Abstract
Background
HOXA cluster antisense RNA 2 (lncRNA HOXA-AS2) is a long noncoding RNA (lncRNA) that aberrantly expressed in various cancers and is closely associated with cancer progression. To overcome the limitation of small sample sizes that are inherent to single studies, a meta-analysis was conducted to explore the relationship between the expression level of HOXA-AS2 and cancer prognosis.
Methods
Correlational studies were retrieved by searching the databases of PubMed, Embase and Web of Science (up to August 10, 2022). The survival and prognosis data included overall survival (OS), and clinical parameters were gathered and analyzed.
Results
Eighteen publications with 1181 patients who were diagnosed with solid tumors were ultimately included. The results showed that, compared with patients with low HOXA-AS2 expression, patients with high HOXA-AS2 expression tended to have poorer overall survival (OS) (HR= 2.52, 95% CI 1.87-3.38, P < 0.01) and shorter disease-free survival (DFS) (HR=7.19, 95% CI 3.20-16.17, P < 0.01). In addition, elevated HOXA-AS2 expression indicated a larger tumor size (OR =2.43, 95% CI 1.53–3.88,P < 0.01), more advanced TNM stage (OR=3.85, 95% CI 2.79-5.31, P < 0.01), earlier lymph node metastasis (LNM) (OR = 4.41, 95% CI 3.05-6.39, P < 0.01) and distant metastasis (DM) (OR= 2.96, 95% CI 1.87-4.7, P < 0.01). Furthermore, HOXA-AS2 expression was notassociated with age (OR=1.15, 95% CI 0.90-1.47), gender (OR=1.16, 95% CI 0.89-1.53), or tumor differentiation (OR=1.21, 95% CI 0.56-2.63). Moreover, aberrant HOXA-AS2 expression was related to drug sensitivity in various types of cancers.
Conclusion
The overexpression of HOXA-AS2 predicted poor cancer prognosis in the Chinese population, including poor OS, DFS, TNM, LNM, and DM. HOXA-AS2 could serve as a promising prognostic biomarker and therapeutic target.
Systematic Review Registration
https://www.crd.york.ac.uk/prospero/, identifier CRD42022352604.
Keywords: lncRNA, HOXA-AS2, cancer, prognosis, meta-analysis, bioinformatics analysis
1 Highlights
Long noncoding RNA HOXA cluster antisense RNA 2 ( HOXA-AS2 ) was revealed increasing expression in most of solid tumor tissues
Elevated HOXA-AS2 expression predicts poor cancer prognosis.
HOXA-AS2 could be served as potential therapeutic target and prognostic marker.
HOXA-AS2 may take effect in chemotherapy and may be correlated with chemoresistance.s
2 Introduction
Cancer is one of the leading causes of death worldwide. Since its onset is hidden, early unobvious symptoms are not easy to detect; therefore, cancer has become a key issue of global concern and causes a heavy economic burden to the world every year (1, 2). The global cancer burden is predicted to reach 28.4 million cases by 2040 (an increase of 47% compared with 2020) according to cancer statistics (1). In recent decades, cancer treatment has evolved from exclusively surgical treatment to multidisciplinary treatment involving chemotherapy, radiotherapy, immunotherapy, and targeted therapy (3). However, the survival rate of cancer patients is still not optimal. Many researchers are beginning to explore new treatment options, and with the rapid advances in molecular medicine, cancer research has reached the level of genes and molecules (4, 5).
Over the past decade, many noncoding RNAs have been closely linked to cancer occurrence and development (6, 7). DNA transcription products include coding RNA and noncoding RNA; the latter occupies more than 90% of the RNA in cells (8). Due to the inability to encode proteins, noncoding RNA was once viewed as an “useless product” of DNA transcription. Nevertheless, there is growing evidence that noncoding RNA plays a role in many processes of cancer progression, including cell proliferation, invasion, autophagy, apoptosis, and epithelial-mesenchymal transition (9, 10).
HOXA-AS2 is a 1048-bp long noncoding RNA located on human chromosome 7p15.2 between the HOXA3 and HOXA4 genes in the HOXA cluster (11). In recent years, an increasing number of publications have shown that HOXA-AS2 is highly expressed in diverse malignancies, including oral squamous cell carcinoma (12, 13), nasopharyngeal carcinoma (14), thyroid cancer (15, 16), breast cancer (17, 18), lung cancer (19–22), gallbladder carcinoma (23), gastric cancer (24, 25), hepatocellular carcinoma (26, 27), pancreatic cancer (28), colorectal cancer (29), bladder cancer (30), prostate cancer (31), ovarian cancer (32), endometrial cancer (33), cervical cancer (34, 35), osteosarcoma (36, 37) and glioblastoma (38). Cell function tests showed that high HOXA-AS2 expression can promote the proliferation and invasion of cancer cells, inhibit apoptosis, and affect the cancer cell cycle (21, 32). HOXA-AS2 could function as a competitive endogenous RNA (ceRNA) affecting the distribution of microRNAs on their targets, which could contribute to tumor progression and occurrence (37, 39). Furthermore, an increasing amount of data have indicated a correlation between abnormal HOXA-AS2 expression and clinicopathological features and prognosis, indicating its potential as a prognostic and therapeutic marker (22, 40, 41). However, owing to the small number of cases in a single study, the results have been inconclusive, as the prognosis reported in such studies do not represent all cancer patients equally. Therefore, this meta-analysis systematically evaluated the prognostic value of HOXA-AS2 in cancer patients.
3 Materials and methods
3.1 Literature search
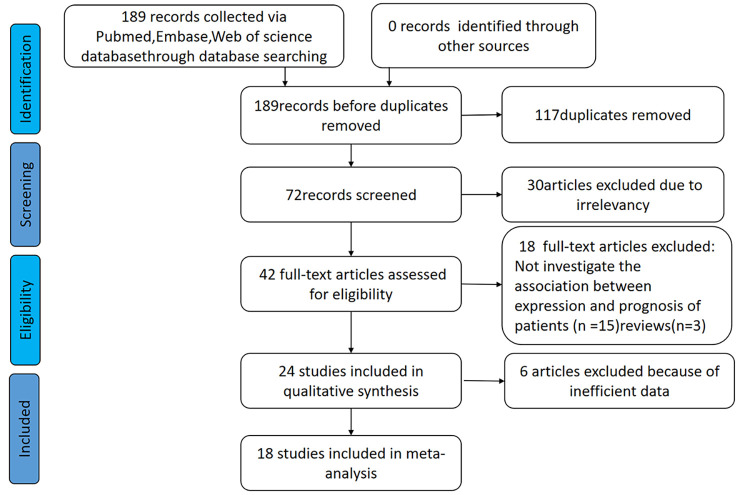
This meta-analysis was conducted in accordance with the statement of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (42). The Embase, PubMed, and Web of Science databases were searched from inception to August 10, 2022. The search terms consisted of different subject terms, including: (“HOXA-AS2” or “Long noncoding RNA HOXA-AS2” or “LncRNA HOXA-AS2” or “HOXA cluster antisense RNA 2”) and (“neoplasm” or “cancer” or “carcinoma” or “tumor” or “malignancy”). The literature was limited to English-language publications and human studies ( Figure 1 ). The study was registered on PROSPERO (registration number: CRD42022352604).
Figure 1.
The flow diagram of the eligible studies.
3.2 Inclusion and exclusion criteria of the literature
The inclusion criteria were as follows: 1) the research mainly focused on the relationship between the expression of HOXA-AS2 and the clinicopathological characterization or prognosis of various solid human malignancies; 2) according to the expression of HOXA-AS2, patients were divided into two groups: a high expression group and a low expression group; 3) sufficient data were provided for the calculation of odds ratio (OR) or hazard ratio (HR) with 95% confidence interval (CI); and 4) the full text was published. The exclusion criteria were as follows: 1) nonprimary research articles, such as case reports, conferences, abstracts, reviews, or editorials; 2) patients were not divided into a high HOXA-AS2 expression group and a low HOXA-AS2 expression group; 3) data were not available or insufficient; and 4) animal experiments.
3.3 Quality evaluation
The Newcastle-Ottawa Scale (NOS) score (43) was applied to evaluate the quality of the included literature across three dimensions: Selection, Comparability and Outcome. The following aspects were evaluated: Adequate of case definition, Representativeness of the cases, Selection of Controls, Definition of Controls, Comparability of cases and controls, Ascertainment of exposure, Same method of ascertainment, Non-response rate. The total score ranges from 0~9, and studies with scores<6 are considered to be of substandard quality and were not included in this meta-analysis.
3.4 Data extraction and quality evaluation
In this study, two reviewers independently extracted the data (publication data) included in the article, and differences were resolved through discussion with a third reviewer. The data included the author’s name, publication year, country, cancer type, number of patients, sample type and detection method, high expression and number of patients with low expression, cut-off value, clinicopathological features, and follow-up times. The HR (95% CI) of survival was obtained directly from the study or calculated using Engauge Digitizer 11.1 software if Kaplan–Meier curves were available.
3.5 Statistical analysis
STATA 16.0 software was adopted for statistical analysis. We used the HR value and its 95% CI as the effect index of survival data (OS, DFS), and the odds ratio (OR) and its 95% CI as the clinical pathology of patients. When the Chi-squared test had a P < 0.10 or when I2 > 50%, the random effects model was selected, and the subgroup analysis was carried out according to the patient region, cancer type, data source method, etc. If P >0.10 or I2<50%, the fixed effects model was adopted.
3.6 Sensitivity analysis for publication bias
Publication bias of the literature was assessed using Egger’s test by applying STATA 16.0 software. The robustness of the results was tested by sensitivity analysis. P<0.05 was considered statistically significant.
3.7 Signal Pathway Network Construction, target gene and drug sensitivity prediction
Pathway Network Construction Related genes for HOXA-AS2 were downloaded from the RNAInter v4.0 database (https://www.rnainter.org) (44). Next, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted via the R package “clusterprofiler”. Cytoscape software (45) was used to construct a visualized signaling pathway network. Moreover, NCI-60 compound activity data and RNA-seq expression profiles from CallMiner™ were downloaded to analyze the drug sensitivity of HOXA-AS2 in pan-cancer (https://discover.nci.nih.gov/cellminer/home.do). Drugs approved by the FDA or clinical trials were adopted for analysis (46). The “ggplot2”, “impute”, and “ggpubr” R packages were used in R software.
4 Results
4.1 Characteristics of studies
The detailed process of the literature identification and selection is presented in Figure 1 . The initial search resulted in 189 articles, of which 117 were excluded as they were duplicate studies. Another 30 articles were eliminated after screening the title and abstracts because of irrelevant findings. Further screening of the remaining 42 articles led to the exclusion of 18 reports due to a lack of study data regarding prognosis or clinicopathologic characteristics, and six were excluded due to missing data or information. Ultimately, 18 articles were included in the current meta-analysis.
A total of 18 articles including 1181 patients were included, and the cancer types included gastric cancer (24), colorectal cancer (CRC) (29, 40), hepatocellular carcinoma(HCC) (26, 27, 41), breast cancer (18), non-small cell lung cancer (NSCLC) (19, 22), prostate cancer (31), osteosarcoma (36, 37), bladder cancer (30), oral squamous cell carcinoma (OSCC) (13), papillary thyroid cancer (PTC) (15, 16), cervical cancer (35) and glioma (47). The publication years were from 2015 to 2021, and all patients were from China. The number of cases ranged from 27 to 128. Among the eighteen included studies, eleven studies provided overall survival (OS), 2 studies provided disease−free survival (DFS). Only 3 studies provided HR values directly, and other studies only provided K-M survival curves ( Table 1 ). The NOS scores varied from 6-9 ( Supplementary Table S2 ).
Table 1.
Basic features of the publications included in this meta-analysis (n=18).
| Study | Year | Region | Sample size | Cancer type | Detection method | Sample | Cut-off | survival analysis | HR statistics | Hazard ratios(95% CI) | Follow-up (month) | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xie M | 2015 | China | 55 | Gastric cancer | qRT-PCR | tissue | median | OS | indirectly | 6.45(1.92,21.63) | 50 | 7 |
| DFS | indirectly | 8.33(1.94,35.75) | ||||||||||
| Li Q | 2016 | China | 30 | CRC | qRT-PCR | tissue | median | OS | indirectly | 4.7(1.01, 21.87) | 28 | 7 |
| Ding J | 2017 | China | 69 | CRC | qRT-PCR | tissue | not reported | lack off | NA | - | NA | 6 |
| Wang F | 2016 | China | 112 | HCC | qRT-PCR | tissue | median | OS | indirectly | 1.96(1.17,3.28) | 80 | 7 |
| Zhang Y | 2018 | China | 58 | HCC | qRT-PCR | tissue | mean | lack off | NA | – | NA | 6 |
| Lu Q | 2020 | China | 116 | HCC | qRT-PCR | tissue | median | OS | directly | 2.76(1.115,6.848) | 25 | 9 |
| Wang Y | 2018 | China | 66 | Osteosar- coma |
qRT-PCR | tissue | median | lack off | NA | – | NA | 6 |
| Wang L | 2019 | China | 27 | Osteosar- coma |
qRT-PCR | tissue | mean | OS | indirectly | 0.75(0.16,3.45) | 60 | 7 |
| Wu L | 2019 | China | 50 | Glioma | qRT-PCR | tissue | not reported | lack off | NA | – | NA | 6 |
| Fang Y | 2017 | China | 38 | Breast cancer | qRT-PCR | tissue | not reported | OS | indirectly | 1.51(0.53, 4.27) | 90 | 7 |
| Li Y | 2017 | China | 103 | NSCLC | qRT-PCR | tissue | median | OS | directly | 6.711(3.526-17.019) | 75 | 9 |
| DFS | directly | 6.737(2.926-20.527) | ||||||||||
| Cui T J | 2019 | China | 40 | NSCLC | qRT-PCR | tissue | not reported | OS | indirectly | 1.70(0.29,10.48) | 75 | 8 |
| Xia F | 2018 | China | 128 | PTC | qRT-PCR | tissue | mean | lack off | NA | – | NA | 6 |
| Jiang L | 2019 | China | 68 | PTC | qRT-PCR | tissue | mean | OS | indirectly | 1.45(0.53, 3.98) | 60 | 7 |
| Wang F | 2019 | China | 80 | Bladder cancer | qRT-PCR | tissue | not reported | lack off | NA | – | NA | 6 |
| Xiao S | 2020 | China | 68 | Prostate cancer | qRT-PCR | tissue | mean | OS | indirectly | 1.55(0.2, 11.91) | 60 | 7 |
| ChenR (a) |
2021 | China | 46 | OSCC | qRT-PCR | tissue | not reported | lack off | NA | – | NA | 6 |
| ChenR (b) |
2021 | China | 27 | Cervical cancer | qRT-PCR | tissue | mean | OS | directly | 2.80(1.0, 7.9) | 120 | 8 |
CRC, colorectal cancer; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; PTC, papillary thyroid cancer; OSCC, Oral Squamous Cell Carcinoma; NA, not available; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; OS, overall survival; DFS, disease−free survival.
4.2 Association between HOXA-AS2 and prognostic indicators
4.2.1 Association between HOXA-AS2 expression and OS
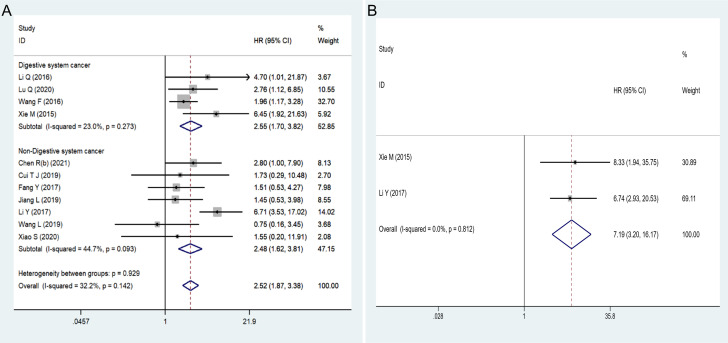
Eleven articles involving 684 patients reported the correlation between the expression level of HOXA-AS2 and the prognosis of cancer patients. No noticeable heterogeneity was observed among the studies (I2 = 32.2%, P=0.142), and therefore, fixed-effects model was utilized to estimate the HR of the studies. The results showed that high HOXA-AS2 expression predicts poor cancer survival (pooled HR=2.52, 95% CI 1.87-3.38, P < 0.01). Subgroup analysis was adopted based on the tumor type, HR estimation method, sample size, follow-up time and tumor tissue origins (mesenchymal tissue, epithelial tissue), which further confirmed that HOXA-AS2 overexpression was associated with poorer OS among cancer patients in all subgroups except for mesenchymal tissue origins (osteosarcoma) (HR=0.75, 95% CI 0.16-3.48) ( Figure 2A , Table 2 ).
Figure 2.
Forest plots showed: (A) the correlation between HOXA-AS2 expression and overall survival (OS). (B) the correlation between HOXA-AS2 expression and disease-free survival (DFS).
Table 2.
Subgroup analysis of the pooled HRs with HOXA-AS2 expression in patients with malignancy.
| No. of studies | No. of patients | Pooled HR (95% CI) | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| Fixed | Random | I2(%) | P-value | ||||
| Overall survival(OS) | 11 | 684 | 2.52 (1.87-3.38) | 2.54 (1.72-3.76) | 32.2 | 0.142 | |
| Cancer type | |||||||
| Digestive system | 4 | 313 | 2.55 (1.70-3.82) | 2.79 (1.67-4.68) | 23 | 0.273 | |
| Non-digestive system | 7 | 371 | 2.48 (1.62-3.81) | 2.17 (1.18-3.99) | 44.7 | 0.093 | |
| HR estimation method | |||||||
| Indirectly | 8 | 438 | 2.00 (1.39-2.86) | 2.00 (1.38-2.88) | 1.5 | 0.418 | |
| Directly | 3 | 246 | 4.06(2.42-6.79) | 3.96 (2.15-7.28) | 27.3 | 0.253 | |
| Sample size | |||||||
| ≥50 | 6 | 522 | 2.75 (1.95-3.87) | 2.94 (1.69-5.09) | 51.7 | 0.066 | |
| <50 | 5 | 162 | 1.97 (1.11-3.51) | 1.97 (1.11-3.51) | 0 | 0.484 | |
| Follow-up time(month) | |||||||
| ≥60 | 8 | 483 | 2.25 (1.62-3.13) | 2.17 (1.36-3.47) | 38.2 | 0.125 | |
| <60 | 3 | 201 | 3.91 (2.03-7.53) | 3.91 (2.03-7.53) | 0 | 0.529 | |
| Tumor Tissue origins | |||||||
| Mesenchymal tissue | 1 | 27 | 0.75 (0.16-3.48) | 0.75 (0.16-3.48) | – | – | |
| Epithelial tissue | 10 | 657 | 2.64 (1.95-3.56) | 2.72 (1.86-3.96) | 26.6 | 0.199 | |
| DiseaseFree Survival (DFS) | 2 | 158 | 7.19 (3.20-16.17) | 7.19 (3.20-16.17) | 0 | 0.812 | |
4.2.2 Association of HOXA-AS2 expression with DFS
Only two studies reported DFS data that could be used to assess the prognostic value of HOXA-AS2. Our analysis suggested that HOXA-AS2 overexpression was associated with DFS (pooled HR = 7.19, 95% CI 3.20-16.17, P < 0.01) ( Figure 2B , Table 2 ). No obvious heterogeneity was observed among the studies (I2 = 0%, P = 0.812).
4.3 Association between HOXA-AS2 and clinicopathological features
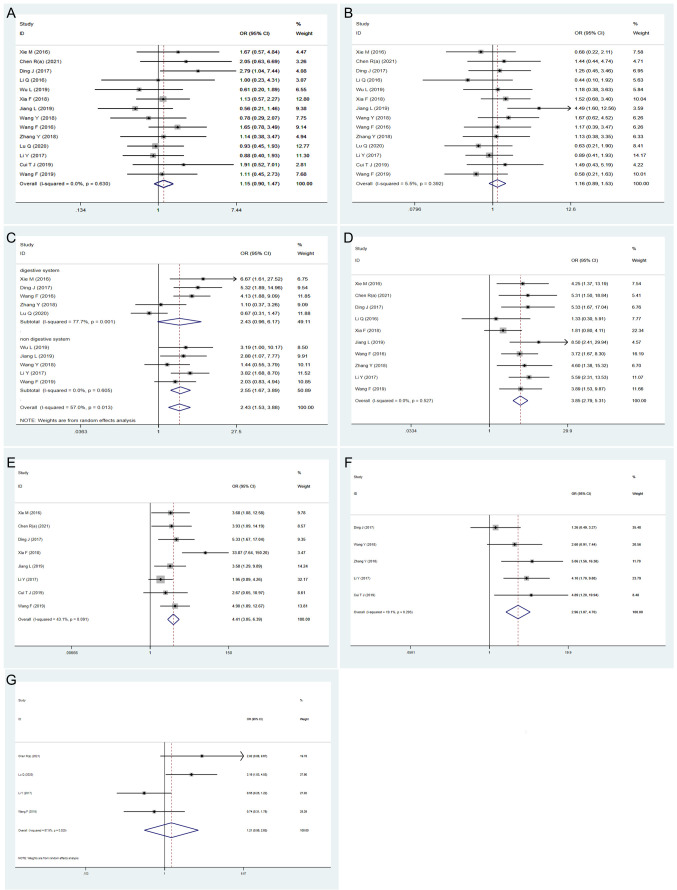
4.3.1 Association between HOXA-AS2 expression and age, gender
Fourteen studies with 1021 patients were examined to determine the association between the expression level of HOXA-AS2 and age ( Figure 3A ). No obvious heterogeneity was observed (I2 = 0%, P=0.630); therefore, the fix-effects model was conducted. We found that there was no significant association between HOXA-AS2 expression and patient age (OR=1.15, 95% CI 0.90-1.47, P > 0.05). Fourteen studies with 1021 patients investigated potential associations between HOXA-AS2 expression and gender. Upon assessment with the fixed model, no obvious heterogeneity was observed amongst the studies (I2 = 5.5%, P=0.392). The expression of HOXA-AS2 was not correlated with patient gender (OR=1.16, 95% CI 0.89-1.53, P > 0.05) ( Figure 3B ).
Figure 3.
Forest plots showing the correlation between HOXA-AS2 expression and clinicopathological parameters. (A) age. (B) gender. (C) tumor size. (D) TNM stage. (E) lymph node metastasis. (F) distant metastasis. (G) differentiation.
4.3.2 Association of HOXA-AS2 expression with tumor size
The relationship between tumor size and HOXA-AS2 expression was evaluated in 10 studies involving 777 patients. There was obvious heterogeneity (I2 = 57%, P=0.013); therefore, the random-effects model was carried out, and subgroup analysis was performed. The combined results indicated a significant positive correlation between high HOXA-AS2 expression and larger tumors (OR=2.43, 95% CI 1.53-3.88, P < 0.01). Subgroup analysis revealed, a strong association between HOXA-AS2 expression and tumor size in non-digestive system malignant tumor (OR =2.55, 95% CI 1.67-7.74, P < 0.01). However, no significant correlation was observed between HOXA-AS2 expression and digestive system malignancies (OR=2.43, 95% CI 0.96–6.17, P > 0.05), indicating that the digestive system tumors may be the source of the heterogeneity ( Figure 3C ).
4.3.3 The relationship between HOXA-AS2 expression and TNM and lymph node metastasis (LNM)
There were 10 and 8 studies reporting the TNM stage and LNM in patients with solid tumors, respectively. The fix-effects model was carried out to show that the expression level of HOXA-AS2 was significantly correlated to TNM stage (OR=3.85, 95% CI 2.79-5.31, P < 0.01) and LNM (OR = 4.41, 95% CI 3.05-6.39, P < 0.01) with no obvious heterogeneity ( Figures 3D, E ).
4.3.4 The relationship between the expression of HOXA-AS2 and distant metastasis (DM), tumor differentiation
A total of 5 qualified studies examined the occurrence of DM in patients with solid tumors. Since there was no significant heterogeneity, we used the fixed-effects model (I2 = 19.1%, P = 0.293) to analyze the DM data. The results suggested that the increased expression of HOXA-AS2 was significantly related to DM (OR=2.96, 95% CI 1.87-4.7, P < 0.01) ( Figure 3F ). Four of the included studies reported the differentiation of tumors, and the random-effects model was used to analyze the relationship between HOXA-AS2 expression and tumor differentiation (I2 = 67.9%, P=0.025). We found that the increased expression of HOXA-AS2 was not significantly correlated with the tumor differentiation (OR=1.21, 95% CI 0.56-2.63, P > 0.05) ( Figure 3G ).
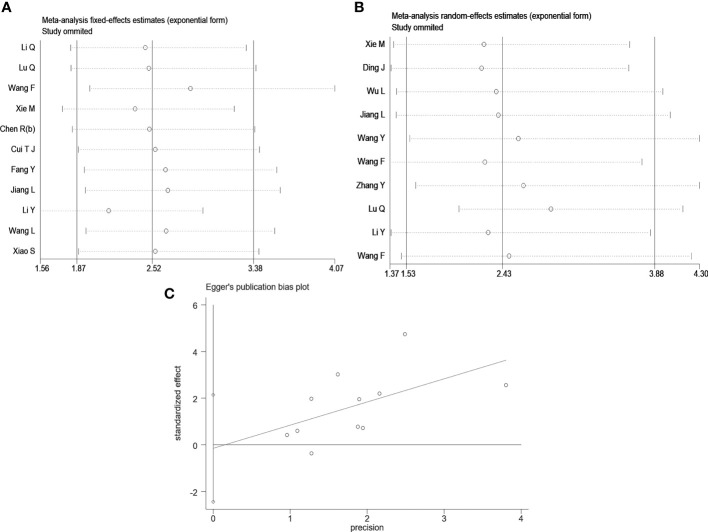
4.4 Sensitivity analysis and publication bias
Sensitivity analysis was conducted by the leave-one-out method to assess the effect of an individual article on the pooled OS and tumor size data. However, no single article significantly altered the results ( Figures 4A, B ). Egger’s test did not reveal evident publication bias for OS (Pr>|t|=0.884) ( Figure 4C ), age (Pr>|t|=0.694), gender (Pr>|t|=0.524), tumor size (Pr>|t|=0.399), TNM stage (Pr>|t|=0.582), LNM (Pr>|t|=0.088), DM (Pr >|t|=0.507), or differentiation (Pr>|t| =0.697) ( Supplementary Figure S1 ).
Figure 4.
Sensitivity analysis and Egger’s test. (A) Sensitivity analysis for OS; (B) Sensitivity analysis for tumor size; (C) The Egger’s test and linear regression plot for the publication bias for OS.
4.5 Prediction of HOXA-AS2 function and drug sensitivity in pan-cancer
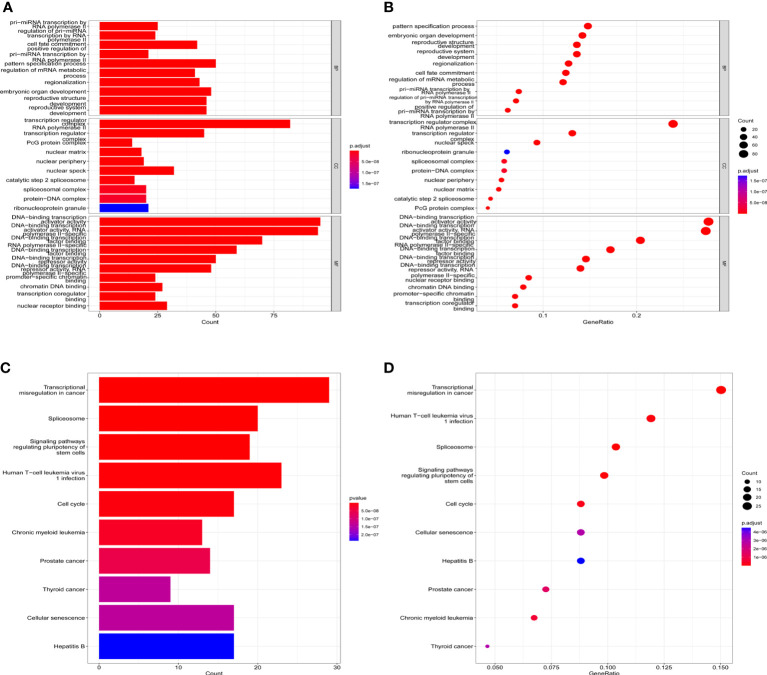
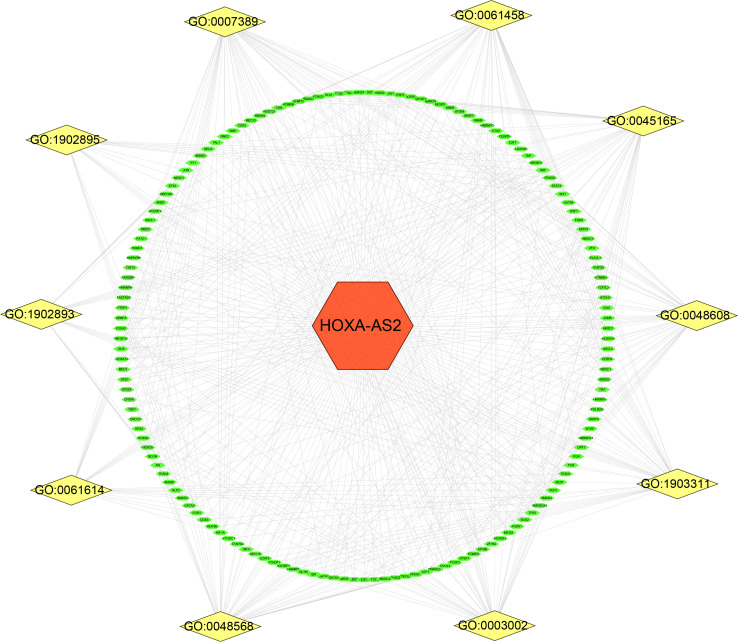
Public databases were used to predict the biological functions and molecular mechanisms of HOXA-AS2. First, we used the RNAInter v4.0 database to screen interacting genes of HOXA-AS2(Confidence Score screening: 0.2-1.0), and 344 protein-coding genes were screened ( Supplementary Table S2 ). Figure 5 shows the underlying molecular mechanisms identified by GO and KEGG pathway analyses. HOXA-AS2 was predicted to be mainly involved in biological processes (BPs), such as pri−miRNA transcription by RNA polymerase II, cell fate commitment, and embryonic organ development; molecular functions (MFs), such as DNA−binding transcription activator activity, RNA polymerase II-specific DNA-binding transcription factor binding; and cellular components (CCs), such as the transcription regulator complex, RNA polymerase II transcription regulator complex ( Figures 5A, B ). Additionally, the KEGG pathway analysis results showed that interacting genes were principally involved in transcriptional misregulation in cancer, the spliceosome, and signaling pathways regulating the pluripotency of stem cells ( Figures 5C, D ). Furthermore, Cytoscape software was applied to construct the top 10 visual signaling pathway networks ( Figure 6 ).
Figure 5.
KEGG and GO term enrichment for HOXA-AS2. (A) Barplots of GO enrichment. (B) Bubble charts of GO enrichment; (C) Barplots of KEGG molecular mechanisms. (D) Bubble charts of KEGG molecular mechanisms.
Figure 6.
Related gene interaction network analysis. Yellow nodes represent the related pathway and green nodes represent target genes. GO:0061614 (pri-miRNA transcription by RNA polymerase II); GO:1902893 (regulation of pri-miRNA transcription by RNA polymerase II); GO:0045165 (cell fate commitment); GO:1902895 (positive regulation of pri-miRNA transcription by RNA polymerase II); GO:0007389 (pattern specification process); GO:1903311 (regulation of mRNA metabolic process); GO:0003002 (regionalization); GO:0048568 (embryonic organ development); GO:0048608 (reproductive structure development); GO:0061458 (reproductive system development).
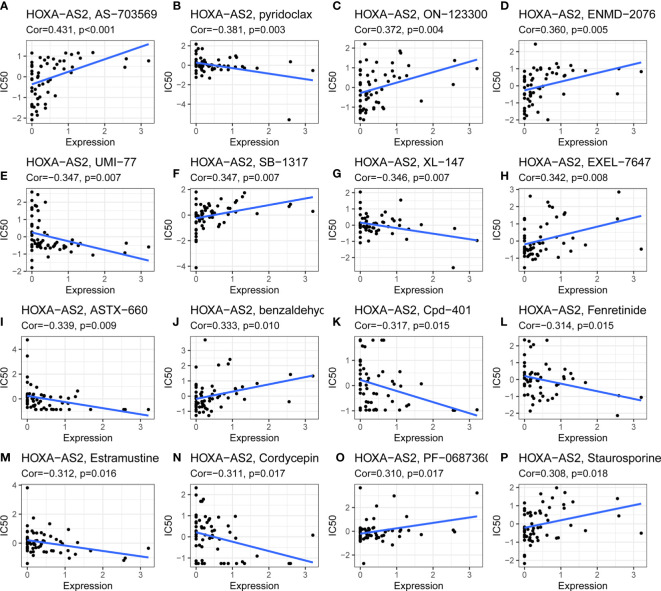
Using CellMiner™, we further investigated the potential correlation between HOXA-AS2 expression and drug sensitivity. Notably, HOXA-AS2 expression was positively correlated with the drug sensitivity of AS-703659, ON-123300, ENMD-2076, SB-1317, EXEL-7647, benzaldehyc, PF-0687360, and staurosporine ( Figures 7A, C, D, F, H, J, O, P ). Our results showed that HOXA-AS2 expression was negatively associated with pyridoclax, UMI-77, XL-147, ASTX-660, Cpd-401, fenretinide, estramustine, and cordycepin ( Figures 7B, E, G, I, K–N ). The data suggested that HOXA-AS2 might contribute to the chemoresistance development. Such as AS-703659 and Staurosporine. Most of the above positively correlated drugs were enzyme inhibitors, indicating that the involvement of HOXA-AS2 in chemoresistance might be related to the metabolism of enzymes and transcriptional misregulation.
Figure 7.
Drug sensitivity analysis of HOXA-AS2. The expression of HOXA-AS2 was associated with the sensitivity of (A) AS-703659, (B) pyridoclax, (C) ON-123300, (D) ENMD-2076, (E) UMI-77, (F) SB-1317, (G) XL-147, (H) EXEL-7647, (I) ASTX-660, (J) benzaldehyc, (K) Cpd-401, (L) Fenretinide, (M) Estramustine, (N) Cordycepin, (O) PF-0687360, (P) Staurosporine.
5 Discussion
In recent years, lncRNAs have attracted increasing attention due to rapid advances in high-throughput sequencing technology (48, 49). Increasing evidence suggests that abnormal lncRNA expression is related to human cancers, and dysregulation of lncRNAs contributes to tumor development by promoting cell proliferation, invasion and metastasis of tumor cells (50, 51). Some studies have reported that lncRNAs,such as LUCAT1 (52), SNHG7 (53), TP73-AS1 (54), are promising to be the potential therapeutic targets and biomarkers for diagnosis, prognostic evaluation or treatment. The prognostic value of HOXA-AS2 expression levels for cancers are unclear, and the underlying mechanism has yet to be elucidated.
HOXA-AS2 was found to be highly expressed in promyelocytic leukemia for the first time (55) and has been confirmed to be dysregulated in diverse tumor cells, such as osteosarcoma (37), glioma (56), NSCLC (20), CRC (57), and cervical cancer (35). It has also been proven to be involved in the biological behavior of many tumor cells as a high-risk factor. In osteosarcoma and cervical cancer, cell function tests show that HOXA-AS2 upregulation promotes tumor cell metastasis, prevents the cell cycle, and epithelial-mesenchymal transition and inhibits apoptosis. Silencing HOXA-AS2 can inhibit the above biological processes (35, 37). In hepatocellular carcinoma, HOXA-AS2 expression was significantly increased in both cell lines and HCC tissues and was associated with poor survival and prognosis (41). Therefore, most studies reported that high expression of HOXA-AS2 carried a higher risk of cancer progression and a poorer survival benefit.
Due to the existing research foundations and the urgent need to improve cancer prognosis, this study evaluated the prognostic value of HOXA-AS2 for various solid tumors. A total of eighteen studies involving 1181 patients and 12 solid tumors were included. The results showed that high expression of HOXA-AS2 was positively correlated with the poor prognosis of patients, including overall survival (HR=2.52, 95% CI 1.87-3.38, P<0.01) and disease-free survival (HR=7.19, 95% CI 3.20-16.17, P<0.01). Given the differential expression of HOXA-AS2 in different tumor tissues, a subgroup analysis for OS was performed based on different tissue sources such as cancer types, different data extraction methods, sample sizes, follow-up times and tumor tissue origins, and there was no noticeable heterogeneity. However, we found no correlation between HOXA-AS2 expression and mesenchymal tissue origins (osteosarcoma) (HR=0.75, 95%CI 0.16-3.48), which may be because only one small-sample study (n=27) was conducted in this subgroup or because of tumor heterogeneity between mesenchymal tumors and epithelial tumors. Therefore, further studies with a larger sample size are needed to validate the prognostic significance of HOXA-AS2 in mesenchymal tumors.
Moreover, we analyzed the relationship between HOXA-AS2 expression levels and clinical attributes. The results of the combined OR showed that elevated HOXA-AS2 expression had no significant correlations with age, sex or tumor differentiation but was positively correlated with advanced TNM stage, earlier lymph node metastasis and distant metastasis. Overall, our data suggested that HOXA-AS2 could serve as a potential reliable tumor prognostic indicator.
Previous studies have demonstrated that HOXA-AS2 is highly expressed in various cancer tissues and closely associated with the prognosis of tumor patients, such as gastric cancer, hepatocellular carcinoma, non-small cell lung cancer, and cervical cancer. Some researchers have suggested that long noncoding RNAs could function as therapeutic targets for cancer and as independent indicators of tumor progression. Liu Y et al. reported that HOXA-AS2 could promote cell proliferation, migration and invasion and inhibit apoptosis in NSCLC via the miR-520a-3p/HOXD8/MAP3K2 axis (58). Zheng et al. revealed that lncHOXA-AS2 expression was increased in NSCLC tissues and that HOXA-AS2 could enhance the migratory and invasive abilities of tumor cells by upregulating IGF2 (20). Wang F et al. revealed that HOXA-AS2 knockdown suppresses the migration, invasion and stemness of bladder cancer cells by regulating miR-125b/Smad2E signaling axis (30). In colorectal cancer, Ding et al. found that silencing HOXA-AS2 could promote apoptosis, inhibit the proliferation and migration by binding to EZH2/LSD1, and upregulated HOXA-AS2 is positively correlated with larger tumor size, advanced tumor stage and early lymph node metastasis. Wu Q et al. reported that HOXA-AS2 may promote the cell proliferation and migration of cervical cancer cells by modulating the Notch pathway (34). Gao Y et al. identified that HOXA-AS2 can play a role in glioblastoma cell viability, invasion, migration, and vasculogenic mimicry by interacting with EFGR via sponging miR-373 (59). Additionally, we explored the mechanisms of HOXA-AS2 expression in ovarian cancer, gallbladder carcinoma, pancreatic cancer, and nasopharyngeal carcinoma and found similar results. These studies demonstrated that HOXA-AS2 is capable of oncogenic activity. It has been shown that HOXA-AS2 is related to various functions, including post-transcriptional regulation (56) and regulation of the pluripotency of stem cells (30). An overview of the molecular mechanisms governing HOX-AS2’s effects on various solid malignancies can be found in Table 3 .
Table 3.
Regulation mechanism of HOXA-AS2 involved in various solid tumors.
| Cancer type | Expression | Micro-RNAs | Targets | Functions | References |
|---|---|---|---|---|---|
| NSCLC | up-regulation | miR-216a-5p | – | Proliferation, migration, invasion | Cui T J (19) |
| up-regulation | miR-520a-3p | HOXD8/MAP3K2 | cell proliferation, migration, invasion | Liu Y (58) | |
| up-regulation | miR-302a-3p | – | invasion, migration, proliferation | Pan X (21) | |
| up-regulation | - | IGF2 | invasion, migration | Zheng (20) | |
| Bladder cancer | up-regulation | miR-125b | Smad2 | migration, invasion, stemness | Wang F (30) |
| Prostate cancer | up-regulation | miR-5590-3p | PBX3 | EMT, proliferation, migration | Xiao S (31) |
| CRC | up-regulation | – | EZH2, LSD1 | cell proliferation, inhibit apoptosis | Ding J (29) |
| HCC | up-regulation | miR-520c-3p | GPC3 | cell proliferation, migration, invasion | Zhang Y (27) |
| hepatoblastoma | up-regulation | miR-217 | HOXA3 | proliferation, migration, invasion | Liu G (60) |
| Ovarian cancer | up-regulation | miR-520a-3p | – | proliferation,migration, invasion | Xie D (32) |
| Endometrial cancer | up-regulation | miR-302c-3p | ZFX,YKL-40 | proliferation, invasion | Song N (33) |
| Breast cancer | up-regulation | miR-520c-3p | TGFBR2, RELA | proliferation, invasion | Fang Y (18) |
| Cervical cancer | up-regulation | miR-509-3p | BTN3A1 | invasion, metastasis | Chen R (35) |
| up-regulation | – | Notch Pathway | cell proliferation, migration | Wu Q (34) | |
| Gastric cancer | up-regulation | – | P21/PLK3/DDIT3 | cell proliferation, tumorigenesis | Xie M (24) |
| Osteosarcoma | up-regulation | miR-520c-3p | – | EMT, migration, invasion | Wang Y (37) |
| up-regulation | miR-124-3p | E2F3 | migration, invasion | Wang L (36) | |
| Gallbladder carcinoma | up-regulation | – | E-cadherin | migration, invasion, EMT | Zhang P (23) |
| Glioma | up-regulation | miR-373 | EFGR | migration, invasion, vasculogenic mimicry | Gao Y (59) |
| up-regulation | – | RND3 | proliferation, invasion, inhibit apoptosis | Wu L (47) | |
| up-regulation | miR-184 | COL6A2 | cell proliferation | Chen P Y (61) | |
| up-regulation | – | E2F8,E2F1,ATF3 | proliferation,inflammation | Le Boiteux (56) | |
| up-regulation | miR-302a | KDM2A/JAG1 | T cell proliferation, immune tolerance | Zhong C (62) | |
| Glioblastoma | up-regulation | miR-885-5p | RBBP4 | cell proliferation | Shou J (38) |
| up-regulation | miR-2116-3p | SERPINA3 | proliferation, invasion, inhibit apoptosis | Sun J (39) | |
| Pancreatic cancer |
up-regulation | – | EZH2, LSD1 | cell proliferation | Lian Y (28) |
| NPC | up-regulation | miR-519 | PD-L1,HIF-1α | proliferation, migration, invasion | Wang S (63) |
| PTC | up-regulation | miR-520c-3p | S100A4 | migration, invasion | Xia F (16) |
| up-regulation | miR-15a-5p | HOXA3 | proliferation, migration, invasion | Jiang L (15) | |
| OSCC | up-regulation | miR-567 | CDK8 | cell proliferation | Chen R (13) |
| up-regulation | – | EZH2 | proliferation, migration, invasion | Zhao Z (12) |
NSCLC, non-small cell lung cancer; CRC, colorectal cancer; PTC, papillary thyroid cancer; NPC, nasopharyngeal carcinoma; OSCC, oral squamous cell carcinoma; HCC, hepatocellular carcinoma.
Based on bioinformatics analysis, we predicted and functionally annotated the target genes of HOXA-AS2. Gene ontology and KEGG pathway enrichment analysis found that target genes were mostly enriched in pri−miRNA transcription by RNA polymerase II, cell fate commitment, regulation of mRNA metabolic process, DNA−binding transcription activator activity, transcription regulator complex, transcriptional misregulation in cancer, and signaling pathways regulating pluripotency of stem cells. These functions mainly included transcriptional regulation and enzyme metabolism. Our results were in consistent with previous reports that HOXA-AS2 was involved in tumor progression by regulating various pathways, which broadened the prospects for further study on HOX-AS2 target therapy.
In addition to the close relationship between lncRNAs and tumor progression and metastasis, an increasing number of studies have also demonstrated that they play an important role in determining cancer cell malignant phenotypes and drug resistance (64–66). To improve the survival of patients with malignant tumors, it is imperative to understand the relationship between lncRNAs and drug resistance. As a next step, we investigated the correlation between HOXA-AS2 expression and drug sensitivity. In acute myeloid leukemia (ALL), Zhao et al. reported that HOXA-AS2 upregulation can reduce glucocorticoid sensitivity by regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway (67). Dong et al. found that HOXA-AS2 overexpression increases the resistance of acute myeloid leukemia cells to adriamycin, perhaps through the miR-520c-3p/S100A4 pathway (68). However, there has been no related research on the relation between HOXA-AS2 and drug sensitivity or resistance in solid cancers until now. We explored the association between HOXA-AS2 expression and drug sensitivity via the CellMiner™ database and found that HOXA-AS2 expression was correlated with many drug sensitivities, such as AS-703569, pyridoclax, ON-123300, and benzaldehyde. Therefore, we deduced that HOXA-AS2 may affect chemotherapy and may be correlated with chemoresistance. Despite of the need for further research into the mechanism of HOXA-AS2 and drug resistance to these drugs, this finding provides us with a potential research avenue for modulating drug resistance and eventually improving cancer prognosis through inhibition of HOXA-AS2 expression.
This large-sample study aims to investigate the correlation between abnormal expression of HOXA-AS2 and tumor prognosis. It also revealed the characteristics of HOXA-AS2 in multiple aspects, including expression pattern, survival prognosis, signaling pathway, and drug sensitivity. HOXA-AS2 might serve as a potential target for cancer treatment since it displayed abnormal expression in multiple cancers and predicted a worse prognosis in cancer patients. To ensure quality and high representativeness, all relevant data should be collected as much as possible using appropriate statistical software and statistical methods. However, this study also has several limitations. 1. Among the included literature, some studies provided HRs and 95% confidence intervals directly, while others provided only survival curves, which caused a certain reporting bias. 2. Due to language limitations, the literature sources used in this study were all English literature, and related literature in other languages was not included, so the conclusion represents only a fraction of the population. 3. The included literature was a retrospective study, and the quality level of the literature may be low. 4. Many studies with positive results are more likely to be published than those with negative results, which leads to certain publication biases. 5. The included studies were all from China, so the results may only be applicable to Chinese or Asian populations. 6. We only predicted the molecular mechanisms and biological functions of HOXA-AS2 via GO enrichment and KEGG analysis, and the drug sensitivity analysis was carried out only by the tumor call line set using online databases. Regulatory mechanisms at a broader level should now be investigated. Overall, studies with larger sample sizes are needed to explore other potential functions of HOXA-AS2 and its role in pro-oncogenic signaling. Using this information, cancer patients might be able to develop new therapeutic strategies.
6 Conclusion
High expression levels of HOXA-AS2 are associated with poor cancer prognosis among Chinese patients, and HOXA-AS2 may serve as an underlying therapeutic target and a promising prognostic biomarker.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author contributions
YZ and QW design the project; CD and SL searched databases and performed literature screen; CD and QW extracted and analyzed the data, analysis; WZ and QW evaluated the quality of included literature; YZ, QW, WZ, and CD contributed to writing the manuscript. Final draft was approved by all the authors. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1030825/full#supplementary-material
The publication bias of HOXA-AS2 related research.
Quality assessment of eligible studies Newcastle-Ottawa scale (NOS).
Interactional Protein-coding Genes sceened by RNAinter database.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 3. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin (2022) 72(5):409–436. doi: 10.3322/caac.21731 [DOI] [PubMed] [Google Scholar]
- 4. Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, et al. Long non-coding RNAs: The regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol (2020) 10:598817. doi: 10.3389/fonc.2020.598817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov (2017) 16(3):167–79. doi: 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet (2018) 19(9):535–48. doi: 10.1038/s41576-018-0017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin C, Yang L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol (2018) 28(4):287–301. doi: 10.1016/j.tcb.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol (2018) 19(3):143–57. doi: 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heydarnezhad Asl M, Pasban Khelejani F, Bahojb Mahdavi SZ, Emrahi L, Jebelli A, Mokhtarzadeh A. The various regulatory functions of long noncoding RNAs in apoptosis, cell cycle, and cellular senescence. J Cell Biochem (2022) 123(6):995–1024. doi: 10.1002/jcb.30221 [DOI] [PubMed] [Google Scholar]
- 10. Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev (2018) 34(2):153–80. doi: 10.1080/02648725.2018.1471566 [DOI] [PubMed] [Google Scholar]
- 11. Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta (2018) 485:229–33. doi: 10.1016/j.cca.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 12. Zhao Z, Xing Y, Yang F, Zhao Z, Shen Y, Song J, et al. LncRNA HOXA-AS2 promotes oral squamous cell proliferation, migration, and invasion via upregulating EZH2 as an oncogene. Technol Cancer Res Treat (2021) 20:15330338211039109. doi: 10.1177/15330338211039109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen R, Wang X, Zhou S, Zeng Z. LncRNA HOXA-AS2 promotes tumor progression by suppressing miR-567 expression in oral squamous cell carcinoma. Cancer Manag Res (2021) 13:5443–55. doi: 10.2147/CMAR.S305946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S, You H, Yu S. Long non-coding RNA HOXA-AS2 promotes the expression levels of hypoxia-inducible factor-1α and programmed death-ligand 1, and regulates nasopharyngeal carcinoma progression via miR-519. Oncol Lett (2020) 20(5):245. doi: 10.3892/ol.2020.12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang L, Wu Z, Meng X, Chu X, Huang H, Xu C. LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 axis. Hum Gene Ther (2019) 30(5):618–31. doi: 10.1089/hum.2018.109 [DOI] [PubMed] [Google Scholar]
- 16. Xia F, Chen Y, Jiang B, Du X, Peng Y, Wang W, et al. Long noncoding RNA HOXA-AS2 promotes papillary thyroid cancer progression by regulating miR-520c-3p/S100A4 pathway. Cell Physiol Biochem (2018) 50(5):1659–72. doi: 10.1159/000494786 [DOI] [PubMed] [Google Scholar]
- 17. Wu J, Li M, Zhang Y. Long noncoding RNA HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in breast cancer. J Cell Biochem (2019) 120(9):14465–75. doi: 10.1002/jcb.28706 [DOI] [PubMed] [Google Scholar]
- 18. Fang Y, Wang J, Wu F, Song Y, Zhao S, Zhang Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget (2017) 8(28):46090–103. doi: 10.18632/oncotarget.17552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui TJ, Lin GS, Dai YM, Zheng JP, Chen Z, Chen Q, et al. LncRNA HOXA-AS2 regulates microRNA-216a-5p to promote malignant progression of non-small cell lung cancer. Eur Rev Med Pharmacol Sci (2019) 23(3 Suppl):264–73. doi: 10.26355/eurrev_201908_18656 [DOI] [PubMed] [Google Scholar]
- 20. Zheng FX, Wang XQ, Zheng WX, Zhao J. Long noncoding RNA HOXA-AS2 promotes cell migration and invasion via upregulating IGF-2 in non-small cell lung cancer as an oncogene. Eur Rev Med Pharmacol Sci (2019) 23(11):4793–9. doi: 10.26355/eurrev_201906_18064 [DOI] [PubMed] [Google Scholar]
- 21. Pan X, Ma D. MiR-302a-3p, sponged by HOXA-AS2, suppresses cell proliferation and invasion in lung cancer cells. J Biomater Tissue Eng (2019) 9(12):1644–52. doi: 10.1166/jbt.2019.2202 [DOI] [Google Scholar]
- 22. Li Y, Jiang H. Up-regulation of long non-coding RNA HOXA-AS2 in non-small cell lung cancer is associated with worse survival outcome. Int J Clin Exp Pathol (2017) 10(9):9690–6. [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P, Cao P, Zhu X, Pan M, Zhong K, He R, et al. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget (2017) 8(20):33137–43. doi: 10.18632/oncotarget.16561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding J, et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget (2015) 6(32):33587–601. doi: 10.18632/oncotarget.5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajabi A, Riahi A, Shirabadi-Arani H, Moaddab Y, Haghi M, Safaralizadeh R. Overexpression of HOXA-AS2 LncRNA in patients with gastric cancer and its association with helicobacter pylori infection. J Gastrointest Cancer (2022) 53(1):72–7. doi: 10.1007/s12029-020-00549-y [DOI] [PubMed] [Google Scholar]
- 26. Wang F, Yang H, Deng Z, Su Y, Fang Q, Yin Z. HOX antisense lincRNA HOXA-AS2 promotes tumorigenesis of hepatocellular carcinoma. Cell Physiol Biochem (2016) 40(1-2):287–96. doi: 10.1159/000452545 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Xu J, Zhang S, An J, Zhang J, Huang J, et al. HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition via the miR-520c-3p/GPC3 axis in hepatocellular carcinoma. Cell Physiol Biochem (2018) 50(6):2124–38. doi: 10.1159/000495056 [DOI] [PubMed] [Google Scholar]
- 28. Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu W, et al. The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex promotes cell proliferation in pancreatic cancer. Am J Transl Res (2017) 9(12):5496–506. [PMC free article] [PubMed] [Google Scholar]
- 29. Ding J, Xie M, Lian Y, Zhu Y, Peng P, Wang J, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis (2017) 6(1):e288. doi: 10.1038/oncsis.2016.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F, Wu D, Chen J, Chen S, He F, Fu H, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res (2019) 375(1):1–10. doi: 10.1016/j.yexcr.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 31. Xiao S, Song B. LncRNA HOXA-AS2 promotes the progression of prostate cancer via targeting miR-509-3p/PBX3 axis. Biosci Rep (2020) 40(8):BSR20193287. doi: 10.1042/BSR20193287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie D, Ling L, Jin X. lncRNA HOXA-AS2 regulates proliferation, migration and invasion of ovarian cancer cells via targeting miR-520a-3p. Chin J Cancer Biother (2020) 27(5):501–7. doi: 10.3872/j.issn.1007-385X.2020.05.005 [DOI] [Google Scholar]
- 33. Song N, Zhang Y, Kong F, Yang H, Ma X. HOXA-AS2 promotes type I endometrial carcinoma via miRNA-302c-3p-mediated regulation of ZFX. Cancer Cell Int (2020) 20:359. doi: 10.1186/s12935-020-01443-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Q, Lu S, Zhang L, Zhao L. LncRNA HOXA-AS2 activates the notch pathway to promote cervical cancer cell proliferation and migration. Reprod Sci (Thousand Oaks Calif) (2021) 28(10):3000–9. doi: 10.1007/s43032-021-00626-y [DOI] [PubMed] [Google Scholar]
- 35. Chen R, He P. Long noncoding RNA HOXA-AS2 accelerates cervical cancer by the miR-509-3p/BTN3A1 axis. J Pharm Pharmacol (2021) 73(10):1387–96. doi: 10.1093/jpp/rgab090 [DOI] [PubMed] [Google Scholar]
- 36. Wang L, Wang L, Zhang X. Knockdown of lncRNA HOXA-AS2 inhibits viability, migration and invasion of osteosarcoma cells by miR-124-3p/E2F3. Onco Targets Ther (2019) 12:10851–61. doi: 10.2147/OTT.S220072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Zhang R, Cheng G, Xu R, Han X. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle (2018) 17(13):1637–48. doi: 10.1080/15384101.2018.1489174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shou J, Gao H, Cheng S, Wang B, Guan H. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int (2021) 21(1):39. doi: 10.1186/s12935-020-01690-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J, Wang L. HOXA-AS2 enhances GBM cell malignancy by suppressing miR-2116-3p thereby upregulating SERPINA3. BMC Cancer (2022) 22(1):366. doi: 10.1186/s12885-022-09462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma (2016) 63(6):977–83. doi: 10.4149/neo_2016_617 [DOI] [PubMed] [Google Scholar]
- 41. Lu Q, Gao J, Tang S, Li Z, Wang X, Deng C, et al. Integrated RNA sequencing and single-cell mass cytometry reveal a novel role of LncRNA HOXA-AS2 in tumorigenesis and stemness of hepatocellular carcinoma. Onco Targets Ther (2020) 13:10901–16. doi: 10.2147/OTT.S272717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ (Clinical Res ed) (2021) 372:n160. doi: 10.1136/bmj.n160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol (2010) 25(9):603–5. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 44. Kang J, Tang Q, He J, Li L, Yang N, Yu S, et al. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res (2022) 50(D1):D326–d32. doi: 10.1093/nar/gkab997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol (Clifton NJ) (2011) 696:291–303. doi: 10.1007/978-1-60761-987-1_18 [DOI] [PubMed] [Google Scholar]
- 46. Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res (2012) 72(14):3499–511. doi: 10.1158/0008-5472.CAN-12-1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu L, Zhu X, Song Z, Chen D, Guo M, Liang J, et al. Long non-coding RNA HOXA-AS2 enhances the malignant biological behaviors in glioma by epigenetically regulating RND3 expression. Onco Targets Ther (2019) 12:9407–19. doi: 10.2147/OTT.S225678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Núñez-Martínez HN, Recillas-Targa F. Emerging functions of lncRNA loci beyond the transcript itself. Int J Mol Sci (2022) 23(11):6258. doi: 10.3390/ijms23116258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rao A, Rajkumar T, Mani S. Perspectives of long non-coding RNAs in cancer. Mol Biol Rep (2017) 44(2):203–18. doi: 10.1007/s11033-017-4103-6 [DOI] [PubMed] [Google Scholar]
- 50. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett (2018) 419:152–66. doi: 10.1016/j.canlet.2018.01.053 [DOI] [PubMed] [Google Scholar]
- 51. Kwok ZH, Tay Y. Long noncoding RNAs: lincs between human health and disease. Biochem Soc Trans (2017) 45(3):805–12. doi: 10.1042/BST20160376 [DOI] [PubMed] [Google Scholar]
- 52. Xing C, Sun SG, Yue ZQ, Bai F. Role of lncRNA LUCAT1 in cancer. BioMed Pharmacother (2021) 134:111158. doi: 10.1016/j.biopha.2020.111158 [DOI] [PubMed] [Google Scholar]
- 53. Wang J, Du S, Wang C, Zhu Z, Xie B, Zhang B. Clinicopathological and prognostic value of long noncoding RNA SNHG7 in cancers: a meta-analysis and bioinformatics. Aging (2021) 13(20):23796–809. doi: 10.18632/aging.203650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen C, Wang J, Feng Y, Liang Y, Huang Y, Zou W. TP73-AS1 as a predictor of clinicopathological parameters and prognosis in human malignancies: a meta and bioinformatics analysis. BMC Cancer (2022) 22(1):581. doi: 10.1186/s12885-022-09658-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao H, Zhang X, Frazao JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem (2013) 114(10):2375–83. doi: 10.1002/jcb.24586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Le Boiteux E, Guichet P-O, Masliantsev K, Montibus B, Vaurs-Barriere C, Gonthier-Gueret C, et al. The long non-coding RNA HOXA-AS2 promotes proliferation of glioma stem cells and modulates their inflammation pathway mainly through post-transcriptional regulation. Int J Mol Sci (2022) 23(9):4743. doi: 10.3390/ijms23094743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tong G, Wu X, Cheng B, Li L, Li X, Li Z, et al. Knockdown of HOXA-AS2 suppresses proliferation and induces apoptosis in colorectal cancer. Am J Transl Res (2017) 9(10):4545–52. [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Y, Lin X, Zhou S, Zhang P, Shao G, Yang Z. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci Rep (2019) 39(5):BSR20190283. doi: 10.1042/BSR20190283 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Gao Y, Yu H, Liu Y, Liu X, Zheng J, Ma J, et al. Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the MiR-373/EGFR axis. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 45(1):131–47. doi: 10.1159/000486253 [DOI] [PubMed] [Google Scholar]
- 60. Liu G, Liu B, Liu X, Xie L, He J, Zhang J, et al. ARID1B/SUB1-activated lncRNA HOXA-AS2 drives the malignant behaviour of hepatoblastoma through regulation of HOXA3. J Cell Mol Med (2021) 25(7):3524–36. doi: 10.1111/jcmm.16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen P-Y, Li X-D, Ma W-N, Li H, Li M-M, Yang X-Y, et al. Comprehensive transcriptomic analysis and experimental validation identify lncRNA HOXA-AS2/miR-184/COL6A2 as the critical ceRNA regulation involved in low-grade glioma recurrence. Oncotargets Ther (2020) 13:4999–5016. doi: 10.2147/OTT.S245896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhong C, Tao B, Li X, Xiang W, Peng L, Peng T, et al. HOXA-AS2 contributes to regulatory T cell proliferation and immune tolerance in glioma through the miR-302a/KDM2A/JAG1 axis. Cell Death Dis (2022) 13(2):160. doi: 10.1038/s41419-021-04461-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang S, You H, Yu S. Long non-coding RNA HOXA-AS2 promotes the expression levels of hypoxia-inducible factor-1 alpha and programmed death-ligand 1, and regulates nasopharyngeal carcinoma progression via miR-519. Oncol Lett (2020) 20(5):245. doi: 10.3892/ol.2020.12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Y, Sun W, Qin Z, Guo S, Kang Y, Zeng S, et al. LncRNA regulation: New frontiers in epigenetic solutions to drug chemoresistance. Biochem Pharmacol (2021) 189:114228. doi: 10.1016/j.bcp.2020.114228 [DOI] [PubMed] [Google Scholar]
- 65. Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy (2021) 17(12):4083–101. doi: 10.1080/15548627.2021.1901204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis (2020) 11(4):233. doi: 10.1038/s41419-020-2426-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhao Q, Zhao S, Li J, Zhang H, Qian C, Wang H, et al. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. BioMed Pharmacother (2019) 109:1640–9. doi: 10.1016/j.biopha.2018.10.046 [DOI] [PubMed] [Google Scholar]
- 68. Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X, et al. Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 axis. Cell Physiol Biochem (2018) 51(2):886–96. doi: 10.1159/000495387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The publication bias of HOXA-AS2 related research.
Quality assessment of eligible studies Newcastle-Ottawa scale (NOS).
Interactional Protein-coding Genes sceened by RNAinter database.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.