Abstract
Background
Young children are frequently exposed to antibiotics, with the potential for collateral consequences to the gut microbiome. The impact of antibiotic exposures to off-target microbes (i.e., bacteria not targeted by treatment) and antibiotic resistance genes (ARGs) is poorly understood.
Methods
We used metagenomic sequencing data from paired stool samples collected prior to antibiotic exposure and at 1 year from over 200 infants and a difference-in-differences approach to assess the relationship between subsequent exposures and the abundance or compositional diversity of microbes and ARGs while adjusting for covariates.
Results
By 1 year, the abundance of multiple species and ARGs differed by antibiotic exposure. Compared to infants never exposed to antibiotics, Bacteroides vulgatus relative abundance increased by 1.72% (95% CI: 0.19, 3.24) while Bacteroides fragilis decreased by 1.56% (95% CI: −4.32, 1.21). Bifidobacterium species also exhibited opposing trends. ARGs associated with exposure included class A beta-lactamase gene CfxA6. Among infants attending day care, Escherichia coli and ARG abundance were both positively associated with antibiotic use.
Conclusion
Novel findings, including the importance of day care attendance, were identified through considering microbiome data at baseline and post-intervention. Thus, our study design and approach have important implications for future studies evaluating the unintended impacts of antibiotics.
Impact
The impact of antibiotic exposure to off-target microbes and antibiotic resistance genes in the gut is poorly defined.
We quantified these impacts in two cohort studies using a difference-in-differences approach. Novel to microbiome studies, we used pre/post-antibiotic data to emulate a randomized controlled trial.
Compared to infants unexposed to antibiotics between baseline and 1 year, the relative abundance of multiple off-target species and antibiotic resistance genes was altered.
Infants who attended day care and were exposed to antibiotics within the first year had a higher abundance of Escherichia coli and antibiotic resistance genes; a novel finding warranting further investigation.
Introduction
Infants and young children are exposed to more antibiotics than any other age group1,2 with an estimated 30–40% of children receiving at least one course of antibiotics within their first year of life.3–7 Although most of these antibiotics are prescribed for acute respiratory tract infections including acute otitis media,8,9 an unintended consequence of antibiotic use is its impact on microbes other than the targeted pathogen.10,11 Understanding antibiotics’ impacts on off-target or “bystander” microbes in the infant gut, in particular, is significant for two main public health reasons.11 First, systemic antibiotics can result in microbial dysbiosis as the developing infant gut is sensitive to perturbation.12 Encompassing both taxonomic and metabolic changes, antibiotics and resulting dysbiosis have been associated with negative health outcomes in young children including overweight/obesity,13–16 asthma,7,17 and celiac disease.18 Second, antibiotic exposures can lead to antimicrobial resistance. This is a particular concern in the human gut, a known reservoir for antimicrobial resistance genes (ARGs).19,20 Indeed, multiple studies have found that antibiotic exposures lead to the proliferation of ARGs that confer resistance to the antibiotic prescribed and to other antibiotics.21–24
Mitigation of antimicrobial resistance is a global health priority as the incidence of infections requiring second or third-line broad-spectrum antibiotics for treatment is on the rise,25 leading to further risk of antibiotic-resistant microbial infections. Integrated strategies that consider the microbiome are increasingly becoming useful for improving the surveillance of ARGs and providing clinical recommendations to prevent antimicrobial-resistant infections.26 Commensal bacteria can harbor ARGs and carriage prevalence of potentially pathogenic microbes can lead to antimicrobial-resistant infections. Thus, there is an urgent need to conduct epidemiologic studies and build quantitative models to understand how antibiotics affect off-target microbes, antimicrobial resistance, and unintended health outcomes.
The goal of the current study was to quantify the population-level effects of antibiotics on the abundance and diversity of off-target microbes and ARGs. Specifically, we use pre/post-antibiotic data to estimate average population-level changes to individual microbes, ARGs, and compositional diversity metrics emulating a randomized controlled trial. This difference-in-differences approach is frequently used in health services and policy research but has not been previously applied to microbiome studies.27 The advantage of this design is that it uses baseline information (i.e., data from before the intervention) to account for intra-subject variation over time and a comparison group to assess differences among those exposed and unexposed to an intervention. This approach is equivalent to assessing the interaction effect between the exposed group and the post-intervention time point.28 As antibiotics are often prescribed for respiratory illnesses and ear infections unnecessarily,8,9,29 this study offers insight that can support antibiotic stewardship practices in light of the effect of antibiotics on commensal gut microbes and ARGs.
Methods
Study cohorts
The New Hampshire Birth Cohort Study (NHBCS) is an ongoing prospective cohort study of over 2250 pregnant women and their young children from New Hampshire and Vermont. A detailed description of the cohort has been published,30–32 but, in brief, enrollment began in 2009 to study the effects of environmental exposures on pregnant women and young children. Extensive data are available for the first year of life and include delivery and pediatric medical records, as well as interview data for 4, 8, and 12 months post-delivery. Stool samples have been collected and undergone shotgun sequencing from a subset of children throughout the first year of life but predominantly at the 6-week and 1-year timepoints. Pregnant women and their children were recruited using the NHBCS’s current Dartmouth Institutional Review Board-approved procedures by the Center for the Protection of Human Subjects.
The DIABIMMUNE Study is a cohort of infants in Finland, Estonia, and Russia focused on the hygiene or microbiota hypothesis and its potential role in the development of autoimmune diseases.33 We used data from infants in a DIABIMMUNE sub-study that aimed to examine the role antibiotics play in microbiome development over the first 3 years of life.23 The sub-study included 39 infants from Finland having either 0 or at least 9 antibiotic exposures within their first 3 years of life. Children in this cohort had stool samples collected throughout their early life starting at about 2 months of age. We accessed quality-controlled shotgun sequencing FASTQ files and covariate data through their publicly available website.34
Antibiotic exposure classification
NHBCS
We were interested in systemic antibiotic exposures between the 6-week and 1-year timepoints (to match available stool samples) with a goal of studying both exposure (yes/no) and the frequency of antibiotic courses given to all infants. A unique advantage of the NHBCS is that antibiotic prescriptions, including indication, are captured in both medical records and caregiver questionnaires administered at 4, 8, and 12 months of age.
As outlined in Supplementary Fig. S1, not all infants with stool samples available had full medical record or interview data over the first year of life. Therefore, we considered two sub-cohorts for assessing antibiotic exposure. The first cohort, the NHBCS Antibiotic Exposure Cohort, classifies antibiotic exposure from medical record and/or interview data. This cohort was used to assess differences between antibiotic exposure versus non-exposure. In this dataset, we have overall less precise knowledge of the timing of antibiotics (i.e., the exact day of antibiotic prescription) but were able to maximize the sample size. Our second cohort, the NHBCS Antibiotic Frequency Cohort, only uses antibiotic prescription data from medical records enabling us to know the number and exact timing of antibiotics prescribed. The schema, sensitivity analysis, assumptions, and sample sizes for these two sub-cohorts are available in the Supplementary Notes.
DIABIMMUNE Study
To increase the external validity of this study and include subjects that were known to have a relatively high number of antibiotic exposures, infants from the DIABIMMUNE Study cohort were included. Antibiotic exposures in the DIABIMMUNE Study were collected through parental reporting of antibiotic use, duration, and reason for use as described previously.33 Infants from the DIABIMMUNE Study were added to NHBCS sub-cohorts to make combined cohorts, referred to respectively as the Antibiotic Exposure and Antibiotic Frequency Cohorts (Supplementary Fig. S1).
Stool microbiome assessment
Details on stool collection and metagenomic DNA shotgun sequencing have previously been described for the NHBCS cohort31,35 and the DIABIMMUNE Study group.23,36 For all samples, taxonomy down to the species level was assigned using MetaPhlAn337 and quantified using ShortBRED38 with ARGs derived from the Comprehensive Antibiotic Resistance Database (CARD) v3.1.1.39 In CARD, model names are sometimes annotated by microbes that they have been identified in, but this may not necessarily reflect the microbial origin. Information on the ARGs profiled is available in Supplementary Table S1. Additional details are discussed in the Supplementary Notes.
Statistical analyses
All statistical analyses were performed using R version 3.6.0,40 with additional details on overall ARG abundance and correlation analysis in the Supplementary Notes.
Assessing individual off-target microbes and antibiotic resistance genes
The difference-in-differences approach is frequently used in health services and policy research to estimate the effect of change due to an intervention or policy on a population when a randomized controlled trial is not possible or ethical.27 Assumptions required for the difference-in-differences approach27,28 were assessed prior to use.
The difference-in-difference estimate was performed using Microbiome Multivariable Associations with Linear Models (MaAsLin2).41 Our general formula was:
Where abundance is the relative abundance of the microbe or ARG in each sample, sample age is the time in days of the sample collection, antibiotic exposure is a dichotomous indicator of if the infant was exposed or not exposed to an antibiotic ever, and sampling interval is a binary variable used to denote that the measurement was from baseline (~ 6 weeks or 2 months) or 1-year. Antibiotic exposure × sampling interval represents the difference-in-difference estimate or interaction between exposure and time interval. We considered intercept random effects at the infant level (1|infant).
Results
Study groups
We used rigorous classification rules (Supplementary Notes) to retrospectively assess antibiotic exposure and frequency in 238 infants in the NHBCS that had shotgun sequencing data from stool samples collected at both ~6-week and ~1-year timepoints (Supplementary Fig. S1). Ultimately, we assigned 183 infants from the NHBCS as exposed or unexposed to antibiotics for a specific condition based on interview and/or medical record data (the NHBCS Antibiotic Exposure Cohort). A group of 99 infants could be classified by antibiotic frequency based on medical record data (NHBCS Antibiotic Frequency Cohort). Sensitivity analyses were used to evaluate exposure misclassification between medical record and interview data, but we found high concordance between the two methods (Supplementary Notes). In addition to infants participating in the NHBCS, we were able to classify the antibiotic exposure and frequency profiles of 33 infants with stool samples collected at approximately 2 months and 1 year from the DIABIMMUNE Study.23
Descriptive characteristics of infants and samples
Comparisons between the DIABIMMUNE and NHBCS Antibiotic Exposure and Antibiotic Frequency sub-cohorts indicated that the proportions of infants that were female and that were born full-term were similar (Table 1, χ2 p value > 0.1). Infants from the DIABIMMUNE Study were more likely than infants from at least one NHBCS sub-cohort to be vaginally delivered (χ2 p value < 0.1). The average baseline age and days breastfeeding of infants from the DIABIMMUNE Study was approximately 20 days greater than that of infants from the NHBCS (Kruskal–Wallis p value < 0.1).
Table 1.
Descriptive overview of infants in the study.
| New Hampshire Birth Cohort Study (NHBCS) Antibiotic Exposure Cohort (n = 183) | NHBCS Antibiotic Frequency Cohort (n = 99) | DIABIMMUNE Study (n = 33) | |
|---|---|---|---|
| Antibiotic classification derivation | Combination of medical record review and parental recall | Medical record review | Parental recall |
| Mean (SD) sample age in days | Baseline: 46 (15) | Baseline: 45 (16) | Baseline: 64 (15) |
| 1 year: 371 (29) | 1 year: 376 (29) | 1 year: 366.6 (15) | |
| Female | 70 (38.3%) | 40 (40.4%) | 14 (42.4%) |
| Vaginal delivery | 131 (71.6%) | 67 (67.7%) | 29 (87.9%) |
| Mean (SD) days breastfeeding by sample collection | Baselinea: 43.0 (17.7) | Baselinea: 41.8 (18.9) | Baseline: 64 (15) |
| 1 year: 247 (133) | 1 year: 238 (140) | 1 year: 281 (99) | |
| Attend day care by 1 year | Yes: 76 (41.5%) | Yes: 38 (38.4%) | N/A |
| No: 91 (49.7%) | No: 45 (45.5%) | ||
| Missing: 16 (8.7%) | Missing: 16 (16.2%) | ||
| Full-term (at least 37 weeks’ gestation) | 171 (93.4%) | 91 (92.0%) | 31 (94.0%) |
| Parity | 0: 89 (48.6%) | 0: 44 (44.4%) | N/A |
| 1: 62 (33.9%) | 1: 41 (41.4%) | ||
| 2+: 26 (14.2%) | 2+: 13 (13.1%) | ||
| Missing: 6 (3.3%) | Missing: 1 (1.0%) | ||
| Maternal education level | High school or equivalent: 16 (8.7%) | High school or equivalent: 9 (9.1%) | N/A |
| Junior college, technical school, or some college: 26 (14.2%) | Junior college, technical school, or some college: 15 (15.2%) | ||
| College: 67 (36.6%) | College: 38 (38.4%) | ||
| Post-graduate: 71 (38.8%) | Post-graduate: 36 (36.4%) | ||
| Missing: 3 (1.6%) | Missing: 1 (1.0%) | ||
| Antibiotic exposure immediately following birth | 6 (3.3%) | 3 (3.0%) | 1 (3.0%) |
| Missing: 3 (1.6%) | |||
| Number of antibiotic prescriptions between 6 weeks and 1 year | N/A | For otitis media: 42 | For otitis media: 36 |
| Not otitis mediab: 5 | Not otitis media: 0 | ||
| Children with antibiotic exposure between 6 weeks and 1 year | For otitis media: 58 (31.7%) | For otitis media: 27 (27.3%) | For otitis media or any condition: 13 (39.4%) |
| For any conditionc: 63 (34.4%) | For any condition: 29 (29.3%) | ||
| Number of antibiotics for otitis media given to child | N/A | 0: 72 | 0: 20 |
| 1: 15 | 1: 3 | ||
| 2+: 12 | 2+: 10 | ||
| Number of antibiotics for any reason given to child between baseline and 1-year time point | N/A | 0: 70 | 0: 20 |
| 1: 15 | 1: 3 | ||
| 2+: 14 | 2+: 10 | ||
| Number of prescriptions by antibiotic type between baseline and 1-year time point | N/A | Amoxicillin-type: 37 | Amoxicillin-type: 20 |
| Cephalosporin-type: 8 | Cephalosporin-type: 7 | ||
| Azithromycin: 2 | Azithromycin: 5 | ||
| Trimetoprime and sulfadiazine: 3 | |||
| Unknown: 1 |
aOne infant included in both NHBCS sub-cohorts did not have available breastfeeding information at baseline.
bOther reasons included: upper respiratory infection (2), pneumonia (1), bronchitis (1), and reaction to pneumococcal vaccine (1).
cOther reasons included: upper respiratory infection (2), eye infection (1), pneumonia (1), and strep throat (1).
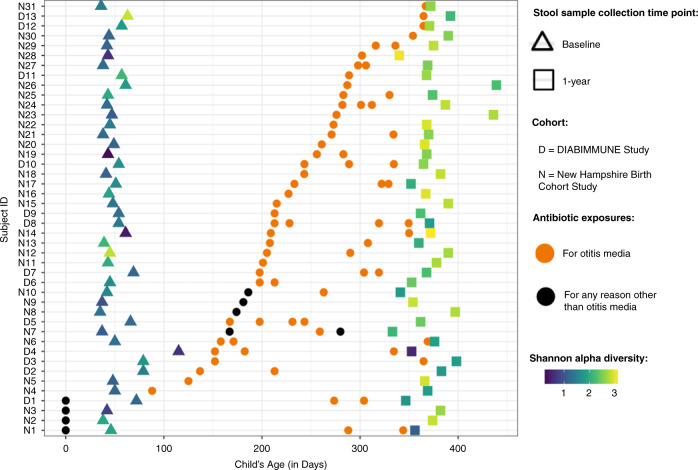
The majority of antibiotic exposures occurred after 6 months of age and for otitis media (Fig. 1). As expected based on the inclusion criteria that DIABIMMUNE Study infants would have high exposure to antibiotics over the first 3 years of life, infants in the DIABIMMUNE Study were more frequently exposed to antibiotics and, when exposed, were given more antibiotics than infants in the NHBCS. Amoxicillin-type antibiotics (e.g., amoxicillin, amoxicillin with clavulanate) were the most commonly prescribed antibiotics among all infants. The alpha diversity of samples did vary by antibiotic exposure both before and after exposure (Supplementary Notes).
Fig. 1. Descriptive overview of antibiotic exposures among New Hampshire Birth Cohort and DIABIMMUNE Study infants exposed to antibiotics in the Antibiotic Frequency Cohort.
Figure only includes antibiotic exposures occurring before the 1-year stool sample collection. Samples are ordered by first antibiotic exposure. Baseline samples were collected at approximately 6 weeks for New Hampshire Birth Cohort Study participants and 2 months for DIABIMMUNE Study infants. Age of antibiotic exposure reflects the start day of antibiotic exposure. Infants with antibiotic exposures prior to the baseline microbiome collection received them immediately following birth (age 0 days).
Abundance of off-target microbes and antibiotic resistance genes
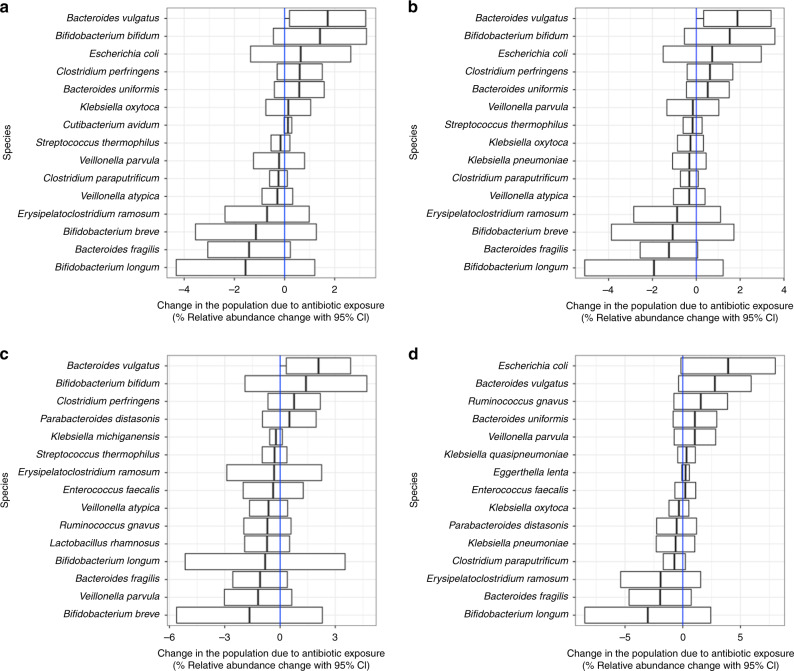
Since all antibiotic exposures were for non-gut-associated infections, we classified all microbes as off-target. Looking specifically at antibiotic-exposed versus unexposed infants from the NHBCS and the DIABIMMUNE Studies (Fig. 2a and Supplementary Table S2), we found the relative abundance change for two microbes was significantly different than zero. In particular, the relative abundance of Bacteroides vulgatus across the two timepoints increased by 1.72% (95% CI: 0.19, 3.24) more than it would have if the infant was never exposed to an antibiotic, while the relative abundance of Actinomyces odontolyticus decreased by 0.008% (95% CI: 0.015, −0.001). Relative abundance changes based on antibiotic exposure went in opposing directions for Bacteroides and Bifidobacterium species. While B. vulgatus abundance increased, Bacteroides fragilis decreased by 1.42% (95% CI: −3.06, 0.23). Similarly, Bifidobacterium bifidum relative abundance increased 1.41% (95% CI: −0.44, 3.27) more than it would have if the infant was never exposed, but Bifidobacterium longum and Bifidobacterium breve decreased by 1.56% (95% CI: −4.32, 1.21) and 1.14% (95% CI: −3.55, 1.27) respectively.
Fig. 2. Off-target species changes in the population due to any antibiotic exposure.
a Change in the population due to antibiotic exposure in the Antibiotic Exposure Cohort (n = 216). The difference-in-difference model for the relative abundance change between exposed and unexposed infants across the two timepoints was adjusted for sample age, duration of breastfeeding, delivery mode, sex, gestational age, antibiotic use immediately following birth, study cohort, and a random effect for each subject. b Change in the population due to antibiotic exposure in NHBCS infants (n = 183). c Change in the population due to antibiotic exposure in NHBCS infants that did not attend day care by 1 year (n = 91). d Change in the population due to antibiotic exposure in NHBCS infants that did attend day care by 1 year (n = 76). b was adjusted for the same variables as a with the exception of study cohort and addition of day care attendance by 1 year. c and d models were adjusted for the same variables as b except for day care attendance by 1 year. For all plots, the 15 species with the greatest absolute change are shown. Species are ordered by the point estimate for the relative abundance change.
As day care attendance increases an infant’s exposure to infections42–44 and is time variant (see Supplementary Notes), we stratified NHBCS infants in the Antibiotic Exposure Cohort by day care attendance by 1 year to attempt to separate the effect of day care attendance and antibiotic exposure on off-target microbes (Fig. 2b–d). Some microbe associations were consistent in both day care strata (i.e., no evidence of effect modification). However, among infants who attended day care, the relative abundance of Escherichia coli and Veillonella parvula was considerably higher among infants exposed to antibiotics compared to those who were unexposed, suggesting a synergistic interaction between antibiotic exposure and day care attendance towards the abundance of certain microbes.
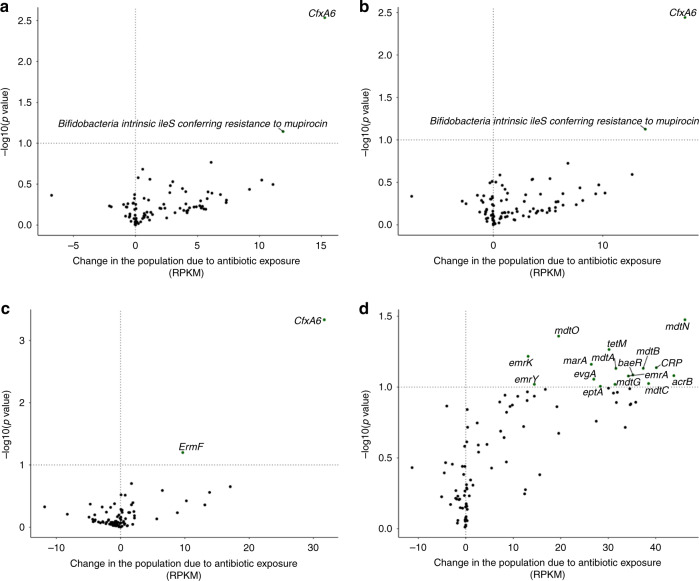
We also assessed the impact of antibiotic exposure on changes in ARGs over the follow-up period (Fig. 3). The most significant change occurred for the Antibiotic Resistance Ontology marker (ARO): 3003097 CfxA6, which increased on average by 15.26 reads per kilobase of reference sequence per million samples reads (RPKM) (95% CI: 5.33, 25.20) more than if the infant was not exposed to antibiotics (Fig. 3a and Supplementary Table S3). Similar results were found among infants only in the NHBCS (Fig. 3b). Results stratified by day care attendance were discordant for many ARGs. Among infants that did not attend day care by 1 year (Fig. 3c), CfxA6 was increased among infants exposed to antibiotics, whereas a different trend in ARG abundance change was identified among infants that attended day care by 1 year. While CfxA6 was no longer associated with antibiotic exposure, many other ARGs were positively associated such as mdtN (ARO: 3003548), tetM (ARO: 3000186), and mdtO (ARO: 3003549) (Fig. 3d).
Fig. 3. Volcano plots demonstrating abundance of antibiotic resistance gene changes in the population due to any antibiotic exposure in RPKM.
Line marks p value < 0.1. a Change in the population due to antibiotic exposure in the Antibiotic Exposure Cohort (n = 216). The difference-in-difference model for the relative abundance change between exposed and unexposed infants across the two timepoints was adjusted for sample age, breastfeeding duration, delivery mode, sex, gestational age, antibiotic use immediately following birth, study cohort, and a random effect for each subject. b Change in the population due to antibiotic exposure in NHBCS infants (n = 183). c Change in the population due to antibiotic exposure in NHBCS infants that did not attend day care by 1 year (n = 91). d Change in the population due to antibiotic exposure in NHBCS infants that did attend day care by 1 year (n = 76). b was adjusted for the same variables as a with the exception of study cohort and addition of day care attendance by 1 year. c and d models were adjusted for the same variables as b except for day care attendance by one year. Only antibiotic resistance genes with a baseline prevalence greater than 20% within each population are included in the plot. RPKM reads per kilobase of reference sequence per million samples reads.
To investigate this further, we looked at overall ARG abundance in the same infants (Supplementary Table S4). In the Antibiotic Exposure Cohort, there was no statistically significant mean ARG difference by antibiotic exposure between timepoints (516.16 RPKM increase in overall RPKM load among infants exposed to an antibiotic, 95% CI: −904.61, 1936.92) after adjustment for covariates. However, among infants who attended day care in the NHBCS, ARGs increased on average by 2305.54 RPKM (95% CI: −379.80, 4990.87) more in infants exposed to antibiotics than those who were not. For all analyses, while only a few ARGs changed significantly across the two timepoints, we found that the majority of the estimated mean differences for individual ARGs was greater than 0 suggesting a greater net RPKM trend increase among infants exposed to antibiotics.
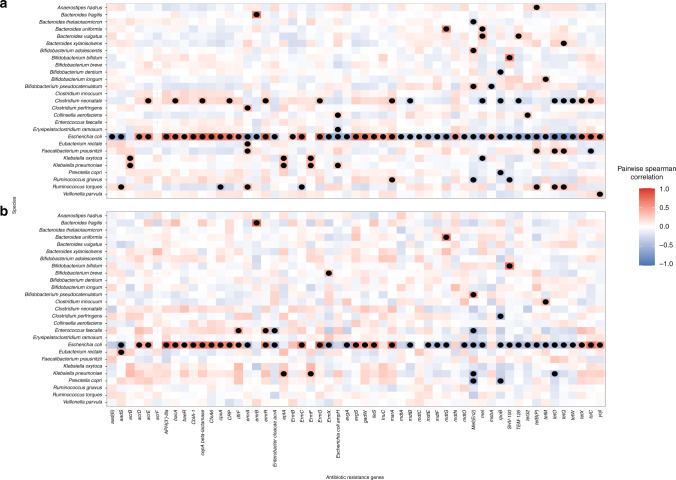
To jointly evaluate associations noted in the off-target microbe and ARG analyses, we conducted a block correlation analysis using Hierarchical All-against-All association (HAllA) testing. Across the 41 species and 55 ARGs analyzed, 111 pairs and 47 clusters were identified as having a statistically significant correlation (Benjamini–Hochberg q value < 0.05) among infants unexposed to antibiotics and 60 pairs and 19 clusters for exposed infants with 49 pairwise associations overlapping (Supplementary Tables S5 and S6). Across infants both unexposed and exposed to antibiotics, E. coli was highly associated positively and negatively with many ARGs (Fig. 4). Results further stratified by day care attendance were similar (Supplementary Fig. S2).
Fig. 4. Pairwise spearman correlation between species and antibiotic resistance genes by antibiotic exposure.
a Infants not exposed (n = 140) and b exposed to antibiotics (n = 76). HAllA was used to determine pairwise associations that were statistically significant. Only species and antibiotic resistance genes with at least one statistically significant pairwise association were included in the plot. Dots represent statistically significant associations (Benjamini–Hochberg adjusted p value < 0.05). Due to length of gene-associated model names, ileS is used as an abbreviation for Bifidobacteria intrinsic ileS conferring resistance to mupirocin and rpoB is used for Bifidobacterium rpoB conferring resistance to rifampicin.
Discussion
Overall findings
We evaluated the off-target effects of antibiotic exposures on the infant gut microbiome and resistome among a large number of infants from the United States and Finland. Using metagenomic sequencing of pre/post-antibiotic stool samples combined with detailed covariate information from two separate cohorts, we identified and quantified the population-level impact antibiotics have on off-target microbes and ARGs within the infant gut. We found that even with a broad categorization of antibiotic exposures between approximately 6 weeks and 1 year of life, antibiotics were associated with changes to off-target microbe and ARG abundance.
In the NHBCS Antibiotic Exposure Cohort, we found that 34.4% of infants were exposed to antibiotics between approximately 6 weeks and 1 year of life with 58 out of the 63 (92.1%) antibiotic exposures prescribed for otitis media. This largely correlates with other studies from similar demographic groups that have estimated approximately 30–40% of infants are exposed to antibiotics in the first year.3–7 Current protocols in the United States recommend antibiotics for most otitis media and lower respiratory tract infections but not for upper respiratory infections.45,46 Due to challenges in diagnosis, gastrointestinal consequences of antibiotics, and concern for the growing threat of antimicrobial resistance, many country-level antibiotic stewardship protocols encourage watchful waiting.47 Accordingly, randomized controlled trials29,48 and population-level cross-sectional studies49,50 have examined delayed, shortened, or no antibiotic therapy for uncomplicated otitis media and respiratory infections.29,48 While these studies are critical, cohort studies with combined covariate and microbiome data also yield valuable evidence in support of antibiotic stewardship practices because they reflect current treatment practices. Thus, while this study’s main goal was to provide a population-level overview of the impact of antibiotics to infant gut microbiota and the resistome, it also provides public health professionals and clinicians with quantifiable results that may be applicable to antibiotic stewardship recommendations.
Antibiotic exposure differentially impacts commensal microbes
Commensal microbes have many roles in the infant gut microbiome that include: (1) degrading polysaccharides into useable sugars and short-chain fatty acids for other gut microbes51,52 and (2) inhibiting pathogen colonization directly through nutrient competition and bacteria-bacteria attacks53 as well as indirectly by stimulating the development of cells necessary for innate and adaptive immunity.54 Thus, decreased presence or abundance due to antibiotic exposure is a major concern. However, previous studies inconsistently identify particular commensal microbial abundances that have increased or decreased as a result of antibiotic exposure and often only profile results at the genus level.55 While the majority of our results did not indicate statistically significant differences between antibiotic-exposed and unexposed infants, our results justified the need to evaluate differences below the genus level especially for Bifidobacterium and Bacteroides.
Bifidobacterium species exhibited conflicting trends in our study. The relative abundance of two species, B. longum and B. breve, consistently decreased in the antibiotic-exposed population. However, B. bifidum increased in relative abundance after antibiotic exposure. This trend suggests that antibiotic exposure may lead to long-term selection changes to favor certain species of Bifidobacterium. Limited studies have specifically assessed Bifidobacterium species,55 but a prior study in 1.5-year-old children found that B. bifidum concentration decreased by 23% in amoxicillin exposed children compared to 54.5% in unexposed children while B. longum concentration remained consistent immediately following exposure.56 Differences identified between studies may be a result of differences in population demographics including the age of the children as well as the time period post antibiotic exposure.
We consistently identified that B. vulgatus relative abundance increased in antibiotic-exposed children but B. fragilis decreased. These results are interesting in context to the DIABIMMUNE Study that identified B. fragilis as a “single-colonization species” and B. vulgatus as a “multiple-colonization species” based on the dominance of one vs. multiple strains across time.23 Species with multiple strains in an environment are thought to contribute to a more resilient microbiome.57,58 While we did not assess strain diversity and at least one previous study suggests these species may have a similar sensitivity to amoxicillin-based antibiotics,59 our results underscore the need to consider differential sensitivity to antibiotics as a result of strain-level diversity, differences in biogeographical niches,60 and variation in ARG carriage by species. Regarding ARG carriage, the CARD continually tracks the prevalence of antibiotic resistance phenotypic profiles in reference sequences for many non-pathogenic microbes. According to the CARD prevalence 3.0.9 update, there is wide variability in ARG phenotypes between commensals from the same and different genera.39 These data, in combination with the results from this study and others assessing antimicrobial susceptibility in bacteria often considered off-target,61,62 provide further evidence that intra- and inter-species variation is important to consider.
Describing differential patterns of antimicrobial resistance abundance
Multiple studies have assessed how antibiotic exposure after the neonatal period impact the resistome21–23,63–67 but have been conducted in predominantly preterm infants21,22 or contained relatively small cohorts (<30 infants) of antibiotic-exposed infants.23,63–66 We found two distinct patterns in our study, but, regardless of antibiotic exposure, found that E. coli was the microbe most frequently correlated with ARGs (Fig. 4 and Supplementary Fig. S2).
In our Antibiotic Exposure Cohort, only ARO: 3003730 Bifidobacteria intrinsic ileS conferring resistance to mupirocin and CfxA6 were statistically significantly increased among antibiotic-exposed infants (Fig. 3a). According to the current version of CARD resistomes (v3.0.9), ARO: 3003730, works through antibiotic target alteration and has only been found in whole-genome sequences of B. longum.39 As we found that B. longum abundance decreased when this ARG increased (Fig. 2), this provides some evidence that this ARG may be associated with multiple species and requires follow-up studies. While we could not identify specific species hosts for CfxA6, three studies21,23,68 also identified CfxA6 beta-lactamase gene and one study67 identified CfxA2 (another CfxA class A beta-lactamase gene69) as ARG markers of interest. CfxA genes work via antibiotic inactivation39 and have been identified in a wide range of microbes from Bacillota (formally the phylum Firmicutes) and Bacteroidota (Bacteroidetes) including Bacteroides, Prevotella, and Capnocytophaga with point mutations differing between CfxA2 and CfxA6.67,69–71 The DIABIMMUNE Study found the CfxA6 gene marker persisted in children long after antibiotic exposure.23 Although some of these infants were included in our study, we independently found that the gene increased in abundance in infants in the NHBCS only (Fig. 3b). The most comparable study to ours demonstrated in 662 Danish children that antibiotic exposures during the first year of life impacted the gut resistome of healthy infants.67 In particular, they found time since antibiotic exposure and number of antibiotics influential to overall ARG abundance. In addition, they identified specific ARGs including CfxA2 and APH(3’)-llla to be increased among infants exposed to a beta-lactam inhibitor compared to infants not exposed to any antibiotic. For CfxA2, they found that it was also increased among infants given another antibiotic compared to no antibiotic exposure. Although they did not have resistome measurements taken before antibiotic exposures to establish baseline ARG abundances, the results of our study aligned with these results and those presented previously in our cohort.35 In addition, a study of healthy adults exposed to a 7-day treatment of cefprozil found specific point mutations in CfxA6 were enriched in multiple participants68 and another study identified CfxA6 to be an important discriminator of gestational age.21 Given all this data, further detailed investigations of the CfxA6 gene at the sequence level are warranted.
Among infants who attended day care by 1 year, we found that multiple ARGs and the overall resistance load were comparatively much higher among infants exposed to antibiotics (Fig. 3d). E. coli relative abundance increased in antibiotic-exposed infants (Fig. 2d) suggesting a synergistic interaction between antibiotic exposure and day care attendance. Although we were unable to identify other studies that assessed both antibiotic exposure and day care attendance, given E. coli’s role in horizontal gene transfer72,73 and previous studies in the NHBCS and others noting their association with harboring antibiotic resistance genes,21,35,67,74,75 the corresponding changes to resistome composition aligned with changes to E. coli relative abundance.
Strengths and limitations
While this study is one of the largest to consider the population-level impact of antibiotic exposures to the infant gut over a follow-up period, we note some important limitations. Our main limitation was the precise timing and dosage of antibiotic exposures in the NHBCS Antibiotic Exposure Cohort. To mitigate this, we utilized stool samples from the DIABIMMUNE Study as a validation cohort since it obtained both precise timing and dose of antibiotic exposures (Supplementary Notes). We also used exposure data with more precision on a smaller sample group of infants via a medical record review to assess timing and type of antibiotic exposure. Although statistical tools have used mixed-effects models to assess microbiome composition longitudinally,41,76 the difference-in-differences approach offers a straight-forward interpretation of the effect of an intervention in a population through its strong counterfactual estimate of the effect of eliminating unnecessary antibiotic exposures on the infant gut microbiome. Moreover, the epidemiologic framework provides the flexibility for researchers to estimate the difference-in-differences using a variety of transformations, statistical packages, and tools. A major consideration, however, in formally applying the difference-in-difference approach to microbiome analyses is fully addressing inherent difference-in-difference assumptions.27,28 For instance, in this analysis, we were unable to fully account for the common trend assumption due to only having two timepoints to assess metagenomic data. However, in the process of validating our assumptions, sensitivity analysis revealed that antibiotic exposure did differentially impact some microbes and ARGs in infants depending on their day care status; a novel finding. Lastly, an important caveat to much microbiome data is inherent limitations in accurately reflecting the composition of gut microbiota and ARGs using stool samples. Thus, while we did identify the relative abundance of microbes and ARGs changing in our differential abundance analyses, we cannot rule out that the change would affect the absolute abundance, rule out unmeasured confounding, nor fully predict changes in function or the gut itself.77 Regardless, misclassification likely would be non-differential by antibiotic exposure and we still can conclude that the infant gut microbiome of antibiotic-exposed and unexposed infants is different.
Conclusion
Our results support the notion that microbiota and the resistome should be considered when weighing the costs and benefits of antibiotic interventions. Moreover, we apply an established framework used to emulate a randomized controlled trial to the microbiome space. This enabled quantification of the magnitude of the population-level effects of antibiotic exposure in the context of other pressures impacting the developing microbiome in early life. We imagine that this novel application can be used to evaluate the potential impact of new or current antibiotic prescription practices on gut microbiota and residual impacts on ARGs.
Supplementary information
Acknowledgements
We have the utmost gratitude to participants, family members, coordinators, clinicians, and researchers involved in the New Hampshire Birth Cohort. We thank Dr Scot Zens, Terran Campbell, Dr Weston Viles, and Dr Jie Zhou for their thoughtful comments and contributions.
Author contributions
R.M.L., J.C.M., M.R.K., and A.G.H. designed the research. J.C.M. and M.R.K. designed the study protocols and provided oversight for cohort and the collection, processing, and analyses of samples. R.M.L. performed the data analysis. R.M.L. and A.G.H. wrote the initial manuscript draft. H.G.M. provided technical and methodological support from the Marine Biological Laboratory. Y.M., T.J.P., and E.F.D. managed the analytic samples. R.M.L., J.C.M., Q.P.N., M.O.C., B.D.R., M.M.P., M.R.K., and A.G.H. interpreted the results. All authors provided consent for publication.
Funding
This work was funded in part by US National Institute Health under award numbers NIGMS P20GM104416, NIEHS P01ES022832 and P20ES018175, and NLM R01LM012723. Additional funds were provided by the US Environmental Protection Agency grants RD83459901 and RD83544201. R.M.L. was funded under the Host-Microbe Interactions Training Grant at Dartmouth NIAID T32AI007519.
Data availability
The whole metagenomic shotgun sequencing samples are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive: https://www.ncbi.nlm.nih.gov/sra (accession number: PRJNA296814). Analytic R markdown scripts are available upon request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
For participants from the New Hampshire Birth Cohort Study, pregnant women, and their children were recruited using the New Hampshire Birth Cohort Study’s current Dartmouth Institutional Review Board-approved procedures by the Center for the Protection of Human Subjects.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-022-02104-w.
References
- 1.Lee GC, et al. Outpatient antibiotic prescribing in the United States: 2000 to 2010. BMC Med. 2014;12:1–8. doi: 10.1186/1741-7015-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicks LA, et al. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 2015;60:1308–1316. doi: 10.1093/cid/civ076. [DOI] [PubMed] [Google Scholar]
- 3.Stam J, et al. Antibiotic use in infants in the first year of life in five European countries. Acta Paediatr. Int. J. Paediatr. 2012;101:929–934. doi: 10.1111/j.1651-2227.2012.02728.x. [DOI] [PubMed] [Google Scholar]
- 4.Dawson-Hahn EE, Rhee KE. The association between antibiotics in the first year of life and child growth trajectory. BMC Pediatr. 2019;19:1–9. doi: 10.1186/s12887-018-1363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinlaw AC, et al. Trends in antibiotic use by birth season and birth year. Pediatrics. 2017;140:e20170441. doi: 10.1542/peds.2017-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, et al. Prescription drug dispensing profiles for one million children: A population-based analysis. Eur. J. Clin. Pharmacol. 2013;69:581–588. doi: 10.1007/s00228-012-1343-1. [DOI] [PubMed] [Google Scholar]
- 7.Patrick DM, et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir. Med. 2020;8:1094–1105. doi: 10.1016/S2213-2600(20)30052-7. [DOI] [PubMed] [Google Scholar]
- 8.Fleming-Dutra KE, et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA. 2016;315:1864–1873. doi: 10.1001/jama.2016.4151. [DOI] [PubMed] [Google Scholar]
- 9.Kronman MP, Zhou C, Mangione-Smith R. Bacterial prevalence and antimicrobial prescribing trends for acute respiratory tract infections. Pediatrics. 2014;134:e956–e965. doi: 10.1542/peds.2014-0605. [DOI] [PubMed] [Google Scholar]
- 10.Tedijanto C, Olesen SW, Grad YH, Lipsitch M. Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc. Natl Acad. Sci. USA. 2018;115:E11988–E11995. doi: 10.1073/pnas.1810840115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morley VJ, Woods RJ, Read AF. Bystander selection for antimicrobial resistance: implications for patient health. Trends Microbiol. 2019;27:864–877. doi: 10.1016/j.tim.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vatanen T, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korpela K, et al. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome. 2017;5:1–9. doi: 10.1186/s40168-017-0245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uzan-Yulzari, A. et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat. Commun. 12, 443 (2021). [DOI] [PMC free article] [PubMed]
- 15.Vallianou N, Dalamaga M, Stratigou T, Karampela I, Tsigalou C. Do antibiotics cause obesity through long-term alterations in the gut microbiome? A review of current evidence. Curr. Obes. Rep. 2021;10:244–262. doi: 10.1007/s13679-021-00438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan S, et al. Impact of exposure to antibiotics during pregnancy and infancy on childhood obesity: a systematic review and meta-analysis. Obesity. 2020;28:793–802. doi: 10.1002/oby.22747. [DOI] [PubMed] [Google Scholar]
- 17.Ong MS, Umetsu DT, Mandl KD. Consequences of antibiotics and infections in infancy: Bugs, drugs, and wheezing. Ann. Allergy, Asthma Immunol. 2014;112:441–445.e1. doi: 10.1016/j.anai.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Dydensborg Sander S, et al. Association between antibiotics in the first year of life and celiac disease. Gastroenterology. 2019;156:2217–2229. doi: 10.1053/j.gastro.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 19.Moore AM, et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One. 2013;8:e78822. doi: 10.1371/journal.pone.0078822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Gasparrini AJ, et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 2019;4:2285–2297. doi: 10.1038/s41564-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson MK, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 2016;1:1–25. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yassour M, et al. Natural history of the infant gut microbiome and impact of antibiotic treatments on strain-level diversity and stability. Sci. Trans. Med. 2016;8:1173–1178. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doan T, et al. Macrolide and nonmacrolide resistance with mass azithromycin distribution. N. Engl. J. Med. 2020;383:1941–1950. doi: 10.1056/NEJMoa2002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. Antimicrobial Resistance Global Report on Surveillance. 10.1007/s13312-014-0374-3 (2014).
- 26.Relman DA, Lipsitch M. Microbiome as a tool and a target in the effort to address antimicrobial resistance. Proc. Natl Acad. Sci. USA. 2018;115:12902–12910. doi: 10.1073/pnas.1717163115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wing C, Simon K, Bello-Gomez RA. Designing difference in difference studies: best practices for public health policy research. Annu. Rev. Public Health. 2018;39:453–469. doi: 10.1146/annurev-publhealth-040617-013507. [DOI] [PubMed] [Google Scholar]
- 28.Caniglia EC, Murray EJ. Difference-in-difference in the time of cholera: a gentle introduction for epidemiologists. Curr. Epidemiol. Rep. 2020;7:203–211. doi: 10.1007/s40471-020-00245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mas-Dalmau, G. et al. Delayed antibiotic prescription for children with respiratory infections: a randomized trial. Pediatrics147, e20201323 (2021). [DOI] [PubMed]
- 30.Gilbert-Diamond D, et al. Rice consumption contributes to arsenic exposure in US women. Proc. Natl Acad. Sci. USA. 2011;108:20656–20660. doi: 10.1073/pnas.1109127108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coker MO, et al. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: a prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2019;127:217–227. doi: 10.1111/1471-0528.15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coker MO, et al. Infant feeding alters the longitudinal impact of birth mode on the development of the gut microbiota in the first year of life. Front. Microbiol. 2021;12:1–11. doi: 10.3389/fmicb.2021.642197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mustonen N, et al. Early childhood infections and the use of antibiotics and antipyretic-analgesics in Finland, Estonia and Russian Karelia. Acta Paediatr. Int. J. Paediatr. 2019;108:2075–2082. doi: 10.1111/apa.14874. [DOI] [PubMed] [Google Scholar]
- 34.Diabimmune. DIABIMMUNE Antibiotics Cohort. https://diabimmune.broadinstitute.org/diabimmune/antibiotics-cohort. Accessed 15 Jan 2021.
- 35.Lebeaux RM, et al. The infant gut resistome is associated with E. coli and early-life exposures. BMC Microbiol. 2021;21:1–18. doi: 10.1186/s12866-021-02129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yassour M, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24:146–154. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminski J, et al. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput. Biol. 2015;486:207–214. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcock BP, et al. CARD 2020: antibiotic resistome surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: a language and environment for statistical computing (2017).
- 41.Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021). [DOI] [PMC free article] [PubMed]
- 42.Kaur R, Morris M, Pichichero ME. Epidemiology of acute otitis media in the postpneumococcal conjugate vaccine era. Pediatrics. 2017;140:1–11. doi: 10.1542/peds.2017-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kansen HM, et al. Risk factors for atopic diseases and recurrent respiratory tract infections in children. Pediatr. Pulmonol. 2020;55:3168–3179. doi: 10.1002/ppul.25042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kørvel-Hanquist A, Koch A, Lous J, Olsen SF, Homøe P. Risk of childhood otitis media with focus on potentially modifiable factors: a Danish follow-up cohort study. Int. J. Pediatr. Otorhinolaryngol. 2018;106:1–9. doi: 10.1016/j.ijporl.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 45.Lieberthal AS, et al. The diagnosis and management of acute otitis media. Pediatrics. 2013;131:e964–e999. doi: 10.1542/peds.2012-3488. [DOI] [PubMed] [Google Scholar]
- 46.Wong DM, Blumberg DA, Lowe LG. Guidelines for the use of antibiotics in acute upper respiratory tract infections. Am. Fam. Physician. 2006;74:956–966. [PubMed] [Google Scholar]
- 47.Suzuki HG, Dewez JE, Nijman RG, Yeung S. Clinical practice guidelines for acute otitis media in children: a systematic review and appraisal of European national guidelines. BMJ Open. 2020;10:e035343. doi: 10.1136/bmjopen-2019-035343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spiro DM, et al. Wait-and-see prescription for the treatment of acute otitis media: a randomized controlled trial. J. Am. Med. Assoc. 2006;296:1235–1241. doi: 10.1001/jama.296.10.1235. [DOI] [PubMed] [Google Scholar]
- 49.Tedijanto C, Grad YH, Lipsitch M. Potential impact of outpatient stewardship interventions on antibiotic exposures of bacterial pathogens. Elife. 2020;9:1–12. doi: 10.7554/eLife.52307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chua KP, Fischer MA, Linder JA. Appropriateness of outpatient antibiotic prescribing among privately insured US patients: ICD-10-CM based cross sectional study. BMJ. 2019;364:1–8. doi: 10.1136/bmj.k5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawson MAE, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–648. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 2017;279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonnell L, et al. Association between antibiotics and gut microbiome dysbiosis in children: systematic review and meta-analysis. Gut Microbes. 2021;13:1–18. doi: 10.1080/19490976.2020.1870402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mangin I, Suau A, Gotteland M, Brunser O, Pochart P. Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe. 2010;16:433–438. doi: 10.1016/j.anaerobe.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Vatanen T, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol. 2019;4:470–479. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz DJ, Langdon AE, Dantas G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 2020;12:1–12. doi: 10.1186/s13073-020-00782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Betriu, C., Sánchez, A., Gómez, M. & Picazo, J. J. In-vitro susceptibilities of species of the. J. Antimicrob. Chemother. 43, 133–136 (1999). [DOI] [PubMed]
- 60.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vázquez-López R, et al. The beta-lactam resistome expressed by aerobic and anaerobic bacteria isolated from human feces of healthy donors. Pharmaceuticals. 2021;14:1–15. doi: 10.3390/ph14060533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duranti S, et al. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl. Environ. Microbiol. 2017;83:1–14. doi: 10.1128/AEM.02894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sosa-Moreno A, et al. Perinatal risk factors for fecal antibiotic resistance gene patterns in pregnant women and their infants. PLoS One. 2020;15:1–21. doi: 10.1371/journal.pone.0234751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bäckhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Moore AM, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015;3:1–10. doi: 10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahman SF, Olm MR, Morowitz MJ, Banfield JF. Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems. 2018;3:1–12. doi: 10.1128/mSystems.00123-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, et al. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe. 2021;29:1–13. doi: 10.1016/j.chom.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 68.Raymond F, et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016;10:707–720. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parker AC, Smith CJ. Genetic and biochemical analysis of a novel ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 1993;37:1028–1036. doi: 10.1128/AAC.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamanai-Shacoori Z, et al. CfxA expression in oral clinical Capnocytophaga isolates. Anaerobe. 2015;35:68–71. doi: 10.1016/j.anaerobe.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 71.Fernández-Canigia L, Cejas D, Gutkind G, Radice M. Detection and genetic characterization of β-lactamases in Prevotella intermedia and Prevotella nigrescens isolated from oral cavity infections and peritonsillar abscesses. Anaerobe. 2015;33:8–13. doi: 10.1016/j.anaerobe.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 72.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 74.Pärnänen K, et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casaburi G, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob. Resist. Infect. Control. 2019;8:1–18. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen EZ, Li H. A two-part mixed-effects model for analyzing longitudinal microbiome compositional data. Bioinformatics. 2016;32:2611–2617. doi: 10.1093/bioinformatics/btw308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montassier E, et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021;6:1043–1054. doi: 10.1038/s41564-021-00920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole metagenomic shotgun sequencing samples are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive: https://www.ncbi.nlm.nih.gov/sra (accession number: PRJNA296814). Analytic R markdown scripts are available upon request.