therefore, a synthetic human miRNA mimic library can be screened with relative ease for therapeutic leads. Cell-produced mature miRNAs are single-stranded RNA molecules generated from a hairpin that folds back to form partially double-stranded regions. In principle, either hairpin strand can be used by RISC for RNA interference. In contrast, miRNA mimics have the same sequence as the desired miRNA strand, while the complementary passenger strand, which usually is a perfect complement to the guide strand, is chemically modified to prevent RISC loading.27 Three miRNA mimics have been tested in cancer trials. The miR-34 mimic, MRX34, for advanced solid tumours28 showed serious immune-mediated adverse events.29 MesomiR-1 is a miR-16 mimic for patients with mesothelioma,30 while remlarsen is a miR-29 mimic administered via intradermal injection for treatment of keloid.31 Several other therapeutic miRNA applications are likely to follow.
Aptamers
The SELEX (systematic evolution of ligands by exponential enrichment) method developed in the 1990s permits selection of RNAs that specifically bind proteins or small organic molecules, starting from libraries of oligoribonucleotides with random sequence.32–34 The selected RNA molecules (or aptamers) exploit the secondary structure of nucleic acids rather than sequence complementarity for binding (Figure 1). Aptamers are usually short (25–40 nt) RNA segments that bind their targets at relatively high affinity.35
Pegaptanib, a 28-mer modified RNA aptamer conjugated with two polyethylene glycol (PEG) molecules, was the first aptamer to reach clinical approval in 2004.36 It binds the 165aa isoform of the vascular endothelial growth factor (VEGF) and has an indication by intravitreal injection in age-related neovascular macular degeneration. After initial clinical success, however, pegaptanib has faced competition with the anti-VEGF monoclonal antibodies ranibizumab and bevacizumab.37
To date, eight other RNA aptamers and five DNA aptamers have undergone clinical trials in various conditions35 but most molecules have so far failed to meet clinical safety and efficacy standards.
Delivery vehicles for small, non-coding RNA therapeutics
A major challenge remains the efficiency with which these molecules cross cellular membranes, with the foremost reason for clinical trial termination so far being lack of efficacy.3 Whether delivered as naked nucleic acid, ligand–oligonucleotide conjugates, or via various nanocarriers, all small RNAs enter cells by endocytosis and need to exit endocytic vesicles to gain access to the inner cellular compartments.38–40
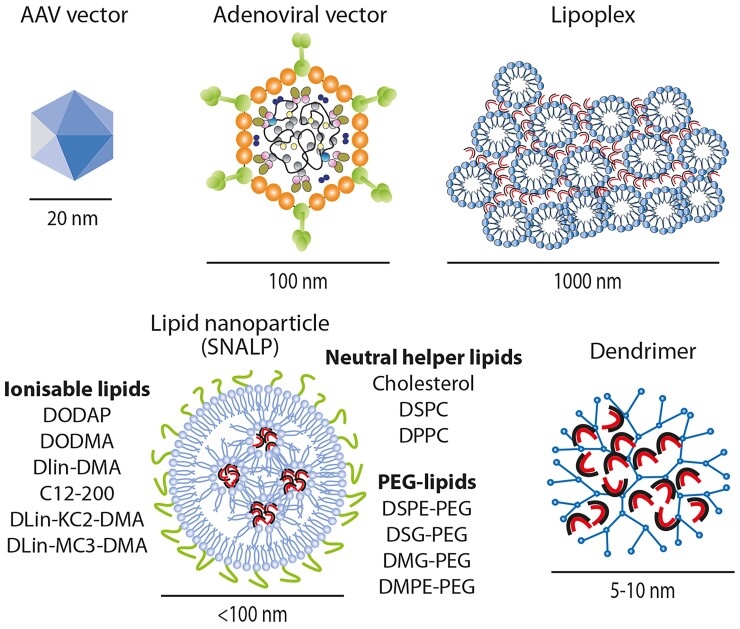
Antisense oligonucleotides with a PS backbone enter the endocytic pathway as naked nucleic acids as they can associate with different uptake molecules on the plasma membrane.41,42 Indeed, all the ASOs approved to date are administered as naked molecules that are injected subcutaneously or intravenously (Table 1). Instead, natural nucleic acids with a phosphodiester backbone, including double-stranded RNAs targeting the RNAi pathway (i.e. siRNAs, miRNA mimics), require either conjugation with a ligand or inclusion within a carrier complex for efficient cellular uptake. Figure 3 shows some of the carrier molecules frequently used for small nucleic acid delivery, with their size compared with that of two viral vectors (AAV and adenovirus).
Figure 3.
Main delivery vehicles for short interfering RNAs and microRNAs, with the indication of their approximate size. The size of the two most used vectors for cardiovascular applications (based on adeno-associated virus [AAV] and adenovirus) are shown for comparison. For stable nucleic acid lipid particles, some of the commonly used lipids for each of the three lipid categories (ionisable lipids, neutral helper lipids, and polyethylene glycol-lipids) are reported. The RNA molecules are schematically shown as curved duplexes.
Uptake ligands
Initial studies indicated that conjugation of ncRNAs with cholesterol favours association with circulating lipoproteins and hepatic uptake, with reduction in exposure to the kidney.43,44 Cholesterol-conjugated ASOs targeting specific miRNAs were originally named ‘antagomirs’.45 Remlarsen, a miRNA that has reached clinical trials, is a cholesterol conjugate.31
The breakthrough technology for naked RNA therapies targeting the liver has been the development of RNA conjugates carrying a trimer of N-acetylgalactosamine (GalNAc), which avidly binds the cellular asialoglycoprotein receptor (ASGPR; reviewed in Springer and Dowdy46). As this receptor is expressed on hepatocyte plasma membrane, this strategy permits specific liver targeting. Of the 4 approved siRNAs, 3 are GalNac-conjugates (Table 1 and Figure 2D), while >10 other GalNac-conjugated ASOs and siRNAs are in late phase clinical development for liver-directed therapy.3
Polymers
Small RNAs can be formulated with synthetic or natural polymers. The former include poly(ethylenimine) (PEI), which binds nucleic acids due to its cationic nature. The resulting large size polyplexes are still positive, interact with negatively charged cell surface polysaccharides and are endocytosed. A pH buffer effect then facilitates endosomal escape. Poly(ethylenimine) is efficient for miRNA delivery47,48 but its toxicity limits clinical application. Other synthetic polymers used for ncRNA delivery are poly-L-lysine and poly(lactic-co-glycolic acid) (PLGA).49–51 Among natural polymers, the positively charged chitosan, which is formed of glucosamine and N-acetylglucosamine, remains attractive because of its biocompatibility and biodegradability.52,53
Several miRNAs have been delivered in animal models using polymers.3 In the cardiovascular field, intracoronary administration of an antagomiR-92 encapsulated in PLGA microspheres of >9 µm diameter was shown to promote angiogenesis and improve cardiac function after MI in pigs.54
Lipids
The most effective methods for small nucleic acid delivery to date are based on lipids (reviewed in Hou et al.55). A first generation of lipofection reagents was based on nucleic acid entrapment by mixtures of a cationic lipid (e.g. DOTMA or DOTAP) formulated with a neutral co-lipid helper (e.g. DOPE). These lipoplexes are very efficient for cell transfection but their large size (often >1 µm; Figure 3) and positive charge results in rapid plasma clearance, toxicity, and inflammation.56–58 Improved biodistribution can be obtained using neutral lipids, such as the commercial preparation MaxSuppressor In Vivo RNA-LANCEr II—composed of DOPC, squalene oil, polysorbate 20, and an antioxidant—which has been used to deliver miRNAs for cancer and cardiovascular applications.59,60 The first miRNA to reach clinical experimentation for solid cancer, miR-34, was formulated in a nanoparticle called Smarticle, composed of amphoteric lipids with an overall anionic charge at physiological pH, while becoming cationic in the acidic tumour environment. Drug development was however halted due to serious side effects.28
Two breakthrough improvements in lipid-mediated delivery were first, ionisable cationic lipids which are positively charged at acidic pH and complex tightly with negatively charged RNA, while becoming neutral at physiological pH, and second, the development of methods to use these molecules to form small lipid nanoparticles (LNPs) that entrap the RNA.61 Patisiran (mentioned earlier) is formulated in an LNP.20 The two other LNPs clinically approved in 2020 were the COVID-19 vaccines pioneered by Moderna (mRNA-127362) and Pfizer/BioNTech (BNT162b263) for the administration of the SARS-CoV-2 Spike mRNA.
Stable nucleic acid lipid particles (SNALPs) are 80–120 nm diameter nanoparticles in which the RNA is surrounded by a lipid bilayer64 (Figure 3). Examples of ionisable lipids essential for SNALP formation include DLin-MC3-DMA (used in patisiran), ALC-0315, and SM102, which are used in the BioNTech/Pfizer and Moderna COVID-19 vaccines, respectively. Other ionisable lipids for LNP formulation that are used experimentally are reviewed in Han et al.65 To form SNALPs, the ionisable lipid is mixed with three other lipids. Two of these helper lipids are usually a phosphocholine-containing lipid (e.g. distearoylphosphatidylcholine [DSPC], dipalmitoylphosphatidylcholine [DPPC]) and cholesterol which, as in cell membranes, provides stability and assists in membrane fusion to facilitate endosomal escape.66 The fourth lipid is a PEG-derivatized lipid which provides stability to the nanoparticle and prolongs its circulatory half-life. Examples of PEGgylated lipids are distearoyl-rac-glycerol-PEG or 1,2-dimyristoyl-rac-glycero-3-methoxy-PEG.
Dendrimers
These are branched polymers carrying cationic groups that interact with nucleic acids. Similar to PEI, endosomal escape is facilitated by a proton sponge effect after endocytosis. Polyamidoamine (PAMAM) dendrimers in particular were capable of delivering an RNA-triple-helix structure comprising two miRNAs and an antagomiR to breast cancer cells in a mouse model.67 As with cationic polymers, the main limitation remains toxicity.
Biological carriers
Exosomes68 are endogenously produced 30–150 nm diameter vesicles spontaneously released by cells that participate in intercellular communication by transferring bioactive material, including small nucleic acids.69 Methods under development include the enrichment of selected miRNAs into exosomes70,71 and the inclusion of specific ligands within exosome membranes for cell targeting.72
Other non-coding RNA delivery methods
Different inorganic carriers (including gold nanoparticles, quantum dots, carbon nanotubes, nanocrystals, silica- and calcium-based nanoparticles) have also been considered for ASO and RNAi therapeutics delivery.73 These systems, however, are significantly less efficient than lipid- or polymer-mediated delivery and to date have undergone limited study in animal models.
Clinically approved non-coding RNAs therapies for cardiovascular disease
Several ncRNA therapies for hyperlipidaemia have recently entered the clinical arena (Table 1), with a potential advantage of increased patient compliance due to an infrequent dosing regimen and higher efficacy either as a standalone or combination treatment.
The siRNA inclisiran targets PCSK9, leading to an up-regulation of hepatocyte low-density lipoprotein (LDL) receptors and consequent reduction in plasma LDL-cholesterol levels.74 A series of randomized placebo-controlled clinical trials (the ORION studies75) tested its efficacy in patients with familial hypercholesterolaemia (FH), CVD, or a high risk of CVD, and demonstrated substantial reduction in LDL cholesterol (∼50%), total cholesterol, and lipoprotein(a) [Lp(a)]. Inclisiran was subcutaneously injected 3–6 monthly for ≥18 months and had a good safety profile and prolonged effects. While potentially a major advance, clinical studies to determine its effects on CVD endpoints and any long-term adverse consequences are still required. Inclisiran was approved for use in the EU and UK in 2020–21.
Volanesorsen is an ASO-targeting ApoC3 which effectively reduces triglyceride levels in familial chylomicronaemia syndrome.76 It was tested as a once-weekly subcutaneous injection for 52 weeks in this randomized placebo-controlled trial in 66 patients. Volanesorsen was approved in the EU in 2019 but not by the US Food and Drug Administration (FDA), at least in part due to the high incidence of thrombocytopenia in treated patients. Mipomersen is an ASO administered by weekly subcutaneous injection that targets ApoB-100 and was approved by the FDA in 2013 as an orphan drug to reduce severe hypercholesterolaemia in patients with homozygous FH. It effectively lowers LDL cholesterol77 but failed to receive EU approval due to a risk of serious liver toxicity and has not been widely used. In the USA, it was withdrawn in 2019. Patisiran, while initially approved for the treatment of HTT-amyloid with neurological manifestations,78 has significant cardiac relevance. The mutant hepatic transthyretin targeted by patisiran leads to accumulation of amyloid deposits in peripheral nerves, the heart, and other organs. In the initial landmark APOLLO study in HTT-amyloid, patisiran was intravenously injected every 3 weeks for 18 months and led to a slight clinical improvement and marked prevention of disease progression when compared with placebo.20 Follow-up data indicate safety at ≥5 years in treated patients. An analysis of cardiac structure and function in a subgroup of patients in this study demonstrated significant reductions in left-ventricular wall thickness, improved longitudinal strain, and reduced pro-BNP levels, hospitalization or mortality79—strongly suggesting efficacy against cardiac disease. These findings are even more impactful, since it is now recognized that HTT-amyloid is quite prevalent and commonly misdiagnosed as idiopathic HF with preserved ejection fraction (HFpEF).
Another approved transthyretin-targeting ncRNA, inotersen, has shown efficacy against neurological HTT-amyloid in a randomized clinical trial involving weekly subcutaneous injection for 15 months.80 Its value in cardiac HTT-amyloid remains to be established.
Small non-coding RNA therapies in the clinical pipeline
A series of novel small ncRNA therapeutics have entered clinical study for different CVD applications; Table 2 lists the most advanced of these.
Table 2.
Main non-coding RNA therapies in the pipeline for cardiovascular applications
| Product (developer/manufacturer) | Type (modifications) | Route of administration | Indication | Target organ | Target mRNA or miRNA | Latest clinical studies |
|---|---|---|---|---|---|---|
| Pelacarsen (Ionis Pharmaceuticals) | ASO (PS, 2′-MOE, GalNAc-conjugated) | Subcutaneous | Elevated Lp(a) | Liver | LPA mRNA | Phase 3 (NCT04023552) |
| Olpasiran (Amgen) | siRNA (GalNAc-conjugated) | Subcutaneous | Elevated Lp(a) | Liver | LPA mRNA | Phase 2 (NCT04270760) |
| SLN360 (Silence Therapeutics) | siRNA (GalNAc-conjugated) | Subcutaneous | Elevated Lp(a) | Liver | LPA mRNA | Phase 1 (NCT04606602) |
| Vupanorsen (Ionis Pharmaceuticals, Pfizer) | ASO (GalNAc-conjugated) | Subcutaneous | Hypertriglyceridemia; Familial Chylomicronemia Syndrome (FCS) | Liver | ANGPTL3 mRNA | Phase 2 (NCT03371355) Phase 2 (NCT04516291) Phase 2 (NCT03360747) Discontinued in January 2022 |
| ARO-ANG3 (Arrowhead Pharmaceuticals) | siRNA (liver targeted) | Subcutaneous | Mixed dyslipidaemia | Liver | ANGPTL3 mRNA | Phase 2 (NCT04832971) |
| MRG-110 (S95010) (miRagen, Servier) | ASO (LNA) | Intradermal | Neovascularization, wound healing | Vasculature | miR-92a-3p | Phase 1 (NCT03494712; NCT03603431) |
| CDR132L (Cardior) | ASO (LNA) | Intravenous | Heart failure | Heart | miR-132-3p | Phase 1b (NCT04045405) |
| Teprasiran (Quark Pharmaceuticals) | siRNA | Intravenous | Acute kidney injury | Kidney | p53 mRNA | Phase 2 (NCT02610283) Phase 3 (NCT03510897) |
| Vutrisiran (Anylam Pharmaceuticals) | siRNA (GalNAc-conjugated) | Subcutaneous | HTT-amyloid with polyneuropathy; TT-amyloid with cardiomyopathy | Liver | Transthyretin mRNA | Phase 3 (NCT03759379) Phase 3 (NCT04153149) |
ASO, antisense oligonucleotide; ds, double stranded; GalNAc, N-acetylgalactosamine; siRNA, short interfering RNA.
The ability to readily target the liver has led to a large pipeline of ncRNA therapies to target different aspects of hyperlipidaemia. An important target isLp(a), a major carrier of oxidized phospholipid in human plasma and a causal risk factor for atherosclerotic CVD and aortic stenosis,81 but not targeted by current small molecule drugs. Lipoprotein(a) contains Apo(a)—the product of the LPA gene—covalently bound to ApoB-100. Pelacarsen (formerly AKCEA-APO(a)-LRx), an ASO-targeting LPA mRNA which is subcutaneously injected 2–4 weekly, lowers Lp(a) levels by up to 80%, and is well tolerated apart from injection-site reactions.82 It is currently in Phase 3 trials in patients with CVD (NCT04023552). Olpasiran, a GalNAc-conjugated siRNA to lower Lp(a), is in Phase 2 studies (NCT04270760), while SLN360 is another GalNAc-conjugated siRNA in Phase 1 trials (NCT04606602).
Another promising hyperlipidaemia target is angiopoietin-like 3 (ANGPTL3), based on findings that loss-of-function variants are associated with substantially lower LDL-cholesterol and triglyceride levels and a reduced CHD risk.83,84 The ANGPTL3 is mainly produced in the liver and is a circulating inhibitor of lipoprotein lipase and endothelial lipase, thereby modulating muscle-free fatty acid uptake, adipose tissue lipogenesis, and hepatic uptake of LDL-cholesterol and remnant cholesterol.85 Importantly, the latter effects are LDL-receptor-independent, meaning that ANGPTL3 lowering should be effective in FH patients with LDL-receptor mutations. Vupanorsen, a GalNAc-modified ASO-targeting ANGPTL3 mRNA, has been investigated in a Phase 2b study (NCT04516291) in diabetic patients with hepatic steatosis and hypertriglyceridaemia where it achieved 38–58% reductions in triglycerides, ApoC3, and remnant cholesterol.86 However, despite this trial meeting its primary objectives, at the end of January 2022, drug development was discontinued due to the perceived insufficient efficacy and liver toxicity (https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-ionis-announce-discontinuation-vupanorsen). Finally, ARO-ANG3, a GalNAc-conjugated siRNA also targeting ANGPTL3, has been assessed in a small open-label study in heterozygous FH87 and is now in a larger Phase 2 trial (NCT04832971).
Two other ASOs in clinical experimentation target endogenous miRNAs, namely miR-92a and miR-132-3p. Antisense oligonucleotide–mediated miR-92a inhibition increases vascularization after MI and hind limb ischaemia,88 accelerates wound healing,89,90 improves re-endothelialization,91 and prevents endothelial dysfunction and atherosclerosis,92 in murine and porcine models. Catheter-based delivery of an anti-miR-92a LNA ASO in pigs effectively reduced infarct size and improved cardiac function.93 Analogous results in pigs were also obtained by intracoronary injection of a miR-92a antagomir encapsulated in PLGA microspheres.54 Based on these findings, a placebo-controlled, dose-escalating study using an intravenously administered anti-miR-92a LNA ASO (MRG-110) was performed in 49 healthy volunteers (NCT03494712). This study demonstrated safety and that a single dose of ASO de-repressed miR-92a target mRNAs in different human peripheral blood cells for 2 weeks.94 Another Phase 1 study (NCT03603431) investigated intradermal injection of 3-weekly MRG-110 doses to promote angiogenesis and improve wound healing. The results indicate safety and preliminary evidence of neoangiogenesis (https://www.globenewswire.com/en/news-release/2019/10/16/1930277/31683/en/miRagen-Announces-New-Clinical-Data-Showing-MRG-110-Positively-Impacted-Tissue-Repair-and-New-Blood-Vessel-Growth.html).
First-in-human data were also reported for an LNA ASO–targeting miR-132-3p for patients with HF.95 Pre-clinical studies had indicated that inhibition of miR-132-3p alone is sufficient to prevent HF in mice and pigs by improving contractility and metabolism, and reducing hypertrophy.96,97 Based on these data, a 16-mer LNA ASO with a PS backbone (named CDR132L) was developed and tested by intravenous injection in a placebo-controlled, dose-escalation Phase 1b study in 28 HF patients with NYHA Classes I–III and left-ventricular ejection fraction >30% (NCT04045405). The study met safety objectives and provided evidence of efficacy based on cardiac function and HF biomarkers.95 More broadly, the study provided evidence of favourable pharmacokinetics upon a 4-weekly ASO-dosing regimen, which has relevance for future applications of ASOs with a similar chemistry.98
Other disease indications being clinically tested include acute kidney injury (AKI) after high-risk cardiac surgery. Teprasiran, a synthetic chemically stabilized siRNA that targets p53, has been tested in a randomized multicentre Phase 2 study in 360 patients (NCT02610283). Teprasiran achieved a 58% relative risk reduction in AKI, with no significant safety issues.99 A larger Phase 3 trial in >1000 patients completed enrolment (NCT03510897) but was terminated early due to results not meeting efficacy outcomes at Day 90 (NCT03510897).
Finally, a subcutaneously administered GalNAc-conjugated siRNA, vutrisiran, is being developed by the manufacturers of patisiran for the same indication of down-regulating transthyretin mRNA for HTT-amyloid. Vutrisiran is in two large Phase 3 trials in patients with HTT-amyloid with polyneuropathy (NCT03759379) and patients with hereditary or non-hereditary TT-amyloid with cardiomyopathy (NCT04153149).
Novel candidate non-coding RNA therapies from pre-clinical studies
Non-coding RNA therapies for heart failure
Cardiac remodelling is an obvious therapeutic target in HF. The underlying processes, such as CM hypertrophy, altered excitation–contraction coupling, cell death, interstitial fibrosis, microvascular rarefaction, and others, are underpinned by substantial changes in the expression of protein-coding genes, ncRNAs, and re-expression of foetal genes. As such, the targeting of gene expression is an attractive approach to tackle remodelling.
A common approach has been to use 2′-OMe-modified antagomiRs or LNAs to inhibit miRNAs involved in cardiac hypertrophy, with early proof-of-concept for in vivo silencing originally reported in mice.100 The therapeutic antagonism of miRNAs to inhibit hypertrophy and remodelling during experimental HF in mice has been shown for numerous miRNAs, which act on either CMs or fibroblasts. These include miR-133,100 mir-29,101 miR-21,102 miR-23a,103 miR-199a-5p,104 miR-24,105 miR-34a,106 and miR-25.107 Other studies reported success with LNA ASOs, e.g. against miR-208a,108 mir-34,109 miR-652,110 miR-154,111 miR-29,112 and for miR-21 in pigs.113 Most of these approaches also reduced fibrosis and cardiac dysfunction. Other anti-miR oligonucleotide applications are reviewed in De Majo and De Windt.114
Approaches to target other cell types may also be effective. The inhibition of leucocyte-expressed miR-155 in mice reduced cardiac inflammation, hypertrophy, and dysfunction after chronic pressure overload.115 Icli et al.116 found that intravenous administration of an LNA targeting miR-26a rapidly induced angiogenesis, reduced MI size, and improved heart function. An RNA aptamer-targeting osteopontin (which drives both fibrosis and hypertrophy) prevented or reversed pressure overload–induced HF in mice.117 Finally, antagonism of endothelial miR-24 limited MI size by preventing endothelial apoptosis.118
Long ncRNAs are also potential targets. Nuclear lncRNAs can modify chromatin structure close to their own transcribed regions or at a different site, act as decoys for proteins or regulate mRNA alternative splicing.119 Cytoplasmic lncRNAs regulate mRNA stability and translation, interact with cytoplasmic proteins and act as miRNA sponges.120 As most aspects of cardiac function are governed by one or multiple lncRNAs,121–124 targeting lncRNAs is an attractive therapeutic option. The first examples of therapeutic lncRNA targeting in the cardiovascular field include GapmeR-mediated silencing of the lncRNA Chast, which effectively prevented or attenuated pressure overload–induced pathological cardiac remodelling,125 lncRNA Meg3, to reduce cardiac fibrosis and improve diastolic function after chronic pressure overload,126 lncRNA Wisper to inhibit MI-induced fibrosis and cardiac dysfunction,127 and lncRNA H19 to inhibit aortic aneurysm development128 or pulmonary arterial hypertension.129 Other applications of anti-lncRNA antisense technology are reviewed in Hobuss et al.,121 Poller et al.,122 Salamon et al.,123 and Lucas et al.124 It is worth noting, however, that the degree of sequence conservation between lncRNAs of different species is relatively limited,130–132 rendering the pre-clinical development of effective therapies more problematic.
Non-coding RNAs for cardiac regeneration
A specific therapeutic area gaining momentum at pre-clinical level is the stimulation of endogenous CM proliferation to achieve cardiac regeneration after MI. As CM proliferation is controlled by both the miRNA and lncRNA networks,133 a few studies over the last years have explored the possibility of delivering synthetic ncRNAs to achieve cardiac regeneration after MI. Several miRNAs that control CM proliferation were identified in two large screenings that analysed whole-genome miRNA libraries in rodent and human cells.24,134 Anti-miR LNAs against miR-15b135 and miR-34a,136 which both inhibit CM proliferation, were shown to exert a beneficial effect after MI in rodents.
The efficacy of antisense approaches, however, strictly depends on the level of expression of the inhibitory ncRNAs and their relative importance in controlling regeneration. A more tempting pharmacological approach is to impart a proliferative phenotype to CMs with small RNAs, irrespective of whether the administered molecules participate in normal cardiac physiology. A single intramyocardial injection of miR-199a-3p or miR-590-3p mimics using a cationic lipid formulation was sufficient to induce cardiac regeneration after MI in mice.137 Similar results were also reported with the intramyocardial administration of miR-19a/19b mimics using a neutral lipid delivery reagent,59 using the same formulation for the daily intravenous administration of miR-302b/c,60 miR-19a/19b59 or miR-708138 mimics, or the intracardiac delivery of cholesterol-modified miR-302b/c mimics using a hydrogel.139 Of note, the administration of miRNA mimics exerts a transient effect lasting approximately 10 days with miR-199a-3p.137 This can overcome the adverse effects and safety concerns potentially caused by the continuous expression of pro-proliferative RNAs upon gene transfer of their coding genes using AAV vectors.140
Non-coding RNA delivery to the heart
Cardiac-specific delivery may be achieved either through cell targeting upon systemic administration or via direct injection through selective catheterization. While the former approach is still in its infancy, the latter appears readily achievable.
Cardiac cell targeting
An ideal ncRNA therapeutic should fulfil the mandate of the so-called Ehrlich’s magic bullet, namely be administered systemically to then reach the target organ in a specific manner. This is achievable for the liver by the conjugation of ASOs and siRNAs with GalNAc, which specifically binds the hepatocyte ASGPR as discussed earlier. No such targeting system yet exists for any cardiac cell type. Virus-mediated gene transfer can achieve tissue-specific efficacy by using CM-specific vectors (e.g. AAV Serotype 9 or 6) and by including CM-specific promoters for transcriptional targeting.141 Neither possibility is available to synthetic ncRNAs.
After intravenous injection, polymer:RNA or lipid:RNA complex is generally phagocytosed by mononuclear cells in the liver, spleen, lymph nodes, and bone marrow.142 A first step towards tissue specificity is therefore to reduce interaction with phagocytic cells and increase circulatory half-life, e.g. by PEG surface modification of LNPs and other nanocarriers to provide a hydrophilic shield. Polyethylene glycol-lipids in LNPs also reduce nanoparticle opsonization by ApoE and other circulating lipoproteins, which results in uptake by lipoprotein receptors.66
Active cellular targeting could be achieved through inclusion in the LNPs or other nanocarriers of ligands that bind specifically expressed surface molecules. Several peptides have been identified through phage display panning techniques to target CMs143–148 or the infarcted myocardium.144,149,150 Additional possibilities for cardiac targeting are a ligand for the angiotensin II Type 1 receptor,151 antibodies against cardiac troponin T152,153 or myosin,154 ligands for activated endothelial cells,155,156 carbohydrates,157 or substrates for enzymes present in the infarct area.158 Despite abundant data in cell culture and rodents, none of these strategies has yet progressed to large animal or clinical testing.
In cancer settings, nanosized particles may accumulate in tumour tissue due to enhanced vascular permeability and retention (EPR effect).159 A similar EPR effect after MI could allow preferential accumulation of LNPs in the damaged myocardium. While the post-MI EPR effect begins to diminish after 24–48 h and thus might be too short for treatments aimed at preventing adverse remodelling (reviewed in Prajnamitra et al.160), this time frame may be sufficient for nanoparticles carrying ncRNA payloads that induce acute cardioprotection or stimulate post-MI cardiac regeneration.
Direct cardiac-specific delivery
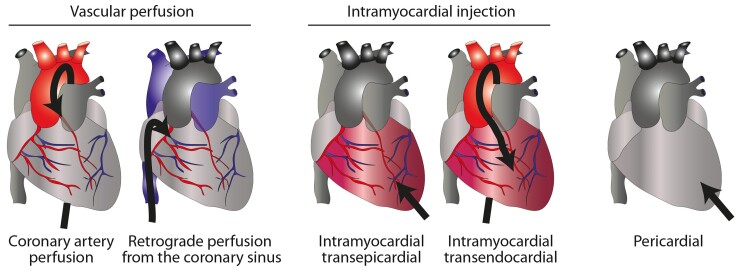
The heart is amenable to physical targeting by direct injection or catheterization of its main vessels (Figure 4).
Figure 4.
Routes for cardiac delivery. These include injection into the coronary artery as during standard percutaneous coronary intervention or retrograde into the coronary sinus; or intramyocardial, through either direct injection after mini-thoracotomy or during bypass surgery, or after percutaneous catheterization to reach the left ventricle, followed by transendocardial delivery. A fifth route includes administration into the pericardial sac.
Intramyocardial injection
This may be performed surgically (mini-thoracotomy or open-chest surgery) through transepicardial injection or percutaneously via transendocardial injection. The surgical approach allows precise delivery and in multiple locations (e.g. the infarct border zone) but is limited by its invasiveness and difficulty in accessing areas such as the interventricular septum. Percutaneous transendocardial delivery via the left-ventricular cavity is less invasive and safe in expert hands.161,162 Different imaging techniques (e.g. intravascular ultrasound) or peri-procedural electromechanical mapping (e.g. NOGA; Biosense Webster) are required to inject nanomedicine products.163,164
Intracoronary administration
The simplest and clinically most applicable route for direct delivery of a therapeutic remains antegrade coronary catheterization, which can be safely performed even in patients with marked cardiac dysfunction.165,166 However, the transit time in the coronary circulation is very short and the endothelial cell barrier needs to be crossed. The latter is normally impermeable to particles >20 nm diameter,167 much smaller than typical LNPs (80–100 nm) and even larger lipoplexes. Numerous approaches have been proposed to improve antegrade delivery efficacy, such as increasing permeability or coronary sinus occlusion,168,169 but delivery may also be enhanced early after MI due to the aforementioned EPR effect or the use of closed recirculation systems, by which coronary sinus blood is collected and re-delivered to the coronary artery through an extracorporeal membrane oxygenation system.170–172
Higher efficacy may be achieved by retrograde administration via the coronary sinus,173–176 a relatively simple interventional procedure usually accompanied by transient antegrade blockade of coronary artery flow.164 Maintenance of high pressure increases efficacy of this approach.173,177 Potential issues are the presence of cardiac resynchronization devices and the risk of coronary sinus rupture.
Pericardial administration
Percutaneous intra-pericardial delivery could offer broad access to the entire heart except the septum.178 The safety of this approach for epicardial catheter electrophysiological ablations has progressively improved but a major issue for ncRNA therapy may be restricted delivery to the epicardial surface and removal of agent by the pericardial lymphatic system.164,179
Conclusions and future perspectives
Small nucleic acid therapies offer an exciting new therapeutic modality for CVD and an opportunity to tackle disease pathways that are not targeted by current small molecule approaches. Despite this perceived potential, there are several challenges that need to be overcome before wider clinical application of ncRNA therapies. A major issue that remains is improvements in the carriers and RNA modifications that would allow efficient cellular uptake and intracellular stability of the agents; lipid-based nanocarriers currently appear the most promising approach, at least until efficient CM-targeting systems are developed.
An additional problem relates to toxicity following the undesired activation of the innate immune system. As part of the antiviral defence mechanism, cells recognize both single- and double-stranded RNA via pathogen-associated molecular pattern receptors.180 The predominant pathway that recognizes small RNAs starts with the detection of RNA structures by Toll-like receptors 3, 7, and 8 in endosomes.181 This can result in the production of pro-inflammatory cytokines in dendritic cells and macrophages, or the stimulation of a Type I interferon response.182 In addition, double-stranded molecules are recognized by intracytoplasmic receptors, such as retinoic acid-inducible gene I (RIG1) and melanoma differentiation-associated protein 5 (MDA5).183 Further improvements in the design of ncRNA therapeutics should aim to reduce immune recognition. Already available evidence indicates that innate immune system activation by siRNAs is sequence specific184 and that immunogenicity can be markedly decreased by the incorporation of 2′-OMe or 2′-F modifications in the ribose ring of nucleotides.185–187
Potential off-target effects of ncRNA therapies also need to be borne in mind. For example, siRNAs can induce off-target effects due to their miRNA-like activity, by which other cellular genes can also be down-regulated.188–191 Analogous problems can occur with miRNA mimics, in which the passenger strand, which is usually chemically modified, can accumulate in transfected cells and exert effects on other cellular mRNAs.192 This can be even more relevant considering that these off-target effects can be cell-type specific and thus difficult to predict or reproduce in cell culture.193
Notwithstanding these hurdles, the class of ncRNA therapies may truly portend a paradigm shift in our approaches to the management of many CVD.
Contributor Information
Ajay M Shah, King’s College London, British Heart Foundation Centre of Research Excellence, School of Cardiovascular and Metabolic Medicine and Sciences, The James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Mauro Giacca, King’s College London, British Heart Foundation Centre of Research Excellence, School of Cardiovascular and Metabolic Medicine and Sciences, The James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Funding
A.M.S. is supported by the British Heart Foundation (CH/1999001/11735, RE/18/2/34213, RM/17/3/33381) and the Fondation Leducq (17CVD04). M.G. is supported by the European Research Council (Advanced Grant 787971), the British Heart Foundation (RG/19/11/34633), and the European Commission Horizon 2020 grants 825670 and 874764. Both authors are supported by the National Institute for Health Research Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust with King’s College London (IS-BRC-543 1215-20006).
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol 2018;9:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203–222. [DOI] [PubMed] [Google Scholar]
- 3. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics—challenges and potential solutions. Nat Rev Drug Discov 2021;20:629–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A 1978;75:285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest 2017;127:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bobbin ML, Rossi JJ. RNA Interference (RNAi)-based therapeutics: delivering on the promise? Annu Rev Pharmacol Toxicol 2016;56:103–122. [DOI] [PubMed] [Google Scholar]
- 7. Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 2011;18:1111–1120. [DOI] [PubMed] [Google Scholar]
- 8. van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov 2012;11:860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896–899. [DOI] [PubMed] [Google Scholar]
- 10. Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 2004;43:13233–13241. [DOI] [PubMed] [Google Scholar]
- 11. Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, et al. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J Biol Chem 2004;279:36317–36326. [DOI] [PubMed] [Google Scholar]
- 12. Sorensen MD, Kvaerno L, Bryld T, Hakansson AE, Verbeure B, Gaubert G, et al. Alpha-L-ribo-configured locked nucleic acid (alpha-L-LNA): synthesis and properties. J Am Chem Soc 2002;124:2164–2176. [DOI] [PubMed] [Google Scholar]
- 13. Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res 2002;30:1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, et al. Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem 1993;268:14514–14522. [PubMed] [Google Scholar]
- 15. Braasch DA, Liu Y, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res 2002;30:5160–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fluiter K, Mook OR, Vreijling J, Langkjaer N, Hojland T, Wengel J, et al. Filling the gap in LNA antisense oligo gapmers: the effects of unlocked nucleic acid (UNA) and 4'-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol Biosyst 2009;5:838–843. [DOI] [PubMed] [Google Scholar]
- 17. Neil CR, Seiler MW, Reynolds DJ, Smith JJ, Vaillancourt FH, Smith PG, et al. Reprogramming RNA processing: an emerging therapeutic landscape. Trends Pharmacol Sci 2022;43:437–454. [DOI] [PubMed] [Google Scholar]
- 18. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 20. Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 2018;379:11–21. [DOI] [PubMed] [Google Scholar]
- 21. Balwani M, Sardh E, Ventura P, Peiro PA, Rees DC, Stolzel U, et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent Porphyria. N Engl J Med 2020;382:2289–2301. [DOI] [PubMed] [Google Scholar]
- 22. Ray KK, Landmesser U, Leiter LA, Kallend D, Dufour R, Karakas M, et al. Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med 2017;376:1430–1440. [DOI] [PubMed] [Google Scholar]
- 23. Scott LJ, Keam SJ. Lumasiran: first approval. Drugs 2021;81:277–282. [DOI] [PubMed] [Google Scholar]
- 24. Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376–381. [DOI] [PubMed] [Google Scholar]
- 25. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 2012;110:1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 2012;19:937–954. [DOI] [PubMed] [Google Scholar]
- 28. Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 2017;35:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 2020;122:1630–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 2017;18:1386–1396. [DOI] [PubMed] [Google Scholar]
- 31. Gallant-Behm CL, Piper J, Lynch JM, Seto AG, Hong SJ, Mustoe TA, et al. A microRNA-29 mimic (remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol 2019;139:1073–1081. [DOI] [PubMed] [Google Scholar]
- 32. Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990;344:467–468. [DOI] [PubMed] [Google Scholar]
- 33. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990;346:818–822. [DOI] [PubMed] [Google Scholar]
- 34. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990;249:505–510. [DOI] [PubMed] [Google Scholar]
- 35. Byun J. Recent progress and opportunities for nucleic acid aptamers. Life (Basel) 2021;11:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siddiqui MA, Keating GM. Pegaptanib: in exudative age-related macular degeneration. Drugs 2005;65:1571–1577. [DOI] [PubMed] [Google Scholar]
- 37. Solomon SD, Lindsley K, Vedula SS, Krzystolik MG, Hawkins BS. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. Cochrane Database Syst Rev 2019;3:CD005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Juliano RL. Intracellular trafficking and endosomal release of oligonucleotides: what we know and what we don't. Nucleic Acid Ther 2018;28:166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J. Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J Control Release 2015;203:1–15. [DOI] [PubMed] [Google Scholar]
- 40. Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet 2014;15:541–555. [DOI] [PubMed] [Google Scholar]
- 41. Crooke ST, Wang S, Vickers TA, Shen W, Liang XH. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol 2017;35:230–237. [DOI] [PubMed] [Google Scholar]
- 42. Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 2017;35:222–229. [DOI] [PubMed] [Google Scholar]
- 43. Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004;432:173–178. [DOI] [PubMed] [Google Scholar]
- 44. Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol 2017;57:81–105. [DOI] [PubMed] [Google Scholar]
- 45. Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689. [DOI] [PubMed] [Google Scholar]
- 46. Springer AD, Dowdy SF. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther 2018;28:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 1995;92:7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem 2008;19:1448–1455. [DOI] [PubMed] [Google Scholar]
- 49. Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A 2012;109:E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devulapally R, Sekar NM, Sekar TV, Foygel K, Massoud TF, Willmann JK, et al. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 2015;9:2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang S, Zhang J, Wang Y, Chen M. Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542-3p for triple negative breast cancer therapy. Nanomedicine 2016;12:411–420. [DOI] [PubMed] [Google Scholar]
- 52. Ragelle H, Vandermeulen G, Preat V. Chitosan-based siRNA delivery systems. J Control Release 2013;172:207–218. [DOI] [PubMed] [Google Scholar]
- 53. Kaban K, Salva E, Akbuga J. In vitro dose studies on chitosan nanoplexes for microRNA delivery in breast cancer cells. Nucleic Acid Ther 2017;27:45–55. [DOI] [PubMed] [Google Scholar]
- 54. Bellera N, Barba I, Rodriguez-Sinovas A, Ferret E, Asin MA, Gonzalez-Alujas MT, et al. Single intracoronary injection of encapsulated antagomir-92a promotes angiogenesis and prevents adverse infarct remodeling. J Am Heart Assoc 2014;3:e000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater 2021;6:1078–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Filion MC, Phillips NC. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochim Biophys Acta 1997;1329:345–356. [DOI] [PubMed] [Google Scholar]
- 57. Ito Y, Kawakami S, Charoensit P, Higuchi Y, Hashida M. Evaluation of proinflammatory cytokine production and liver injury induced by plasmid DNA/cationic liposome complexes with various mixing ratios in mice. Eur J Pharm Biopharm 2009;71:303–309. [DOI] [PubMed] [Google Scholar]
- 58. Litzinger DC. Limitations of cationic liposomes for antisense oligonucleotide delivery in vivo. J Liposome Res 1997;7:51–61. [Google Scholar]
- 59. Gao F, Kataoka M, Liu N, Liang T, Huang ZP, Gu F, et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun 2019;10:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med 2015;7:279ra238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kulkarni JA, Cullis PR, van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther 2018;28:146–157. [DOI] [PubMed] [Google Scholar]
- 62. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021;384:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med 2020;383:2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kulkarni JA, Darjuan MM, Mercer JE, Chen S, van der Meel R, Thewalt JL, et al. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 2018;12:4787–4795. [DOI] [PubMed] [Google Scholar]
- 65. Han X, Zhang H, Butowska K, Swingle KL, Alameh MG, Weissman D, et al. An ionizable lipid toolbox for RNA delivery. Nat Commun 2021;12:7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv Drug Deliv Rev 2016;99:129–137. [DOI] [PubMed] [Google Scholar]
- 67. Conde J, Oliva N, Atilano M, Song HS, Artzi N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat Mater 2016;15:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 69. Jiang L, Vader P, Schiffelers RM. Extracellular vesicles for nucleic acid delivery: progress and prospects for safe RNA-based gene therapy. Gene Ther 2017;24:157–166. [DOI] [PubMed] [Google Scholar]
- 70. Mathiyalagan P, Sahoo S. Exosomes-based gene therapy for microRNA delivery. Methods Mol Biol 2017;1521:139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Abreu RC, Ramos Cristiana V, Becher C, Lino M, Jesus C, Costa Martins PA, et al. Exogenous loading of miRNAs into small extracellular vesicles. J Extracell Vesicles 2021;10:e12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ohno SI, Kuroda M. Exosome-mediated targeted delivery of miRNAs. Methods Mol Biol 2016;1448:261–270. [DOI] [PubMed] [Google Scholar]
- 73. Boca S, Gulei D, Zimta AA, Onaciu A, Magdo L, Tigu AB, et al. Nanoscale delivery systems for microRNAs in cancer therapy. Cell Mol Life Sci 2020;77:1059–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cupido AJ, Kastelein JJP. Inclisiran for the treatment of hypercholesterolaemia: implications and unanswered questions from the ORION trials. Cardiovasc Res 2020;116:e136–e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Raal FJ, Kallend D, Ray KK, Turner T, Koenig W, Wright RS, et al. Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N Engl J Med 2020;382:1520–1530. [DOI] [PubMed] [Google Scholar]
- 76. Witztum JL, Gaudet D, Freedman SD, Alexander VJ, Digenio A, Williams KR, et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N Engl J Med 2019;381:531–542. [DOI] [PubMed] [Google Scholar]
- 77. Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet 2010;375:998–1006. [DOI] [PubMed] [Google Scholar]
- 78. Titze-de-Almeida SS, Brandao PRP, Faber I, Titze-de-Almeida RPRP. Leading RNA interference therapeutics part 1: silencing hereditary transthyretin amyloidosis, with a focus on patisiran. Mol Diagn Ther 2020;24:49–59. [DOI] [PubMed] [Google Scholar]
- 79. Solomon SD, Adams D, Kristen A, Grogan M, Gonzalez-Duarte A, Maurer MS, et al. Effects of patisiran, an RNA interference therapeutic, on cardiac parameters in patients with hereditary transthyretin-mediated amyloidosis. Circulation 2019;139:431–443. [DOI] [PubMed] [Google Scholar]
- 80. Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med 2018;379:22–31. [DOI] [PubMed] [Google Scholar]
- 81. Swerdlow DI, Rider DA, Yavari A, Lindholm MW, Campion GV, Nissen SE. Treatment and prevention of lipoprotein(a)-mediated cardiovascular disease: the emerging potential of RNA interference therapeutics. Cardiovasc Res 2022;118:1218–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsimikas S, Karwatowska-Prokopczuk E, Gouni-Berthold I, Tardif JC, Baum SJ, Steinhagen-Thiessen E, et al. Lipoprotein(a) reduction in persons with cardiovascular disease. N Engl J Med 2020;382:244–255. [DOI] [PubMed] [Google Scholar]
- 83. Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med 2010;363:2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stitziel NO, Khera AV, Wang X, Bierhals AJ, Vourakis AC, Sperry AE, et al. ANGPTL3 deficiency and protection against coronary artery disease. J Am Coll Cardiol 2017;69:2054–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Graham MJ, Lee RG, Brandt TA, Tai LJ, Fu W, Peralta R, et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med 2017;377:222–232. [DOI] [PubMed] [Google Scholar]
- 86. Gaudet D, Karwatowska-Prokopczuk E, Baum SJ, Hurh E, Kingsbury J, Bartlett VJ, et al. Vupanorsen, an N-acetyl galactosamine-conjugated antisense drug to ANGPTL3 mRNA, lowers triglycerides and atherogenic lipoproteins in patients with diabetes, hepatic steatosis, and hypertriglyceridaemia. Eur Heart J 2020;41:3936–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Watts G, Schwabe C, Scott R, Gladding P, Sullivan D, Baker J, et al. Abstract 15751: pharmacodynamic effect of ARO-ANG3, an investigational RNA interference targeting hepatic angiopoietin-like protein 3, in patients with hypercholesterolemia. Circulation 2020;142:15751. [Google Scholar]
- 88. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009;324:1710–1713. [DOI] [PubMed] [Google Scholar]
- 89. Gallant-Behm CL, Piper J, Dickinson BA, Dalby CM, Pestano LA, Jackson AL. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen 2018;26:311–323. [DOI] [PubMed] [Google Scholar]
- 90. Lucas T, Schafer F, Muller P, Eming SA, Heckel A, Dimmeler S. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat Commun 2017;8:15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Daniel JM, Penzkofer D, Teske R, Dutzmann J, Koch A, Bielenberg W, et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc Res 2014;103:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Loyer X, Potteaux S, Vion AC, Guerin CL, Boulkroun S, Rautou PE, et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res 2014;114:434–443. [DOI] [PubMed] [Google Scholar]
- 93. Hinkel R, Penzkofer D, Zuhlke S, Fischer A, Husada W, Xu QF, et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 2013;128:1066–1075. [DOI] [PubMed] [Google Scholar]
- 94. Abplanalp WT, Fischer A, John D, Zeiher AM, Gosgnach W, Darville H, et al. Efficiency and target derepression of anti-miR-92a: results of a first in human study. Nucleic Acid Ther 2020;30:335–345. [DOI] [PubMed] [Google Scholar]
- 95. Taubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J 2021;42:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Foinquinos A, Batkai S, Genschel C, Viereck J, Rump S, Gyongyosi M, et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Commun 2020;11:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Batkai S, Genschel C, Viereck J, Rump S, Bar C, Borchert T, et al. CDR132L Improves systolic and diastolic function in a large animal model of chronic heart failure. Eur Heart J 2021;42:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Baker AH, Giacca M. Antagonism of miRNA in heart failure: first evidence in human. Eur Heart J 2021;42:189–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Thielmann M, Corteville D, Szabo G, Swaminathan M, Lamy A, Lehner LJ, et al. Teprasiran, a small interfering RNA, for the prevention of acute kidney injury in high-risk patients undergoing cardiac surgery: a randomized clinical study. Circulation 2021;144:1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618. [DOI] [PubMed] [Google Scholar]
- 101. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980–984. [DOI] [PubMed] [Google Scholar]
- 103. Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci U S A 2009;106:12103–12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 2010;12:1220–1227. [DOI] [PubMed] [Google Scholar]
- 105. Li RC, Tao J, Guo YB, Wu HD, Liu RF, Bai Y, et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ Res 2013;112:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013;495:107–110. [DOI] [PubMed] [Google Scholar]
- 107. Wahlquist C, Jeong D, Rojas-Munoz A, Kho C, Lee A, Mitsuyama S, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature 2014;508:531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011;124:1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A 2012;109:17615–17620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bernardo BC, Nguyen SS, Winbanks CE, Gao XM, Boey EJ, Tham YK, et al. Therapeutic silencing of miR-652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J 2014;28:5097–5110. [DOI] [PubMed] [Google Scholar]
- 111. Bernardo BC, Nguyen SS, Gao XM, Tham YK, Ooi JY, Patterson NL, et al. Inhibition of miR-154 protects against cardiac dysfunction and fibrosis in a mouse model of pressure overload. Sci Rep 2016;6:22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sassi Y, Avramopoulos P, Ramanujam D, Gruter L, Werfel S, Giosele S, et al. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating wnt signaling. Nat Commun 2017;8:1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hinkel R, Ramanujam D, Kaczmarek V, Howe A, Klett K, Beck C, et al. AntimiR-21 prevents myocardial dysfunction in a pig model of ischemia/reperfusion injury. J Am Coll Cardiol 2020;75:1788–1800. [DOI] [PubMed] [Google Scholar]
- 114. De Majo F, De Windt LJ. RNA therapeutics for heart disease. Biochem Pharmacol 2018;155:468–478. [DOI] [PubMed] [Google Scholar]
- 115. Heymans S, Corsten MF, Verhesen W, Carai P, van Leeuwen RE, Custers K, et al. Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation 2013;128:1420–1432. [DOI] [PubMed] [Google Scholar]
- 116. Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res 2013;113:1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Li J, Yousefi K, Ding W, Singh J, Shehadeh LA. Osteopontin RNA aptamer can prevent and reverse pressure overload-induced heart failure. Cardiovasc Res 2017;113:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011;124:720–730. [DOI] [PubMed] [Google Scholar]
- 119. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Geisler S, Coller J. RNA In unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013;14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hobuss L, Bar C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol 2019;10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J 2018;39:2704–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Salamon I, Saccani Jotti G, Condorelli G. The long noncoding RNA landscape in cardiovascular disease: a brief update. Curr Opin Cardiol 2018;33:282–289. [DOI] [PubMed] [Google Scholar]
- 124. Lucas T, Bonauer A, Dimmeler S. RNA therapeutics in cardiovascular disease. Circ Res 2018;123:205–220. [DOI] [PubMed] [Google Scholar]
- 125. Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, et al. Long noncoding RNA chast promotes cardiac remodeling. Sci Transl Med 2016;8:326ra322. [DOI] [PubMed] [Google Scholar]
- 126. Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res 2017;121:575–583. [DOI] [PubMed] [Google Scholar]
- 127. Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, et al. The long noncoding RNA wisper controls cardiac fibrosis and remodeling. Sci Transl Med 2017;9:eaai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation 2018;138:1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Omura J, Habbout K, Shimauchi T, Wu WH, Breuils-Bonnet S, Tremblay E, et al. Identification of long noncoding RNA H19 as a new biomarker and therapeutic target in right ventricular failure in pulmonary arterial hypertension. Circulation 2020;142:1464–1484. [DOI] [PubMed] [Google Scholar]
- 130. Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014;1840:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ramirez-Colmenero A, Oktaba K, Fernandez-Valverde SL. Evolution of genome-organizing long non-coding RNAs in metazoans. Front Genet 2020;11:589697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019;571:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Braga L, Ali H, Secco I, Giacca M. Non-coding RNA therapeutics for cardiac regeneration. Cardiovasc Res 2021;117:674–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Diez-Cunado M, Wei K, Bushway PJ, Maurya MR, Perera R, Subramaniam S, et al. miRNAs that induce human cardiomyocyte proliferation converge on the Hippo pathway. Cell Rep 2018;23:2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 2012;110:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, et al. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res 2015;117:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lesizza P, Prosdocimo G, Martinelli V, Sinagra G, Zacchigna S, Giacca M. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ Res 2017;120:1298–1304. [DOI] [PubMed] [Google Scholar]
- 138. Deng S, Zhao Q, Zhen L, Zhang C, Liu C, Wang G, et al. Neonatal heart-enriched miR-708 promotes proliferation and stress resistance of cardiomyocytes in rodents. Theranostics 2017;7:1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang LL, Liu Y, Chung JJ, Wang T, Gaffey AC, Lu M, et al. Sustained miRNA delivery from an injectable hydrogel promotes cardiomyocyte proliferation and functional regeneration after ischaemic injury. Nat Biomed Eng 2017;1:983–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019;569:418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zacchigna S, Zentilin L, Giacca M. Adeno-associated virus vectors as therapeutic and investigational tools in the cardiovascular system. Circ Res 2014;114:1827–1846. [DOI] [PubMed] [Google Scholar]
- 142. Weissleder R, Nahrendorf M, Pittet MJ. Imaging macrophages with nanoparticles. Nat Mater 2014;13:125–138. [DOI] [PubMed] [Google Scholar]
- 143. Zhang H, Li N, Sirish P, Mahakian L, Ingham E, Curry FR, et al. The cargo of CRPPR-conjugated liposomes crosses the intact murine cardiac endothelium. J Control Release 2012;163:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mentkowski KI, Lang JK. Exosomes engineered to express a cardiomyocyte binding peptide demonstrate improved cardiac retention in vivo. Sci Rep 2019;9:10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang X, Chen Y, Zhao Z, Meng Q, Yu Y, Sun J, et al. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc 2018;7:e008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kanki S, Jaalouk DE, Lee S, Yu AY, Gannon J, Lee RT. Identification of targeting peptides for ischemic myocardium by in vivo phage display. J Mol Cell Cardiol 2011;50:841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. McGuire MJ, Samli KN, Johnston SA, Brown KC. In vitro selection of a peptide with high selectivity for cardiomyocytes in vivo. J Mol Biol 2004;342:171–182. [DOI] [PubMed] [Google Scholar]
- 148. Dasa SSK, Suzuki R, Gutknecht M, Brinton LT, Tian Y, Michaelsson E, et al. Development of target-specific liposomes for delivering small molecule drugs after reperfused myocardial infarction. J Control Release 2015;220:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Huang Z, Song Y, Pang Z, Zhang B, Yang H, Shi H, et al. Targeted delivery of thymosin beta 4 to the injured myocardium using CREKA-conjugated nanoparticles. Int J Nanomedicine 2017;12:3023–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhang Y, Chen X, Liu L, Tian J, Hao L, Ran HT. Photoacoustic imaging of myocardial infarction region using non-invasive fibrin-targeted nanoparticles in a rat myocardial ischemia-reperfusion model. Int J Nanomedicine 2021;16:1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Dvir T, Bauer M, Schroeder A, Tsui JH, Anderson DG, Langer R, et al. Nanoparticles targeting the infarcted heart. Nano Lett 2011;11:4411–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Li M, Tang X, Liu X, Cui X, Lian M, Zhao M, et al. Targeted miR-21 loaded liposomes for acute myocardial infarction. J Mater Chem B 2020;8:10384–10391. [DOI] [PubMed] [Google Scholar]
- 153. Liu M, Li M, Sun S, Li B, Du D, Sun J, et al. The use of antibody modified liposomes loaded with AMO-1 to deliver oligonucleotides to ischemic myocardium for arrhythmia therapy. Biomaterials 2014;35:3697–3707. [DOI] [PubMed] [Google Scholar]
- 154. Khaw BA, DaSilva J, Hartner WC. Cytoskeletal-antigen specific immunoliposome-targeted in vivo preservation of myocardial viability. J Control Release 2007;120:35–40. [DOI] [PubMed] [Google Scholar]
- 155. Scott RC, Rosano JM, Ivanov Z, Wang B, Chong PL, Issekutz AC, et al. Targeting VEGF-encapsulated immunoliposomes to MI heart improves vascularity and cardiac function. FASEB J 2009;23:3361–3367. [DOI] [PubMed] [Google Scholar]
- 156. Wang B, Cheheltani R, Rosano J, Crabbe DL, Kiani MF. Targeted delivery of VEGF to treat myocardial infarction. Adv Exp Med Biol 2013;765:307–314. [DOI] [PubMed] [Google Scholar]
- 157. Yamada Y, Kobayashi H, Iwasa M, Sumi S, Ushikoshi H, Aoyama T, et al. Postinfarct active cardiac-targeted delivery of erythropoietin by liposomes with sialyl Lewis X repairs infarcted myocardium in rabbits. Am J Physiol Heart Circ Physiol 2013;304:H1124–H1133. [DOI] [PubMed] [Google Scholar]
- 158. Nguyen MM, Carlini AS, Chien MP, Sonnenberg S, Luo C, Braden RL, et al. Enzyme-responsive nanoparticles for targeted accumulation and prolonged retention in heart tissue after myocardial infarction. Adv Mater 2015;27:5547–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Fang J, Islam W, Maeda H. Exploiting the dynamics of the EPR effect and strategies to improve the therapeutic effects of nanomedicines by using EPR effect enhancers. Adv Drug Deliv Rev 2020;157:142–160. [DOI] [PubMed] [Google Scholar]
- 160. Prajnamitra RP, Chen HC, Lin CJ, Chen LL, Hsieh PC. Nanotechnology approaches in tackling cardiovascular diseases. Molecules 2019;24:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Krause KT, Jaquet K, Geidel S, Schneider C, Mandel C, Stoll HP, et al. Percutaneous endocardial injection of erythropoietin: assessment of cardioprotection by electromechanical mapping. Eur J Heart Fail 2006;8:443–450. [DOI] [PubMed] [Google Scholar]
- 162. Tao Z, Chen B, Zhao Y, Chen H, Wang L, Yong Y, et al. HGF percutaneous endocardial injection induces cardiomyocyte proliferation and rescues cardiac function in pigs. J Biomed Res 2010;24:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Dib N, Menasche P, Bartunek JJ, Zeiher AM, Terzic A, Chronos NA, et al. Recommendations for successful training on methods of delivery of biologics for cardiac regeneration: a report of the International Society for Cardiovascular Translational Research. JACC Cardiovasc Interv 2010;3:265–275. [DOI] [PubMed] [Google Scholar]
- 164. Ishikawa K, Aguero J, Naim C, Fish K, Hajjar RJ. Percutaneous approaches for efficient cardiac gene delivery. J Cardiovasc Transl Res 2013;6:649–659. [DOI] [PubMed] [Google Scholar]
- 165. Lourenco AP, Leite-Moreira AF, Balligand JL, Bauersachs J, Dawson D, de Boer RA, et al. An integrative translational approach to study heart failure with preserved ejection fraction: a position paper from the Working Group on Myocardial Function of the European Society of Cardiology. Eur J Heart Fail 2018;20:216–227. [DOI] [PubMed] [Google Scholar]
- 166. Losordo DW, Dimmeler S. Therapeutic angiogenesis and vasculogenesis for ischemic disease: part II: cell-based therapies. Circulation 2004;109:2692–2697. [DOI] [PubMed] [Google Scholar]
- 167. Claesson-Welsh L, Dejana E, McDonald DM. Permeability of the endothelial barrier: identifying and reconciling controversies. Trends Mol Med 2021;27:314–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Emani SM, Shah AS, Bowman MK, Emani S, Wilson K, Glower DD, et al. Catheter-based intracoronary myocardial adenoviral gene delivery: importance of intraluminal seal and infusion flow rate. Mol Ther 2003;8:306–313. [DOI] [PubMed] [Google Scholar]
- 169. Hayase M, Del Monte F, Kawase Y, Macneill BD, McGregor J, Yoneyama R, et al. Catheter-based antegrade intracoronary viral gene delivery with coronary venous blockade. Am J Physiol Heart Circ Physiol 2005;288:H2995–H3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Byrne MJ, Power JM, Preovolos A, Mariani JA, Hajjar RJ, Kaye DM. Recirculating cardiac delivery of AAV2/1SERCA2a improves myocardial function in an experimental model of heart failure in large animals. Gene Ther 2008;15:1550–1557. [DOI] [PubMed] [Google Scholar]
- 171. Kaye DM, Preovolos A, Marshall T, Byrne M, Hoshijima M, Hajjar R, et al. Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals. J Am Coll Cardiol 2007;50:253–260. [DOI] [PubMed] [Google Scholar]
- 172. Fargnoli AS, Katz MG, Yarnall C, Sumaroka MV, Stedman H, Rabinowitz JJ, et al. A pharmacokinetic analysis of molecular cardiac surgery with recirculation mediated delivery of betaARKct gene therapy: developing a quantitative definition of the therapeutic window. J Card Fail 2011;17:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Boekstegers P, von Degenfeld G, Giehrl W, Heinrich D, Hullin R, Kupatt C, et al. Myocardial gene transfer by selective pressure-regulated retroinfusion of coronary veins. Gene Ther 2000;7:232–240. [DOI] [PubMed] [Google Scholar]
- 174. Hou D, Youssef EA, Brinton TJ, Zhang P, Rogers P, Price ET, et al. Radiolabeled cell distribution after intramyocardial, intracoronary, and interstitial retrograde coronary venous delivery: implications for current clinical trials. Circulation 2005;112:I150–I156. [DOI] [PubMed] [Google Scholar]
- 175. Raake PW, Schlegel P, Ksienzyk J, Reinkober J, Barthelmes J, Schinkel S, et al. AAV6.betaARKct cardiac gene therapy ameliorates cardiac function and normalizes the catecholaminergic axis in a clinically relevant large animal heart failure model. Eur Heart J 2013;34:1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Tuma J, Fernandez-Vina R, Carrasco A, Castillo J, Cruz C, Carrillo A, et al. Safety and feasibility of percutaneous retrograde coronary sinus delivery of autologous bone marrow mononuclear cell transplantation in patients with chronic refractory angina. J Transl Med 2011;9:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Raake P, von Degenfeld G, Hinkel R, Vachenauer R, Sandner T, Beller S, et al. Myocardial gene transfer by selective pressure-regulated retroinfusion of coronary veins: comparison with surgical and percutaneous intramyocardial gene delivery. J Am Coll Cardiol 2004;44:1124–1129. [DOI] [PubMed] [Google Scholar]
- 178. Ladage D, Turnbull IC, Ishikawa K, Takewa Y, Rapti K, Morel C, et al. Delivery of gelfoam-enabled cells and vectors into the pericardial space using a percutaneous approach in a porcine model. Gene Ther 2011;18:979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Sahoo S, Kariya T, Ishikawa K. Targeted delivery of therapeutic agents to the heart. Nat Rev Cardiol 2021;18:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol 2011;30:16–34. [DOI] [PubMed] [Google Scholar]
- 181. Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 2003;5:834–839. [DOI] [PubMed] [Google Scholar]
- 182. Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science 2003;300:1524–1525. [DOI] [PubMed] [Google Scholar]
- 183. Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev 2007;18:363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 2005;23:457–462. [DOI] [PubMed] [Google Scholar]
- 185. Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2'-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther 2007;15:1663–1669. [DOI] [PubMed] [Google Scholar]
- 186. Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 2006;13:494–505. [DOI] [PubMed] [Google Scholar]
- 187. Lee Y, Urban JH, Xu L, Sullenger BA, Lee J. 2'Fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Ther 2016;26:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev 2003;17:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Seok H, Lee H, Jang ES, Chi SW. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol Life Sci 2018;75:797–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, et al. 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 2006;3:199–204. [DOI] [PubMed] [Google Scholar]
- 191. Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 2006;12:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Sokilde R, Newie I, Persson H, Borg A, Rovira C. Passenger strand loading in overexpression experiments using microRNA mimics. RNA Biol 2015;12:787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Goldgraben MA, Russell R, Rueda OM, Caldas C, Git A. Double-stranded microRNA mimics can induce length- and passenger strand-dependent effects in a cell type-specific manner. RNA 2016;22:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.