Abstract
Warmer climatic conditions have been associated with fewer COVID-19 cases. Herein we infected K18-hACE2 mice housed at the standard animal house temperature of ∼22 °C, or at ∼31 °C, which is considered to be thermoneutral for mice. On day 2 post infection, RNA-Seq analyses showed no significant differential gene expression lung in lungs of mice housed at the two temperatures, with almost identical viral loads and type I interferon responses. There was also no significant difference in viral loads in lungs on day 5, but RNA-Seq and histology analyses showed clearly elevated inflammatory signatures and infiltrates. Thermoneutrality thus promoted lung inflammation. On day 2 post infection mice housed at 31 °C showed reduced viral loads in nasal turbinates, consistent with increased mucociliary clearance at the warmer ambient temperature. These mice also had reduced virus levels in the brain, and an ensuing amelioration of weight loss and a delay in mortality. Warmer air temperatures may thus reduce infection of the upper respiratory track and the olfactory epithelium, resulting in reduced brain infection. Potential relevance for anosmia and neurological sequelae in COVID-19 patients is discussed.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ARDS, acute respiratory distress syndrome; K18-hACE2, transgenic mice where the keratin 18 promoter drives expression of human angiotensin converting enzyme 2; CCID50, 50 % cell culture infectivity dose; RT-qPCR, quantitative reverse transcription polymerase chain reaction; RNA-Seq, RNA sequencing (whole transcriptome); IFN, interferon; TPM, transcripts per million; DEGs, differentially expressed genes; GSEA, gene set enrichment analysis; IPA, Ingenuity pathway analysis; NES, normalized enrichment score; ImmuneSigDB, immune signatures database; H&E, hematoxylin and eosin; vs., versus
Keywords: SARS-CoV-2, COVID-19, Temperature, Climate, Thermoneutrality, Mucociliary clearance, Brain, Anosmia
Graphical abstract
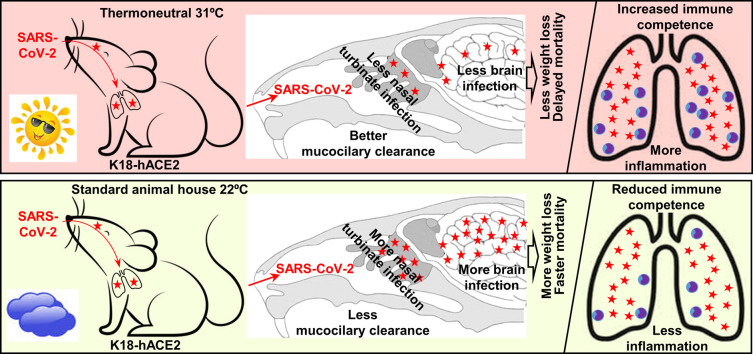
Interpretation of the data presented herein; warmer ambient temperatures improve mucocilary clearance thereby reducing nasal turbinate infection, which reduces infection of the olfactory epithelium and brain infection in K18-hACE2 mice. Warmer temperatures also improve immune competence leading to promotion of inflammation in the lungs.
1. Introduction
Ambient temperature can have important effects on virus replication in a diversity of settings and for a range of different viruses (Lane et al., 2018; Prow et al., 2017; Wimalasiri-Yapa et al., 2021; Zhang et al., 2017). In particular, for respiratory viruses, colder climatic conditions have traditionally been associated with promotion of infection, with, for instance, clear seasonal fluctuations in the northern hemisphere seen for influenza and common cold viruses (Maciorowski et al., 2021; Madaniyazi et al., 2021). Epidemiological studies on SARS-CoV-2 infections and COVID-19 disease have similarly associated lower climatic temperatures with an increase in the number of infections and an increase in the mortality rate (Burra et al., 2021; Chen et al., 2021; Christophi et al., 2021; Mu et al., 2021; Sobral et al., 2020; Solmaz et al., 2021; Wang et al., 2021b; Wu et al., 2020; Yuan et al., 2021). A range of mechanisms may be involved such as virus stability, humidity and human behavior (Maciorowski et al., 2021). However, once infected, increased air temperature (over a 2–11 °C range) in field hospitals in China correlated with improved survival (Cai et al., 2020). Perhaps in contrast, mild hypothermia (cooling the blood to 33.5 °C) was recently used to treat a COVID-19 patient (Cruces et al., 2021), with mild hypothermia therapy also shown to reduce markers of injury and inflammation in experimental models of acute respiratory distress syndrome (ARDS) (Akyol et al., 2022; Angus et al., 2022).
Mouse models have been used to explore the role of ambient temperature on infection and disease caused by a number of different viruses. For instance, survival from rabies virus challenge was higher for mice housed at 35 °C when compared with 21 °C (Bell and Moore, 1974). Similarly, survival after herpes simplex virus and Coxsackie A-21 infections was higher in mice held at 36 °C when compared with 25 °C (Underwood et al., 1966). These results have been attributed to increased immune cell competence at the warmer temperatures, with the energy needed to drive immune processes not being diverted towards heat generation (Vialard and Olivier, 2020). Humans are deemed to be thermoneutral at ≈20–22 °C, whereas for mice the thermoneutral temperature is believed to be ≈29–32 °C (Seeley and MacDougald, 2021). Arguably, experimental mice kept at temperatures below thermoneutrality (usually 20–22 °C) are permanently cold stressed, potentially complicating interpretation of mouse experiments and their relevance to human disease (Ganeshan and Chawla, 2017; Seeley and MacDougald, 2021; Speakman and Keijer, 2012). However, others have argued that cold stress may be ameliorated by provision of nesting materials and co-housing with other mice allowing huddling (Gaskill and Garner, 2014), with such housing conditions now generally met in most animal houses.
The innate anti-viral type I interferon (IFN) system represents a key response to many acute viral infections, and can provide rapid early protection against explosive viral replication (Munoz-Moreno et al., 2021; Rudd et al., 2012). Tissues and cells kept at only a few degrees below normal mammalian core body temperatures (≈36–37 °C) have been shown to exhibit suboptimal type I IFN responses after infection with respiratory (Boonarkart et al., 2017; Eccles, 2021; Foxman et al., 2015; Foxman et al., 2016) and non-respiratory viruses (Lane et al., 2018; Prow et al., 2017; Zhang et al., 2017) resulting in increased viral replication. This effect can also be observed in vivo; for instance, mice infected with chikungunya virus and housed at 30 °C, rather than 22 °C, showed reduced virus replication in feet with a subsequent reduction in arthritis (Prow et al., 2017). However, SARS-CoV-2 infection of IFNAR−/− mice indicated that endogenously induced type I IFN responses do not significantly suppress SARS-CoV-2 replication (Rawle et al., 2021), with SARS-CoV-2 proteins able to inhibit various steps in type I IFN production and the ensuing responses (Bastard et al., 2022; Guo et al., 2022; Setaro and Gaglia, 2021). Type I IFN responses are, nevertheless, induced and contribute to immunopathology (Kim and Shin, 2021; Lu et al., 2022; Rawle et al., 2021), with inhibition of type I IFN production using a cGAS-STING inhibitor (H-151) able to inhibit SARS-CoV-2 lung immunopathology in K18-hACE2 mice (Andreakos, 2022; Domizio et al., 2022).
K18-hACE2 mice have been widely used to evaluate interventions against SARS-CoV-2 infection and disease (Alsoussi et al., 2020; Garcia-Arriaza et al., 2021; Hassan et al., 2020; Mills et al., 2021; Rosenfeld et al., 2021; Yan et al., 2022; Zheng et al., 2021) and (when housed at 22 ± 1 °C) develop lung infection and respiratory disease resembling severe COVID-19-associated ARDS (Arce and Costoya, 2021; Yinda et al., 2021). RNA-Seq and bioinformatic analyses of SARS-CoV-2 infected lungs of K18-hACE2 mice also show a high level of concordance in proinflammatory cytokine responses (the key drivers of ARDS) to those seen in infected human lungs/lung tissues (Bishop et al., 2022). K18-hACE2 mice infected via the intranasal route also develop a fulminant brain infection, which is associated with mortality (Carossino et al., 2022; Fumagalli et al., 2022; Kumari et al., 2021). Fulminant brain infection is, however, not a feature of human disease (Butowt et al., 2021; Carossino et al., 2022; Fernandez-Castaneda et al., 2022), although evidence for some level of brain infection in COVID-19 patients has emerged (Balasubramanian et al., 2022; Chertow et al., 2021; Fu et al., 2022; Gagliardi et al., 2021; Liu et al., 2021; Serrano et al., 2021; Song et al., 2021; Wang et al., 2021a; Zhou et al., 2021), and may play an important part in the neurological manifestations of COVID-19 and long-COVID (Bauer et al., 2022; None, 2021). Infection of the olfactory epithelium (Bryche et al., 2020; de Melo et al., 2021; Zheng et al., 2021), perhaps also involving infection of olfactory neurons (Lempriere, 2021; Piras et al., 2021; Ye et al., 2021; Zhang et al., 2021), has been reported for animal models and humans and is likely associated with anosmia (loss of smell), a common COVID-19 symptom (Kumar et al., 2021). In K18-hACE2 mice, virus likely enters the brain via the olfactory epithelium (Carossino et al., 2022; Kumari et al., 2021; Yu et al., 2022), with high virus titers seen in the nasal turbinates after intranasal delivery of virus (Carossino et al., 2022; Guimond et al., 2022; Kumari et al., 2021; van Oosten et al., 2022; Yu et al., 2022; Zheng et al., 2021). Brain infection is also seen in hamsters (de Melo et al., 2021) and in a mouse model where hACE2 is driven from the mouse ACE2 promoter (Sun et al., 2020). Infection with a number of viruses in the family Coronaviridae have also been associated with neurological involvement (Alluwaimi et al., 2020).
The K18-hACE2 mouse model was used herein to explore the role of ambient temperature on SARS-CoV-2 infection and disease. Mice were housed at standard animal house temperature of 22 ± 1 °C or were housed at 31 ± 1 °C; the latter considered thermoneutral for mice (Seeley and MacDougald, 2021). The mice were then infected with SARS-CoV-2 to ascertain whether the increased temperature would influence infection, the innate immune responses and/or immunopathology. Mice housed at 31 °C showed decreased infection of nasal turbinates and brain, but also showed increased lung inflammation.
2. Materials and methods
2.1. Ethics statement and regulatory compliance
All mouse work was conducted in accordance with the Australian code for the care and use of animals for scientific purposes as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute Animal Ethics Committee (P3600). All infectious SARS-CoV-2 work was conducted in a dedicated suite in a biosafety level 3 (PC3) facility at the QIMR Berghofer MRI (Australian Department of Agriculture, Water and the Environment certification Q2326 and Office of the Gene Technology Regulator certification 3445). Breeding and use of GM mice was approved under a Notifiable Low Risk Dealing (NLRD) Identifier: NLRD_Suhrbier_Oct2020: NLRD 1.1(a). Mice were euthanized using carbon dioxide.
2.2. SARS-CoV-2
The SARS-CoV-2 isolate (hCoV-19/Australia/QLD02/2020) (GISAID accession EPI_ISL_407896) (original strain) was kindly provided by Dr. Alyssa Pyke (Queensland Health Forensic & Scientific Services, Queensland Department of Health, Brisbane, Australia). Virus stocks were generated in Vero E6 cells as described (Rawle et al., 2021). The virus was determined to be mycoplasma free using co-culture with a non-permissive cell line (i.e. HeLa) and Hoechst staining as described (La Linn et al., 1995). The Fetal Bovine Serum (Gibco) used to propagate cells and virus was determined to be endotoxin free using RAW264-HIV-LTR-luc indicator cells (Johnson et al., 2005). The virus was titered using CCID50 assays (see below).
2.3. K18-hACE2 mice, housing and infection
K18-hACE2 mice (strain B6.Cg-Tg(K18-ACE2)2Prlmn/J, JAX Stock No: 034860) were purchased from The Jackson Laboratory, USA, and were maintained in-house as heterozygotes by backcrossing to C57BL/6J mice (Bishop et al., 2022). Mice were genotyped using hACE2 Primers: Forward: 5′-CTT GGT GAT ATG TGG GGT AGA -3′; Reverse: 5′-CGC TTC ATC TCC CAC CAC TT -3′ (recommended by NIOBIOHN, Osaka, Japan) (Amarilla et al., 2021; Bishop et al., 2022).
The housing conditions for the mice were as follows; light = 12:12 h dark/light cycle, 7:45 a.m. sunrise and 7:45 p.m. sunset, 15 min light dark and dark light ramping time. Enclosures, M.I.C.E cage (Animal Care Systems, Colorado, USA); up to 6 mice per cage. Ambient temperature, 22 ± 1 °C. Humidity, ≈55 %. Ventilation, 100 % fresh air, 16 complete air exchange/h/room, with cages on continuous air flow sourced from the room. Environmental enrichment, paper cups (Impact-Australia), tissue paper, cardboard rolls. Bedding, PuraChips (Able Scientific) (aspen fine). Food, double bagged Norco rat and mouse pellet (AIRR, Darra, Qld.). Drinking water was deionized and acidified with HCl (pH ≈ 3.2). Although acidification can influence the microbiome (Whipple et al., 2021), this is standard practice to prevent infection of the water supply; a pertinent feature especially at the higher housing temperature.
For each experiment mice were sorted into groups with a similar age distribution and the same gender in each group. Mice were then moved to the PC3 facility and housed under the same conditions as above but at 22 ± 1 °C or 31 ± 1 °C for 24 h prior to being infected, and were housed at these temperatures until they were euthanized. Mice were infected intrapulmonarily via the nasal route with 5 × 104 CCID50 of virus in 50 μl medium while under light anaesthesia; 3 % isoflurane (Piramal Enterprises Ltd., Andhra Pradesh, India) delivered using The Stinger, Rodent Anaesthesia System (Advanced Anaesthesia Specialists/Darvall, Gladesville, NSW, Australia). Mice were euthanized using CO2, and tissues were homogenized using four ceramic beads at 6000 rpm twice for 15 s (Precellys 24 Homogenizer, Bertin Instruments, Montigny-le-Bretonneux, France). After centrifugation for 10 min, 9400 ×g at 4 °C, virus titers in supernatants were determined by CCID50 assays using Vero E6 cells.
2.4. Disease scores
Overt clinical signs of mice were scored on a scale of 0–3 (Diseases scores) according to posture, activity, and fur ruffling. For all criteria, the normal condition was designated as 0. For posture, hunching only while at rest was designated as 1, moderate hunching with some impairment of normal movement was designated as 2, and severe hunching with difficulty in maintaining upright posture was designated as 3. For activity, a mild to moderate decrease was designated as 1, stationary unless stimulated was designated as 2, and reluctant to move even if stimulated was designated as 3. For fur ruffling, mild to moderate fur ruffling was designated as 1, severe ruffling was designated as 2, and any sign of shivering was designated as 3. Any animal reaching a level of 3 in any single criterion was euthanized, and any animal reaching a level of 2 in two or more criteria was euthanized.
2.5. CCID50 assays
CCID50 assays were undertaken using Vero E6 cells using 10 fold serial dilution in duplicate as described (Rawle et al., 2021; Yan et al., 2021).
2.6. Histology
Lungs were fixed in 10 % formalin, embedded in paraffin, and sections stained with H&E (Sigma-Aldrich, Darmstadt, Germany). Slides were scanned using Aperio AT Turbo (Aperio, Vista, CA, USA) and images extracted using Aperio ImageScope software v12.3.2.8013 (Leica Biosystems, Wetzlar, Germany). Leukocyte infiltrates were quantified by measuring nuclear (strong purple staining) / cytoplasmic (total red staining) pixel ratios in scanned H&E stained images (n = 2 whole lung sections per mouse), and was undertaken using Aperio Positive Pixel Count Algorithm (Leica Biosystems) (Prow et al., 2017). Quantitation of white space in scanned images of H&E stained lung parenchyma (with areas greater than ≈100 μm set as a threshold) was undertaken using PixelClassifierTools in QuPath v0.3.2.
2.7. RT-qPCR
Quantitative reverse transcriptase PCR was undertaken as described (Rawle et al., 2021) using the following primers: SARS-CoV-2 E, F 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′, R 5′-ATATTGCAGCAGTACGCACACA; and mouse Rpl13a F 5′-GAGGTCGGGTGGAAGTACCA-3′, R 5′-TGCATCTTGGCCTTTTCCTT-3′.
2.8. RNA-Seq and bioinformatics
In-house RNA-Seq was undertaken as described using female mice (n = 6 mice per cage) using Illumina Nextseq 550 platform generating 75 bp paired end reads (Bishop et al., 2022; Rawle et al., 2021). The per base sequence quality for >90 % bases was above Q30 for all samples. Subsequent analyses were undertaken as described (Bishop et al., 2022; Rawle et al., 2021), with the mouse reference genome GRCm39 primary assembly and GENCODE M27 used in the combined reference that included SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2; 29,903 bp). In brief, counts for mouse genes and for SARS-CoV-2 were generated using RSEM and differentially expressed genes were determined using EdgeR. To avoid missing type I IFN genes, which have low read counts (Wilson et al., 2017), a low filter of row sum normalized read count >1 was used.
DEGs in direct and indirect interactions were analyzed using Ingenuity Pathway Analysis (IPA) (QIAGEN) using the Canonical pathways, Up-Stream Regulators (USR) and Diseases and Functions features (Rawle et al., 2022). Enrichment for biological processes, molecular functions, KEGG pathways, and other gene ontology categories in DEG lists was elucidated using the STRING database (Szklarczyk et al., 2019) in Cytoscape (v3.7.2) (Shannon et al., 2003). GSEAs were undertaken using MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/) and ImmuneSigDB (https://immunespace.org/announcements/home/thread.view?rowId=50).
2.9. Statistics
Statistical analyses of experimental data were performed using IBM SPSS Statistics for Windows, Version 19.0 (IBM Corp., Armonk, NY, USA). The t-test was used when the difference in variances was <4, skewness was > minus 2 and kurtosis was <2 (deemed normally distributed). Otherwise, for non-parametric data the Mann Whitney U test was used if the difference in variances was <4, otherwise the Kolmogorov-Smirnov test was used.
3. Results
3.1. Nasal turbinates on day 2 post infection showed lower viral loads at 31 °C vs. 22 °C
In this K18-hACE2 mouse model of SARS-CoV-2 infection and disease, virus is generally inoculated into the lungs of lightly anesthetized mice via the intranasal route, with the nasal turbinates (part of the upper respiratory tract) becoming infected and showing high viral loads (Carossino et al., 2022; Guimond et al., 2022; Kumari et al., 2021; van Oosten et al., 2022; Yu et al., 2022; Zheng et al., 2021), which peak around day 2 post infection (Oladunni et al., 2020; Rathnasinghe et al., 2020).
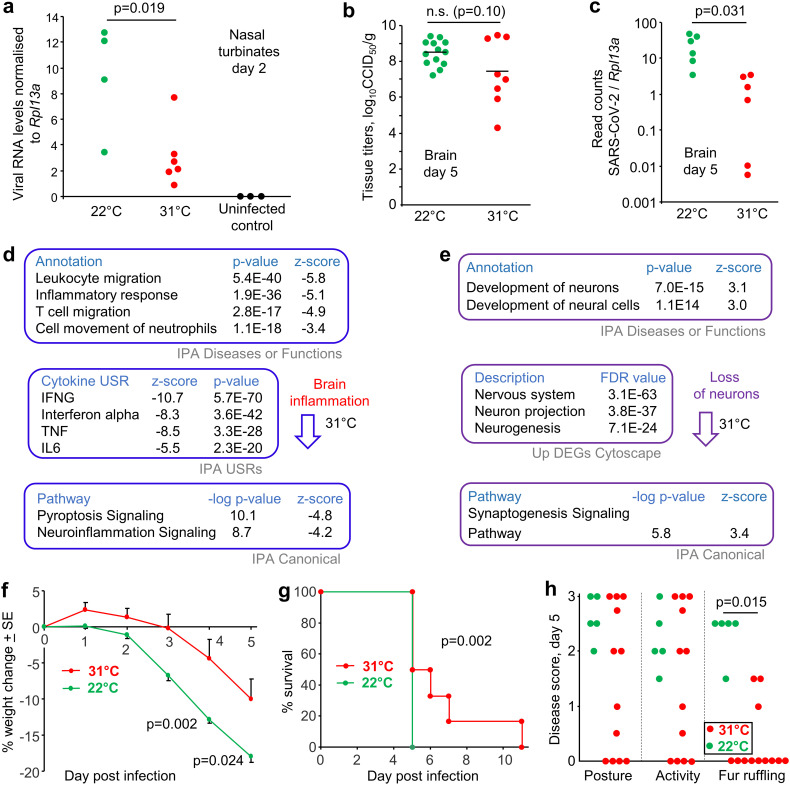
K18-hACE2 mice were housed at standard animal house temperatures (22 ± 1 °C) and were then moved to rooms held at 31 ± 1 °C or 22 ± 1 °C, hereafter referred to as 31 °C or 22 °C, respectively. A day later, all mice were inoculated with 50 μl of 5 × 104 CCID50 SARS-CoV-2 (original strain, QLD02) into the lungs via the intranasal route. On day 2 post infection, mice housed at 31 °C showed significantly lower levels of viral RNA in their nasal turbinates than mice housed at 22 °C, as measure by RT-qPCR and normalized to the house keeping gene Rpl13a (Mogal and Abdulkadir, 2006) (Fig. 1a).
Fig. 1.
Nasal turbinate and brain infection for mice housed at 31 °C vs. 22 °C. a K18-hACE2 mice were housed at 31 °C or 22 °C and were infected with SARS-CoV-2 intrapulmonarily via the intranasal route. Nasal turbinates were harvested on day 2 post infection from mice infected with SARS-CoV-2 and viral load determined by RT-qPCR normalized to Rpl13a (House-keeping gene). Statistics by Mann Whitney U test. b Brains were harvested on day 5 post infection and tissue titers determined by CCID50 assays. Data from 2 independent experiments. c. Brains were harvested on day 5 post infection and analyzed by RNA Seq. TPM normalized viral read counts normalized to TPM normalized Rpl13a read counts are shown. Statistics by Kolmogorov Smirnov test. d Brains were harvested on day 5 post infection and were analyzed by RNA Seq to provide 2022 DEGs. DEGs were analyzed by IPA and selected top inflammation annotations shown (full data sets are available in Supplementary Table 1). e As for d but showing selected neuron-associated signatures identified in IPA, and by Cytoscape in up-regulated DEGs. f Percent body weight change relative to day 0 determined on the indicated days; n = 12 mice per group, data derived from two independent experiments. Statistics by Kolmogorov-Smirnov tests. g Survival; time until mice reached ethically defined end points for euthanasia; n = 6 for 31 °C and n = 17 for 22 °C (the latter derived from 3 independent experiments). Significance by log rank statistic. h Disease scores (data for 31 °C taken from 2 independent experiments). Statistics by Kolmogorov-Smirnov test.
Nasal turbinate and brain infection for mice housed at 31 °C vs. 22 °C. a K18-hACE2 mice were housed at 31 °C or 22 °C and were infected with SARS-CoV-2 intrapulmonarily via the intranasal route. Nasal turbinates were harvested on day 2 post infection from mice infected with SARS-CoV-2 and viral load determined by RT-qPCR normalized to Rpl13a (House-keeping gene). Statistics by Mann Whitney U test. b Brains were harvested on day 5 post infection and tissue titers determined by CCID50 assays. Data from 2 independent experiments. c. Brains were harvested on day 5 post infection and analyzed by RNA Seq. TPM normalized viral read counts normalized to TPM normalized Rpl13a read counts are shown. Statistics by Kolmogorov Smirnov test. d Brains were harvested on day 5 post infection and were analyzed by RNA Seq to provide 2022 DEGs. DEGs were analyzed by IPA and selected top inflammation annotations shown (full data sets are available in Supplementary Table 1). e As for d but showing selected neuron-associated signatures identified in IPA, and by Cytoscape in up-regulated DEGs. f Percent body weight change relative to day 0 determined on the indicated days; n = 12 mice per group, data derived from two independent experiments. Statistics by Kolmogorov-Smirnov tests. g Survival; time until mice reached ethically defined end points for euthanasia; n = 6 for 31 °C and n = 17 for 22 °C (the latter derived from 3 independent experiments). Significance by log rank statistic. h Disease scores (data for 31 °C taken from 2 independent experiments). Statistics by Kolmogorov-Smirnov test.
3.2. Brains on day 5 post infection showed lower viral loads and inflammation at 31 °C vs. 22 °C
Infection of K18-hACE2 mice with SARS-CoV-2QLD02 is associated with a fulminant brain infection (Guimond et al., 2022; van Oosten et al., 2022) that is also seen with other SARS-CoV-2 strains, with the virus likely reaching the olfactory bulb and then the brain via the olfactory neuroepithelium (Carossino et al., 2022; Fumagalli et al., 2022; Ye et al., 2021; Yu et al., 2022). The olfactory epithelium is located at the back of the nasal turbinates, separated from the olfactory bulb by the cribriform plate (Gardner et al., 2015), with axons of the olfactory neurons passing through the plate to the olfactory bulb (Bilinska et al., 2020).
The mean infectious virus titers in brain on day 5 post infection were lower for mice housed at 31 °C; but this did not reach significance (Fig. 1b). However, RNA-Seq did show a significantly lower level of viral RNA (read counts normalized to Rpl13a) in the brains of these mice on day 5 (Fig. 1c), consistent with lower viral RNA levels in the nasal turbinates on day 2 (Fig. 1a). RNA-Seq provides viral RNA reads from both infectious virus, as well as non-infectious viral RNA that has accumulated over several days of infection (Martin and Griffin, 2018).
Consistent with the reduced viral load (Fig. 1c), the brains of infected mice housed at 31 vs. 22 °C showed a series of inflammation-associated annotations with negative z-scores (Fig. 1d; Supplementary Table 1). Neurons are infected in K18-hACE2 brains (Carossino et al., 2022; Dong et al., 2022), with neuron-associated signatures identified with positive z-scores and neuron-associated genes found in up-regulated DEGs, indicating reduced infection and disruption of these cells at 31 °C (Fig. 1e, Supplementary Table 1).
3.3. Improved survival of SARS-CoV-2 infected K18-hACE2 mice housed at 31 °C vs. 22 °C
In the K18-hACE2 model the fulminant brain infection is associated with weight loss and mortality (Carossino et al., 2022; Fumagalli et al., 2022; Yu et al., 2022), with ethically defined end points for euthanasia usually met by day 5 post infection (Amarilla et al., 2021; Bishop et al., 2022; Guimond et al., 2022).
Mice held at 31 °C showed significantly less weight loss on days 4 and 5 post infection (Fig. 1f) and survived slightly, but significantly, longer (Fig. 1g), consistent with the reduced level of brain infection and inflammation. Although disease score for activity and posture were not significantly different, fur ruffling was significantly less prominent in mice housed at 31 °C (Fig. 1h). Ruffled fur is generally considered to be a sign of a febrile response (Groeneveld et al., 1988; Smith et al., 2010), with shivering and chills likely to be ameliorated at warmer temperatures. The febrile response has recently been shown to involve the brain (Eskilsson et al., 2021).
3.4. No significant differential gene expression for lungs on day 2 for 31 °C vs. 22 °C
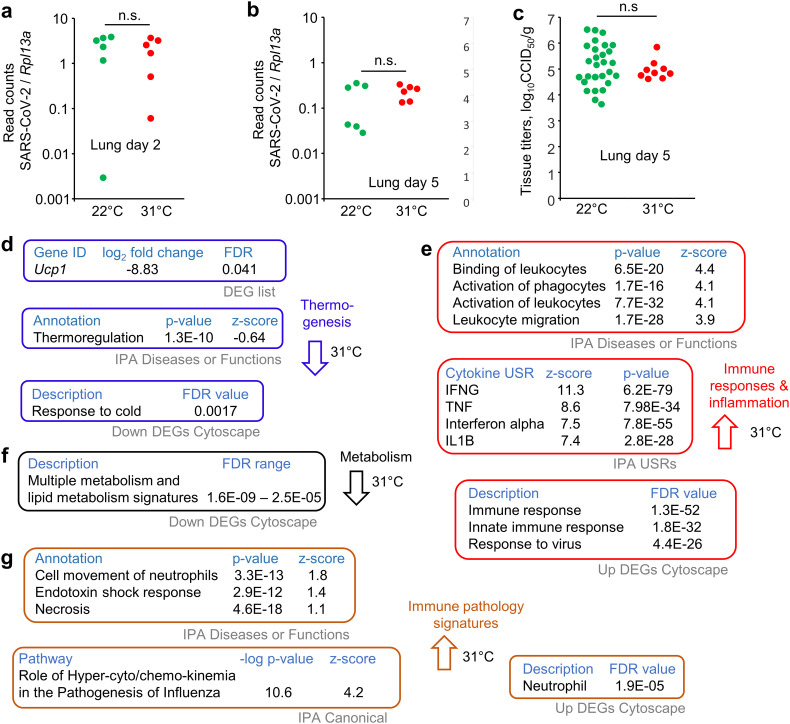
Although in humans SARS-CoV-2 infection generally travels from the upper to the lower respiratory track, infection of lungs in the K18-hACE2 mouse model requires direct inoculation of virus into the lungs of anesthetized mice, with an estimated ≈40 μl of the inoculum reaching the lower respiratory track. Lungs were harvested on day 2 post infection and were analyzed by RNA-Seq, with SARS-CoV-2 read counts not significantly different for mice held at the two temperatures (Fig. 2a).
Fig. 2.
Lung RNA-Seq for mice housed at 31 °C vs. 22 °C. a Lungs were taken day 2 post infection and were analyzed by RNA-Seq. TPM normalized read counts for SARS-CoV-2 divided by TPM normalized read counts for Rpl13a (house keeping gene) are shown for each mouse. n.s. – not significant, Kolmogorov Smirnov test. b As for a but for lungs taken on day 5 post infection. Statistics as in a. c Lung virus titers on day 5 post infection determined by CCID50 assays; data derived from 4 independent experiments. Limit of detection ≈2 log10CCID50/g. Statistics as in a. d RNA-Seq of mouse lungs taken day 5 post infection comparing mice held at 31 °C vs. 22 °C. The 841 DEGs were analyzed by IPA and Cytoscape. Thermogenesis associated genes and annotations are shown (full data sets are available in Supplementary Table 3). e As for d but showing selected dominant immune response and inflammation-associated annotations. f The DEGs down-regulated at 31 °C in lungs were analyzed by Cytoscape, with multiple metabolism signatures identified (Supplementary Table 3f). g As for d but showing dominant immune pathology associated annotations.
Lung RNA-Seq for mice housed at 31 °C vs. 22 °C. a Lungs were taken day 2 post infection and were analyzed by RNA-Seq. TPM normalized read counts for SARS-CoV-2 divided by TPM normalized read counts for Rpl13a (house keeping gene) are shown for each mouse. n.s. – not significant, Kolmogorov Smirnov test. b As for a but for lungs taken on day 5 post infection. Statistics as in a. c Lung virus titers on day 5 post infection determined by CCID50 assays; data derived from 4 independent experiments. Limit of detection ≈2 log10CCID50/g. Statistics as in a. d RNA-Seq of mouse lungs taken day 5 post infection comparing mice held at 31 °C vs. 22 °C. The 841 DEGs were analyzed by IPA and Cytoscape. Thermogenesis associated genes and annotations are shown (full data sets are available in Supplementary Table 3). e As for d but showing selected dominant immune response and inflammation-associated annotations. f The DEGs down-regulated at 31 °C in lungs were analyzed by Cytoscape, with multiple metabolism signatures identified (Supplementary Table 3f). g As for d but showing dominant immune pathology associated annotations.
RNA-Seq comparison of the transcriptome of infected lungs for mice held at the two temperatures provided no significantly differentially expressed genes (DEGs) on day 2 post infection. Consistent with this finding, type I IFN responses and type I IFN read counts were essentially identical (Supplementary Fig. 1). Thus, although substantial type I IFN activity was induced by SARS-CoV-2 infection on day 2 (as would be expected (Rawle et al., 2021)), neither the induction of type I IFN mRNA, nor the type I IFN-induced transcriptional responses showed any significant differences for 31 °C vs. 22 °C. Although type I IFN responses generally operate more effectively at temperatures closer to core body temperatures (Boonarkart et al., 2017; Eccles, 2021; Foxman et al., 2015; Foxman et al., 2016; Lane et al., 2018; Prow et al., 2017; Zhang et al., 2017), this would appear not to manifest during acute SARS-CoV-2 infection in this model.
To ascertain if there was any indications of a temperature effect, Gene Set Enrichment Analyses (GSEAs) were undertaken using ranked gene list (Supplementary Table 2a) and gene sets from the Molecular Signatures Database (MSigDB), a collection of ≈30,000 gene sets. Two dominant themes emerged from the signatures, muscle and epithelial differentiation (Supplementary Table 2b). These might be interpreted as a mild increase at 31 °C of smooth muscle hyperplasia and squamous metaplasia of lung epithelium, both of which have been described as inflammatory responses in SARS-CoV-2 infected lungs (Boszormenyi et al., 2021; Margaroli et al., 2021; Ramasamy et al., 2022; Recalde-Zamacona et al., 2020; Woolsey et al., 2021).
3.5. Increased inflammation signatures in lungs on day 5 post infection for 31 °C vs. 22 °C
RNA-Seq analysis of infected lungs on day 5 post infection for mice housed at 31 °C showed no significant difference in viral read counts (Fig. 2b). Viral titers in the lungs were also not significantly different (Fig. 2c). Nevertheless, RNA-Seq analysis of SARS-CoV-2 infected lungs on day 5 post infection for mice housed at 31 °C vs. 22 °C provided 841 DEGs (q < 0.05) (Supplementary Table 3a, b). The gene expression profile and bioinformatic analyses did, to some extent, reflect the different housing temperatures (Fig. 2d). Specifically, the most down-regulated gene at 31 °C was the mitochondrial uncoupling protein, Ucp1 (Fig. 2d; Supplementary Table 3b), with Ucp1 up-regulated by cooler ambient temperatures to promote thermogenesis (Cui et al., 2016; Denjean et al., 1999). The Thermoregulation annotation in IPA Disease or Functions showed a slight, but significant, negative z-score for mice housed at 31 °C (Fig. 2d; Supplementary Table 3d). The GO Process term Response to cold was identified as significant in the DEGs down-regulated at 31 °C (Fig. 2d; Supplementary Table 3f).
IPA and Cytoscape analyses of the DEGs showed a series of annotations illustrating that for mice housed at 31 °C, leukocyte activity and migration was increased, and cytokine responses were higher (Fig. 2e; Supplementary Table 3c–e). This observation is consistent with reports showing enhanced immune function and immune cell metabolism for mice housed at thermoneutral temperatures when compared with ≈21 °C (Carpenter et al., 2020; Rubin, 2017; Seeley and MacDougald, 2021; Vialard and Olivier, 2020). Mounting an immune response incurs a considerable energy burden, which may be harder to meet when energy allocations are diverted to thermogenesis (Derting Terry and Compton, 2003; Ganeshan et al., 2019). Such an interpretation is supported by the prominence of signatures associated with metabolic processes and metabolism of lipids in the down-regulated DEGs for infected lungs at 31 °C vs. 22 °C (Fig. 2f; Supplementary Table 3f). For mice held at 31 °C the requirement to burn fat to supply energy for thermogenesis is reduced, with an ensuing increase in energy availability for immune responses.
The acute respiratory distress syndrome (ARDS) that is associated with severe COVID-19 is generally viewed as a pro-inflammatory immunopathology with inter alia excessive IL-6, TNF, IL-1β (Wong and Perlman, 2022) and neutrophils (Cui et al., 2021). Although IL-6 was not identified (Supplementary Table 3c), the following inflammation-associated signatures emerged with positive z-scores for mice infected at 31 °C; (i) TNF and IL-1 β (Fig. 2e), (ii) neutrophils (Fig. 2g; Supplementary Table 3d, e), as well as (iii) shock, necrosis and influenza pathogenesis (Fig. 2g; Supplementary Table 3d, e, g). These data illustrate that at the warmer housing temperature, SARS-CoV-2-associated lung inflammation was more severe, suggestive of an increase in ARDS severity.
3.6. Lungs on day 5 post infection showed more cellular infiltrates at 31 °C vs. 22 °C
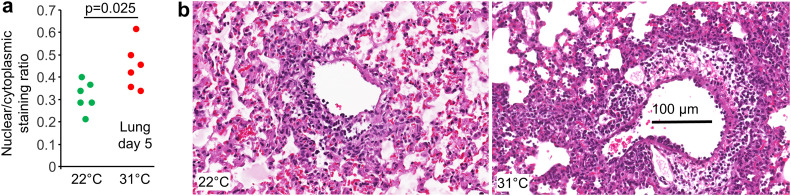
The bioinformatic signatures identified by IPA (e.g. Fig. 2e, leukocyte migration) were supported by histology, with Aperio image analysis of H&E stained lung sections showing an increase in the ratio of nuclear to cytoplasmic staining for mice housed at 31 °C (Fig. 3a). As leukocytes have a higher ratio of nuclear to cytoplasmic staining than resident tissue cells, an overall increase in nuclear/cytoplasmic staining ratios indicates increased levels of leukocyte infiltration (Prow et al., 2019; Prow et al., 2017). Examples of H&E staining are shown in Fig. 3b (an uninfected control is shown in Supplementary Fig. 2a). Lung consolidation (loss of alveolar airspaces) was measured as the proportion of white space in H&E stained lung parenchyma (Amarilla et al., 2021), but showed no significant differences (Supplementary Fig. 2b, c).
Fig. 3.
Lung histology for mice housed at 31 °C vs. 22 °C. a Aperio Positive Pixel Count Algorithm (Aperio ImageScope analysis) of H&E stained lung sections showing strong purple pixels (nuclear staining) over total red (cytoplasmic staining) ratios, which indicates the level of leukocyte infiltration. Statistics by t-test. b Example of H&E staining showing increased leucocyte infiltrates in infected lungs on day 5 for mice housed at 31 °C.
3.7. GSEAs indicated that inflammatory infiltrates and processes were similar for 31 °C vs. 22 °C
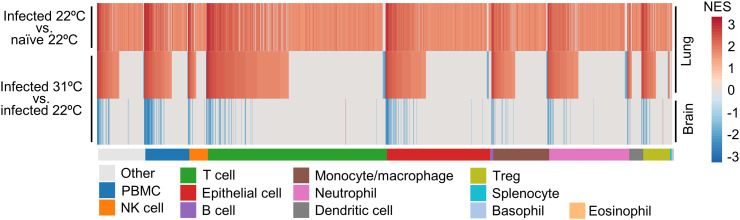
ImmuneSigDB provides ≈5000 immunology-specific gene sets that can be used to interrogate gene lists (herein ranked by fold change) using GSEAs (Godec et al., 2016). We have previously reported results of such an analysis for infected lungs vs. naïve lungs for this model at 22 °C, with significant GSEAs grouped by the cell type mentioned in the gene set annotation and ranked by Normalized Enrichment Scores (NES) in each group (Bishop et al., 2022). Here we compare the GSEAs for infected vs. naïve lungs (Fig. 4, top heat map) with the same analysis for infected lungs for mice housed at 31 °C vs. 22 °C (Fig. 4, middle heat map). The results show a high level of concordance, with gene sets showing significant enrichment and high NES overlapping considerably for the two data sets (Fig. 4; Supplementary Table 4). As might be expected, a greater number of significant GSEAs were identified for infected lungs vs. naïve lungs (n = 1967 significant GSEAs) than for infected lungs 31 °C vs. 22 °C (n = 997). The major cellular infiltrates and associated immune processes would thus appear to be largely similar at 22 °C and 31 °C.
Fig. 4.
GSEAs using ImmuneSigDB. GSEAs were undertaken using gene sets from ImmuneSigDB and ranked gene lists. Significant GSEAs were grouped by the indicated cell type (mentioned in the GSEA annotation) and ranked by NES and plotted as heat maps. Top, previously published results for lungs from Infected vs. naïve K18-hACE2 mice. Middle, lungs of infected mice 31 °C vs. 22 °C. Bottom, brains of infected mice 31 °C vs. 22 °C.
The same process was undertaken for infected brains for mice housed at 31 °C vs. 22 °C (Fig. 4, bottom heat map). Generally, annotations with positive NES in lung (increased at 31 °C) showed negative NES in brain (decreased at 31 °C) (Fig. 4; Supplementary Table 4), suggesting the cellular infiltrates and processes in infected lungs and brains are largely comparable, but increased in lung and decreased in brain for mice housed at 31 °C.
4. Discussion
We show herein that the improved ability of the early innate type I IFN anti-viral response to operate at temperatures closer to normal mammalian core body temperatures (≈36–37 °C) (Boonarkart et al., 2017; Eccles, 2021; Foxman et al., 2015; Foxman et al., 2016; Lane et al., 2018; Prow et al., 2017; Zhang et al., 2017) did not appear to manifest significantly for SARS-CoV-2 infection of lungs of K18-hACE2 mice housed at 31 °C. The overriding ability of SARS-CoV-2 to evade and inhibit early anti-viral type I IFN responses (Beyer and Forero, 2022; Guo et al., 2022; Oh and Shin, 2022; Setaro and Gaglia, 2021; Tay et al., 2022; Wong and Perlman, 2022) and for lung viral loads to be largely unaffected by endogenous type I IFN responses (Boudewijns et al., 2020; Rawle et al., 2021), likely provide an explanation for these observations.
Both RNA-Seq and histology analyses illustrated that mice housed at 22 °C had decreased lung inflammation and leukocyte infiltrates, consistent with a reduction in the severity of ARDS. Immune functions are often decreased for mice housed under standard animal house temperatures, which are considered to be below thermoneutrality (Carpenter et al., 2020; Rubin, 2017; Seeley and MacDougald, 2021; Vialard and Olivier, 2020). Herein, such reduced immune function would appear also to result in decreased lung immunopathology. The observation is consistent with the general contention that cryotherapy (e.g. icing) reduces inflammation and tissue damage (Kwiecien and McHugh, 2021), with mild hypothermia also able to ameliorate lung inflammation in animal models of non-viral ARDS (Akyol et al., 2022; Angus et al., 2022). In humans ARDS is the main cause of morbidity and mortality for COVID-19 patients; however, there is no compelling data suggesting ARDS in humans is less severe during the winter, or in geographic regions with cooler climates. Quite the contrary, transmission and mortality is generally seen to increase during cooler climatic conditions. However, such epidemiological correlations may be unable to detect any reduction in ARDS severity at cooler temperatures, given that other overarching mechanisms are in play. For instance, during winter in northern climates there is less UV-inactivation and dehydration of virus in droplets/aerosols, and people spend more time indoors (Burra et al., 2021; Chen et al., 2021; Christophi et al., 2021; Mu et al., 2021; Sobral et al., 2020; Wang et al., 2021b; Wu et al., 2020). Increased air temperature, over a 2 °C to 11 °C range, in COVID-19 field hospitals also decreased mortality (Cai et al., 2020); however, other factors may again come into play at these low temperatures, such as impaired tissue regeneration (Kwiecien and McHugh, 2021). Our data is consistent with the notion that mild hypothermia (housing at 22 °C) might reduce the severity of lung inflammation (Fig. 2e, g) and thus by implication also ARDS (Cruces et al., 2021; Dos Reis Ururahy and Park, 2021). Human data supporting such a contention may be limited (Solmaz et al., 2021) because, unlike laboratory mice, COVID-19 patients with severe ARDS are generally not kept at temperatures below thermoneutrality. Although mild hypothermia has been suggested as a potential treatment option (Cruces et al., 2021; Dos Reis Ururahy and Park, 2021), any advantage over anti-inflammatory corticosteroid treatments (Griesel et al., 2022; Wagner et al., 2021) might need to be established, given the latter is arguably simpler, cheaper and safer.
The reduced level of brain infection and inflammation for mice housed at 31 °C likely underpins the reduced weight loss and delayed mortality (Carossino et al., 2022; Fumagalli et al., 2022; Kumari et al., 2021; Yu et al., 2022). Reduced brain infection likely arises from reduced infection of the nasal turbinates at 31 °C vs. 22 °C (Fig. 1a), given the virus probably enters the brain via the olfactory epithelium (Carossino et al., 2022; Kumari et al., 2021). Reduced infection of the upper respiratory track (nasal turbinates) at 31 °C may be due to the more than doubling of the beat frequency of cilia from nasal epithelium held at for 31 °C vs. 22 °C (Green et al., 1995). Mucociliary clearance is the first line of defense against airway infection and is driven by the beating of 200–300 cilia present in every airway epithelial cell (Gallo et al., 2021; Kawaguchi et al., 2022). The beating cilia propel mucus and associated pathogens out of the respiratory track to the laryngopharynx, where it is ultimately swallowed (Robinot et al., 2021). Mucociliary clearance is believed to mitigate against SARS-CoV-2 infection (Chatterjee et al., 2020; Courtney and Bax, 2021; Ferreira et al., 2022), although ultimately ciliated cells are targeted by viral infection in humans (Robinot et al., 2021). Increased mucociliary clearance may be one of the factors that contribute to reduced respiratory infections during warmer climatic conditions, although its relative importance given other factors is unclear (Moriyama et al., 2020).
For the K18-hACE2 mouse model described herein, 50 μl of medium containing the SARS-CoV-2 inoculum is delivered intranasally, and whatever fluid remains in the nasal cavity would likely be rapidly and largely cleared to allow breathing, given mice are obligatory nasal breathers. Then more efficient mucociliary clearance at 31 °C would explain the lower viral RNA levels in the nasal turbinates of mice house at this temperature (Fig. 1a); although it should be noted that mucociliary clearance was not measured in our study and other factors may be involved (Horstmann et al., 1977; Tippe et al., 1998). Humidity can effect mucocillary clearance (Kudo et al., 2019), and humidity was similar for mice held at the two temperatures (≈55 %). Reduced infection of the nasal turbinates and thus the olfactory epithelium, would delay brain infection, and the ensuing weight loss and mortality (Fumagalli et al., 2022). Why increased mucociliary clearance did not significantly reduce lungs titers for mice housed at 31 °C is unclear. However, this may simply reflect the difficulty in shifting, via mucociliary clearance, the ≈40 μl of viral inoculum that is delivered directly into the lungs, before significant viral infection has taken place. In humans, infection tends to migrate from the upper to the lower respiratory track, a feature not recapitulated by mouse models. While initial infection of the upper respiratory track in humans may be modulated by mucocilary clearance (Chatterjee et al., 2020; Courtney and Bax, 2021; Kumar et al., 2021), to what extent warmer ambient air temperatures would increase mucociliary clearance in human lungs and thereby lead to ameliorated lung infection remains unclear (Bridges et al., 2022).
The value of the K18-hACE2 mouse model for understanding human disease has been extensively discussed (Arce and Costoya, 2021; Bishop et al., 2022; Dong et al., 2022; Oladunni et al., 2020; Yinda et al., 2021; Yu et al., 2022; Zheng et al., 2021). To what extent the fulminant brain infection seen in this model is informative for neurological manifestations of COVID-19 in humans (Dangayach et al., 2022) remains controversial (Butowt et al., 2021; Carossino et al., 2022). However, olfactory disorders (including anosmia) appear to be considerably more common in cooler Western countries (33.4 %) than in warmer East Asian countries (8.3 %) (Kumar et al., 2021), an observation consistent with Fig. 1a. Importantly, a significant associated between persistent anosmia and long-lasting cognitive problems was recently reported at a conference (RADC, 2022), suggesting that in humans there may also be a link between infection of the olfactory epithelium and brain involvement.
5. Conclusion
When compared with standard animal house temperature of ≈22 °C, at thermoneutral housing temperature (≈31 °C) the K18-hACE2 mouse model of SARS-CoV-2 infection and disease showed reduced infection levels in the nasal turbinates and the brain (leading to reduced weight loss and delayed mortality), but increased inflammation in the lung. The data support the view that warmer climatic ambient air temperatures lead to an increase in mucociliary clearance of the viral inoculum in the nasal turbinates. The reduced infection of the nasal turbinates is associated with reduced infection of the brain, with brain infection fulminant and associated with mortality in this mouse model. A growing body of evidence suggests brain infection can occur in humans and may give rise to neurological manifestations of COVID-19 and long-COVID; however, brain infection is neither fulminant nor lethal. The K18-hACE2 mouse model recapitulates many aspects of severe ARDS, and at 31 °C, inflammatory cytokines and leukocyte infiltrates were significantly elevated in lungs, arguing that, once infected, warmer ambient temperatures may worsen lung inflammation. The observation is consisted with the general contention that mild hypothermia can reduce inflammation; although the suggested use of mild hypothermia as a treatment for COVID-19 ARDS may not be germane, given inter alia the availability of corticosteroids as an anti-inflammatory treatment.
The following are the supplementary data related to this article.
Supplementary figures
Brain
Lungs 2 dpi.
Lungs 5 dpi.
ImmuneSigDB.
CRediT authorship contribution statement
Conceptualization, A.S.; Methodology, T.T.L., T.D., D.J.R. and A.S.; Formal analysis, T.D., D.J.R., A.S. and C.B.; Investigation, T.T.L., W.N., K·Y, and B.T.; Data curation, T.D., D.J.R., A.S.; Writing – original draft, A.S. Writing – review and editing, T.D. and D.J.R.; Visualization, A.S.; Supervision, D.J.R. and A.S.; Project administration, A.S.; Funding acquisition, A.S., D.J.R.
Funding
The authors thank the Brazil Family Foundation (and others) for their generous philanthropic donations that helped set up the PC3 (BSL3) SARS-CoV-2 research facility at QIMR Berghofer MRI, as well as ongoing research into SARS-CoV-2, COVID-19 and long-COVID. A.S. is supported by the National Health and Medical Research Council (NHMRC) of Australia (Investigator grant APP1173880). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the following QIMRB staff; Dr. I. Anraku for management of the PC3 facility at QIMR Berghofer MRI, Dr. Viviana Lutzky for proof reading, Drs Clay Winterford and Crystal Chang for histology services, the animal house staff for mouse breeding and agistment, and Dr. Gunter Hartel for assistance with statistics.
Editor: Scott Sheridan
Data availability
All data is provided in the manuscript and accompanying supplementary files. Raw sequencing data (fastq files) generated for this publication for RNA-Seq have been deposited in the NCBI SRA, BioProject: PRJNA813692 and are publicly available at the date of publication.
References
- Akyol O., Demirgan S., Sengelen A., Guneyli H.C., Oran D.S., Yildirim F., et al. Mild hypothermia via external cooling improves lung function and alleviates pulmonary inflammatory response and damage in two-hit rabbit model of acute lung injury. J. Investig. Surg. 2022;35:1472–1483. doi: 10.1080/08941939.2022.2064010. [DOI] [PubMed] [Google Scholar]
- Alluwaimi A.M., Alshubaith I.H., Al-Ali A.M., Abohelaika S. The coronaviruses of animals and birds: their zoonosis, vaccines, and models for SARS-CoV and SARS-CoV2. Front. Vet. Sci. 2020;7 doi: 10.3389/fvets.2020.582287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., et al. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilla A.A., Sng J.D.J., Parry R., Deerain J.M., Potter J.R., Setoh Y.X., et al. A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 2021;12:3431. doi: 10.1038/s41467-021-23779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreakos E. STINGing type I IFN-mediated immunopathology in COVID-19. Nat. Immunol. 2022;23:478–480. doi: 10.1038/s41590-022-01174-6. [DOI] [PubMed] [Google Scholar]
- Angus S.A., Henderson W.R., Banoei M.M., Molgat-Seon Y., Peters C.M., Parmar H.R., et al. Therapeutic hypothermia attenuates physiologic, histologic, and metabolomic markers of injury in a porcine model of acute respiratory distress syndrome. Physiol. Rep. 2022;10 doi: 10.14814/phy2.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce V.M., Costoya J.A. SARS-CoV-2 infection in K18-ACE2 transgenic mice replicates human pulmonary disease in COVID-19. Cell. Mol. Immunol. 2021;18:513–514. doi: 10.1038/s41423-020-00616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian N., James T.D., Pushpavathi S.G., Marcinkiewcz C.A. bioRxiv; 2022. Repeated Ethanol Exposure and Withdrawal Alters ACE2 Expression in Discrete Brain Regions: Implications for SARS-CoV-2 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Zhang Q., Zhang S.Y., Jouanguy E., Casanova J.L. Type I interferons and SARS-CoV-2: from cells to organisms. Curr. Opin. Immunol. 2022;74:172–182. doi: 10.1016/j.coi.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer L., Laksono B.M., de Vrij F.M.S., Kushner S.A., Harschnitz O., van Riel D. The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. 2022;45:358–368. doi: 10.1016/j.tins.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.F., Moore G.J. Effects of high ambient temperature on various stages of rabies virus infection in mice. Infect. Immun. 1974;10:510–515. doi: 10.1128/iai.10.3.510-515.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer D.K., Forero A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2021.167265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.R., Dumenil T., Rawle D.J., Le T.T., Yan K., Tang B., et al. Mouse models of COVID-19 recapitulate inflammatory pathways rather than gene expression. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonarkart C., Suptawiwat O., Sakorn K., Puthavathana P., Auewarakul P. Exposure to cold impairs interferon-induced antiviral defense. Arch. Virol. 2017;162:2231–2237. doi: 10.1007/s00705-017-3334-0. [DOI] [PubMed] [Google Scholar]
- Boszormenyi K.P., Stammes M.A., Fagrouch Z.C., Kiemenyi-Kayere G., Niphuis H., Mortier D., et al. The post-acute phase of SARS-CoV-2 infection in two macaque species is associated with signs of ongoing virus replication and pathology in pulmonary and extrapulmonary tissues. Viruses. 2021;13(8):1673. doi: 10.3390/v13081673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges J.P., Vladar E.K., Huang H., Mason R.J. Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax. 2022;77:203–209. doi: 10.1136/thoraxjnl-2021-217561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryche B., St Albin A., Murri S., Lacote S., Pulido C., Ar Gouilh M., et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav. Immun. 2020;89:579–586. doi: 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burra P., Soto-Díaz K., Chalen I., Gonzalez-Ricon R.J., Istanto D., Caetano-Anollés G. Temperature and latitude correlate with SARS-CoV-2 epidemiological variables but not with genomic change worldwide. Evol. Bioinforma. 2021;17 doi: 10.1177/1176934321989695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowt R., Meunier N., Bryche B., von Bartheld C.S. The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models. Acta Neuropathol. 2021;141:809–822. doi: 10.1007/s00401-021-02314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Huang T., Liu X., Xu G. The effects of "Fangcang, huoshenshan, and Leishenshan" hospitals and environmental factors on the mortality of COVID-19. PeerJ. 2020;8 doi: 10.7717/peerj.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carossino M., Kenney D., O'Connell A.K., Montanaro P., Tseng A.E., Gertje H.P., et al. Fatal neurodissemination and SARS-CoV-2 tropism in K18-hACE2 mice is only partially dependent on hACE2 expression. Viruses. 2022;14:535. doi: 10.3390/v14030535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K.C., Zhou Y., Hakenjos J.M., Fry C.D., Nemzek J.A. Thermoneutral housing temperature improves survival in a murine model of polymicrobial peritonitis. Shock. 2020;54:688–696. doi: 10.1097/SHK.0000000000001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M., van Putten J.P.M., Strijbis K. Defensive properties of mucin glycoproteins during respiratory infections-relevance for SARS-CoV-2. mBio. 2020;11 doi: 10.1128/mBio.02374-20. e02374-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Prettner K., Kuhn M., Geldsetzer P., Wang C., Barnighausen T., et al. Climate and the spread of COVID-19. Sci. Rep. 2021;11:9042. doi: 10.1038/s41598-021-87692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow D., Stein S., Ramelli S., Grazioli A., Chung J.-Y., Singh M., et al. SARS-CoV-2 infection and persistence throughout the human body and brain. Res. Square. 2021 doi: 10.21203/rs.3.rs-1139035/v1. [DOI] [Google Scholar]
- Christophi C.A., Sotos-Prieto M., Lan F.Y., Delgado-Velandia M., Efthymiou V., Gaviola G.C., et al. Ambient temperature and subsequent COVID-19 mortality in the OECD countries and individual United States. Sci. Rep. 2021;11:8710. doi: 10.1038/s41598-021-87803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney J.M., Bax A. Hydrating the respiratory tract: an alternative explanation why masks lower severity of COVID-19. Biophys. J. 2021;120:994–1000. doi: 10.1016/j.bpj.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruces P., Cores C., Casanova D., Pizarro F., Diaz F. Successful use of mild therapeutic hypothermia as compassionate treatment for severe refractory hypoxemia in COVID-19. J. Crit. Care. 2021;63:260–263. doi: 10.1016/j.jcrc.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Nguyen N.L., Zarebidaki E., Cao Q., Li F., Zha L., et al. Thermoneutrality decreases thermogenic program and promotes adiposity in high-fat diet-fed mice. Physiol. Rep. 2016;4 doi: 10.14814/phy2.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S.N., Tan H.Y., Fan G.C. Immunopathological roles of neutrophils in virus infection and COVID-19. Shock. 2021;56:345–351. doi: 10.1097/SHK.0000000000001740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangayach N.S., Newcombe V., Sonnenville R. Acute neurologic complications of COVID-19 and postacute sequelae of COVID-19. Crit. Care Clin. 2022;38:553–570. doi: 10.1016/j.ccc.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo G.D., Lazarini F., Levallois S., Hautefort C., Michel V., Larrous F., et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denjean F., Lachuer J., Geloen A., Cohen-Adad F., Moulin C., Barre H., et al. Differential regulation of uncoupling protein-1, -2 and -3 gene expression by sympathetic innervation in brown adipose tissue of thermoneutral or cold-exposed rats. FEBS Lett. 1999;444:181–185. doi: 10.1016/s0014-5793(99)00056-3. [DOI] [PubMed] [Google Scholar]
- Derting Terry L., Compton S. Immune response, not immune maintenance, is energetically costly in wild white-footed mice (Peromyscus leucopus) Physiol. Biochem. Zool. 2003;76:744–752. doi: 10.1086/375662. [DOI] [PubMed] [Google Scholar]
- Domizio J.D., Gulen M.F., Saidoune F., Thacker V.V., Yatim A., Sharma K., et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Mead H., Tian L., Park J.G., Garcia J.I., Jaramillo S., et al. The K18-human ACE2 transgenic mouse model recapitulates non-severe and severe COVID-19 in response to an infectious dose of the SARS-CoV-2 virus. J. Virol. 2022;96 doi: 10.1128/JVI.00964-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Reis Ururahy R., Park M. Cheap and simple, could it get even cooler? Mild hypothermia and COVID-19. J. Crit. Care. 2021;63:264–268. doi: 10.1016/j.jcrc.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles R. Why is temperature sensitivity important for the success of common respiratory viruses? Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskilsson A., Shionoya K., Engblom D., Blomqvist A. Fever during localized inflammation in mice is elicited by a humoral pathway and depends on brain endothelial interleukin-1 and interleukin-6 signaling and central EP3 receptors. J. Neurosci. 2021;41:5206. doi: 10.1523/JNEUROSCI.0313-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castaneda A., Lu P., Geraghty A.C., Song E., Lee M.H., Wood J., et al. Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell. 2022;185(2452–2468) doi: 10.1016/j.cell.2022.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L.C., Gomes C.E.M, Rodrigues-Neto J.F., Jeronimo S.M.B. Genome-wide association studies of COVID-19: Connecting the dots. Infect. Gen. Evol. 2022;106:105379. doi: 10.1016/j.meegid.2022.105379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman E.F., Storer J.A., Fitzgerald M.E., Wasik B.R., Hou L., Zhao H., et al. Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:827–832. doi: 10.1073/pnas.1411030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman E.F., Storer J.A., Vanaja K., Levchenko A., Iwasaki A. Two interferon-independent double-stranded RNA-induced host defense strategies suppress the common cold virus at warm temperature. Proc. Natl. Acad. Sci. 2016;113:8496. doi: 10.1073/pnas.1601942113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.W., Xu H.S., Liu S.J. COVID-19 and neurodegenerative diseases. Eur. Rev. Med. Pharmacol. Sci. 2022;26:4535–4544. doi: 10.26355/eurrev_202206_29093. [DOI] [PubMed] [Google Scholar]
- Fumagalli V., Rava M., Marotta D., Di Lucia P., Laura C., Sala E., et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abl9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi S., Poloni E.T., Pandini C., Garofalo M., Dragoni F., Medici V., et al. Detection of SARS-CoV-2 genome and whole transcriptome sequencing in frontal cortex of COVID-19 patients. Brain Behav. Immun. 2021;97:13–21. doi: 10.1016/j.bbi.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo O., Locatello L.G., Mazzoni A., Novelli L., Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K., Chawla A. Warming the mouse to model human diseases. Nat. Rev. Endocrinol. 2017;13:458–465. doi: 10.1038/nrendo.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K., Nikkanen J., Man K., Leong Y.A., Sogawa Y., Maschek J.A., et al. Energetic trade-offs and hypometabolic states promote disease tolerance. Cell. 2019;177(2):399–413.e12. doi: 10.1016/j.cell.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arriaza J., Garaigorta U., Perez P., Lazaro-Frias A., Zamora C., Gastaminza P., et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021;95(7):e02260–20. doi: 10.1128/JVI.02260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J., Rudd P.A., Prow N.A., Belarbi E., Roques P., Larcher T. Infectious chikungunya virus in the saliva of mice, monkeys and humans. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill B.N., Garner J.P. Letter-to-the-editor on "Not so hot: optimal housing temperatures for mice to mimic the thermal environment of humans". Mol. Metab. 2014;3:335–336. doi: 10.1016/j.molmet.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godec J., Tan Y., Liberzon A., Tamayo P., Bhattacharya S., Butte A.J., et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A., Smallman L.A., Logan A.C., Drake-Lee A.B. The effect of temperature on nasal ciliary beat frequency. Clin. Otolaryngol. Allied Sci. 1995;20:178–180. doi: 10.1111/j.1365-2273.1995.tb00040.x. [DOI] [PubMed] [Google Scholar]
- Griesel M., Wagner C., Mikolajewska A., Stegemann M., Fichtner F., Metzendorf M.I., et al. Inhaled corticosteroids for the treatment of COVID-19. Cochrane Database Syst. Rev. 2022;3(3) doi: 10.1002/14651858.CD015125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld P.H., Claassen E., Kuper C.F., Van Rooijen N. The role of macrophages in LPS-induced lethality and tissue injury. Immunology. 1988;63:521–527. [PMC free article] [PubMed] [Google Scholar]
- Guimond S.E., Mycroft-West C.J., Gandhi N.S., Tree J.A., Le T.T., Spalluto C.M., et al. Synthetic heparan sulfate mimetic pixatimod (PG545) potently inhibits SARS-CoV-2 by disrupting the spike-ACE2 interaction. ACS Cent. Sci. 2022;8:527–545. doi: 10.1021/acscentsci.1c01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K., Barrett B.S., Morrison J.H., Mickens K.L., Vladar E.K., Hasenkrug K.J., et al. Interferon resistance of emerging SARS-CoV-2 variants. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182(3):744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann G., Iravani J., Norris Melville G., Richter H.G. Influence of temperature and decreased water content of inspired air on the ciliated bronchial epithelium. A physiological and electron microscopical study. Acta Otolaryngol. 1977;84:124–131. doi: 10.3109/00016487709123950. [DOI] [PubMed] [Google Scholar]
- Johnson B.J., Le T.T., Dobbin C.A., Banovic T., Howard C.B., Flores Fde M., et al. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J. Biol. Chem. 2005;280:4037–4047. doi: 10.1074/jbc.M411569200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K., Nakayama S., Saito D., Kogiso H., Yasuoka K., Marunaka Y., et al. Ezrin knockdown reduces procaterol-stimulated ciliary beating without morphological changes in mouse airway cilia. J. Cell Sci. 2022;135(6) doi: 10.1242/jcs.259201. [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Shin E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021;53:750–760. doi: 10.1038/s12276-021-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo E., Song E., Yockey L.J., Rakib T., Wong P.W., Homer R.J., et al. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc. Natl. Acad. Sci. U. S. A. 2019;116:10905–10910. doi: 10.1073/pnas.1902840116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.A., Lee S.W.Y., Lock C., Keong N.C. Geographical variations in host predisposition to COVID-19 related anosmia, ageusia, and neurological syndromes. Front. Med. 2021;8 doi: 10.3389/fmed.2021.661359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P., Rothan H.A., Natekar J.P., Stone S., Pathak H., Strate P.G., et al. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses. 2021;13:132. doi: 10.3390/v13010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecien S.Y., McHugh M.P. The cold truth: the role of cryotherapy in the treatment of injury and recovery from exercise. Eur. J. Appl. Physiol. 2021;121:2125–2142. doi: 10.1007/s00421-021-04683-8. [DOI] [PubMed] [Google Scholar]
- La Linn M., Bellett A.J., Parsons P.G., Suhrbier A. Complete removal of mycoplasma from viral preparations using solvent extraction. J. Virol. Methods. 1995;52:51–54. doi: 10.1016/0166-0934(94)00136-5. [DOI] [PubMed] [Google Scholar]
- Lane W.C., Dunn M.D., Gardner C.L., Lam L.K.M., Watson A.M., Hartman A.L. The efficacy of the interferon Alpha/Beta response versus arboviruses is temperature dependent. MBio. 2018;9 doi: 10.1128/mBio.00535-18. e00535-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempriere S. SARS-CoV-2 detected in olfactory neurons. Nat. Rev. Neurol. 2021;17:63. doi: 10.1038/s41582-020-00449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.M., Tan B.H., Wu S., Gui Y., Suo J.L., Li Y.C. Evidence of central nervous system infection and neuroinvasive routes, as well as neurological involvement, in the lethality of SARS-CoV-2 infection. J. Med. Virol. 2021;93:1304–1313. doi: 10.1002/jmv.26570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.Y., Feng P.H., Yu M.S., Chen M.C., Lin A.J., Chen J.L., et al. Current utilization of interferon alpha for the treatment of coronavirus disease 2019: a comprehensive review. Cytokine Growth Factor Rev. 2022;63:34–43. doi: 10.1016/j.cytogfr.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciorowski D., Sharma D., Kunamneni A. Environmental factors and their role in the transmission of SARS-CoV-2. Biosaf Health. 2021;3:235–237. doi: 10.1016/j.bsheal.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaniyazi L., Ng C.F.S., Seposo X., Toizumi M., Yoshida L.M., Honda Y., et al. Role of temperature, influenza and other local characteristics in seasonality of mortality: a population-based time-series study in Japan. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-044876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaroli C., Benson P., Sharma N.S., Madison M.C., Robison S.W., Arora N., et al. Spatial mapping of SARS-CoV-2 and H1N1 lung injury identifies differential transcriptional signatures. Cell. Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N.M., Griffin D.E. Interleukin-10 modulation of virus clearance and disease in mice with alphaviral encephalomyelitis. J. Virol. 2018;92 doi: 10.1128/JVI.01517-17. e01517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Humphrey S.J., Fortuna P.R.J., Lor M., Foster S.R., Quaife-Ryan G.A., et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell. 2021;184(8):2167–2182. doi: 10.1016/j.cell.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogal A., Abdulkadir S.A. Effects of histone deacetylase inhibitor (HDACi); trichostatin-a (TSA) on the expression of housekeeping genes. Mol. Cell. Probes. 2006;20:81–86. doi: 10.1016/j.mcp.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Moriyama M., Hugentobler W.J., Iwasaki A. Seasonality of respiratory viral infections. Annu. Rev. Virol. 2020;7:83–101. doi: 10.1146/annurev-virology-012420-022445. [DOI] [PubMed] [Google Scholar]
- Mu Y., Shao M., Zhong B., Zhao Y., Leung K.M.Y., Giesy J.P., et al. Transmission of SARS-CoV-2 virus and ambient temperature: a critical review. Environ. Sci. Pollut. Res. Int. 2021;28:37051–37059. doi: 10.1007/s11356-021-14625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Moreno R., Martinez-Romero C., Garcia-Sastre A. Induction and evasion of type-I interferon responses during influenza a virus infection. Cold Spring Harb. Perspect. Med. 2021;11(10):a038414. doi: 10.1101/cshperspect.a038414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- None T.L.N. Long COVID: understanding the neurological effects. Lancet Neurol. 2021;20:247. doi: 10.1016/S1474-4422(21)00059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Shin O.S. SARS-CoV-2-mediated evasion strategies for antiviral interferon pathways. J. Microbiol. 2022;60:290–299. doi: 10.1007/s12275-022-1525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladunni F.S., Park J.G., Pino P.A., Gonzalez O., Akhter A., Allue-Guardia A., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras I.S., Huentelman M.J., Walker J.E., Arce R., Glass M.J., Vargas D. medRxiv; 2021. Olfactory Bulb and Amygdala Gene Expression Changes in Subjects Dying with COVID-19. [Google Scholar]
- Prow N.A., Tang B., Gardner J., Le T.T., Taylor A., Poo Y.S., et al. Lower temperatures reduce type I interferon activity and promote alphaviral arthritis. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow N.A., Hirata T.D.C., Tang B., Larcher T., Mukhopadhyay P., Alves T.L., et al. Exacerbation of chikungunya virus rheumatic immunopathology by a high fiber diet and butyrate. Front. Immunol. 2019;10:2736. doi: 10.3389/fimmu.2019.02736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADC Persistent loss of smell due to COVID-19 closely connected to long-lasting cognitive problems. Alzheimer’s Association International Conference. 2022. https://aaic.alz.org/downloads2022/COVID-and-Cognition-News-Release-AAIC2022.pdf San Diego.
- Ramasamy S., Kolloli A., Kumar R., Husain S., Soteropoulos P., Chang T.L., et al. Comprehensive analysis of disease pathology in immunocompetent and immunocompromised hosts following pulmonary SARS-CoV-2 infection. Biomedicines. 2022;10(6):1343. doi: 10.3390/biomedicines10061343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., Strohmeier S., Amanat F., Gillespie V.L., Krammer F., Garcia-Sastre A., et al. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9:2433–2445. doi: 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawle D.J., Le T.T., Dumenil T., Yan K., Tang B., Nguyen W., et al. ACE2-lentiviral transduction enables mouse SARS-CoV-2 infection and mapping of receptor interactions. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawle D.J., Dumenil T., Tang B., Bishop C.R., Yan K., Le T.T., et al. Microplastic consumption induces inflammatory signatures in the colon and prolongs a viral arthritis. Sci. Total Environ. 2022;809 doi: 10.1016/j.scitotenv.2021.152212. [DOI] [PubMed] [Google Scholar]
- Recalde-Zamacona B., Garcia-Tobar L., Argueta A., Alvarez L., De Andrea C.E., Fernandez Alonso M., et al. Histopathological findings in fatal COVID-19 severe acute respiratory syndrome: preliminary experience from a series of 10 Spanish patients. Thorax. 2020;75:1116–1118. doi: 10.1136/thoraxjnl-2020-215577. [DOI] [PubMed] [Google Scholar]
- Robinot R., Hubert M., de Melo G.D., Lazarini F., Bruel T., Smith N., et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 2021;12:4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld R., Noy-Porat T., Mechaly A., Makdasi E., Levy Y., Alcalay R., et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021;12:944. doi: 10.1038/s41467-021-21239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R.L. Mice housed at elevated vivarium temperatures display enhanced T-cell response and survival to francisella tularensis. Comp. Med. 2017;67:491–497. [PMC free article] [PubMed] [Google Scholar]
- Rudd P.A., Wilson J., Gardner J., Larcher T., Babarit C., Le T.T., et al. Interferon response factors 3 and 7 protect against chikungunya virus hemorrhagic fever and shock. J. Virol. 2012;86:9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley R.J., MacDougald O.A. Mice as experimental models for human physiology: when several degrees in housing temperature matter. Nat. Metab. 2021;3:443–445. doi: 10.1038/s42255-021-00372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G.E., Walker J.E., Arce R., Glass M.J., Vargas D., Sue L.I. medRxiv; 2021. Mapping of SARS-CoV-2 Brain Invasion and Histopathology in COVID-19 Disease. [Google Scholar]
- Setaro A.C., Gaglia M.M. All hands on deck: SARS-CoV-2 proteins that block early anti-viral interferon responses. Curr. Res. Virol. Sci. 2021;2 doi: 10.1016/j.crviro.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.R., Steele K.E., Shamblin J., Honko A., Johnson J., Reed C., et al. The pathogenesis of Rift Valley fever virus in the mouse model. Virology. 2010;407:256–267. doi: 10.1016/j.virol.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Sobral M.F.F., Duarte G.B., da Penha Sobral A.I.G., Marinho M.L.M., de Souza Melo A. Association between climate variables and global transmission oF SARS-CoV-2. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmaz I., Ozcaylak S., Alakus O.F., Kilic J., Kalin B.S., Guven M., et al. Risk factors affecting ICU admission in COVID-19 patients; could air temperature be an effective factor? Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.13803. [DOI] [PubMed] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R., Keijer J. Not so hot: optimal housing temperatures for mice to mimic the thermal environment of humans. Mol. Metab. 2012;2:5–9. doi: 10.1016/j.molmet.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay D.J.W., Lew Z.Z.R., Chu J.J.H., Tan K.S. Uncovering novel viral innate immune evasion strategies: what has SARS-CoV-2 taught us? Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.844447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippe A., Korbel R., Ziesenis A., Heyder J. Viscoelastic properties of canine tracheal mucus: effects of structural inhomogeneities and storage. Scand. J. Clin. Lab. Invest. 1998;58:259–264. doi: 10.1080/00365519850186652. [DOI] [PubMed] [Google Scholar]
- Underwood G.E., Baker C.A., Weed S.D. Protective effect of elevated temperature on mice infected with coe virus. J. Immunol. 1966;96:1006–1012. [PubMed] [Google Scholar]
- van Oosten L., Yan K., Rawle D.J., Le T.T., Altenburg J.J., Fougeroux C., et al. An S1-nanoparticle vaccine protects against SARS-CoV-2 challenge in K18-hACE2 mice. J. Virol. 2022;12(5):e0181321. doi: 10.1128/jvi.00844-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard F., Olivier M. Thermoneutrality and immunity: how does cold stress affect disease? Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.588387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Griesel M., Mikolajewska A., Mueller A., Nothacker M., Kley K., et al. Systemic corticosteroids for the treatment of COVID-19. Cochrane Database Syst. Rev. 2021;8 doi: 10.1002/14651858.CD014963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Z., Zhou J., Han S., Kang Z., Chuang H., et al. Next-generation sequencing and proteomics of cerebrospinal fluid from COVID-19 patients with neurological manifestations. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.782731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tang K., Feng K., Lin X., Lv W., Chen K., et al. Impact of temperature and relative humidity on the transmission of COVID-19: a modelling study in China and the United States. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-043863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple B., Agar J., Zhao J., Pearce D.A., Kovacs A.D. The acidified drinking water-induced changes in the behavior and gut microbiota of wild-type mice depend on the acidification mode. Sci. Rep. 2021;11:2877. doi: 10.1038/s41598-021-82570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.A., Prow N.A., Schroder W.A., Ellis J.J., Cumming H.E., Gearing L.J., et al. RNA-seq analysis of chikungunya virus infection and identification of granzyme a as a major promoter of arthritic inflammation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasiri-Yapa B., Barrero R.A., Stassen L., Hafner L.M., McGraw E.A., Pyke A.T., et al. Temperature modulates immune gene expression in mosquitoes during arbovirus infection. Open Biol. 2021;11(1) doi: 10.1098/rsob.200246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.R., Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy? Nat. Rev. Immunol. 2022;22:47–56. doi: 10.1038/s41577-021-00656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., et al. Establishment of an african green monkey model for COVID-19 and protection against re-infection. Nat. Immunol. 2021;22:86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Jing W., Liu J., Ma Q., Yuan J., Wang Y., et al. Effects of temperature and humidity on the daily new cases and new deaths of COVID-19 in 166 countries. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.139051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Rawle D.J., Le T.T., Suhrbier A. Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol. J. 2021;18:123. doi: 10.1186/s12985-021-01587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Dumenil T., Tang B., Le T.T., Bishop C.R., Suhrbier A. Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2. Virus Evol. 2022;8(2):veac063. doi: 10.1093/ve/veac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Zhou J., He Q., Li R.T., Yang G., Zhang Y., et al. SARS-CoV-2 infection in the mouse olfactory system. Cell Discov. 2021;7:49. doi: 10.1038/s41421-021-00290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Port J.R., Bushmaker T., Offei Owusu I., Purushotham J.N., Avanzato V.A., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Deng W., Bao L., Qu Y., Xu Y., Zhao W., et al. Comparative pathology of the nasal epithelium in K18-hACE2 tg mice, hACE2 tg mice, and hamsters infected with SARS-CoV-2. Vet. Pathol. 2022;59(4):602–612. doi: 10.1177/03009858211071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Wu Y., Jing W., Liu J., Du M., Wang Y., et al. Association between meteorological factors and daily new cases of COVID-19 in 188 countries: a time series analysis. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tang X., Sheng X., Xing J., Zhan W. The influence of temperature on viral replication and antiviral-related genes response in hirame rhabdovirus-infected flounder (Paralichthys olivaceus) Fish Shellfish Immunol. 2017;68:260–265. doi: 10.1016/j.fsi.2017.07.029. [DOI] [PubMed] [Google Scholar]
- Zhang A.J., Lee A.C., Chu H., Chan J.F., Fan Z., Li C., et al. Severe acute respiratory syndrome coronavirus 2 infects and damages the mature and immature olfactory sensory neurons of hamsters. Clin. Infect. Dis. 2021;73:e503–e512. doi: 10.1093/cid/ciaa995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Xu J., Hou Y., Leverenz J.B., Kallianpur A., Mehra R., et al. Network medicine links SARS-CoV-2/COVID-19 infection to brain microvascular injury and neuroinflammation in dementia-like cognitive impairment. Alzheimers Res. Ther. 2021;13:110. doi: 10.1186/s13195-021-00850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Brain
Lungs 2 dpi.
Lungs 5 dpi.
ImmuneSigDB.
Data Availability Statement
All data is provided in the manuscript and accompanying supplementary files. Raw sequencing data (fastq files) generated for this publication for RNA-Seq have been deposited in the NCBI SRA, BioProject: PRJNA813692 and are publicly available at the date of publication.